- 1Affiliated Hospital of Nanjing University of Chinese Medicine, Nanjing, China
- 2Jiangsu Key Laboratory of Pediatric Respiratory Disease, Institute of Pediatrics, Nanjing University of Chinese Medicine, Nanjing, China
- 3Children’s Hospital of Soochow University, Suzhou, China
- 4College of Life Sciences, Nanjing Agricultural University, Nanjing, China
- 5Department of Chemistry and Institute of Biomedical Sciences of Shanghai Medical School, Fudan University, Shanghai, China
Gubenfangxiao decoction (GBFXD) is a traditional Chinese medicine based on a combination of Yu-Ping-Feng-San and Erchen decoctions. GBFXD has been widely used for decades in treating asthma at the Affiliated Hospital of Nanjing University of Chinese Medicine. Previously, GBFXD was found to reduce lung inflammation and airway remodeling; however, the underlying mechanism remains unknown. In this study, the effects of GBFXD on asthmatic mice were evaluated based on pathology and lung function; airway hyperresponsiveness (AHR) and pathology were compared among the two different mouse models utilized. Furthermore, the mechanism of action of GBFXD on asthmatic mice was analyzed using iTRAQ labeling technology combined with ingenuity pathway analysis (IPA). Modeling analysis of pre- and post-treatment proteins identified 75 differentially expressed proteins. These proteins were related to B-cell development, calcium-induced lymphocyte apoptosis, antigen presentation, and Th1 and Th2 activation pathways. Moreover, 68 differentially expressed proteins were identified in the GBFXD treatment group compared with the model group. Upstream regulatory factors predicted that interleukin (IL)-4 (necessary for inducing polarization of type 2 [M2] macrophages), cyclooxygenase, and prostaglandin E2 are significantly elevated in the model group. Based on IPA analysis, it was concluded that several pathways, including mitochondrial dysfunction and oxidative phosphorylation, are closely associated with the therapeutic effects of GBFXD in asthma. Moreover, the differential expression of several proteins, including the M2 markers, MRC1, ARG1, Retnla, Chil3, and CHIA, were validated by western blotting, confirming that GBFXD can reduce airway inflammation, which fits the pattern of an alternative M2 activation state, and attenuate AHR. Overall, our findings indicate that GBFXD significantly suppresses M2 macrophage polarization, providing further insights into the mechanism underlying the protective effects of GBFXD.
Introduction
Asthma is one of the most common chronic, non-infectious airway diseases in children, with a global prevalence of 1–18% (Ebmeier et al., 2017; Papi et al., 2018). As a developing country, with the change of industrialization and lifestyle, the incidence of asthma in Chinese children is rising (Guo et al., 2018). Asthma is characterized by chronic airway inflammation, airway hyperresponsiveness and airway remodeling (Grainge et al., 2011). However, asthma in childhood and adult asthma is significantly different in epidemiology. Respiratory syncytial virus (RSV) is one of the main causes of recurrent asthma in children (Jartti and Gern, 2017; William et al., 2018). Moreover, epidemiological studies have shown that early infection with RSV can increase the risk of asthma in children (Carroll et al., 2017). Experiments have confirmed that the increased eosinophils can be found in airway of infants and mice after infection with RSV, which is associated with the involvement of viral prion-induced Th2 type cytokines as well as eosinophil chemotactic factors during RSV replication (Phipps et al., 2007). In addition, RSV infection can also cause damage to the airway surface (Zhang et al., 2002). At present, research on the role of RSV infection in asthma via proteomics has ever been yet reported.
The prevalence of asthma in children in China is 3.01% (Liu, 2019). However, the long-term use rate of ICS is only 57.8% (Liu and Chen, 2013). As a traditional medication, herbal compound has become a common alternative treatment for asthma in children in China and other Asian countries (Huang et al., 2013; Geng et al., 2016). At the molecular level, traditional Chinese medicine formulations are multi-targeted and multi-component. Combinatorial systems biology and omics technology currently provide powerful tools for addressing the complexities and mechanisms of traditional Chinese medicine formulations (Yue et al., 2017; Chun et al., 2018).
Gubenfangxiao decoction (GBFXD) has been used clinically for decades. It is consisted of traditional Chinese medicine compound Yupingfeng powder (YPF-P) and Ershen Tang (ECD). Previous clinical trials have confirmed the efficacy and safety for long-term prevention of asthma recurrence (Yuan et al., 2010). Subsequent studies have shown that GBFXD can reduce lung inflammation in OVA-sensitized mice, inhibit the expression of asthma susceptibility genes ORMDL3 and ADAM33, and reduce endoplasmic reticulum stress (ERS) (Huang et al., 2016; Lu et al., 2016). In this study, as a model of chronic persistant asthma, RSV-OVA-sensitized mice was established. iTRAQ-based proteomics studies and ingenuity pathway analysis (IPA) were performed to reveal the protein features of RSV-OVA-sensitized mice and investigate the underlying mechanisms of GBFXD.
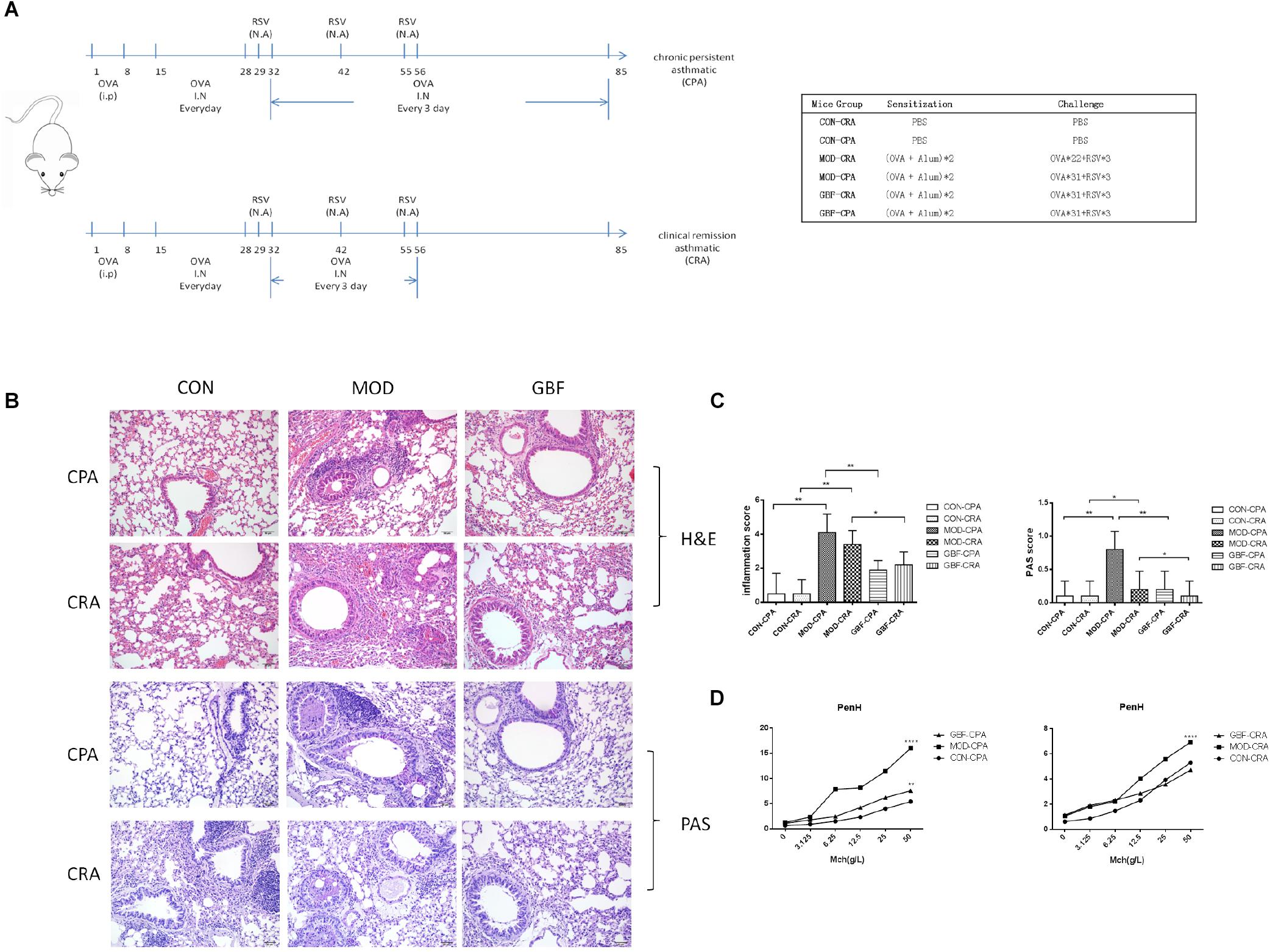
Figure 1. Effect of GBFXD treatment on airway hyperresponsiveness in ovalbumin-challenged mice and histological examination of lung tissue for airway inflammation (H&E and PAS staining). (A) Experimental scheme for the induction of airway inflammation in a mouse model. (B) H&E staining showing asthmatic inflammation. PAS staining identified epithelial goblet cells. (C) Total inflammation scores in all animal groups. The percentage of PAS-positive cells per bronchiole was calculated. (D) Airway responsiveness to aerosolized methacholine was measured with WBP. Mice were placed in the main chamber and nebulized first with PBS and then with increasing doses (3.125–50 mg/mL) of methacholine. Data represent the mean ± SEM of five independent experiments (two-way ANOVA by Tukey’s multiple comparisons test; ∗∗p < 0.01; ∗∗∗p < 0.0001; ∗∗∗∗p < 0.0001).
Materials and Methods
Animal Experiments and Drug Treatments
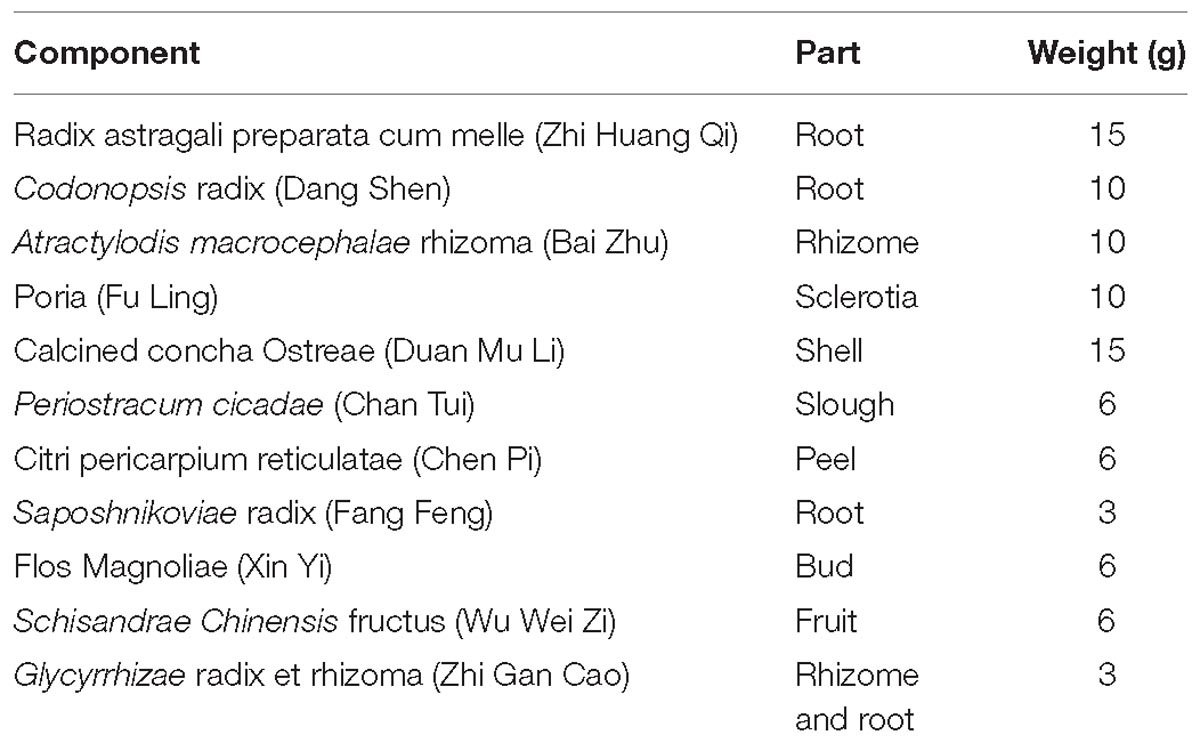
GBFXD Preparation
GBFXD, which consists of 11 components (Table 1), was purchased from the Jiangsu Province Hospital of Chinese Medicine (Jiangsu, China) and authenticated by Dr. Sheng-Jin Liu of the College of Pharmacy, Nanjing University of Chinese Medicine, Nanjing, China. The principal components of GBFXD were prepared and identified by high-performance liquid chromatography-ultraviolet in a previous study (Lu et al., 2016).
Animal Model Establishment
Four-week-old, female, specific-pathogen-free BALB/c mice (16–18 g) were purchased from Suzhou JOINN Laboratories Co., Ltd. (Jiangsu, China). The animals were housed in the experimental animal center of Nanjing University of Chinese Medicine and maintained under a 12 h light/dark cycle at a constant temperature of 22 ± 2°C. Cages, bedding, food, and water were sterilized before use. Mice were sensitized with 20 μg intraperitoneal OVA injections (grade II; Sigma-Aldrich, St. Louis, MO, United States), after which the clinical remission asthmatic (CRA) and chronic persistent asthmatic (CPA) models were established at two different challenge frequencies. The excitation models included 2.5% OVA atomization and RSV in nasal drop form with a titer of 1.0 × 10 TCID50/mL (Figure 1A). The mice were randomly divided into six groups as follows, CON-CRA control group, MOD-CRA model group, GBF-CRA (36 g/kg/d) treatment group, CON-CPA control group, MOD-CPA model group, and GBF-CPA (36 g/kg/d) treatment group. Prior to the experiments, there were no significant differences among the groups in terms of animal weight. All experimental procedures were performed in accordance with the National Institutes of Health Guidelines for Laboratory Animals and approved by the Animal Ethics Committee of Nanjing University of Chinese Medicine [no. SYXK (Su) 2014–0001].
Proteomics
Protein Extraction and Digestion
Lungs were excised, immediately frozen at −80°C, and ground in liquid N2. Cold RIPA extraction buffer (Beyotime, Haimen, China) was added to the pulverized tissue and then the mixture was sonicated. Next, 1 mM phenylmethanesulfonyl fluoride (Beyotime), 2 mM ethylenediaminetetraacetic acid, 10 mM dithiothreitol, and protease inhibitor cocktails (Roche, Basel, Switzerland) were added, after which the mixture was centrifuged at 4°C and 30,000 × g for 15 min. The supernatant was collected and added to five volumes of cold acetone containing 10% (v/v) trichloroacetic acid, thoroughly mixed, and incubated at −20°C overnight. The mixture was centrifuged again at 4°C and 30,000 × g and the supernatant was discarded. The precipitate was then washed three times with chilled acetone, dissolved in RIPA buffer, and air-dried. Proteins were quantified with a BCA kit (Thermo Fisher Scientific, Waltham, MA, United States), after which 300 μg of total protein was mixed with sequencing-grade trypsin (Promega, Madison, WI, United States) at an enzyme-to-protein ratio of 1:50 and incubated at 37°C for 16 h. Peptides obtained from the digestion were dried by vacuum centrifugation.
iTRAQ Labeling and High-pH Reverse-Phase (RP) Fractionation
Peptides were processed using 4-plex iTRAQ reagent (AB Sciex, Framingham, MA, United States) according to manufacturer’s instructions. Control samples were labeled with 116 iTRAQ tags, model samples were labeled with 115 iTRAQ tags, GBFXD samples were labeled with 114 iTRAQ tags, and the mixtures were labeled with 117 iTRAQ tags. High pH RP fractionation was then performed with the U3000 HPLC chromatography system (Thermo Fisher Scientific). The iTRAQ-labeled peptide mixtures were reconstituted with 100 μL of high pH RP buffer A (98% H2O, 2% acetonitrile; pH 10.0) and loaded onto a C18 column with a particle size of 1.7 μm (2.1 mm × 100 mm; Waters Corporation, Milford, MA, United States). The column was eluted with the following gradient program, 3–18% buffer B (2% H2O, 98% acetonitrile; pH 10.0) for 30 min; 18–32% B for 15 min; 32–98% B for 6 min; and holding at 98% B for 15 min. The flow rate was 0.2 mL/min and elution was monitored by measuring the absorbance at 214 nm.
Liquid Chromatography-Tandem Mass Spectrometry (LC-MS/MS) Analysis
Peptides were re-dissolved in buffer A (2% acetonitrile, 0.1% formate) and centrifuged at 4°C and 20,000 × g for 10 min. The final peptide concentration of each fraction was ∼0.2 μg/μL. The peptides (10 μL) were then loaded onto a 2-cm C18 trap column using the Nano LC System autosampler (Thermo Fisher Scientific) and eluted onto a 15-cm analytical C18 column with an inner diameter of 75 μm using a 3–55% buffer B (84% acetonitrile, 0.1% formate) gradient. The elution process ran for 112 min at a flow rate of 300 nL/min. The peptides were ionized via nano-electrospray ionization at a voltage of 2.2 kV. Data-dependent MS/MS was performed using an LTQ-Orbitrap XL mass spectrometer (Thermo Fisher Scientific). Using the MS1 full scan at a resolution of 60,000, the five most abundant precursor ions surpassing the 5,000 count threshold were selected for MS/MS analysis at a dynamic exclusion duration of 60 s. Normalized collision energy for high-energy collision dissociation (HCD) was set to 40.0. Finally, the ion fragments were detected in the Orbitrap at a resolution of 7,500 m/z scan ranges were 350–1800 and 100–1800 Da for the MS1 and the MS2 scans, respectively.
Protein Identification and Quantification
Raw MS data were converted into.MGF files using Proteome Discoverer v.1.4 (Thermo Fisher Scientific). Proteins were identified with the SEQUEST search engine and considered differentially expressed when their fold changes were > 1.5 or <0.66 and p < 0.05. Parameters required for qualification are listed in Table 2.
Bioinformatic Analysis of Differentially Expressed Proteins (DEPs)
Protein-encoding genes were functionally categorized according to biological process, molecular function, and cellular components by the PANTHER gene classification system. Proteomic data were analyzed by Ingenuity® Pathways Analysis (IPA; Qiagen, Hilden, Germany) to elucidate the hidden biological significance of the experimental data. Biological information obtained from data analysis is presented as biological function/disease, canonical pathways, networks, and upstream regulators.
Histopathology
Lung tissues were infused with 4% paraformaldehyde, embedded in paraffin, and processed for histology. Paraffin-embedded samples were sectioned (5-mm thick) and stained with hematoxylin and eosin (H&E) to examine the extent of peribronchial inflammation. Glandular hyperplasia was analyzed in randomly selected samples using the periodic acid Schiff (PAS) staining method.
Airway Hyperresponsiveness (AHR) Testing
BABL/c mice in their natural state were placed in Whole Body Plethysmography(WBP) and allowed to stabilize for 3 min to adapt to the environment. Basic physiological respiratory parameters were measured for 3 min, after which normal saline was added to the inhalation tubes in the boxes. Diluted methacholine (300 μL) was then injected into the boxes at a concentration of 0, 3.125, 6.25, 12.5, and 25 g/L. The mist generation rate was adjusted to 50%. For every minute of nebulization and inhalation, Enhanced pause (Penh) values were calculated using the software of the associated computer interface; the average Penh value at each challenge dose was calculated 3 min after inhalation.
Validation
Total RNA from the lung samples was isolated using TRIzol reagent (Takara Bio, Kusatsu, Japan) according to manufacturer’s instructions. Reverse transcription master mix (G490) and EvaGreen qPCR master mix (MasterMix-ER) were purchased from Applied Biological Materials Inc. (Richmond, BC, Canada). ARG1, MRC1, Retnla, CHIL3, TNF, and IFNG mRNA levels were determined by quantitative reverse transcription (qRT)-PCR using GAPDH as a reference gene (Table 3). Expression levels of ARG1, CHIA, and Retnla (Abcam, Cambridge, United Kingdom) in lung tissue were also measured by western blotting using β-actin as loading control. Protein samples from the lungs were fractionated via SDS-PAGE. Three independent experiments were performed.
Data Analysis
Data are expressed as the mean ± standard error of the mean (SEM). All statistical analyses were performed using GraphPad Prism 6.0 (GraphPad Software Inc., San Diego, CA, United States). Statistical analysis was performed using one-way analysis of variance (ANOVA) followed by Dunnett’s post hoc test to determine the statistical significance. Differences were considered significant when p < 0.05.
Results
GBFXD Alleviates Airway Inflammation and AHR Caused by Persistent Challenge
We improved the previous method (Lu et al., 2016) and established a CPA model that more closely simulates chronic persistent asthmatic characteristics than the one previously tested. H&E staining, PAS staining, and AHR of the two models tested were compared. In both models, the mice developed typical inflammatory changes including bronchial congestion and inflammatory cell infiltration. Mucus expression in the airways was evaluated by quantifying and comparing PAS-positive cells between the two models; the CPA model presented with more prominent goblet cell hyperplasia and mucus overproduction. After GBFXD treatment, however, the average number of PAS-stained goblet cells was greatly reduced (Figures 1B,C). Asthma is an airway obstruction often accompanied by varying degrees of AHR. As the Penh metric can reflect airway responsiveness and Penh values are accepted AHR indicators, we calculated Penh values after induction with a methacholine concentration gradient. We found higher AHR in the CPA model than in the CRA model, with GBFXD showing significantly reduced AHR (Figure 1D).
Proteomic Expression Patterns
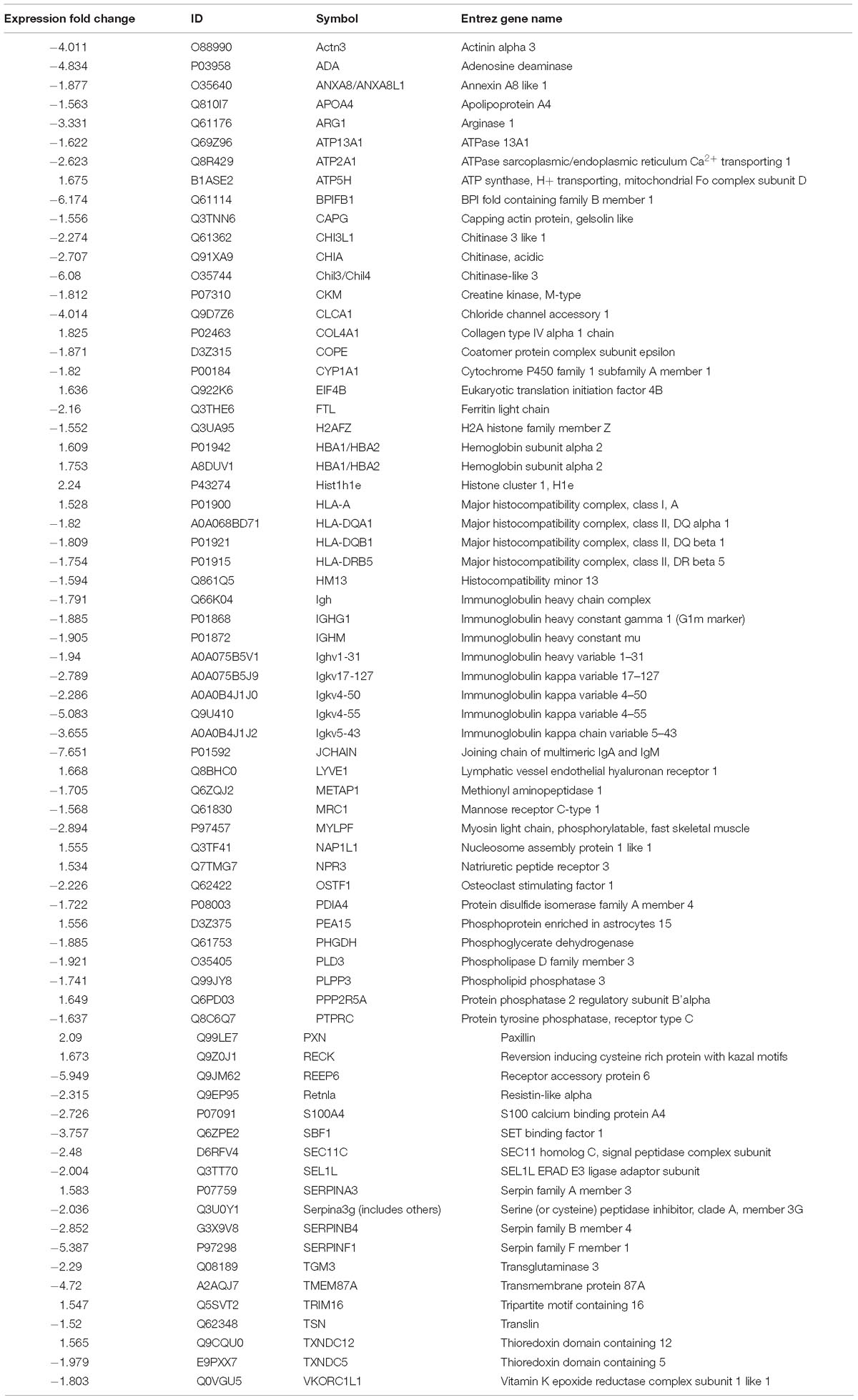
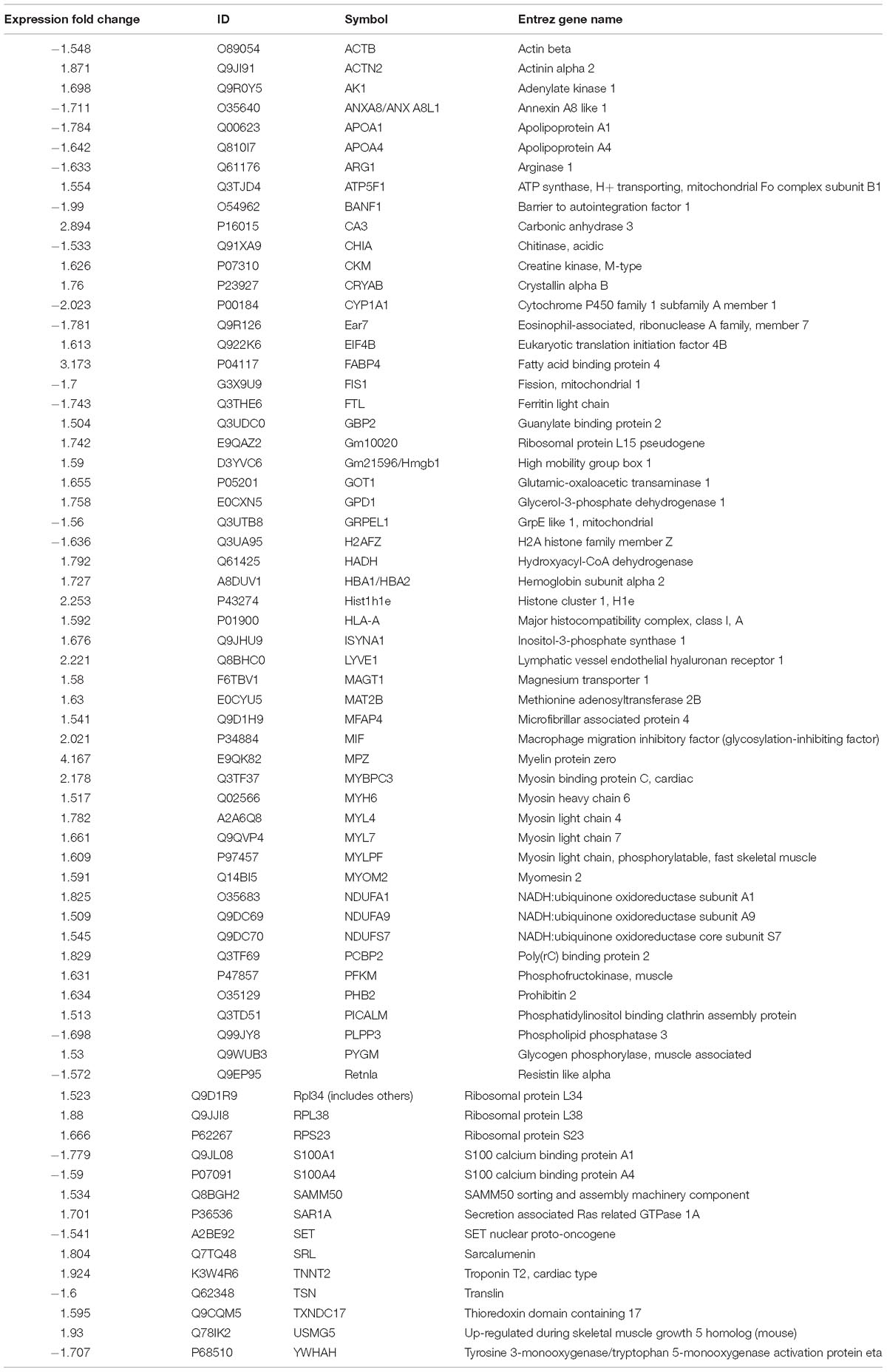
To compare the pathological and pulmonary function results, we selected the CPA model for further proteomic experiments. A total of 1,956 proteins were identified by iTRAQ with a < 1% false discovery rate (FDR) after analyzing three biological replicates of mouse lung tissue. Proteins with significantly altered expression levels at a fold change cut-off of > 1.5 relative to the model group were considered DEPs (p < 0.05). We found 71 DEPs in the model group compared with control; 17 proteins were downregulated while 54 proteins were upregulated (Figure 2A and Table 4). Moreover, a total of 68 proteins were differentially expressed after GBFXD treatment compared with that of the model group; 47 proteins were upregulated and 20 were downregulated (Figure 2B and Table 5). EIF4B, HBA1/HBA2, Hist1h1e, HLA-A, and LYVE1 were repressed in the model group but were found upregulated after GBFXD treatment. In contrast, ANXA8/ANXA8L1, APOA, ARG1, CHIA, CYP1A1, FTL, H2AFZ, PLPP3, Retnla, S100A4, and TSN were induced in the model group but downregulated in response to GBFXD treatment (Figure 2C). Furthermore, DEPs were classified using the PANTHER gene classification systems and the enriched gene ontology (GO) terms were found enriched compared with the control group, hydrolase activity (GO:0016787), protein binding (GO:0005515), metabolic process (GO:0008152), and response to stimuli (GO:0050896). DEPs between the GBFXD and model groups were mainly involved in metabolic processes (GO:0008152), cellular processes (GO:0009987), cellular component organization or biogenesis (GO:0071840), intracellular processes (GO:0005622), and protein binding (GO:0005515; Figures 2D–I).
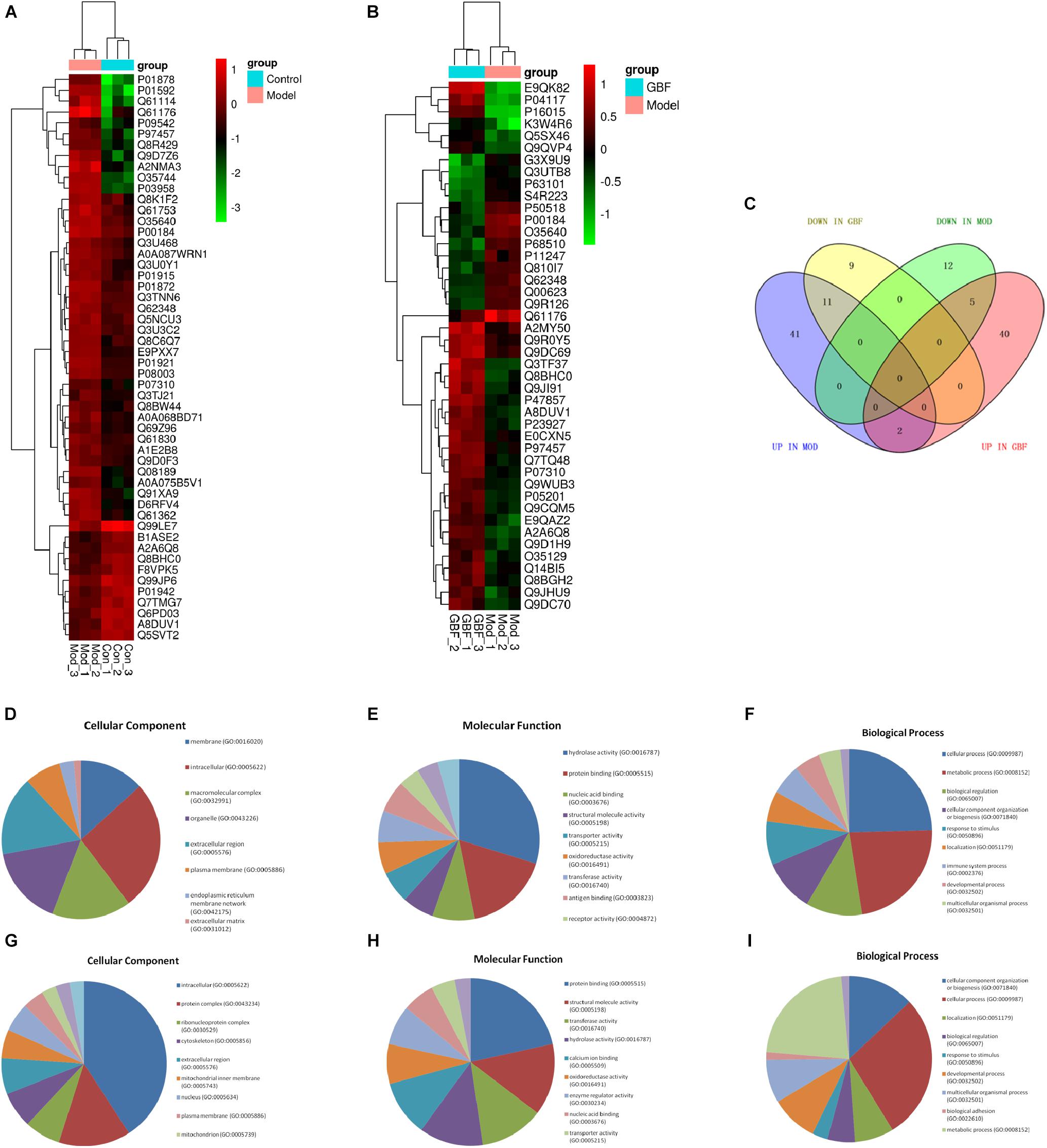
Figure 2. Protein expression profiles. (A,B) Heatmap of the expression levels of differentially expressed proteins (DEPs) compared with those of the model group quantified by iTRAQ-based stable isotope tagging. (C) Venn diagrams showing the distribution of shared DEPs. Blue represents upregulated proteins in the model group (up in MOD), green represents downregulated proteins in the model group (down in MOD), pink represents upregulated proteins in the GBFXD group (up in GBF), and yellow represents downregulated proteins in the GBFXD group (down in GBF). Overlapping sections indicate the number of common proteins across different groups. GO, gene ontology enrichment analysis. GO analysis of DEPs after modeling; (D) biological process, (E) cellular component, and (F) molecular function. GO analysis of DEPs after GBFXD treatment; (G) biological process, (H) cellular component, and (I) molecular function.
Analysis of Proteomic Data via IPA
Proteomic Analysis of the Model
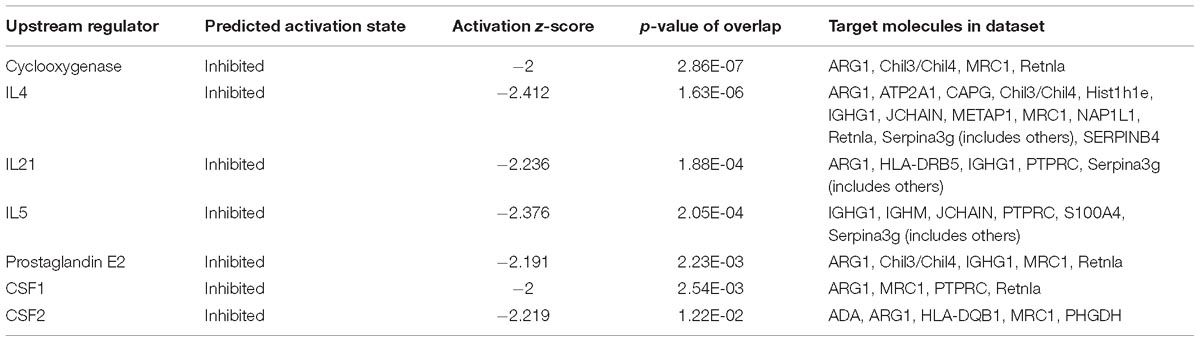
Proteomic data were analyzed via IPA, in which proteins are analyzed as a network using canonical pathways, disease networks, and predicted upstream regulators. Bioinformatic analysis showed that the model was involved in 165 pathways and 80 biological functions (Figure 3). Functional enrichments in inflammatory response, respiratory diseases, and inflammatory disease suggest that this model exhibits significant pulmonary inflammation. The high degree of enrichment in organismal injury and abnormalities, connective tissue disorders, and tissue morphology also suggests substantial changes in histomorphology. Humoral immune response and immune cell trafficking were also highly involved. The top 20 enriched disease and function terms are shown in Figure 3A. Canonical pathway analysis of the data revealed that our established model significantly activated multiple pathways in the mouse immune system; these included B-cell development, calcium-induced lymphocyte apoptosis, antigen presentation, iCOS-iCOSL signaling in T-helper cells, dendritic cell maturation, PKCθ signaling in T-lymphocytes, and Th1 and Th2 activation pathways. We also found that the DEPs were associated with 30 metabolic pathways. Upstream regulator predictions suggested that six inflammatory cytokines were upregulated and one downregulated (Z-Score > 1 or <−1); among the upregulated cytokines, the classical Th2 cytokines interleukin (IL)-4 and IL-5 drive M2 macrophage polarization in asthma pathogenesis (Table 6). Moreover, ARG1, MRC1, Chil3/Chil4, CHIA, and Retnla are typical cytokines secreted by M2 macrophages typical M2 cytokines that were found among the DEPs. In addition, GSF1, GSF2, cyclooxygenase, and prostaglandin E2 were significantly predicted upstream. These findings suggest that macrophages play an important role in this asthma model (Table 6). We further edited and expanded the functional network according to the molecular relationship most strongly correlated with the DEPs. The results showed that the DEPs constitute a reliable network and participate in inflammation and immune cell trafficking. The expression of the M2-type macrophage factors ARG1, MRC1, Chil3/Chil4, and Retnla is related to serum amyloid A (SAA) and cyclooxygenase. HBA1/HBA2 is a common biochemical indicator in the network. Therefore, there is abnormal blood oxygen saturation in the reaction state (Figure 3F).
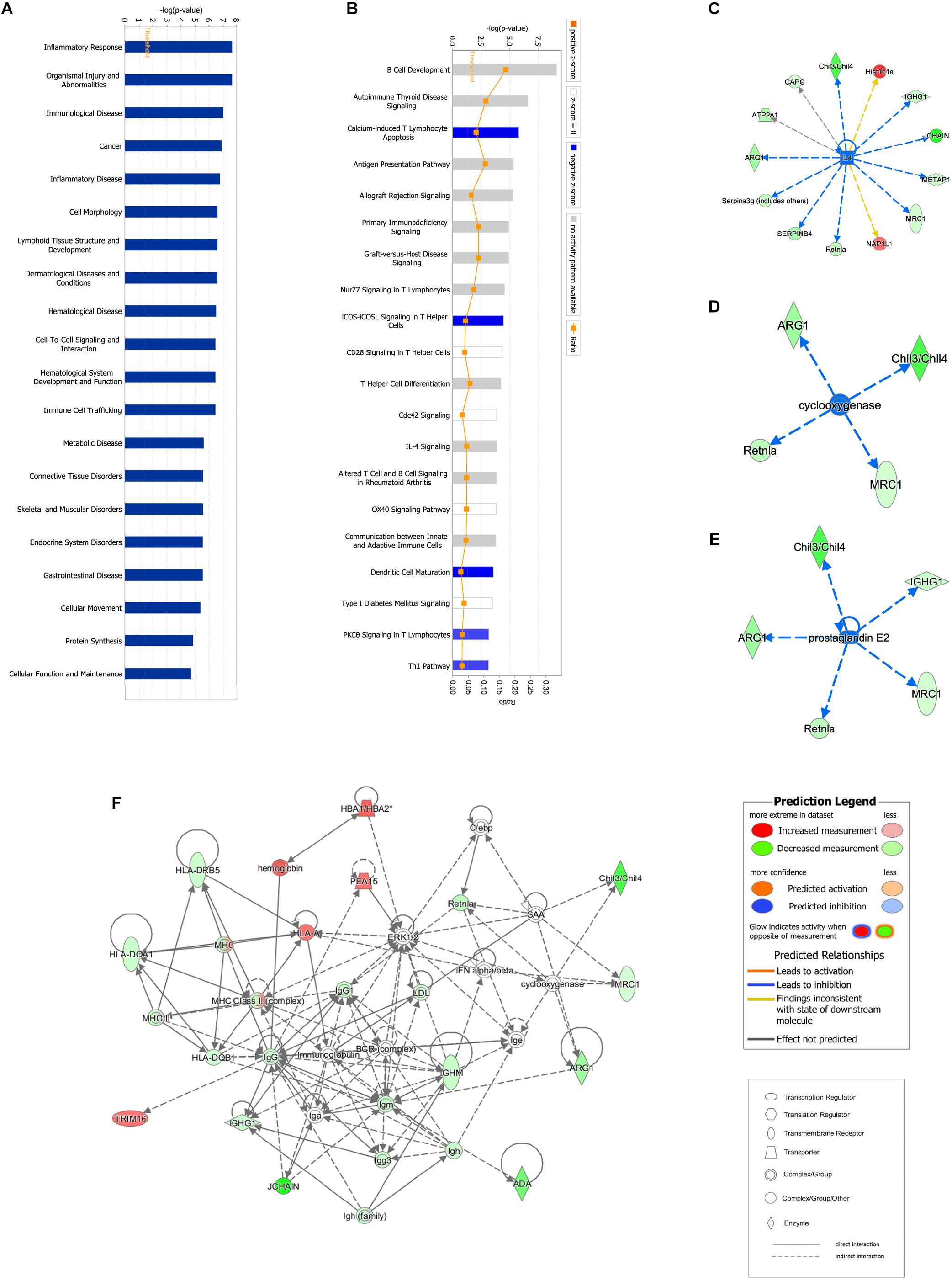
Figure 3. Bioinformatic analysis of DEPs between the control and model groups using Ingenuity Pathway Analysis (IPA) and z-score algorithms. (A) Biological function enrichment. (B) Canonical pathways enrichment (p-values for all functions and pathways presented are < 0.05). (C–E) Upstream regulatory factors predicted based on DEPs. (F) Relationships between DEPs and proteins predicted to be involved in interactions, as well as relevant signaling pathways, are presented. Statistical predictions of the networks were based on the Ingenuity Knowledge base. Red nodes represent upregulated proteins and green nodes represent downregulated proteins.
Proteomic Analysis Revealed the Therapeutic Effects of GBFXD
Ingenuity pathway analysis correlation analysis was performed to compare the GBFXD treatment and model groups and a total of 159 pathways were found to be involved. Pathways with significant regulation included actin cytoskeleton signaling, rho regulation of actin-based motility, ILK signaling, and sirtuin signaling. In addition, highly enriched pathways included mitochondrial dysfunction and oxidative phosphorylation. DEPs were found associated with 76 biological programs, 6 metabolic pathways, the immune program, and morphologically related programs; lipid and carbohydrate metabolism were the most enriched. Networks constructed with DEPs suggested that GBFXD treatment may have increased mitochondrial respiratory function and oxidative phosphorylation (Figure 4). Furthermore, we compared the highest scoring internetworks predicted by the two protein groups. Seven points were coincident (ARG1, cyclooxygenase, ERK1/2, HBA1/HBA2, hemoglobin, Retnla, and SAA; Figure 4); among them, ARG1, Retnla, and HBA1/HBA2 were common DEPs to both groups. Network analysis suggested that GBFXD regulates mitochondrial energy metabolism and corrects M2 factor expression. Moreover, it was predicted that IFNG is significantly activated after GBFXD treatment, which supports our previous experimental findings. Upstream regulatory factor prediction also suggested that GBFXD significantly inhibits RICTOR. Taken together, the two significant upstream predictions indicate that GBFXD inhibits M2 macrophages in our model.
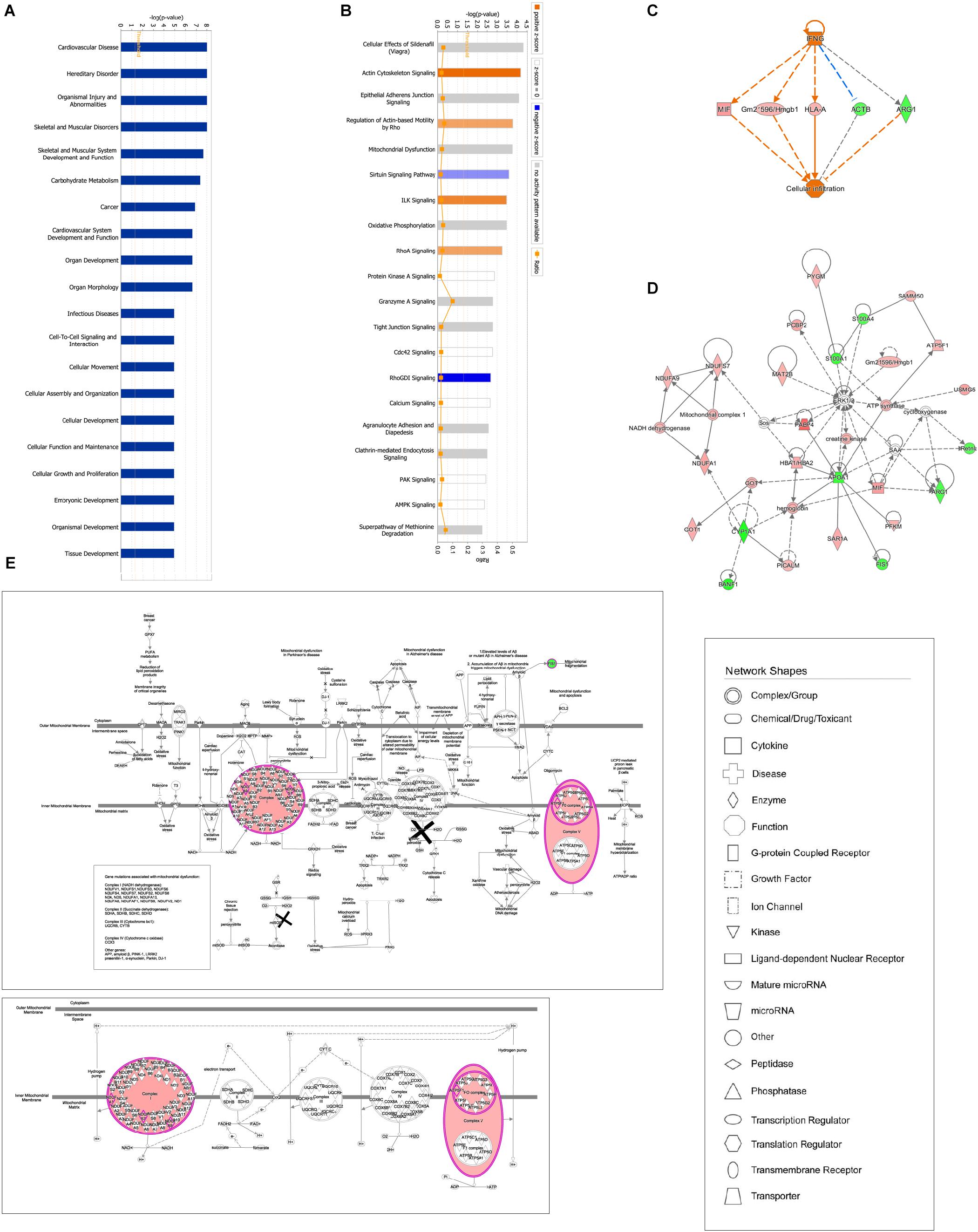
Figure 4. Bioinformatic analysis of DEPs in the lung after GBFXD treatment. (A) Biological function enrichment. (B) The most enriched pathways as a summary of DEPs between the GBFXD and model groups. (C) Regulatory effects resulting from integrating target molecules, upstream factors, and downstream biological functions. (D) The most credible interaction networks. (E) IPA of mitochondrial function and the oxidative phosphorylation pathway.
Validation
We further performed western blot analysis and found that expression levels of ARG1, CHIA, and Retnla in lung tissue were significantly decreased in the GBFXD group (Figure 5, Supplementary Data Sheet S1). This result is in agreement with the protein levels measured by iTRAQ analysis and the changing trends in transcription indicated by qRT-PCR. Therefore, our proteomic analysis output was deemed reliable.
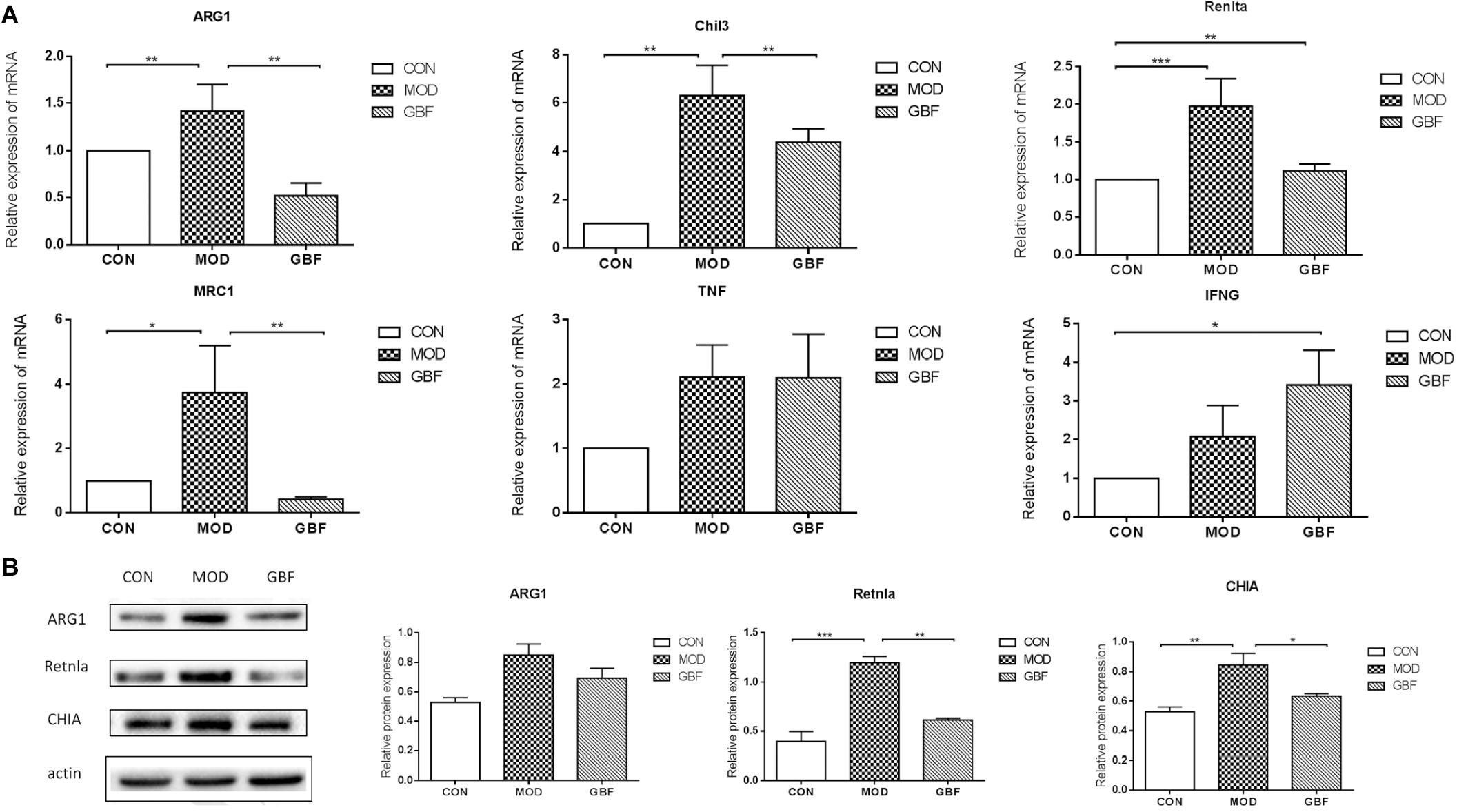
Figure 5. Validation of differentially expressed candidate proteins identified using proteomics. (A) qRT-PCR of six selected genes presented relative to control samples and normalized against GAPDH in each sample. (B) Western blot analysis shows decreased ARG1, Retnla, and CHIA levels in the GBFXD group (B). Values represent the means ± SE of three independent experiments with ≥3 replicates per experiment. ∗∗p < 0.01; ∗∗∗p < 0.0001; ∗∗∗∗p < 0.0001.
Discussion
Asthma is the most common chronic airway inflammation disorder whose incidence continues to increase worldwide. The pathophysiological features of asthma include continuous chronic airway inflammation, AHR, and airway remodeling. Moreover, there are dramatic epidemiological differences between childhood and adult asthma. As a first-line treatment for asthma, ICS is not well tolerated in children. Moreover, it may be difficult to diagnose asthma in certain children under the age of five (Pando et al., 2010; Klok et al., 2011; Bush, 2018; Ferrante and Grutta, 2018). As an alternative to ICS, several Asian countries employ GBFXD against asthma. Previous studies have shown that GBFXD reduces pulmonary inflammation during clinical remission in asthmatic mice (Huang et al., 2016; Lu et al., 2016) however, the mechanism remains unknown. Therefore, to understand the therapeutic role of GBFXD, we established a clinically consistent chronic asthmatic model and performed proteomic analysis. The study considerated the epidemiological characteristics of asthma in children; this subpopulation often presents with persistent allergen exposure inevitably (Szentpetery et al., 2017). Therefore, we increased the frequency of OVA challenges compared with that of our previous model. We compared both models and found that mice in the continuous OVA-induced model exhibit remarkable pathological features that were corroborated by lung function tests.
Alveolar macrophages, airway surface mucus, microorganisms, and airway epithelium constitute the first line of airway defense (Hussell and Bell, 2014). Macrophages can be adaptively polarized according to the metabolic characteristics of the tissue and immune environments. Macrophage polarization is categorized as the classical M1 type and the alternative M2 type. M1 polarization is pro-inflammatory and is initiated by adaptive immunity, whereas M2 polarization is anti-inflammatory and is critical for tissue damage and recovery during the late phases of infection (Wynn et al., 2013; Holtzman et al., 2014). M2 polarization mediates airway remodeling, matrix deposition, and other pathophysiological mechanisms in asthma (Wynn and Vannella, 2016). Th2 inflammatory factors are required to polarize macrophages toward the M2 phenotype (Dyken and Locksley, 2013). As OVA is often used to trigger Th2-type immune responses, the combination of OVA and RSV can aggravate Th2-type responses. Furthermore, excessive M2 macrophages are activated in lung tissue after RSV infection (Keegan et al., 2016; Naessens et al., 2016). Therefore, the increased M2 macrophage polarization exhibited by our model is credible.
Clinical studies have found that CD206+ macrophage is highly expressed in the trachea and bronchoalveolar lavage fluid in asthmatic patients. The degree of its expression is correlated with biological sex, asthma severity, and lung function. However, CD206+ macrophage may be insensitive to ICS treatment (Melgert et al., 2010, 2011; Draijer et al., 2016). Markers of alternatively activated macrophages include MRC1, ARG1, Retnla, and Chil3 and these are upregulated in the lungs. Herein, proteomic analysis indicated the presence of M2 macrophage markers, MRC1, ARG1, Retnla, Chil3, and CHIA in asthmatic. GBFXD treatment inhibited these factors (Supplementary Figure S1A). The inhibitory effect on M2 macrophages by GBFXD may result from its therapeutic effect in attenuating airway remodeling and ARH, which interact with each other and constitute the hidden pathogen inducing susceptibility to asthma. Therefore, GBFXD may represent an alternative therapeutic agent against asthma.
The upstream predictive function of our experiment indicated that IL-4/IL-5/IL-13 are significantly activated in this model. Arg1 is induced by IL-4/IL-13 in macrophages and counterbalances inflammatory signals. L-arginine metabolism in macrophages is a defining feature of alternative versus classical macrophage activation (Duan et al., 2011). Moreover, chitinase-like protein is elevated in serum and mainly in alveolar cells and macrophages of the lungs of asthmatic patients (Chupp et al., 2007). ARG1 and CHIL3 are well-recognized susceptibility genes closely associated with childhood asthma that increase AHR and weaken lung function; additionally, the polymorphic and epigenetic states of ARG1 are associated with childhood asthma progression (Li et al., 2006; Ober et al., 2008; Salam et al., 2009; Cunningham et al., 2011). Retnla (resistin-like molecule-α), also known as FIZZ 1 (found in inflammatory zone 1), is also highly induced in allergic lung inflammation and bleomycin-induced lung fibrosis (Holcomb et al., 2000; Liu et al., 2014). Furthermore, macrophages can strongly express Retnla in chronic type 2 inflammation (Nair et al., 2003). In agreement, our proteomic analysis identified changes in these proteins that led to clinical outcomes. At the same time, lung function and pathology results also support these conclusions. Therefore, we believe that our model would be valuable in the study of chronic persistent asthma in children.
Mitochondria are the energy centers of cellular activity. They also maintain cellular ion homeostasis and lipid metabolism. Clinical studies have detected dysfunction and structural changes in the mitochondria of airway epithelial cells of asthmatic patients. Similar changes observed in asthmatic animal models indicated that abnormal oxidative stress, mitochondrial membrane potential, and energy metabolism occur (Mabalirajan et al., 2008; Aguilera and Bacsi, 2009; Zifa et al., 2012; Flaquer et al., 2014; Girodet et al., 2015; Wang et al., 2017). Th1/Th2 imbalance also adversely affects mitochondrial function (Chang et al., 2009; Pattnaik et al., 2016). Furthermore, arginine and chitinase metabolism affect mitochondrial oxidative stress (Zhang et al., 2009; Chao and Carter, 2015; Xu et al., 2016). In the present study, proteomic analysis showed that after GBFXD treatment, NDUFA1, NDUFA9, NDUFS7, ATP5F1, and HADH were upregulated while FIS1 was downregulated. Pathway and network analyses also suggested that GBFXD increases ATP synthesis and that mitochondrial complex 1 interacts with mitigating M2 macrophage markers (Supplementary Table S1 and Supplementary Figure S1B).
Conclusion
Treatment with GBFXD can improve Th1/Th2 balance, inhibit alternatively activated macrophages, and reduce AHR, mucus secretion, and airway remodeling. We believe that GBFXD shows efficacy due to its mechanism of action, specifically the improvement in macrophage polarization and restoration of mitochondrial function.
Ethics Statement
All experimental procedures were performed in accordance with the National Institutes of Health Guidelines for Laboratory Animals and approved by the Animal Ethics Committee of Nanjing University of Chinese Medicine (201810A026).
Author Contributions
XZ conceived the project and designed the experiments. L-wL performed the data analysis and interpretation and Q-qX helped to write the manuscript. Q-qX, YL, MT, and Y-mD provided the experimental assistance. CD and YZ contributed to reagents, materials, and analysis tools.
Funding
This work was supported by the Major Research Plan of Natural Science Foundation of the Jiangsu Higher Education Institutions of China (Grant No. 15KJA360003), the Natural Science Foundation of China (Grant No. 81774367), the Innovation Program for Graduate Students of Jiangsu Province, China (Grant Nos. KYCX18_1553 and KYCX18_1555), the leading academics training program of Chinese Medicine in Jiangsu Province, China (Grant No. SLJ0224) and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2019.00441/full#supplementary-material
FIGURE S1 | Effect of GBFXD on expression of M2 and mitochondrial complex 1 marker in macrophages in mouse models. (A) Lung tissue was fixed and sections were processed for Immunofluorescence detection of M2 macrophage (CD206+f4/80+). (B) After macrophage collected from Balf, total mRNA was isolated and examined for transcript levels of the mitochondrial complex 1 markers (NDUFA1, NDUFA9, NDUFS7, ATP5F1) by RT-PCR analysis. Values represent the means ± SE of three independent experiments with 3 replicates per experiment. ∗∗p < 0.01; ∗∗∗p < 0.0001; ∗∗∗∗p < 0.0001.
TABLE S1 | Primer sequences of NDUFA1, NDUFA9, NDUFS7, ATP5F1.
References
Aguilera, A. L., and Bacsi, A. M. A. (2009). Mitochondrial dysfunction increases allergic airway inflammation. J. Immunol. 183, 5379–5387. doi: 10.4049/jimmunol.0900228
Bateman, E. D., Boushey, H. A., Bousquet, J., Busse, W. W., Clark, T. J., Pauwels, R. A., et al. (2004). Can guideline-defined asthma control be achieved? the gaining optimal asthma control study. Am. J. Respir. Crit. Care Med. 170, 836–844. doi: 10.1164/rccm.200401-033oc
Bush, A. (2018). Management of asthma in children. Minerva Pediatr. 70, 444–457. doi: 10.23736/S0026-4946.18.05351-3
Carroll, K. N., Gebretsadik, T., Escobar, G. J., Wu, P., Li, S. X., Walsh, E. M., et al. (2017). Respiratory syncytial virus immunoprophylaxis in high-risk infants and development of childhood asthma. J. Allergy Clin. Immunol. 139, 66–71. doi: 10.1016/j.jaci.2016.01.055
Chang, H. H., Chen, C. S., and Lin, J. Y. (2009). High dose vitamin C supplementation increases the Th1/Th2 cytokine secretion ratio, but decreases eosinophilic infiltration in bronchoalveolar lavage fluid of ovalbumin-sensitized and challenged mice. J. Agric. Food Chem. 57, 10471–10476. doi: 10.1021/jf902403p
Chao, H., and Carter, A. B. (2015). The metabolic prospective and redox regulation of macrophage polarization. J. Clin. Cell. Immunol. 6:371.
Chun, W., Qing, R., Xue-Tong, C., Song, Z. Q., Ning, Z. C., and Gan, J. H. (2018). System pharmacology-based strategy to decode the synergistic mechanism of zhi-zhu wan for functional dyspepsia. Front. Pharmacol. 9:841. doi: 10.3389/fphar.2018.00841
Chupp, G. L., Lee, C. G., Jarjour, N., Shim, Y. M., Holm, C. T., He, S., et al. (2007). A chitinase-like protein in the lung and circulation of patients with severe asthma. N. Engl. J. Med. 357, 2016–2027. doi: 10.1056/nejmoa073600
Cunningham, J., Basu, K., Tavendale, R., Palmer, C. N., Smith, H., and Mukhopadhyay, S. (2011). The CHI3L1 rs4950928 polymorphism is associated with asthma-related hospital admissions in children and young adults. Ann. Allergy Asthma Immunol. 106, 381–386. doi: 10.1016/j.anai.2011.01.030
Draijer, C., Boorsma, C. E., Robbe, P., Timens, W., Hylkema, M. N., Ten Hacken, N. H., et al. (2016). Human asthma is characterized by more IRF5+ M1 and CD206+ M2 macrophages and less IL-10+ M2-like macrophages around airways compared with healthy airways. J. Allergy Clin. Immunol. 140, 280–283. doi: 10.1016/j.jaci.2016.11.020
Duan, Q. L., Gaume, B. R., Hawkins, G. A., Himes, B. E., Bleecker, E. R., Klanderman, B., et al. (2011). Regulatory haplotypes in ARG1 are associated with altered bronchodilator response. Am. J. Respir. Crit. Care Med. 183, 449–454. doi: 10.1164/rccm.201005-0758OC
Dyken, S. J. V., and Locksley, R. M. (2013). Interleukin-4- and interleukin-13-mediated alternatively activated macrophages: roles in homeostasis and disease. Annu. Rev. Immunol. 31, 317–343. doi: 10.1146/annurev-immunol-032712-095906
Ebmeier, S., Thayabaran, D., Braithwaite, I., Bénamara, C., Weatherall, M., and Beasley, R. (2017). Trends in international asthma mortality: analysis of data from the who mortality database from 46 countries (1993–2012). Lancet 390, 935–945. doi: 10.1016/S0140-6736(17)31448-4
Ferrante, G., and Grutta, S. L. (2018). The burden of pediatric asthma. Front. Pediatr. 6:186. doi: 10.3389/fped.2018.00186
Flaquer, A., Heinzmann, A., Rospleszcz, S., Mailaparambil, B., Dietrich, H., Strauch, K., et al. (2014). Association study of mitochondrial genetic polymorphisms in asthmatic children. Mitochondrion 14, 49–53. doi: 10.1016/j.mito.2013.11.002
Gao, B. Z., Chen, J. C., Liao, L. H., Xu, J. Q., Lin, X. F., and Ding, S. S. (2015). Erchen decoction prevents high-fat diet induced metabolic disorders in C57BL/6 mice. Evid. Based Complement. Altern. Med. 2015:501272.
Geng, Y. Y., Wang, W. G., Zhang, J. H., Bi, S. Y., Li, H. X., and Lin, M. J. (2016). Effects of Traditional Chinese Medicine herbs for tonifying Qi and kidney, and replenishing spleen on intermittent asthma in children aged 2 to 5 years old. J. Tradit. Chin. Med. 36, 32–38. doi: 10.1016/s0254-6272(16)30005-x
Girodet, P. O., Allard, B., Thumerel, M., Begueret, H., Dupin, I., Ousova, O., et al. (2015). Bronchial smooth muscle remodeling in non-severe asthma. Am. J. Respir. Crit. Care Med. 193, 627–633.
Grainge, C. L., Lau, L. C., Ward, J. A., Dulay, V., Lahiff, G., and Wilson, S., et al. (2011). Effect of bronchoconstriction on airway remodeling in asthma. N. Engl. J. Med. 364, 2006–2015. doi: 10.1056/NEJMx180023
Guo, X., Li, Z., Ling, W., Long, J., Su, C., Li, J., et al. (2018). Epidemiology of childhood asthma in mainland china (1988-2014): a meta-analysis. Allergy Asthma Proc. 39, 15–29. doi: 10.2500/aap.2018.39.4131
Holcomb, I. N., Kabakoff, R. C., Chan, B., Baker, T. W., Gurney, A., Henzel, W., et al. (2000). FIZZ1, a novel cysteine-rich secreted protein associated with pulmonary inflammation, defines a new gene family. EMBO J. 19, 4046–4055. doi: 10.1093/emboj/19.15.4046
Holtzman, M. J., Byers, D. E., Alexander-Brett, J., and Wang, X. (2014). The role of airway epithelial cells and innate immune cells in chronic respiratory disease. Nat. Rev. Immunol. 14, 686–698. doi: 10.1038/nri3739
Huang, T. P., Liu, P. H., Lien, A. Y., Yang, S. L., Chang, H. H., and Yen, H. R. (2013). Characteristics of traditional Chinese medicine use in children with asthma: a nationwide population-based study. Allergy 68, 1610–1613. doi: 10.1111/all.12273
Huang, Z., Gao, L., Zhao, X., Ling, H., and Chen, W. (2016). Effect of Gubenfangxiao decoction on respiratory syncytial virus-induced asthma and expression of asthma susceptibility gene orosomucoid 1-like protein 3 in mice. J. Tradit. Chin. Med. 36, 101–106. doi: 10.1016/s0254-6272(16)30015-2
Hussell, T., and Bell, T. J. (2014). Alveolar macrophages: plasticity in a tissue-specific context. Nat. Rev. Immunol. 14, 81–93. doi: 10.1038/nri3600
Jartti, T., and Gern, J. E. (2017). Role of viral infections in the development and exacerbation of asthma in children. J. Allergy Clin. Immunol. 140, 895–906. doi: 10.1016/j.jaci.2017.08.003
Keegan, A. D., Shirey, K. A., Bagdure, D., Blanco, J., Viscardi, R. M., and Vogel, S. N. (2016). Enhanced allergic responsiveness after early childhood infection with respiratory viruses: are long-lived alternatively-activated macrophages the missing link? Pathog. Dis. 74:ftw047. doi: 10.1093/femspd/ftw047
Klok, T., Kaptein, A., Duiverman, E., and Brand, P. (2011). Poor adherence to inhaled corticosteroids in childhood asthma: don’t blame (only) the patient. Eur. Respir. J. 38(Suppl. 55):4818.
Li, H., Romieu, I., Sienra-Monge, J. J., Ramirez-Aguilar, M., Estela Del Rio-Navarro, B., Kistner, E. O., et al. (2006). Genetic polymorphisms in Arginase I and II and childhood asthma and atopy. J. Allergy Clin. Immunol. 117, 119–126. doi: 10.1016/j.jaci.2005.09.026
Liu, C. H. (2019). The prevalence, diagnosis and management of asthma in children in China. J. Otolaryngol. Ophthalmol. Shandong University, 33, 28–32.
Liu, C. H., and Chen, Y. Z. (2013). Analysis of the epidemiology and the present situation of prevention and treatment of asthma in chilren in China. Chinese J. Pract. Pediatr. 28, 809–811.
Liu, T., Yu, H., Ullenbruch, M., Jin, H., Ito, T., Wu, Z., et al. (2014). The in vivo fibrotic role of FIZZ1 in pulmonary fibrosis. PLoS One 9:e88362. doi: 10.1371/journal.pone.0088362
Lu, Y., Xu, J. Y., Zhang, X. H., and Zhao, X. (2016). Gu-Ben-Fang-Xiao decoction attenuates sustained airway inflammation by suppressing ER stress response in a murine asthma remission model of respiratory syncytial virus infection. J. Ethnopharmacol. 192, 496–509. doi: 10.1016/j.jep.2016.09.039
Mabalirajan, U., Dinda, A. K., Kumar, S., Roshan, R., Gupta, P., Sharma, S. K., et al. (2008). Mitochondrial structural changes and dysfunction are associated with experimental allergic asthma. J. Immunol. 181, 3540–3548. doi: 10.4049/jimmunol.181.5.3540
Melgert, B. N., Oriss, T. B., Qi, Z., Dixon-McCarthy, B., Geerlings, M., Hylkema, M. N., et al. (2010). Macrophages: regulators of sex differences in asthma? Am. J. Respir. Cell Mol. Biol. 42, 595–603. doi: 10.1165/rcmb.2009-0016OC
Melgert, B. N., ten Hacken, N. H., Rutgers, B., Timens, W., Postma, D. S., and Hylkema, M. N. (2011). More alternative activation of macrophages in lungs of asthmatic patients. J. Allergy Clin. Immunol. 127, 831–833. doi: 10.1016/j.jaci.2010.10.045
Naessens, T., Schepens, B., Smet, M., Pollard, C., Van Hoecke, L., De Beuckelaer, A., et al. (2016). GM-CSF treatment prevents respiratory syncytial virus-induced pulmonary exacerbation responses in postallergic mice by stimulating alveolar macrophage maturation. J. Allergy Clin. Immunol. 137, 700–709. doi: 10.1016/j.jaci.2015.09.031
Nair, M. G., Cochrane, D. W., and Allen, J. E. (2003). Macrophages in chronic type 2 inflammation have a novel phenotype characterized by the abundant expression of Ym1 and Fizz1 that can be partly replicated in vitro. Immunol. Lett. 85, 173–180. doi: 10.1016/s0165-2478(02)00225-0
Ober, C., Tan, Z., Sun, Y., Possick, J. D., Pan, L., and Nicolae, R. (2008). Effect of variation in CHI3L1 on serum YKL-40 level, risk of asthma, and lung function. N. Engl. J. Med. 358, 1682–1691. doi: 10.1056/NEJMoa0708801
Pando, S., Lemière, C., Beauchesne, M. F., Perreault, S., Forget, A., and Blais, L. (2010). Suboptimal use of inhaled corticosteroids in children with persistent asthma: inadequate prescription, poor drug adherence, or both? Pharmacotherapy 30, 1109–1116. doi: 10.1592/phco.30.11.1109
Papi, A., Brightling, C., Pedersen, S. E., and Reddel, H. K. (2018). Asthma. Lancet 391, 783–800. doi: 10.1016/S0140-6736(17)33311-1
Pattnaik, B., Bodas, M., Bhatraju, N. K., Ahmad, T., Pant, R., Guleria, R., et al. (2016). IL-4 promotes asymmetric dimethylarginine accumulation, oxo-nitrative stress, and hypoxic response-induced mitochondrial loss in airway epithelial cells. J. Allergy Clin. Immunol. 138, 130.e9–141.e9. doi: 10.1016/j.jaci.2015.11.036
Phipps, S., Lam, C. E., Mahalingam, S., Newhouse, M., Ramirez, R., Rosenberg, H. F., et al. (2007). Eosinophils contribute to innate antiviral immunity and promote clearance of respiratory syncytial virus. Blood 110, 1578–1586. doi: 10.1182/blood-2007-01-071340
Salam, M. T., Islam, T., Gauderman, W. J., and Gilliland, F. D. (2009). Roles of arginase variants, atopy, and ozone in childhood asthma. J. Allergy Clin. Immunol. 123, 596.e8–602.e8. doi: 10.1016/j.jaci.2008.12.020
Szentpetery, S. S., Gruzieva, O., Forno, E., Han, Y. Y., Bergström, A., Kull, I., et al. (2017). Combined effects of multiple risk factors on asthma in school-aged children. Respir. Med. 133, 16–21. doi: 10.1016/j.rmed.2017.11.002
Wang, C. M., Zhang, X. J., Ying, J. M. A., and Li, X. (2017). Mutational analysis of mitochondrial tRNA genes in patients with asthma. Iran. J. Public Health 46, 620–625.
Wang, D., Zhang, B. B., Qu, X. X., Gao, F., and Yuan, M. Y. (2015). Microwave-assisted extraction of polysaccharides from Yupingfeng powder and their antioxidant activity. Pharmacogn. Mag. 11, 546–554. doi: 10.4103/0973-1296.160468
William, D., Pingsheng, W., Tebeb, G., and Tina, H. (2018). Rsv prevention in infancy and asthma in later life. Lancet Respir. Med. 6:e32. doi: 10.1016/s2213-2600(18)30231-5
Wynn, T. A., Chawla, A., and Pollard, J. W. (2013). Macrophage biology in development, homeostasis and disease. Nature 496, 445–455. doi: 10.1038/nature12034
Wynn, T. A., and Vannella, K. M. (2016). Macrophages in tissue repair, regeneration, and fibrosis. Immunity 44, 450–462. doi: 10.1016/j.immuni.2016.02.015
Xu, W., Ghosh, S., Comhair, S. A., Asosingh, K., Janocha, A. J., Mavrakis, D. A., et al. (2016). Increased mitochondrial arginine metabolism supports bioenergetics in asthma. J. Clin. Invest. 126, 2465–2481. doi: 10.1172/JCI82925
Yuan, X. J., Sun, Y. Q., Wang, S. M., Li, Y. N., Wang, M. Q., and Liu, X. R. (2010). Clinical research of Gubenfangxiao decoction combined with point application on chronic asthmatic children in 100 cases. Zhong Hua Zhong Yi Yao Za Zhi 21, 2306–2309
Yue, S. J., Xin, L. T., Fan, Y. C., Li, S. J., Tang, Y. P., Duan, J. A., et al. (2017). Herb pair Danggui-Honghua: mechanisms underlying blood stasis syndrome by system pharmacology approach. Sci. Rep. 7:40318. doi: 10.1038/srep40318
Zhang, L., Peeples, M. E., Boucher, R. C., Collins, P. L., and Pickles, R. J. (2002). Respiratory syncytial virus infection of human airway epithelial cells is polarized, specific to ciliated cells, and without obvious cytopathology. J. Virol. 76, 5654–5666. doi: 10.1128/JVI.76.11.5654-5666.2002
Zhang, L., Wang, M., Kang, X., Boontheung, P., Li, N., Nel, A. E., et al. (2009). Oxidative stress and asthma: proteome analysis of chitinase-like proteins and FIZZ1 in lung tissue and bronchoalveolar lavage fluid. J. Proteome Res. 8, 1631–1638. doi: 10.1021/pr800685h
Keywords: GU-BEN-FANG-XIAO decoction, iTRAQ, chronic persistent asthma, macrophage, IPA
Citation: Liu L-w, Xing Q-q, Zhao X, Tan M, Lu Y, Dong Y-m, Dai C and Zhang Y (2019) Proteomic Analysis Provides Insights Into the Therapeutic Effect of GU-BEN-FANG-XIAO Decoction on a Persistent Asthmatic Mouse Model. Front. Pharmacol. 10:441. doi: 10.3389/fphar.2019.00441
Received: 23 October 2018; Accepted: 08 April 2019;
Published: 07 May 2019.
Edited by:
Aiping Lu, Hong Kong Baptist University, Hong KongReviewed by:
Wei Song, Peking Union Medical College Hospital (CAMS), ChinaSongxiao Xu, Artron BioResearch Inc., Canada
Copyright © 2019 Liu, Xing, Zhao, Tan, Lu, Dong, Dai and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xia Zhao, zhaoxiahy@hotmail.com
 Li-wei Liu
Li-wei Liu Qiong-qiong Xing1,2
Qiong-qiong Xing1,2 Xia Zhao
Xia Zhao