- 1Carol Davila University of Medicine and Pharmacy, Bucharest, Romania
- 2Center of Excellence in Robotics and Autonomous Systems, Military Technical Academy Ferdinand I, Bucharest, Romania
- 3Gastroenterology Department, Dr. Carol Davila Central Military Emergency University Hospital, Bucharest, Romania
Celiac disease (CD) is a chronic autoimmune disease that occurs in genetically predisposed individuals in whom the ingestion of gluten leads to damage of the small bowel. It is estimated to affect 1 in 100 people worldwide, but is severely underdiagnosed. Currently available guidelines require CD-specific serology and atrophic histology in duodenal biopsy samples for the diagnosis of adult CD. In pediatric CD, but in recent years in adults also, nonbioptic diagnostic strategies have become increasingly popular. In this setting, in order to increase the diagnostic rate of this pathology, endoscopy itself has been thought of as a case finding strategy by use of digital image processing techniques. Research focused on computer aided decision support used as database video capsule, endoscopy and even biopsy duodenal images. Early automated methods for diagnosis of celiac disease used feature extraction methods like spatial domain features, transform domain features, scale-invariant features and spatio-temporal features. Recent artificial intelligence (AI) techniques using deep learning (DL) methods such as convolutional neural network (CNN), support vector machines (SVM) or Bayesian inference have emerged as a breakthrough computer technology which can be used for computer aided diagnosis of celiac disease. In the current review we summarize methods used in clinical studies for classification of CD from feature extraction methods to AI techniques.
Introduction
Celiac disease (CD) is a systemic autoimmune disease driven by gluten ingestion in genetically susceptible individuals. At some point during their lifetime, some of the DQ2/DQ8 positive individuals become gluten intolerant and develop an autoimmune reaction in response to dietary gluten, leading to small bowel injury consisting in villous atrophy (VA) and crypt hyperplasia. Although it is one of the most common chronic digestive disorders, with prevalence rate of 1% worldwide (Ludvigsson et al., 2016), CD is severely underdiagnosed. This is due to the frequently mislabeling patients with irritable bowel syndrome, lack of awareness among medical professionals about the extra-intestinal presentations of the disease (Jinga et al., 2018) and missed opportunities to screen for CD such as first-grade relatives, high-risk groups and not least scoping the upper gastrointestinal tract for unrelated reasons. Un-diagnosed CD bears the risk of several complications (nutritional, fertility-related and even malignancy) and reduced quality of life (Fuchs et al., 2018). Although the diagnosis of adult CD is very clear cut (CD-specific serology and sampling of duodenal mucosa by upper gastrointestinal endoscopy) and access to diagnostic tools has improved considerably, CD remains heavily underdiagnosed, with only 15%–20% of patients being detected through current strategies.
In the setting of open-access endoscopy and increasing number of examinations worldwide (including on-demand procedures), some have considered using endoscopy as an opportunity for detection of unsuspected CD, by careful analysis of the small bowel mucosa and recognition of subtle markers of VA. In fact, a study has shown that up to 5% of CD patients have undergone a previous endoscopy examination in the years before the diagnosis, and this could be considered a missed opportunity to diagnose it earlier (Lebwohl et al., 2012). Thus, endoscopy can be viewed not only as a diagnostic tool to confirm the disease by tissue sampling, but also as a case-finding tool for CD. Some have even proposed random duodenal biopsies during all upper endoscopy examinations, but this has proven a low diagnostic yield for CD with a high burden for endoscopists and pathologists and lack of cost-effectiveness (Herrod and Lund, 2018). Thus, the interest has been changed over to a better selection of patients in whom biopsy sampling should be carried out and the way to do it is by detection of markers of VA during endoscopy. However, recognition of changes in the duodenal mucosa can be challenging, especially in the setting of patchy or mild disease (Balaban et al., 2015); in order to overcome the subjectiveness in detecting these endoscopic markers of VA and to better delineate the subtle mucosal changes seen in early CD, some have proposed the use of computer-based processing of endoscopic images for the detection of VA, which could trigger the examiner to perform biopsies for the diagnostic protocol of CD.
Endoscopy with biopsy is currently considered the gold standard for the diagnosis of adult CD. Computer-assisted systems for the diagnosis of CD could improve the whole diagnostic work-up, by saving costs, time and manpower and at the same time increase the safety of the procedure by avoiding biopsy sampling and prolonged sedation associated with the multiple biopsy protocol. Not least, this nonbiopsy protocol could translate into a longer life for the endoscope, by avoiding warn of the working channel of the scope. Also, the histological staging of biopsies is subject to a significant degree of intra- and interobserver variability (Weile et al., 2000; Corazza et al., 2007; Mubarak et al., 2011; Arguelles-Grande et al., 2012). A further limitation of the endoscopy biopsies for the diagnosis of CD is due to the possibly patchy distribution of CD (Bonamico et al., 2004; Hopper et al., 2007), areas affected by CD can be in the midst of normal mucosa. So, given the case that the biopsies would be targeted from areas of healthy mucosa, CD could be missed due to a sampling error. Therefore, observer independent diagnostic methods such as computer-assisted diagnosis systems are very useful to improve the accuracy of diagnosis.
From the first research focused on computer-assisted system in the context of automated diagnosis of CD which has started in 2008 (Vécsei et al., 2008), over 50 publications on the topic that are using spatial domain, transform domain, scale-invariant and and spatio-temporal features have appeared (Hegenbart et al., 2015) but artificial intelligence (AI), machine learning (ML) and deep learning (DL) have emerged as a breakthrough computer technology in this world of big data and computational power based on graphics processing units. In the field of medical images, the accumulation of enormous digital images and medical records drove a need for the utilization of AI to efficiently deal with these data, which also become fundamental resources for the machine to learn by itself. ML and AI techniques have played an important role in the medical field, supporting such activities as medical image processing, computer-aided diagnosis, image interpretation, image fusion, image registration, image segmentation, image-guided therapy, and image retrieval and analysis (Razzak et al., 2018; Yang and Bang, 2019). In this work, we try to give a comprehensive overview of the research focused on computer-assisted diagnosis of CD from classical features extraction to AI. Several image-processing techniques have been reported so far in the literature, with good diagnostic performance in discriminating CD patients from healthy controls. Applying these image-processing techniques could be used to select in real-time, during endoscopy, patients with high probability of CD, who warrant a full diagnostic work-up including small bowel biopsies. The purpose of our review is to summarize current evidence of computerized methods in detecting CD, according to their diagnostic accuracy.
Methods
We conducted the present research according to the principles of the preferred reporting items for meta-analysis protocol (PRISMA) (Moher et al., 2015).
A systematic search of the literature was carried out in September 2019 in PubMed (Medline) database, using the following search criteria: CD (Mesh) and terms referring to computer-aided detection by image processing – computer, digital, image processing, AI, DL, neural network, quantitative assessment or texture features. There were no restrictions set on the search with regard to article type, text availability or publication date.
Publications revealed through this search were assessed for consistency with the topic, according to their title and abstract, by the two first authors. Conflicts resulted from independent data extraction according to inclusion and exclusion criteria were resolved by consensus.
Studies included in our systematic review were required to meet the following inclusion criteria: (i) full-text paper available in English, (ii) original papers describing image-processing techniques for computer-aided diagnosis of CD. We excluded case-reports, reviews and descriptive papers without validation of methods described on CD patients. We also excluded papers referring to digital processing of histology images in diagnosing CD. References from the retrieved articles were also checked for possible match with the review topic, in order to identify potentially relevant publications that could have been missed on the initial search.
For each study included in the systematic review we recorded the following data: first author, year of publication, type of endoscopic image used, study population (CD cases and controls), method tested and diagnostic performance (sensitivity, specificity, diagnostic accuracy).
Results
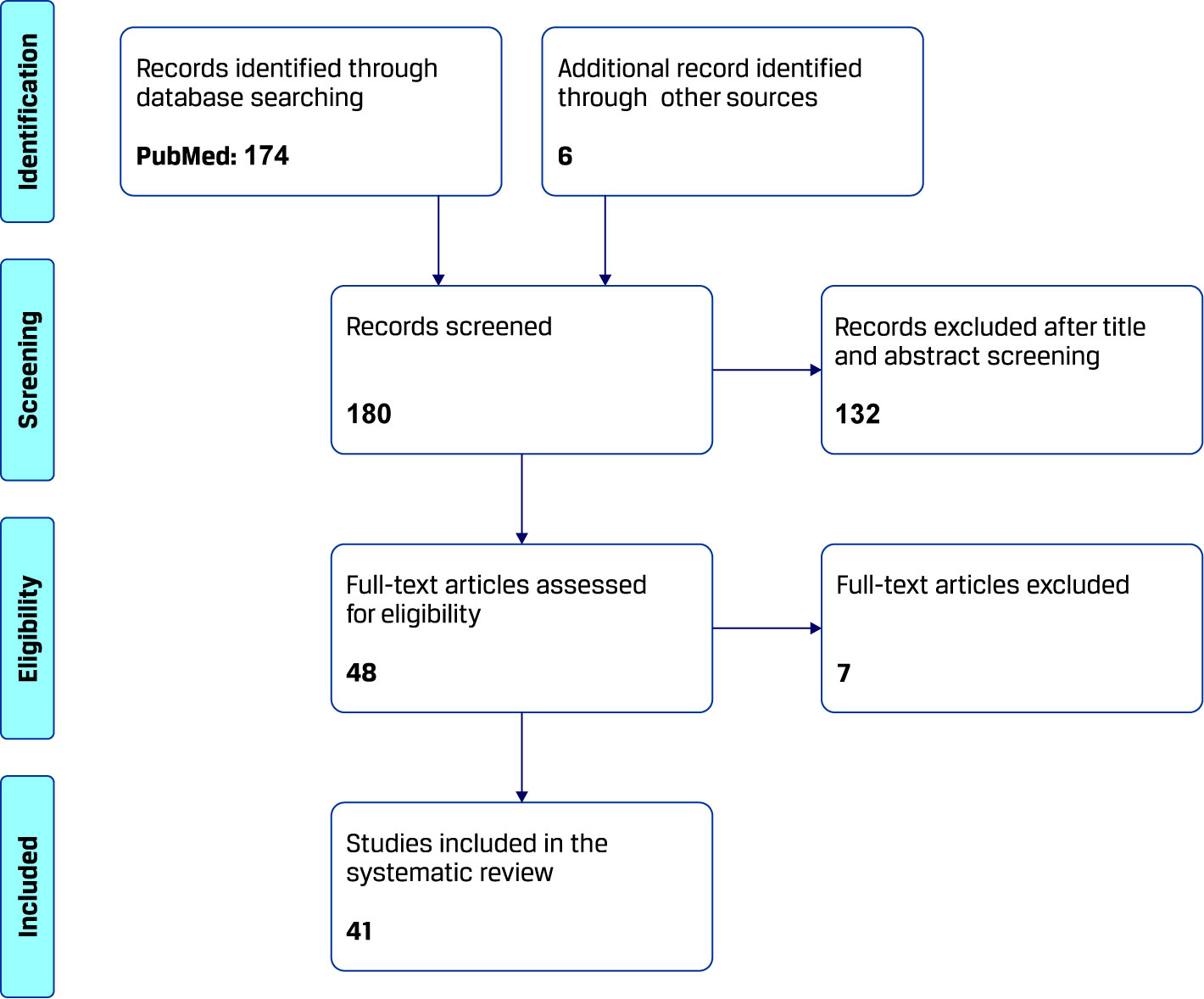
The process of study selection for this systematic review is summarized in Figure 1. The search yielded 174 results from 1970 onward, which were evaluated according to the above described methodology. Another six papers were found through other sources. A total of 139 papers were excluded because of irrelevance to the topic (confounding use of search words in the papers) or type of article (review/editorial) and the remaining 41 publications, consisting in original work describing image processing techniques for computer-aided diagnosis of CD, were analyzed for this review.
Among published papers, several techniques have been validated for the diagnosis of CD—first attempts were focused on features extraction methods used for classification such as spatial domain features, transform domain features, scale-invariant features and spatio-temporal features. More recent AI techniques DL methods such as convolutional neural network (CNN) have emerged as a breakthrough computer technology which can be used for computer aided diagnosis of CD. A statistic of common methods is presented, as well as an evaluation of their use in CD diagnosis.
Feature Extraction Methods
Table 1 summarizes the feature extraction methods used in clinical studies for the classification of the CD and the overall classification rates (OCR) [sensitivity (sens), specificity (spec) and accuracy (acc)]. Features can be classified into four main categories: spatio-domain features, transform domain features, scale-invariant features, and spatio-temporal features. Images used for feature extraction are obtained either by standard endoscopy or using video-capsule. While some of the studies reported the number of subjects analyzed (healthy and with CD), others have reported the number of full images and the number of subimages obtained as patches from the full images that were used for training and testing.
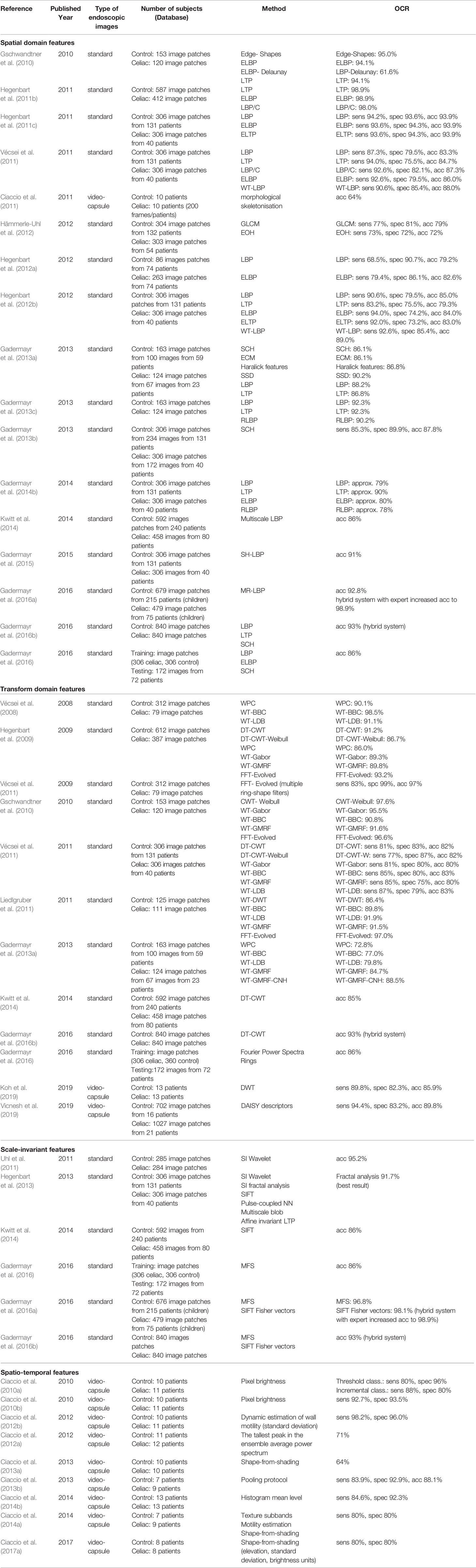
Table 1 Summary of features extraction methods used in clinical studies for classification of celiac disease (CD).
The spatial domain features that were used for CD classification are the edge shapes, shape curvature histograms (SCH), gray-level cooccurence matrix (GLCM), edge orientation histogram (EOH), local binary patterns (LBP) and extended LBP (ELBP), local ternary patterns (LTP) and extended LTP (ELTP), LBP in wavelet subbands (WT-LBP), rotation-invariant LBP (RLBP), LBP combined with a contrast measure (LBP/C), soft histogram LBP (SH-LBP), and multiresolution LBP (MR-LBP).
Transform domain features used in studies were pyramidal Wavelet transform (WPC), best-basis centroids base (WT-BBC), local discriminant basis (WT-LDB), Wavelet-based Gaussian Markov random fields (WT-GMRF), WT-GMRF with custom neighborhoods (WT-GMRF-CNH), dual-tree complex Wavelet transform correlation signature (DT-CWT), DT-CWT-Weibull, Gabor Wavelet transform (GWT), best-basis decomposition of Wavelet transform (WT-BBC), Fourier power spectra rings, and DAISY descriptors.
Common scale-invariant features are scale invariant wavelet based features (SI-WTF), scale invariant methods based on fractal analysis (SI-FA), scale-invariant feature transform (SIFT), multiscale blob features (MBF), affine invariant LTP (AILTP), multifractal spectrum (MFS), and SIFT Fisher vectors.
Among spatio-temporal features one can mention the pixel brightness, dynamic estimate of wall motility (Bassotti et al., 1994; Tursi, 2004) (standard deviation), periodicity in brightness, histogram mean level, shape-from-shading, pooling protocol, and statistical and syntactical measurements. Ciaccio et al. proposed some quantitative measurement (statistical and syntactical measurement, motility estimation) in video capsule endoscopy images in order to detect and measure the presence of VA in CD patients (Ciaccio et al., 2016a; Ciaccio et al., 2016b). Also in (Ciaccio et al., 2017b), Ciaccio et al. described and discussed methods used for quantitative detection and analysis of VA in the small intestinal mucosa of CD patients using video capsule endoscopy images but these remain to be further validated in larger samples (Ciaccio et al., 2017b).
Best results obtained using spatial domain features were obtained by Hegenbart et al. (Hegenbart et al., 2011b) by extracting various LBP from standard endoscopic images. In a database consisting of 999 image patches (587 control and 412 CD), overall classification rates varied between 98.04% and 98.93%.
In terms of transform-domain features best results were obtained by Vécsei et al. (2009) using FFT-evolved multiple ring-shape filters applied on standard endoscopic images. Database consisted of 390 image patches (312 control and 79 CD). The algorithm proved 97% accuracy, 83% sensitivity and 99% specificity in diagnosing CD.
Best results for scale-invariant features were obtained by Gadermayr et al. (2016a) using multifractal spectrum features and SIFT Fisher vectors extracted from standard endoscopic images. Database consisted of 676 control image patches and 479 CD image patches from 290 patients (all children). Performance of the method was 98.1% for SIFT Fisher vectors and 96.8% for multifractal spectrum features. When human knowledge was incorporated, performance increased to 98.9%.
Best results for spatio-temporal features were obtained by Ciaccio et al. (2012b) using dynamic estimate of wall motility (standard deviation) computed on video-capsule endoscopic images. Database consisted of 200 frames per patient extracted from 10 control patients and 11 CD patients. Diagnostic performance was high, with 98.2% sensitivity and 96% specificity.
Gadermayr et al. in (2015) compared different endoscopic image configuration [white-light imaging (WLI) (Gasbarrini et al., 2003) and narrow-band imaging (NBI) (Emura et al., 2008; Valitutti et al., 2014)] to find which image data are most accurate in case of computer aided CD diagnosis but in (Gadermayr et al., 2016a; Gadermayr et al., 2016b) same authors et al. showed that an hybrid system which incorporated expert knowledge in automated CD diagnosis increased the accuracy with 6% (see Table 1).
Spatial domain features were based on the similarity of specific features also observed by a human analyst; they are robust and fast in terms of computation, making them suitable for real time classification. Transform domain features have the advantage of analysing endoscopic images on multiscale and multiorientation levels. Common tools are based on Wavelet transform, Fourier transform and Gabor transform. Scale-invariant features are suitable to analyse characteristics that are affected by scaling, but are more demanding in terms of computation than previous features. Spatio-temporal features have higher robustness to features that are not visible directly on the acquired images. In terms of computation, the more complex the feature extraction algorithm is, the more are suitable to an offline analysis. All extracted features presented herein are further used as inputs to various classifiers, such as: support vector machine (SVM), k-nearest neighbors (kNN), Bayes classifiers, and random forests. All these classifiers are standard pattern recognition techniques.
AI Techniques
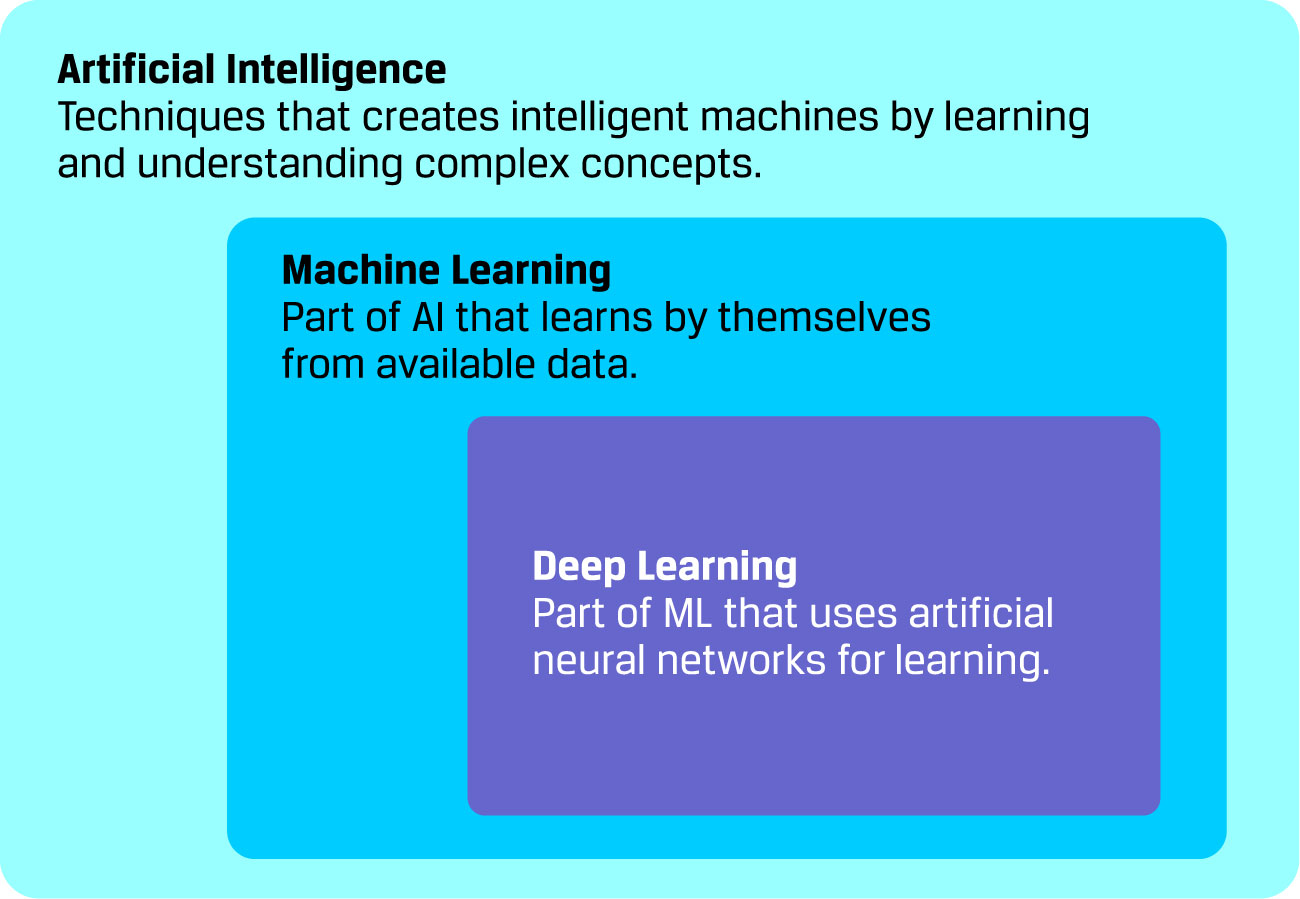
The AI is the field of computer science that aims to create intelligent machines by learning and understanding complex concepts. As an AI branch, ML deals with intelligent machines that learns by themselves from available data. Moreover, DL refers to a family of ML methods that uses neural networks for learning (see Figure 2). These artificial neural networks (ANN) are biological-inspired computing systems that allows computers to learn.
In field of medical image processing there are some application that use AI. In Yang and Bang’s review (Yang and Bang, 2019) about applications of AI in gastroenterology, which summarizes clinical studies that are using AI in the upper and lower gastrointestinal field, there is a sole mention of CD. The review presented only Zhou’s study which has achieved a sensitivity and specificity of 100%. Seguí et al. in (Seguí et al., 2016) presented a generic feature descriptor for the classification of video capsule endoscopy images. In order to build the system they created a large database containing only color images, designed a CNN architecture and performed an exhaustive validation of the proposed method. They achieved very good results: 96% accuracy. Gadermayr et al. in (Gadermayr et al., 2018) investigated the capability of state-of-the-art neural network approaches for diagnosis of CD and proposed pipelines for fully-automated patient-wise diagnosis as well as for integrating expert knowledge into the automated decision process.
The availability of big data and computational power have led to the use of AI in medical applications on a large scale. Table 2 summarizes AI techniques used in clinical studies for classification of CD. Common neural networks used in studies made for CD diagnosis are the AlexNet (Wimmer et al., 2016b; Yang and Bang, 2019), GoogLeNet (Zhou et al., 2017), VGGf net (Wimmer et al., 2016b; Gadermayr et al., 2017; Wimmer et al., 2017; Wimmer et al., 2018), and VGG16 net (Wimmer et al., 2016b; Wimmer et al., 2018). Only a single study used video-capsule images (Zhou et al., 2017), while all the others researches used standard endoscopic images. Comparing to the feature extraction, databases used in AI are based on a much larger number of patients [e.g., 353 patients in (Wimmer et al., 2016b)].
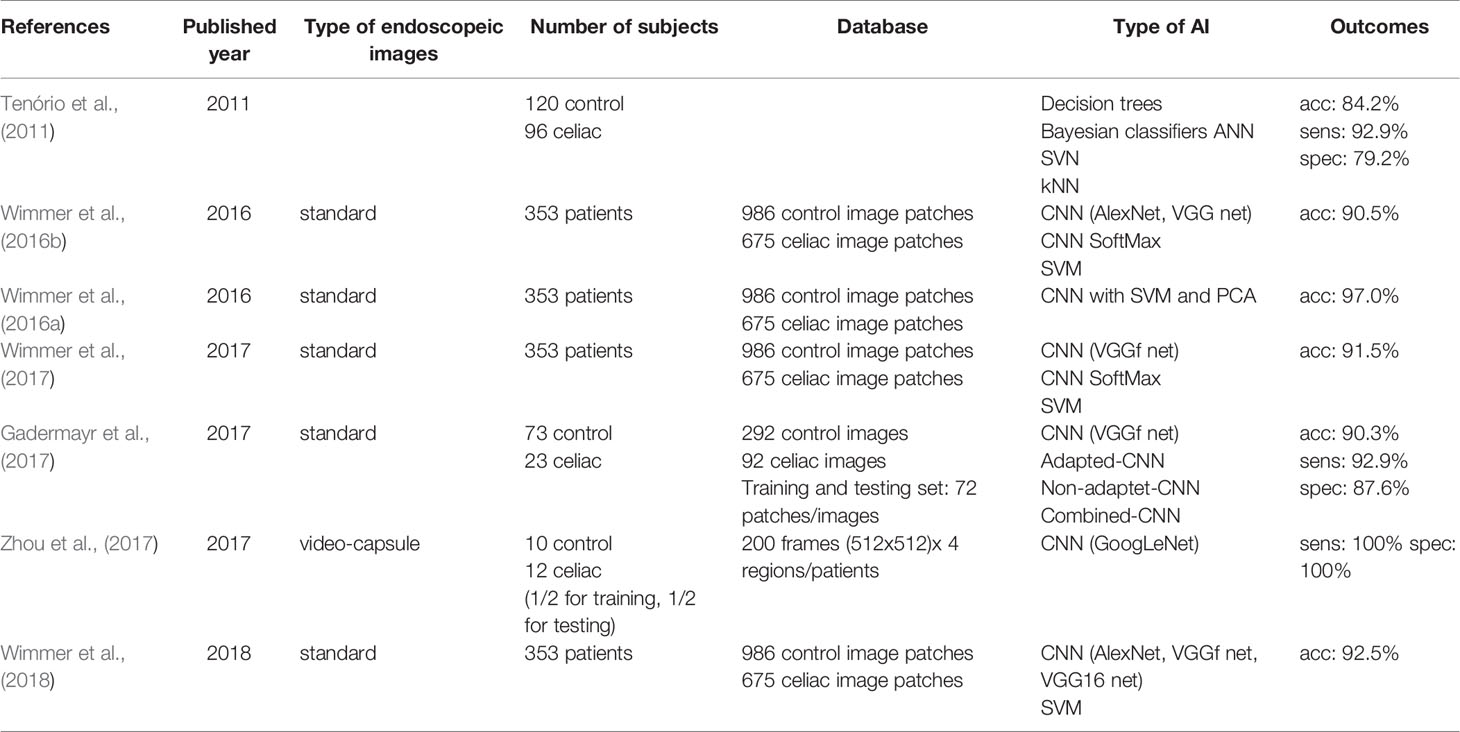
Table 2 Summary of AI techniques used in clinical studies for classification of celiac disease (CD).
Best results have been obtained on video-capsule images by Zhou et al. using GoogLeNet (Zhou et al., 2017). Although the database was reduced in terms of number of patients, 400 images were used for training that led to 100% specificity and 100% sensitivity. Best results on standard endoscopy images were obtained by Wiemer et al. using CNN with SVM and principal component analysis (PCA) (Wimmer et al., 2016a). They obtained a 97% good classification rate based on 1661 image patches (986 control and 675 CD).
Discussion and Limitations
Even if the gold standard for the diagnosis of CD is considered to be the duodenal biopsy, advanced endoscopic techniques such as chromoendoscopy and water-immersion have been researched as enhanced tools to detect VA. The most notable techniques include the modified immersion technique (MIT) (Gasbarrini et al., 2003) under traditional white-light illumination (denoted as WLMIT), as well as MIT under narrow band imaging (Emura et al., 2008; Valitutti et al., 2014) (denoted as NBIMIT). These endoscopic techniques were specifically designed for improving the visual confirmation of CD during endoscopy. Other studies have proposed the use of video capsule images processing in detecting CD (Ciaccio et al., 2010a; Ciaccio et al., 2010b; Ciaccio et al., 2012a; Ciaccio et al., 2012b; Ciaccio et al., 2013b; Ciaccio et al., 2014b; Ciaccio et al., 2017a). Although it is considered as a noninvasive technique, its use is relatively low due to high cost and low resolution of image samples (de Bruaene et al., 2015).
One of the most important issues that are encountered in endoscopic image analysis is related to degradations such as noise, reflections, blurring and scaling due by weak illumination and downsized sensors. Some of the papers proposed some methods of improving these degradations (Hegenbart et al., 2011a; Hegenbart et al., 2011b; Gadermayr et al., 2014a).
Database construction is a critical subject in the AI-based classification of CD using endoscopic imagery. When large databases are not available, one can use data augmentation to artificially increase the number of samples of the database (Wimmer et al., 2017).
A major limitation of using automated-processing of duodenal images captured during endoscopy for diagnosing CD is the wide differential diagnosis of VA. Several other diseases other than CD can manifest as VA in the small bowel—giardiasis, Helicobacter pylori infection, Whipple’s disease, tropical sprue, collagenous sprue, eosinophilic gastroenteritis, common variable immunodeficiency, intestinal lymphoma, Crohn’s disease, HIV-enteropathy, or drug-induced enteropathy (Jinga et al., 2017; Schiepatti et al., 2019). Upon detection of VA by digitally processing of endoscopy images, CD-specific serology will discriminate CD from other causes of seronegative VA.
Another limit of using computerized automation methods for image processing is that of artifacts due to the presence of air bubbles, residues or secretions in the duodenum. This could represent an issue for selection of images frames which are processed for assessment of VA. Not least, selection of point of interest (POI) regions from the image captured during endoscopy is yet to be automatized; this could be considered a selection bias in current studies, as it was done manually during image processing. Also, a special concern is to be raised for cases with mild enteropathy, when changes in the duodenum are subtle, and those with patchy disease, when selection of the POI could be nonrepresentative.
Another limitation of the current review is the heterogeneity of studies with respect to cases analyzed, as some of the studies report the number of patients included, while others the number of images processed.
Conclusion
In the last decades, there’s been a growing interest in image processing techniques for detection of VA. Computer-aided diagnosis of CD by processing images of the small bowel captured during endoscopy is feasible and warrants further development for integration into endoscopy consoles.
Author Contributions
AM and DB discussed the research idea. C-CM and MJ proposed the research design. AM and DB performed the literature search and selection of articles. AM and DB drafted the first manuscript. MJ and C-CM critically reviewed the manuscript, acting as guarantors of the paper. All authors approved the final version of the manuscript.
Funding
This paper was financially supported by “Carol Davila” University of Medicine and Pharmacy through Contract no. 23PFE/17.10.2018 funded by the Ministry of Research and Innovation within PNCDI III, Program 1 – Development of the National RD system, Subprogram 1.2 – Institutional Performance – RDI excellence funding projects.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Arguelles-Grande, C., Tennyson, C. A., Lewis, S. K., Green, P. R., Bhagat, G. (2012). Variability in small bowel histopathology reporting between different pathology practice settings: impact on the diagnosis of coeliac disease. J. Clin. Pathol. 65 (3), 242–247. doi: 10.1136/jclinpath-2011-200372
Balaban, D., Popp, A., Vasilescu, F., Haidautu, D., Purcărea, R., Jinga, M. (2015). Diagnostic yield of endoscopic markers for celiac disease. J. Med. Life. 8 (4), 452–457.
Bassotti, G., Castellucci, G., Betti, C., Fusaro, C., Cavalletti, M. L., Bertotto, A., et al. (1994). Abnormal gastrointestinal motility in patients with celiac sprue. Digestive Dis. Sci. 39, 1947–1954. doi: 10.1007/BF02088130
Bonamico, M., Mariani, P., Thanasi, E., Ferri, M., Nenna, R., Tiberti, C., et al. (2004). Patchy villous atrophy of the duodenum in childhood celiac disease. J. Pediatr. Gastroenterol. Nutr. 38 (2), 204–207. doi: 10.1097/00005176-200402000-00019
Ciaccio, E. J., Tennyson, C. A., Bhagat, G., Lewis, S. K., Green, P. H. R. (2010a). Classification of videocapsule endoscopy image patterns: comparative analysis between patients with celiac disease and normal individuals. Biomed. Eng. Online. 9 (1), 44. doi: 10.1186/1475-925X-9-44
Ciaccio, E. J., Tennyson, C. A., Lewis, S. K., Krishnareddy, S., Bhagat, G., Green, P. H. R. (2010b). Distinguishing patients with celiac disease by quantitative analysis of videocapsule endoscopy images. Comput. Methods Programs Biomed. 100 (1), 39–48. doi: 10.1016/j.cmpb.2010.02.005
Ciaccio, E., Bhagat, G., Tennyson, C., Lewis, S., Hernandez, L., Green, P. (2011). Quantitative assessment of endoscopic images for degree of villous atrophy in celiac disease. Digestive Dis. Sci. 56, 805–811. doi: 10.1007/s10620-010-1371-6
Ciaccio, E. J., Tennyson, C. A., Bhagat, G., Lewis, S. K., Green, P. H. R. (2012a). Transformation of videocapsule images to detect small bowel mucosal differences in celiac versus control patients. Comput. Methods Programs Biomed. 108 (1), 28–37. doi: 10.1016/j.cmpb.2011.12.008
Ciaccio, E. J., Tennyson, C. A., Bhagat, G., Lewis, S. K., Green, P. R. (2012b). Quantitative estimates of motility from videocapsule endoscopy are useful to discern celiac patients from controls. Digestive Dis. Sci. 57, 2936–2943. doi: 10.1007/s10620-012-2225-1
Ciaccio, E. J., Tennyson, C. A., Bhagat, G., Lewis, S. K., Green, P. H. R. (2013a). Use of shape-from-shading to estimate three-dimensional architecture in the small intestinal lumen of celiac and control patients. Comput. Methods Programs Biomed. 111 (3), 676–684. doi: 10.1016/j.cmpb.2013.06.002
Ciaccio, E. J., Tennyson, C. A., Bhagat, G., Lewis, S. K., Green, P. R. (2013b). Implementation of a polling protocol for predicting celiac disease in videocapsule analysis. World J. Gastrointestinal Endoscopy 5 (7), 313–322. doi: 10.4253/wjge.v5.i7.313
Ciaccio, E. J., Tennyson, C. A., Bhagat, G., Lewis, S. K., Green, P. H. R. (2014a). Methods to quantitate videocapsule endoscopy images in celiac disease. Bio-med. Mater. Eng. 24 (6), 1895–1911. doi: 10.3233/BME-140999
Ciaccio, E. J., Tennyson, C. A., Bhagat, G., Lewis, S. K., Green, P. H. R. (2014b). Use of basis images for detection and classification of celiac disease. Bio-med. Mater. Eng. 24 (6), 1913–1923. doi: 10.3233/BME-141000
Ciaccio, E. J., Bhagat, G., Lewis, S. K., Green, P. H. R. (2016a). Extraction and processing of videocapsule data to detect and measure the presence of villous atrophy in celiac disease patients. Comput. Biol. Med. 78, 97–106. doi: 10.1016/j.compbiomed.2016.09.009
Ciaccio, E. J., Bhagat, G., Lewis, S. K., Green, P. R. (2016b). Recommendations to quantify villous atrophy in video capsule endoscopy images of celiac disease patients. World J. Gastrointestinal Endoscopy. 8 (18), 653–662. doi: 10.4253/wjge.v8.i18.653
Ciaccio, E. J., Bhagat, G., Lewis, S. K., Green, P. R. (2017a). Use of shape-from-shading to characterize mucosal topography in celiac disease videocapsule images. World J. Gastrointestinal Endoscopy. 9 (7), 310–318. doi: 10.4253/wjge.v9.i7.310
Ciaccio, E. J., Lewis, S. K., Bhagat, G., Green, P. H. R. (2017b). Coeliac disease and the videocapsule: what have we learned till now. Ann. Trans. Med. 5 (9), 197. doi: 10.21037/atm.2017.05.06
Corazza, G. R., Villanacci, V., Zambelli, C., Milione, M., Luinetti, O., Vindigni, C., et al. (2007). Comparison of the interobserver reproducibility with different histologic criteria used in celiac disease. Clin. Gastroenterol. Hepatol. 5 (7), 838–843. doi: 10.1016/j.cgh.2007.03.019
de Bruaene, C. V., de Looze, D., Hindryckx, P. (2015). Small bowel capsule endoscopy: Where are we after almost 15 years of use? World J. Gastrointestinal Endoscopy 7 (1), 13–36. doi: 10.4253/wjge.v7.i1.13
Emura, F., Saito, Y., Ikematsu, H. (2008). Narrow-band imaging optical chromocolonoscopy: advantages and limitations. World J. Gastroenterol. 14 (31), 4867–4872. doi: 10.3748/wjg.14.4867
Fuchs, V., Kurppa, K., Huhtala, H., Mäki, M. J., Kekkonen, L., Kaukinen, K. (2018). Delayed celiac disease diagnosis predisposes to reduced quality of life and incremental use of health care services and medicines: A prospective nationwide study. U. Eur. Gastroenterol. J. 6 (4), 567–575. doi: 10.1177/2050640617751253
Gadermayr, M., Liedlgruber, M., Uhl, A., Vécsei, A. (2013a). Evaluation of different distortion correction methods and interpolation techniques for an automated classification of celiac disease. Comput. Methods Programs Biomed. 112 (3), 694–712. doi: 10.1016/j.cmpb.2013.07.001
Gadermayr, M., Liedlgruber, M., Uhl, A., Vécsei, A. (2013b). “Shape curvature histogram: A shape feature for celiac disease diagnosis,” in MCV. Springer, 175–184. doi: 10.1007/978-3-319-05530-5
Gadermayr, M., Uhl, A., Vécsei, A. (2013c). “Feature extraction with intrinsic distortion correction in celiac disease imagery: No need for rasterization,” in MCV. Springer. 196–204. doi: 10.1007/978-3-319-05530-5
Gadermayr, M., Uhl, A., Vécsei, A. (2014a). “Degradation adaptive texture classification: A case study in celiac disease diagnosis brings new insight,” in ICIAR. Springer. 263–273. doi: 10.1007/978-3-319-11755-3
Gadermayr, M., Uhl, A., Vécsei, A. (2014b). Is a precise distortion estimation needed for computer aided celiac disease diagnosis?,” in ICISP, Springer. 620–628. doi: 10.1007/978-3-319-07998-1
Gadermayr, M., Kogler, H., Uhl, A., Vécsei, A. (2015). “Comparing endoscopic imaging configurations in computer-aided celiac disease diagnosis,” in 2015 International Conference on Image Processing Theory, Tools and Applications (IPTA), IEEE, 446–451.
Gadermayr, M., Uhl, A., Vécsei, A. (2015). “Boosting small-data performance of lbp: A case study in celiac disease diagnosis,” in SCIA. Springer. 224–233. doi: 10.1007/978-3-319-19665-7
Gadermayr, M., Uhl, A., Vécsei, A. (2016). Fully automated decision support systems for celiac disease diagnosis. IRBM. 37 (1), 31–39. doi: 10.1016/j.irbm.2015.09.009
Gadermayr, M., Kogler, H., Karla, M., Merhof, D., Uhl, A., Vécsei, A. (2016a). Computer-aided texture analysis combined with experts’ knowledge: Improving endoscopic celiac disease diagnosis. World J. Gastroenterol. 22 (31), 7124–7134. doi: 10.3748/wjg.v22.i31.7124
Gadermayr, M., Kogler, H., Karla, M., Vécsei, A., Uhl, A., Merhof, D. (2016b). “Incorporating human knowledge in automated celiac disease diagnosis,” in 2016 Sixth International Conference on Image Processing Theory, Tools and Applications (IPTA), IEEE. doi: 10.1109/IPTA.2016.7821009
Gadermayr, M., Wimmer, G., Uhl, A., Kogler, H., Vécsei, A., Merhof, D. (2017). “Fully-automated cnn-based computer aided celiac disease diagnosis,” in ICIAP. Springer. 467–478. doi: 10.1007/978-3-319-68548-9
Gadermayr, M., Wimmer, G., Kogler, H., Vécsei, A., Merhof, D., Uhl, A. (2018). Automated classification of celiac disease during upper endoscopy: Status quo and quo vadis. Comput. Biol. Med. 102, 221–226. doi: 10.1016/j.compbiomed.2018.04.020
Gasbarrini, A., Ojetti, V., Cuoco, L., Cammarota, G., Migneco, A., Armuzzi, A., et al. (2003). Lack of endoscopic visualization of intestinal villi with the immersion technique in overt atrophic celiac disease. Gastrointestinal Endoscopy 57 (3), 348–351. doi: 10.1067/mge.2003.116
Gschwandtner, M., Liedlgruber, M., Uhl, A., Vécsei, A. (2010). “Experimental study on the impact of endoscope distortion correction on computer-assisted celiac disease diagnosis,” in Proceedings of the 10th IEEE International Conference on Information Technology and Applications in Biomedicine, 1–6. doi: 10.1109/itab.2010.5687708
Hämmerle-Uhl, J., Höller, Y., Uhl, A., Vécsei, A. (2012). “Endoscope distortion correction does not (easily) improve mucosa-based classification of celiac disease,” in Medical image computing and computer-assisted intervention: MICCAI International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer. 574–581. doi: 10.1007/978-3-642-33454-2
Hegenbart, S., Kwitt, R., Liedlgruber, M., Uhl, A., Vécsei, A. (2009). “Impact of duodenal image capturing techniques and duodenal regions on the performance of automated diagnosis of celiac disease,” in 2009 Proceedings of 6th International symposium on Image and Signal Processing and Analysis, IEEE. 718–723. doi: 10.1109/ISPA.2009.5297637
Hegenbart, S., Uhl, A., Vécsei, A. (2011a). “Impact of endoscopic image degradations on lbp based features using one-class svm for classification of celiac disease,” in 2011 7th International Symposium on Image and Signal Processing and Analysis (ISPA), IEEE. 715–720.
Hegenbart, S., Uhl, A., Vécsei, A. (2011b). “Impact of histogram subset selection on classification using multi-scale lbp-operators,” in Bildverarbeitung für die Medizin. Springer. 359–363. doi: 10.1007/978-3-642-19335-4
Hegenbart, S., Uhl, A., Vécsei, A. (2011c). “Systematic assessment of performance prediction techniques in medical image classification a case study on celiac disease,” in Information processing in medical imaging: proceedings of the conference. Springer. 22, 498–509. doi: 10.1007/978-3-642-22092-0
Hegenbart, S., Uhl, A., Vécsei, A. (2012a). “On the implicit handling of varying distances and gastrointestinal regions in endoscopic video sequences with indication for celiac disease,” in 2012 25th IEEE International Symposium on Computer-Based Medical Systems (CBMS), IEEE. doi: 10.1109/CBMS.2012.6266354
Hegenbart, S., Maimone, S., Uhl, A., Vécsei, A., Wimmer, G. (2012b). “Customised frequency pre-filtering in a local binary pattern-based classification of gastrointestinal images,” in MCBR-CDS. Springer. 99–109. doi: 10.1007/978-3-642-36678-9
Hegenbart, S., Uhl, A., Vécsei, A., Wimmer, G. (2013). “Scale invariant texture descriptors for classifying celiac disease,” in Medical Image Analysis. 17 (4). 458–474. doi: 10.1016/j.media.2013.02.001
Hegenbart, S., Uhl, A., Vécsei, A. (2015). Survey on computer aided decision support for diagnosis of celiac disease. Comput. Biol. Med. 65, 348–358. doi: 10.1016/j.compbiomed.2015.02.007
Herrod, P. J. J., Lund, J. N. (2018). “Random duodenal biopsy to exclude coeliac disease as a cause of anaemia is not cost-efective and should be replaced with universally performed pre-endoscopy serology in patients on a suspected cancer pathway,” in Techniques in Coloproctology. 22 (2), 121–124. doi: 10.1007/s10151-018-1756-7
Hopper, A. D., Cross, S. S., Sanders, D. S. (2007). Patchy villous atrophy in adult patients with suspected gluten-sensitive enteropathy: is a multiple duodenal biopsy strategy appropriate? Endoscopy 40 (3), 219–224. doi: 10.1055/s-2007-995361
Jinga, M., Balaban, D. V., Peride, I., Niculae, A., Monica, D., Florina, V., et al. (2017). Crypt hyperplastic enteropathy in distal duodenum in helicobacter pylori infection-report of two cases without evidence of celiac disease. Rom. J. Morphol. Embryol. = Rev. Roum. Morphol. Embryol. 58, 685–688.
Jinga, M., Popp, A., Balaban, D. V., Dima, A. M., Jurcut, C. (2018). Physicians’ attitude and perception regarding celiac disease: A questionnaire-based study. Turkish J. Gastroenterol. 29 (4), 419–426. doi: 10.5152/tjg.2018.17236
Koh, J. E. W., Hagiwara, Y., Oh, S. L., Tan, J. H., Ciaccio, E. J., Green, P. H. R., et al. (2019). Automated diagnosis of celiac disease using dwt and nonlinear features with video capsule endoscopy images. Future Gener. Comp. Syst. 90, 86–93. doi: 10.1016/j.future.2018.07.044
Kwitt, R., Hegenbart, S., Rasiwasia, N., Vécsei, A., Uhl, A. (2014). “Do we need annotation experts? a case study in celiac disease classification,” in Medical image computing and computer-assisted intervention : MICCAI International Conference on Medical Image Computing and Computer-Assisted Intervention, vol. 17, 454–461. doi: 10.1007/978-3-319-10470-6
Lebwohl, B., Bhagat, G., Markoff, S., Lewis, S. K., Smukalla, S. M., Neugut, A. I., et al. (2012). Prior endoscopy in patients with newly diagnosed celiac disease: A missed opportunity? Digestive Dis. Sci. 58, 1293–1298. doi: 10.1007/s10620-012-2551-3
Liedlgruber, M., Uhl, A., Vécsei, A. (2011). “Statistical analysis of the impact of distortion (correction) on an automated classification of celiac disease,” in 2011 17th International Conference on Digital Signal Processing (DSP), IEEE, doi: 10.1109/ICDSP.2011.6004900
Ludvigsson, J. F., Card, T. R., Kaukinen, K., Bai, J. C., Zingone, F., Sanders, S., et al. (2016). Screening for coeliac disease in the general population and in 1 high-risk groups 2. United Eur. Gatroenterol. J. 3 (2), 106–120. doi: 10.1177/2050640614561668
Moher, D., Shamseer, L., Clarke, M., Ghersi, D., Liberati, A., Petticrew, M., et al. (2015). Preferred reporting items for systematic review and meta-analysis protocols (prisma-p) 2015 statement. Syst. Rev. 4 (1). doi: 10.1186/2046-4053-4-1
Mubarak, A., Nikkels, P. G. J., Houwen, R. H. J., Kate, F. J. W. T. (2011). Reproducibility of the histological diagnosis of celiac disease. Scand. J. Gastroenterol. 46 (9), 1065–1073. doi: 10.3109/00365521.2011.589471
Razzak, M. I., Naz, S., Zaib, A. (2018). Deep learning for medical image processing: Overview, challenges and future. Classification in BioApps, Springer, 323–350. doi: 10.1007/978-3-319-65981-7
Schiepatti, A., Sanders, D. S., Zuffada, M., Luinetti, O., Iraqi, A. M. E., Biagi, F. (2019). Overview in the clinical management of patients with seronegative villous atrophy. Eur. J. Gastroenterol. Hepatol. 31 (4), 409–417. doi: 10.1097/MEG.0000000000001340
Seguí, S., Drozdzal, M., Pascual, G., Radeva, P., Malagelada, C., Azpiroz, F., et al. (2016). Generic feature learning for wireless capsule endoscopy analysis. Comput. Biol. Med. 79, 163–172. doi: 10.1016/j.compbiomed.2016.10.011
Tenório, J. M., Hummel, A. D., Cohrs, F. M., Sdepanian, V. L., Pisa, I. T., Marin, H. F. (2011). Artificial intelligence techniques applied to the development of a decision-support system for diagnosing celiac disease. Int. J. Med. Inf. 80 (11), 793–802. doi: 10.1016/j.ijmedinf.2011.08.001
Tursi, A. (2004). Gastrointestinal motility disturbances in celiac disease. J. Clin. Gastroenterol. 38 (8), 642–645. doi: 10.1097/01.mcg.0000118792.58123.c1
Uhl, A., Vécsei, A., Wimmer, G. (2011). “Complex wavelet transform variants in a scale invariant classification of celiac disease,” in IbPRIA. doi: 10.1007/978-3-642-21257-4
Vécsei, A., Fuhrmann, T., Uhl, A. (2008). Towards automated diagnosis of celiac disease by computer-assisted classification of duodenal imagery in 4th IET International Conference on Advances in Medical, Signal and Information Processing (MEDSIP 2008), IET. doi: 10.1049/cp:20080465
Vécsei, A., Fuhrmann, T., Liedlgruber, M., Brunauer, L., Payer, H., Uhl, A. (2009). Automated classification of duodenal imagery in celiac disease using evolved fourier feature vectors. Comput. Methods Programs Biomed. 95 (2 Suppl), S68–S78. doi: 10.1016/j.cmpb.2009.02.017
Vécsei, A., Amann, G., Hegenbart, S., Liedlgruber, M., Uhl, A. (2011). Automated marsh-like classification of celiac disease in children using local texture operators. Comput. Biol. Med. 41 6, 313–325. doi: 10.1016/j.compbiomed.2011.03.009
Valitutti, F., Oliva, S., Iorfida, D., Aloi, M., Gatti, S., Trovato, C. M., et al. (2014). Narrow band imaging combined with water immersion technique in the diagnosis of celiac disease. Digestive liver Dis. : Off. J. Ital. Soc. Gastroenterology Ital. Assoc. Study Liver 46 (12), 1099–1102. doi: 10.1016/j.dld.2014.08.039
Vicnesh, J., Wei, J. K. E., Ciaccio, E. J., Oh, S. L., Bhagat, G., Lewis, S. K., et al. (2019). Automated diagnosis of celiac disease by video capsule endoscopy using daisy descriptors. J. Med. Syst. 43, 1–9. doi: 10.1007/s10916-019-1285-6
Weile, B., Hansen, B. F., Hägerstrand, I., Hansen, J. P., Krasilnikoff, P. A. (2000). Interobserver variation in diagnosing coeliac disease. a joint study by danish and swedish pathologists. APMIS : Acta Pathol. Microbiol. Immunol. Scand. 108 (5), 380–384. doi: 10.1034/j.1600-0463.2000.d01-72.x
Wimmer, G., Hegenbart, S., Vécsei, A., Uhl, A. (2016a). “Convolutional neural network architectures for the automated diagnosis of celiac disease,” in CARE@MICCAI. Springer. 104–113. doi: 10.1007/978-3-319-54057-3
Wimmer, G., Vécsei, A., Uhl, A. (2016b). “Cnn transfer learning for the automated diagnosis of celiac disease,” in 2016 Sixth International Conference on Image Processing Theory, Tools and Applications (IPTA), IEEE, doi: 10.1109/IPTA.2016.7821020
Wimmer, G., Uhl, A., Vécsei, A. (2017). “Evaluation of domain specific data augmentation techniques for the classification of celiac disease using endoscopic imagery,” in 2017 IEEE 19th International Workshop on Multimedia Signal Processing (MMSP), IEEE. doi: 10.1109/MMSP.2017.8122221
Wimmer, G., Vécsei, A., Häfner, M., Uhl, A. (2018). Fisher encoding of convolutional neural network features for endoscopic image classification. J. Med. Imaging. 5 (3), 034504. doi: 10.1117/1.JMI.5.3.034504
Yang, Y. J., Bang, C. S. (2019). Application of artificial intelligence in gastroenterology. World J. Gastroenterol. 25 (14), 1666–1683. doi: 10.3748/wjg.v25.i14.1666
Keywords: celiac disease, computer aided diagnosis, artificial intelligence, endoscopy, feature extraction
Citation: Molder A, Balaban DV, Jinga M and Molder C-C (2020) Current Evidence on Computer-Aided Diagnosis of Celiac Disease: Systematic Review. Front. Pharmacol. 11:341. doi: 10.3389/fphar.2020.00341
Received: 01 December 2019; Accepted: 09 March 2020;
Published: 16 April 2020.
Edited by:
Jean-Marie Boeynaems, Université libre de Bruxelles, BelgiumReviewed by:
Michael Thiede, IUBH University of Applied Sciences, GermanyKurt Neumann, Independent Researcher, Kerékteleki, Hungary
Copyright © 2020 Molder, Balaban, Jinga and Molder. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Daniel Vasile Balaban, dmFzaWxlLmJhbGFiYW5AdW1mY2Qucm8=
 Adriana Molder
Adriana Molder Daniel Vasile Balaban
Daniel Vasile Balaban Mariana Jinga
Mariana Jinga Cristian-Constantin Molder
Cristian-Constantin Molder