
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Pharmacol. , 04 August 2020
Sec. Pharmacogenetics and Pharmacogenomics
Volume 11 - 2020 | https://doi.org/10.3389/fphar.2020.00936
This article is part of the Research Topic Precision Psychiatry from a Pharmacogenetics Perspective View all 10 articles
The aim of this study was to provide dose recommendations for risperidone in Asian people based on cytochrome P450 enzyme CYP2D6 genotype. First, we investigated the influence of CYP2D6 polymorphism on the pharmacokinetics of risperidone in Chinese patients with schizophrenia. Then, we performed a search for studies covering the relationship between pharmacokinetic parameters of risperidone and CYP2D6 genotype. Pooled pharmacokinetic parameters were meta-analyzed using a random-effects model. Lastly, we calculated the dose adjustment for risperidone based on CYP2D6 genotype for white and Asian people. Significant differences between the extensive metabolizer and intermediate metabolizer groups were observed for dose-adjusted risperidone level, 9-hydroxyrisperidone level, and risperidone/9-hydroxyrisperidone ratio, but not for the total active moiety. Meta-analysis showed that significant differences were observed among the four phenotype groups, including steady state concentration, peak risperidone concentration, and the area under the curve, using the Kruskal-Wallis test. No differences were found in oral clearance. For risperidone, dose recommendations for poor and ultrarapid metabolizers of CYP2D6 for Asians were different compared to that for white people for poor metabolizers (dose adjustment around 45% for white people, while for Asians the risperidone dose should be reduced by 26%). For ultrarapid metabolizers, risperidone dose should be increased by about 33% for white people and 30% for Asians. This was a first attempt to apply pharmacogenetics to suggest dose-regimens for Asian people; further research to replicate and extend these findings is recommended.
Risperidone (RIS) is an atypical antipsychotic (AAP) drug that is prescribed for the treatment of autism, schizophrenia, and acute bipolar mania, which is metabolized by the CYP2D6 enzyme in the liver to its major active metabolite, 9-hydroxyrisperidone (9-OH-RIS, also known as paliperidone). Over 100 allelic variants of the CYP2D6 gene have been reported (https://www.pharmvar.org/gene/CYP2D6). These give rise to four different metabolizer phenotypes (enzyme activity levels) based on the sum of an activity score that is attributed to each allele including: (1) poor metabolizers (PM) without enzyme activity (activity score 0); (2) intermediate metabolizers (IM) with reduced enzyme activity (activity score 0–1.25); (3) normal metabolizers (NM) with normal activity (activity score 1.25–2.25); and (4) ultrarapid metabolizers (UM) with increased enzyme activity (activity score >2.25) (Caudle et al., 2020). Although it was believed that the sum of the plasma risperidone and 9-hydroxyrisperidone concentration (known as the “active moiety”) is responsible for the therapeutic response to, and adverse effects of, risperidone, there are prior data that indicate switching from risperidone to another drug that is higher in UMs and PMs (Jukic et al., 2019). The plasma concentration of risperidone may range from subtherapeutic levels in the UM group to supratherapeutic and potentially toxic concentrations in the PM group, thereby (if RIS and 9-OH-RIS differ in adverse effect profile) potentially increasing the possibility of adverse effects.
In previous studies, the plasma concentration of risperidone and the risperidone/9-hydroxyrisperidone ratio in patients varied by CYP2D6 predicted phenotype group, suggesting that the determination of an accurate CYP2D6 genotype-predicted phenotype may be helpful for the individualization of drug therapy in a clinical setting (van der Weide and van der Weide, 2015; Vanwong et al., 2016).
Risperidone dose adjustments were recommended based on CYP2D6 metabolic phenotype (Kirchheiner et al., 2001; Kirchheiner et al., 2004; Kirchheiner et al., 2005). As significant differences in CYP2D6 allele frequencies have been observed between different populations (Gaedigk et al., 2017), dose recommendations presented in previous studies may not be applicable to East Asian populations, such as the Chinese. In a study by Stingl et al. (Stingl et al., 2013), an updated version of pharmacogenetic-based therapeutic recommendations for risperidone was presented, integrating previous reviews (Kirchheiner et al., 2001; Kirchheiner et al., 2004; Kirchheiner et al., 2005) of studies on the pharmacogenetics of psychotropics that were published before 2010. In their analysis, nine studies were included (eight Caucasian, and one Korean study). Pharmacokinetic studies often have small sample sizes with limited precision in parameter estimates; it is therefore necessary to perform systematic reviews and meta-analyses to provide more robust estimates. We aimed to contribute to the data available on Asian people (specifically, Chinese) and to review data to date in order to develop risperidone dose recommendations for Asians.
The outpatients who were followed up at Peking University Sixth Hospital (Beijing, China) were recruited in 2010-2011. In addition, the present study was a part of the Chinese Antipsychotics Pharmacogenomics Consortium (CAPOC) project (Yu et al., 2018). Individuals included in our study were diagnosed with schizophrenia based on the Structured Clinical Interview for DSM-IV, were of North Han Chinese ancestry, scored more than 60 on the Positive and Negative Syndrome Scale (PANSS) (and scored more than four on at least three positive items), were physically healthy, with all laboratory parameters within normal limits, had a condition that could be treated with oral medication, and were able to provide informed consent. Both first-episode and relapsed patients with schizophrenia were enrolled from inpatient departments of psychiatric hospitals affiliated with CAPOC. Patients were excluded from the study if they were diagnosed with schizoaffective disorder, delusional disorder, brief psychotic disorder, schizophreniform disorder, psychosis associated with substance use or medical conditions, learning disability, pervasive developmental disorder, delirium, dementia, amnesia, or other cognitive disorders. In addition, patients were excluded if they had severe, unstable physical diseases (such as diabetes, thyroid diseases, hypertension, and cardiac diseases), malignant syndrome or acute dystonia, documented histories of epilepsy or febrile convulsions, a DSM-IV diagnosis of alcohol or drug dependence, a history of drug-induced neuroleptic malignant syndrome, required long-acting injectable medication to maintain treatment adherence, were regularly treated with clozapine for treatment resistance during the past month (patients who had taken clozapine for reasons other than treatment resistance were eligible), were treated with electroconvulsive therapy (ECT) during the previous month, had previously attempted suicide, or had experienced symptoms of severe excitement and agitation, had abnormal liver or renal function (i.e., aspartate aminotransferase ≥80 U/L, alanine aminotransferase ≥80 U/L, blood urea nitrogen ≥9.75 mmol/L, urine creatinine ≥21.6 mmol per day), did not have a legal guardian (there was a hospital stipulation that written informed consent was required from the patient’s legal guardian), had QTc prolongation, a history of congenital QTc prolongation, or recent (i.e., within the past six months) myocardial infarction, were pregnant or breastfeeding women, or had a contraindication to any of the drugs to which they could be assigned. Patients who were already taking antipsychotic medications were obliged to switch to risperidone within one week, and adjusted drug dosages based on treatment effectiveness within two weeks (risperidone from 2 mg to 6 mg per day). The dosage then remained unchanged throughout the study. Patients receiving a risperidone-based regimen for more than four weeks were enrolled. Patients were excluded if they were unable to take medicine regularly or if they needed to change the medication. No combination of any other antipsychotics, antidepressants, antianxiety medications, or mood stabilizers was allowed during the study period. ECT therapy was prohibited. Participants were advised not to take drugs known to induce liver enzymes (e.g., rifampicin, carbamazepine, phenobarbital, phenytoin, etc.) for two weeks prior to enrollment and throughout the study period. Participants had to minimize usage of aspirin and non-steroidal anti-inflammatory drugs throughout the study period. Systematic psychotherapy was not permitted for the duration of the study. The study was approved by the institutional ethics review boards of the Peking University Sixth Hospital (Beijing, China), and written informed consent was obtained.
Blood samples were collected from a total of 130 patients after 4-6 weeks of treatment with a stable dose of risperidone, at 8.00 a.m., before the antipsychotic morning dose, 12 hours after the bedtime dose. Blood was collected in EDTA tubes for genotypic analysis and measurement of steady-state plasma concentrations of risperidone and 9-hydroxyrisperidone.
DNA was extracted using the Wizard Genomic DNA purification Kit (Promega). Genotyping was performed using the iPLEX™ Assay with a Sequenom MassARRAY®-SNP platform (CapitalBio Technology Co., Ltd.). The polymorphisms detected were as follows: 1708delT, 886C>T, 1022C>T, 1847G>A, 2540delAACT, 100C>T, 1612T>A, 2989G>A, 77G>A, 3202C>T, 882G>C, 2951G>C, 1977_1978insG, 2550delAl, 1759G>A, 2616delAAG, 2936A>C, representing the following alleles: normal or uncertain function *1, *2,*43; decrease of function *9,*10, *14,*17, *41, *49; loss of function *3, *4, *6, *7, *11, *19, *20, *44, *56. CYP2D6 gene copy number variation was derived from Illumina Human OmniZhongHua BeadChips, which were designed for the Chinese population, using the cnvpartition-cnv-analysis-plugin-v3.2.0 for GenomeStudio 2.0.
The CYP allele designations refer to those defined by the Pharmacogene Variation Consortium (https://www.pharmvar.org/gene/CYP2D6) (Gaedigk et al., 2019). The activity score was computed and the phenotype was extrapolated based on the genotypic data (Supplement 1, Tables S1-1, and S1-2).
The elimination half-lives of risperidone and 9-hydroxyrisperidone have been reported to be 3–29 h and 20–29 h, respectively (Huang et al., 1993; Yasui-Furukori et al., 2003). Therefore, plasma concentrations of these compounds had already reached a steady state in all participants before the blood samples were taken.
Plasma concentrations of risperidone and 9-hydroxyrisperidone were measured by a high-performance liquid chromatography tandem mass spectrometry coupled with an online solid-phase extraction method that could be used to measure risperidone, paliperidone, and olanzapine using small (40uL) samples as described (Ruan et al., 2018). The intra- and inter-assay coefficients of variation (CV) were 2.2% and 4.8% respectively. We first extracted the analytes from plasma samples and then pre-concentrated and purified it by C8 (5 μm, 2.1 × 30 mm) solid-phase extraction cartridges. Then the analytes were chromatographed on an Xbidge™ C18 column (3.5 μm, 100 × 2.1 mm), and analyzed by tandem mass spectrometry.
We searched the following databases: PubMed, Embase, the Cochrane Library, the China National Knowledge Infrastructure (CNKI), and the Wanfang database, including all articles published up to Oct 8, 2019. The search strategy was based on combinations of the keywords “CYP2D6” AND “polymorphism” AND “risperidone OR ris.” Reference lists of retrieved articles or previous meta-analyses were manually checked to identify any further eligible studies.
Two independent investigators screened titles and abstracts for relevance to the topic independently. The remaining potential publications were evaluated by reading the full text. Retrieved studies were included if they met the following criteria: 1) observational or clinical trial design, excluding abstracts and conference literature; 2) conducted in individuals (healthy volunteers or patients); 3) included measurements of the pharmacokinetic parameters of risperidone, such as the area under the curve (AUC), total clearance(Cl), and steady-state concentration (Css); and 4) determined CYP2D6 polymorphism. Review articles and studies involving nonhuman participants and in vitro experiments were excluded. When the results of a study were reported in further interim analyses, only the most recent, complete, and updated data were included in this meta-analysis.
For each article, the following information was extracted from the article using a systematic extraction form: the first author’s name, year of publication, country or geographical origin of investigation, study design, follow-up, treatment, comparison, number of patients, and the outcome measurements. Any disagreements were resolved by discussion with a third investigator. In trials in which data were presented as the median (IQR) or median (range), we contacted the authors to request the raw data. In cases where we did not receive a response, the mean (SD) was calculated according to the 68–95–99.7 rule (Pukelsheim, 1994), also known as the Empirical rule, which states that the so-called three-sigma rule of thumb expresses a conventional heuristic that nearly all values may be taken to lie within three standard deviations of the mean. Even for non-normally distributed variables, at least 88.8% of cases should fall within a properly calculated three-sigma interval. For unimodal distributions, the probability of being within the three-sigma interval is at least 95% (Vysochanskij, 1980). The data in our meta-analysis were pharmacokinetic parameters adjusted by dosage, such as steady state dose-normalized plasma risperidone concentration (Css/dose), peak dose-normalized plasma risperidone concentration (Cmax/dose), and dose-normalized area under the plasma concentration (AUC/dose). If the extracted data were not presented as dose-adjusted, data were divided by the dose presented in the paper. For some raw data, the units were adjusted to facilitate consistency in the analysis.
Phenotypes, or the metabolizer group classification of CYP2D6, were assigned to participants depending on the combination of the two identified alleles. The “allele and gene activity score” system is one of the most popular classification systems (Caudle et al., 2020). Table 1 shows the details of our data using this system. Individuals were classified into four different metabolic phenotypes according to predictive enzymatic activity, as described above. Based on the algorithm presented by Gaedigk et al. (2008, 2017) and Borges et al. (2006), participants were defined as normal metabolizers (NM) if two fully functional alleles were present or if they carried one reduced-function allele in combination with a fully functioning allele (AS:1.25-2.25), or intermediate metabolizers (IM) if they carried two reduced-function alleles or one fully functional and one nonfunctional allele, or one reduced and one nonfunctional allele (AS:0-1.25). In addition, poor metabolizers (PM) carried two nonfunctional alleles (AS=0); ultrarapid metabolizers (UM) were defined as having at least one extra copy of a functional allele (AS>2.25) (Caudle et al., 2020). No UMs or other structural variants were detected.
Data are presented as the median (IQR). Descriptive statistics were used to describe the clinical characteristics of all participants. After testing for normality, a Mann-Whitney U test was employed for between group comparisons. P values < 0.05 were considered nominally statistically significant (with no adjustment for multiple testing). Statistical analyses were performed using SPSS version 21.0 (SPSS Inc., Chicago, IL, USA).
Dose adjustments were computed according to a method described previously (Kirchheiner et al., 2004; Stingl et al., 2013). In brief, dose adjustments were based on differences in pharmacokinetic parameters, including oral clearance, area under the concentration time curve, and concentration at steady state, observed between phenotype groups.
For CYP2D6, about 0.5% of the Chinese population are poor metabolizers, 32.5% are intermediate, and 66.5% are normal metabolizers (Gaedigk et al., 2017). Thus, in general, the average dose (Dav) that is recommended in Asian populations is as follows:
where DPM, DIM, and DNM represent the optimal dose for the groups of poor metabolizers, intermediate metabolizers, and normal metabolizers, respectively.
In white people, about 7% are poor metabolizers of CYP2D6, 9% are intermediate, 82% normal metabolizers, and 2% ultrarapid metabolizers (Del Tredici et al., 2018). Thus, the average dose (Dav) that is recommended in white people may be calculated as follows:
It is hypothesized that the relationship between relative dose adjustments and relative changes of pharmacokinetic parameters observed in the phenotype groups is linear; therefore, dose adjustments for the intermediate and the poor metabolizer groups DIM and DPM were calculated according to equations 2 and 3 of Kirchheiner et al. (2004):
Assuming the average dose (Dav) is 100%, then the adjusted dose for the NM group, DNM, for CYP2D6 in Asian people is as follows:
where ClPM, ClIM, and ClNM are the empirically determined clearances for the respective phenotype groups.
For the computation of the dose adjustments in ultrarapid metabolizers (DUM) - if PK data were available - the percent dose was calculated as follows:
If no data on the UM group were available, the UM dose was computed based on the following linear relationship assumption:
For AUC or concentration of steady state, the calculation of dose recommendations for each phenotype group was similar to the above.
Statistical analysis was performed using STATA version 12.0 (StataCorp LLC, College Station, TX, USA). Due to substantial heterogeneity between studies, pooled pharmacokinetic parameters were calculated using a random-effects model. The inverse variance method was used for weighting studies. The heterogeneity between studies was formally assessed by the I2 statistic. Pre-planned subgroup analyses were conducted to examine whether effects of CYP2D6 phenotype differed comparing Asian to white people. Comparison of the means of pooled pharmacokinetic parameters between metabolizer phenotype groups was performed by a Kruskal-Wallis test, as the data were not normally distributed, using GraphPad Prism 7.00 (GraphPad, La Jolla, CA, USA) software. A P-value of <0.05 was considered nominally significant.
The total number of north Han Chinese patients included in this study was 130, including 78 males and 52 females with schizophrenia aged 30.6 (18-46) years old, not known to be related to each other. The allele frequencies of CYP2D6*1, *2, *10, *19, and *41 in the 130 participants analyzed were 25.0%, 12.7%, 51.5%, 4.2%,and 2.3%, respectively. These findings were similar to the data presented in previous studies (Lee et al., 2009; Yoo et al., 2011). The CYP2D6*3, *7, *9, *11, *14, *20, *44, and *56 alleles were not detected in any of the participants included in the current study. Phenotype extrapolation from CYP2D6 genotype resulted in 71 (54.6%) and 59 (45.4%) patients classified into the NM and IM groups, respectively. Frequencies of each CYP2D6 genotype and the corresponding inferred phenotype are presented in Table 2.
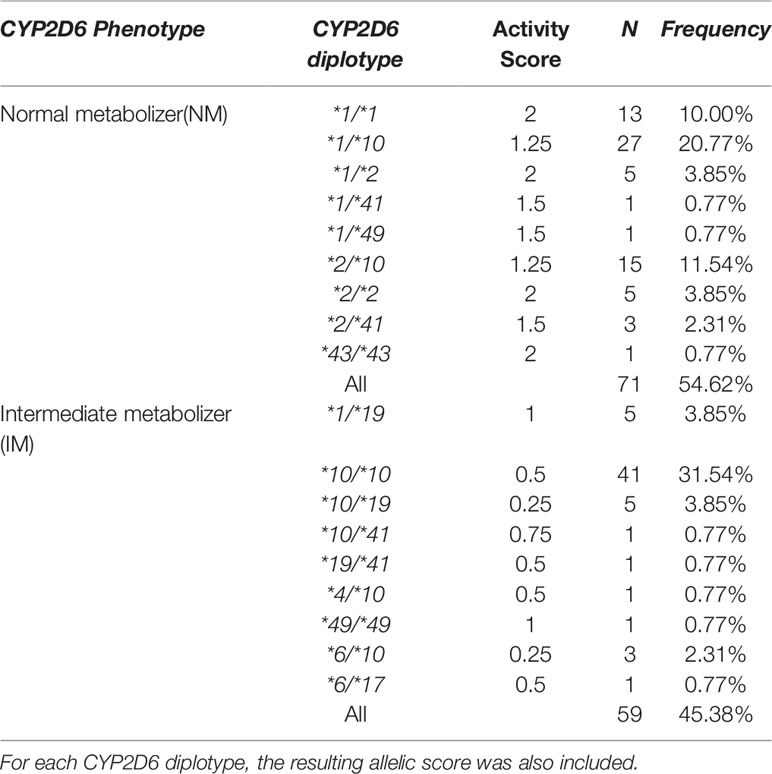
Table 2 Frequencies of CYP2D6 phenotypes and corresponding CYP2D6 diplotypes identified in the present study.
Of the 130 participants, for 38 there were no plasma risperidone levels available. There was no significant difference between the demographic characteristics of these missing participants and the other 92 participants (data not shown). The median daily baseline dosages of risperidone were 6mg and 5mg in the NM and IM group, respectively, as shown in Table 3, with no significant between-group difference(p=0.275). The nonparametric test for pairwise comparisons between groups identified significant differences between the NM and IM groups for dose-adjusted risperidone concentration (p=0.003), 9-hydroxyrisperidone concentration (p=0.001), and risperidone/9-hydroxyrisperidone ratio (p<0.001), as shown in Figure 1 and Table 3. As CYP2D6*10 has a high frequency in Asian people and is associated with decreased enzyme activity, an additional head to head comparison was made between *1/*1, *1/*10, and *10/*10 patients for risperidone pharmacokinetic parameters (Table S1-3 and Figure S1).
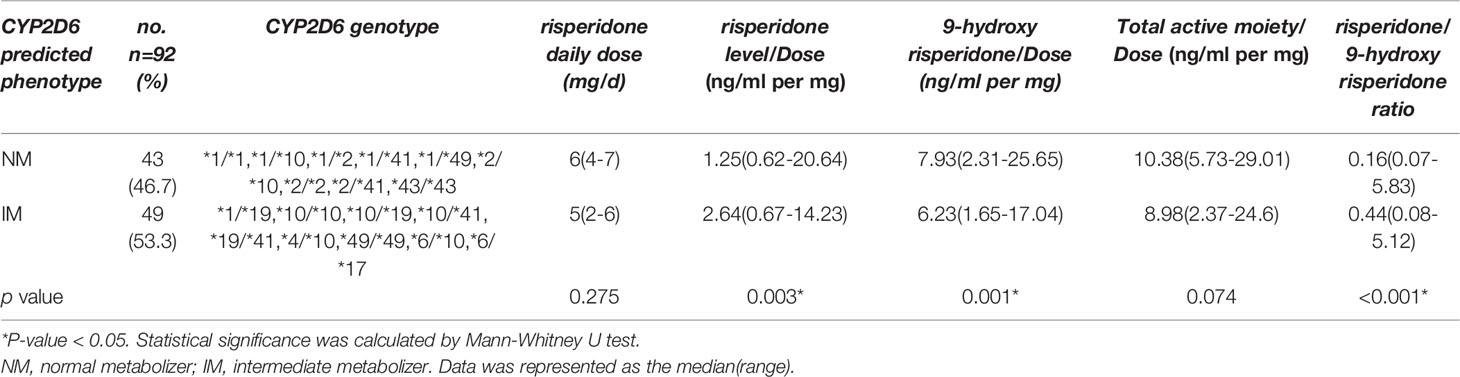
Table 3 The relationship between CYP2D6 predicted phenotype and risperidone daily dose(mg/d), and dose-adjusted concentrations of risperidone, 9-hydroxyrisperidone, total active moiety, and risperidone/9-hydroxyrisperidone ratio (n =92).
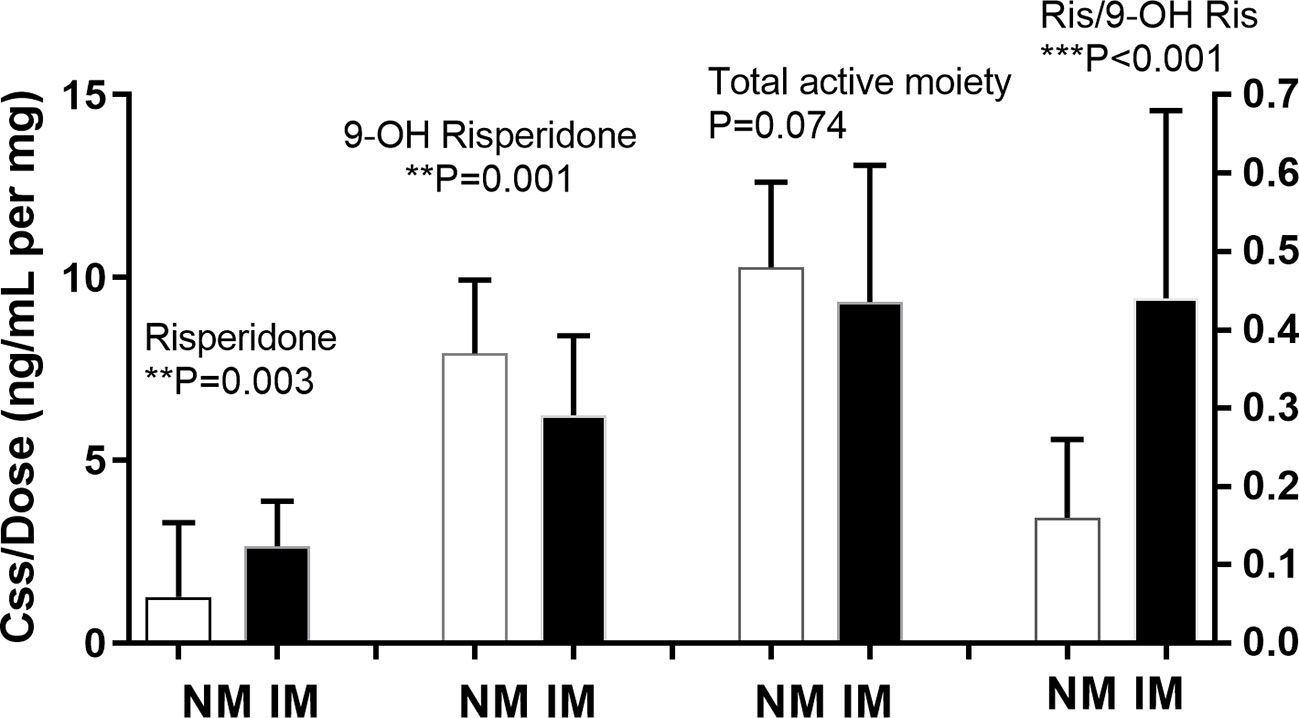
Figure 1 Relationship between CYP2D6 predicted phenotype and risperidone level, 9-hydroxyrisperidone level, total active moiety level, and risperidone/9-hydroxyrisperidone level. Bars represent the median (interquartile range). The metabolic ratio was plotted on the right Y-axis. NM, normal metabolizer; IM, intermediate metabolizer; RIS, risperidone; 9-OH RIS,9-hydroxyrisperidone; RIS/OHRIS, risperidone/9-hydroxyrisperidone ratio. **means p < 0.01, ***means p < 0.001.
Among the 130 patients, 77 patients were prescribed benzhexol to alleviate extrapyramidal-related symptoms, and 73 patients were given benzodiazepines, such as clonazepam or alprazolam, as hypnotics. No other potential CYP inhibitors were taken.
A systematic review of the literature identified 28 independent studies that met the pre-defined search criteria. Twenty-nine studies were included in our meta-analysis, with a total sample size of 2624 participants (Figure 2).
The pooled pharmacokinetic data are shown in Table 4 and Figure 3. For risperidone, significant differences were observed among the four phenotype groups for the following parameters: the steady state concentration [Css, p<0.0001], peak risperidone concentration [Cmax, p=0.0054], and area under the curve [AUC, p=0.0003], using a Kruskal-Wallis test. No significant between group difference was observed for the oral clearance [CL/F, p=0.5100]. For 9-OH-RIS, a significant difference was found only in the Cmax [p=0.0005]. For the total active moiety, a significant difference was found in the Css[p=0.0488], with no significant difference for the Cmax or AUC (there was only one paper reporting the CL/F of the active moiety of risperidone, therefore it could not be pooled). Subgroup analyses were conducted to examine whether the effects of CYP2D6 phenotype differed between Asian and white people. However, owing to limited data available for Cmax, AUC, and CL/F, subgroup analysis by ethnicity was conducted only for Css. For pooled analysis of the pharmacokinetics of 9-OH-RIS and the activity moiety stratified by CYP2D6 phenotype, see Supplement 2 (Table S2 and Figure S2).
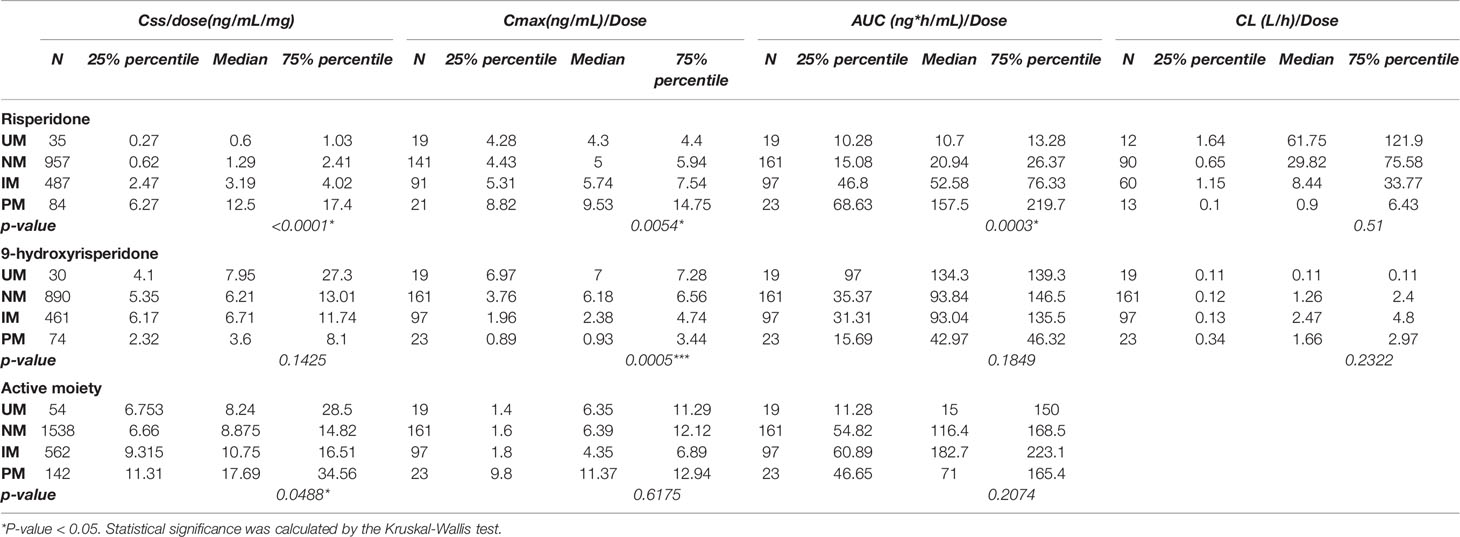
Table 4 Pooled analysis for pharmacokinetics of risperidone, 9-hydroxyrisperidone, and the activity moiety stratified by the CYP2D6 phenotype.
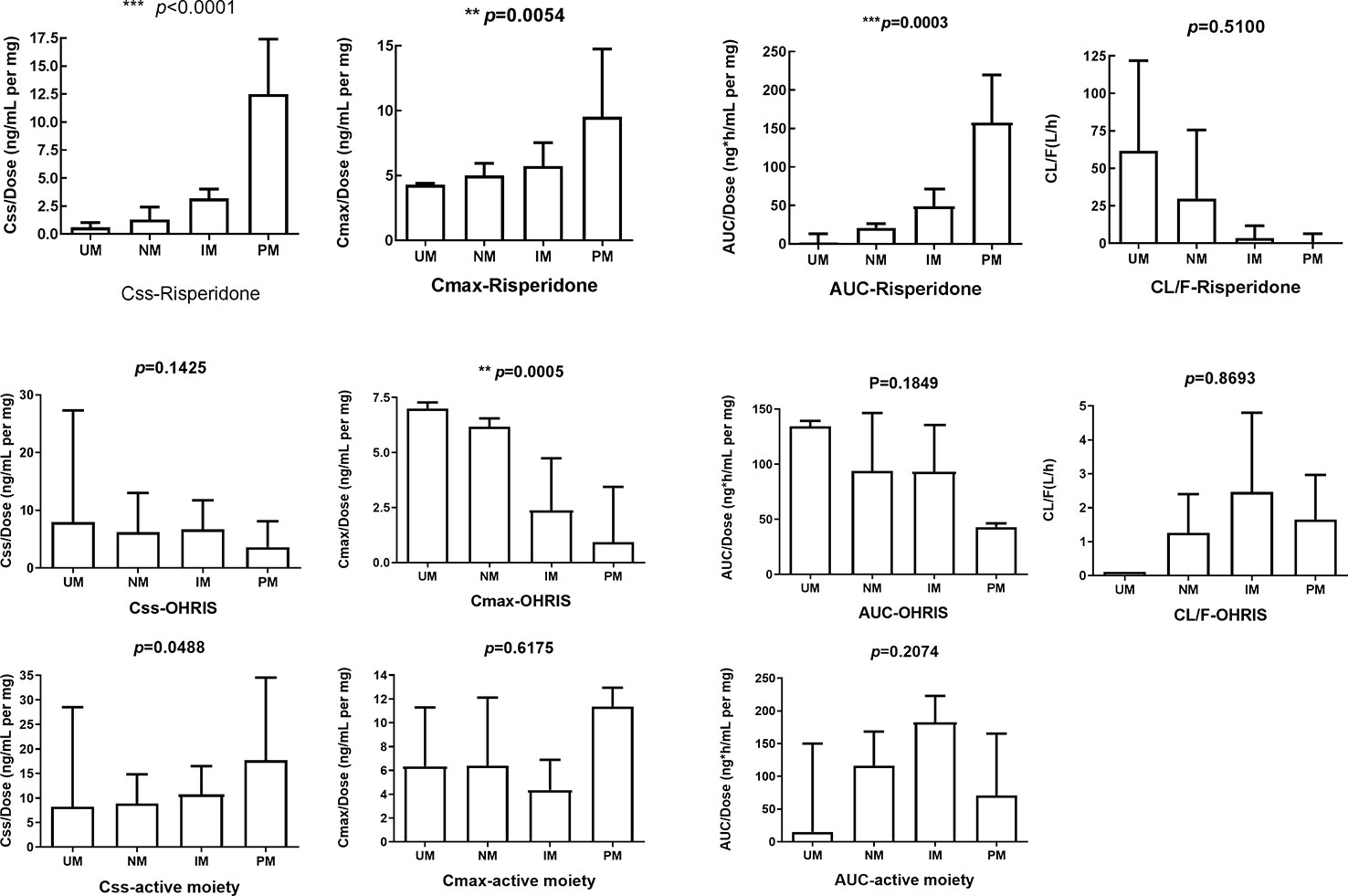
Figure 3 Effects of CYP2D6 metabolizer phenotype on selected pharmacokinetic parameters. All parameters, except apparent oral clearance, were dose-adjusted. The first row represents pharmacokinetic parameters for risperidone, the second for 9-hydroxyrisperidone, and the third for active moiety of risperidone. There was only one paper reporting the CL/F of the active moiety of risperidone, therefore, this was not pooled. Css, Steady state dose-normalized; Cmax, peak dose-normalized plasma; AUC, dose-normalized area under the plasma concentration; CL/F, apparent oral clearance. Error bars represent standard deviations. **means p < 0.01, ***means p < 0.001.
A meta-analysis of the data presented in Table 4 demonstrated a significant effect in all pharmacokinetic (PK) parameters except for clearance. Because the normal metabolizers (NM) represented the majority of the population, NM was set as the reference group. When comparing the total active moiety of risperidone between ultrarapid metabolizers (UM) and NMs, a significant difference was found in Css/dose (mean difference[90% CI]: -4.12[-7.50, -0.75], p=0.02). Moreover, when comparing intermediate metabolizers(IM) and NM, significant differences were found in Css/dose (mean difference[90% CI]:-2.48[-3.70,-1.27], p<0.0001) and AUC/dose (mean difference[90% CI]:-20.68[-23.50,-17.86, p<0.00001). In addition, when comparing poor metabolizers (PM) with NM, the same trend was observed and nominally significant differences were found in the Css/dose (mean difference[90% CI]:- 7.17[-13.48,-0.87], p=0.03) and AUC/dose (mean difference[90% CI]:-19.85[-36.69,-3.01], p=0.02). For Forest plots, see Figures S3-1–S3-9).
Information on the four CYP2D6 phenotypic groups for all human studies found for risperidone is shown in Table S4 (Supplement 4), with the percent dose adjustment calculated based on pharmacokinetic parameters (oral clearance, area under the concentration time curve, and concentration at steady state) observed by phenotype group. For the UM group, extrapolation was performed assuming a linear gene–dose effect according to the calculation methods given above. If possible, data on the active moiety of the drug (sum of parent drug and active metabolite) after multiple dosing were taken, and in cases where more studies existed, mean dose adjustments were calculated. Good concordance of the quantitative effects on pharmacokinetic parameters existed between various studies. If the concentration of the active moiety was known, dose recommendations were based on this. Thus, in summary for Asian people, we suggest the risperidone dose should be reduced by 26% for PM, and increased by 30% for UM, while for white people, for PMs the dose should be reduced by 45% and for UMs increased by 33%.
Risperidone is an active compound that is mainly metabolized in the liver by cytochrome P450 enzymes such as CYP2D6. An active metabolite in the main hydroxylation pathway is 9-hydroxyrisperidone. Another minor metabolic pathway involves N-dealkylation (Fang et al., 1999). In a previous in vitro study, three CYP enzymes were identified, CYP2D6, CYP3A4, and CYP3A5, which showed risperidone to 9-hydroxyrisperidone metabolizing activities, at 7.5, 0.4, and 0.2 pmol pmol–1 CYP min–1, respectively (Fang et al., 1999). In in vivo studies, Yasui- Furukori et al. (Yasui-Furukori et al., 2001) demonstrated that CYP2D6 played a predominant role in the formation of (+)-9-hydroxyrisperidone, while CYP3A4 appeared to be primarily involved in the formation of (-)-9-hydroxyrisperidone. These findings were confirmed by using inhibitors of CYP2D6 (quinidine) and CYP3A4 (ketoconazole) to inhibit the formation of 9-hydroxyrisperidone (Fang et al., 1999). Thus, 9-hydroxyrisperidone is the major active metabolite of risperidone, and CYP2D6 is the major enzyme involved in its formation, although CYP3A4 also seems to contribute.
In this study, we first investigated the influence of CYP2D6 polymorphism on the pharmacokinetics of risperidone in 130 Chinese outpatients. According to results presented previously (Johansson et al., 1994; Yoo et al., 2011), Asian populations show a high frequency (about 50%) for the CYP2D6*10 allele and a low frequency of nonfunctional CYP2D6 alleles. In our study, the allele frequency of CYP2D6*10 was 51.5%, which is similar to that presented previously. As shown in a meta-analysis (Gaedigk et al., 2017), over 99% of Asians are normal metabolizers (66.5%) and intermediate metabolizers (32.5%). In our study, 71 out of 130 (54.62%) patients were NM and 59 out of 130 (45.38%) were IM, with no participants in the PM or UM groups. However, it should be noted that we did not use a method of directly identifying Ums but rather inferred these from array data, and therefore it is possible that there were patients who were UMs or with other structural variants of CYP2D6 that were not identified in our sample.
For risperidone pharmacokinetics in the Chinese sample, significant differences were observed between the NM and IM groups for the dose-adjusted concentrations of risperidone (p=0.003), 9-hydroxyrisperidone (p=0.001), and the risperidone/9-hydroxyrisperidone ratio (p<0.001). Our meta-analysis results were consistent with this. In risperidone-treated patients, the RIS/9-OH-RIS concentration ratio is an index of CYP2D6 activity and the concentration-to-dose (C/D) ratio, in which C includes RIS+9-OH-RIS, is an index of the active moiety and its total clearance from the body (de Leon et al., 2010). Our results indicated that the NM and IM groups had different CYP2D6 activity but did not show significant differences in total clearance from the body. The pooled analysis and meta-analysis provide more robust data regarding the effects of different CYP2D6 metabolizer phenotypes on the pharmacokinetics of risperidone in white and Asian people.
The US Food and Drug Administration (FDA) (https://www.accessdata.fda.gov/scripts/cder/daf/index.cfm?event=overview.process&varApplNo=210655) appears to have considered that, as the active moiety was the sum of risperidone and 9-hydroxyrisperidone, and CYP2D6 metabolizer status would be predicted to merely change the ratio between these two but not the total of the two, there was no need for a recommendation for patients to undergo cytochrome P450 (CYP) PGx testing when initiating risperidone. Similarly, the Royal Dutch Pharmacists Association - Pharmacogenetics Working Group (DPWG) 2018 update did not recommend that any action was required for this gene-drug interaction (https://www.pharmgkb.org/chemical/PA451257/guidelineAnnotation/PA166104943) Although Swen et al. (2011) concluded that there were “insufficient data to allow calculation of dose adjustment,” they also suggested selection of an alternative drug, vigilance for adverse drug events, and/or adjustment of doses according to clinical response for patients who were CYP2D6 poor metabolizers, intermediate metabolizers, or ultrarapid metabolizers (Swen et al., 2011). Subsequent works (Stingl and Viviani, 2015; Jukic et al., 2019) showed that patients in the PM or UM groups had a significantly higher rate of switching from risperidone to another medication. Jukic et al. (Jukic et al., 2019) analyzed data from 1288 patients treated with risperidone and found that CYP2D6 IMs and PMs had 1.4- and 1.6-fold higher exposure, respectively, to the active moiety for risperidone (i.e., risperidone plus 9-hydroxyrisperidone). The rate of switching to another medication was also higher in UMs and PMs than in NMs. Roos van Westrhenen et al. (2020) recommended reduction of risperidone dose by 33% in PMs and IMs on the basis of the results of Jukic et al. (2019). Our meta-analytic data support dose adjustments for risperidone based on CYP2D6 genotype for white people, and also suggest a greater reduction (45%) for PMs. This data also extends to Asian populations. We have computed dose adjustments as follows. In Asian populations, for PMs reduce the recommended dose by 26% and for UMs increase the dose by 30%. In white people, for PMs reduce the recommended dose by 45% and for UMs increase it by 33%.
As mentioned above, a limitation of our study was the method of derivation of CYP2D6 copy number. How UMs and those with structural variants are assessed is likely to be a limitation common to previous studies in this field. Therefore, the dose recommendation for Asian people who are UMs should be considered provisional, pending replication.
In this study, we also measured the clinical response for the 130 patients by Positive and Negative Syndrome Scale (PANSS) at the baseline and the end of two weeks, four weeks, and six weeks after taking the assigned antipsychotic medication. We did not observe any association between CYP2D6 polymorphism and the PANSS percent change in our sample of 130 Asian patients with schizophrenia (t=0.561, p=0.576 at 2 weeks; t=0.196, p=0.845 at 4 weeks; t=-0.129, p=0.898 at six weeks).
In this study, dose adjustments were computed assuming a linear relationship between relative dose adjustments and relative changes of pharmacokinetic parameters observed in the phenotype groups. While this may not be valid (Jukic et al., 2019), as we did not observe any UMs, the assumption of linearity could not be tested.
In our pooled analysis, we incorporated pharmacokinetic parameters, whether from a single-dose study (SD) data or from multiple dosing (MD), as well as from healthy volunteers or patients. Given that saturable pharmacokinetics, irreversible enzyme blockade, and enzyme up- or downregulation might change the outcome under multiple dosing or under different physical conditions, it is inappropriate for data from single-dose experiments to be extrapolated to long-term drug therapy, or, likewise, to extrapolate data from healthy individuals to patients (Kirchheiner et al., 2004). Several factors affect risperidone dosage, including patient-related factors (i.e. gender, age, ethnicity, and body mass index (BMI)), factors related to psychiatric illness (i.e. acute vs. chronic phase, illness severity) (Gardner et al., 2010) and concomitant administration of medication (i.e. CYP2D6 inhibitors, such as fluoxetine or paroxetine, and CYP3A4 inducers such as carbamazepine or phenytoin) (Spina and de Leon, 2015). Moreover, multigenic interactions and gene-environment interactions will have to be considered. Therefore, in the future, haplotypic analyses and multigenetic analyses with large sample sizes, and high-quality phenotypic data should be included. Furthermore, the raw data in our meta-analysis, presented as the median (range), were used to infer the mean (SD), and in some cases the units had to be converted for data harmonization. We acknowledge that this conversion might result in some measurement bias. For CYP2D6 phenotypic grouping, in a previous study, African‐American subjects with CYP2D6*1/*2 and *2/*2 genotypes had significantly lower activity compared with white people. It is also possible that a subgroup of CYP2D6*2 alleles are associated with reduced enzyme activity (Wang et al., 2014; Wang et al., 2015). A potential difference in activity of the *2 allele between Asian and white people might contribute to some of the differences among the enzyme activity of these ethnic groups. This might also result in a degree of dose adjustment being required in CYP2D6 phenotypes derived from CYP2D6*2.
Further research aiming to address the limitations that we have outlined above, particularly in regard to the UM group, is indicated. In order to implement routine pre-treatment genetic testing, additional parameters would need to be considered, including the effects of concomitant CYP2D6 inhibiting medications and CYP3A inducers, clinical utility, and cost-benefit analyses. In regard to clinical utility, we did not observe any association between CYP2D6 polymorphism and clinical response in our sample of 130 Asian patients with schizophrenia (as measured by the PANSS).
The original contributions presented in the study are publicly available. This data can be found here: https://doi.org/10.18170/DVN/MQB2Y3.
This study was carried out in accordance with the recommendations of the institutional ethics review boards, Peking University Sixth Hospital with written informed consent from all subjects. All subjects gave written informed consent in accordance with the Declaration of Helsinki. The protocol was approved by the institutional ethics review boards, Peking University Sixth Hospital.
YC and WY presented the ideas. YC and YS developed the theory and performed the computations. HY, LW, and TL collected the samples and carried out the experiment. DZ and WY supervised the project.
The study was funded by National Key R&D Program of China (2016YFC1307000, 2017YFC1311100); National Natural Science Foundation of China (81825009, 81901358, 81221002); Peking University Clinical Scientist Program supported by “the Fundamental Research Funds for the Central Universities”(BMU2019LCKXJ012); Academy of Medical Sciences Research Unit (2019-I2M-5-006); PKUHSC-KCL Joint Medical Research (BMU2020KCL001); Beijing Science and Technology Commission (D171100007017002).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We are greatly indebted to all the study participants without whom this study would not have been possible.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2020.00936/full#supplementary-material
Borges, S., Desta, Z., Li, L., Skaar, T. C., Ward, B. A., Nguyen, A., et al. (2006). Quantitative effect of CYP2D6 genotype and inhibitors on tamoxifen metabolism: implication for optimization of breast cancer treatment. Clin. Pharmacol. Ther. 80, 61–74. doi: 10.1016/j.clpt.2006.03.013
Caudle, K. E., Sangkuhl, K., Whirl Carrillo, M., Swen, J. J., Haidar, C. E., Klein, T. E., et al. (2020). StandardizingCYP 2D6 Genotype to Phenotype Translation: Consensus Recommendations from the Clinical Pharmacogenetics Implementation Consortium and Dutch Pharmacogenetics Working Group. Clin. Trans. Sci. 13, 116–124. doi: 10.1111/cts.12692
de Leon, J., Wynn, G., Sandson, N. B. (2010). The pharmacokinetics of paliperidone versus risperidone. Psychosomatics 51, 80–88. doi: 10.1176/appi.psy.51.1.80
Del Tredici, A. L., Malhotra, A., Dedek, M., Espin, F., Roach, D., Zhu, G., et al. (2018). Frequency of CYP2D6 Alleles Including Structural Variants in the United States. Front. Pharmacol. 9, 305. doi: 10.3389/fphar.2018.00305
Fang, J., Bourin, M., Baker, G. B. (1999). Metabolism of risperidone to 9-hydroxyrisperidone by human cytochromes P450 2D6 and 3A4. Naunyn-Schmiedeberg’s Arch. Pharmacol. 359, 147–151. doi: 10.1007/PL00005334
Gaedigk, A., Sangkuhl, K., Whirl-Carrillo, M., Klein, T., Leeder, J. S. (2017). Prediction of CYP2D6 phenotype from genotype across world populations. Genet. Med. 19, 69–76. doi: 10.1038/gim.2016.80
Gaedigk, A., Sangkuhl, K., Whirl-Carrillo, M., Twist, G. P., Klein, T. E., Miller, N. A. (2019). The Evolution of PharmVar. Clin. Pharmacol. Ther. 105, 29–32. doi: 10.1002/cpt.1275
Gardner, D. M., Murphy, A. L., O'Donnell, H., Centorrino, F., Baldessarini, R. J. (2010). International consensus study of antipsychotic dosing. Am. J. Psychiatry 167 (6), 686–693. doi: 10.1176/appi.ajp.2009.09060802
Huang, M. L., Van Peer, A., Woestenborghs, R., De Coster, R., Heykants, J., Jansen, A. A., et al. (1993). Pharmacokinetics of the novel antipsychotic agent risperidone and the prolactin response in healthy subjects. Clin. Pharmacol. Ther. 54, 257–268. doi: 10.1038/clpt.1993.146
Johansson, I., Oscarson, M., Yue, Q. Y., Bertilsson, L., Sjoqvist, F., Ingelman-Sundberg, M. (1994). Genetic analysis of the Chinese cytochrome P4502D locus: characterization of variant CYP2D6 genes present in subjects with diminished capacity for debrisoquine hydroxylation. Mol. Pharmacol. 46, 452–459. doi: 10.1002/med.2610140504
Jukic, M., Smith, R., Haslemo, T., Molden, E., Ingelman-Sundberg, M. (2019). Effect of CYP2D6 genotype on exposure and efficacy of risperidone and aripiprazole: a retrospective, cohort study. Lancet Psychiatry 6, 418–426. doi: 10.1016/S2215-0366(19)30088-4
Kirchheiner, J., Brosen, K., Dahl, M. L., Gram, L. F., Kasper, S., Roots, I., et al. (2001). CYP2D6 and CYP2C19 genotype-based dose recommendations for antidepressants: a first step towards subpopulation-specific dosages. Acta Psychiatr. Scand. 104, 173–192. doi: 10.1034/j.1600-0447.2001.00299.x
Kirchheiner, J., Nickchen, K., Bauer, M., Wong, M. L., Licinio, J., Roots, I., et al. (2004). Pharmacogenetics of antidepressants and antipsychotics: the contribution of allelic variations to the phenotype of drug response. Mol. Psychiatry 9, 442–473. doi: 10.1038/sj.mp.4001494
Kirchheiner, J., Fuhr, U., Brockmoller, J. (2005). Pharmacogenetics-based therapeutic recommendations–ready for clinical practice? Nat. Rev. Drug Discovery 4, 639–647. doi: 10.1038/nrd1801
Lee, S. J., Lee, S. S., Jung, H. J., Kim, H.-S., Park, S.-J., Yeo, C.-W., et al (2009). Discovery of novel functional variants and extensive evaluation of CYP2D6 genetic polymorphisms in Koreans. Drug Metab. Dispos. 37 (7), 1464–1470. doi: 10.1124/dmd.108.022368
Pukelsheim, F. (1994). The Three Sigma Rule, Vol. 48, No. 2 (May 1994), pp. 88-91 Published by: Taylor & Francis, Ltd. on behalf of the American Statistical Association Stable URL: Accessed: 09-10-2019 08:51 UTC. Am. Stat. 2, 88–91. doi: 10.2307/2684253
Ruan, C., Guo, W., Zhou, M., Guo, G., Wang, C., Li, W., et al. (2018). Quantitative determination of risperidone, paliperidone and olanzapine in human serum by liquid chromatography-tandem mass spectrometry coupled with on-line solid-phase extraction. Biomed. Chromatogr. 32 (7), e4209. doi: 10.1002/bmc.4209
Spina, E., de Leon, J. (2015). Clinical applications of CYP genotyping in psychiatry. J. Neural. Transm. (Vienna) 122 (1), 5–28. doi: 10.1007/s00702-014-1300-5
Stingl, J., Viviani, R. (2015). Polymorphism in CYP2D6 and CYP2C19, members of the cytochrome P450 mixed-function oxidase system, in the metabolism of psychotropic drugs. J. Intern. Med. 277, 167–177. doi: 10.1111/joim.12317
Stingl, J. C., Brockmoller, J., Viviani, R. (2013). Genetic variability of drug-metabolizing enzymes: the dual impact on psychiatric therapy and regulation of brain function. Mol. Psychiatry 18, 273–287. doi: 10.1038/mp.2012.42
Swen, J. J., Nijenhuis, M., de Boer, A., Grandia, L., Maitland-van der Zee, A. H., Mulder, H., et al (2011). Pharmacogenetics: from bench to byte--an update of guidelines. Clin. Pharmacol. Ther. 89 (5), 662–673. doi: 10.1038/clpt.2011.34
van der Weide, K., van der Weide, J. (2015). The Influence of the CYP3A4*22 Polymorphism and CYP2D6 Polymorphisms on Serum Concentrations of Aripiprazole, Haloperidol, Pimozide, and Risperidone in Psychiatric Patients. J. Clin. Psychopharm. 35, 228–236. doi: 10.1097/JCP.0000000000000319
van Westrhenen, R., Aitchison, K. J., Ingelman-Sundberg, M., Jukić, M. M. (2020). Pharmacogenomics of antidepressant and antipsychotic treatment: how far have we got and where are we going? Front. Psychiatry 11, 94. doi: 10.3389/fpsyt.2020.00094
Vanwong, N., Ngamsamut, N., Hongkaew, Y., Nuntamool, N., Puangpetch, A., Chamnanphon, M., et al. (2016). Detection of CYP2D6 polymorphism using Luminex xTAG technology in autism spectrum disorder: CYP2D6 activity score and its association with risperidone levels. Drug Metab. Pharmacok. 31, 156–162. doi: 10.1016/j.dmpk.2016.01.005
Vysochanskij, Y. I. P. (1980). Justification of the 3σ rule for unimodal distributions. Theory Probab. Math. Stat 21, 25–36.
Wang, D., Papp, A. C., Sun, X. (2015). Functional characterization of CYP2D6 enhancer polymorphisms. Hum. Mol. Genet. 24, 1556–1562. doi: 10.1093/hmg/ddu566
Wang, D., Poi, M. J., Sun, X., Gaedigk, A., Leeder, J. S., Sadee, W. (2014). Common CYP2D6 polymorphisms affecting alternative splicing and transcription: long-range haplotypes with two regulatory variants modulate CYP2D6 activity. Hum. Mol. Genet. 23, 268–278. doi: 10.1093/hmg/ddt417
Yasui-Furukori, N., Hidestrand, M., Spina, E., Facciola, G., Scordo, M. G., Tybring, G. (2001). Different enantioselective 9-hydroxylation of risperidone by the two human CYP2D6 and CYP3A4 enzymes. Drug Metab. Dispos. 29, 1263–1268.
Yasui-Furukori, N., Mihara, K., Kondo, T., Kubota, T., Iga, T., Takarada, Y., et al. (2003). Effects of CYP2D6 genotypes on plasma concentrations of risperidone and enantiomers of 9-hydroxyrisperidone in Japanese patients with schizophrenia. J. Clin. Pharmacol. 43, 122–127. doi: 10.1177/0091270002239819
Yoo, H., Lee, S., Kang, H., Cho, H., Lee, I., Lee, Y. (2011). Influence of ABCB1 genetic polymorphisms on the pharmacokinetics of risperidone in healthy subjects withCYP2D6 *10/*10. Brit. J. Pharmacol. 164, 433–443. doi: 10.1111/j.1476-5381.2011.01385.x
Keywords: cytochrome P-450, CYP2D6, polymorphism, pharmacogenetics, risperidone
Citation: Cui Y, Yan H, Su Y, Wang L, Lu T, Zhang D and Yue W (2020) CYP2D6 Genotype-Based Dose Recommendations for Risperidone in Asian People. Front. Pharmacol. 11:936. doi: 10.3389/fphar.2020.00936
Received: 27 April 2019; Accepted: 08 June 2020;
Published: 04 August 2020.
Edited by:
Katherine J. Aitchison, University of Alberta, CanadaReviewed by:
Vangelis G. Manolopoulos, Democritus University of Thrace, GreeceCopyright © 2020 Cui, Yan, Su, Wang, Lu, Zhang and Yue. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dai Zhang, ZGFpemhhbmdAYmptdS5lZHUuY24=; Weihua Yue, ZHJ5dWVAYmptdS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.