
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Pharmacol., 17 August 2018
Sec. Drug Metabolism and Transport
Volume 9 - 2018 | https://doi.org/10.3389/fphar.2018.00927
Pigs are commonly used as an animal model to evaluate the toxic effects of exogenous compounds. Cytochrome P450 1A1 (CYP1A1) metabolizes numerous exogenous compounds and is abundantly expressed in the liver, kidneys, and intestines. The high amino acid similarity between human and porcine CYP1A1 indicates that they probably have the same metabolic characteristics. Therefore, understanding the regulatory mechanism of CYP1A1 expression in pigs is particularly important for predicting the toxicology and metabolic kinetics of exogenous chemicals. Currently, the transcriptional regulation of porcine CYP1A1 has rarely been studied, especially regarding basal transcription. In this study, we first confirmed that the key regulatory elements of porcine CYP1A1 basal transactivation are in the proximal promoter region using promoter truncation analysis via a dual luciferase assay in a porcine kidney cell line LLC-PK1. Two overlapping cis-elements, the xenobiotic response element (XRE) and GC box, in this proximal region potentially play key roles in the basal transactivation of porcine CYP1A1. Furthermore, using electrophoretic mobility shift assay and chromatin immunoprecipitation, the GC box binding protein Sp1 was confirmed to bind to the proximal promoter of porcine CYP1A1, instead of AhR, the XRE binding protein. In LLC-PK1 cells, by knocking down either Sp1 or AhR, the expression of porcine CYP1A1 at the mRNA level and protein level was significantly downregulated, suggesting both proteins are important for porcine CYP1A1 expression. However, promoter activity analysis in LLC-PK1 cells treated with an AhR agonist and antagonist confirmed that AhR does not participate in the basal regulation of porcine CYP1A1 at the proximal promoter. In conclusion, our study revealed that the proximal promoter is the key regulatory region for porcine CYP1A1 basal expression. Although AhR plays an important role in the transactivation of porcine CYP1A1 expression, the key determinant transcription factor for its basal transactivation is Sp1 at the proximal promoter of porcine CYP1A1.
CYP1A1 is one of the most important CYP450 family members for metabolizing multiple exogenous and endogenous substrates such as drugs, carcinogens, steroids, and toxicants (Guengerich et al., 1991; Gray and Squires, 2013). The expression of CYP1A1 is correlated with carcinogen bioactivation (Guengerich, 1988) and the biotransformation of various chemical substrates by oxygenation, demethylation, or dealkylation. The pig is a valuable animal model as a substitute for humans in pharmacological and toxicological studies (Kojima and Degawa, 2016). Porcine CYP1A1 (pCYP1A1) is the orthologous protein to human CYP1A1, which plays a key role in the metabolism of various toxic and carcinogenic compounds (Ma and Lu, 2007). Therefore, clarifying the regulation of CYP1A1 expression in pigs is instructive in understanding the metabolism of various drugs in humans.
CYP1A1 and 1A2 are highly induced by many chemicals at the mRNA and protein levels in different cells. However, the basal transcription mechanism remains to be further elucidated. It was first found in Drosophila SL2 cells that specific protein 1 (Sp1), aryl hydrocarbon receptor (AhR) and AhR receptor nuclear translocator (Arnt) interacted with each other and bound to the GC box at the promoter to enhance the transcription of CYP1A1 (Wang et al., 1999). In human cells, the GC box is a key cis-element that activates CYP1A1 promoter activity (Zhang et al., 1998). Decreased expression of AhR in human hepatocytes inhibits the expression of CYP1A1 (Le et al., 2010). The mechanisms underlying CYP1A1 basal transcription remain unresolved, especially concerning the functions of Sp1 and AhR in mammalian cells.
Sp1 is expressed ubiquitously in cells and transactivates many genes by binding to the GC box at promoters through three C2H2-type zinc fingers at the C-terminus (Wierstra, 2008). The pathways via which Sp1 acts in the regulation of gene expression are versatile. One or more Sp1 molecules bind to a single site at the promoter of a gene to activate gene transcription; additionally, multiple Sp1 molecules are recruited to multiple Sp1 binding sites at the promoter to synergistically activate gene expression (Deniaud et al., 2009). Many studies confirmed that Sp1 participates in the regulation of CYP450 gene expression. As an example, Sp1 is critical for the regulation of CYP3A5 basal expression in humans and pigs (Iwano et al., 2001; Saito et al., 2001; Bombail et al., 2004). However, few studies have focused on Sp1 function in the regulation of CYP1A1 in human cells and none have focused on porcine cells.
CYP1A1 expression has been confirmed to be inducible by many exogenous compounds. AhR is regarded as the target transcription factor to induce CYP1A1. AhR is abundant in the cytoplasm, is transported to the nucleus by heterodimerization with ARNT and then binds to the xenobiotic response element (XRE) at the promoter of the gene, thereby activating gene expression (Whitelaw et al., 1993). Many exogenous compounds, such as polycyclic aromatic hydrocarbons (PAHs), biphenyls (PCBs), and halogenated aromatic hydrocarbons (HAHs), induce CYP1A1 expression by AhR-dependent pathways (Hanlon et al., 2005; Fazili et al., 2010; Xie et al., 2018). However, several studies have found that AhR-independent pathways regulate the induction of CYP1A1. For example, α-naphthoflavone, an AhR-interfering agent, does not antagonize the induction of CYP1A1 in HepG2 cells treated with TCDD (Kikuchi et al., 1998). Retinoids induced CYP1A1 via binding to the retinoic acid response element in its promoter region (Vecchini et al., 1994). These studies indicate that the regulatory pattern of CYP1A1 is coordinated by multiple pathways. However, the regulation of porcine CYP1A1 remains unclear presently.
In our study, initially, we confirmed that the key regulatory elements for its basal transcription are in the proximal promoter region using truncated promoter analysis by a dual luciferase assay. Two overlapped cis-elements, the GC box and XRE, possibly recruiting Sp1 and AhR, respectively, transactivate basal transcription in the proximal promoter of pCYP1A1. Knockdown of Sp1 and AhR suggested that only Sp1 is involved in the regulation of the proximal promoter of pCYP1A1. Furthermore, in LLC-PK1 cells treated with an agonist and antagonist of AhR, neither activation nor inhibition of AhR activity affected the basal transcription of pCYP1A1 in the proximal promoter. Electrophoretic mobility shift assay (EMSA) and chromatin immunoprecipitation (ChIP) confirmed that Sp1 binds to the proximal promoter of pCYP1A1.
A 3.5-kb fragment (-3424 to +157; +1 indicates the transcriptional start site) from the 5′-flanking region of porcine CYP1A1 (GenBank Accession No. NC_010449.5) was amplified by PCR from the genomic DNA of porcine liver tissues. This fragment was inserted into the pGL3-Basic vector at Xho I and Mlu I sites to generate the -3424-Luc plasmid. Using this plasmid as a template and the same downstream primer, a series of upstream truncated primers were used to amplify fragments of different lengths. These fragments were inserted into the pGL3-Basic vector at Xho I and Mlu I sites to generate the truncated plasmids. Using primers designed for different mutation patterns, ΩPCR was used to construct different mutation vectors using the -43-LUC plasmid as a template. The open reading frames for the expression of Arnt (GenBank Accession No. NC_010446.5), AhR (GenBank Accession No. NC_010451.4), and Sp1 (GenBank Accession No. NC_010447.4) were amplified with the corresponding primers from cDNA that was reversely transcribed from total RNA extracted from porcine liver cells. The open reading frames were inserted into the Xba I and Hind III sites of pcDNA3.1/myc-His(-) A vectors (Invitrogen). The primers used are listed in Supplementary Table S1.
LLC-PK1 cells (ATCC, CL-101) were cultured in M199 medium, and COS7 cells (ATCC, CRL-1651) were cultured in DMEM medium. All media were supplemented with 10% fetal bovine serum (PAN-Biotech, Aidenbach, Germany), 1% penicillin G/streptomycin (Invitrogen), 1× SITE (Sigma), 0.1 μM dexamethasone and 0.1 μM insulin (Invitrogen). All cell lines were cultivated at 37°C, 5% CO2.
Total RNA was isolated using TRIZOL reagent (Invitrogen). Total RNA was treated with DNase I (M0303S, New England BioLabs) to remove the genomic DNA before performing reverse transcription. Thereafter, 1 μg of RNA was reversely transcribed using M-MLV Reverse Transcriptase (Promega), random primer (6mer) and 10 mM dNTPs (TaKaRa).
Real-time PCR was performed using the BioRad CFX96 real-time PCR detection system (Bio-Rad, Hercules, CA, United States) according to the manufacturer’s instructions. RT-PCR was performed in 20-μL volumes containing SYBR Green I Dye. The following parameters were used: 94.0°C for 2 min, followed by 40 cycles of 94.0°C for 20 s, 50.0°C for 30 s, and 72.0°C for 30 s. The gene expression levels were normalized using GAPDH as a control. The 2-ΔΔCT method was used to process data and calculate the relative expression of genes. The primers used are listed in Supplementary Table S1.
According to the manufacturer’s instructions (Beyotime, Shanghai, China), RIPA lysis buffer containing 1 mM phenylmethylsulfonyl fluoride (PMSF) was used to extract total cellular protein. The protein concentration was adjusted using the BCA Protein Assay Kit (Pierce). Next, 30 μg of total protein was subjected to 10% polyacrylamide gel electrophoresis, followed by transfer to a PVDF membrane (Millipore). The membranes were blocked in a buffer containing 5% skim milk and 0.1% Tween 20 in Tris-buffered saline (0.15 M sodium chloride and 20 mM Tris base, pH 8.0) for 1 h at room temperature. Next, the membranes were incubated with antibodies overnight at 4°C. Secondary antibodies were incubated for 1 h at room temperature. The bands were detected using the ChemiDoc MP Imaging system (Bio-Rad, United States). The antibodies used in this work included anti-GAPDH (Santa Cruz, sc-32233), anti-Sp1 (Abcam, ab13370), anti-AhR (Cell Signaling Technology, D5S6H), and anti-CYP1A1 (Santa Cruz, sc-101828).
Cells were grown in 24-well plates for 24 h to 70–90% confluence. Next, 0.5 μg of the pGL3-basic plasmid containing the promoter and 2.5 ng of the pRL-CMV plasmid were transfected into cells with 1.5 μL of Lipofectamine 3000 (Invitrogen). The transfection reagent was removed, and then 500 μL of medium with 10% FBS was added to each well after 5 h. After 24 h, the cells were lysed, and the luciferase activities were measured using the Dual Luciferase Reporter Assay System (Promega) on a Turner Designs TD-20/20n luminometer (Promega). The dual luciferase reporter assay system was purchased from Promega (Madison, WI, United States). The ratio of the firefly luciferase activity to the Renilla luciferase activity represents the strength of the promoter.
All siRNAs were synthesized by GenePharma (Suzhou). The cells were grown in 24-well plates for 24 h to 70–90% confluence, and then 40 pmol of siRNA and plasmid were transfected into the cells with 1.5 μL of Lipofectamine 3000 (Invitrogen). Non-targeting siRNA (scramble) was used as the negative control. The transfection reagent was removed, and 500 μL of medium with 10% FBS was added to each well after 5 h. After 24 h, the cells were lysed, and the luciferase activities were measured. The siRNA sequences used are listed in Supplementary Table S1.
For overexpression experiments, COS-7 cells were transfected with the constructs of the Sp1 or AhR gene. The Nuclear Extraction Kit (Beyotime, Haimen, China) was used to extract overexpressed nuclear protein from COS7 cells. EMSA was performed using the EMSA Assay Kit (Beyotime, Haimen, China). The mixtures containing unlabeled probe, nuclear extract (4 μg) and binding buffer were preincubated at 23°C for 10 min. Next, 0.1 pmol of the biotinylated probe was added and incubated at 23°C for 20 min in a total volume of 9 μL. The mixture was subjected to electrophoresis in 4.5% non-denaturing polyacrylamide [acrylamide/bisacrylamide 29:1 (v/v)] gels, followed by the addition of bromophenol blue in a blank hole as an indicator. The gel was transferred to a nylon membrane (+) after the bromophenol blue appeared at approximately 2 cm from the bottom of the gel. The nylon membrane (+) was crosslinked for 2 min under UV light (254 nm). The nylon membrane (+) was then processed with the EMSA Assay Kit (Beyotime, Haimen, China), followed by the detection by chemiluminescence (Millipore, Bedford, MA, United States). For competition experiments, 100-fold molar excess of unlabeled probe was used. For supershift experiments, 1 μL of anti-Sp1 (Abcam, ab13370), and anti-AhR (Cell Signaling Technology, D5S6H) were used.
According to the manufacturer’s instructions (Cell Signaling Technology, #9003, United States), the enrichment of Sp1 and AhR in the CYP1A1 promoter in LLC-PK1 cells was analyzed. The antibodies used in this work included anti-histone H3 (Cell Signaling Technology, D2B12), anti-Sp1 (Abcam, ab13370), and anti-AhR (Cell Signaling Technology, D5S6H). The primers used are listed in Supplementary Table S1.
All experiments were performed independently at least three times. All the data are represented as the means ± standard deviation (SD). Statistical significance was assessed by ANOVA. Values of P < 0.05 were considered statistically significant.
To identify the cis-elements that regulate CYP1A1 expression in the porcine CYP1A1 promoter, DNA fragments with different lengths, spanning from the ATG of the pCYP1A1 gene to -3424 bp upstream were fused upstream of the firefly luciferase gene. Each promoter plasmid and reference plasmid, pRL-CMV harboring the Renilla luciferase gene, were co-transfected into LLC-PK1 cells. Normalized luciferase activity indicates the strength of the promoter for transcription activities. As shown in Figure 1A, the removal of DNA fragments from -3424 to -1981 bp led to the apparent incremental change in firefly luciferase activity, suggesting there are negative regulatory elements in this region to inhibit the basal transcription of porcine pCYP1A1. More importantly, the firefly luciferase activity was apparently decreased when DNA fragments from -43 to -28 bp were deleted, from 27- to 1.7-fold that of the normalized basic control. This suggested that the proximal promoter is the key region for pCYP1A1 transactivation and contains positive regulatory elements between -43 and -28 bp (Figure 1A).
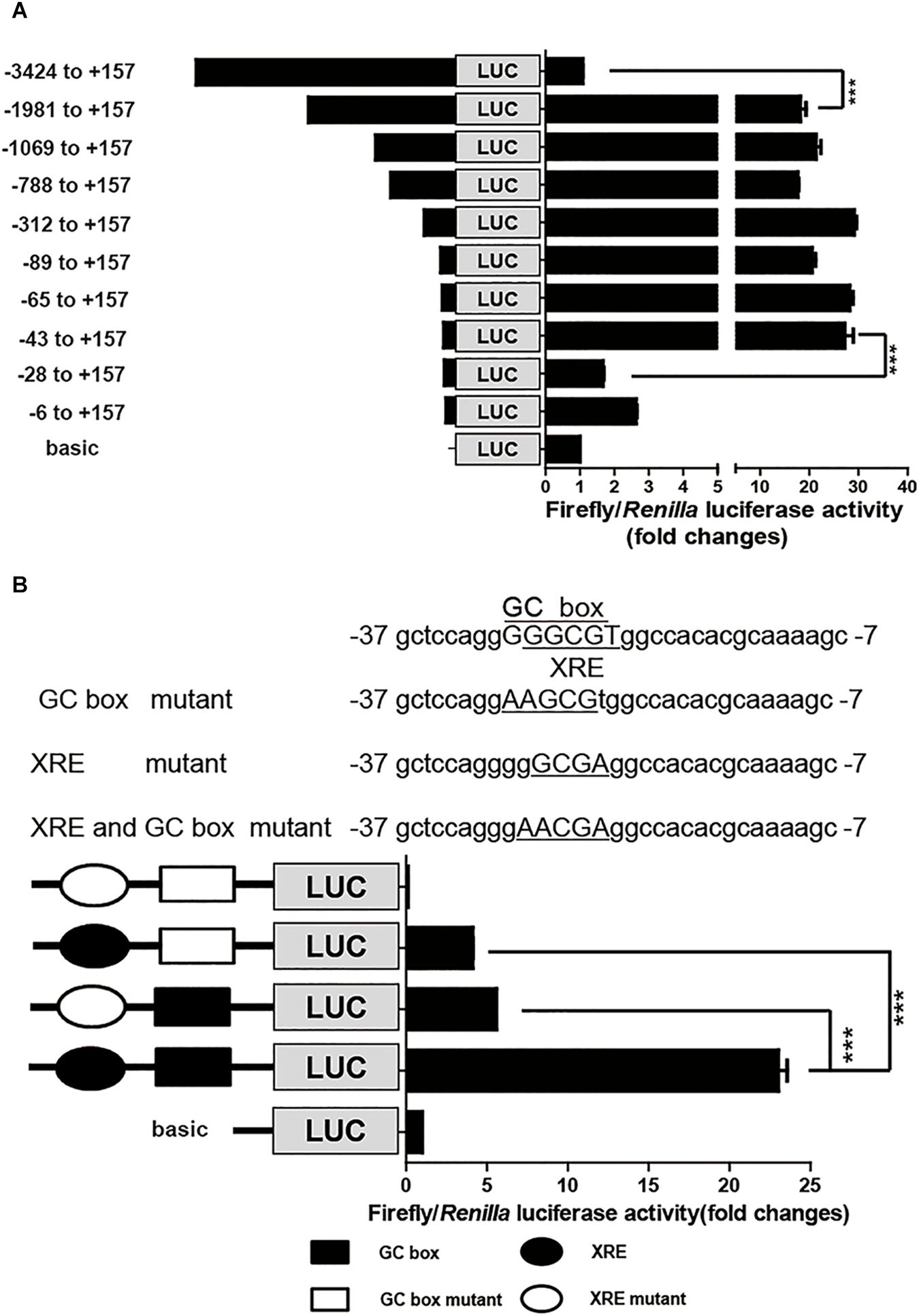
FIGURE 1. The proximal GC box and XRE determine pCYP1A1 basal expression. (A) Deletion analysis of the pCYP1A1 promoter in LLC-PK1 cells. pGL3-basic containing different lengths of the pCYP1A1 promoter fragments and pRL-CMV plasmid were transfected into LLC-PK1 cells, and the luciferase activities were measured, as described in section “Materials and Methods.” Basic represents empty vector pGL3-basic, which served as the control. The values on the right show the ratio of the luciferase activity of the promoter fragment to the luciferase activity of pGL3-basic. The left diagrams show the deletion pattern of the pCYP1A1 promoter. The number indicates the position of the fragment (base pair) at the transcription start site. (B) Mutation analysis of the proximal pCYP1A1 promoter in LLC-PK1 cells. The mutation pattern is shown above and represent the mutations that are introduced into the –37 to –7 promoter region to destroy the XRE and GC box. The mutated bases are underlined. The left diagram shows the mutation pattern of the pCYP1A1 promoter. The value on the right shows the ratio of the luciferase activity of the promoter fragment to the luciferase activity of pGL3-basic. The experiment was repeated three times. Error bars represent the standard deviation of three replicates. ∗∗∗P < 0.001.
The MatInspector program1 was used to analyze the candidate cis-elements in the proximal promoter DNA sequence regarding transcriptional regulation. Two cis-elements, GC box and XRE, were found in this -43 to -28 bp region, and they probably play important roles in pCYP1A1 expression. These two cis-elements overlapped in the region, and the core DNA sequences only have one base difference. Therefore, direct deletion of one cis-element to confirm its role in the pCYP1A1 promoter transactivation is not applicable. To verify the critical roles for the two cis-elements in basal transcription, the key binding sites were mutated, and then the strengths of the mutated promoters were analyzed by the dual luciferase assay. The relative luciferase activity was decreased by 4.5-fold in the plasmid with the mutated GC box and 5.6-fold in the plasmid with mutated XRE (Figure 1B). This implied either that both cis-elements were essential for the basal transcription or the mutation abolished the function of non-mutated cis element. The double mutations of the GC box and XRE in the promoter region caused the complete loss of basal transactivity because the relative luciferase activity was even lower than that of the reference control. Our data suggest that the mutation of key binding sites may lead to a combination of repressor in this region. Therefore, a mutation assay cannot confirm whether either one of these two cis elements or both are the key players.
Next, we further confirmed whether Sp1 or AhR plays a key transactivation role in the proximal promoter region by measuring the relative luciferase activity of the (-43/+157) promoter in the cell lines with the knocking down of Sp1 or AhR. As shown in Figures 2A,B, siRNA successfully repressed the expression of Sp1 or AhR, respectively. The protein and mRNA levels of pCYP1A1 following the knocking down of Sp1 or AhR were apparently decreased (Figures 2C,D). This suggested that both transcription factors play important regulatory roles in pCYP1A1 expression. However, when only measuring the transcription in the proximal promoter using the relative luciferase assay, a twofold decrease in transactivity was observed in the Sp1-knockdown cells but not in AhR-knockdown cells (Figures 2E,F). These data demonstrated that Sp1, rather than AhR, affects the regulation of the pCYP1A1 proximal promoter. Taken together, the data proved that, in the proximal region of the pCYP1A1 promoter, Sp1 transactivates the basal expression of pCYP1A1, and AhR does not contribute to the proximal DNA transactivation of pCYP1A1.
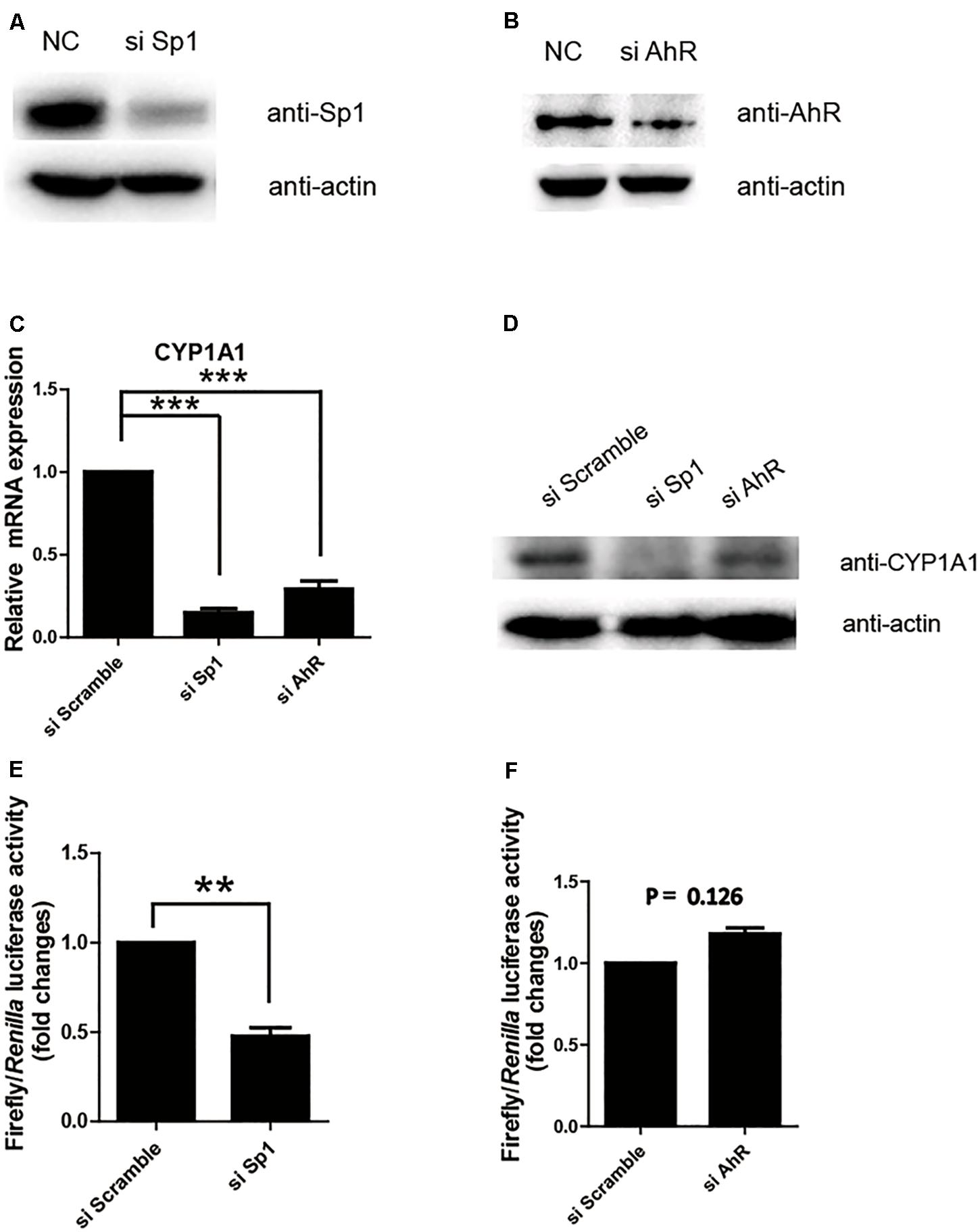
FIGURE 2. Sp1 is required for the basal transactivation of the proximal promoter of pCYP1A1. (A,B) Western blotting confirmed Sp1 or AhR protein levels after siRNA transfection in LLC-PK1 cells. (C,D) mRNA and protein levels of pCYP1A1 in the cells with knocked down Sp1 or AhR. RT-PCR confirmed the pCYP1A1 mRNA levels after transfection of siRNA. Western blotting confirmed the pCYP1A1 protein levels after transfection of siRNA. (E,F) Sp1 but not AhR affects the proximal promoter activity of pCYP1A1. The siRNA, pRL-CMV plasmid and -43-Luc plasmid were co-transfected into LLC-PK1 cells. Non-targeting siRNA (NC) was used as the negative control. The promoter activity of the -43-Luc plasmid was measured. The value on the ordinate indicates the ratio of the value of the promoter activity transferred to the si Sp1 or si AhR to the value of the initiation activity transferred to the NC. The experiment was repeated three times. Error bars represent the standard deviation of three replicates. ∗∗P < 0.01 and ∗∗∗P < 0.001.
As mentioned above, AhR expression in cells does not affect the proximal promoter activity of pCYP1A1, which is contradicted to the previous reported that AhR affects the activity of the CYP1A1 proximal promoter (Kobayashi et al., 1996). Two drugs, β-naphthoflavone and resveratrol, were used to further address the function of AhR in the proximal promoter of pCYP1A1. β-Naphthoflavone, as an agonist for AhR, has been reported to induce pCYP1A1 expression via the translocation of cytoplasmic AhR to the nucleus (Hankinson, 1995; Kikuchi et al., 1998). In β-naphthoflavone-treated LLC-PK1 cells, pCYP1A1 mRNA was increased by 2000-fold relative to the level in untreated cells (Figure 3B). Nevertheless, the (-43/+157) promoter was not changed significantly compared with the relative luciferase activity in β-naphthoflavone-treated LLC-PK1 cells with the activity in untreated control at 24 h (Figure 3A). Resveratrol inhibits the binding of AhR to the promoter DNA, thereby AhR activation of gene expression is inhibited. When (-43/+157) promoter plasmids were transfected into cells, similarly, there was no significant change in the relative luciferase activity in resveratrol-treated cells for 24 h (Figure 3C). In resveratrol-treated LLC-PK1 cells, pCYP1A1 mRNA was decreased by 18-fold relative to that in untreated cells (Figure 3D).
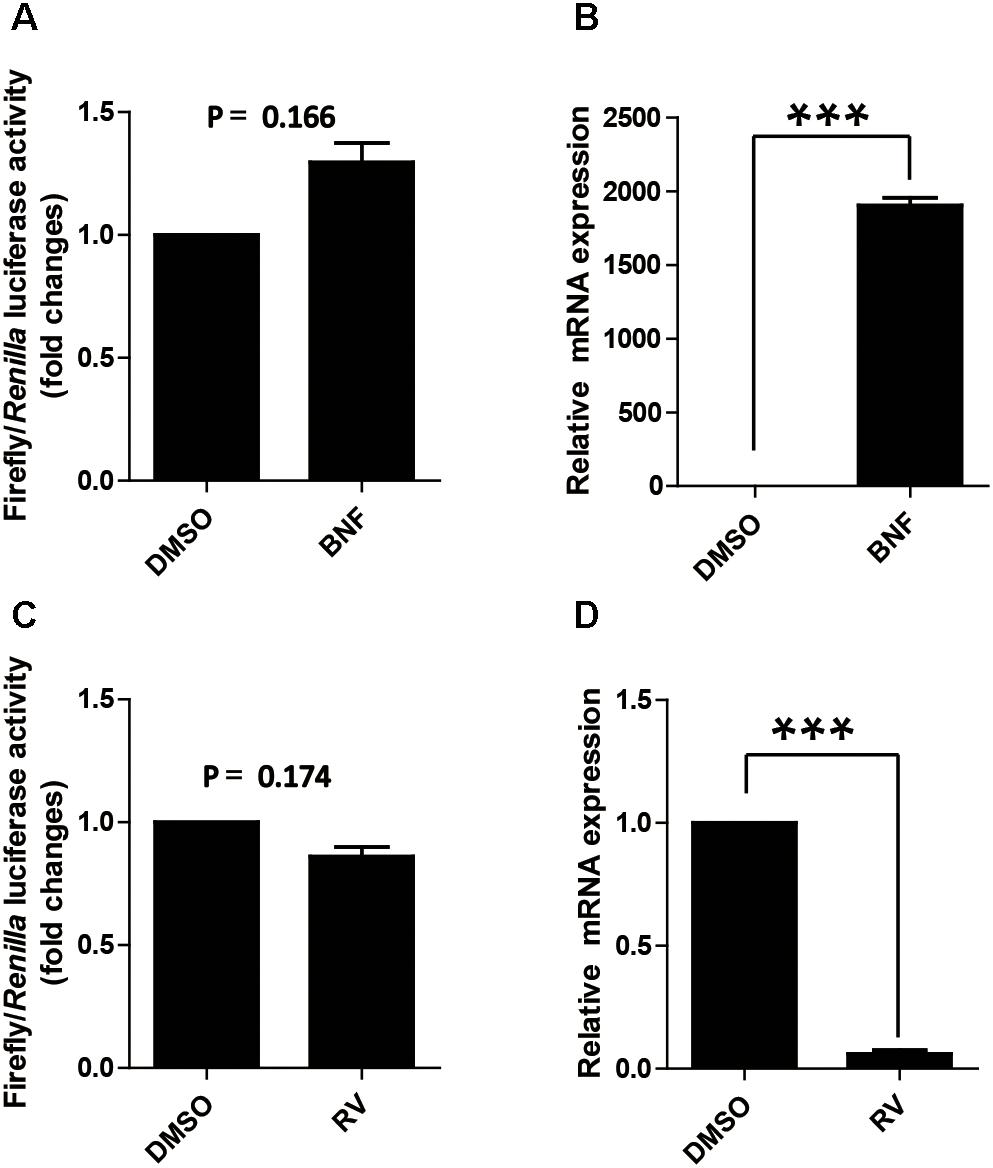
FIGURE 3. Effect of the agonist and antagonist for AhR to treat cells on promoter activity of pCYP1A1. (A) Effect of β-naphthoflavone on -43-Luc plasmid promoter activity. “BNF” indicates β-naphthoflavone treatment at a concentration of 2 μM for 24 h. DMSO treated cells were used as the control. The values on the ordinate represent the ratio of the activity of the promoter after BNF treatment to the value of the activity of the DMSO treated promoter. (B) The relative quantified mRNA level of CYP1A1 in LLC-PK1 Cells treated with BNF for 24 h. DMSO-treated cells were used as the control. In the control cells, pCYP1A1 relative to glyceraldehydes-3-phosphate dehydrogenase (GADPH) mRNA level was set to 1. (C) Effect of resveratrol on -43-Luc plasmid promoter activity. “RV” means resveratrol used at a concentration of 180 μM for 24 h. DMSO-treated cells were used as the control. The values on the ordinate represent the ratio of the activity of the promoter after RV treatment to the value of the activity of the DMSO-treated promoter. (D) The relative quantified mRNA level of CYP1A1 in LLC-PK1 cells treated with 180 μM RV for 24 h. DMSO-treated cells served as the control. In the control cells, pCYP1A1 relative to the mRNA level of GAPDH was set to 1. The experiment was repeated three times. Error bars represent the standard deviation of three replicates. ∗∗∗P < 0.001.
The agonist and antagonist for AhR do not significantly affect the relative luciferase activity of the (-43/+157) promoter plasmid. These data further confirmed that the transcription factor that affects the (-43/+ 157) promoter activity is not AhR.
Sp1 and AhR bind to the GC box and XRE, respectively, which are located in the proximal promoter region of pCYP1A1. As mentioned above, it is not applicable to verify the key role of these two cis-elements in the regulation of pCYP1A1 by deletion or mutation of the cis-elements. Therefore, we initially confirmed the binding of Sp1 or AhR to the DNA motif in vitro by EMSA. Sp1 or AhR was overexpressed in COS-7 cells. For Sp1, when the probe was incubated with the nuclear protein, probe migration would lag to form a band. A supershift occurred after the addition of the Sp1 antibody, and the band disappeared after the addition of a 100-fold molar excess of the unlabeled probe, but not after the addition of a 100-fold molar excess of unlabeled and mutated probe (Figure 4A). For AhR, it is dubious that AhR bound with the probe, and no supershift band was observed (Figure 4B). These data indicated that Sp1 binds at the position of -37 to -7 bp of the pCYP1A1 promoter and then transactivates the expression of pCYP1A1. ChIP was used to verify whether Sp1 and AhR bind to the pCYP1A1 proximal promoter. The binding of Sp1 to the porcine CYP1A1 proximal promoter was fourfold greater than that of the negative control, while the binding of AhR to the porcine CYP1A1 proximal promoter was not significantly different from that of the negative control (Figure 4C). Taken together, those data proved that Sp1, but not AhR, binds to the proximal region of the pCYP1A1 promoter.
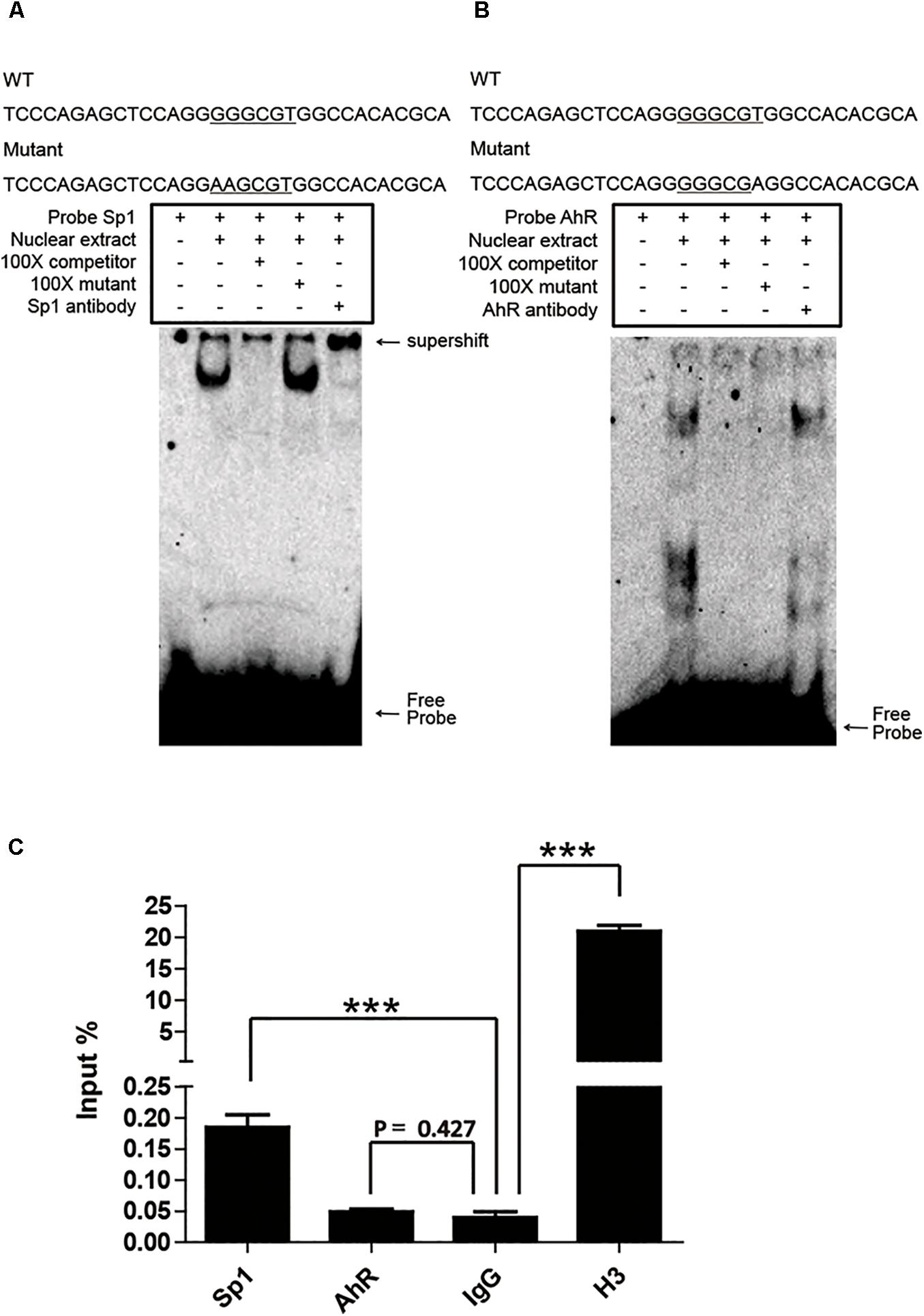
FIGURE 4. Sp1 binds to the GC box and then affects the activity of the pCYP1A1 promoter. (A) EMSA was used to confirm that Sp1 binds to the GC box at the proximal pCYP1A1 promoter. (B) EMSA was used to confirm that AhR does not bind to XRE at the proximal pCYP1A1 promoter. The mutated bases are underlined. For the competition experiments, 100-fold molar excess of the unlabeled probe was used. The DNA protein complexes and supershifts are indicated by arrows. (C) ChIP was used to confirm that Sp1 or AhR binds to cis elements at the proximal pCYP1A1 promoter. IgG was used as the negative control. Histone H3 antibody served as the positive control. The experiment was repeated three times. Error bars represent the standard deviation of three replicates.∗∗∗P < 0.001.
Cytochrome P450 is a superfamily of enzymes known to metabolize exogenous compounds. Among them, CYP1A1 is well-known for its ability to metabolize drugs and other xenobiotic substances (Guengerich et al., 1991). The pig is an animal model that can substitute for humans in pharmacological and toxicological studies. The amino acid sequence similarity of CYP1A1 between pigs and humans exceeds 80%, indicating that they have the same metabolic characteristics. Presently, the regulatory mechanism of CYP1A1 in pigs is not clear. To analyze the conservation of the regulatory mechanisms of CYP1A1 in humans and pigs, it is necessary to clarify the expression regulation mechanism of pCYP1A1.
Truncation analysis of the pCYP1A1 promoter DNA showed that the proximal region (-43 to -28) is the key determinant region for pCYP1A1 expression. In this region, two cis-elements were identified, but the core binding sequence is overlapped. We also found that these two cis-elements at the human CYP1A1 proximal promoter overlap and play important roles in human CYP1A1 basal transcription (Internal communication). Furthermore, by knocking down either Sp1 or AhR via siRNA, the expression levels of pCYP1A1 at the mRNA and protein levels were significantly decreased, suggesting that both are important in pCYP1A1 basal transcription. However, using EMSA and ChIP, it was confirmed that only Sp1 binds to the GC box in the proximal promoter. Instead, AhR did not bind to this region, although the expression of pCYP1A1 was drastically decreased in si-AhR cells. This finding suggested that, in the proximal promoter region, Sp1 determines the strength of transcription. AhR probably affected the expression of pCYP1A1 in this proximal region, which harbors other functional XRE sites.
Studies have shown that AhR is an important transcription factor that participates in the expression of CYP1A1. The high induction of CYP1A1 expression by many exogenous compounds like PAHs, PCBs, and HAHs relies on the AhR-dependent pathway (Ma et al., 2000; Monk et al., 2003; Uno et al., 2004). Among them, the mechanism by which TCDD induces the expression of CYP1A1 through AhR is clear. TCDD increases the level of AhR in the nucleus and induces the expression of CYP1A1 in humans and pigs (Xu et al., 2000; Hombachklonisch et al., 2006). In addition, AhR is a methylation-sensitive transcription factor. TCDD induces a decrease in DNA methylation in the Cyp1a1 promoter region of mice, thereby enhancing the expression of Cyp1a1 via an AhR-dependent pathway (Amenya et al., 2016).
In humans, AhR is important for the basal regulation of CYP1A (Le et al., 2010), but how AhR activates the pCYP1A1 basal promoter activity is unclear. In our study, AhR was also important for the basal regulation of CYP1A1 in pigs. Intriguingly, we found that the DNA region that regulates pCYP1A1 expression is mainly located proximal to the transcription start site, whereas XRE in this region does not participate in the regulation of pCYP1A1. These data are inconsistent with previous studies. Therefore, we performed promoter strength analysis under the condition that AhR was regulated by its agonist or antagonist to further address the effect of AhR on the proximal promoter of pCYP1A1. β-Naphthoflavone has been reported to enhance gene expression by activating AhR, and resveratrol reduces gene expression by inhibiting AhR (Lee and Safe, 2001; Fujita and Mtetsuhashi, 2010). Indeed, β-naphthoflavone and resveratrol treatment of LLC-PK1 cells increased or decreased the expression of pCYP1A1 mRNA, suggesting that AhR binds to the promoter of pCYP1A1. Using the MatInspector program to analyze the 3.5-kb promoter DNA, 12 XREs were predicted to be in the promoter region of pCYP1A1, and 10 of them were located in the region of -819 to -2494 nt upstream of the transcription start site. However, when proximal promoter analysis was applied, it showed that neither activation nor deactivation of AhR showed significant effects on the activity of the pCYP1A1 proximal promoter. Our results demonstrate that AhR regulates the expression of pCYP1A1, but it is not involved in the regulation of the proximal promoter of pCYP1A1.
Considering only the regulation of this proximal promoter, the luciferase assay clearly suggested that only Sp1 transactivates the basal expression of pCYP1A1 rather than AhR. Sp1 participates in the expression of many CYP450 genes, such as CYP1A1, CYP1B1, CYP2J2, CYP3A29, and CYP3A46 (Zhang et al., 1998; Tsuchiya et al., 2003; Spiecker et al., 2004; Dong et al., 2015; Liu et al., 2016). The amino acid sequence for Sp1 in pigs and humans has more than 90% identity similarity, leading us to believe that Sp1 might have a conserved regulatory function in CYP1A1 expression. In humans, Sp1 activates the CYP1A1 promoter (Zhang et al., 1998), and phosphorylation of Sp1 regulates CYP1A1 expression (Shimoyama et al., 2014). We also found that the GC box at the proximal promoter of pCYP1A1 is the main cis-element that plays a regulatory role by recruiting Sp1.
CYP1A1 not only functions to metabolize exogenous compounds but also functions in the metabolism of endogenous chemicals, such as polyunsaturated fatty acids (Westphal et al., 2011; Chi et al., 2016). Comparing the pattern of CYP1A1 expression regulation in pigs and humans, we can see that Sp1 is very conservative in the regulation of CYP1A1. This shows the importance of Sp1 for pigs and humans in the gene regulation such as CYP1A1.
In summary, by analyzing the porcine CYP1A1 promoter activity, we found that the proximal promoter is very important for the regulation of pCYP1A1 expression. Two cis elements were found in the proximal promoter region: XRE and GC box with the binding of Sp1 and AhR, respectively. However, only Sp1 plays a transactivation role in the proximal promoter region of pCYP1A1. AhR also regulates the expression of pCYP1A1, but it does not function at the proximal promoter.
XX, JJ, WY, and RC conducted the experiments and data analysis. XX and JW wrote this article. YD and JW conceived the study.
This work was supported by the Natural Science Foundation of Guangdong Province (2015A030312005), the Science and Technology Program of Guangzhou (201804020067 and 201607010177), and the Project of Innovation Capacity Promotion for Higher Education in Guangdong Province (2017KCXTD001 and 2014GKXM020).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2018.00927/full#supplementary-material
Amenya, H. Z., Tohyama, C., and Ohsako, S. (2016). Dioxin induces Ahr-dependent robust DNA demethylation of the Cyp1a1 promoter via Tdg in the mouse liver. Sci. Rep. 6:34989. doi: 10.1038/srep34989
Bombail, V., Taylor, K., Gibson, G. G., and Plant, N. (2004). Role of Sp 1, C/EBP alpha, HNF 3, and PXR in the basal- and xenobiotic-mediated regulation of the CYP3A4 gene. Drug Metab. Dispos. 32, 525–535. doi: 10.1124/dmd.32.5.525
Chi, H. N., Brenner, S., Huttary, N., Atanasov, A. G., Dirsch, V. M., Chatuphonprasert, W., et al. (2016). AHR/CYP1A1 interplay triggers lymphatic barrier breaching in breast cancer spheroids by inducing 12(S)-HETE synthesis. Hum. Mol. Genet. 25, 5006–5016. doi: 10.1093/hmg/ddw329
Deniaud, E., Baguet, J., Chalard, R., Blanquier, B., Brinza, L., Meunier, J., et al. (2009). Overexpression of transcription factor Sp1 leads to gene expression perturbations and cell cycle inhibition. PLoS One 4:e7035. doi: 10.1371/journal.pone.0007035
Dong, L., Chen, Q., Liu, X., Wen, J., Jiang, J., and Deng, Y. (2015). Role of specificity protein 1, hepatocyte nuclear factor 1α, and pregnane x receptor in the basal and rifampicin-induced transcriptional regulation of porcine cytochrome p450 3a46. Drug Metab. Dispos. 43, 1458–1467. doi: 10.1124/dmd.115.065565
Fazili, I. S., Jiang, W., Wang, L., Felix, E. A., Khatlani, T., Coumoul, X., et al. (2010). Persistent induction of cytochrome p4501a1 in human hepatoma cells by 3-methylcholanthrene: evidence for sustained transcriptional activation of the CYP1A1 promoter. J. Pharmacol. Exp. Ther. 333, 99–109. doi: 10.1124/jpet.109.162222
Fujita, Y., and Mtetsuhashi, Y. (2010). beta-Naphthoflavone analogs as potent and soluble aryl hydrocarbon receptor agonists: improvement of solubility by disruption of molecular planarity. Bioorg. Med. Chem. 18, 1194–1203. doi: 10.1016/j.bmc.2009.12.036
Gray, M. A., and Squires, E. J. (2013). Effects of nuclear receptor transactivation on boar taint metabolism and gene expression in porcine hepatocytes. J. Steroid Biochem. Mol. Biol. 133, 110–119. doi: 10.1016/j.jsbmb.2012.09.025
Guengerich, F. P. (1988). Roles of cytochromes P-450 enzymes in chemical carcinogenesis and cancer chemotherapy. Cancer Res. 48, 2946–2954.
Guengerich, F. P., Brian, W. R., Iwasaki, M., Sari, M. A., Bäärnhielm, C., and Berntsson, P. (1991). Oxidation of dihydropyridine calcium channel blockers and analogs by human liver cytochrome P-450 IIIA4. J. Med. Chem. 34, 1838–1844. doi: 10.1021/jm00110a012
Hankinson, O. (1995). The aryl hydrocarbon receptor complex. Annu. Rev. Pharmocol. Toxicol. 35, 307–340. doi: 10.1146/annurev.pa.35.040195.001515
Hanlon, P. R., Zheng, W., Ko, A. Y., and Jefcoate, C. R. (2005). Identification of novel TCDD-regulated genes by microarray analysis. Toxicol. Appl. Pharmacol. 202, 215–228. doi: 10.1016/j.taap.2004.06.018
Hombachklonisch, S., Pocar, P., Kauffold, J., and Klonisch, T. (2006). Dioxin exerts anti-estrogenic actions in a novel dioxin-responsive telomerase-immortalized epithelial cell line of the porcine oviduct (TERT-OPEC). Toxicol. Sci. 2, 519–528. doi: 10.1093/toxsci/kfj102
Iwano, S., Saito, T., Takahashi, Y., Fujita, K., and Kamataki, T. (2001). Cooperative regulation of CYP3A5 gene transcription by NF-Y and Sp family members. Biochem. Biophys. Res. Commun. 286, 55–60. doi: 10.1006/bbrc.2001.5352
Kikuchi, H., Hossain, A., Yoshida, H., and Kobayashi, S. (1998). Induction of cytochrome P-450 1A1 by omeprazole in human HepG2 cells is protein tyrosine kinase-dependent and is not inhibited by alpha-naphthoflavone. Arch. Biochem. Biophys. 358, 351–358. doi: 10.1006/abbi.1998.0869
Kobayashi, A., Sogawa, K., and Fujii-Kuriyama, Y. (1996). Cooperative interaction between ahr.arnt and sp1 for the drug-inducible expression of CYP1A1 gene. J. Biol. Chem. 271, 12310–12316. doi: 10.1074/jbc.271.21.12310
Kojima, M., and Degawa, M. (2016). Sex differences in constitutive mRNA levels of CYP2B 22, CYP2C 33, CYP2C 49, CYP3A 22, CYP3A29 and CYP3A46 in the pig liver: comparison between meishan and landrace pigs. Drug Metab. Pharmacokinet. 31, 185–192. doi: 10.1016/j.dmpk.2016.02.00
Le, V. M., Jouan, E., and Fardel, O. (2010). Involvement of aryl hydrocarbon receptor in basal and 2,3,7,8-tetrachlorodibenzo-p-dioxin-induced expression of target genes in primary human hepatocytes. Toxicol. In vitro 24, 1775–1781. doi: 10.1016/j.tiv.2010.07.001
Lee, J. E., and Safe, S. (2001). Involvement of a post-transcriptional mechanism in the inhibition of CYP1A1 expression by resveratrol in breast cancer cells. Biochem. Pharmacol. 62, 1113–1124. doi: 10.1016/S0006-2952(01)00763-8
Liu, X., Wen, J., Chen, R., Zhang, T., Jiang, J., and Deng, Y. (2016). T-2 toxin induces the expression of porcine CYP3A22 via the upregulation of the transcription factor, NF-Y. Biochim. Biophys. Acta 1860, 2191–2201. doi: 10.1016/j.bbagen.2016.05.009
Ma, Q., and Lu, A. Y. H. (2007). CYP1A induction and human risk assessment: an evolving tale of in vitro and in vivo studies. Drug Metab. Dispos. 35, 1009–1016. doi: 10.1124/dmd.107.015826
Ma, Q., Renzelli, A. J., Baldwin, K. T., and Antonini, J. M. (2000). Superinduction of CYP1A1 gene expression. J. Biol. Chem. 275, 12676–12683. doi: 10.1074/jbc.275.17.12676
Monk, S. A., Denison, M. S., and Rice, R. H. (2003). Reversible stepwise negative regulation of CYP1A1 in cultured rat epidermal cells. Arch. Biochem. Biophys. 419, 158–169. doi: 10.1016/j.abb.2003.09.004
Saito, T., Takahashi, Y., Hashimoto, H., and Kamataki, T. (2001). Novel transcriptional regulation of the human CYP3A7 gene by Sp1 and Sp3 through nuclear factor kappa B-like element. J. Biol. Chem. 276, 38010–38022. doi: 10.1074/jbc.M106130200
Shimoyama, S., Kasai, S., Kahn-Perlès, B., and Kikuchi, H. (2014). Dephosphorylation of Sp1 at Ser-59 by protein phosphatase 2A (PP2A) is required for induction of CYP1A1 transcription after treatment with 2,3,7,8-tetrachlorodibenzo-p-dioxin or omeprazole. Biochim. Biophys. Acta 1839, 107–115. doi: 10.1016/j.bbagrm.2013.12.004
Spiecker, M., Darius, H., Hankeln, T., Soufi, M., Sattler, A. M., Schaefer, J. R., et al. (2004). Risk of coronary artery disease associated with polymorphism of the cytochrome P450 epoxygenase CYP2J2. Circulation. 110, 2132–2136. doi: 10.1161/01.cir.0000143832.91812.60
Tsuchiya, Y., Nakajima, M., and Yokoi, T. (2003). Critical enhancer region to which AhR/ARNT and Sp1 bind in the human CYP1B1 gene. J. Biochem. 133, 583–592. doi: 10.1093/jb/mvg075
Uno, S., Dalton, T. P., Sinclair, P. R., Gorman, N., Wang, B., Smith, A. G., et al. (2004). CYP1A1(-/-) male mice: protection against high-dose TCDD-induced lethality and wasting syndrome, and resistance to intrahepatocyte lipid accumulation and uroporphyria. Toxicol. Appl. Pharmacol. 196, 410–421. doi: 10.1016/j.taap.2004.01.014
Vecchini, F., Lenoirviale, M. C., Cathelineau, C., Magdalou, J., Bernard, B. A., and Shroot, B. (1994). Presence of a retinoid responsive element in the promoter region of the human cytochrome P4501A1 gene. Biochem. Biophys. Res. Commun. 201, 1205–1212. doi: 10.1006/bbrc.1994.1833
Wang, F., Wang, W., and Safe, S. (1999). Regulation of constitutive gene expression through interactions of Sp1 protein with the nuclear aryl hydrocarbon receptor complex. Biochemistry 38, 11490–11500. doi: 10.1021/bi982578f
Westphal, C., Konkel, A., and Schunck, W. H. (2011). CYP-eicosanoids—A new link between omega-3 fatty acids and cardiac disease? Prostaglandins Other Lipid Mediat. 96, 99–108. doi: 10.1016/j.prostaglandins.2011.09.001
Whitelaw, M., Pongratz, I., Wilhelmsson, A., Gustafsson, J. A., and Poellinger, L. (1993). Ligand-dependent recruitment of the Arnt coregulator determines DNA recognition by the dioxin receptor. Mol. Cell. Biol. 4, 2504–2514. doi: 10.1128/MCB.13.4.2504
Wierstra, I. (2008). Sp1: emerging roles–beyond constitutive activation of TATA-less housekeeping genes. Biochem. Biophys. Res. Commun. 372, 1–13. doi: 10.1016/j.bbrc.2008.03.074
Xie, S., Junaid, M., Bian, W., Luo, J., Syed, J. H., Wang, C., et al. (2018). Generation and application of a novel transgenic zebrafish line Tg(cyp1a:mCherry) as an in vivo assay to sensitively monitor PAHs and TCDD in the environment. J. Hazard. Mater. 344, 723–732. doi: 10.1016/j.jhazmat.2017.11.021
Xu, L., Li, A. P., Kaminski, D. L., and Ruh, M. F. (2000). 2,3,7,8 Tetrachlorodibenzo- p -dioxin induction of cytochrome P4501A in cultured rat and human hepatocytes. Chem. Biol. Interact. 124, 173–189. doi: 10.1016/S0009-2797(99)00149-0
Keywords: porcine, CYP1A1, Sp1, AhR, basal transactivation
Citation: Xie X, Jiang J, Ye W, Chen R, Deng Y and Wen J (2018) Sp1, Instead of AhR, Regulates the Basal Transcription of Porcine CYP1A1 at the Proximal Promoter. Front. Pharmacol. 9:927. doi: 10.3389/fphar.2018.00927
Received: 27 May 2018; Accepted: 30 July 2018;
Published: 17 August 2018.
Edited by:
Petr Pavek, Charles University, CzechiaReviewed by:
Su-Jun Lee, Inje University, South KoreaCopyright © 2018 Xie, Jiang, Ye, Chen, Deng and Wen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yiqun Deng, eXFkZW5nQHNjYXUuZWR1LmNu Jikai Wen, amt3ZW5Ac2NhdS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.