
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Pharmacol. , 03 October 2019
Sec. Drug Metabolism and Transport
Volume 10 - 2019 | https://doi.org/10.3389/fphar.2019.01146
 Yu-Wei Lin1†
Yu-Wei Lin1† Mei-Ling Han1†
Mei-Ling Han1† Jinxin Zhao1
Jinxin Zhao1 Yan Zhu1
Yan Zhu1 Gauri Rao2
Gauri Rao2 Alan Forrest2
Alan Forrest2 Jiangning Song3
Jiangning Song3 Keith S. Kaye4
Keith S. Kaye4 Paul Hertzog5,6
Paul Hertzog5,6 Anthony Purcell3
Anthony Purcell3 Darren Creek7
Darren Creek7 Qi Tony Zhou8
Qi Tony Zhou8 Tony Velkov9*
Tony Velkov9* Jian Li1*
Jian Li1*Polymyxins are used as a last-resort class of antibiotics against multidrug-resistant (MDR) Gram-negative Pseudomonas aeruginosa. As polymyxin monotherapy is associated with potential development of resistance, combination therapy is highly recommended. This study investigated the mechanism underlying the synergistic killing of polymyxin B and enrofloxacin against extensive drug-resistant (XDR) P. aeruginosa. An XDR isolate P. aeruginosa 12196 was treated with clinically relevant concentrations of polymyxin B (2 mg/L) and enrofloxacin (1 mg/L) alone or in combination. Metabolome profiles were investigated from bacterial samples collected at 1-and 4-h posttreatment using liquid chromatography with tandem mass spectrometry (LC-MS/MS), and data were analyzed using univariate and multivariate statistics. Significantly perturbed metabolites (q < 0.05, fold change ≥ 2) were subjected to pathway analysis. The synergistic killing by polymyxin B–enrofloxacin combination was initially driven by polymyxin B as indicated by the perturbation of lipid metabolites at 1 h in particular. The killing was subsequently driven by enrofloxacin via the inhibition of DNA replication, resulting in the accumulation of nucleotides at 4 h. Furthermore, the combination uniquely altered levels of metabolites in energy metabolism and cell envelope biogenesis. Most importantly, the combination significantly minimized polymyxin resistance via the inhibition of lipid A modification pathway, which was most evident at 4 h. This is the first study to elucidate the synergistic mechanism of polymyxin B–enrofloxacin combination against XDR P. aeruginosa. The metabolomics approach taken in this study highlights its power to elucidate the mechanism of synergistic killing by antibiotic combinations at the systems level.
Extensive drug-resistant (XDR) Pseudomonas aeruginosa is a major burden to the global health-care system and has been highlighted by the World Health Organization as a priority pathogen with “Serious” threat to human health (Mcphee et al., 2006; Gales et al., 2011; World Health Organization, 2014). Due to the dry discovery pipeline, few novel classes of antibiotics will become available in the near future (Boucher et al., 2013). “Old” polymyxins (i.e., polymyxin B and colistin, also known as polymyxin E) are a last-line therapy that are increasingly used for life-threatening infections caused by XDR P. aeruginosa (Falagas et al., 2005; Li et al., 2006; Nation and Li, 2009; Yahav et al., 2012; Nation et al., 2014; Cheah et al., 2016a). Although polymyxins remain effective against XDR P. aeruginosa, recent pharmacokinetic/pharmacodynamic (PK/PD) studies suggest that polymyxin monotherapy is potentially associated with increased emergence of resistance (Tam et al., 2005; Cheah et al., 2015; Cheah et al., 2016b). Moreover, reports of infections caused by XDR P. aeruginosa, including polymyxin-resistant XDR isolates, are on the rise (Hsueh et al., 1998; Adams et al., 2009; Gales et al., 2012; Goli et al., 2016). In a recent in vitro PK/PD study, we demonstrated that polymyxin B in combination with enrofloxacin is highly effective against XDR P. aeruginosa, which is resistant to both, and significantly minimizes the emergence of polymyxin resistance (Lin et al., 2018). However, the underlying mechanism of the synergistic killing by this novel drug combination remains unknown.
Metabolomics investigates dynamic global metabolite levels in biological systems in response to biological stimuli or perturbations (Chen et al., 2007; Kaddurah-Daouk and Weinshilboum, 2014; Mastrangelo et al., 2014; Vincent et al., 2016). To date, metabolomics is increasingly employed in drug discovery and development to elucidate the mechanism of drug action (Peng et al., 2015). In the present study, we investigated the synergistic killing mechanism of polymyxin B and enrofloxacin combination against a clinical isolate of XDR P. aeruginosa using metabolomics. The mechanistic findings provide important pharmacological information for optimizing this promising combination in patients.
A solution of polymyxin B (sulfate, Sigma-Aldrich, Castle Hill, NSW, Australia; batch number BCBD1065V) was freshly prepared in sterile Milli-Q water (Millipore Australia, North Ryde, NSW, Australia). Enrofloxacin (Sigma-Aldrich) was first dissolved in dimethyl sulfoxide (DMSO; Sigma-Aldrich) and subsequently diluted in sterile Milli-Q water to obtain a final DMSO concentration of ≤10% (v/v) (Tran et al., 2016a).
An XDR P. aeruginosa 12196 with a polymyxin MIC of 64 mg/L and enrofloxacin MIC of 4 mg/L was examined (Lin et al., 2018). P. aeruginosa 12196 was stored in tryptone soy broth with 20% glycerol at −80°C and sub-cultured onto nutrient agar plates before each experiment (Maifiah et al., 2016; Maifiah et al., 2017). Overnight culture was subsequently prepared in 10-ml cation-adjusted Mueller-Hinton broth (CAMHB) and diluted 100-fold using fresh media to prepare 200 ml of mid-logarithmic culture with a starting inoculum of approximately 108 CFU/ml (Maifiah et al., 2016; Maifiah et al., 2017). To each bacterial culture, polymyxin B (2 mg/L), enrofloxacin (1 mg/L), or a combination was added. Bacterial culture without any antibiotics served as a control. Three biological replicates were prepared independently from different colonies of XDR P. aeruginosa 12196 on three consecutive days. Bacterial cultures were incubated at 37°C in a shaking incubator (180 rpm). Samples were collected at 0, 1, and 4 h and immediately quenched in a dry ice–ethanol bath for 30 s to halt the metabolism. Subsequently, the OD600 value of each sample was measured and normalized to 0.50 ± 0.02 with fresh CAMHB. Subsequently, 15 ml of each normalized sample culture was transferred to 15-ml Falcon tubes (Thermo Fisher Scientific, Melbourne, Australia) for metabolite extraction.
Metabolite sample preparation was carried out as reported previously (Han et al., 2018). Briefly, 15 ml of each bacterial culture was centrifuged at 3,200 × g at 4 C. The supernatant was discarded, and cell pellets were resuspended in cold 0.9% sodium chloride solution. Samples were centrifuged at 3,200 × g for 5 min to remove extracellular metabolites and media components. Following the washing step, bacterial pellets were resuspended in 0.5 ml of chloroform/methanol/water (1:3:1, v/v/v) containing 1 µM internal standards (CHAPS, CAPS, PIPES, and TRIS). Subsequently, bacterial samples were frozen in liquid nitrogen and thawed on ice to release intracellular metabolites. The samples were then centrifuged at 14,000 × g for 10 min, and 200 µL of supernatants was transferred into ultra-performance liquid chromatography (UPLC) vials for liquid chromatography with tandem mass spectrometry (LC-MS/MS) analysis. QC samples were prepared by mixing equal amounts of all tested samples and processed as a “real” sample outlined above.
Metabolite samples were analyzed on a Q-exactive Orbitrap mass spectrometer coupled with a Dionex U3000 high-performance LC (HPLC; Thermo Fisher) with a ZIC-pHILIC column (5 µm, polymeric, 150 × 4.6 mm; SeQuant, Merck). The MS system was operated at 35,000 resolution in both positive and negative electrospray ionization modes with a detection range of 85–1,275 m/z. Column temperature was maintained at 25 C, and the mobile phase consisted of 20 mM of ammonium carbonate (solvent A) and acetonitrile (solvent B). Metabolites were eluted in a step gradient, starting with 80% solvent B at a flow rate of 0.3 ml/min followed by a linear gradient to 50% solvent B over 15 min (Maifiah et al., 2016; Maifiah et al., 2017). All samples were randomized and analyzed in a single LC-MS batch. Analytical reproducibility was monitored on the basis of pooled QC samples throughout the batch, which were periodically analyzed after groups of six samples. Analyses of a mixture of pure standards containing >250 metabolites were performed to assist in the identification of metabolites.
IDEOM (http://mzmatch.sourceforge.net/ideom.php) and mzMatch were employed for metabolomics analysis (Scheltema et al., 2011; Creek et al., 2012). Raw mass spectrometric data files were processed as described previously based on the intensity (>100,000 counts), shape (codadw > 0.8), and reproducibility (RSD < 0.5) of the LC-MS peaks. Elemental composition and exact mass were used for open-source database searching, including MassBank (http://www.massbank.jp). Putative metabolites were identified by accurate mass (±5 ppm) and retention time with authentic standards (<50%) as indicated by IDEOM confidence score of 9 or 10, or by accurate mass (±5 ppm) and predicted retention time to achieve an IDEOM confidence scores of ≥5 (Creek et al., 2012). Several different databases [e.g., PseudoCyc, Kyoto Encyclopedia of Genes and Genomes (KEGG), BioCyc HMDB, and LipidMaps] were used to map the metabolite, and global metabolomics profiles of samples were analyzed using univariate and multivariate analyses in MetaboAnalyst 4.0 (Xia et al., 2015). Data were filtered using interquartile range (IQR), normalized relative to the median, log2 transformed, and auto-scaled. PCA was performed to identify and remove outliers that were defined as samples outside of ±2 standard deviations (SDs) along the principal component 1 axis (PC1). Statistical significance of differences between metabolites was determined using one-way analysis of variance (ANOVA), Benjamini–Hochberg multiple testing correction (q < 0.05), Fisher’s least significant difference (LSD) test, and fold change (FC; log2FC ≥ 1). FC values were calculated using raw intensity and geometric mean of the biological replicates. BioCyc (Karp et al., 2005), iPath (Letunic et al., 2008), and the KEGG (Kanehisa and Goto, 2000) were employed for pathway analysis.
The intra-experimental variability was assessed based on the median relative standard deviations (RSDs) of the samples, which ranged from 15% to 24% (Figure S1) and were well within the acceptable limits for metabolomics studies (Kirwan et al., 2014). Furthermore, the principal component analysis (PCA) plots showed that all samples [including six quality control (QC) samples] were tightly clustered together, demonstrating an excellent reproducibility of our analytical methods (Figure S1). Univariate and multivariate analyses revealed that over 500 putative metabolites were identified in the metabolome of XDR P. aeruginosa 12196 induced by polymyxin B (2 mg/L) and enrofloxacin (1 mg/L) alone or in combination at 1- and 4-h postdrug treatment. The nature of these metabolites indicated that a wide range of pathways were perturbed (Figures 1, 2 and S2; Supplementary Tables 1–2). Univariate analysis showed that polymyxin B alone induced 6.3% (36) and 5.3% (30) metabolic changes at 1 and 4 h, respectively. Likewise, the combination induced 11.4% (65) and 21.8% (124) metabolic changes at 1 and 4 h, respectively. On the other hand, enrofloxacin alone induced minimal metabolic changes at 1 h (Figures 1, 2 and S2). PCA demonstrated that polymyxin B alone and in combination with enrofloxacin induced significant global metabolic changes as early as 1 h (Figure 1A). Perturbations in the metabolome of XDR P. aeruginosa 12196 induced by polymyxin B (2 mg/L) and enrofloxacin (1 mg/L) alone or in combination were evident at 1 and 4 h post drug treatment. Many metabolic features were shared between the two monotherapies and the combination with more significant changes at 4 h, demonstrating a time-dependent antibacterial effect by the drug combination (Figure 1B). At 1 h, the number of perturbed metabolites that were common between polymyxin B alone and the combination was much higher than that between enrofloxacin alone and the combination. Interestingly, at 4 h, the metabolic alterations were largely caused by the combination (Figure 1B). Overall, the combination of polymyxin B and enrofloxacin produced significantly greater perturbations in the metabolomes at 1 and 4 h than either polymyxin B or enrofloxacin alone.
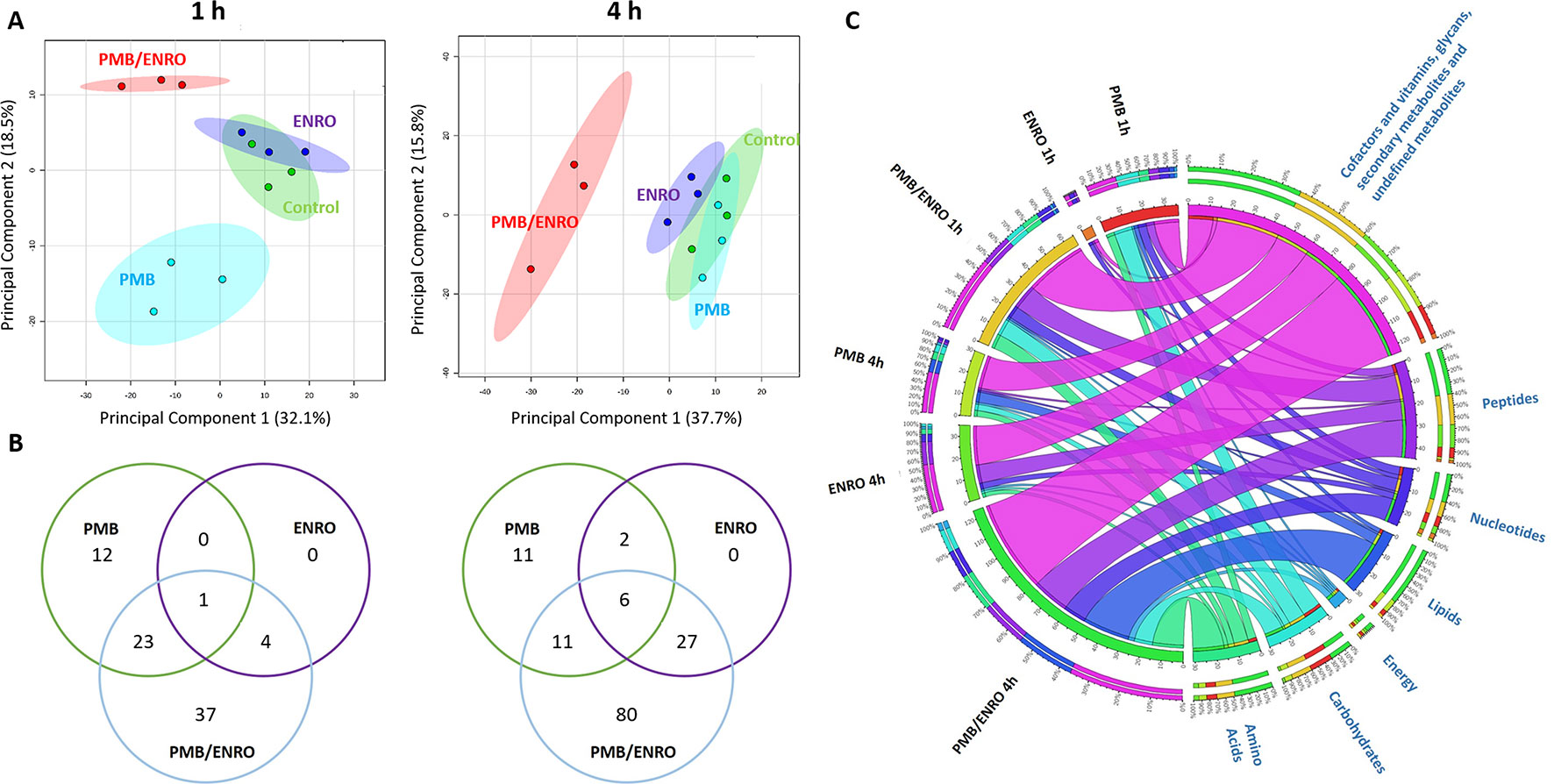
Figure 1 Metabolomics analyses of XDR P. aeruginosa 12196 treated with polymyxin B (PMB) and enrofloxacin (ENRO) alone or in combination (PMB/ENRO). (A) Principal component analysis (PCA) score plots for the metabolites from bacterial cultures treated with polymyxin B, enrofloxacin, and the combination at 1 and 4 h. Green, light blue, purple, and red represent untreated control, polymyxin B alone (PMB), enrofloxacin alone (ENRO), and the combination (COMB), respectively. (B) Venn diagram for the comparison of the numbers of metabolites that were significantly altered by each treatment at each time point. Significant metabolites were selected based on log2 fold change (FC) ≥ 1 and q < 0.05. (C) Bipartite graph connected different treatment groups with significantly altered metabolites in major classes.
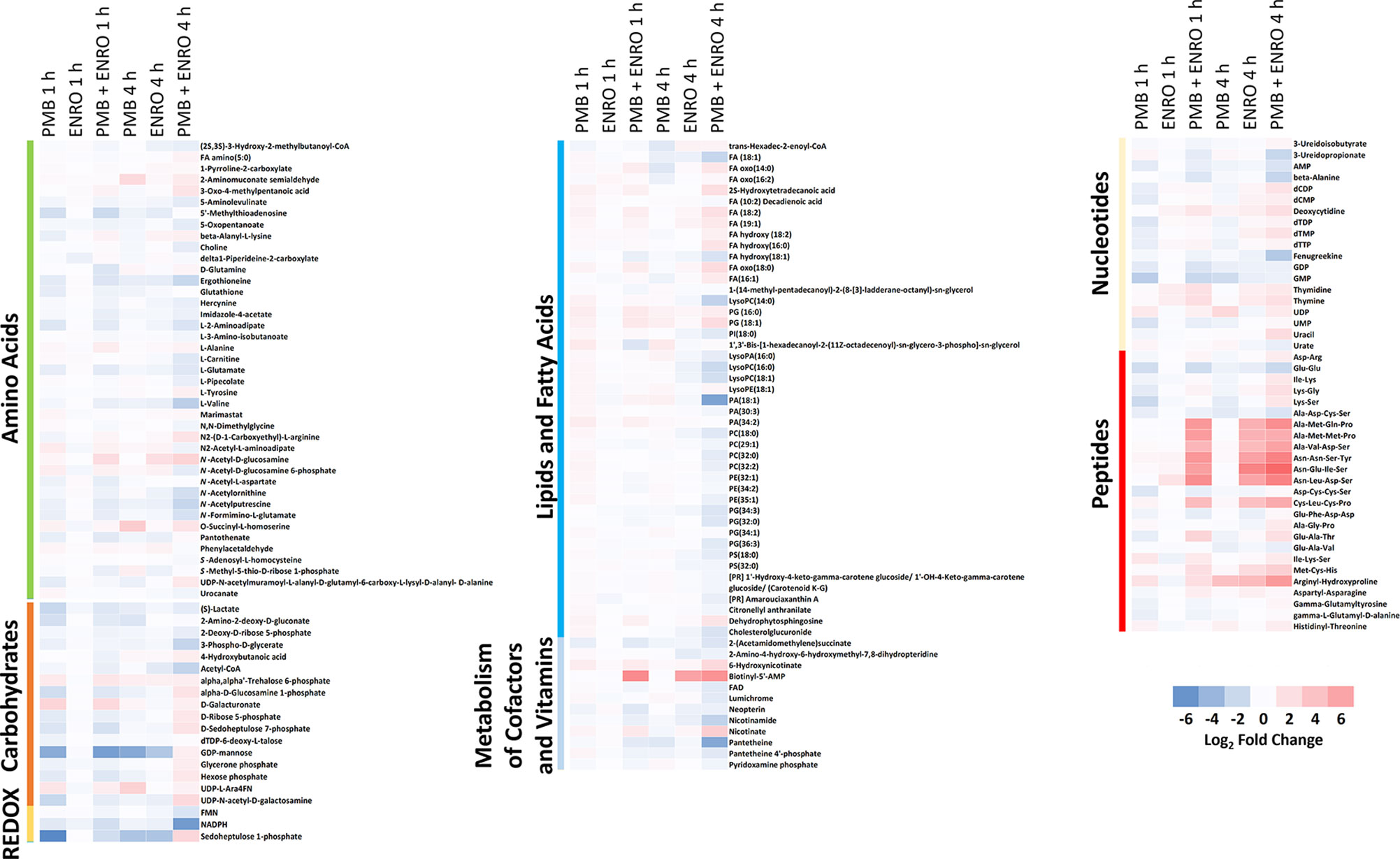
Figure 2 Heatmap profiles of the relative abundance of putative metabolites in XDR P. aeruginosa 12196 following treatment with polymyxin B alone (PMB), enrofloxacin alone (ENRO), and its combination (PMB + ENRO) at 1 and 4 h. Metabolites were grouped into different classes: amino acids, carbohydrates, energy, lipids (lipid metabolism, fatty acids, and phospholipids), co-factors and vitamins, nucleotides, and peptides. Only the significantly perturbed metabolites in the major perturbed pathways are included. Data represent log2FC compared with the untreated control samples at respective time points.
Pathway analyses revealed that multiple metabolic pathways were affected by antibiotic treatments. In details, at 1 and 4 h polymyxin B alone significantly perturbed phospholipid and fatty acid metabolisms, whereas enrofloxacin alone had minimal metabolic perturbations on both metabolic pathways (Figures 1, 2 and S2). On the contrary, the combination therapy significantly perturbed a greater number of key metabolic pathways, including lipid, carbohydrate, nucleotide, and energy metabolism (Figures 1, 2 and S2). The levels of perturbed metabolites are provided in Supplementary Tables 1–2.
Polymyxin B alone and its combination with enrofloxacin significantly perturbed phospholipid and fatty acid levels at 1- and 4-h posttreatment (Figures 2 and 3). More specifically, at 4 h, the drug combination significantly decreased the levels of phospholipids, phosphatidylserine (PS), phosphatidylethanolamine (PE), and phosphatidylglycerol (PG) (Figure 3). Enrofloxacin alone did not have a significant impact on phospholipid levels. Interestingly, the decreased phospholipid levels were accompanied with an accumulation of a large number of fatty acids intracellularly at 4 h (Figure 3). Importantly, at 4 h, polymyxin B alone led to significantly increased levels of uridine 5′-diphospho-beta-(4-deoxy-4-formamido-l-arabinose) (UDP-l-Ara4FN) (log2FC = 2.95), a key precursor of 4-amino-4-deoxy-l-arabinose (l-Ara4N)-modified lipid A but not in the groups of enrofloxacin alone (log2FC = 0.11) or the combination (log2FC = 0.79) (Figure 2).
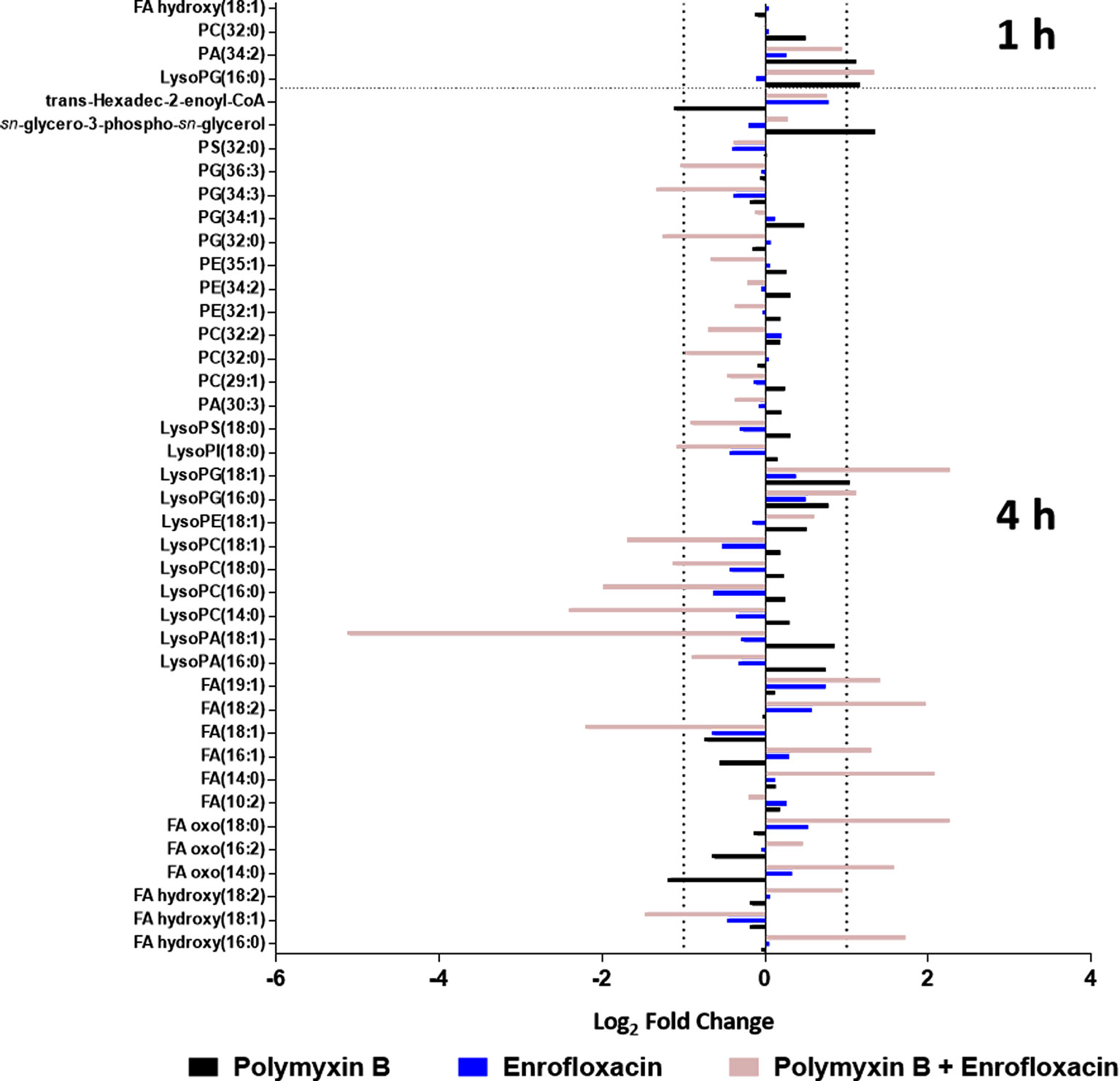
Figure 3 Metabolic perturbations in the phospholipid and fatty acid pathway following treatments with polymyxin B, enrofloxacin, and the combination against XDR P. aeruginosa 12196. Data represent log2FC compared with the untreated control samples at respective time points.
Several intermediate metabolites in pyrimidine metabolism were significantly enriched at 4 h by the combination. In particular, the levels of deoxycytidine (log2FC = 2.04), deoxycytidine monophosphate (dCMP; log2FC = 1.10), deoxycytidine diphosphate (dCDP; log2FC = 1.78), thymine (log2FC = 1.95), thymidine (log2FC = 1.43), deoxythymidine monophosphate (dTMP; log2FC = 1.82), and uridine diphosphate (UDP; log2FC = 1.27) were all significantly increased at 4 h by the combination (Figure 4). For each monotherapy, only UDP (log2FC = 2.48) was elevated following polymyxin B alone at 4 h, while levels of deoxycytidine (log2FC = 1.06), thymine (log2FC = 1.24), and thymidine (log2FC = 1.02) were significantly increased by enrofloxacin alone at 4 h (Figure 4).
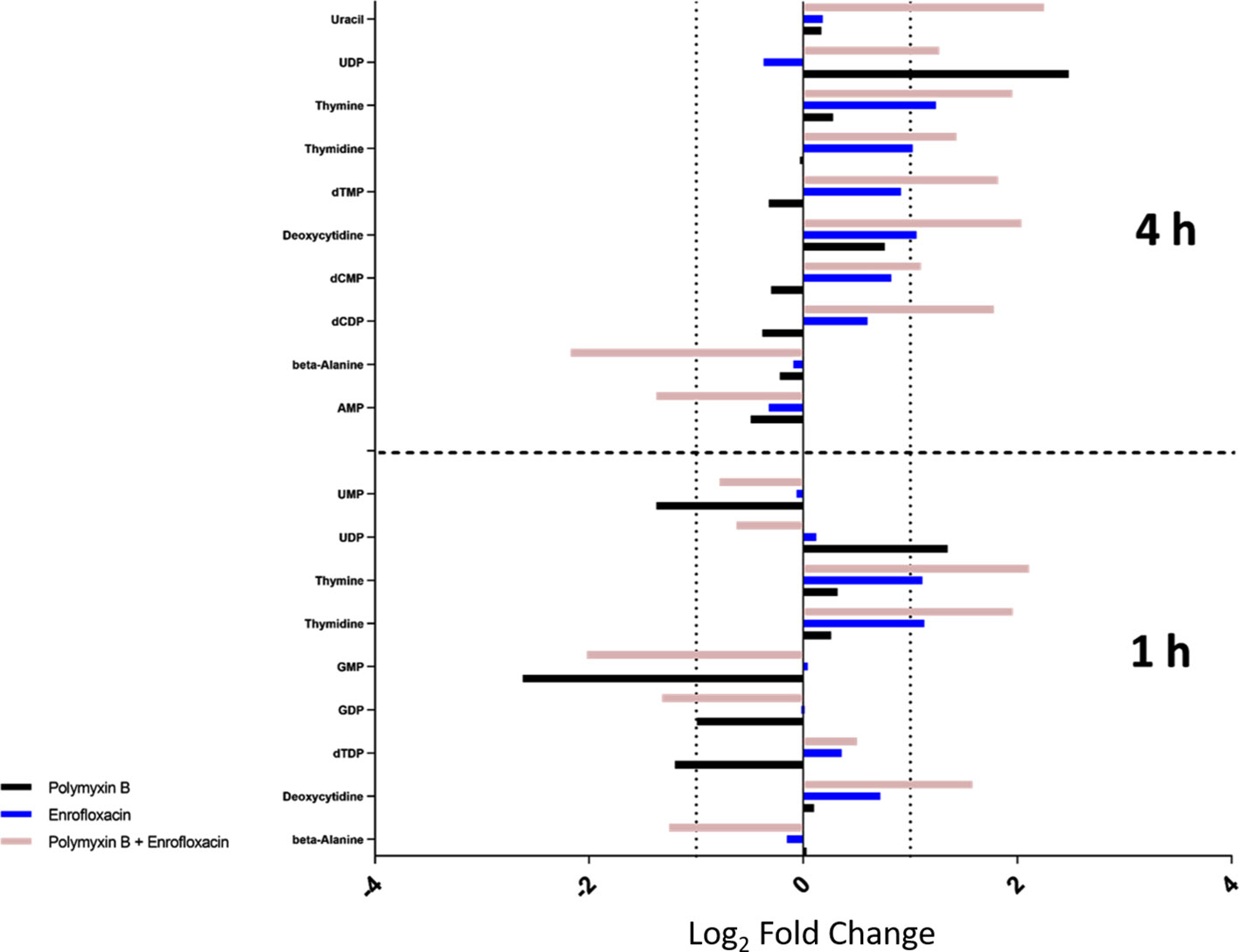
Figure 4 Metabolic perturbations in nucleotide metabolism at 1 and 4 h. Data represent log2FC relative to untreated control samples at respective time points.
In contrast, the combination significantly decreased the levels of the metabolites in purine metabolism at 1 or 4 h. In details, the levels of guanosine monophosphate (GMP; log2FC = −2.02) and guanosine diphosphate (GDP; log2FC = −1.32) were significantly decreased by the combination at 1 h, whereas at 4 h, adenosine monophosphate (AMP; log2FC = −1.37) was significantly decreased (Figure 4). Polymyxin B alone also significantly perturbed GMP (log2FC = −2.62) at 1 h but had a minimal effect on purine nucleotide metabolism at 4 h (Figures 4 and S2; Supplementary Tables 1–2). At the examined concentration, enrofloxacin alone had a minimal effect on purine metabolism at 1 and 4 h.
In addition to nucleotide metabolism, metabolites related to energy metabolism were significantly depleted by the combination but not the monotherapies (except for sedoheptulose 1-phosphate). Specifically, at 1 and 4 h, two important redox co-factors, flavin mononucleotide (FMN; log2FC = −0.31 and −1.37, respectively) and nicotinamide adenine dinucleotide phosphate (NADPH; log2FC = −1.63 and −5.57, respectively), were significantly depleted by the combination (Figure 5 and Supplementary Tables 1–2). Neither polymyxin B nor enrofloxacin alone exhibited significant effects on the levels of redox co-factors at 1 and 4 h.
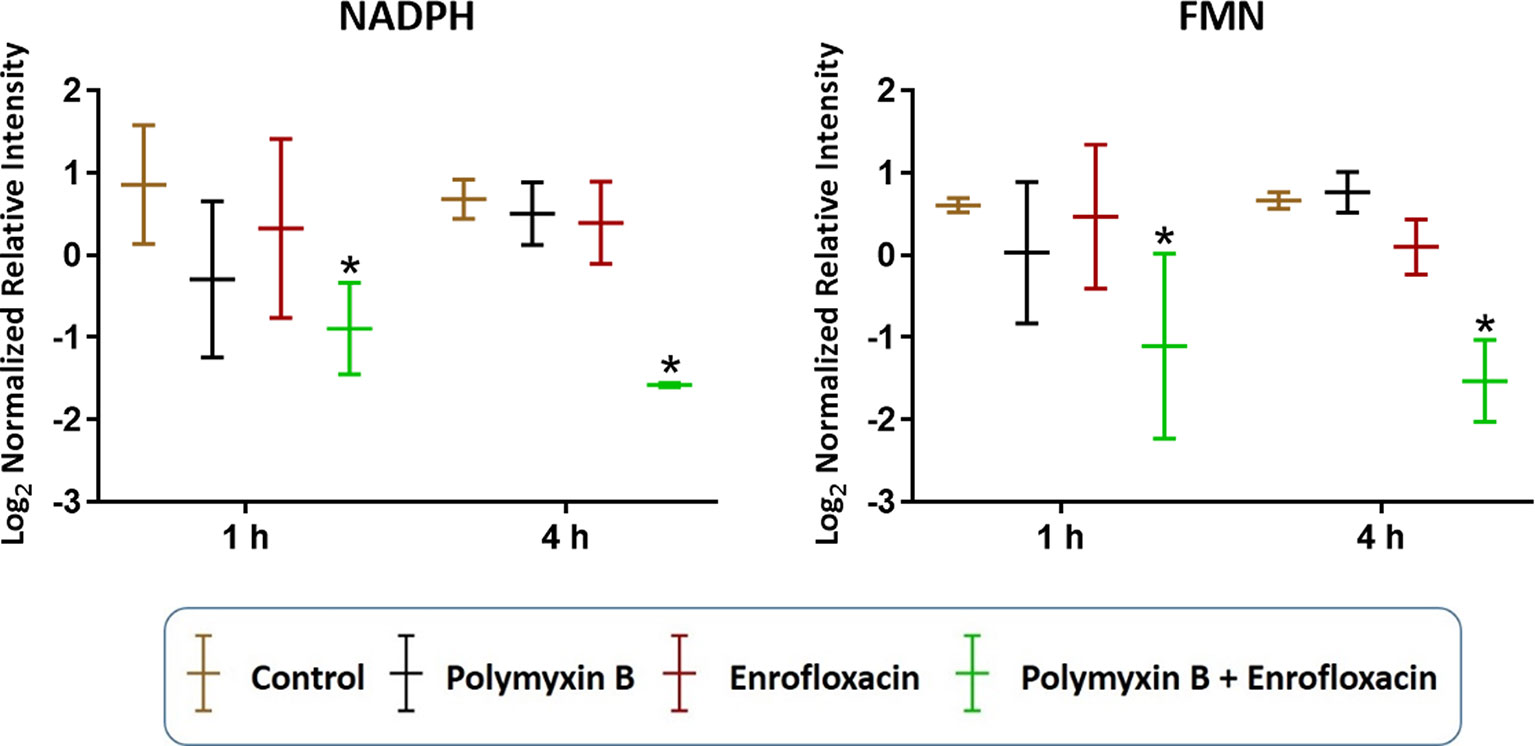
Figure 5 Depletion of key redox co-factors following treatments with polymyxin B, enrofloxacin, and the combination against XDR P. aeruginosa 12196. Bars labeled with an asterisk indicate significant changes in the abundance of metabolites (q < 0.05; ≥1 − log2FC). Data represent means of normalized intensity ± standard deviation (n = 3). Intensity was normalized relative to the median, log2 transformed, and auto-scaled.
The combination of polymyxin B and enrofloxacin significantly perturbed key intermediates in the pentose phosphate pathway (PPP) (Figure 6). Two key intermediate metabolites, d-ribose 5-phosphate (log2FC = −1.02 and −0.72, respectively) and d-sedoheptulose 7-phosphate (log2FC = −1.29 and −1.52, respectively), were significantly decreased at 1 h by polymyxin B alone and the combination. Interestingly, d-ribose 5-phosphate (log2FC = 1.47) and d-sedoheptulose 7-phosphate (log2FC = 1.75) were significantly enriched by the combination at 4 h (log2FC ≥ 1, q < 0.05) (Figure 6). Notably, neither glycolysis nor citric acid cycle was significantly perturbed by each monotherapy or the combination at the tested concentrations (Figure S2 and Supplementary Tables 1–2).
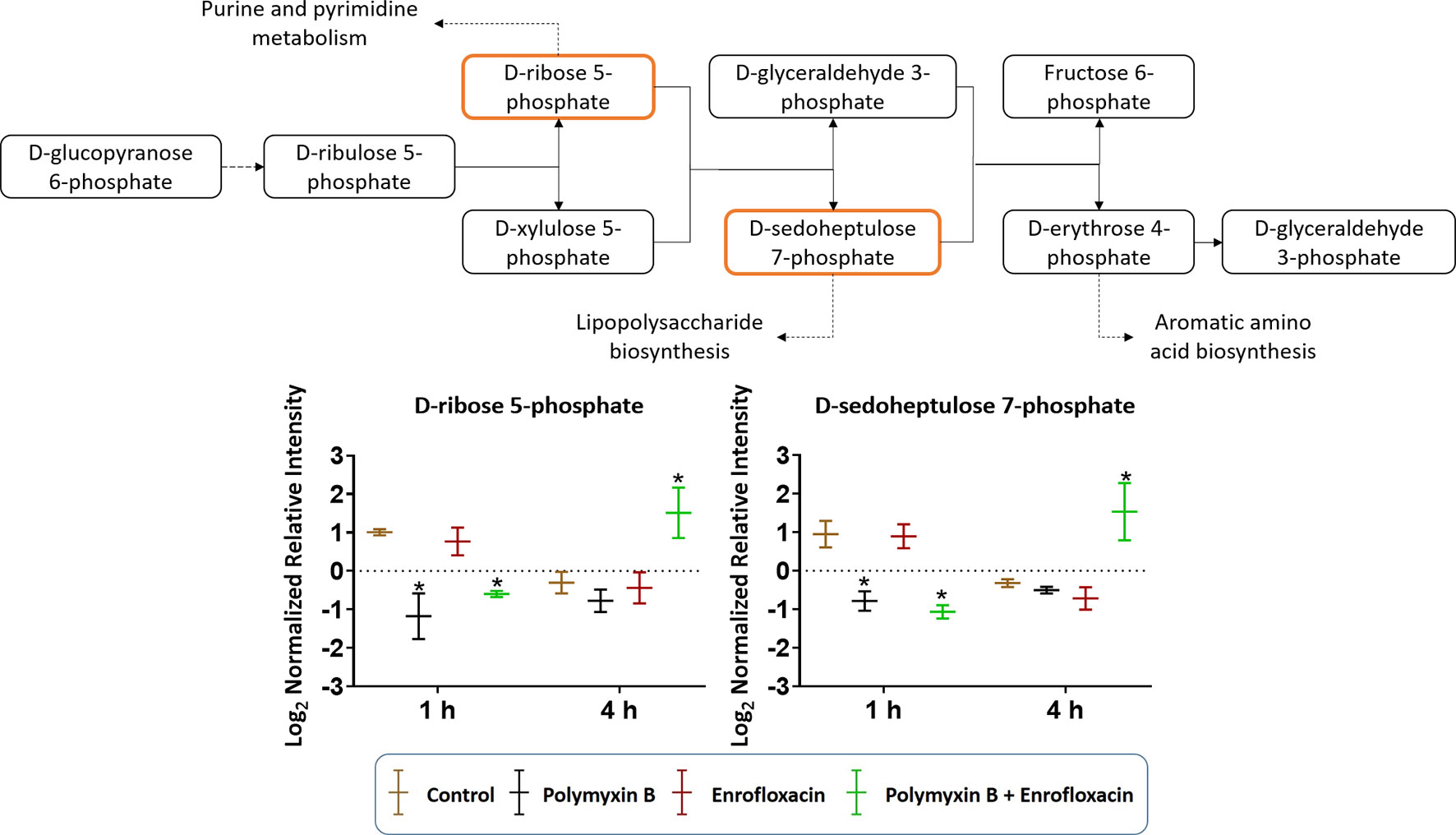
Figure 6 Metabolic perturbations in the pentose phosphate pathway in XDR P. aeruginosa 12196. Orange boxes indicate the metabolites that were significantly perturbed. Bar charts show the raw intensity at respective time points (1 and 4 h), and bars labelled with an asterisk indicate significant changes in the abundance of metabolites (q < 0.05; ≥1 − log2FC). Raw intensity was normalized relative to the median, log2 transformed, and auto-scaled. Data represent geometric means of normalized intensity ± standard deviation (n = 3).
Cell envelope biogenesis was significantly perturbed following treatments with the polymyxin B–enrofloxacin combination (Figure 7). The level of UDP-N-acetyl-d-glucosamine (log2FC = −2.07) was decreased at 1 h in response to polymyxin B alone but increased at 4 h following treatment with the combination (log2FC = 2.45). Interestingly, a similar trend was also observed for UDP-N-acetylmuramoyl-l-alanyl-d-glutamyl-6-carboxyl-lysyl-d-alanyl-d-alanine at 4 h in response to the combination (log2FC = 1.45), whereas enrofloxacin alone had minimal effects on the cell envelope biogenesis pathway at either time point (Figure 7).
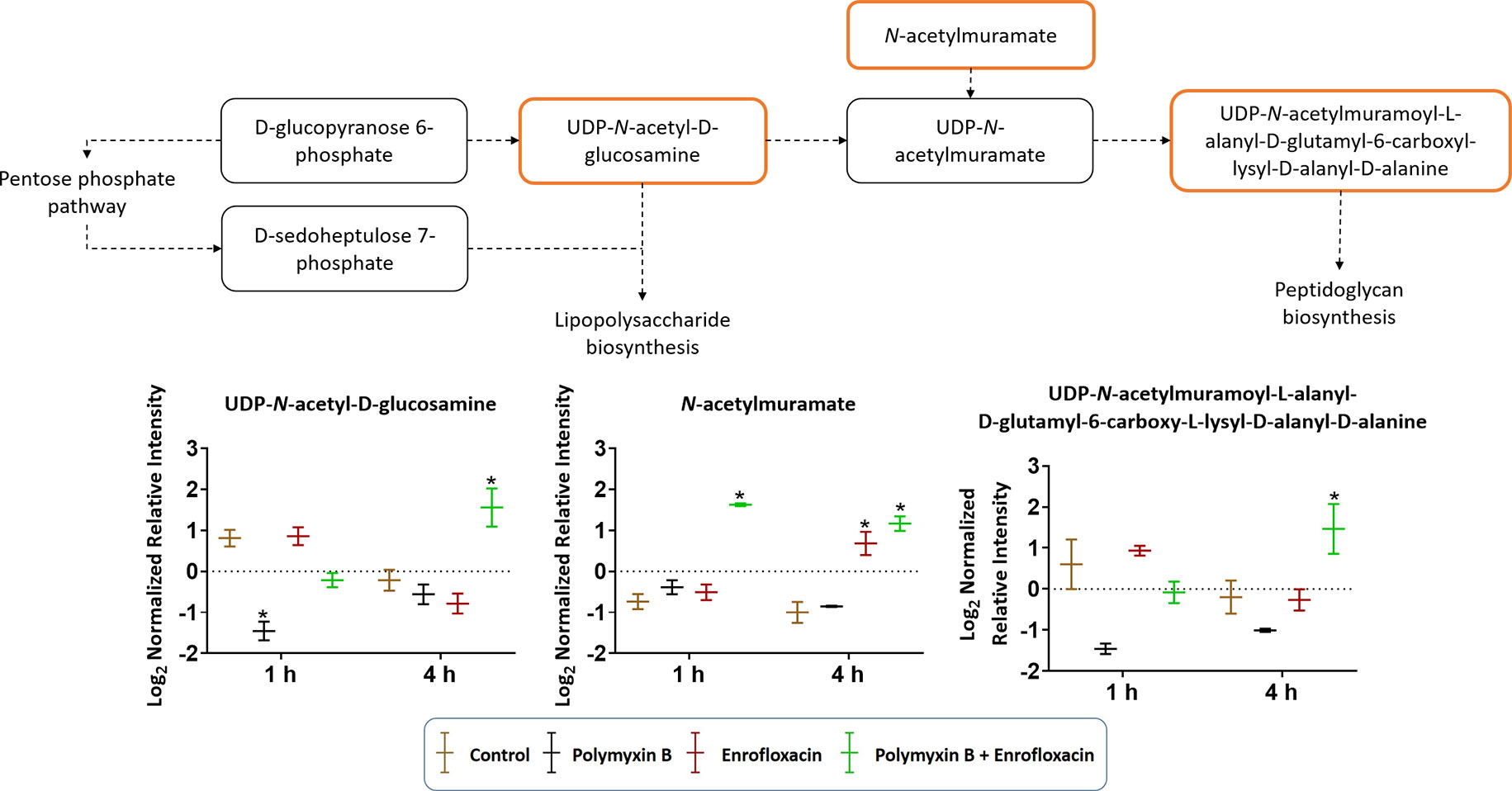
Figure 7 Metabolic perturbations in cell envelope biogenesis in XDR P. aeruginosa 12196. Orange boxes indicate the metabolites that were significantly increased. Bar charts show raw intensity at respective time points (1 and 4 h). Bars labelled with an asterisk indicate significant changes in the abundance of metabolites (q < 0.05; ≥1 − log2FC). Raw intensity was normalized relative to the median, log2 transformed, and auto-scaled. Data represent geometric means of normalized intensity ± standard deviation (n = 3).
Resistance to the last-resort polymyxins can emerge after monotherapy (Bergen et al., 2010; Abdul Rahim et al., 2015; Schneider et al., 2016; Tran et al., 2016b); therefore, rational polymyxin combinations with other antibiotics have been strongly recommended from the PK/PD perspective (Nation et al., 2015). The use of polymyxin B in combination with enrofloxacin, a fluoroquinolone, is highly effective against polymyxin- and enrofloxacin-resistant XDR Pseudomonas aeruginosa (Lin et al., 2018). To our knowledge, the present study is the first to demonstrate that the synergistic killing of polymyxin B in combination with enrofloxacin was time dependent, which was initially facilitated by polymyxin B and subsequently driven by enrofloxacin.
Enrofloxacin is partially metabolized into ciprofloxacin in vivo by the cytochrome P450 enzymes (Küng et al., 1993; Kaartinen et al., 1995; Giguere et al., 1996; Mengozzi et al., 1996; Salvadori et al., 2015), and its PK profile in humans is currently undetermined; therefore, clinically achievable concentrations of ciprofloxacin were used for enrofloxacin (1 mg/L) in the present study (Sánchez Navarro et al., 2002). Polymyxin B was examined at 2 mg/L to ensure the clinical relevance of our findings (Sandri et al., 2013). With an inoculum of 108 CFU/ml, neither 2 mg/L of polymyxin B nor 1 mg/L of enrofloxacin had a significant killing effect on XDR P. aeruginosa 12196; surprisingly, the combination of polymyxin B and enrofloxacin exhibited synergistic killing at 4 h. In order to understand the molecular basis of the dynamic extensive killing, bacterial metabolic profiles were examined at 1 and 4 h following antibiotic treatment (Figure 1).
Consistent with the phenotypical synergistic killing observed in our previous PK/PD studies, our metabolomics results showed that the combination was clearly separated from each monotherapy alone at both time points (Figure 1). Metabolic pathway analyses revealed that the synergistic killing of polymyxin B–enrofloxacin combination was attributed to the perturbations of key metabolic pathways, including lipid, carbohydrate, nucleotide, and energy metabolism (Figures 1, 2 and S2; Supplementary Tables 1–2). A large number of metabolites associated with fatty acids and lipids were significantly perturbed following polymyxin B alone and the combination at 1 and 4 h (Figure 3). These findings are in agreement with the primary mode of action of polymyxins via the disruption of the bacterial outer membrane (Li et al., 2006). Notably, this finding is also consistent with previous results in Acinetobacter baumannii, in which colistin significantly perturbed the gene expression and metabolites in fatty acid and lipid metabolism (Henry et al., 2015; Maifiah et al., 2017). On the contrary, enrofloxacin alone did not affect fatty acid and lipid metabolisms at 1 and 4 h (Figures 1, 3 and S2), which is consistent with its mode of action by the inhibition of topoisomerase II (DNA gyrase) and prevention of the replication of DNA (Küng et al., 1993; Kaartinen et al., 1995; Giguere et al., 1996; Mengozzi et al., 1996; Salvadori et al., 2015). DNA damage activates the SOS gene network that results in the production of DNA repair proteins (Power and Phillips, 1992) and the accumulation of nucleotides (Dörries et al., 2014). Furthermore, transcriptomic analysis of Staphylococcus aureus showed that fluoroquinolone treatment up-regulates the expression of ribonucleotide reductases and several genes involved in the DNA repair pathways (Cirz et al., 2007). This finding is consistent with the metabolomics data obtained in the present study with P. aeruginosa. Our results showed that SOS responses induced by enrofloxacin alone and the combination resulted in the accumulation of pyrimidine metabolites to cope with the inhibition of DNA replication at 4 h (Figure 4). Consistently, the level of d-ribose 5-phosphate in PPP, a key precursor in nucleotide metabolism, was also significantly increased following the treatment with the combination at 4 h, but not at 1 h when polymyxin killing predominated (Figure 6). Overall, our metabolomics results highlight the dominant effect of enrofloxacin on the synergy observed with the combination at 4 h.
Despite the largely overlapping metabolic perturbations between polymyxin B or enrofloxacin alone and the combination, the present data revealed that the synergistic combination induced several unique metabolic alterations in energy metabolism (Figures 2 and 5) and cell envelope biogenesis (Figure 7). It is evident that the combination uniquely caused a significant depletion of key redox co-factors, including FMN and NADPH, at 1- and 4-h posttreatment (Figure 5). It is likely that P. aeruginosa diverted the energy required to synthesize nucleotides toward DNA repair, as a result of the activation of the SOS gene network. The observed decrease in energy metabolism coupled with significant perturbations in PPP suggests an imbalanced redox state due to the treatment with this combination. Interestingly, at 4 h, the combination led to significant increase in fatty acids levels and decrease in phospholipid levels (Figure 3). These alterations might be attributed to the reduced utilization as an energy source and membrane remodelling. Overall, our findings indicate that the inhibition of energy metabolism plays a key role in the mechanism of synergistic bacterial killing by the combination at 4 h.
Moreover, the combination displayed significant and persistent effects on the cell envelope biogenesis in XDR P. aeruginosa (Figure 7). At 4 h, a significant increase in the levels of a peptidoglycan biosynthesis metabolite UDP-N-acetylmuramoyl-l-alanyl-d-glutamyl-6-carboxy-l-lysyl-d-alanyl-d-alanine was observed with the combination (Figure 7). In Streptococcus faecalis, the inhibition of DNA replication by a fluoroquinolone resulted in the formation of thicker cell wall (Higgins et al., 1974). It is very likely that the synergistic killing at 4 h by the combination is driven by the secondary antibiotic, enrofloxacin. In addition to the inhibition of cell wall biogenesis, the combination synergistically inhibited the lipopolysaccharide (LPS) modification pathway (Figure 2). Polymyxin B alone against P. aeruginosa leads to the development of resistance most commonly via lipid A modification with aminoarabinose (Miller et al., 2011). UDP-l-Ara4FN, a key precursor of lipid A modification (Breazeale et al., 2005; Gatzeva-Topalova et al., 2005), was significantly enriched following the treatment with polymyxin B alone at both 1 and 4 h; however, this effect was not observed with the combination (Figure 2). Our results clearly demonstrated that polymyxin resistance via the lipid A modification can emerge rapidly (e.g., as early as in 1 h) even in resistant isolates. Importantly, the inhibition of polymyxin resistance by enrofloxacin plays a key role in the synergistic antibacterial killing. From a PK/PD perspective, our metabolomics results are clinically significant and highlight the importance of a combination therapy in minimizing the development of resistance to the last-line polymyxins.
The development of effective polymyxin combination therapy is of utmost importance in response to the increasing incidence of infections caused by XDR Gram-negative “superbugs.” To the best of our knowledge, this is the first systems pharmacology study to investigate the synergistic effect of polymyxins with a fluoroquinolone antibiotic against XDR P. aeruginosa, which is resistant to all antibiotics, including polymyxins and fluoroquinolones. Importantly, co-administration of enrofloxacin reduced the emergence of polymyxin resistance by inhibiting lipid A modification. These results provide important mechanistic insights into optimizing the clinical use of this promising combination using PK/PD approaches.
This article is dedicated to the memory of Professor Alan Forrest, a friend of many and an inspiring researcher.
All datasets generated for this study are included in the article/Supplementary Files.
JL and TV conceived the project, and all authors were involved in the design of the experiments. Y-WL and M-LH performed the experiments, and Y-WL, M-LH, JZ, YZ, GR, AF, JS, KK, PH, AP, DC, and QZ analyzed the results. All authors reviewed the manuscript.
JL, TV, GR, AF, JS, PH, AP, DC, and KK are supported by a research grant from the National Institute of Allergy and Infectious Diseases of the National Institutes of Health (R01 AI111965). Y-WL and M-LH are recipients of the 2018–2019 Faculty Bridging Fellowship, Monash University. GR, KK, QZ, TV and JL are supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under Award Numbers R01AI132681 and R01AI146160.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors are grateful to Monash Biomedical Proteomics Facility (MBPF) for technical assistance with the metabolomics study. JL and AP are Australian National Health and Medical Research Council (NHMRC) Principal Research Fellows, PH is an Australian NHMRC Senior Principal Research Fellow, and TV and DC are Australian NHMRC Career Development Level 2 Research Fellows. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Allergy and Infectious Diseases or the National Institutes of Health.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2019.01146/full#supplementary-material
Abdul Rahim, N., Cheah, S. E., Johnson, M. D., Yu, H., Sidjabat, H. E., Boyce, J., et al. (2015). Synergistic killing of NDM-producing MDR Klebsiella pneumoniae by two ‘old’ antibiotics—polymyxin B and chloramphenicol. J. Antimicrob. Chemother. 70, 2589–2597. doi: 10.1093/jac/dkv135
Adams, M. D., Nickel, G. C., Bajaksouzian, S., Lavender, H., Murthy, A. R., Jacobs, M. R., et al. (2009). Resistance to colistin in Acinetobacter baumannii associated with mutations in the PmrAB two-component system. Antimicrob. Agents Chemother. 53, 3628–3634. doi: 10.1128/AAC.00284-09
Bergen, P. J., Bulitta, J. B., Forrest, A., Tsuji, B. T., Li, J., Nation, R. L. (2010). Pharmacokinetic/pharmacodynamic investigation of colistin against Pseudomonas aeruginosa using an in vitro model. Antimicrob. Agents Chemother. 54, 3783–3789. doi: 10.1128/AAC.00903-09
Boucher, H. W., Talbot, G. H., Benjamin, D. K., Bradley, J., Guidos, R. J., Jones, R. N., et al. (2013). 10 × ’20 progress—development of new drugs active against gram-negative bacilli: an update from the Infectious Diseases Society of America. Clin. Infect. Dis. 56, 1685–1694. doi: 10.1093/cid/cit152
Breazeale, S. D., Ribeiro, A. A., Mcclerren, A. L., Raetz, C. R. (2005). A formyltransferase required for polymyxin resistance in Escherichia coli and the modification of lipid A with 4-amino-4-deoxy-L-arabinose. Identification and function oF UDP-4-deoxy-4-formamido-L-arabinose. J. Biol. Chem. 280, 14154–14167. doi: 10.1074/jbc.M414265200
Cheah, S.-E., Li, J., Bergen, P. J., Nation, R. L. (2016a). “Polymyxin pharmacokinetics and pharmacodynamics,” in Antibiotic pharmacodynamics. Eds. Rotschafer, J. C., Andes, D. R., Rodvold, K. A. (New York, NY: Springer New York), 221–260. doi: 10.1007/978-1-4939-3323-5_10
Cheah, S.-E., Wang, J., Turnidge, J. D., Li, J., Nation, R. L. (2015). New pharmacokinetic/pharmacodynamic studies of systemically administered colistin against Pseudomonas aeruginosa and Acinetobacter baumannii in mouse thigh and lung infection models: smaller response in lung infection. J. Antimicrob. Chemother. 70, 3291–3297. doi: 10.1093/jac/dkv267
Cheah, S. E., Li, J., Tsuji, B. T., Forrest, A., Bulitta, J. B., Nation, R. L. (2016b). Colistin and polymyxin B dosage regimens against Acinetobacter baumannii: differences in activity and the emergence of resistance. Antimicrob. Agents Chemother. 60, 3921–3933. doi: 10.1128/AAC.02927-15
Chen, C., Gonzalez, F. J., Idle, J. R. (2007). LC-MS-based metabolomics in drug metabolism. Drug Metab. Rev. 39, 581–597. doi: 10.1080/03602530701497804
Cirz, R. T., Jones, M. B., Gingles, N. A., Minogue, T. D., Jarrahi, B., Peterson, S. N., et al. (2007). Complete and SOS-mediated response of Staphylococcus aureus to the antibiotic ciprofloxacin. J. Bacteriol. 189, 531–539. doi: 10.1128/JB.01464-06
Creek, D. J., Jankevics, A., Burgess, K. E., Breitling, R., Barrett, M. P. (2012). IDEOM: an Excel interface for analysis of LC–MS-based metabolomics data. Bioinformatics 28, 1048–1049. doi: 10.1093/bioinformatics/bts069
Dörries, K., Schlueter, R., Lalk, M. (2014). Impact of antibiotics with various target sites on the metabolome of Staphylococcus aureus. Antimicrob. Agents Chemother. 58, 7151–7163. doi: 10.1128/AAC.03104-14
Falagas, M. E., Kasiakou, S. K., Saravolatz, L. D. (2005). Colistin: the revival of polymyxins for the management of multidrug-resistant gram-negative bacterial infections. Clin. Infect. Dis. 40, 1333–1341. doi: 10.1086/429323
Gales, A. C., Castanheira, M., Jones, R. N., Sader, H. S. (2012). Antimicrobial resistance among gram-negative bacilli isolated from Latin America: results from SENTRY Antimicrobial Surveillance Program (Latin America, 2008–2010). Diagn. Microbiol. Infect. Dis. 73, 354–360. doi: 10.1016/j.diagmicrobio.2012.04.007
Gales, A. C., Jones, R. N., Sader, H. S. (2011). Contemporary activity of colistin and polymyxin B against a worldwide collection of Gram-negative pathogens: results from the SENTRY Antimicrobial Surveillance Program (2006–09). J. Antimicrob. Chemother. 66, 2070–2074. doi: 10.1093/jac/dkr239
Gatzeva-Topalova, P. Z., May, A. P., Sousa, M. C. (2005). Structure and mechanism of ArnA: conformational change implies ordered dehydrogenase mechanism in key enzyme for polymyxin resistance. Structure 13, 929–942. doi: 10.1016/j.str.2005.03.018
Giguere, S., Sweeney, R., Belanger, M. (1996). Pharmacokinetics of enrofloxacin in adult horses and concentration of the drug in serum, body fluids, and endometrial tissues after repeated intragastrically administered doses. Am. J. Vet. Res. 57, 1025–1030. doi: 10.1016/j.str.2005.03.018
Goli, H. R., Nahaei, M. R., Ahangarzadeh Rezaee, M., Hasani, A., Samadi Kafil, H., Aghazadeh, M. (2016). Emergence of colistin resistant Pseudomonas aeruginosa at Tabriz hospitals, Iran. Iran. J. Microbiol. 8, 62–69.
Han, M.-L., Liu, X., Velkov, T., Lin, Y.-W., Zhu, Y., Li, M., et al. (2018). Metabolic analyses revealed time-dependent synergistic killing by colistin and aztreonam combination against multidrug-resistant Acinetobacter baumannii. Front. Microbiol. 9, 2776. doi: 10.3389/fmicb.2018.02776
Henry, R., Crane, B., Powell, D., Deveson Lucas, D., Li, Z., Aranda, J., et al. (2015). The transcriptomic response of Acinetobacter baumannii to colistin and doripenem alone and in combination in an in vitro pharmacokinetics/pharmacodynamics model. J. Antimicrob. Chemother. 70, 1303–1313. doi: 10.1093/jac/dku536
Higgins, M., Daneo-Moore, L., Boothby, D., Shockman, G. (1974). Effect of inhibition of deoxyribonucleic acid and protein synthesis on the direction of cell wall growth in Streptococcus faecalis. J. Bacteriol. 118, 681–692.
Hsueh, P. R., Teng, L. J., Yang, P. C., Chen, Y. C., Ho, S. W., Luh, K. T. (1998). Persistence of a multidrug-resistant Pseudomonas aeruginosa clone in an intensive care burn unit. J. Clin. Microbiol. 36, 1347–1351.
Kaartinen, L., Salonen, M., Älli, L., Pyörälä, S. (1995). Pharmacokinetics of enrofloxacin after single intravenous, intramuscular and subcutaneous injections in lactating cows. J. Vet. Pharmacol. Ther. 18, 357–362. doi: 10.1111/j.1365-2885.1995.tb00604.x
Kaddurah-Daouk, R., Weinshilboum, R. M. (2014). Pharmacometabolomics: implications for clinical pharmacology and systems pharmacology. Clin. Pharmacol. Ther. 95, 154–167. doi: 10.1038/clpt.2013.217
Kanehisa, M., Goto, S. (2000). KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic. Acids Res. 28, 27–30. doi: 10.1093/nar/28.1.27
Karp, P. D., Ouzounis, C. A., Moore-Kochlacs, C., Goldovsky, L., Kaipa, P., Ahrén, D., et al. (2005). Expansion of the BioCyc collection of pathway/genome databases to 160 genomes. Nucleic. Acids Res. 33, 6083–6089. doi: 10.1093/nar/gki892
Kirwan, J. A., Weber, R. J. M., Broadhurst, D. I., Viant, M. R. (2014). Direct infusion mass spectrometry metabolomics dataset: a benchmark for data processing and quality control. Sci. Data 1, 140012. doi: 10.1038/sdata.2014.12
Küng, K., Riond, J. L., Wanner, M. (1993). Pharmacokinetics of enrofloxacin and its metabolite ciprofloxacin after intravenous and oral administration of enrofloxacin in dogs. J. Vet. Pharmacol. Ther. 16, 462–468. doi: 10.1111/j.1365-2885.1993.tb00212.x
Letunic, I., Yamada, T., Kanehisa, M., Bork, P. (2008). iPath: interactive exploration of biochemical pathways and networks. Trends Biochem. Sci. 33, 101–103. doi: 10.1016/j.tibs.2008.01.001
Li, J., Nation, R. L., Turnidge, J. D., Milne, R. W., Coulthard, K., Rayner, C. R., et al. (2006). Colistin: the re-emerging antibiotic for multidrug-resistant Gram-negative bacterial infections. Lancet Infect. Dis. 6, 589–601. doi: 10.1016/S1473-3099(06)70580-1
Lin, Y.-W., Yu, H. H., Zhao, J., Han, M.-L., Zhu, Y., Akter, J., et al. (2018). Polymyxin B in combination with enrofloxacin exerts synergistic killing against extensive drug-resistant Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 62, e0028–e0018. doi: 10.1128/AAC.00028-18
Maifiah, M. H. M., Cheah, S.-E., Johnson, M. D., Han, M.-L., Boyce, J. D., Thamlikitkul, V., et al. (2016). Global metabolic analyses identify key differences in metabolite levels between polymyxin-susceptible and polymyxin-resistant Acinetobacter baumannii. Sci. Rep. 6, 22287. doi: 10.1038/srep22287
Maifiah, M. H. M., Creek, D. J., Nation, R. L., Forrest, A., Tsuji, B. T., Velkov, T., et al. (2017). Untargeted metabolomics analysis reveals key pathways responsible for the synergistic killing of colistin and doripenem combination against Acinetobacter baumannii. Sci. Rep. 7, 45527. doi: 10.1038/srep45527
Mastrangelo, A., Armitage, G., García, A., Barbas, C. (2014). Metabolomics as a tool for drug discovery and personalised medicine. A review. Curr. Top. Med. Chem. 14, 2627–2636. doi: 10.2174/1568026614666141215124956
Mcphee, J. B., Bains, M., Winsor, G., Lewenza, S., Kwasnicka, A., Brazas, M. D., et al (2006). Contribution of the PhoP-PhoQ and PmrA-PmrB two-component regulatory systems to Mg2+-induced gene regulation in Pseudomonas aeruginosa. J. Bacteriol. 188, 3995–4006. doi: 10.1128/JB.00053-06
Mengozzi, G., Intorre, L., Bertini, S., Soldani, G. (1996). Pharmacokinetics of enrofloxacin and its metabolite ciprofloxacin after intravenous and intramuscular administrations in sheep. Am. J. Vet. Res. 57, 1040–1043.
Miller, A. K., Brannon, M. K., Stevens, L., Johansen, H. K., Selgrade, S. E., Miller, S. I., et al. (2011). PhoQ mutations promote lipid A modification and polymyxin resistance of Pseudomonas aeruginosa found in colistin-treated cystic fibrosis patients. Antimicrob. Agents Chemother. 55, 5761–5769. doi: 10.1128/AAC.05391-11
Nation, R. L., Li, J. (2009). Colistin in the 21st century. Curr. Opin. Infect. Dis. 22, 535. doi: 10.1097/QCO.0b013e328332e672
Nation, R. L., Li, J., Cars, O., Couet, W., Dudley, M. N., Kaye, K. S., et al. (2015). Framework for optimisation of the clinical use of colistin and polymyxin B: the Prato polymyxin consensus. Lancet Infect. Dis. 15, 225–234. doi: 10.1016/S1473-3099(14)70850-3
Nation, R. L., Velkov, T., Li, J. (2014). Colistin and polymyxin B: peas in a pod, or chalk and cheese? Clin. Infect. Dis. 59, 88–94. doi: 10.1093/cid/ciu213
Peng, B., Li, H., Peng, X.-X. (2015). Functional metabolomics: from biomarker discovery to metabolome reprogramming. Protein Cell 6, 628–637. doi: 10.1007/s13238-015-0185-x
Power, E., Phillips, I. (1992). Induction of the SOS gene (umuC) by 4-quinolone antibacterial drugs. J. Med. Microbiol. 36, 78–82. doi: 10.1099/00222615-36-2-78
Salvadori, M., De Vito, V., Owen, H., Giorgi, M. (2015). Pharmacokinetics of enrofloxacin and its metabolite ciprofloxacin after intracoelomic administration in Tortoises (Testudo hermanni). Isr. J. Vet. Med. 70, 45–48.
Sánchez Navarro, M. D., Sayalero Marinero, M. L., Sánchez Navarro, A. (2002). Pharmacokinetic/pharmacodynamic modelling of ciprofloxacin 250 mg/12 h versus 500 mg/24 h for urinary infections. J. Antimicrob. Chemother. 50, 67–72. doi: 10.1093/jac/dkf079
Sandri, A. M., Landersdorfer, C. B., Jacob, J., Boniatti, M. M., Dalarosa, M. G., Falci, D. R., et al. (2013). Population pharmacokinetics of intravenous polymyxin B in critically ill patients: implications for selection of dosage regimens. Clin. Infect. Dis. 57, 524–531. doi: 10.1093/cid/cit334
Scheltema, R. A., Jankevics, A., Jansen, R. C., Swertz, M. A., Breitling, R. (2011). PeakML/mzMatch: a file format, Java library, R library, and tool-chain for mass spectrometry data analysis. Anal. Chem. 83, 2786–2793. doi: 10.1021/ac2000994
Schneider, E. K., Azad, M. A., Han, M.-L., Zhou, Q., Wang, J., Huang, J. X., et al. (2016). An “unlikely” pair: the antimicrobial synergy of polymyxin B in combination with the cystic fibrosis transmembrane conductance regulator drugs KALYDECO and ORKAMBI. ACS Infect. Dis. 2, 478–488. doi: 10.1021/acsinfecdis.6b00035
Tam, V. H., Schilling, A. N., Vo, G., Kabbara, S., Kwa, A. L., Wiederhold, N. P., et al. (2005). Pharmacodynamics of polymyxin B against Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 49, 3624–3630. doi: 10.1128/AAC.49.9.3624-3630.2005
Tran, T. B., Cheah, S.-E., Yu, H. H., Bergen, P. J., Nation, R. L., Creek, D. J., et al. (2016a). Anthelmintic closantel enhances bacterial killing of polymyxin B against multidrug-resistant Acinetobacter baumannii. J. Antibiot. 69, 415–421. doi: 10.1038/ja.2015.127
Tran, T. B., Velkov, T., Nation, R. L., Forrest, A., Tsuji, B. T., Bergen, P. J., et al. (2016b). Pharmacokinetics/pharmacodynamics of colistin and polymyxin B: are we there yet? Int. J. Antimicrob. Agents 48, 592–597. doi: 10.1016/j.ijantimicag.2016.09.010
Vincent, I. M., Ehmann, D. E., Mills, S. D., Perros, M., Barrett, M. P. (2016). Untargeted metabolomics to ascertain antibiotic modes of action. Antimicrob. Agents Chemother. 60, 2281–2291. doi: 10.1128/AAC.02109-15
World Health Organization. (2014). Antimicrobial resistance: global report on surveillance. World Health Organization. Available at: https://apps.who.int/iris/bitstream/handle/10665/112642/9789241564748_eng.pdf;jsessionid = 7D47E40CA5F5ADBE0369B26442B9BCD4?sequence = 1 (Accessed August 9, 2019).
Xia, J., Sinelnikov, I. V., Han, B., Wishart, D. S. (2015). MetaboAnalyst 3.0—making metabolomics more meaningful. Nucleic. Acids Res. 43, W251–W257. doi: 10.1093/nar/gkv380
Keywords: polymyxin, extensive drug-resistant, Pseudomonas aeruginosa, enrofloxacin, metabolomics
Citation: Lin Y-W, Han M-L, Zhao J, Zhu Y, Rao G, Forrest A, Song J, Kaye KS, Hertzog P, Purcell A, Creek D, Zhou QT, Velkov T and Li J (2019) Synergistic Combination of Polymyxin B and Enrofloxacin Induced Metabolic Perturbations in Extensive Drug-Resistant Pseudomonas aeruginosa. Front. Pharmacol. 10:1146. doi: 10.3389/fphar.2019.01146
Received: 13 June 2019; Accepted: 04 September 2019;
Published: 03 October 2019.
Edited by:
Yurong Lai, Gilead, United StatesReviewed by:
Zhihao Liu, University of Illinois at Chicago, United StatesCopyright © 2019 Lin, Han, Zhao, Zhu, Rao, Forrest, Song, Kaye, Hertzog, Purcell, Creek, Zhou, Velkov and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tony Velkov, dG9ueS52ZWxrb3ZAdW5pbWVsYi5lZHUuYXU=; Jian Li, amlhbi5saUBtb25hc2guZWR1
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.