- 1Laboratory of Biotechnology, Institute of Chemical Biology and Fundamental Medicine, Siberian Branch of the Russian Academy of Sciences, Novosibirsk, Russia
- 2Laboratory of Genome Editing, Institute of Chemical Biology and Fundamental Medicine, Siberian Branch of the Russian Academy of Sciences, Novosibirsk, Russia
- 3Department of Natural Sciences, Novosibirsk State University, Novosibirsk, Russia
- 4Laboratory of Biomedical Chemistry, Institute of Chemical Biology and Fundamental Medicine, Siberian Branch of the Russian Academy of Sciences, Novosibirsk, Russia
Cell penetrating peptides (CPP) are promising agents for transporting diverse cargo into the cells. The amino acid sequence and the mechanism of lactaptin entry into the cells allow it to be included into CPP group. Lactaptin, the fragment of human milk kappa-casein, and recombinant lactaptin (RL2) were initially discovered as molecules that induced apoptosis of cultured cancer cells and did not affect non-malignant cells. Here, we analyzed the recombinant lactaptin potency to form complexes with nucleic acids and to act as a gene delivery system. To study RL2-dependent delivery, three type of nucleic acid were used as a models: plasmid DNA (pDNA), siRNA, and non-coding RNA which allow to detect intracellular localization through their functional activity. We have demonstrated that RL2 formed positively charged noncovalent 110-nm-sized complexes with enhanced green fluorescent protein (EGFP)-expressing plasmid DNA. Ca2+ ions stabilized these complexes, whereas polyanion heparin displaced DNA from the complexes. The functional activity of delivered nucleic acids were assessed by fluorescent microscopy using A549 lung adenocarcinoma cells and A431 epidermoid carcinoma cells. We observed that RL2:pDNA complexes provided EGFP expression in the treated cells and that strongly confirmed the entering pDNA into the cells. The efficiency of cell transformation by these complexes increased when RL2:pDNA ratio increased. Pre-treatment of the cells with anti-RL2 antibodies partly inhibited the entry of pDNA into the cells. RL2-mediated delivery of siRNA against EGFP was analyzed when A549 cells were co-transfected with EGFP-pDNA and RL2:siRNA complexes. siRNA against EGFP efficiently inhibited the expression of EGFP being delivered as RL2:siRNA complexes. We have previously demonstrated that non-coding U25 small nucleolar RNA (snoRNA) can decrease cell viability. Cancer cell transfection with RL2-snoRNA U25 complexes lead to a substantial decrease of cell viability, confirming the efficiency of snoRNA U25 delivery. Collectively, these findings indicate that recombinant lactaptin is able to deliver noncovalently associated nucleic acids into cancer cells in vitro.
Introduction
Successful delivery of exogenous DNA and RNA molecules into cells has major implications for gene-based technologies and therapeutic approaches. Since free DNA and RNA are not readily taken up by cells, various delivery systems were developed during the last twenty years (Bolhassani, 2011). Delivery molecules or vesicles help nucleic acids to cross biological barriers and prevent enzymatic degradation by endogenous nucleases. Besides the lipid-based delivery system, the polymer-based delivery system or inorganic nanoparticles, peptide carriers can be also used for nucleic acids transport into the cells (Zorko and Langel, 2005; Rathnayake et al., 2017). Among peptide carriers, the most promising are short amphipathic and cationic peptides, which translocate themselves across membranes and are collectively named as cell-penetrating peptides (CPP) (Hoyer and Neundorf, 2012; Morris and Labhasetwar, 2015). There is no unique classification for CPPs yet, and usually, they are classified according to their origin, specificity to cargo, and amino acid composition. In 1997, Morris et al. demonstrated that 27-residue CPP peptide vector being premixed with single- or double-stranded deoxyoligonucleotides efficiently delivered them into cultured mammalian cells (Morris et al., 1997). Later, two CPPs—transportan and penetratin—successfully delivered 21-mer peptide nucleic acid (PNA) complementary to the human galanin receptor type 1 mRNA resulted in the suppression of galanin receptor (Pooga et al., 1998). Thereby, CPPs deliver functionally active nucleic acids into the cells.
Lactaptin, an 8.6-kDa proteolytic fragment of human milk kappa-casein, selectively kills various tumor cells while it is used in micromole concentration (Semenov et al., 2010; Koval et al., 2012). We have earlier demonstrated that recombinant lactaptin (RL2) penetrates into cells partly through lipid raft-mediated dynamin-independent pinocytosis and partly through direct penetration across the plasma membrane (Chinak et al., 2016). The comparison of RL2 primary structure and the mechanism of its penetration into the cell suggests that it can be assigned to the CPP.
In the present work, we analyzed if lactaptin analog forms complexes with nucleic acids and if it acts as a delivery system with no cytotoxic effects.
Results
RL2 Forms Non-Covalent Complexes With Plasmid DNA
CPPs, like other cationic proteins or peptides, can condense DNA through their cationic residues by non-covalent electrostatic interactions (Tecle et al., 2003). Since RL2 has the properties of cell-penetrating peptides, its ability to deliver cargo molecules into the cells was studied using a plasmid DNA (pEGFP). This plasmid contains a coding sequence of the enhanced green fluorescent protein (EGFP) under the control of the cytomegalovirus (CMV) immediate early promoter, allowing expression of EGFP in eukaryotic cells.
The charge ratio N/P or peptide nitrogen per nucleic acid phosphate of RL2:pEGFP complexes was calculated as described in the Materials and Methods section for the analysis of the efficient ratio of RL2 and pDNA. Here, to optimize empirically the ratio of RL2 and pEGFP for the non-covalent complexes (RL2:pEGFP) formation, we have varied the RL2 amount, whereas pDNA amount was constant within N/P ratio of 0.5 to 5. UV-vis spectroscopy analysis of complexes (Supplementary Figure 1) demonstrates that RL2:pDNA complexes have maximum absorbance in the range of 260 to 280 nm, whereas pDNA has an absorption maximum at 260 nm and RL2 at 280 nm.
The efficiency of complex formation was analyzed by agarose gel retardation assay. Data obtained (Figure 1A) shows the conditions when pDNA movement retards in gel indicating complex formation. The increase of RL2 amount leads to the decrease of free plasmid DNA in a reaction mixture. Complete retardation was reached at a N/P charge ratio of five (Figure 1A). It indicates that all pDNA was totally involved into RL2:pEGFP complexes which had no mobility under the conditions of electrophoresis due to their larger size compared to the pores size, or the neutral charge.
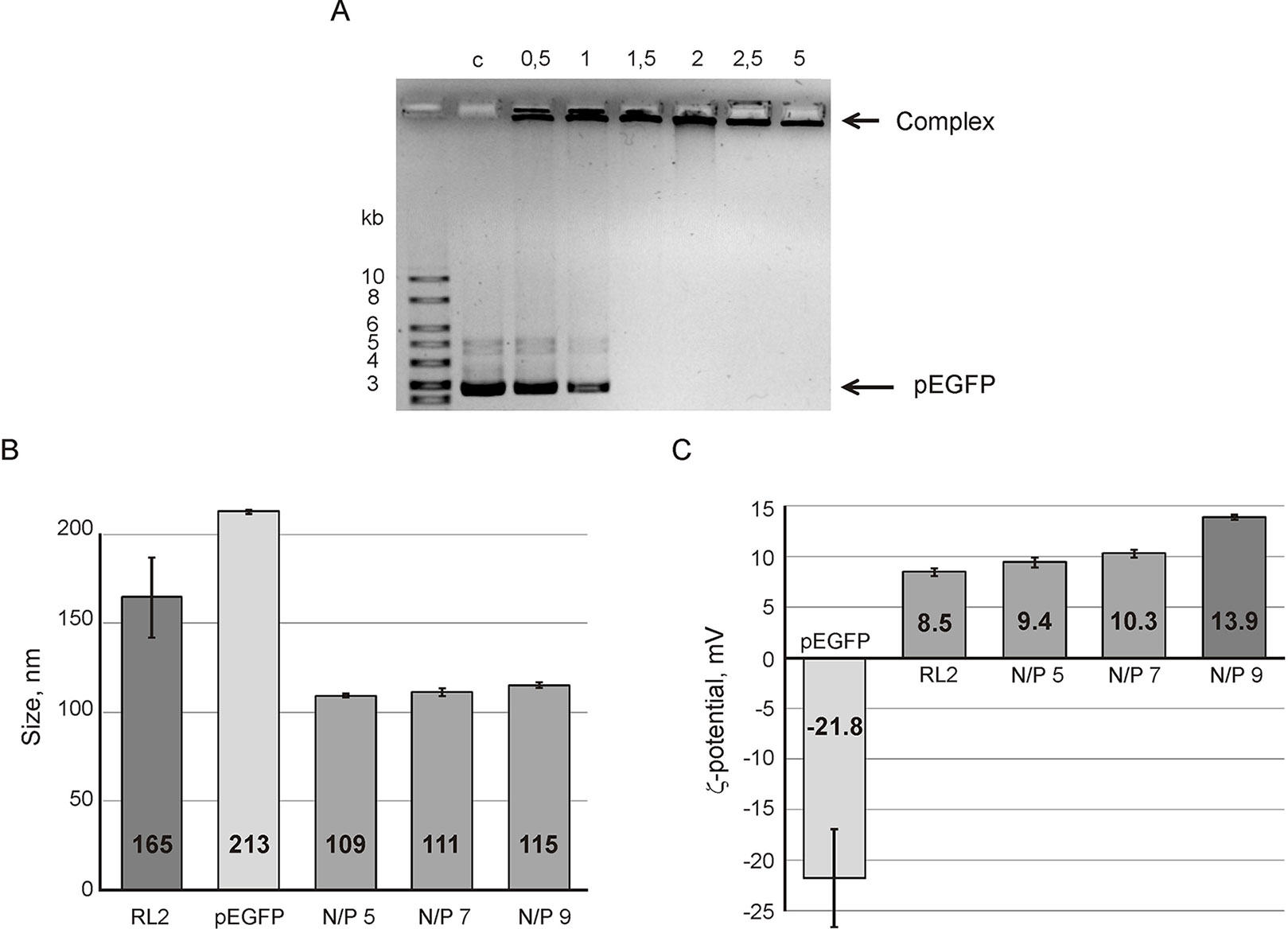
Figure 1 Analysis of RL2:pEGFP complex formation. (A) Gel retardation assays of the RL2:pEGFP complex. N/P ratios of RL2/pDNA charge have varied from 0.5 to 5 in reaction mixtures; C- control (free pDNA). Particle size (B) and ζ potential (C) of RL2:pEGFP complexes were analyzed for N/P ratios 5, 7, and 9. Reactions were performed in MES buffer for 5 min at 37°C. Particle size was estimated by the DLS technique. The data are representative of three independent repeats and are shown as the mean ± SD.
To characterize RL2:pEGFP complexes, the hydrodynamic size and ζ-potential of the complexes were estimated using the dynamic light scattering (DLS) technique. Figure 1B shows that the complex size is near 110 nm with no significant differences for the charge ratio range from 5 to 9. The size of the RL2:pEGFP complex was smaller than the sizes of RL2 or pEGFP alone. Thus, when complexes form, both molecules condense.
Electrokinetic potential (ζ-potential) of pDNA and RL2 in a solution demonstrated a negative charge for DNA and a positive charge for RL2. For complexes, ζ-potential gradually increases with the N/P increasing from 5 to 9 (Figure 1C). We suppose that the increasing of ζ-potential is specified by the rising amount of surface-exposed cationic groups of RL2 molecules in RL2:pDNA complexes.
Ca2+ and Heparin Change the Stability of RL2:pEGFP Complexes
Since RL2 originates from milk k-casein, it can be sensitive to Ca2+ ions concentration. It is known that environmental Ca2+ can stimulate aggregation or dissociation of casein’s micelles (Ye and Harte, 2013). To reveal the Ca2+ effects on the stability of RL2:pEGFP complex, complexes were formed under semi-extreme N/P ratios. Under N/P = 7, there was no free pDNA and Ca2+ did not release pDNA from the RL2:pEGFP complex (Figure 2A). Under N/P = 1, only half of pDNA molecules were in the RL2:pEGFP complex, and these conditions allow us to analyze Ca2+ concentration when total pDNA is incorporated in complex with RL2. The increase of Ca2+ concentration rises the amount of pDNA incorporated in the complex with RL2 (Figure 2A).
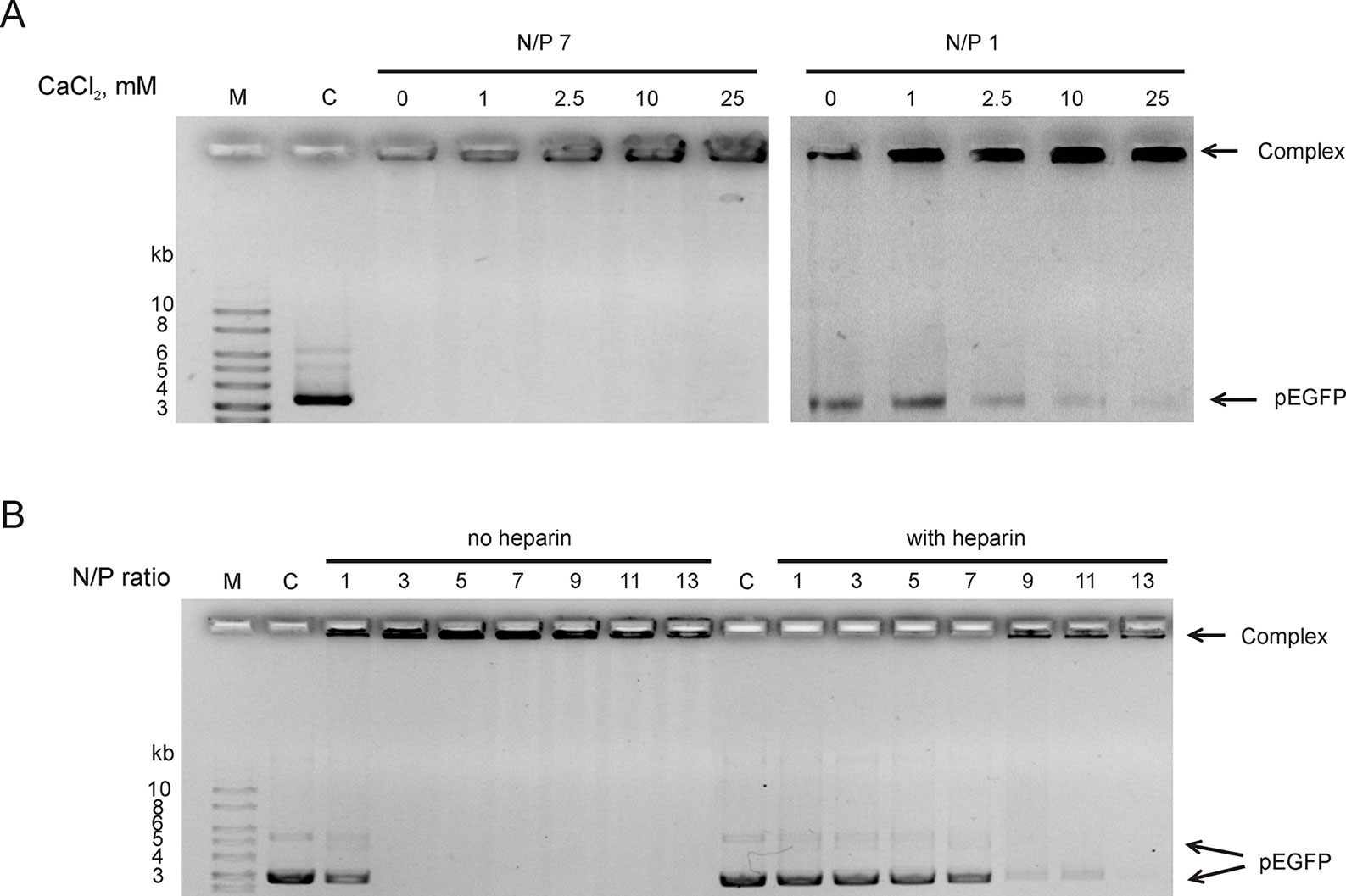
Figure 2 RL2:pEGFP complex analysis in the presence of Ca2+ (A) and heparin in ratio 2.4 Units/1 mkg pEGFP (B). Gel retardation assays. C—free pDNA (150 ng); M—DNA ladder. Complexes were pre-formed and after that Ca2+ or heparin were added. Representative picture of three independent experiments.
Heparin is a stronger polyanion than DNA; therefore, it can competitively bound to RL2, releasing plasmid from RL2:pEGFP complexes. Heparin sodium salt (2.4 units per 1 µg DNA) was added to pre-formed RL2:pEGFP complexes, and the appearance of free pDNA was analyzed. We showed that heparin releases pEGFP from the complexes with N/P ≤ 11 (Figure 2B). Complexes with high N/P ratio were more resistant to heparin.
Thus, Ca2+ divalent cations stabilize complexes and polyanion heparin destabilizes RL2:pEGFP complexes.
Plasmid DNA Delivery
To examine whether RL2 delivers pEGFP into human cancer cells the functional-based method was used. A549 and A431 cells were incubated with RL2:pEGFP complexes with various N/P ratios. After that, a reporter EGFP protein was analyzed by fluorescence microscopy. EGFP fluorescence was observed in the cells treated with complexes characterized by the high N/P ratio (Figure 3A). No fluorescence signal was seen when cells were treated with naked pEGFP, indicating that plasmid DNA itself did not enter into the cells under these conditions. The increase of RL2 content in complexes led to the increase of the number of cells with EGFP, and the highest EGFP fluorescence was in cells incubated with a RL2:pEGFP complexes with N/P = 9.
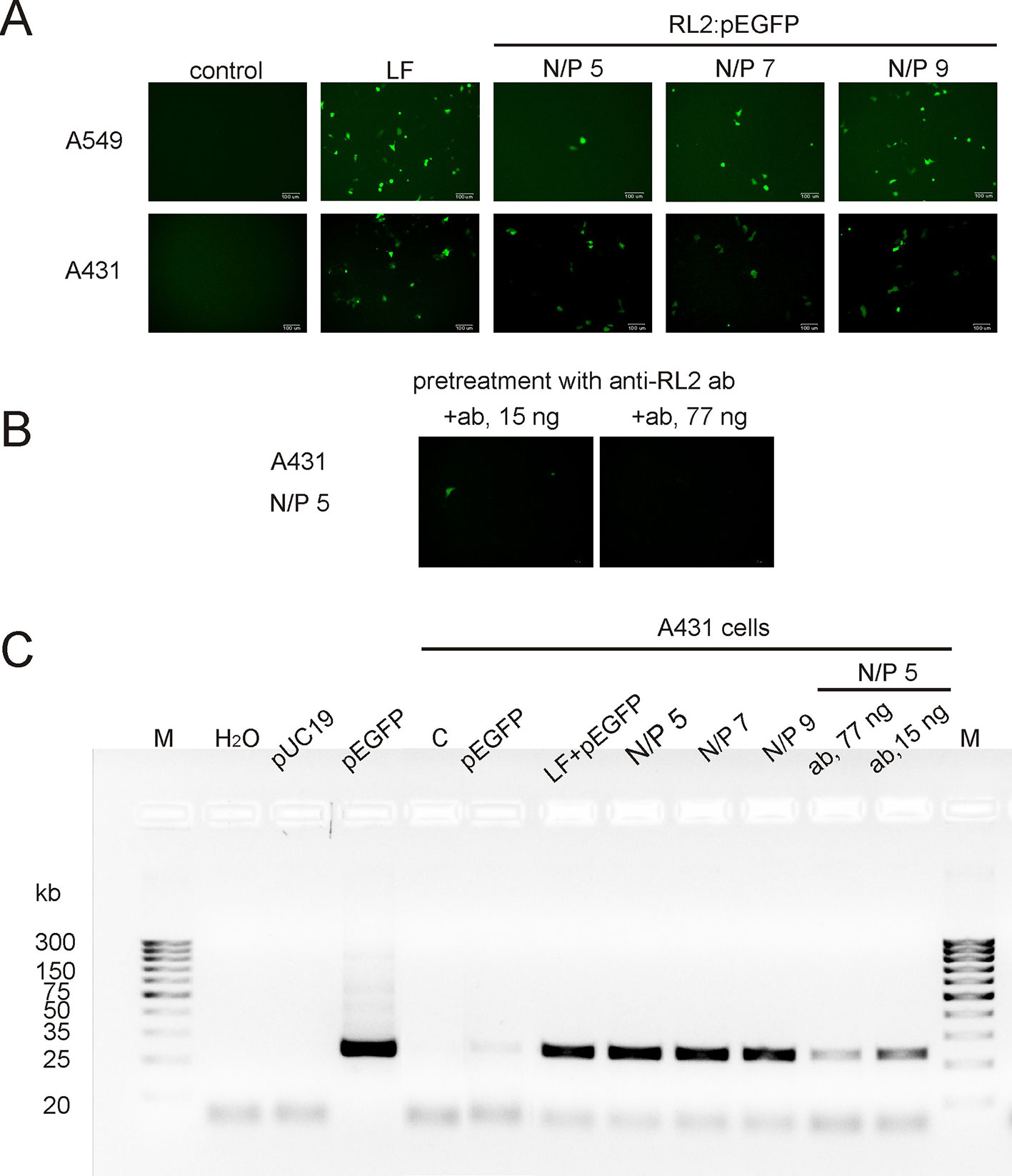
Figure 3 Transfection of cancer cells by RL2:pEGFP at charge ratios from 5 to 9. (A) Cells were incubated with LF—Lipofectamine 3000 and pEGFP, free pEGFP or RL2-pEGFP complexes with various N/P ratios and 24 h after EGFP was analyzed by fluorescent microscopy. (B) Cells were pre-treated with anti-RL2 antibodies (ab) for 3 h and next transected with a complex RL2:pEGFP. (C) Electrophoretic analysis of the products of RT-PCR reaction with EGFP-specific primers. Free pUC19 and pEGFP were used as matrixes in PCR reaction for the negative and positive controls, respectively. Representative pictures of three independent experiments.
The entering of RL2:pEGFP complexes into the A431 cells was also evaluated by RT-PCR analysis with EGFP-specific primers using whole cellular RNAs as a matrixes. EGFP-specific PCR products were detected in cells treated with RL2:pEGFP complexes within N/P ratio of 5 to 9 (Figure 3C). No PCR product was seen when cells were transfected with pEGFP alone or with control pDNA.
Thus, RL2 provides translocation of plasmid DNA into the cells and saves pEGFP functional activity upon internalization. Pre-treatment of the cells with anti-RL2 antibodies partly inhibited the entry of pEGFP into the cells (Figures 3B, C).
RL2:siRNA Complex Formation
To analyze RL2:siRNA complex formation, the well-described anti-EGFP siRNA, which targets the EGFP gene coding region, was used (Nagy et al., 2003). The efficiency of complex formation was analyzed with N/P ratios of RL2:siRNA from 1 to 15. Gel retardation assay revealed that the free form of siRNA in the samples disappeared when the N/P ratio was 10 and higher (Figure 4). Thus, RL2 efficiently forms complexes with siRNA with N/P more than 5.
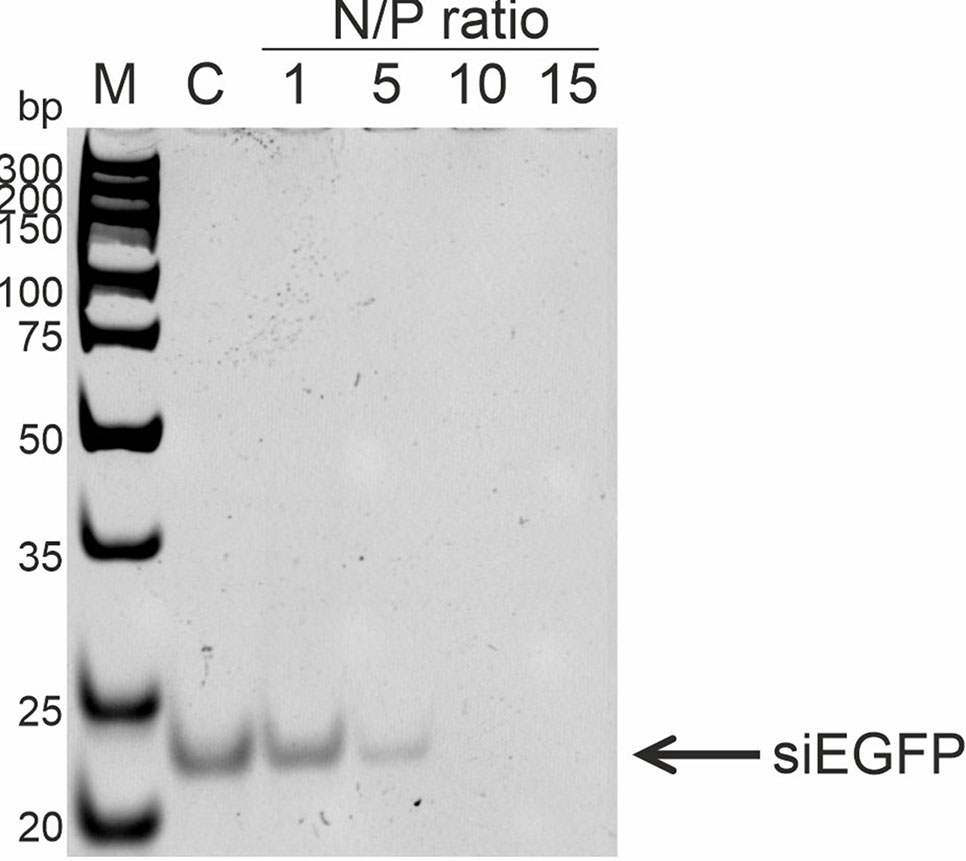
Figure 4 Analysis of RL2:siRNA complex formation. Gel retardation assays. c—free siRNA; M—DNA ladder. Representative picture of two independent experiments.
siRNA Delivery
To study RL2-based siRNA delivery, at first, A549 cells were transfected with pEGFP, and then cells were incubated with pre-forming RL2:siRNA complexes. For the stage of pEGFP transfection, the Lipofectamine 3000 was used to avoid excessive RL2 amount for the cell treatment (positive control). Cells transected with naked pEGFP (negative control) demonstrate no EGFP fluorescent signal (Figure 5A). EGFP was down-regulated in the RL2:siRNA-treated cells when N/P was 25 or higher. Flow cytometry analysis of transfected cells confirmed the data obtained and showed approximately a 50% decrease of EGFP for the complex with N/P = 35 (Figures 5B, C). These experiments demonstrate the specific effects of anti-EGFP siRNA, allowing us to conclude that siRNA efficiently penetrates into the cells treated with RL2:siRNA complexes.
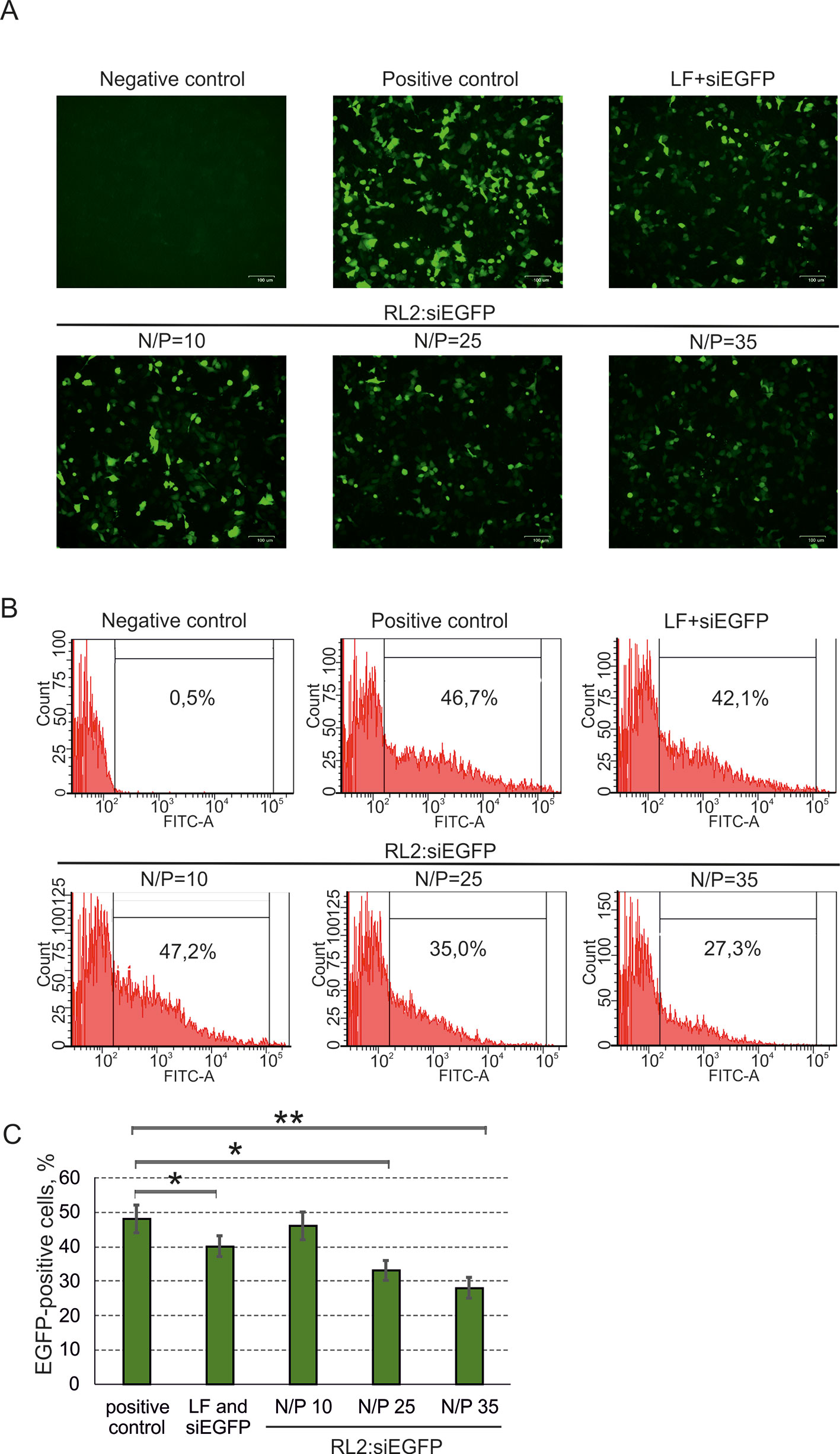
Figure 5 Specific activity of anti-EGFP siRNA delivered into A549 cells as a RL2:siRNA complex. LF—Lipofectamine 3000. Negative control—cells were treated with naked pEGFP; positive control—cells treated with pEGFP and LF. (A) Representative images of fluorescent microscopy data; (B) Representative images of flow cytometry analysis. Representative images of EGFP-positive gate excludes EGFP-negative cells (relative to negative control). (C) Quantitative evaluation of EGFP down-regulation. The data are representative of three independent repeats and are shown as the mean ± SD. *P < 0.05; **P < 0.03.
Cytotoxic Activity of RL2:U25 snoRNA Complex
Non-coding RNAs have been shown to modulate various cellular responses as well as induce death of cancer cells in vitro (Van et al., 2012; Stepanov et al., 2016). Earlier, we demonstrated that non-coding artificial analogue of U25 box C/D snoRNA (snoRNA U25) decreased the viability of various cancer cells in vitro (Nushtaeva et al., 2018). The cytotoxicity U25 snoRNA was in part due to the activation of inflammation-involved genes. U25 snoRNA is a single-stranded molecule in comparison with double-stranded pDNA or siRNA. The complex of RL2 with snoRNA was performed as described in the Materials and Methods section. For the experiments, RL2 was used in low-cytotoxic concentration. A549 cells were treated with RL2:snoRNA complex, and 48 h after MTT analysis revealed the decrease of cell viability (Figure 6A). Figure 6A demonstrates that Lipofectamine-delivered U25 snoRNA decreased the viability of treated cells more substantially than RL2-delivered. Concentration-response curves show the rise of cytotoxic effects with the rising of RL2 concentration only for RL2:snoRNA complexes, but not for free RL2 (Figure 6B). The increase of RL2 quantity in the complex elevates the penetration potency of such complexes, this seems to be the same as the other investigated complex of nucleic acids with RL2. Analyzing the data obtained, we conclude that the decrease of viability of treated cells was induced by snoRNA activity, and this efficiency correlated with the penetration potency of RL2:snoRNA complex.
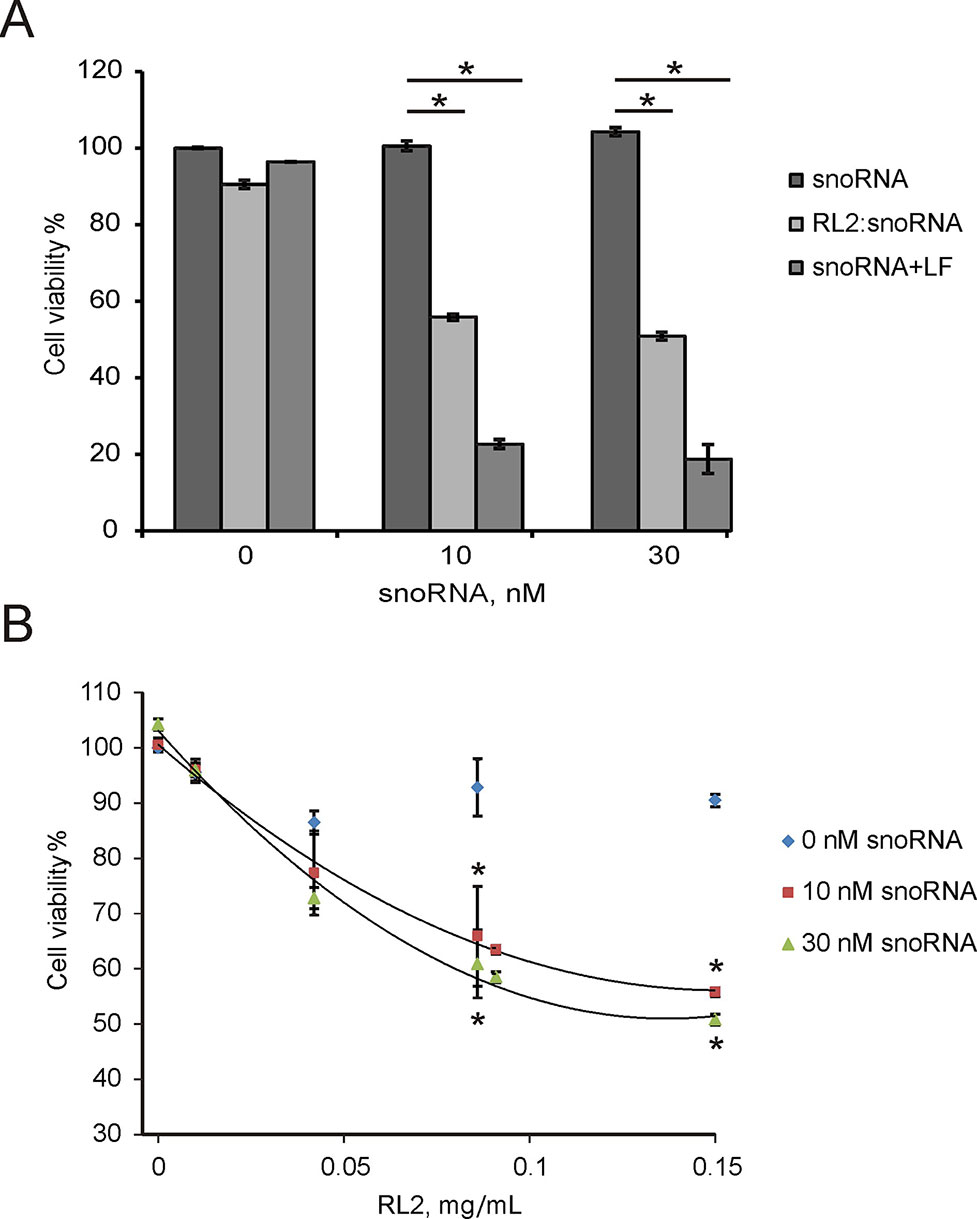
Figure 6 Cytotoxic activity of non-coding artificial analogue of U25 box C/D snoRNA in A549 cells. Cells were treated with RL2:snoRNA complexes, and after 48 h of incubation, cells viability was analyzed by MTT. LF—Lipofectamine 3000. (A) Comparison with LF-dependent delivery. *P < 0.05 compared with control (free RNA); (B) Concentration-response curves for the different RL2 concentration. The data are representative of three independent repeats. *P < 0.05 compared with control (only RL2 treatment).
Discussion
Negative charge of DNA and RNA molecules hampers their movement across cellular membranes. To reach intracellular space, DNA or RNA molecules usually need to be packaged in vesicles or carriers. Usually, peptides are used as cell-targeting compounds or cell-specific ligands being conjugated with vesicles or carriers. Among various carriers, the CPP peptides are the potential delivery system for nucleic acids (Ignatovich et al., 2003; Järver et al., 2012; Shukla et al., 2014; Kizil et al., 2015). Forming protein-nucleic acid complex, CPPs can absorb on the surface of the double-stranded helix generally by electrostatic interaction, and such complexes can aggregate with size of up to 100 nm and have a positive charge (Rathnayake et al., 2017). CPPs were demonstrated as potential siRNA delivery systems for the suppression of gene expression. Low-cytotoxic arginine-rich CPPs were successfully used for the gene delivery to the brain (Morris and Labhasetwar, 2015).
Recombinant analog of lactaptin RL2 originates from human k-casein and demonstrates CPP-like properties (Chinak et al., 2016). Due to amphiphilic characteristics, the caseins themselves can self-assemble into micelles (Portnaya et al., 2008). In drug delivery on the organism level, casein spheres showed a superior biocompatibility being introduced orally and were successfully used as carriers of doxorubicin (Chen et al., 1987). Therefore, here we analyzed if lactaptin analog RL2 translocates nucleic acids into the cancer cells.
We observed that double stranded as well as single stranded nucleic acids were efficiently translocated into the cancer cells being in complex with RL2. The delivery of EGFP reporter gene-containing plasmid into the cells was tested for EGFP-expressing activity. Since the transcription of mRNAs in eukaryotic cells takes place strictly in the nucleus, we suggest that at least pEGFP enters the nucleus. Whether pEGFP releases from the complex in the cytoplasm and then free pEGFP is transported through the nuclear membrane or RL2-pEGFP complex enters the nucleus—this requires more investigations.
The efficiency of delivery was dependent on the RL2 amount in the complexes:RL2-rich complexes were more efficient according to the cargo functional activity. The actual process of DNA or RNA condensation with CPPs has not yet been fully understood. Our finding that pDNA : RL2 complex size is smaller than the sizes of RL2 or pEGFP alone indicates the condensation of DNA in the complex. Nowadays, the most used DNA-condensing cationic peptides in gene delivery are lysine-reach, like RPC or arginine-reach, like viral Tat peptides (Wender et al., 2000; Read et al., 2003). RL2 contains five lysines and eight arginines (Supplementary Figure 2), and this is likely enough to act as a DNA-condensing molecule. We suppose that the increasing of ζ-potential is specified by the rising amount of surface-exposed cationic groups of RL2 molecules in RL2:pDNA complexes.
Although stability of RL2:pDNA complexes has a few differences in the N/P range of 1.5 to 13 (Figures 1A and 2B), the EGFP ectopic expression level increased with N/P increasing from 5 to 9. This could explain that RL2:pEGFP complexes with high amount of RL2 exert higher resistance to the nucleases. The same results were obtained for the siRNA delivery: despite of the findings that strong RL2:siRNA complexes were formed with N/P range from ten and higher, the efficiency of delivery increased with the increasing of RL2 in the complex. This could mean that a high RL2 concentration is needed for the successful delivery or alternatively, that complexes with high N/P value are fully shielded from extracellular enzymes or that the high RL2 concentration provides early endosome escape of siRNA also. Moreover, we suppose that RL2-mediated nucleic acids transport is non-dependent on the cargo size, but only on the RL2 amount that was confirmed by the effective delivery of big plasmid DNA and small siRNA.
We have also observed that Ca2+ stabilizes complexes of RL2 with nucleic acids. It is known that at low concentration divalent cations like Ca2+ and Mg2+ promote the condensation of CPP particles into agglomerates (McIntyre et al., 2017). Next, to study the plasmid protection outside the cells and intracellular release of cargo, we confirmed the pDNA release from the complexes by heparin displacement. Polyanion heparin is a suitable molecule for the modeling of nucleic acid displacement in the complexes because before the complexes reach the plasma membrane phospholipids, the delivering complexes interact with proteoglycans of the extracellular matrix, such as heparin. High heparin concentration induced total release of pDNA from the RL2:pDNA complexes.
Taken together, these findings indicate that recombinant lactaptin is able to deliver noncovalently associated nucleic acids into cancer cells in vitro.
Materials and Methods
Materials
Heparin sodium salt (H4784-1G) and CaCl2 were from Sigma-Aldrich. DNA Gel Loading Dye (R0611) and Lipofectamine 3000 were from ThermoFisher, molecular weight markers Sky High (10 and 500 bp) were from BioLabMix, Russia. MTT (3-(4,5-dimethyl-2-thiazolyl) -2,5-diphenyl-2H-tetrazolium bromide was from Sigma-Aldrich. Plasmid pEGFP-C1 (GenBank Accession U55763; cat 6084-1) was purchased from BD Biosciences Clontec, monoclonal anti-RL2 mouse IgG (clone F14; Biosan, Russia).
The recombinant analogue of lactaptin RL2 was obtained from E. coli and purified as described previously (Semenov et al., 2010). The 98% purity of the isolated protein was confirmed by RP-HPLC chromatography on C5 reverse phase column (Discovery BIO Wide Pore C5; Sigma) in water (0.5% TFA)-acetonitil solvent system using HPLC Station (Bio-RAD Laboratories) as well as by RP-HPLC on C18 (ProntosSIL) using Milichrom A-02 station (EcoNova, Russia) (Supplementary Figure 3A) and by SDS-PAAG electrophoresis under reduction conditions (Supplementary Figure 3B).
Cell Culture
A549 human lung carcinoma cells (ATCC CCL-185) and A431 squamous carcinoma cells (ATCC CRL-1555) were grown in DMEM, supplemented with 10% fetal bovine serum (FBS), 2 mM l-glutamine, 100 U/mL penicillin, 100 mg/mL streptomycin, and 0.25 μg/mL amphotericin B, all from Gibco BRL Co. USA, at 37°C, 5% CO2.
N/P Calculation
N/P values were calculated by the formula:
where C is the concentration (mole/L).
Amino-groups and imino-groups of amino acids in RL2 were estimated as fully protonated for lysine, arginine, and histidine under experimental conditions. Phosphate groups of nucleic acids were accounted as totally deprotonated with negative charge.
Complexes Formation
RL2:pEGFP complexes were prepared by varying a charge ratios (N/P). RL2 (1.1 × 10−8 ? to 9.7 × 10−8 ?) and plasmid DNA pEGFP-C1 (2.9 × 10−8 ?) were dissolved in 25 mM MES buffer, pH 5.5, and then they were mixed and incubated for 5 min at 37°C.
For the RL2:siRNA complex formation, at first siRNA duplexes were formed. Chemically synthesized oligoribonucleotides 5′-GAACGGCAUCAAGGUGAACTT-3′ (sense) and 5′-GUUCACCUUGAUGCCGUUCTT-3′ (antisense) were from Institute of Chemical Biology and Fundamental Medicine, SB RAS, Russia. Oligoribonucleotides (1 × 10−5 М) were incubated in the buffer containing 50 мМ potassium acetate, 15 мМ HEPES-KOH, pH 7.4 and 1 мМ magnesium acetate for 3 min at 90°C, and then mixture was cooled to 37°C. RL2 was mixed with anti-EGFP siRNA at N/P ratios of 10 to 35 in 25 mM MES buffer, pH 5.5 for 5 min at 37°С.
For the RL2:snoRNA complex formation, artificial analogue of U25 box C/D snoRNA was synthesized as was described in the study of Stepanov et al. (2018). RL2 (0.05–0.2 mg/mL) and snoRNA (10 or 30 nM) were dissolved in 25 mM MES buffer, pH 5.5, and then were mixed and incubated for 5 min at 37°С.
Gel Retardation Assays
Aliquots of noncovalent complexes (reaction mixture, 10 µL) were loaded onto 0.5% agarose gel (for RL2:pDNA) or onto 15% PAAG (RL2:siRNA) with DNA Gel Loading Dye. Retardation was analyzed by electrophoresis in Tris-Acetate-EDTA (TAE) buffer or Tris-Borate-EDTA (TBE) buffer. DNA was visualized with ethidium bromide by Gel Doc XR+ Image system (BioRad, USA), and siRNA was visualized with SYBR Green I Nucleic Acid Gel Stain by BioRad GelDoc XR+ Image system (BioRad, USA).
Dynamic Light Scattering (DLS)/Determination of Particle Size, ξ-Potential, and UV-vis
The hydrodynamic diameter and zeta potential of RL2:pEGFP were determined by dynamic laser light scattering using a Zetasizer Nano ZS (Malvern Instruments Ltd, UK) at a wavelength of 623 nm and 25°C. All DLS results were calculated as the average of at least triplicate measurements and presented as mean ± SD.
UV-vis spectroscopy analysis of complexes was performed using NanoDrop 2000c (Thermo Scientific, USA).
pDNA Delivery Microscopic Assays
A549 and A431 cells were seeded (2 x 104 cells/cm2) on 24-well plates 24 h prior the experiment. Cell media was replaced with fresh Opti-MEM (ThermoFisher, USA) containing 2 mM l-glutamine, and pre-formed complexes were added to the cells to the final concentration of pEGFP 1 ng/µL. After 3 h, medium was replaced with complete fresh DMEM media supplemented with 10% FBS, and cells were incubated for additional 24 h. EGFP fluorescence was analyzed using fluorescent microscope (ZOE Bio-Rad, USA).
pDNA Delivery RT-PCR Assays
After microscopic analysis, cells were washed three times with PBS and total RNA was isolated by phenol-chloroform extraction using the Lira reagent (Biolabmix Ltd, Novosibirsk, Russia) according to the manufacturer’s protocol. The quality of total RNA was assessed by agarose gel electrophoresis or capillary electrophoresis with an Agilent 2100 Bioanalyzer, using 28S/18S > 2 or RIN > 8.0 criterion. RT-PCR were performed in the one-tube reaction mixture BioMaster RT-PCR SYBR Blue (Biolabmix Ltd, Novosibirsk, Russia, www.biolabmix.ru) with gene-specific primers: EGFP 5′-GTAAACGGCCACAAGTTCAG-3′ and 5′-GGTGCGCTCCTGGACGTAGC-3′; hypoxanthine-guanine phosphoribosyltransferase (HPRT): 5′-CATCAAAGCACTGAATAGAAAT-3′ and 5′-TATCTTCCACAATCAAGACATT-3′. For RT-PCR analysis 1 µg of total RNA was used in the reaction under the following conditions: incubation at 25°C for 5 min followed by 42°C for 30 min and at 95°C for 4 min, next incubation at 59°C for 30 s, and at 72°C for 20 s followed by 30 cycles of denaturation at 95°C for 30 s, annealing at 59°C for 30 s, and elongation at 72°C for 20 s.
Pre-Treatment With Anti-RL2 Antibodies
Cells were grown in 12-well plates up to 80% confluence before the replacement for the fresh DMEM:F12 medium supplemented with l-glutamine. Monoclonal anti-RL2 IgG (15 and 77 ng) were added to the cells and incubated for 15 min. Next, cells were treated with RL2:pEGFP complexes and incubated for 3 h. Subsequently, culture medium was replaced with fresh DMEM:F12 medium supplemented 10% FBS and l-glutamine and cells were incubated for 24 h at 37°C in CO2 incubator. Cells were analyzed by fluorescence microscopy (ZOE Bio-Rad, USA), and cellular lysates were used for subsequent RT-PCR analysis.
siRNA Delivery Assay
A549 cells were seeded (2 x 104 cells/cm2) on 12-well plates. After 24 h, cell media was replaced with fresh Opti-MEM (ThermoFisher, USA) containing 2 mM l-glutamine. Plasmid pEGFP-C1 with Lipofectamine 3000 (1 µg DNA/well) and RL2:siRNA complex were added to the cells. Cells were incubated 3 h at 37°C, and after that, FBS and antibiotics were added to the cells. Cells were incubated for 48 h and analyzed by fluorescence microscopy (ZOE Bio-Rad, USA) or by flow cytometry (BD FACSCantoII). For flow cytometry analysis cells were fixed in 5% formaldehyde for 2 h. Fluorescence was detected in the FITC channel (emission 535 nm, excitation 488 nm). Data Analysis was performed using the software package BD FACSDiva software v6.1.3.
snoRNA Delivery Assay (MTT)
A549 cells were seeded in 96-well plates at density of 2 x 103 cells per well. After 24 h cell media was replaced with 100 µL DMEM, containing 2 mM l-glutamine, and 10.5 µL RL2:U25 complexes per well. After 2 h, 50 µL DMEM supplemented with 30% FBS, 300 U/ mL penicillin, 300 mg/mL streptomycin, 0.75 μg/mL amphotericin B, and 2-mM l-glutamine was added, and cells were incubated for 48 h, and MTT analysis was performed as was described in the study of Koval et al. (2014). Cell viability was expressed as a means percentage of control ± SD for triplicate independent experiments.
Statistical Analysis
All experiments were repeated three times independently, and data are presented as mean ± SD. The Mann-Whitney U-test was used to define statistically significant differences between groups. All error bars represent standard error of the mean. P value that was less or equal 0.05 was considered as significant.
Data Availability
The data that support the findings of this study are available from the authors upon reasonable request.
Author Contributions
OK designed the study and analyzed the data. OC and EG performed the experiments. IP performed determination of particle size and zeta potential. GS and EZ performed the experiments with snoRNA. The draft manuscript was prepared by OK with input from VR. All authors agreed the final version. All authors read and approved the final manuscript.
Funding
This work funded by the Russian State funded budget project of ICBFM SB RAS AAAA-A17-117020210023-1.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We acknowledge Russian State funded budget project of ICBFM SB RAS AAAA-A17-117020210023-1 for supporting this work.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2019.01043/full#supplementary-material
Supplementary Figure 1 | Analysis of formation of RL2:pEGFP complexes with N/P 5 – 11 by UV-vis spectroscopy.
Supplementary Figure 2 | Amino acid sequence of RL2.
Supplementary Figure 3 | Purity analysis of RL2. A – HPLC analysis of RL2 comparing with blanking chromatography on ProntoSIL 120-5-B – Electrophoretic analysis of RL2 in 13% SDS-PAGE in reducing conditions.
References
Bolhassani, A. (2011). Potential efficacy of cell-penetrating peptides for nucleic acid and drug delivery in cancer. Biochim. Biophys. Acta Rev. Cancer 1816, 232–246. doi: 10.1016/j.bbcan.2011.07.006
Chen, Y., Willmott, N., Anderson, J., Florence, A. T. (1987). Comparison of albumin and casein microspheres as a carrier for doxorubicin. J. Pharm. Pharmacol. 39, 978–985. doi: 10.1111/j.2042-7158.1987.tb03144.x
Chinak, O. A., Fomin, A. S., Nushtaeva, A. A., Koval, O. A., Savelyeva, A. V., Kuligina, E. V., et al. (2016). Penetration of the peptide lactaptin into human cancer cells. Russ. J. Bioorg. Chem. 42, 361–371. doi: 10.1134/S1068162016040063
Futaki, S. (2005). Membrane-permeable arginine-rich peptides and the translocation mechanisms. Adv. Drug Delivery Rev. 57, 547–558. doi: 10.1016/j.addr.2004.10.009
Hoyer, J., Neundorf, I. (2012). Peptide vectors for the nonviral delivery of nucleic acids. Acc. Chem. Res. 45, 1048–1056. doi: 10.1021/ar2002304
Ignatovich, I. A., Dizhe, E. B., Pavlotskaya, A. V., Akifiev, B. N., Burov, S. V., Orlov, S. V., et al. (2003). Complexes of plasmid DNA with basic domain 47-57 of the HIV-1 Tat protein are transferred to mammalian cells by endocytosis-mediated pathways. J Biol. Chem. 278, 42625–42636. doi: 10.1074/jbc.M301431200
Järver, P., Coursindel, T., Andaloussi, S. E., Godfrey, C., Wood, M. J., Gait, M. J. (2012). Peptide-mediated cell and in vivo delivery of antisense oligonucleotides and siRNA. Mol Ther Nucleic Acids 1, e27. doi: 10.1038/mtna.2012.18
Kizil, C., Iltzsche, A., Thomas, A. K., Bhattarai, P., Zhang, Y., Brand, M. (2015). Efficient cargo delivery into adult brain tissue using short cell-penetrating peptides. PLoS ONE 10, e0124073. doi: 10.1371/journal.pone.0124073
Koval, O. A., Fomin, A. S., Kaledin, V. I., Semenov, D. V., Potapenko, M. O., Kuligina, E. V., et al. (2012). A novel pro-apoptotic effector lactaptin inhibits tumor growth in mice models. Biochimie 94, 2467–2474. doi: 10.1016/j.biochi.2012.08.017
Koval, O. A., Tkachenko, A. V., Fomin, A. S., Semenov, D. V., Nushtaeva, A. A., Kuligina, E. V., et al. (2014). Lactaptin induces p53-independent cell death associated with features of apoptosis and autophagy and delays growth of breast cancer cells in mouse xenografts. PLoS ONE 9, e93921. doi: 10.1371/journal.pone.0093921
McIntyre, I., Sullivan, M., O′Riordan, D. (2017). Altering the level of calcium changes the physical properties and digestibility of casein-based emulsion gels. Food Funct. 8, 1641–1651. doi: 10.1039/C6FO01719C
Morris, M. C., Vidal, P., Chaloin, L., Heitz, F., Divita, G. (1997). A new peptide vector for efficient delivery of oligonucleotides into mammalian cells. Nucleic Acids Res. 25, 2730–2736. doi: 10.1093/nar/25.14.2730
Morris, V. B., Labhasetwar, V. (2015). Arginine-rich polyplexes for gene delivery to neuronal cells. Biomaterials 60, 151–160. doi: 10.1016/j.biomaterials.2015.04.052
Nagy, P., Arndt-Jovin, D. J., Jovin, T. M. (2003). Small interfering RNAs suppress the expression of endogenous and GFP-fused epidermal growth factor receptor (erbB1) and induce apoptosis in erbB1-overexpressing cells. Exp. Cell Res. 285, 39–49. doi: 10.1016/S0014-4827(02)00050-2
Nushtaeva, A. A., Stepanov, G. A., Semenov, D. V., Juravlev, E. S., Balahonova, E. A., Gerasimov, A. V., et al. (2018). Characterization of primary normal and malignant breast cancer cell and their response to chemotherapy and immunostimulatory agents. BMC Cancer 18, 728. doi: 10.1186/s12885-018-4635-8
Pooga, M., Soomets, U., Hällbrink, M., Valkna, A., Saar, K., Rezaei, K., et al. (1998). Cell penetrating PNA constructs regulate galanin receptor levels and modify pain transmission in vivo.. Nat. Biotechnol. 16, 857–861. doi: 10.1038/nbt0998-857
Portnaya, I., Ben-Shoshan, E., Cogan, U., Khalfin, R., Fass, D., Ramon, O., et al. (2008). Self-assembly of bovine β-casein below the isoelectric pH. J Agric Food Chem. 56, 2192–2198. doi: 10.1021/jf072630r
Rathnayake, P. V. G. M., Gunathunge, B. G. C. M., Wimalasiri, P. N., Karunaratne, D. N., Ranatunga, R. J. K. U. (2017). Trends in the binding of cell penetrating peptides to siRNA: A molecular docking study. J. Biophysics 2017, 1–12. doi: 10.1155/2017/1059216
Read, M. L., Bremner, K. H., Oupický, D., Green, N. K., Searle, P. F., Seymour, L. W. (2003). Vectors based on reducible polycations facilitate intracellular release of nucleic acids: intracellular activation of RPC-based gene therapy vectors. J. Gene Med. 5, 232–245. doi: 10.1002/jgm.331
Semenov, D. V., Fomin, A. S., Kuligina, E. V., Koval, O. A., Matveeva, V. A., Babkina, I. N., et al. (2010). Recombinant analogs of a novel milk pro-apoptotic peptide, lactaptin, and their effect on cultured human cells. Protein J. 29, 174–180. doi: 10.1007/s10930-010-9237-5
Shukla, R. S., Qin, B., Cheng, K. (2014). Peptides used in the delivery of small noncoding RNA. Mol. Pharm. 11, 3395–3408. doi: 10.1021/mp500426r
Stepanov, G. A., Filippova, J. A., Nushtaeva, A. A., Kuligina, E. V., Koval, O. A., Richter, V. A., et al, (2016). “Artificial analogues of circulating box C/D RNAs induce strong innate immune response and MicroRNA activation in human adenocarcinoma cells,” in Circulating Nucleic Acids in Serum and Plasma – CNAPS IX. Eds. Gahan, P. B., Fleischhacker, M., Schmidt, B. (Cham: Springer International Publishing), 121–125. doi: 10.1007/978-3-319-42044-8_24
Stepanov, G., Zhuravlev, E., Shender, V., Nushtaeva, A., Balakhonova, E., Mozhaeva, E., et al. (2018). Nucleotide modifications decrease innate immune response induced by synthetic analogs of snRNAs and snoRNAs. Genes 9, 531. doi: 10.3390/genes9110531
Tecle, M., Preuss, M., Miller, A. D. (2003). Kinetic study of DNA condensation by cationic peptides used in nonviral gene therapy: analogy of DNA condensation to protein folding †. Biochemistry 42, 10343–10347. doi: 10.1021/bi034325e
Van, D. N., Roberts, C. F., Marion, J. D., Lépine, S., Harikumar, K. B., Schreiter, J., et al. (2012). Innate immune agonist, dsRNA, induces apoptosis in ovarian cancer cells and enhances the potency of cytotoxic chemotherapeutics. FASEB J. 26, 3188–3198. doi: 10.1096/fj.11-202333
Wender, P. A., Mitchell, D. J., Pattabiraman, K., Pelkey, E. T., Steinman, L., Rothbard, J. B. (2000). The design, synthesis, and evaluation of molecules that enable or enhance cellular uptake: peptoid molecular transporters. Proc. Natl. Acad. Sci. 97, 13003–13008. doi: 10.1073/pnas.97.24.13003
Ye, R., Harte, F. (2013). Casein maps: effect of ethanol, pH, temperature, and CaCl2 on the particle size of reconstituted casein micelles. J. Dairy Sci. 96, 799–805. doi: 10.3168/jds.2012-5838
Keywords: nucleic acids, cell penetrating peptides, lactaptin, gene delivery, snoRNA
Citation: Chinak O, Golubitskaya E, Pyshnaya I, Stepanov G, Zhuravlev E, Richter V and Koval O (2019) Nucleic Acids Delivery Into the Cells Using Pro-Apoptotic Protein Lactaptin. Front. Pharmacol. 10:1043. doi: 10.3389/fphar.2019.01043
Received: 10 May 2019; Accepted: 16 August 2019;
Published: 18 September 2019.
Edited by:
Hector A. Cabrera-Fuentes, University of Giessen, GermanyReviewed by:
Carlo Gaetano, IRCCS Scientific Clinical Institutes Maugeri (ICS Maugeri), ItalyHan Qiao,Shanghai Jiao-Tong , University School of Medicine, China
Copyright © 2019 Chinak, Golubitskaya, Pyshnaya, Stepanov, Zhuravlev, Richter and Koval. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Olga Koval, b19rb3ZhbEBuaWJvY2gubnNjLnJ1
†These authors have contributed equally to this work
 Olga Chinak
Olga Chinak Ekaterina Golubitskaya
Ekaterina Golubitskaya Inna Pyshnaya4
Inna Pyshnaya4 Grigory Stepanov
Grigory Stepanov Olga Koval
Olga Koval