- 1Jiangsu Key Laboratory of Traditional Medicine and Translational Research, China Pharmaceutical University, Nanjing, China
- 2State Key Laboratory of Quality Research in Chinese Medicine, Institute of Chinese Medical Sciences, University of Macau, Taipa, Macau
Schisandra chinensis is a commonly used traditional Chinese medicine, which has been widely used in the treatment of acute myocardial infarction in China. However, it has been difficult to systematically clarify the major pharmacological effect of Schisandra, due to its multi-component complex mechanism. In order to solve this problem, a comprehensive network analysis method was established based-on “component–gene ontology–effect” interactions. Through the network analysis, reduction of cardiac preload and myocardial contractility was shown to be the major effect of Schisandra components, which was further experimentally validated. In addition, the expression of NCOR2 and NFAT in myocyte were experimentally confirmed to be associated with Schisandra in the treatment of AMI, which may be responsible for the preservation effect of myocardial contractility. In conclusion, the three-mode gene ontology network can be an effective network analysis workflow to evaluate the pharmacological effects of a multi-drug complex system.
Introduction
Cardiovascular disease, including atherosclerosis, myocardial infarction, heart failure and stroke, is the leading cause of morbidity and mortality in developed nations. Acute myocardial infarction (AMI) is induced through the narrowing of arteries caused by atherosclerotic plaques or the acute occlusion of the coronary artery by thrombosis, and has received extensive attention due to its high risk and poor outcome among all the symptoms of coronary heart disease (Holmes et al., 2011; Husted and Ohman, 2015).
Traditional Chinese medicine (TCM) is a commonly used therapeutic strategy for the treatment of AMI in China. And Schisandra chinensis is a commonly used TCM that has been clinically proven to alleviate the damage of myocytes after the onset of AMI. Many pharmacological research results have clarified the mechanisms of Schisandra (Li et al., 1996; Panossian and Wikman, 2008; Chang et al., 2013; Chen et al., 2013; Zhan et al., 2014). However, few studies have evaluated the major effects and possible mechanisms responsible for the treatment of AMI, due to the complexity of the multi-mechanisms associated with TCM (Gao et al., 2016; Tang et al., 2016). Thus, in order to research the multi-mechanism complex system in TCM, network pharmacology has been commonly used in recent years to predict the major target proteins or signal pathways of TCM.
In this study, network pharmacology was applied to analyze the major effect of Schisandra. In contrast to protein interaction networks, enriched gene ontology (GO) terms of AMI related genes were used to construct a gene ontology interaction (GOI) network, which can be used to simulate the functional interactions between differential expressed genes of disease. In general, this study aims to identify and validate major mechanism and related pharmacological effects of Schisandra in the treatment of AMI through a GOI network, which may offer a new method of network analysis to evaluate complex bio-systems.
Materials and Methods
Component Identification
For the construction of a “component–gene ontology” network of Schisandra in the treatment of AMI, the HPLC-Q-TOF-MS was used to analyze the Schisandra extract. Targets of Schisandra ingredients were further screened to enrich and construct the “component–gene ontology” network, which was a component for the integration of a three-mode network.
Sample Solution Preparation
To ensure the consistency of network analysis and experimental validation results, Schisandra chinensis Fructus of the same batch was utilized for the extraction, analysis and pharmacological experiment. Schisandra chinensis Fructus from Schisandra chinensis (Turcz.) Baill was obtained from Tianjin Tasly Pride Pharmaceutical Co. (Tianjin, China). The crude drug was extracted through a reflux condenser with 10 times the amount of distilled water at 100°C for 1 h. This procedure was repeated three times. The combined Schisandra extract was then concentrated under reduced pressure and dissolved through distilled water into an appropriate concentration for administration to mice. Deionized water was prepared using a Milli-Q Ultrapure water system (Millipore, Bedford, MA, United States). For analysis of the Schisandra component, dry extract was dissolved in 50% methanol and then centrifuged at 12000 rpm for 15 min. The supernatant was transferred to a 1.5 mL brown HPLC vial (Grace, Chicago, IL, United States) and stored at 4°C for analysis.
HPLC-Q-TOF-MS-MS Analysis Conditions
Chromatographic experiments were conducted on a Shimadzu Shimadzu (Kyoto, Japan) LC-2010 series. Chromatographic separation was performed on a Kromasil 100-5C18 (250 × 4.6 mm, 5 μm particle size) column, with the column temperature maintained at 30°C. The mobile phase was composed of solvent A (acetonitrile containing 0.01% v/v formic acid) and solvent B (ultrapure water containing 0.02% v/v acetic acid). The gradient elution conditions were: 0-15 min, 20% A; 15-25 min, 20-22% A; 25-45 min, 22-32% A; 45-65 min, 32-34% 75 min, 34-42% A; 75-95 min, 42-60% A; 95-110 min, 60-70% A; 110-125 min, 70-100% A; 125-130 min, 100% A. The injection volume was 15 μL. The elution rate was 0.8 mL/min and the detector was set at 203 nm.
The 6520 Q-TOF mass spectrometer (Agilent Technologies, Santa Clara, CA, United States) was equipped with an electrospray ionization (ESI) source. Ultrahigh purity argon was used as the collision gas and high purity nitrogen as the nebulizing gas. The following MS conditions were used: detector voltage was 1.65 kV, capillary voltage was 3.5 kV, heat block temperature was 325°C, nebulizer was 35 psig, nebulizing gas (N2) flow was 8.0 L/min, drying gas pressure (N2) was 72 kPa, ion trap pressure was 1.9 × 102 Pa, TOF pressure was 2.2 × 104 Pa, ion accumulation time was 100 ms. Scan ranges were set at m/z 100–1000 in both the positive and negative modes. The accurate mass determination was corrected by calibration using the sodium trifluoroacetate clusters as a reference. The peak area of molecular ion was then measured and normalized for the rough quantification of identified Schisandra components.
Construction and Analysis of “Component-Gene Ontology-Effect” Three-Mode Network
In order to simulate and analyze the correlation between components and pharmacological effects, a three-mode network model, which included the pathological relationship between component and pharmacological effect, was constructed (Figure 1A).
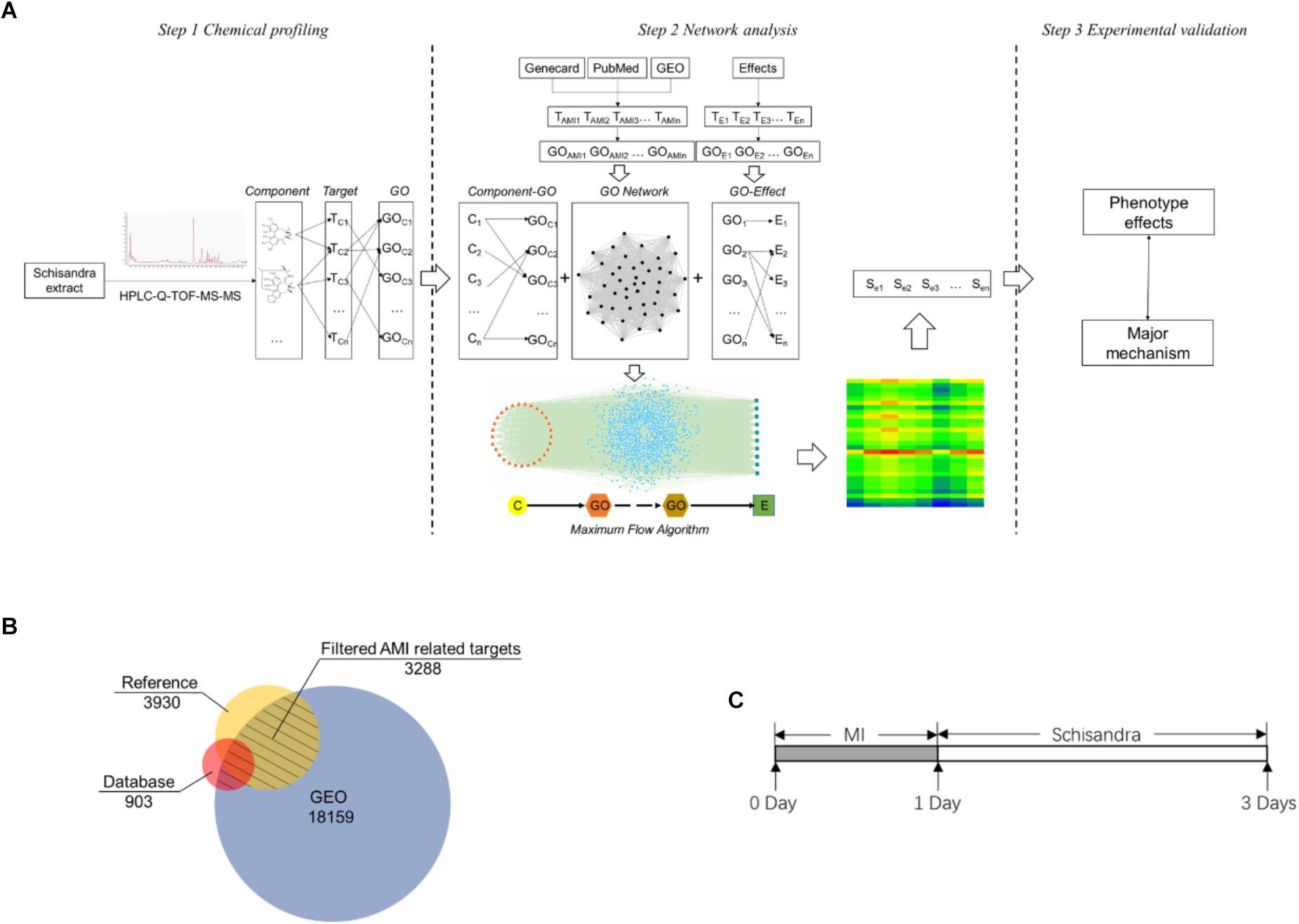
Figure 1. The Schisandra three-mode GOI network analysis workflow. (A) The components in the Schisandra extract were identified through HPLC-Q-TOF-MS-MS. Target proteins relevant to component (TC) were then obtained through PharmMapper. AMI related differential expressed genes (TAMI) and pharmacological effect related proteins (TE) were screened through data mining from three different databases (B). After target data were enriched through the GO annotation database, the relationship between the component and pharmacological effects were connected through GO terms, and the “component–gene ontology–effect” three-mode network was constructed. The relation score between component and effect was calculated using a maximum flow algorithm. In the end, the prediction results were validated through pharmacological experiments (C), with the corresponding drug given to mice one day after ligation of the left anterior descending.
Data Mining of Targets Related to Schisandra and AMI
Screening of identified Schisandra component’s related target
The chemical structures of Schisandra components identified through HPLC-Q-TOF-MS-MS were drawn using the ChemDraw 2004. After the structures were transformed into ∗.mol format, they were uploaded to the PharmMapper1 (Iyer et al., 2015) server to screen for component related drug-like target proteins. The filter species was set as Mus Musculus, and the top 300 drug-related proteins of each component were collected.
Myocardial infarction relevant targets
In order to reduce the bias of datamining, several different data sources were utilized to collect the AMI-related differentially expressed genes. The target data obtained from the public database would be more widely acknowledged. The differential expression genes collected from the GEO database ensures the precision of experimental animal species and organs. Target data screened from references of the PubMed database guarantees the inclusion of the latest research targets related to AMI.
For the screening of public disease databases, three data sources: GeneCard2, PharmGKB3, and the Therapeutic Target Database4 were searched using the keywords “Coronary heart disease OR Coronary artery disease OR Myocardial infarction OR Myocardial ischemia.” Snapshots of these databases were taken in December 2016. The collected target dataset was marked as TDB.
The PubMed database5 was then utilized for reference mining of the latest researched on AMI associated gene datasets, by screening the search terms “Coronary heart disease OR Coronary artery disease OR Myocardial infarction OR Myocardial ischemia.” Abstracts of reference within the past 5 years were downloaded and literature mining was carried out in order to collect the target data. After removing duplicates, the target dataset related to each pharmacological effect could be collected and was marked as TRef.
The next step was datamining for data on differentially expressed genes (DEG) in the Gene Expression Omnibus (GEO6) database. The experimental terms of interest were evaluated by screening experimental designs. Designs that contained a model constructed by ligation of left anterior descending (LAD) surgery on the species of Mus Musculus were included, while raw data terms without enough duplications (>3) were excluded. The collected raw data (GSE775, GSE19322, and GSE49937) were then normalized by the RMA method through the Affy package in Bioconductor7. The significant DEGs between control and AMI model groups were calculated using the Limma (linear models for microarray data) package. Gene sets with an expression of Log2 FC (Fold Change) > 1.5 and FDR (False Discovery Rate) < 0.05 were filtered for and collected. The collected GEO data were marked as TGEO (Barrett et al., 2007; Cappuzzello et al., 2009; Song et al., 2013; Steudemann et al., 2013; Gray et al., 2014; Liu et al., 2014, 2015, 2017; Zhang et al., 2014a,b; Peterson et al., 2015; Wang et al., 2015; Andersen et al., 2017; Gupta et al., 2017; Wu et al., 2017; Fang et al., 2018; Gunawan et al., 2018; Murugesan and Premkumar, 2018; Sabrkhany et al., 2018).
For the final step, the NCBI gene database8 was used to remove duplicates in the collected dataset and to normalize the official symbol in the Mus Musculus species. The union of targets collected from the open source database (TDB) and reference mining (TRef) was then intersected with the DEG data (TGEO) mined from the GEO database (Figure 1B):
In which, TAMI refers to the target dataset of AMI.
Pharmacological effect correlated targets
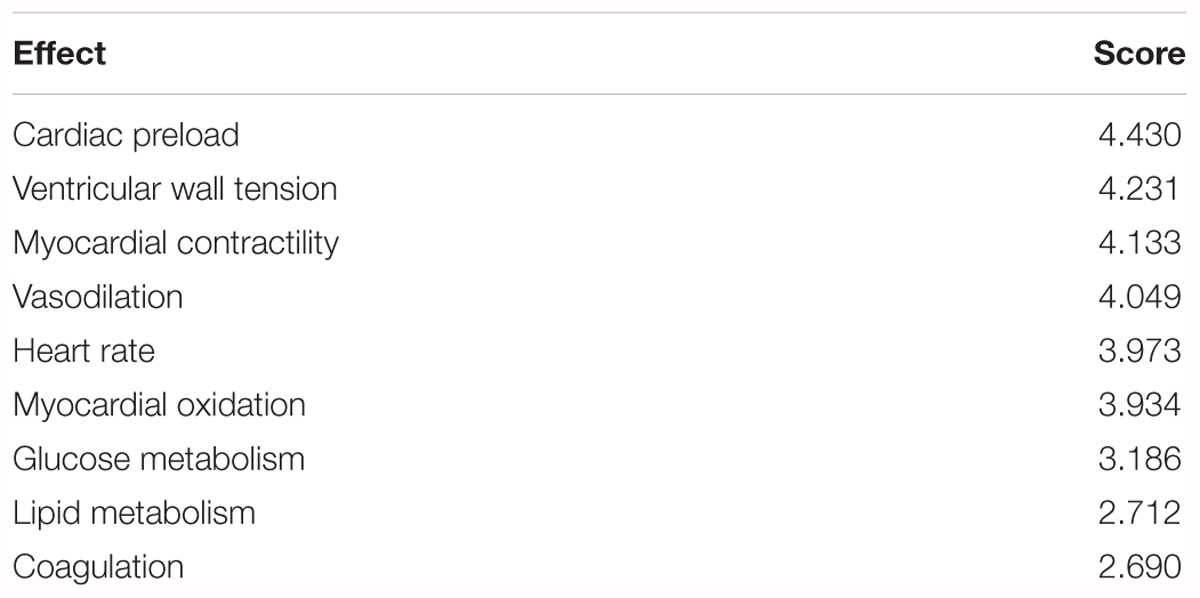
For the construction and analysis of a “gene ontology–effect” network, the targets related to pharmacological effects were screened according to the authoritative classification and references (Figure 2). The treatment mechanisms that were suitable for the network analysis and experimental evaluation can be summarized as anticoagulation, vasodilation, glucose metabolism, lipid metabolism, reduction of heart preload, ventricular wall tension, heart rate and myocardial contractility (Holmes et al., 2011; Husted and Ohman, 2015). Target data of pharmacological effects were screened through literature mining of PubMed references. The effect related target data were enriched by the GO terms for the further construction of a network model.
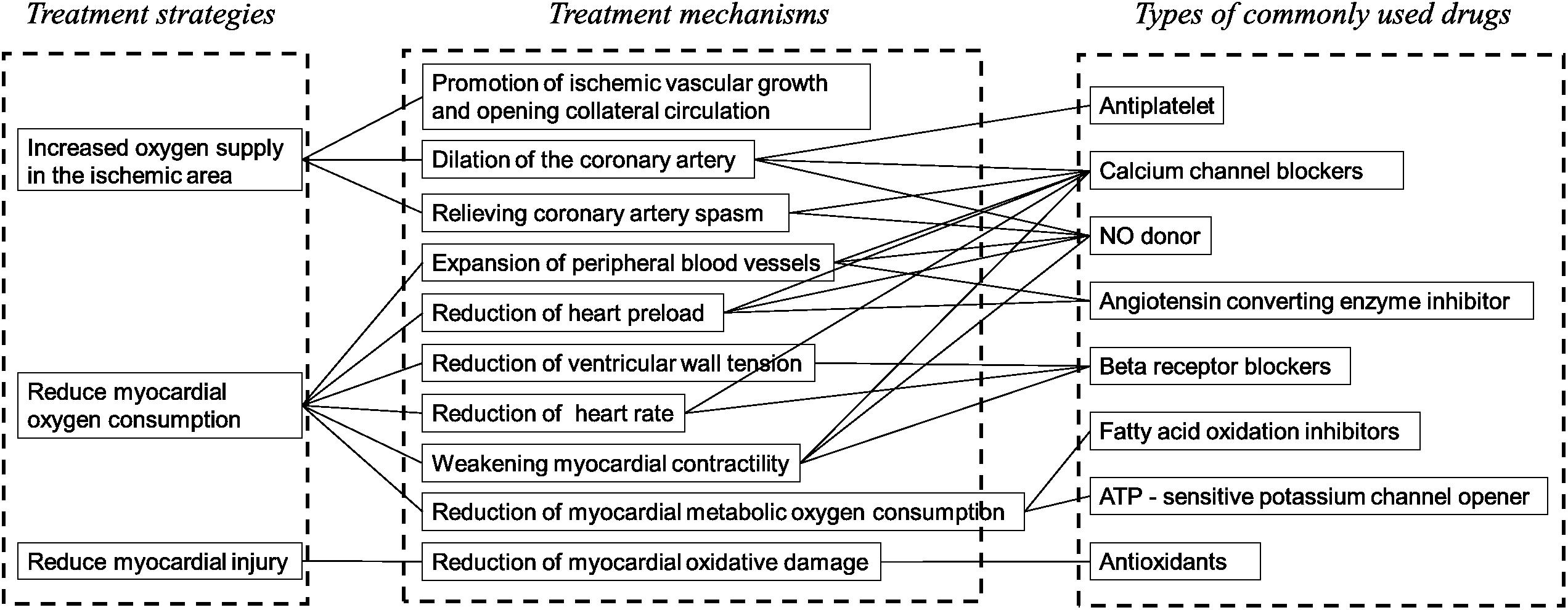
Figure 2. The clinical strategies and related mechanisms for the treatment of AMI. From eight clinically common drugs used in the treatment of AMI, the major therapeutic strategies of AMI can be classified into three categories: increase of the oxygen supply in the ischemic area, reduction of myocardial metabolism and protection of myocyte. The effect for the evaluation of Schisandra in the treatment of AMI can be selected: dilation of the artery, decrease of the heart afterload, decrease of the ventricular wall tension, decrease of the heart rate, weakening of the myocardial contractility, decrease of the myocardial metabolic oxygen consumption.
Construction of Three-Mode Network
Construction of GOI network
Based on screened AMI targets, the GO were enriched through the DAVID database, and further integrated with each other to construct the GOI network (Figure 5A) using the EnrichmentMap9 plugin in Cytoscape version 2.8. Each node represented a gene set corresponding to a GO term, and the edges and its weight were displayed and calculated based on the number of overlapping genes between two GO terms. The overlap similarity coefficient between significant GO terms was calculated and the cutoff was set to 0.5 (Merico et al., 2010; Isserlin et al., 2014; Cui et al., 2015).
Construction of “component-gene ontology” network
To construct the “component–gene ontology” two-mode network, which was based on screened component related targets, the DAVID database10 was used to perform GO enrichment annotation based on screened component related targets. The relationship between components and GO terms (including biological process, metabolic function and cell component) were then constructed into a “component-gene ontology” network (Figure 5B). There are two types of nodes in this network, Schisandra components and GO terms. Also, there are two types of edges in this network, including GO interaction and a component-GO term. In order to cover the scale of genes contained in the GO terms and the significance of these genes, weight scores between the Schisandra component and GO terms were calculated based on the percentage of proteins in each GO term and the significance test p-value. The formula was as follows:
In which, WCm refers to the weighted score of the Schisandra component related GO term m; rm refers to the ratio of protein included in the GO term; pm refers to the significance test p-value of the GO term; norm refers to the min-max normalization method which was used to standardize x within the dataset (Isler and Kuntalp, 2010; Lu et al., 2012; Benaliouche and Touahria, 2014; Dinc et al., 2014; Cao et al., 2016; Zhang et al., 2016). The formulas were as follows:
The pathways of the Schisandra component that were related to the target dataset were enriched through the KEGG and Biocarta database (Sun et al., 2011; Shangguan et al., 2015). Pathway scores were calculated using a significance p-value and the content ratio of the related component (Atobe et al., 2015; Dunkelberger et al., 2017; Babu et al., 2018; Desfontaine et al., 2018). The formulas were as follows:
In which Ri refers to the content ratio of component i compared to all the identified k components. The potential pathway of Schisandra in the treatment of AMI were sequenced and filtered according to the accumulate score.
Construction of “gene ontology–effect” network
Pharmacological effect related targets were also enriched into GO terms, and then integrated into a “gene ontology-effect” two-mode network (Figure 5C). This network contained two types of nodes including GO terms and effects, and two types of edges; GO-GO interaction and GO-effect interaction. Weighted scores between the effect and GO terms were calculated based on the percentage of proteins in each GO term and the p-value. The formula was as follows:
In which, WEn refers to the weighted score of the effect related GO term n; rn refers to the ratio of protein included in the GO term; pn refers to the significance test p-value of the GO term; norm refers to the range method which was used for normalization.
Integration of “component–gene ontology–effect” three-mode network
In order to reduce the prediction bias and highlight the functional relation, the Schisandra components and pharmacological effects were connected through an AMI related GOI network, which represented functional interactions in the condition of myocardial infarction. The “component-gene ontology-effect” three-mode network (Figure 5D) was constructed through the integration of a “component-gene ontology” network and “gene ontology-effect” network. This three-mode network can be utilized to simulate and predict the action level and pharmaceutical mechanisms of the Schisandra component in the treatment of AMI related effects.
Analysis of Three-Mode Network
It would be feasible to evaluate the intensity of pharmacological effects in the disease condition, using a maximum flow algorithm based on the constructed three-mode network. A maximum flow algorithm was therefore carried out using Pajek software to calculate the score of each component related to effects based on the “component-gene ontology-effect” three-mode network (Looijestijn et al., 2015). The result scores were then adjusted using an entropy algorithm (Prado-Prado et al., 2011; Teschendorff et al., 2014; Hayasaka et al., 2015; Zhao et al., 2015), and the information entropy of effect j was defined as Hj:
In which, Zij refers to the score calculated by the maximum flow algorithm, i infers the component i in all m components, Hj refers to the information entropy of effect j in n effects.
The entropy weight ωj of the index j is defined as:
The predicted weight of the Schisandra components correlated to nine effects were calculated as:
The score was then adjusted with the content ratio of each component:Score = Wij×Ri, and the final score of the Schisandra component or the effect, based on the three–mode network, was calculated with the sum of the responsible adjusted score in the matrix:
Wi refers to the score of component i in all m components, Wj refers to the score of effect j in all n effects.
Experimental Validation
Surgery of LAD Ligation Model
The model of AMI was established by ligating the left anterior descending artery (LAD) of ICR mice (Gao et al., 2010; Borst et al., 2011) that weighed approximately 25 g (Laboratory Animal Unit of Qinglongshan, China), and hearts of the Sham group were operated without ligation. All mice that survived the surgery were randomly divided into the Schisandra, and the positive control group. According to the previous research based on “Yiqifumai” (Tasly, China) (Li et al., 2015) in the treatment of coronary heart disease, the experiment doses of Schisandra were set to a low dose of 0.056 g/kg, a medium dose of 0.23 g/kg and a high dose of 0.90 g/kg. In order to evaluate the unknown therapeutic effect of Schisandra on AMI in an unbiased way, five commonly used clinical drugs including Amlodipine (1601001, Yangzijiang, China) 1.52 mg/kg, Metoprolol (1703001, AstraZeneca) 15.17 mg/kg, Captopril (15022511, Changzhou, China) 7.58 mg/kg, Trimetazidine (2008516, Servier) 9.10 mg/kg were provided as the positive controls. Since the pharmacological mechanisms of these five positive drugs are different, they were only appended as the positive control when the pharmacological indicators were directly related to the corresponding mechanism of the positive drugs. All corresponding drugs were administered 24 h after the model was established, and the measurements were carried on 72 h after surgery (Figure 1C).
Echocardiography and Blood Pressure Analysis
Heart echocardiograms and blood pressure were measured 24 h after the mice received the corresponding drugs. Mice were anesthetized with isoflurane, placed on prewarmed trays, and maintained at normothermic levels during the examination. Parasternal long axis view-dependent M-mode and 2-D echocardiographic studies for determination of cardiac left ventricular hypertrophy were performed using a 55 MHz linear array transducer system (Vevo 770). Blood pressure was measured and assessed as the mean value of thirty consecutive measurements using a tail-cuff sphygmomanometer under unstressed conditions (BP-2000; Visitech, United States) (Sakata et al., 2017).
Histological Analyses
The heart tissues were merged into 4% formalin for 3 days and PBS with 30% sucrose for 1 day, then the tissues were embedded by an OCT compound and sliced into 10 μm using a freezing microtome. The slides with heart tissues were stained sequentially with eosin Y and hematoxylin Gill No. 3 for HE staining (201611, Jiancheng Bioengineering Institute, China). The pathological score of each sample was then evaluated based on the degree of necrosis and inflammation in the heart tissue.
Detection of Serum Biomarkers
Mice were sacrificed 48 h after administration. The blood serum of mice in each group were centrifuged at 3000 rpm for 10 min and then stored at room temperature within 20 min. The contents of lactate dehydrogenase (LDH) and free fatty acid (FFA) in the serum were detected through a test kit (201703, Jiancheng Bioengineering Institute, China). Serum noradrenaline (NE), angiotonin II (Ang II), NT and brain natriuretic peptide (NT-BNP), were detected using an enzyme-linked immunosorbent assay (ELISA) kit (201711, Jiancheng Bioengineering Institute, China). Each experiment was performed three times.
Immunocytochemistry
For the immunostaining experiments, slides with heart tissue were rinsed three times through PBS. Tissue was blocked for 1 h at 4°C in blocking buffer (PBS, 5% BSA, and 0.2% Triton). The slides were then incubated for 12 h at 4°C in NFAT or NCOR2(SMRT) antibodies against mouse (Proteintech Technology, China), which was diluted with TBST and mixed with 3% BSA. Slides were rinsed three times in PBS, followed by incubation in Alexa Fluor 488 secondary antibody conjugate (Beyotime, China). After incubation with DAPI for 5 min and the final rinse in PBS, slides were viewed under a fluorescence microscope.
Blood Flux and Clotting Time Measurements
The construction of coagulopathy model
Male ICR mice were randomly divided into six groups, including the Control, Model, Schisandra (Low: 0.056 g/kg, Middle: 0.23 g/kg, High: 0.90 g/kg) and Aspirin (BJ26934, Bayer, Italy) 15.17 mg/kg. Each mouse in the model and treatment group was injected with 0.08 ml/kg Adrenaline (ADR, Sigma–Aldrich). The mice were placed in water (4∼6°C) for 4 min after being injected with Adr, and were then injected again with 0.08 ml/kg Adr for a second time. The corresponding drug was administrated after injection for the second time. All these procedures were repeated for 3 days.
The blood flux measurement
The blood flux was measured 30 min after the last administration. Mice were anesthetized with chloral hydrate and prostrated on the trays. The Moor Laser Doppler Imaging device (Moor LDI, Moor Instruments Ltd, Axminster, Devon, United Kingdom) was utilized to measure blood flow (Blood Flux) in the micro vessels of the ears. The device was calibrated following the manufacturer’s guidelines. A recording of approximately 2 min for each sample was made, using both digital and graphic modes. For each measurement, a noise-free recording of approximately 5 s was used to calculate the average flux. The output data was then analyzed using the manufacturer’s software (Moorsoft v2.0) (Opazo Saez et al., 2005; Clough et al., 2009; Peeters et al., 2012).
Clotting time measurement
For each mouse, 100 μL blood was adopted through capillary from retrobulbar venous plexus, and the clotting time was evaluated at the time an obvious agglutination was identified (Li X. et al., 2013; Chapin and Hajjar, 2015; Chen et al., 2015).
Vasoconstriction Measurements
Animals and tissue preparation
Male Sprague-Dawley rats weighing between 250 and 300 g (Laboratory Animal Unit of Qinglongshan, China) were anesthetized 50 mg/kg, i.p. with Chloral hydrate. Rats were sacrificed through stunning and cervical dislocation, and the thoracic aorta was surgically removed. The aorta was cleaned of any adhering fat and connective tissues and cut into 3 mm wide rings (Li Y. et al., 2013; Baharara et al., 2014; Dai et al., 2017).
Isometric tension measurement
The aortic rings were immediately immersed in 37°C Krebs solution (NaCl 118 mM, KCl 4.7 mM, CaCl2 2.5 mM, MgSO4 1.2 mM, KH2PO4 1.2 mM, NaHCO3 25 mM, and Glucose 11.1 mM) after removal and aerated with 5% CO2/95% O2. Then the aortic rings were connected to a force transducer (Technology Co, Ltd., Chengdu, China) and stretched progressively to 2.0 g. All changes in tension were expressed as a percentage decrease in the contraction activated through 1 μM Adr after being mixed with Schisandra extracts with a dose between 9.0 × 10-20g/mL to 9.0 × 10-04 g/mL.
Statistical Analysis
Statistical differences were assessed by one way-ANOVA. p < 0.05 was considered statistically significant. Data were expressed as the mean ± SD.
Results
Pharmacological Effect Analysis of Schisandra Based on a “Component–Gene Ontology–Effect” Three-Mode Network
Data Mining of Targets Related to Schisandra Components
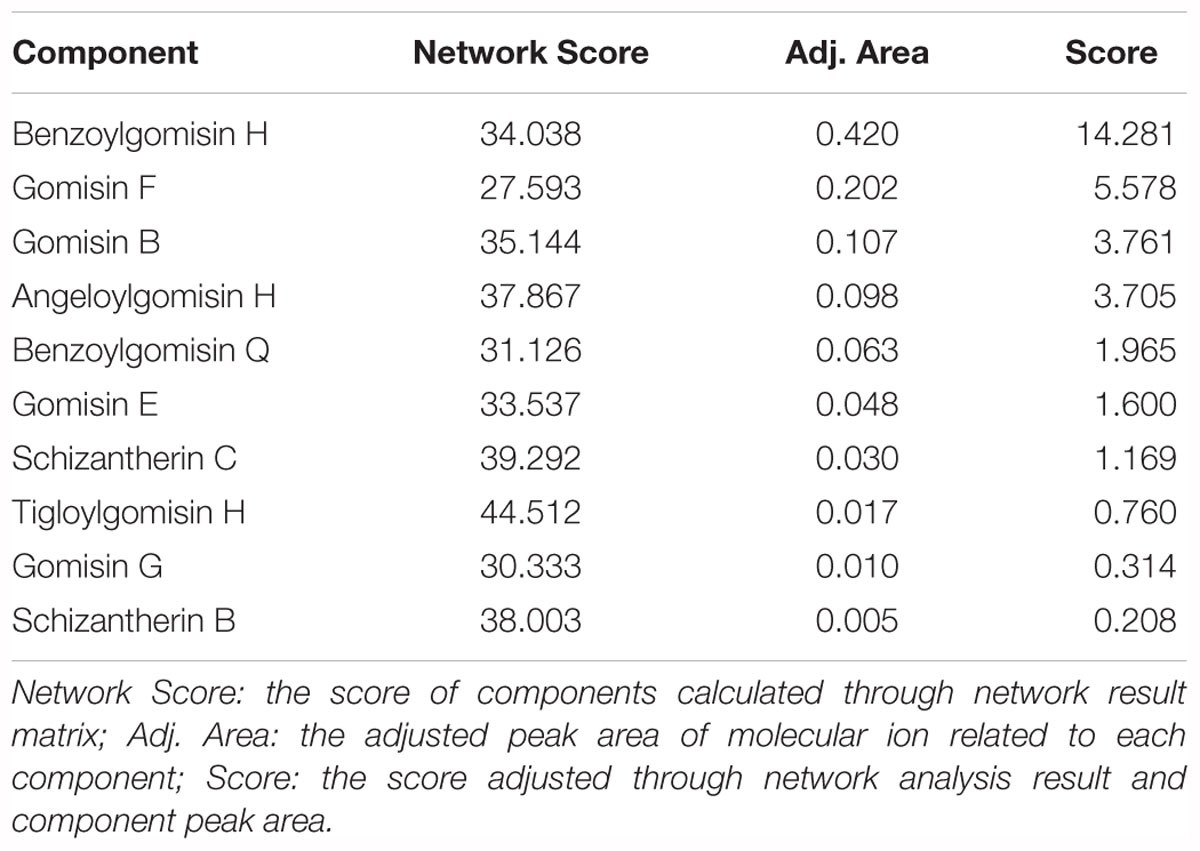
Ten components, including Tigloylgomisin H, Angeloylgomisin H, Benzoylgomisin H, Benzoylgomisin Q, Gomisin G, Schisantherin B, Gomisin B, Gomisin E, Schizantherin C, and Gomisin F, were identified from the water extracts of Schisandra (Figures 3, 4 and Supplementary Table S1) (Li et al., 2006; Wang et al., 2011). Pathway enrichment of target data based on identified components showed that “NFAT and Hypertrophy of the heart” and “Map Kinase Inactivation of SMRT Corepressor” were the top two correlated pathways of Schisandra in the treatment of AMI (Table 1 and Supplementary Tables S2, S3).
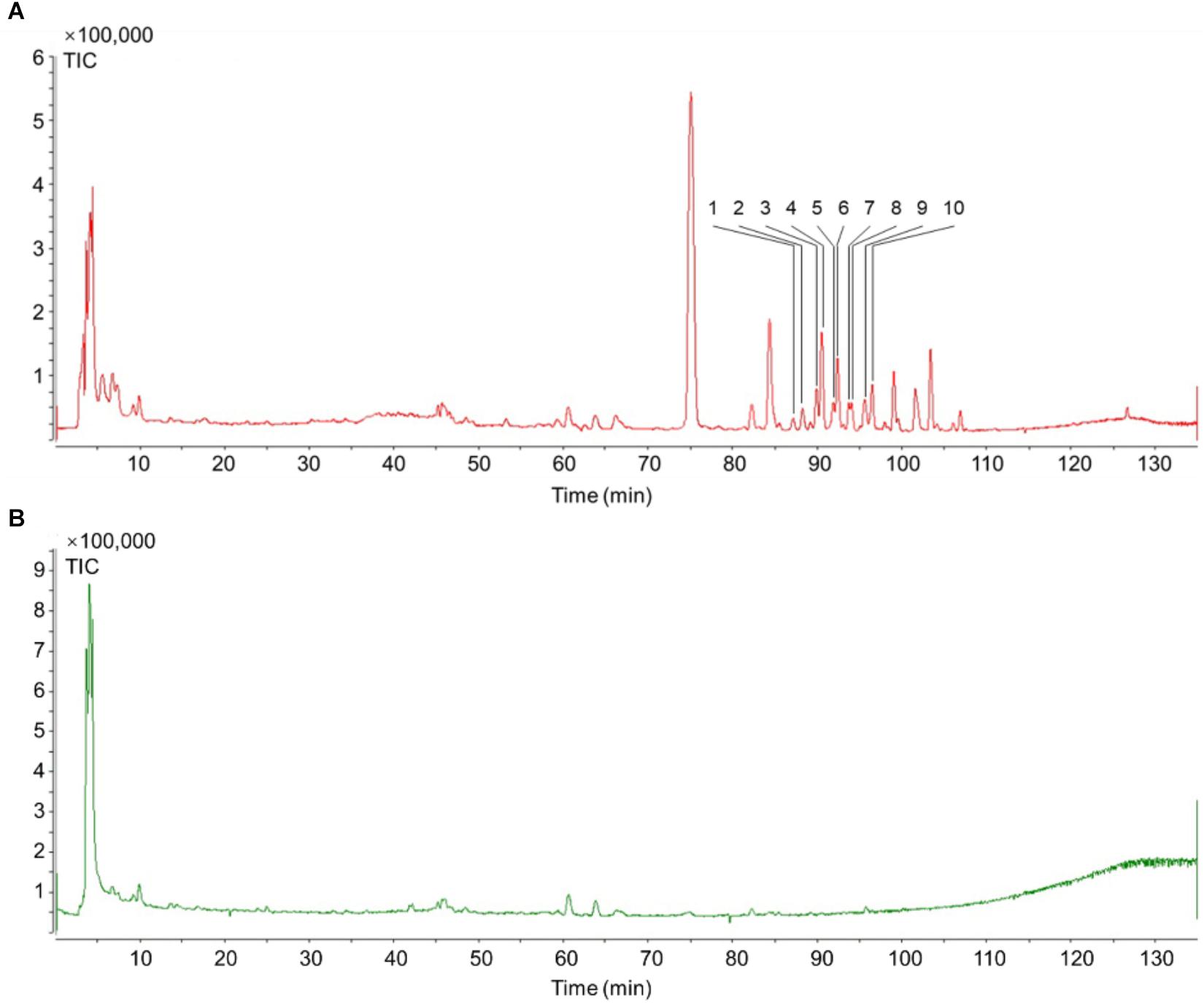
Figure 3. TIC of Schisandra extracts by HPLC-Q-TOF-MS-MS. (A) Positive ion mode MS spectra of SchisandraSchisandra chinensis; (B) Negative ion mode MS spectra of SchisandraSchisandra chinensis.
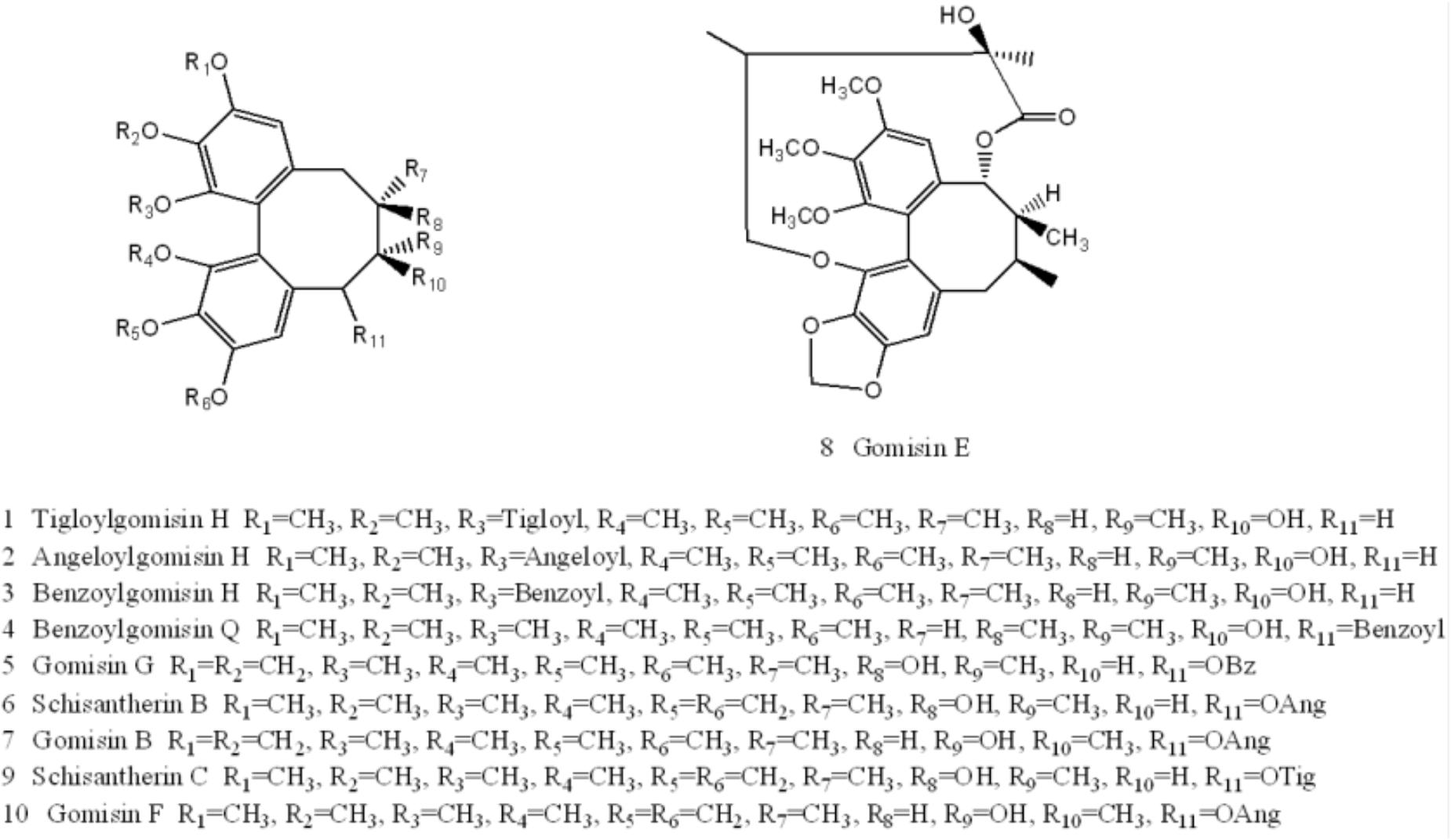
Figure 4. Structures of ten components identified in Schisandra extract: Tigloylgomisin H (1), Angeloylgomisin H (2), Benzoylgomisin H (3), Benzoylgomisin Q (4), Gomisin G (5), Schizantherin B (6), Gomisin B (7), Gomisin E (8), Schizantherin C (9), Gomisin F (10).
Construction and Analysis of “Component–Gene Ontology-Effect” Three-Mode Network
Through mining of AMI related genes from three different data sources, the GEO data displayed the largest scale compared to the other two data sources (Figure 1B). After combining the target data from the database and intersection with GEO data, 3,288 DEG s from the heart tissue of Mus Musculus were confirmed (Figure 1B and Supplementary Tables S4, S5).
The GO terms enriched from collected AMI targets were connected with each other in order to integrate them into the GOI network (Tarnavski et al., 2004; Hunt et al., 2012; Tsukamoto et al., 2013). The “component-gene ontology-effect” three-mode network (Figure 5D) was then constructed by integrating the “component-gene ontology” network (Figure 5A), GOI network (Figure 5B) and the “gene ontology-effect” network (Figure 5C and Supplementary Tables S6, S7). The predicted score matrix of the Schisandra component related to the AMI effect was visualized by a heatmap (Figure 6). And the result indicated that cardiac preload and myocardial contractility were the top two pharmacological effects of the Schisandra components (Table 2), while Benzoylgomisin H was the top active component among all the identified components in the treatment of AMI (Table 3).
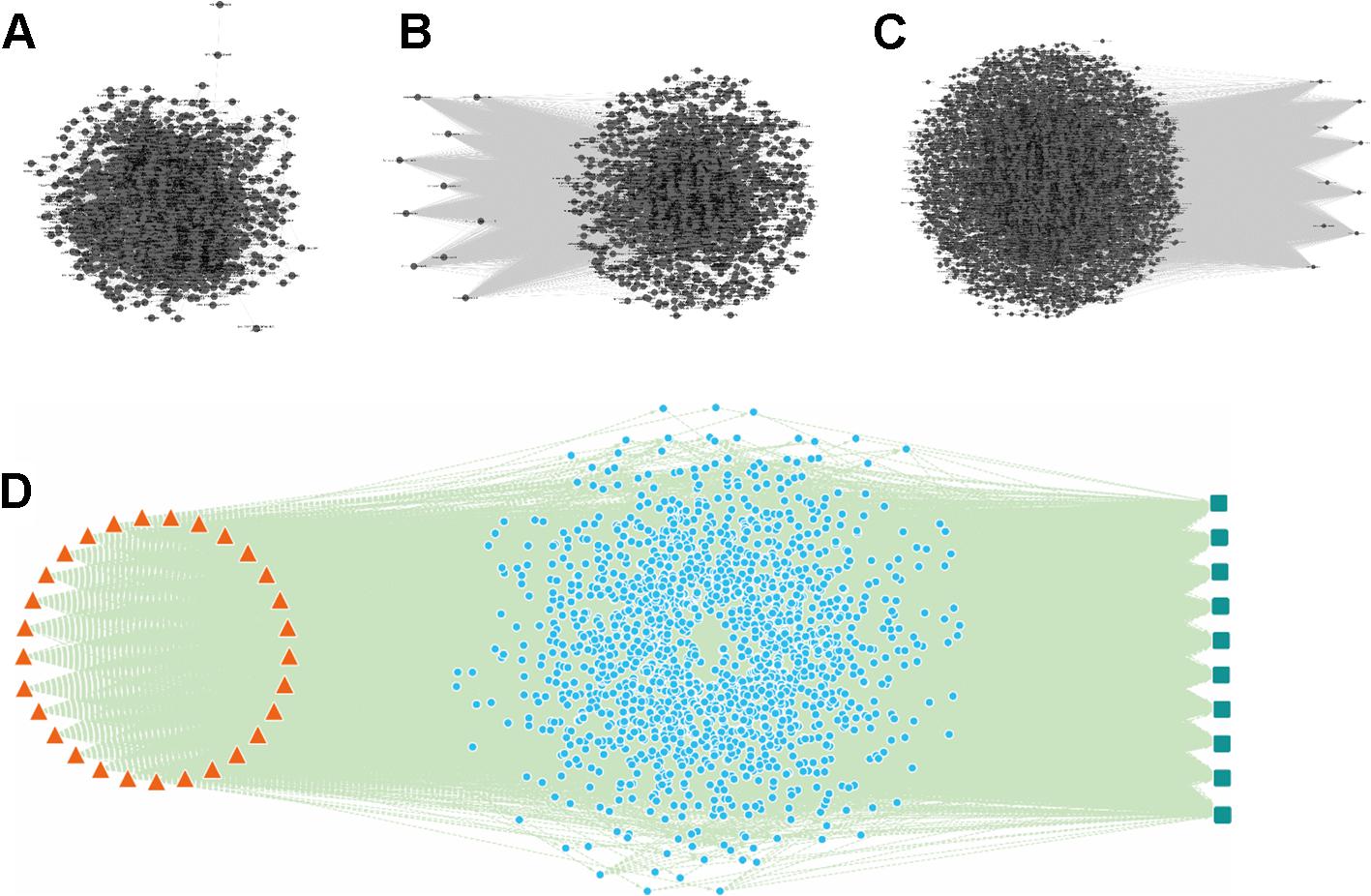
Figure 5. The “component-gene ontology-effect” three-mode network of Schisandra. Firstly, the AMI target related gene ontology interaction (GOI) network (B) was constructed by mining and enrichment of AMI related target genes. Secondly, the target proteins screened through identified Schisandra components were enriched into GO terms. The interactions between GO and the component were constructed and visualized through the “component-gene ontology” two-mode network (A). Then, the “gene ontology-effect” two-mode network (C) for the treatment of AM pharmacological effects was constructed from the target data collected from reference mining. Finally, the three sub-networks were integrated, and the same GO nodes were combined to obtain a “component-gene ontology-effect” three-mode network (D). The triangle in orange represents the components identified from Schisandra, the blue circle represents the common GO which are both relevant to the components and effects (green square).
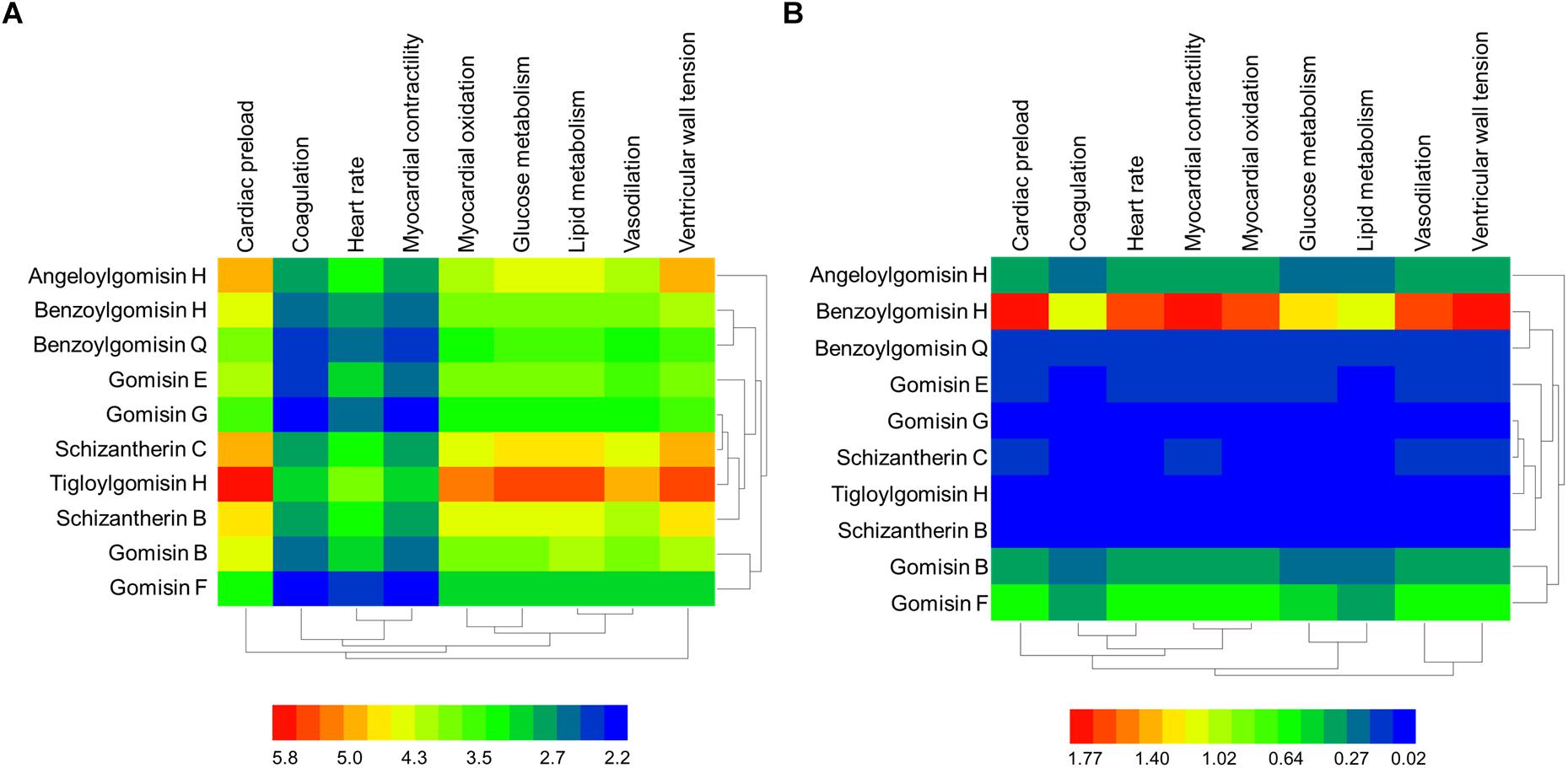
Figure 6. The predicted score heatmap of the Schisandra components related to the effects based on three-mode network. (A) The score matrix calculated from the three-mode network; (B) The score matrix adjusted with the content ratio of Schisandra components.
Experimental Validation of Network Analysis Results
Pharmacological Effect of Schisandra in the Treatment of AMI
The pathological analysis results showed that granulocytes were infiltrated into the heart tissue of the left ventricle and there was significant necrosis after LAD ligation surgery. Schisandra can alleviate the necrosis of the left ventricle in a dose-dependent way (Figures 7A,B). The serum level of LDH also indicated that Schisandra can significantly reduce the necrosis of myocytes after AMI (Figure 7C).
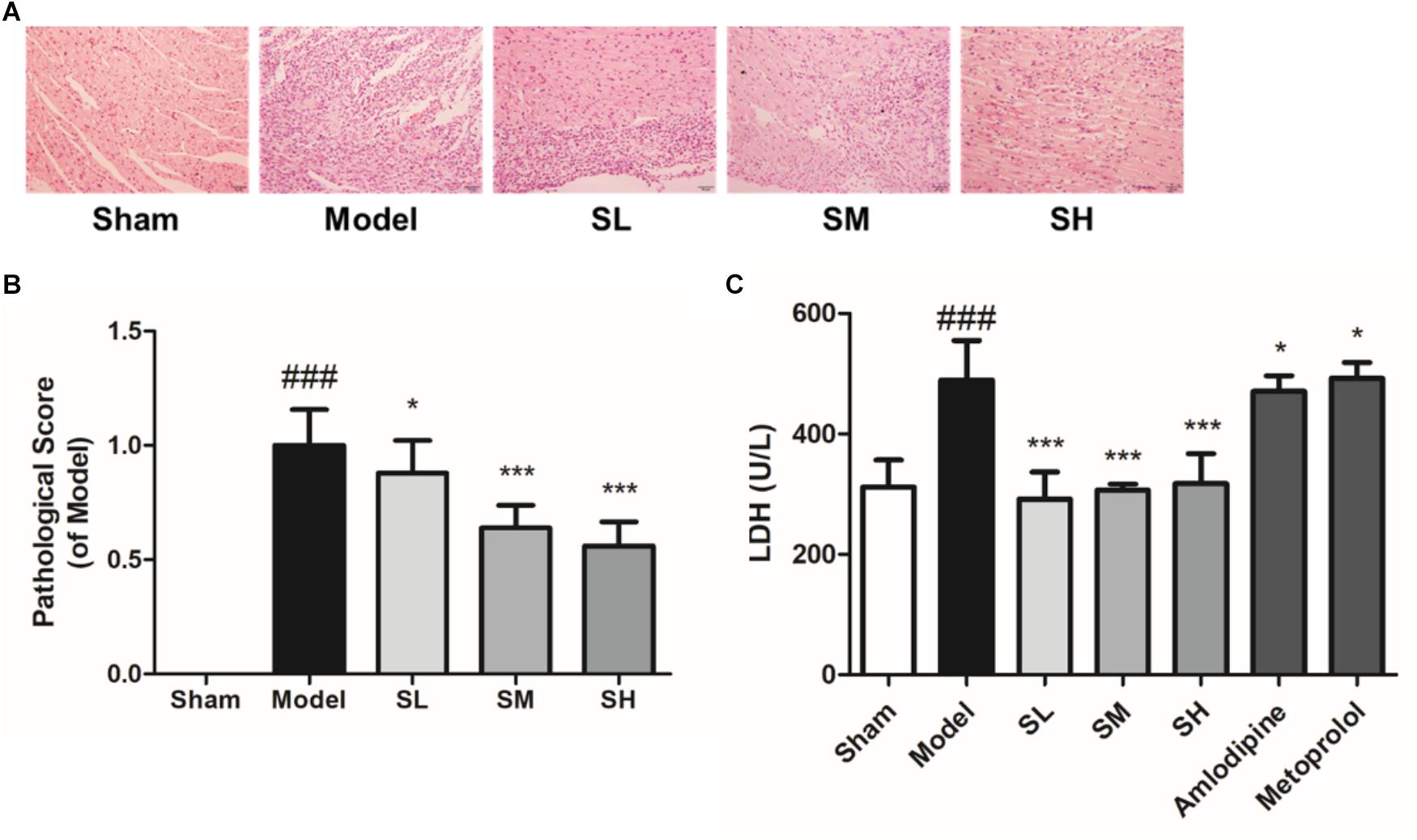
Figure 7. The histopathological staining and serum LDH of AMI mice treated with Schisandra. Sham group: subjected to the same surgical treatment without ligation, SL group: Schisandra extract was given to mice at the dose of 0.056 g/kg, SM group: Schisandra extract was given to mice at the dose of 0.23 g/kg, SH group: Schisandra extract was given to mice at the dose of 0.90 g/kg. (A) Mice heart sections were stained with H&E after given corresponding drug for two days (200×); (B) Pathological scores of H&E staining; (C) Serum LDH was tested after 2 days treatment of Schisandra. All data are shown as the mean ± SD. ###P < 0.001 vs. Sham group; ∗P < 0.05, ∗∗∗P < 0.001 vs. Model group.
Schisandra extract also showed a significant depressor effect both on the systolic blood pressure and diastolic blood pressure (Figures 8A,B). The pulse was elevated after the treatment of Schisandra in a negative feedback (Figure 8C). Studies of an adrenaline-induced artery ring model in vitro showed that Schisandra extract has a vasodilation effect at an EC50 dose of 5.144 × 10-11 g/mL (Figure 8D), which could partly explain the depressor effect of blood pressure.
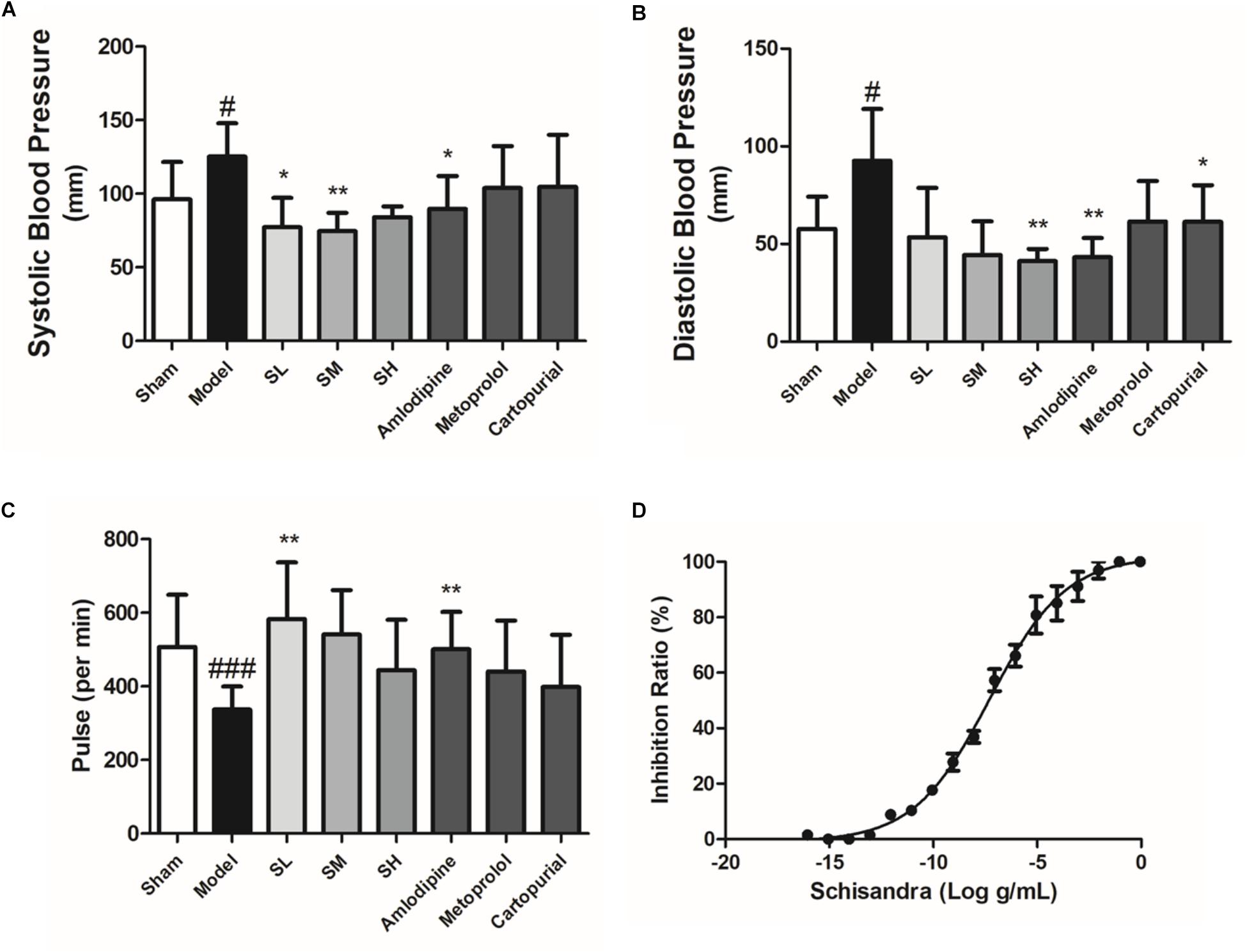
Figure 8. The blood pressure, heart rate and vasodilation ratio of Schisandra. Systolic blood pressure (A), diastolic pressure (B), and pulse (C) was tested 30 min after the last corresponding drug was given. (D) The vasodilation effect of Schisandra. The rat main artery was cut into 3 mm artery rings and were activated through 1 μM Adr. All data are shown as the mean ± SD. #P < 0.05, ###P < 0.001 vs. Sham group; ∗P < 0.05, ∗∗P < 0.01 vs. Model group.
The content of serum Ang II indicated that Schisandra could significantly elevate the content of serum Ang II (Figure 9A), which may be responsible for the vasodilation effect. On the other hand, serum NE was significantly reduced after LAD ligation surgery, whereas Schisandra could significantly reduce serum NE in a dose-dependent way, indicating that Schisandra could inhibit the sympathetic nervous system (Figure 9B). Additionally, the serum NT-BNP was significantly reduced after treatment with Schisandra (Figure 9C), which suggests that left ventricular wall tension and blood pressure can be significantly reduced (Demirtas et al., 2018; Pala et al., 2018; Shen et al., 2018; Xiong et al., 2018). These results indicate that Schisandra could significantly inhibit the activation of the sympathetic nervous system so that the contractility of the left ventricle could also be reduced. Additionally, the serum FFA result showed that Schisandra failed to condition the lipid metabolic function of myocytes during the 3 days of treatment (Figure 9D), and the immunofluorescence result of CPT1 also showed the same trend (Supplementary Figure S1). This result indicates that the conditioning of metabolism is not the major pharmacological effect of Schisandra.
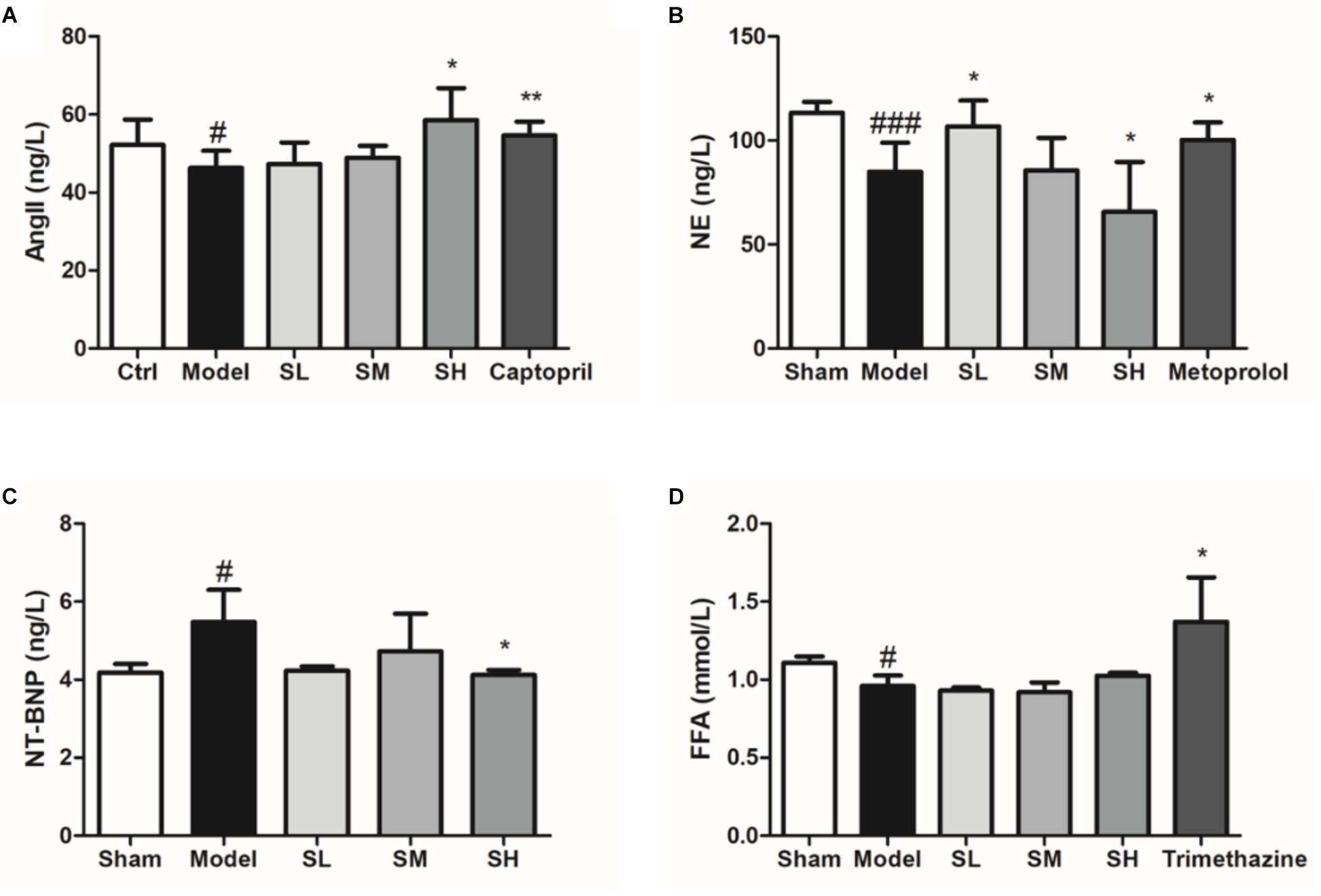
Figure 9. Serum Ang II (A), NE (B), NT-BNP (C), and FFA (D) of Schisandra treated AMI mice. All data are shown as the mean ± SD. #P < 0.05, ###P < 0.001 vs. Sham group; ∗P < 0.05, ∗∗P < 0.05 vs. Model group.
The ultrasonic cardiogram (US) result showed that Schisandra could significantly increase the ejection factor (EF) and fractional shortening (FS) of the AMI heart at a low dose, whereas the heart output and diameter in the left ventricle (LVIDd, LVIDs) was not affected by Schisandra after AMI (Figures 10A–E). It could be inferred from the result of BP and US that Schisandra can release heart burden by reducing myocardial contractility and increasing heart output through vasodilation.
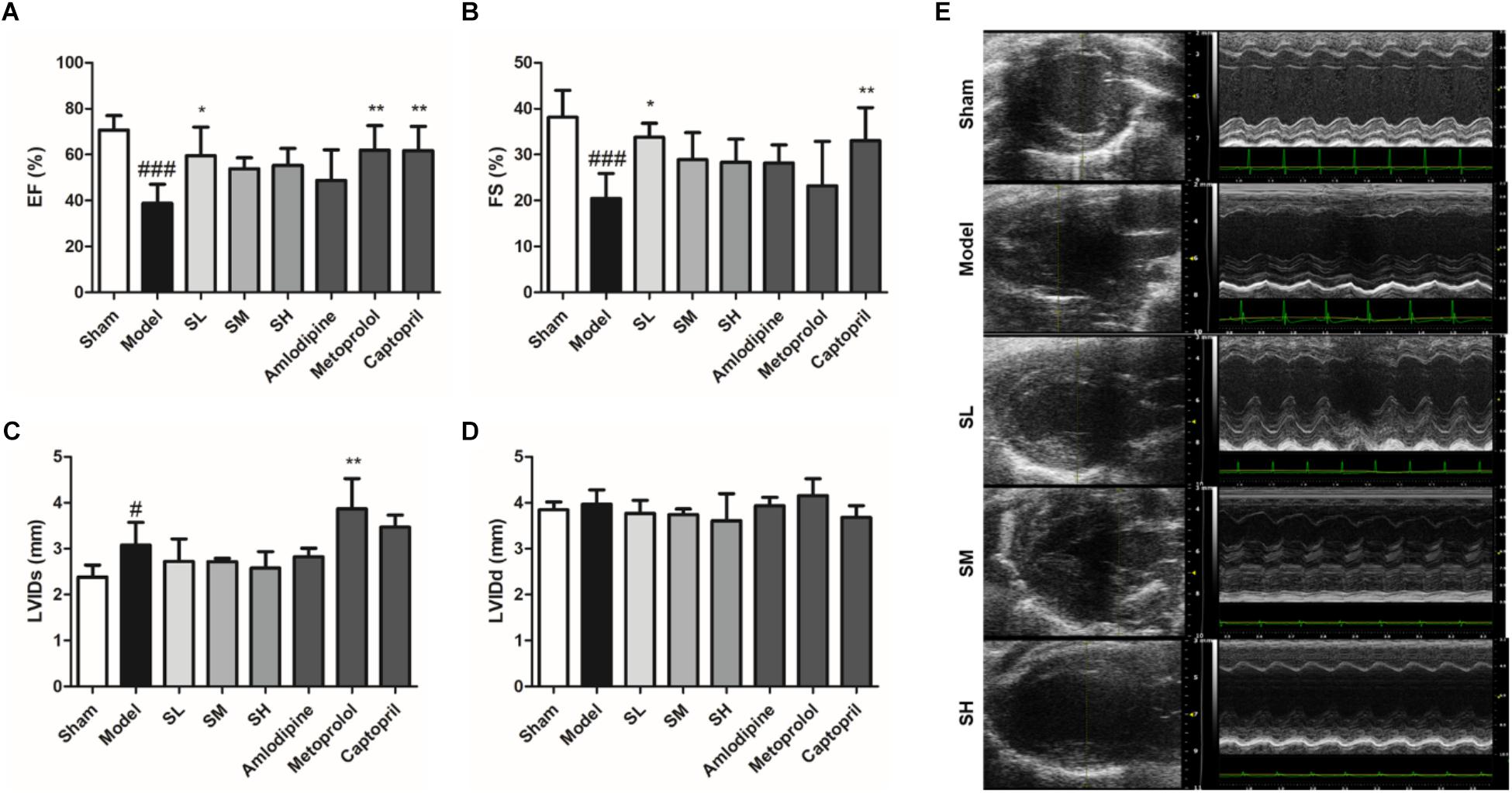
Figure 10. The EF (A), FS (B), LVIDs (C), and LVIDd (D) result of Schisandra treated AMI mice. (E) The Ultrasonic Cardiogram of Schisandra treated mice was tested 30 min after the last corresponding drugs were given. All data are shown as the mean ± SD. #P < 0.05, ###P < 0.001 vs. Sham group; ∗P < 0.05, ∗∗P < 0.01 vs. Model group.
The blood flux results also indicate that Schisandra may induce the inhibition of sympathetic nerves and the subsequent downregulation of blood pressure (Figures 11A,B). On the other hand, Schisandra can significantly prolong the clotting time on the coagulopathy model (Figure 11C), which means it will lead to an increase of blood flow.
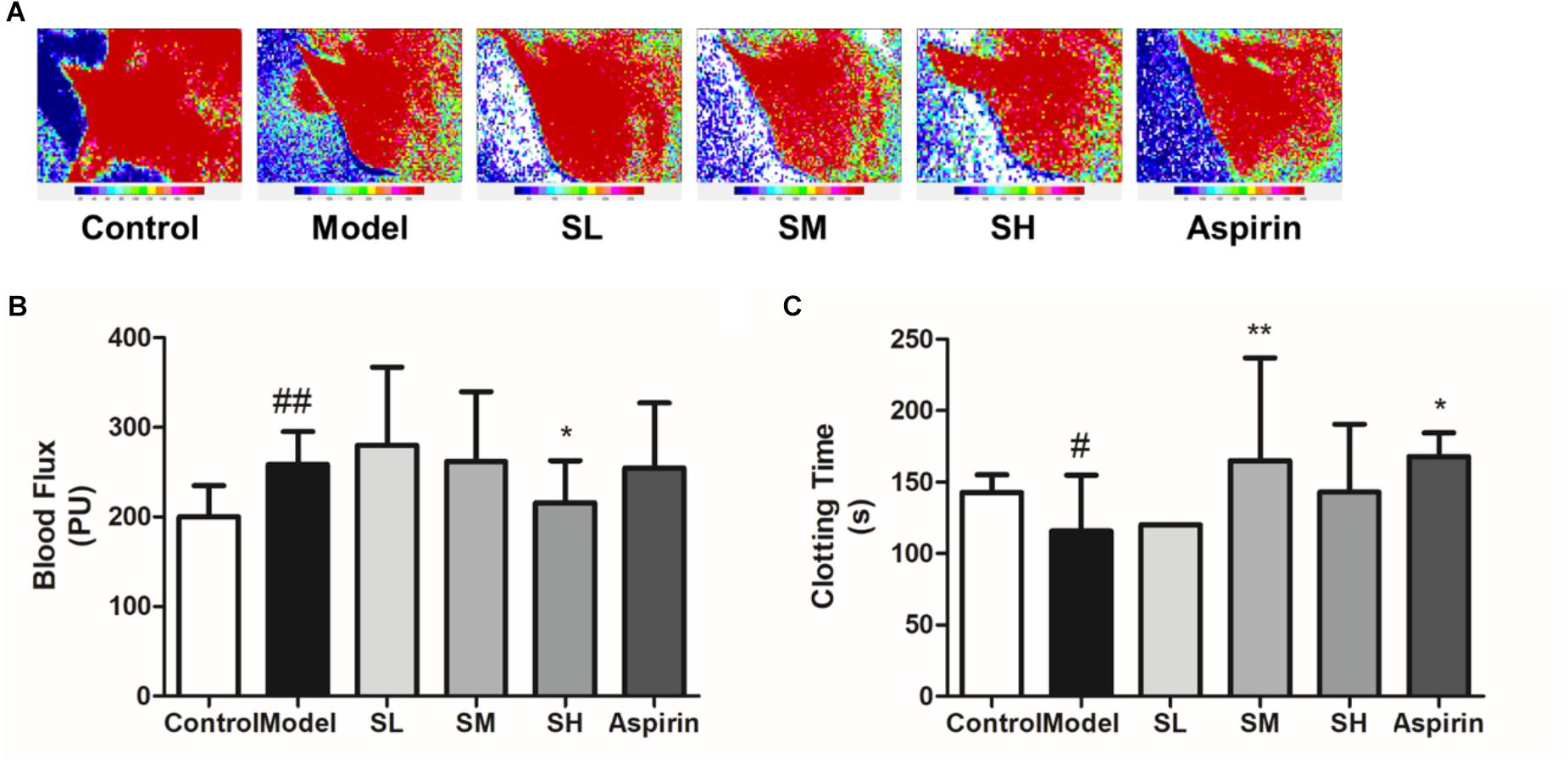
Figure 11. The blood flux (B) and clotting time (C) of AMI mice after treatment with Schisandra. (A) The mice were injected with Adr and then stimulated through 4°C water. After repeating these procedures twice, the FLPI was used to test blood flow in the ear vein. Then one drop of blood was taken for the test of clotting time by counting the seconds when the blood started to coagulate. All data are shown as the mean ± SD. #P < 0.05, ##P < 0.01 vs. Sham group; ∗P < 0.05, ∗∗P < 0.01 vs. Model group.
In conclusion, Schisandra extract showed significant cardiac inhibition and vasodilation effects, which further induces a decrease of heart burden and further preserves the heart contractility. These major pharmacological effects showed the same trend as the predicted results of the three-mode network.
NFAT and NCOR2 in Heart Tissue After Treatment With Schisandra
Immunofluorescence staining result showed that the NFAT expression increased after the onset of AMI, and Schisandra could significantly reduce NFAT expression in myocytes (Figures 12A,B), whereas the inhibition effect was attenuated at a high dose. The expression of NCOR2 in the heart tissues indicated that the pathologically reduced heart output led to an upregulation of NCOR2 expression in myocytes after the onset of AMI (Kawamiya et al., 2010), and Schisandra could efficiently inhibit the over-expression of NCOR2 in myocytes (Figures 13A,B).
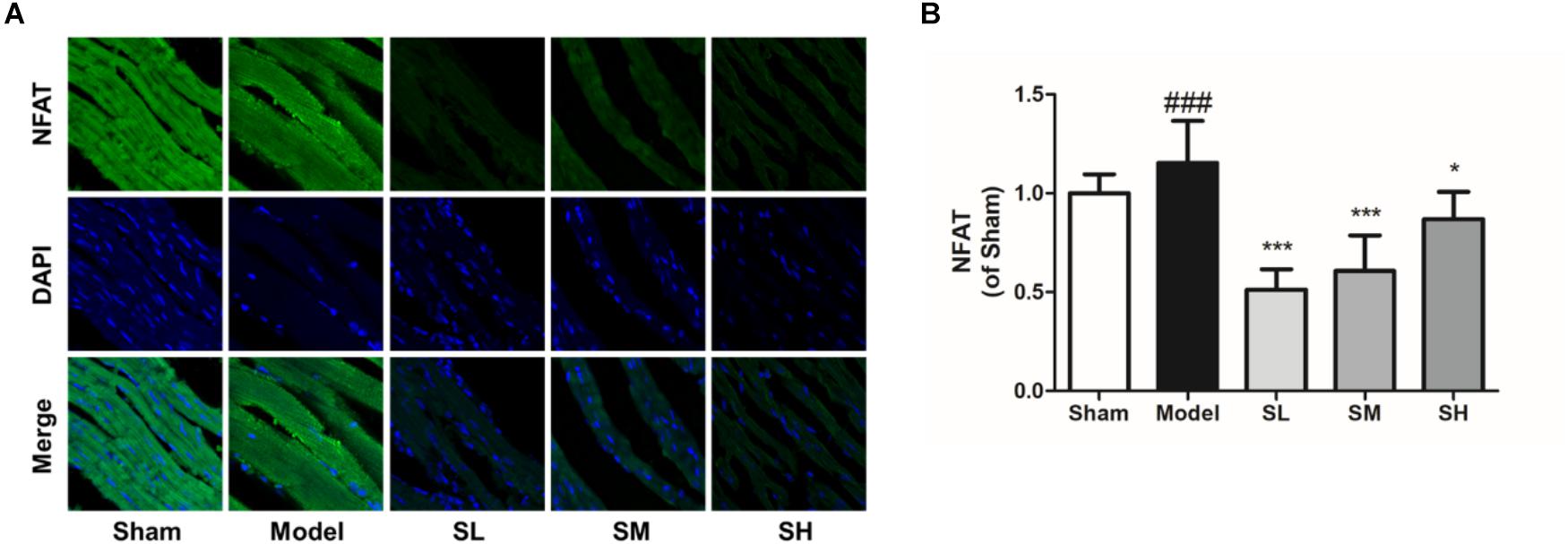
Figure 12. The immunofluorescence of NFAT in the heart tissue after the treatment of Schisandra. (A) The immunofluorescence staining of NFAT in heart tissue (400×); (B) The quantitation of NFAT from immunofluorescence staining results. All data are shown as the mean ± SD, ###P < 0.001 vs. Sham group; ∗P < 0.05, ∗∗∗P < 0.001 vs. Model group.
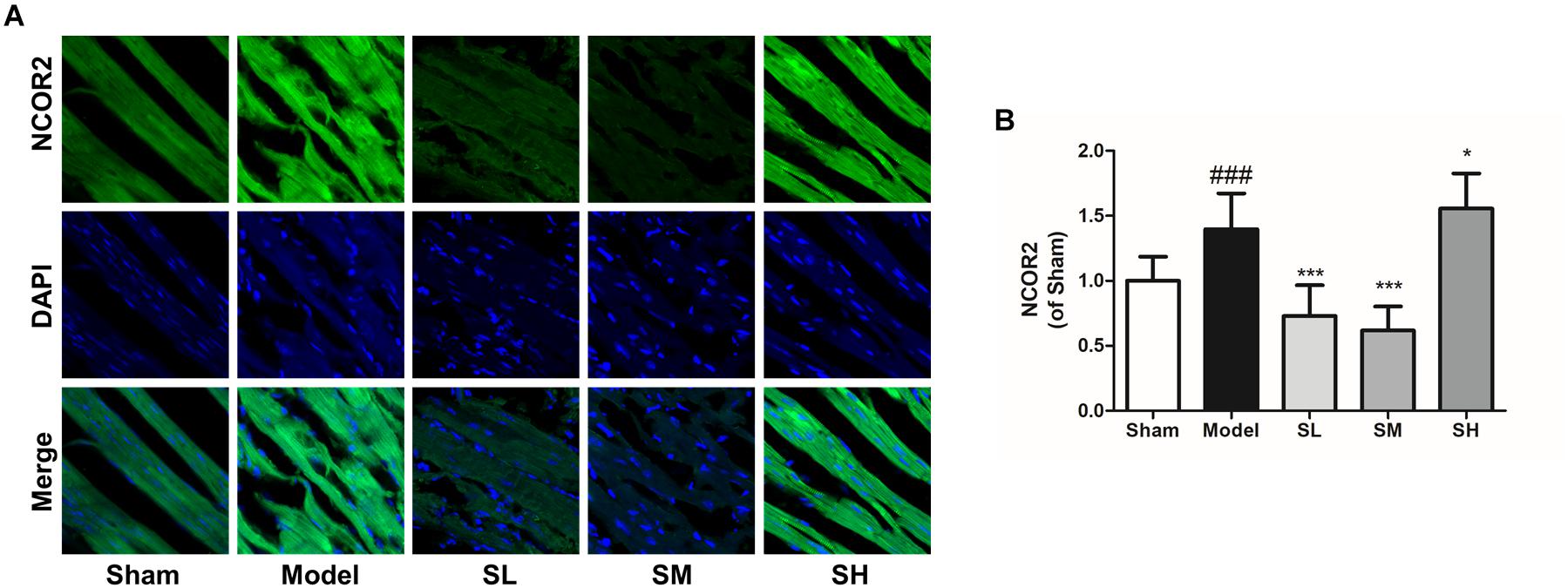
Figure 13. The immunofluorescence of NCOR2 in the heart tissue after the treatment of Schisandra. (A) The immunofluorescence staining of NCOR2 in heart tissue (400×); (B) the quantitation of NCOR2 from immunofluorescence staining results. All data are shown as the mean ± SD, ###P < 0.001 vs. Sham group; ∗P < 0.05, ∗∗∗P < 0.001 vs. Model group.
Discussion
Network pharmacology is an effective way to predict the complex mechanisms of TCM and has widely been used in the prediction of pharmacological mechanisms in order to guide pharmacological research. Thus, the development of a new network analysis methodology with higher accuracy and based on an open source high-throughput dataset, will support the pharmacological research of TCM in a much more convenient and economical way. However, it remained difficult to predict and evaluate the relationship between the component and pharmacological effect of the multi-drug system in a systematical way due to the complex mechanisms between proteins. Thus, it would be more valuable to clarify the pharmacological function of the drug rather than just filtering one target protein to guide further pharmacological research. Generally speaking, it would be more reasonable to highlight the pharmacological functional characteristics of the network model rather than a single target prediction, and the GO enrichment could solve this problem.
In this research, the target data was collected from public databases, reference literature mining and GEO array data to ensure that the collected data included the latest research results and to ensure that the expression difference significance in the heart tissue of Mus Musculus had been experimentally verified. Instead of component related target proteins only, the GO was used for the enrichment of drug-related targets in order to reduce the false positive prediction error of target prediction and to highlight the structural-functional characteristics of the drug components. The “component–gene ontology–effect” three-mode network, based on the GOI network, which represented the pharmacological functional signal pathway stream between the drug component and phenotype effect, was then integrated and the maximum flow algorithm was used to analyze the pharmacological functional message flow between the components and effects. By using this method, the phenotype effect of a complex system could be evaluated.
After the onset of AMI, the necrosis of myocytes in the left ventricle led to a decrease of contraction force. The decreased heart output then induced an increased amount of blood retention in the left ventricle, which further increased the heart burden and oxygen consumption. The hypoxia of myocytes leads to apoptosis and fibration in the long term (Herum et al., 2015; Lighthouse and Small, 2016). According to the results of the network analysis, a reduction of cardiac preload and myocardial contractility was the major effect of Schisandra in the treatment of AMI. The further experimental validation also proved that, compared to other effects, Schisandra could significantly reduce the blood pressure through the inhibition of the sympathetic nerves, which led to a decrease of heart preload and the myocyte ischemia and apoptosis could be reduced.
The immunofluorescence results confirmed that Schisandra components in the treatment of AMI were related to NCOR2 and NFAT. Research has shown that SMRT (NCOR2) can cooperate with thyroid hormone receptor-α (TRα) and up-regulate the expression of CaMK II, which leads to an unregulated sensitivity with sympathetic nerve activation (Kawamiya et al., 2010; Barish et al., 2012; Xie et al., 2015). After the onset of AMI, NCOR2 expression was decreased according to the references. However, experiment results in Mus Musculus showed that the increased expression of NCOR2 after LAD ligation surgery may be induced by negative feedback of reduced heart output at the onset of ischemia. The Schisandra could significantly reduce the expression of NCOR2, which may induce the negative effect of heart burden. However, research has shown that the Ca2+-calcineurin-NFAT pathway was activated due to the activation of the β-receptor after the onset of AMI. The increased expression of NFAT induces the reconstruction of the left ventricle, leading to heart failure in the long term after AMI. Schisandra could inhibit the expression of NFAT (Lee et al., 2003; de Salvi Guimaraes et al., 2017), which may inhibit fibrosis in the long term, so the output function of the heart will be preserved. However, further research needs to be conducted for complete experimental validation of NCOR2 and NFAT pathways related to Schisandra.
In general, the “component-gene ontology-effect” three-mode network could be a proper model to analyze effects, based on a multi-drug complex system, in a systematical way. This network analysis and experiment validation workflow may provide a reasonable method to research multi-drug systems including TCM.
Data Availability
Publicly available datasets were analyzed in this study. This data can be found here: http://www.genecards.org.
Author Contributions
SH carried out this research with the help of HZ and the guidance of JQ, YH, and BY.
Funding
This research was supported by Science and Technology Development Fund of Macao SAR on projects 013-2015-A1.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We sincerely acknowledge Mrs. Qianru Zhang for her technical assistance, and Prof. Yuanjia Hu for valuable suggestions. We would like to thank the Science and Technology Development Fund of Macao SAR for financial support on projects 013-2015-A1 for this research.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2019.00232/full#supplementary-material
FIGURE S1 | The immunofluorescence of CPT1 in the heart tissue after the treatment of Schisandra. (A) The immunofluorescence staining of CPT1 in heart tissue (400 ×); (B) The quantitation of CPT1 from immunofluorescence staining results. All data are shown as the mean ± SD. ###p < 0.001 vs. Sham group.
TABLE S1 | Components identified from Schisandra extract through HPLC-Q-TOF-MS-MS.
TABLE S2 | Target data simulated through Pharmmapper based on identified Schisandra components.
TABLE S3 |Schisandra related pathways enriched through simulated targets.
TABLE S4 | AMI related targets collected through datamining.
TABLE S5 | Gene ontology terms of AMI related targets.
TABLE S6 | Targets related with pharmacological effects.
TABLE S7 | Gene ontology terms of AMI pharmacological effects.
Footnotes
- ^ lilab.ecust.edu.cn/PharmMapper/
- ^ http://www.genecards.org/
- ^ https://www.pharmgkb.org/
- ^ http://bidd.nus.edu.sg/group/ttd/
- ^ http://www.ncbi.nlm.nih.gov/pubmed/
- ^ http://www.ncbi.nlm.nih.gov/geo
- ^ http://www.bioconductor.org/
- ^ https://www.ncbi.nlm.nih.gov/gene/
- ^ http://www.baderlab.org/Software/EnrichmentMap
- ^ https://david-d.ncifcrf.gov
References
Andersen, M. E., Cruzan, G., Black, M. B., Pendse, S. N., Dodd, D., Bus, J. S., et al. (2017). Assessing molecular initiating events (MIEs), key events (KEs) and modulating factors (MFs) for styrene responses in mouse lungs using whole genome gene expression profiling following 1-day and multi-week exposures. Toxicol. Appl. Pharmacol. 335, 28–40. doi: 10.1016/j.taap.2017.09.015
Atobe, T., Mori, M., Yamashita, F., Hashida, M., and Kouzuki, H. (2015). Artificial neural network analysis for predicting human percutaneous absorption taking account of vehicle properties. J. Toxicol. Sci. 40, 277–294. doi: 10.2131/jts.40.277
Babu, N., Advani, J., Solanki, H. S., Patel, K., Jain, A., Khan, A. A., et al. (2018). miRNA and proteomic dysregulation in non-small cell lung cancer in response to cigarette smoke. Microrna 7, 38–53. doi: 10.2174/2211536607666180103165343
Baharara, J., Namvar, F., Ramezani, T., Hosseini, N., and Mohamad, R. (2014). Green synthesis of silver nanoparticles using achillea biebersteinii flower extract and its anti-angiogenic properties in the rat aortic ring model. Molecules 19, 4624–4634. doi: 10.3390/molecules19044624
Barish, G. D., Yu, R. T., Karunasiri, M. S., Becerra, D., Kim, J., Tseng, T. W., et al. (2012). The Bcl6-SMRT/NCoR cistrome represses inflammation to attenuate atherosclerosis. Cell Metabol. 15, 554–562. doi: 10.1016/j.cmet.2012.02.012
Barrett, T., Troup, D. B., Wilhite, S. E., Ledoux, P., Rudnev, D., Evangelista, C., et al. (2007). NCBI GEO: mining tens of millions of expression profiles–database and tools update. Nucleic Acids Res. 35, D760–D765. doi: 10.1093/nar/gkl887
Benaliouche, H., and Touahria, M. (2014). Comparative study of multimodal biometric recognition by fusion of iris and fingerprint. ScientificWorldJournal 2014:829369. doi: 10.1155/2014/829369
Borst, O., Ochmann, C., Schonberger, T., Jacoby, C., Stellos, K., Seizer, P., et al. (2011). Methods employed for induction and analysis of experimental myocardial infarction in mice. Cell Physiol. Biochem. 28, 1–12. doi: 10.1159/000331708
Cao, X. H., Stojkovic, I., and Obradovic, Z. (2016). A robust data scaling algorithm to improve classification accuracies in biomedical data. BMC Bioinform. 17:359. doi: 10.1186/s12859-016-1236-x
Cappuzzello, C., Napolitano, M., Arcelli, D., Melillo, G., Melchionna, R., Di Vito, L., et al. (2009). Gene expression profiles in peripheral blood mononuclear cells of chronic heart failure patients. Physiol. Genomics 38, 233–240. doi: 10.1152/physiolgenomics.90364.2008
Chang, R., Li, Y., Yang, X., Yue, Y., Dou, L., Wang, Y., et al. (2013). Protective role of deoxyschizandrin and schisantherin A against myocardial ischemia-reperfusion injury in rats. PLoS One 8:e61590. doi: 10.1371/journal.pone.0061590
Chapin, J. C., and Hajjar, K. A. (2015). Fibrinolysis and the control of blood coagulation. Blood Rev. 29, 17–24. doi: 10.1016/j.blre.2014.09.003
Chen, J. M., Yuan, Z. L., Wu, C. W., and Li, K. S. (2015). Effects of alcohol extract substance from the aerial roots of Ficus microcarpa on coagulability and cerebral thrombotic activity. Guangxi Kexue 22, 148–152. doi: 10.13656/j.cnki.gxkx.20140610.006.html
Chen, P. S., Pang, S. S., Yang, N. Q., Meng, H. Y., Liu, J., Zhou, N. T., et al. (2013). Beneficial effects of schisandrin B on the cardiac function in mice model of myocardial infarction. PLoS One 8:e79418. doi: 10.1371/journal.pone.0079418
Clough, G., Chipperfield, A., Byrne, C., de Mul, F., and Gush, R. (2009). Evaluation of a new high power, wide separation laser doppler probe: potential measurement of deeper tissue blood flow. Microvasc. Res. 78, 155–161. doi: 10.1016/j.mvr.2009.05.003
Cui, S., Zhang, X. Y., Hai, S., Lu, H., Chen, Y. C., Li, C., et al. (2015). Molecular mechanisms of osteoarthritis using gene microarrays. Acta Histochem. 117, 62–68. doi: 10.1016/j.acthis.2014.11.003
Dai, R., Wang, T., Si, X. Q., Jia, Y. Y., Wang, L. L., Yuan, Y., et al. (2017). Vasodilatory effects and underlying mechanisms of the ethyl acetate extracts from Gastrodia elata. Can. J. Physiol. Pharmacol. 95, 564–571. doi: 10.1139/cjpp-2016-0407
de Salvi Guimaraes, F., de Moraes, W. M., Bozi, L. H., Souza, P. R., Antonio, E. L., Bocalini, D. S., et al. (2017). Dexamethasone-induced cardiac deterioration is associated with both calcium handling abnormalities and calcineurin signaling pathway activation. Mol. Cell. Biochem. 424, 87–98. doi: 10.1007/s11010-016-2846-3
Demirtas, A. O., Karabag, T., and Demirtas, D. (2018). Ischemic modified albumin predicts critical coronary artery disease in unstable angina pectoris and non-ST-elevation myocardial infarction. J. Clin. Med. Res. 10, 570–575. doi: 10.14740/jocmr3417w
Desfontaine, V., Losacco, G. L., Gagnebin, Y., Pezzatti, J., Farrell, W. P., Gonzalez-Ruiz, V., et al. (2018). Applicability of supercritical fluid chromatography - mass spectrometry to metabolomics. I – Optimization of separation conditions for the simultaneous analysis of hydrophilic and lipophilic substances. J. Chromatogr. A 1562, 96–107. doi: 10.1016/j.chroma.2018.05.055
Dinc, I., Sigdel, M., Dinc, S., Sigdel, M. S., Pusey, M. L., and Aygun, R. S. (2014). “Evaluation of normalization and pca on the performance of classifiers for protein crystallization images,” in Proceedings of the IEEE Southeastcon 2014 (Piscataway, NJ: IEEE). doi: 10.1109/SECON.2014.6950744
Dunkelberger, J. R., Serão, N. V. L., Weng, Z., Waide, E. H., Niederwerder, M. C., Kerrigan, M. A., et al. (2017). Genomic regions associated with host response to porcine reproductive and respiratory syndrome vaccination and co-infection in nursery pigs. BMC Genomics 18:865. doi: 10.1186/s12864-017-4182-8
Fang, L., Wang, Y., and Chen, X. (2018). Gene expression profiling and bioinformatics analysis of hereditary gingival fibromatosis. Biomed. Rep. 8, 133–137. doi: 10.3892/br.2017.1031
Gao, E., Lei, Y. H., Shang, X. Y., Huang, Z. M., Zuo, L., Boucher, M., et al. (2010). A novel and efficient model of coronary artery ligation and myocardial infarction in the mouse. Circ. Res. 107, 1445–1453. doi: 10.1161/CIRCRESAHA.110.223925
Gao, L., Hao, J., Niu, Y. Y., Tian, M., Yang, X., Zhu, C. H., et al. (2016). Network pharmacology dissection of multiscale mechanisms of herbal medicines in stage IV gastric adenocarcinoma treatment. Medicine 95:e4389. doi: 10.1097/md.0000000000004389
Gray, W. D., French, K. M., Ghosh-Choudhary, S., Maxwell, J. T., Brown, M. E., Platt, M. O., et al. (2014). Identification of therapeutic covariant MicroRNA clusters in hypoxia-treated cardiac progenitor cell exosomes using systems biology. Circ. Res. 116, 255–263. doi: 10.1161/circresaha.116.304360
Gunawan, A., Jakaria, Listyarini, K., Furqon, A., Sumantri, C., Akter, S. H., and Uddin, M. J. (2018). Transcriptome signature of liver tissue with divergent mutton odour and flavour using RNA deep sequencing. Gene 676, 86–94. doi: 10.1016/j.gene.2018.06.086
Gupta, M. K., Behera, S. K., Dehury, B., and Mahapatra, N. (2017). Identification and characterization of differentially expressed genes from human microglial cell samples infected with Japanese encephalitis virus. J. Vector Borne Dis. 54, 131–138.
Hayasaka, S., Zhao, J., Liang, X., and Xu, K. (2015). Competition between homophily and information entropy maximization in social networks. PLoS One 10:e0136896. doi: 10.1371/journal.pone.0136896
Herum, K. M., Lunde, I. G., Skrbic, B., Louch, W. E., Hasic, A., Boye, S., et al. (2015). Syndecan-4 is a key determinant of collagen cross-linking and passive myocardial stiffness in the pressure-overloaded heart. Cardiovasc. Res. 106, 217–226. doi: 10.1093/cvr/cvv002
Holmes, D. R., Becker, J. A., Granger, C. B., Limacher, M. C., Page, R. L., Sila, C., et al. (2011). ACCF/AHA 2011 health policy statement on therapeutic interchange and substitution. J. Am. Coll. Cardiol. 58, 1287–1307. doi: 10.1016/j.jacc.2011.06.001
Hunt, D. L., Campbell, P. H., Zambon, A. C., Vranizan, K., Evans, S. M., Kuo, H. C., et al. (2012). Early postmyocardial infarction survival in Murphy Roths large mice is mediated by attenuated apoptosis and inflammation but depends on genetic background. Exp. Physiol. 97, 102–114. doi: 10.1113/expphysiol.2011.060269
Husted, S. E., and Ohman, E. M. (2015). Pharmacological and emerging therapies in the treatment of chronic angina. Lancet 386, 691–701. doi: 10.1016/s0140-6736(15)61283-1
Isler, Y., and Kuntalp, M. (2010). Heart rate normalization in the analysis of heart rate variability in congestive heart failure. [Evaluation Studies]. Proc. Inst. Mech. Eng. H 224, 453–463. doi: 10.1243/09544119JEIM642
Isserlin, R., Merico, D., Voisin, V., and Bader, G. D. (2014). Enrichment Map – A Cytoscape app to visualize and explore OMICs pathway enrichment results. F1000Res 3:141. doi: 10.12688/f1000research.4536.1
Iyer, P., Bolla, J., Kumar, V., Gill, M., and Sobhia, M. (2015). In silico identification of targets for a novel scaffold, 2-thiazolylimino-5-benzylidin-thiazolidin-4-one. Mol. Divers. 19, 855–870. doi: 10.1007/s11030-015-9578-2
Kawamiya, T., Kato, K., Horibe, H., Yokoi, K., Oguri, M., Yoshida, T., et al. (2010). Association of genetic variants with myocardial infarction in Japanese individuals with or without metabolic syndrome. Exp. Ther. Med. 1, 969–975. doi: 10.3892/etm.2010.147
Lee, I. S., Lee, H. K., Dat, N. T., Lee, M. S., Kim, J. W., Na, D. S., et al. (2003). Lignans with inhibitory activity against NFAT transcription from SchisandraSchisandra chinensis. Planta Med. 69, 63–64. doi: 10.1055/s-2003-37024
Li, F., Tan, Y. S., Chen, H. L., Yan, Y., Zhai, K. F., Li, D. P., et al. (2015). Identification of schisandrin as a vascular endothelium protective component in YiQiFuMai powder injection using HUVECs binding and HPLC-DAD-Q-TOF-MS/MS analysis. J. Pharmacol. Sci. 129, 1–8. doi: 10.1016/j.jphs.2015.02.003
Li, P. C., Poon, M. K. T., and Ko, K. M. (1996). SchisandraSchisandra chinensis-dependent myocardial protective action of sheng-mai-san in rats. Am. J. Chin. Med. 24, 255–262. doi: 10.1142/s0192415x96000311
Li, S. L., Li, P., Sheng, L. H., Li, R. Y., Qi, L. W., and Zhang, L. Y. (2006). Live cell extraction and HPLC-MS analysis for predicting bioactive components of traditional Chinese medicines. J. Pharm. Biomed. Anal. 41, 576–581. doi: 10.1016/j.jpba.2006.01.014
Li, X., Wei, X. L., Li, K., Tian, L. Q., Shi, Y. B., and Zhang, Z. L. (2013). Effect of carbonized aahesive rehmannia root tuber and prepared rehmannia root on bleeding time and coagulation time in mice. China J. Tradit. Chin. Med. Press 28, 927–929.
Li, Y., Cui, X. Z., Solomon, S. B., Remy, K., Fitz, Y., and Eichacker, P. Q. (2013). B. anthracis edema toxin increases cAMP levels and inhibits phenylephrine-stimulated contraction in a rat aortic ring model. Am. J. Physiol. Heart Circul. Physiol. 305, H238–H250. doi: 10.1152/ajpheart.00185.2013
Lighthouse, J. K., and Small, E. M. (2016). Transcriptional control of cardiac fibroblast plasticity. J. Mol. Cell Cardiol. 91, 52–60. doi: 10.1016/j.yjmcc.2015.12.016
Liu, H., Xu, R., Liu, X., Sun, R., and Wang, Q. (2014). Bioinformatics analysis of gene expression in peripheral blood mononuclear cells from children with type 1 diabetes in 3 periods. Exp. Clin. Endocrinol. Diab. 122, 477–483. doi: 10.1055/s-0034-1372599
Liu, L., Zhu, Q., Wang, J., Xi, Q., Zhu, H., and Gu, M. (2015). Gene expression changes in human mesenchymal stem cells from patients with osteoporosis. Mol. Med. Rep. 12, 981–987. doi: 10.3892/mmr.2015.3514
Liu, P., Jiang, W., Zhou, S., Gao, J., and Zhang, H. (2017). Combined analysis of ChIP sequencing and gene expression dataset in breast cancer. Pathol. Oncol. Res. 23, 361–368. doi: 10.1007/s12253-016-0116-z
Looijestijn, J., Blom, J. D., Aleman, A., Hoek, H. W., and Goekoop, R. (2015). An integrated network model of psychotic symptoms. Neurosci. Biobehav. Rev. 59, 238–250. doi: 10.1016/j.neubiorev.2015.09.016
Lu, H. Y., Wang, S. S., Cai, R., Meng, Y., Xie, X., and Zhao, W. J. (2012). Rapid discrimination and quantification of alkaloids in corydalis tuber by near-infrared spectroscopy. J. Pharm. Biomed. Anal. 59, 44–49. doi: 10.1016/j.jpba.2011.09.037
Merico, D., Isserlin, R., Stueker, O., Emili, A., and Bader, G. D. (2010). Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PLoS One 5:e13984. doi: 10.1371/journal.pone.0013984
Murugesan, M., and Premkumar, K. (2018). Hypoxia stimulates microenvironment in human embryonic stem cell through inflammatory signalling: an integrative analysis. Biochem. Biophys. Res. Commun. 498, 437–444. doi: 10.1016/j.bbrc.2018.02.194
Opazo Saez, A. M., Mosel, F., Nurnberger, J., Rushentsova, U., Gossl, M., Mitchell, A., et al. (2005). Laser doppler imager (LDI) scanner and intradermal injection for in vivo pharmacology in human skin microcirculation: responses to acetylcholine, endothelin-1 and their repeatability. Br. J. Clin. Pharmacol. 59, 511–519. doi: 10.1111/j.1365-2125.2004.02344.x
Pala, S., Sari, M., Kahveci, G., Alizade, E., Arslantas, U., and Uslu, A. (2018). Plasma YKL-40 elevation on admission and follow-up is associated with diastolic dysfunction and mortality in patients with acute myocardial infarction. Cardiol. Res. Pract. 2018:8701851. doi: 10.1155/2018/8701851
Panossian, A., and Wikman, G. (2008). Pharmacology of SchisandraSchisandra chinensis bail.: an overview of Russian research and uses in medicine. J. Ethnopharmacol. 118, 183–212. doi: 10.1016/j.jep.2008.04.020
Peeters, W., Anthonissen, M., Deliaert, A., Van der Hulst, R., and Van den Kerckhove, E. (2012). A comparison between laser-doppler imaging and colorimetry in the assessment of scarring: “a pilot study”. Skin Res. Technol. 18, 188–191. doi: 10.1111/j.1600-0846.2011.00552.x
Peterson, J., Zhang, J., Ahn, J., Suh, Y., Hwang, S., Davis, M. E., et al. (2015). Identification of CTLA2A, DEFB29, WFDC15B, SERPINA1F and MUP19 as Novel tissue-specific secretory factors in mouse. PLoS One 10:e0124962. doi: 10.1371/journal.pone.0124962
Prado-Prado, F., García-Mera, X., Abeijón, P., Alonso, N., Caamaño, O., Yáñez, M., et al. (2011). Using entropy of drug and protein graphs to predict FDA drug-target network: theoretic-experimental study of MAO inhibitors and hemoglobin peptides from Fasciola hepatica. Eur. J. Med. Chem. 46, 1074–1094. doi: 10.1016/j.ejmech.2011.01.023
Sabrkhany, S., Kuijpers, M. J. E., Knol, J. C., Olde Damink, S. W. M., Dingemans, A. C., Verheul, H. M., et al. (2018). Exploration of the platelet proteome in patients with early-stage cancer. J. Proteom. 177, 65–74. doi: 10.1016/j.jprot.2018.02.011
Sakata, F., Ito, Y., Mizuno, M., Sawai, A., Suzuki, Y., Tomita, T., et al. (2017). Sodium chloride promotes tissue inflammation via osmotic stimuli in subtotal-nephrectomized mice. Lab. Invest. 97, 432–446. doi: 10.1038/labinvest.2017.4
Shangguan, H., Tan, S. Y., and Zhang, J. R. (2015). Bioinformatics analysis of gene expression profiles in hepatocellular carcinoma. Eur. Rev. Med. Pharmacol. Sci. 19, 2054–2061.
Shen, Z. X., Yang, Q. Z., Li, C., Du, L. J., Sun, X. N., Liu, Y., et al. (2018). Myeloid peroxisome proliferator-activated receptor gamma deficiency aggravates myocardial infarction in mice. Atherosclerosis 274, 199–205. doi: 10.1016/j.atherosclerosis.2018.05.005
Song, Y., Ahn, J., Suh, Y., Davis, M. E., and Lee, K. (2013). Identification of novel tissue-specific genes by analysis of microarray databases: a human and mouse model. PLoS One 8:e64483. doi: 10.1371/journal.pone.0064483
Steudemann, C., Bauersachs, S., Weber, K., and Wess, G. (2013). Detection and comparison of microRNA expression in the serum of doberman pinschers with dilated cardiomyopathy and healthy controls. BMC Vet. Res. 9:12. doi: 10.1186/1746-6148-9-12
Sun, L. J., Wang, L., Jiang, M. H., Huang, J. X., and Lin, H. (2011). Glycogen debranching enzyme 6 (AGL), enolase 1 (ENOSF1), ectonucleotide pyrophosphatase 2 (ENPP2_1), glutathione S-transferase 3 (GSTM3_3) and mannosidase (MAN2B2) metabolism computational network analysis between chimpanzee and human left cerebrum. Cell Biochem. Biophys. 61, 493–505. doi: 10.1007/s12013-011-9232-2
Tang, H. Q., He, S. B., Zhang, X. Y., Luo, S. L., Zhang, B. X., Duan, X. J., et al. (2016). A Network pharmacology approach to uncover the pharmacological mechanism of xuanhusuo powder on osteoarthritis. Evid. Based Complemen. Alternat. Med. 2016, 1–10. doi: 10.1155/2016/3246946
Tarnavski, O., McMullen, J. R., Schinke, M., Nie, Q., Kong, S., and Izumo, S. (2004). Mouse cardiac surgery: comprehensive techniques for the generation of mouse models of human diseases and their application for genomic studies. Physiol. Genomics 16, 349–360. doi: 10.1152/physiolgenomics.00041.2003
Teschendorff, A. E., Sollich, P., and Kuehn, R. (2014). Signalling entropy: a novel network-theoretical framework for systems analysis and interpretation of functional omic data. Methods 67, 282–293. doi: 10.1016/j.ymeth.2014.03.013
Tsukamoto, K., Mani, D. R., Shi, J., Zhang, S., Haagensen, D. E., Otsuka, F., et al. (2013). Identification of apolipoprotein D as a cardioprotective gene using a mouse model of lethal atherosclerotic coronary artery disease. Proc. Natl. Acad. Sci. U.S.A. 110, 17023–17028. doi: 10.1073/pnas.1315986110
Wang, D., Cai, F., Ge, J., and Yin, L. (2015). Brief exercises affect gene expression in circulating monocytes. Scand. J. Immunol. 82, 429–435. doi: 10.1111/sji.12345
Wang, Y. H., Qiu, C., Wang, D. W., Hu, Z. F., Yu, B. Y., and Zhu, D. N. (2011). Identification of multiple constituents in the traditional Chinese medicine formula Sheng-Mai San and rat plasma after oral administration by HPLC-DAD-MS/MS. J. Pharm. Biomed. Anal. 54, 1110–1127. doi: 10.1016/j.jpba.2010.11.034
Wu, J., Chen, Z. P., Shang, A. Q., Wang, W. W., Chen, Z. N., Tao, Y. J., et al. (2017). Systemic bioinformatics analysis of recurrent aphthous stomatitis gene expression profiles. Oncotarget 8, 111064–111072. doi: 10.18632/oncotarget.22347
Xie, L., Pi, X., Townley-Tilson, W. H. D., Li, N., Wehrens, X. H. T., Entman, M. L., et al. (2015). PHD2/3-dependent hydroxylation tunes cardiac response to β-adrenergic stress via phospholamban. J. Clin. Investig. 125, 2759–2771. doi: 10.1172/jci80369
Xiong, L., Liu, Y., Zhou, M. M., Wang, G. J., Quan, D. J., Shen, C. J., et al. (2018). Targeted ablation of cardiac sympathetic neurons improves ventricular electrical remodelling in a canine model of chronic myocardial infarction. Europace 20, 2036–2044. doi: 10.1093/europace/euy090
Zhan, S. Y., Guo, W. J., Shao, Q., Fan, X. H., Li, Z., and Cheng, Y. Y. (2014). A pharmacokinetic and pharmacodynamic study of drug-drug interaction between ginsenoside Rg1, ginsenoside Rb1 and schizandrin after intravenous administration to rats. J. Ethnopharmacol. 152, 333–339. doi: 10.1016/j.jep.2014.01.014
Zhang, T., Zhao, L. L., Cao, X., Qi, L. C., Wei, G. Q., Liu, J. Y., et al. (2014a). Bioinformatics analysis of time series gene expression in left ventricle (LV) with acute myocardial infarction (AMI). Gene 543, 259–267. doi: 10.1016/j.gene.2014.04.002
Zhang, T., Zhao, L. L., Zhang, Z. R., Fu, P. D., Su, Z. D., Qi, L. C., et al. (2014b). Interaction network analysis revealed biomarkers in myocardial infarction. Mol. Biol. Rep. 41, 4997–5003. doi: 10.1007/s11033-014-3366-4
Zhang, Y. P., Wuyunqiqige, Zheng, W., Liu, S., and Zhao, C. (2016). gDNA-Prot: predict DNA-binding proteins by employing support vector machine and a novel numerical characterization of protein sequence. J. Theor. Biol. 406, 8–16. doi: 10.1016/j.jtbi.2016.06.002
Keywords: Schisandra, myocardial infarction, effect, three-mode network, gene ontology
Citation: Hu S, Zuo H, Qi J, Hu Y and Yu B (2019) Analysis of Effect of Schisandra in the Treatment of Myocardial Infarction Based on Three-Mode Gene Ontology Network. Front. Pharmacol. 10:232. doi: 10.3389/fphar.2019.00232
Received: 16 October 2018; Accepted: 22 February 2019;
Published: 20 March 2019.
Edited by:
Alexander N. Shikov, St. Petersburg Institute of Pharmacy, RussiaReviewed by:
Yanling Zhao, 302 Military Hospital of China, ChinaBrian Yee Hong Lam, University of Cambridge, United Kingdom
Copyright © 2019 Hu, Zuo, Qi, Hu and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuanjia Hu, eXVhbmppYWh1QHVtYWMubW8= Boyang Yu, Ym95YW5neXVjcHVAMTYzLmNvbQ==
 Siyao Hu
Siyao Hu Huali Zuo2
Huali Zuo2 Yuanjia Hu
Yuanjia Hu