
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Pharmacol. , 05 July 2018
Sec. Experimental Pharmacology and Drug Discovery
Volume 9 - 2018 | https://doi.org/10.3389/fphar.2018.00706
 Frank T. Stümpel1*†
Frank T. Stümpel1*† Juliane Stein1†
Juliane Stein1† Kirsten Himmler1
Kirsten Himmler1 Beatrix Scholz1
Beatrix Scholz1 Matthias D. Seidl1
Matthias D. Seidl1 Boris V. Skryabin2
Boris V. Skryabin2 Frank U. Müller1
Frank U. Müller1Background: Atrial fibrillation (AF) is a significant cause of morbidity and mortality with foreseeably increasing prevalence. While large animal models of the disease are well established but resource intensive, transgenic AF mouse models are not yet widely used to develop or validate novel therapeutics for AF. Hemizygous mice with a cardiomyocyte-specific overexpression of the human cAMP response element modulator (CREM) isoform IbΔC-X spontaneously develop AF on grounds of an arrhythmogenic substrate consisting of alterations in structure, conduction, and calcium handling.
Objective: We investigated if homozygous expression of the CREM-IbΔC-X transgene in mice alters the time course of AF development, and if homozygous CREM-IbΔC-X transgenics could be suitable as a disease model of AF.
Methods: Southern Blot, quantitative real-time PCR, and immunoblotting were used to identify and verify homozygous transgenics. Cardiac gravimetry, quantitative real-time RT-PCR, histology, survival analysis, and repeated ECG recordings allowed assessment of phenotypic development and effects of antiarrhythmic drugs.
Results: Homozygous animals could be identified by Southern blot and quantitative PCR, showing a strong trend to increased transgenic protein expression. In homozygous animals, atrial hypertrophy appeared earlier and more pronounced than in hemizygous animals, going along with an earlier onset of spontaneous AF, while no increased early mortality was observed. Application of a rate-controlling drug (esmolol) led to the expected result of a decreased heart rate. Application of a rhythm-controlling drug (flecainide) showed effects on heart rate variability, but did not lead to a definitive conversion to sinus rhythm.
Conclusion: We suggest homozygous CREM-IbΔC-X overexpressing mice as a reliable model of early onset, rapidly progressive AF.
In most developed countries, atrial fibrillation (AF) is a significant cause of morbidity and mortality, often due to its thromboembolic complications (Wolf et al., 1998). Because the prevalence of this disease rises with age its importance as therapeutic challenge is expected to increase further (Chugh et al., 2014). Presently, pharmacological antiarrhythmic treatment options are limited to drugs with significant adverse effects and contraindications. If catheter ablation or surgical options are unavailable, not feasible, or unsuccessful, most patients will ultimately remain in permanent AF and need lifelong antithrombotic therapy, which itself carries risks and remains a balancing act between embolism and bleeding (Gallagher et al., 2014). To gain insights into the mechanisms of AF pathophysiology, as well as to develop and test new antiarrhythmic substances to treat AF, reliable animal models are needed. To date, most AF models used are large animal models (dogs, goats, and pigs) which had their AF caused by intervention (tachypacing, artificial mitral insufficiency, papillary muscle ischemia, etc.) (Nishida et al., 2010). Thus, these models need considerable resources in animal preparation and care.
More recently, several genetic mouse models exhibiting spontaneous AF have been described (Riley et al., 2012). However, many of these models show rapidly progressive phenotypes with ventricular involvement, leading to increased early mortality, while others develop AF only late in the process or with incomplete penetrance.
CREM-IbΔC-X belongs to the CREB/CREM/ATF group of cAMP-dependent transcription factors, which serve important functions in connecting cAMP-dependent signaling pathways to transcriptional activity of the cell (Müller et al., 2000; Kehat et al., 2006a; Altarejos and Montminy, 2011). CREM-IbΔC-X itself is a repressor isoform, initially identified in failing human hearts (Müller et al., 1998). Mice with a heart-directed overexpression of CREM-IbΔC-X revealed a striking phenotype with progressive atrial dilation and development of spontaneous AF (Müller et al., 2005). This phenotype could, at least in part, be attributed to the formation of an arrhythmogenic substrate, consisting of structural alterations, impaired conduction, and increased Ca2+ leak from the sarcoplasmic reticulum (Chelu et al., 2009; Kirchhof et al., 2013; Li et al., 2014; Seidl et al., 2017; Bukowska et al., 2018).
In this study we aimed to identify a possible relationship between the gene dose of CREM-IbΔC-X expression in the heart and the strength of the developed cardiac phenotype, as well as to evaluate a breeding line homozygous for CREM-IbΔC-X as a possible disease model of AF, e.g., for testing antiarrhythmic drugs.
Hemizygous animals transgenic for the HA-tagged CREM-IbΔC-X cDNA under control of the αMHC promoter were created as previously described. All animals used here were offspring of founder line Tg1 (Müller et al., 2005). To obtain animals homozygous for the transgene, two hemizygous transgenic animals were mated, and homozygous transgenics were identified by Southern Blot or quantitative real-time PCR. A breeding colony of homozygous transgenics was then established. Homozygous transgenics (CREMT/T) were compared with age-matched hemizygous (CREMT/W) and wild-type (WT) animals. All experiments were performed conforming to EU and national animal protection laws and regulations and as approved by the responsible authorities of the state of North Rhine-Westphalia.
Approximately 5 μg of genomic DNA purified from (tail) tissue samples was digested with restriction endonuclease EcoRV (Thermo Scientific Fermentas, Waltham, MA, United States), fractionated on 0.8% agarose gels, and transferred to GeneScreen nylon membranes (PerkinElmer, Waltham, MA, United States). The membranes were hybridized with a 32P-labeled 2.2 kb probe containing sequences PCR-amplified from the CREM-IbΔC-X transgene, using primers 5′-TAGCCCACACCAGAAATGACAGAC-3′ (forward) and 5′-GTGGTGTGACATAATTGGACAAACTAC-3′ (reverse), and washed with (final concentrations) 0.5x SSPE (1x SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.7]) and 0.5% (w/v) sodium dodecyl sulfate at 65°C.
Purified genomic DNA was used as template. Real-time qPCR was performed in a LightCycler carousel-type device (Roche, Mannheim, Germany) using the LightCycler FastStart DNA Master SYBR Green I kit (Roche). Primers used to identify CREM-BHIb-cDNA were (forward) 5′-CCAAAATTCAATCCCTGCTT-3′ and (reverse) 5′-TCGACATTCTTTGGCAGCTT-3′. Primers were exon-spanning to exclude the amplification of wild-type CREM DNA containing introns. Quantification was performed using LightCycler software v. 3.5 (Roche), and statistical analysis was done using REST Software v. 2009 (M. Pfaffl, TU Munich, Germany and Qiagen, Hilden, Germany).
Total quantification of mRNA levels was performed as previously described (Seidl et al., 2017) with following modifications: Briefly, RNA was extracted from both atria using TRIzol® (Thermo Fisher) and Direct-zolTM RNA MiniPrep (Zymo Research, Irvine, CA, United States) according to the manufacturer’s instructions. Samples of 0.9 μg RNA were randomly reversely transcribed to cDNA using Transcriptor First Strand cDNA synthesis kit (Roche) and quantitative real time RT-PCR was carried out using a LightCycler 480 System (Roche). Relative quantification was performed by calculating relative expression ratios using the ΔΔCt method and the relative expression software tool (REST Software v. 2009). Hypoxanthine-guanine phosphoribosyltransferase (Hprt) was used as a reference gene. Primers used were: Akap9 (A-kinase anchor protein 9) fwd 5′-AAGGAATGCGAGACCCTGAA-3′, rev 5′-GACAGCCTTCACTAACCCCT-3′, Ank2 (ankyrin 2, brain) fwd 5′-AGGTGGTCAGATTGATGCCA-3′, rev 5′-GCCTTGTATTGGAGCAGGTG-3′, Cacna1c (calcium channel, voltage-dependent, L type, alpha 1C subunit [Cav1.2]) fwd 5′-GCTCTCTTCACCGTCTCCAC-3′, rev 5′-GACGAAACCCACGAAGATGT-3′, Cav3 (caveolin 3) fwd 5′-CAGCCTCACAATGATGACCG-3′, rev 5′-TTCCATACACCGTCGAAGCT-3′, Cxadr (coxsackie virus and adenovirus receptor) fwd 5′-ATGGCTGATATCCCCGTCTG-3′, rev 5′-CTCTTCCGATCCATCCACGA-3′, Fxyd1 (FXYD domain-containing ion transport regulator 1) fwd 5′-GAACCGGATCCATTCACCTA-3′, rev 5′-TTCCCCAGTTCTCTGCTGTT-3′, Gja5 (gap junction protein, alpha 5 [connexin 40]) fwd 5′-AGAGCCTGAAGAAGCCAACT-3′, rev 5′-GGCGTGGACACAAAGATGA-3′, Kcnd2 (potassium voltage-gated channel, Shal-related family, member 2 [Kv4.2]) fwd 5′-CTGCTCACGGAGACACAAAA-3′, rev 5′-CGGCTGTTGGATAGTGGAGT-3′,Kcne1 (potassium voltage-gated channel, Isk-related subfamily, member 1) fwd 5′-TTCTTCACCCTGGGCATCAT-3′, rev 5′-CCGCCTGGTTTTCAATGACA-3′, Kcnv2 (potassium channel, subfamily V, member 2 [Kv11.1]) fwd 5′-TCTCTGCAGCTGTCTACTCG-3′, rev 5′-AATAATGCCAAAAGCGATGC-3′, Mef2c (myocyte enhancer factor 2C) fwd 5′-ACCAGGACAAGGAATGGGAG-3′, rev 5′-GGCGGCATGTTATGTAGGTG-3′, Myl3 (myosin, light polypeptide 3) fwd 5′-CCGAGTTTGATGCCTCCAAG-3′, rev 5′-CTTCCTGTTTTGGCTTCCCC-3′, Scn5a (sodium channel, voltage-gated, type V, alpha [Nav1.5]) fwd 5′-TACCGCATAGTGGAGCACAG-3′, rev 5′-ATCTCGGCAAAGCCTAAGGT-3′.
Heart tissue was homogenized in a buffer containing NaHCO3 and SDS as previously described (Rockman et al., 1996). Proteins were electrophoresed under reducing conditions in SDS-PA gels in Hoefer SE 600 chambers and blotted to nitrocellulose membranes (GE Healthcare, Little Chalfont, United Kingdom). HA-tagged CREM isoforms were detected using an anti-HA-antibody (1:1000; Cell Signaling, Danvers, MA, United States), an HRP-conjugated anti-rabbit-IgG 2nd antibody (1:5000; GE Healthcare), and the enhanced chemiluminescence (ECL) system (Promega, Madison, WI, United States). Images were acquired and quantitatively analyzed using a ChemiDoc MP system and Image Lab software (Bio-Rad, Hercules, CA, United States).
Masson’s trichrome staining was performed on 5 μm sections of paraffin embedded hearts for estimation of interstitial fibrosis as previously described (Bukowska et al., 2018). Whole heart sections were imaged by light microscopy using NIS-Elements software (Nikon, Tokyo, Japan) and fibrotic areas within the ventricle (excluding the cardiac skeleton and the valves) were detected and quantified using Image-Pro Plus software (Media Cybernetics Inc., Rockville, MD, United States).
Mice were anesthetized with isoflurane (∼1.2% v/v) and nitrous oxide (∼66% v/v) while being placed on a warmed pad in supine position. Needle electrodes were attached to obtain limb leads (Einthoven) and augmented limb leads (Goldberger). ECGs were recorded and analyzed electronically using PowerLab hardware and LabChart Pro software (ADInstruments, Bella Vista, Australia). Where required by the experimental protocol, a tube catheter was inserted into the left external jugular vein and drugs, i.e., the β1-adrenoceptor antagonist (class II antiarrhythmic) esmolol (Baxter, Unterschleißheim, Germany) 10 mg/kg bw or the Na+-channel blocker (class Ic antiarrhythmic) flecainide (MEDA Pharma, Bad Homburg, Germany) 5 mg/kg bw, were administered intravenously using an automated syringe pump (B. Braun, Melsungen, Germany). After invasive procedures, animals were euthanized by CO2 inhalation without having regained consciousness. Hearts were excised, photographed, weighed, and stored at -80°C until further analysis.
Animals were monitored weekly by ECG for the occurrence of atrial fibrillation. The first occurrence of AF was noted as event. Animals that could not be followed further were censored. For survival analysis, only deaths from natural causes were counted as events. Survival without AF and overall survival time were estimated using the Kaplan–Meier estimator and compared using the log-rank test.
Data are given as mean ± standard error of the mean (SEM), geometric mean and 95% confidence interval, or median and interquartile range, depending on the type of data, as specified in the “Results” section. “n” indicates the number of independent experiments. Two groups of normally distributed data were compared by student’s t-test. More than two groups of normally distributed data were compared by one-way ANOVA, with post hoc testing done by the Holm–Sidak method. Two groups of data from repeated measurements were compared using the paired t-test. More than two groups of data from repeated measurements were compared by one-way RM ANOVA. Groups of non-normally distributed data were compared by Kruskal–Wallis ANOVA on Ranks, with post hoc testing done by Dunn’s method. Groups of time-to-event data (like survival) were compared using the Kaplan–Meier estimator and the log-rank test, with post hoc testing done by the Holm–Sidak method. Gene expression data were compared using the ΔΔCt randomization method. The SigmaStat package (Systat Software Inc., San Jose, CA, United States) was used to perform all statistical analyses, except for the ΔΔCt randomization method, which was done using REST software (see above).
To identify possible homozygous transgenic animals among the offspring of two hemizygous transgenic parents, genomic tail-tip DNA was digested with the restriction endonuclease EcoRV and hybridized to a 2.2 kb probe containing the ORF of the human CREM-IbΔC-X cDNA, obtained by PCR from the transgenic construct (Figure 1). As the introduced transgene (7868 b) does not contain EcoRV restriction sites, hybridization to DNA fragments larger than ∼8 kb was expected. Indeed, samples of known transgenic animals yielded signals of more than 15 kb size, probably due to multiple consecutive insertions of the transgene. Two of the transgenic animal samples showed much stronger signals than the others, identifying these animals as homozygous transgenics.
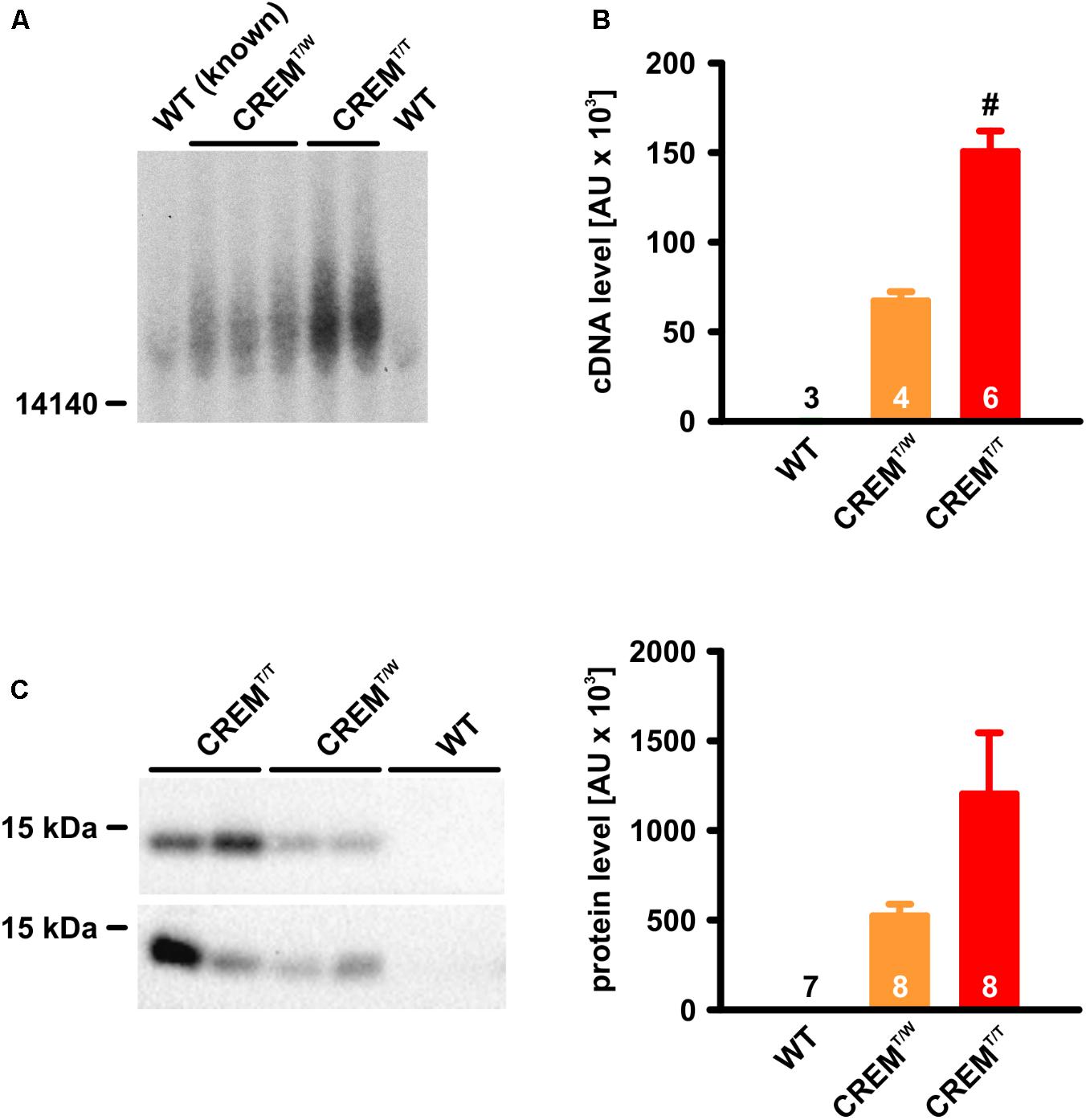
FIGURE 1. Identification and verification of CREMT/T animals. (A) Southern Blot. Note that two of the positive samples yielded much stronger signals than the others. These samples were identified as CREMT/T. (B) Quantitative real-time PCR from genomic DNA. Note that, while the transgenic sequence was virtually undetectable in WT, its detection in CREMT/T was about twice as high as in CREMT/W. (#p < 0.05 vs. CREMT/W). (C) Detection of HA-tagged CREM-IbΔC-X protein expression by western blot. Note that CREMT/T hearts expressed the transgenic product about twice as highly as CREMT/W hearts (CREMT/T vs. CREMT/W p = 0.09). All quantified data are given as mean ± SEM.
(In wild-type animals a much weaker signal was visible, probably due to hybridization of the probe to mouse CREM exons H or Ib [loci chr18:3273422-3273581 and chr18:3267589-3267732, GRCm38/mm10 mouse genome assembly], which are more than 90% homologous with the human sequence. In EcoRV-digested genomic mouse DNA, these loci are expected to be part of a 35828 b fragment).
As a second means of verification of CREMT/T animals, quantitative real-time PCR was employed, using genomic DNA as template. Primers were binding over exon boundaries within the transgene sequence, thus being specific to CREM-IbΔC-X cDNA lacking introns. Calculated transgene DNA content in CREMT/T samples (150600 ± 11365 arbitrary units, AU, n = 6) was about twice as high as in CREMT/W samples (67280 ± 5061 AU, p < 0.05, n = 4, Student’s t-test). In WT samples, the transgene sequence was virtually undetectable (3 ± 1 AU, n = 3).
As a further control of homozygosity, homozygous transgenic animals identified by one of these means were mated with wild-types, which, without exception, only led to transgenic progeny in 13 litters.
To investigate if the homozygous existence of the transgene goes along with an increased expression of its protein products, ventricle homogenates were electrophoresed in SDS-PA-gels and blotted to nitrocellulose membranes, and CREM proteins being translated from the transgene were immunologically detected by their HA-tags (Figure 1). As expected from earlier experiments, a 14 kDa-protein was detected in transgenic samples, putatively corresponding to HIbII (Müller et al., 2005). While no expression was detected in WT (n = 7), expression of this protein in CREMT/T (1203191 ± 340900 AU, n = 8) was about twice as high as in CREMT/W (524054 ± 64820 AU, n = 8, p = 0.09, Student’s t-test).
Overexpression of CREM-IbΔC-X protein products leads to marked atrial hypertrophy (Müller et al., 2005). To assess if the increased transgene expression in homozygous transgenics alters time course and level of hypertrophy, hearts of WT, CREMT/W, and CREMT/T animals were photographed and weighed at three different ages (7, 14, and 26 weeks), and atrial and ventricular weights relative to body weight were calculated (Figure 2). At 7 weeks of age already, relative atrial weights of CREMT/T (0.44 [0.40–0.50] mg/g, median and interquartile range, n = 15) were significantly (p < 0.05, ANOVA on Ranks) increased when compared to CREMT/W (0.25 [0.25–0.26] mg/g, n = 3) or WT (0.27 [0.26–0.31] mg/g, n = 5), while the latter two groups were not different from each other. At 14 and 26 weeks of age, significant (p < 0.05 vs. WT) atrial hypertrophy had developed also in CREMT/W, but CREMT/T showed much more distinct atrial hypertrophy, being significantly increased as compared to both former groups (14 weeks: WT 0.22 [0.21–0.25] mg/g, n = 11, CREMT/W 0.34 [0.24–0.54] mg/g, n = 29, CREMT/T 0.6 [0.48–1.02] mg/g, n = 33; 26 weeks: WT 0.24 [0.22–0.25] mg/g, n = 19, CREMT/W 0.51 [0.27–0.93] mg/g, n = 29, CREMT/T 0.77 [0.57–3.43] mg/g, n = 41). In addition to atrial hypertrophy, CREMT/T also showed significant ventricular hypertrophy, being detectable at 14 and 26 weeks of age, while no significant differences were detected between CREMT/W and WT (7 weeks: WT 3.99 [3.91–4.32] mg/g, n = 5, CREMT/W 3.67 [3.65–3.91] mg/g, n = 3, CREMT/T 4.58 [3.84–4.66] mg/g, n = 15; 14 weeks: WT 3.63 [3.52–3.82] mg/g, n = 11, CREMT/W 3.57 [3.36–4.03] mg/g, n = 29, CREMT/T 4.13 [3.83–4.71] mg/g, n = 33; 26 weeks: WT 3.57 [3.45–3.67] mg/g, n = 19, CREMT/W 3.3 [3.17–3.54] mg/g, n = 29, CREMT/T 4.42 [4.05–4.89] mg/g, n = 41). Body weights were unchanged (1-way ANOVA) between groups at all three ages (7 weeks: WT 27.26 ± 2.22 g, mean ± SEM, n = 5, CREMT/W 24.47 ± 2.28 g, n = 3, CREMT/T 22.91 ± 0.96 g, n = 15; 14 weeks: WT 30.31 ± 1.26 g, n = 11, CREMT/W 29.59 ± 0.87 g, n = 29, CREMT/T 29.88 ± 0.86 g, n = 33; 26 weeks: WT 38.2 ± 1.07 g, n = 19, CREMT/W 35.97 ± 1.09 g, n = 29, CREMT/T 35.17 ± 0.92 g, n = 41). Summing up, in CREMT/T atrial hypertrophy appeared earlier and much more pronounced than in CREMT/W. Ventricular hypertrophy was also detected in CREMT/T, while being absent in CREMT/W and WT.
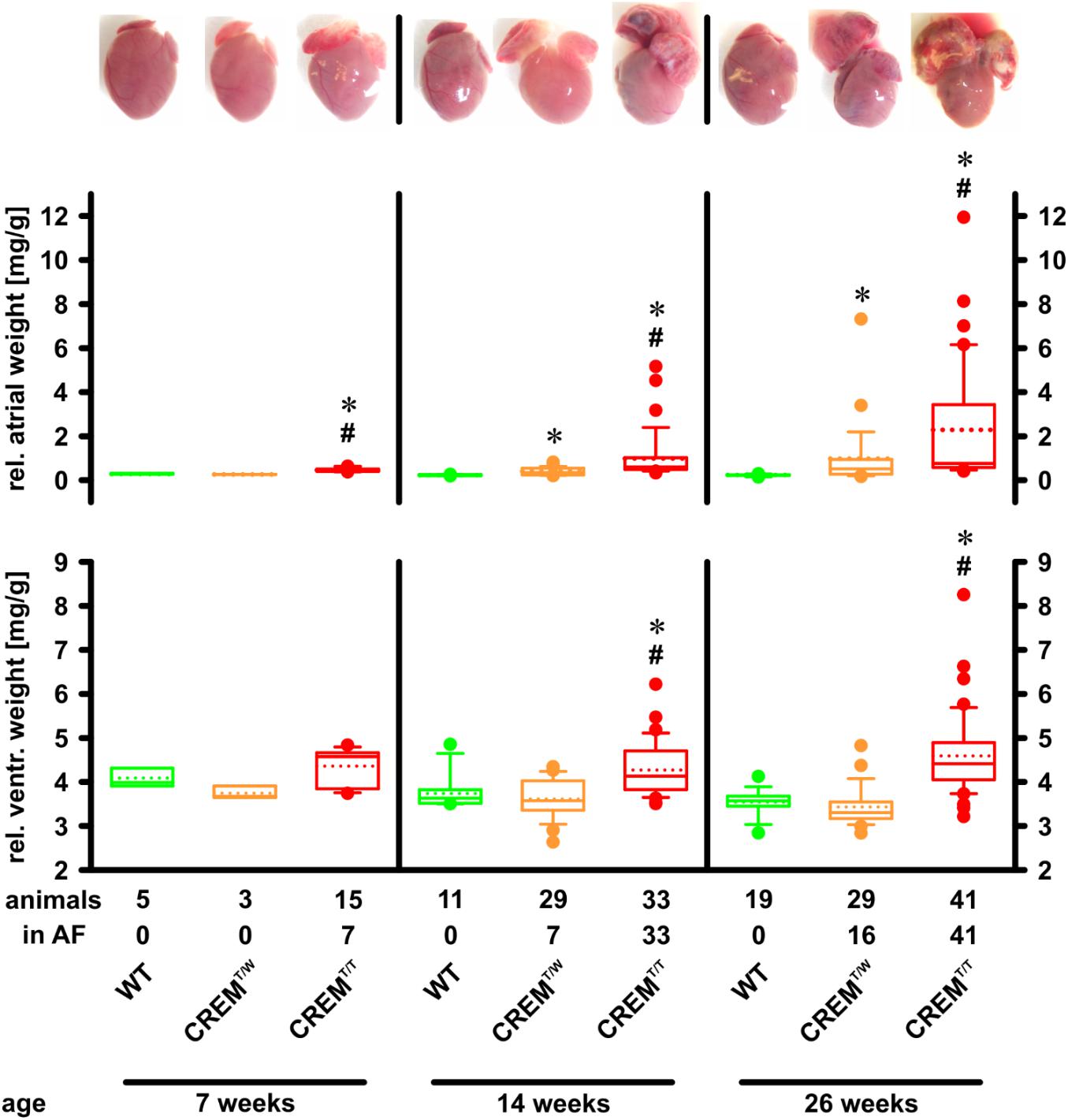
FIGURE 2. Development of atrial and ventricular hypertrophy. Hearts were excised, rinsed, and photographed, and atria and ventricles were weighed. Atrial and ventricular weights are given relative to body weight. Note that atrial hypertrophy developed earlier and was more profound in CREMT/T than in CREMT/W. Further note that in addition to the earlier onset of atrial hypertrophy in CREMT/T at 7 weeks of age, AF had also appeared already at this age, while in CREMT/W it was not yet detected. CREMT/T also developed ventricular hypertrophy, whereas CREMT/W did not. ∗p < 0.05 vs. WT; #p < 0.05 vs. CREMT/W. Boxes represent median and interquartile range; whiskers show the 10th and 90th percentiles; the dotted line is the mean.
Mice overexpressing CREM-IbΔC-X develop spontaneous atrial fibrillation (Müller et al., 2005). A cohort of animals (the same as used to assess atrial and ventricular hypertrophy, with a total of 85 CREMT/T, 65 CREMT/W, and 35 WT) were followed by weekly ECG to identify the onset of AF. We also noted spontaneous deaths in the colony (with a total of 197 CREMT/T, 100 CREMT/W, and 52 WT included into analysis) to estimate overall survival (Figure 3). In CREMT/W, more than 50% had developed AF by the age of 22 weeks (p < 0.05 vs. WT, log-rank test). In CREMT/T, more than 50% had developed AF already by the age of 6 weeks, and all animals had developed AF by the age of 10 weeks (p < 0.05 vs. CREMT/W and WT). No appearances of AF were noted in WT. Overall survival of all three groups was comparable.
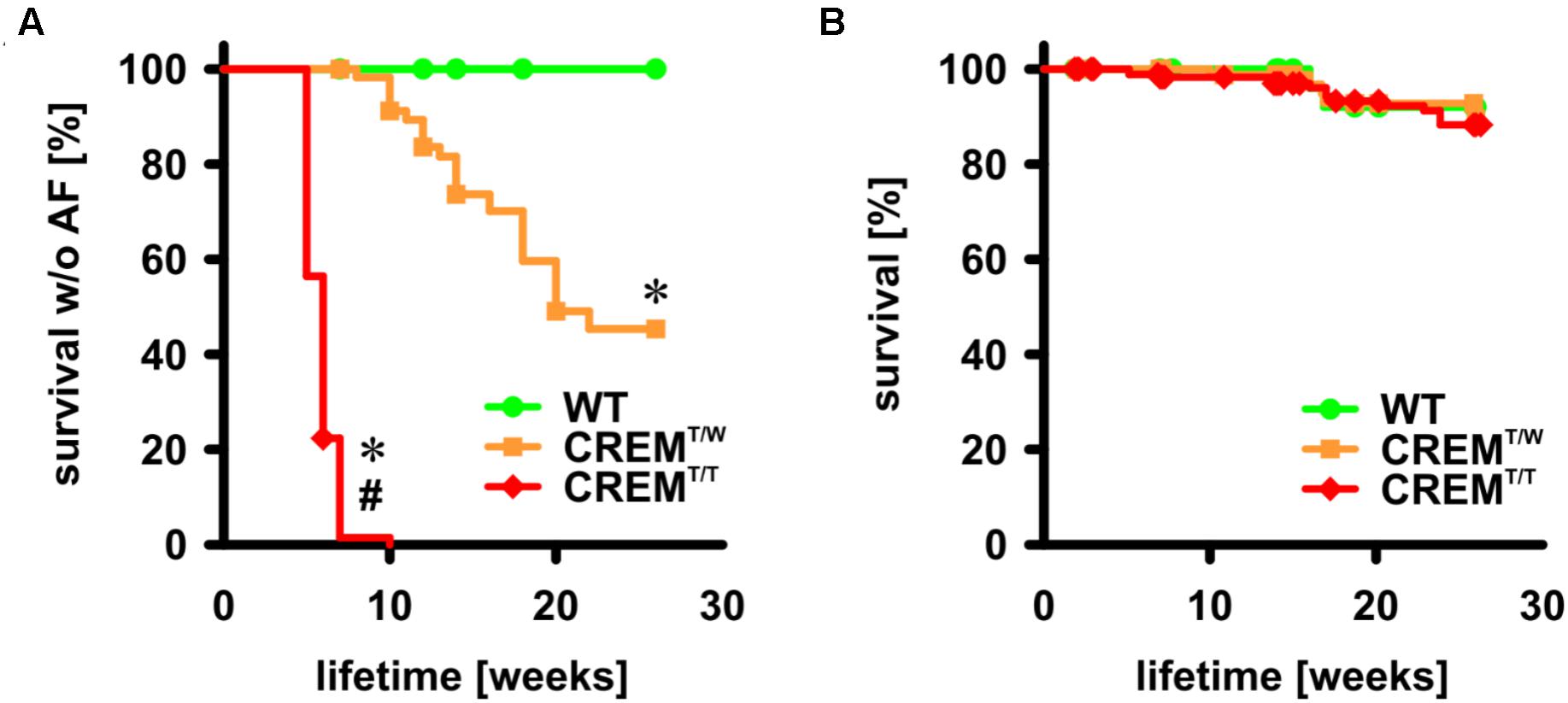
FIGURE 3. Onset of AF and overall survival up to the age of 26 weeks. (A) Animals were followed by once-weekly ECG and the first appearance of spontaneous AF was recorded as event. Animals not available for further examination were censored. Note that all CREMT/T had developed AF by the age of 10 weeks, more than 50% of CREMT/W had developed AF by the age of 26 weeks, and no WT developed AF up to the age of 26 weeks. WT: n = 35, 100% censored; CREMT/W: n = 65, 66% censored; CREMT/T: n = 85, 4% censored ∗p < 0.05 vs. WT, #p < 0.05 vs. CREMT/W. (B) Deaths in the colony from natural causes were recorded as events. Note that there was no difference in survival between groups up to the age of 26 weeks. WT: n = 52, 96% censored, CREMT/W: n = 100, 95% censored, CREMT/T: n = 197, 93% censored.
In atria of hemizygous transgenics, going along with the transition to hypertrophy and AF, a complex pattern and time course of gene regulation changes was found (Seidl et al., 2017). To get a first notion of the effects of the increased transgene expression in homozygous transgenics on gene regulation, mRNA levels of selected genes linked to AF development and identified in the hemizygous model were measured. RNA was extracted from atria of 14 weeks old WT (n = 8), CREMT/W (n = 8), and CREMT/T animals (n = 6), and gene expression was quantified by quantitative real-time RT-PCR relative to the expression of Hprt as reference (Figure 4). Of many of the investigated genes, expression was similar in CREMT/W and CREMT/T, in that there was either no significant difference from WT in both groups (Ank2 WT 1 [0.55–1.84], expression relative to WT and 95% confidence interval, CREMT/W 0.77 [0.35–1.66], CREMT/T 0.79 [0.48–1.42], Cav3 WT 1 [0.41–2.44], CREMT/W 0.73 [0.26–1.54], CREMT/T 0.77 [0.3–1.64], Fxyd1 WT 1 [0.32–3.15], CREMT/W 0.6 [0.21–1.61], CREMT/T 0.57 [0.27–1.07]) or both CREMT/W and CREMT/T were significantly (p < 0.05, ΔΔCt randomization test) different from WT without differences between the former groups (Gja5 WT 1 [0.3–3.36], CREMT/W 0.06 [0.02–0.2], CREMT/T 0.07 [0.03–0.21], Kcnv2 WT 1 [0.56–1.79], CREMT/W 0.03 [0.02–0.06], CREMT/T 0.05 [0.03–0.1], Scn5a WT 1 [0.3–3.34], CREMT/W 0.21 [0.08–0.61], CREMT/T 0.23 [0.09–0.62]). However, several genes were found to be regulated differently between CREMT/W and CREMT/T, insofar as only CREMT/T (and not CREMT/W) was significantly different from WT (Cacna1c WT 1 [0.67–1.5], CREMT/W 0.88 [0.51–1.6], CREMT/T 1.37 [1.01–1.84]) or significant differences were found between CREMT/W and CREMT/T (Akap9 WT 1 [0.57–1.76], CREMT/W 0.7 [0.43–1.21], CREMT/T 1.32 [0.81–2.19], Cxadr WT 1 [0.61–1.64], CREMT/W 0.83 [0.52–1.28], CREMT/T 1.38 [0.81–2.51], Kcnd2 WT 1 [0.72–1.38], CREMT/W 0.43 [0.29–0.64], CREMT/T 0.63 [0.45–0.86], Kcne1 WT 1 [0.11–9.37], CREMT/W 3.45 [0.49–22.24], CREMT/T 11.98 [1.92–68.11], Mef2c WT 1 [0.54–1.84], CREMT/W 0.96 [0.53–1.75], CREMT/T 2.37 [1.48–3.76], Myl3 WT 1 [0.14–7.43], CREMT/W 1.22 [0.21–10.74], CREMT/T 0.26 [0.04–0.96]).
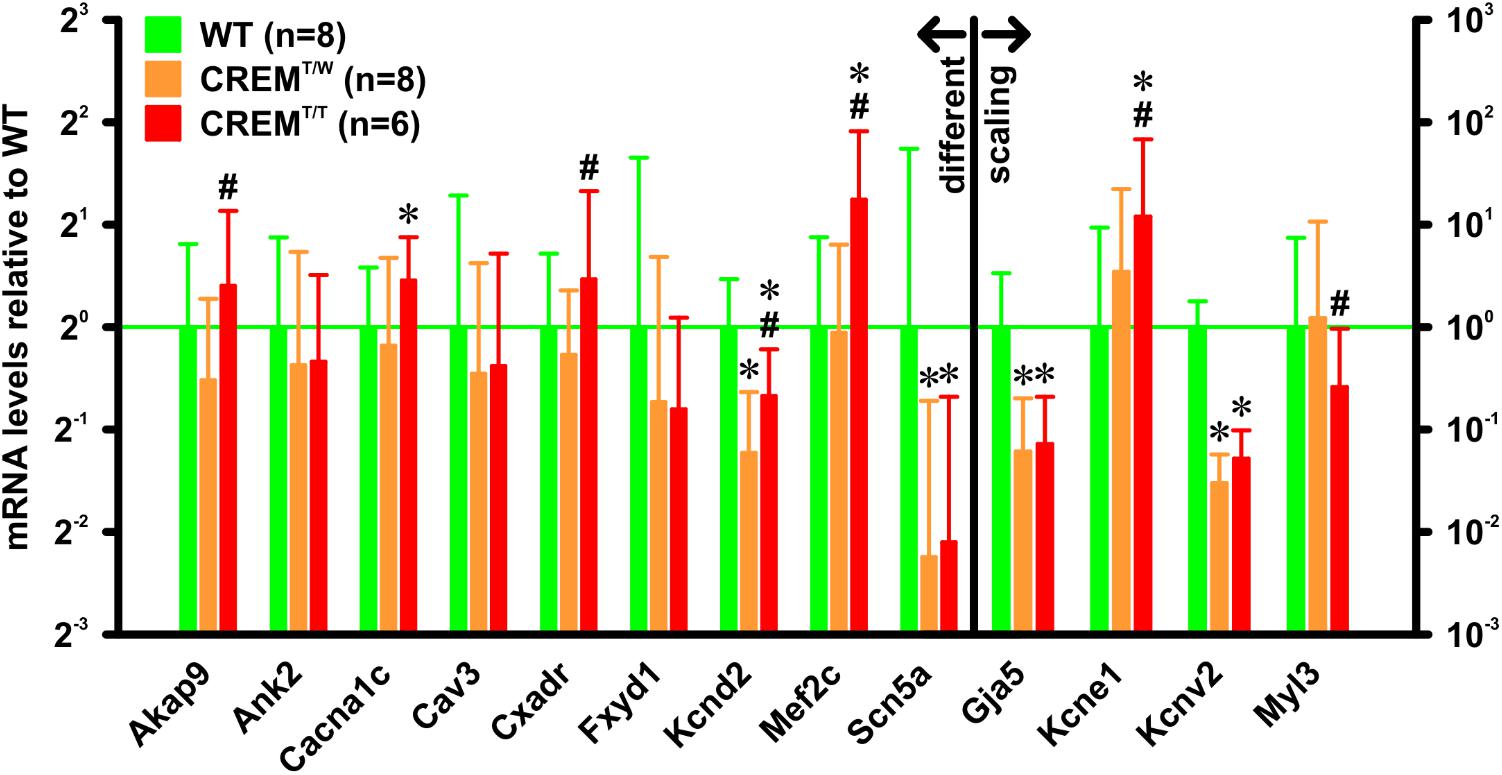
FIGURE 4. mRNA expression levels of selected genes. RNA was extracted from atria from 14 weeks old animals. Gene expression was quantified by quantitative real-time RT-PCR relative to Hprt as a reference gene. All expression levels are given relative to WT. Note that several genes were differently expressed in CREMT/T vs. CREMT/W. ∗p < 0.05 vs. WT; #p < 0.05 vs. CREMT/W. Data given as (geometric) mean and 95% confidence interval.
To assess if homozygous transgenics develop an increased level of ventricular fibrosis, fibrosis (interstitial and perivascular) was measured in Masson’s trichrome stained histologic sections from hearts of 14 weeks old CREMT/T and CREMT/W (three animals per group) by quantifying the blue area (i.e., collagen) within the total ventricular area excl. the cardiac skeleton and the valves (Figure 5). Collagen content of both CREMT/T and CREMT/W was found to be comparable to that of one 14 weeks old WT animal, which was measured for reference, and not significantly different from each other (WT 1.11%, n = 1, CREMT/W 0.9 ± 0.17%, mean ± SEM, n = 3, CREMT/T 1.3 ± 0.15%, n = 3).
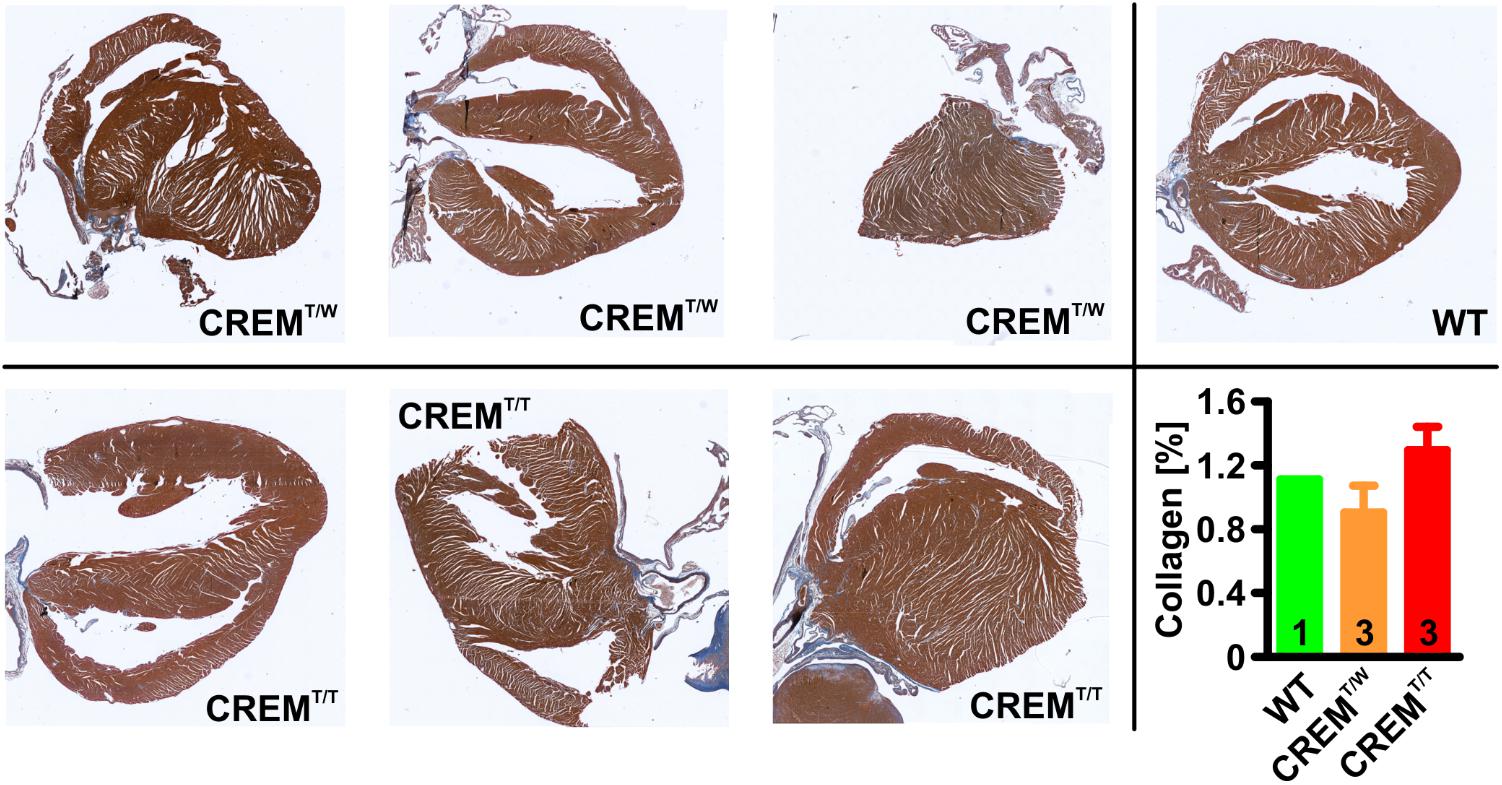
FIGURE 5. Quantification of ventricular fibrosis. 5 μm sections from hearts of 14 weeks old animals were stained with Masson’s trichrome, and interstitial and perivascular fibrosis in ventricles was measured by quantifying the ratio of the blue (i.e., collagen) areas relative to total areas of the ventricles (excl. the cardiac skeleton and the valves). Note that collagen content of both CREMT/T and CREMT/W was found to be comparable to that of one 14 weeks old WT animal, which was measured for reference, and not significantly different from each other. Data given as mean ± SEM.
To examine the potential of the homozygous transgenic model for testing drugs commonly employed in the treatment of human AF, we treated 11 weeks old CREMT/T animals with the β1-adrenoceptor antagonist esmolol (Vaughan Williams class II, 10 mg/kg) and 6–7 weeks old CREMT/T animals with the Na+-channel blocker flecainide (Vaughan Williams class Ic, 5 mg/kg) while simultaneously recording ECGs (Figure 6). During and shortly (2 min) after esmolol infusion, heart rates were reduced (before infusion 435 ± 19 min-1 [mean ± SEM], during inf. 394 ± 24 min-1, after inf. 382 ± 17 min-1, n = 8, p < 0.05, RM ANOVA), while heart rate variability, measured as coefficient of variation of RR internals (CV, i.e., ratio of SD to mean) increased (before infusion 23 ± 2%, during inf. 38 ± 3%, after inf. 33 ± 2%, p < 0.05). Before, during, and after esmolol infusion, all examined animals were (and remained) in AF. To assess effects of flecainide, 90 min of ECG recordings immediately after flecainide administration were compared to (1–5 days) previously recorded control ECGs from the same animals. Over the course of 90 min, flecainide did not change heart rate (flecainide 400 ± 10 min-1, control 408 ± 11 min-1, n = 15). While no animal definitely converted to sinus rhythm during the 90-min follow-up, flecainide significantly reduced beat-to-beat RR variability, measured as the root mean square of successive differences, RMSSD, relative to mean RR (flecainide 26 ± 1%, control 31 ± 2%, p < 0.05, paired t-test). Furthermore, flecainide also tended to reduce RR interval randomness, measured by the turning point rate (Dash et al., 2009). The actual number of turning points tended to differ more from the number expected by randomness after flecainide treatment (16 ± 4 SDs) than without treatment (7 ± 2 SDs, p = 0.08, paired t-test).
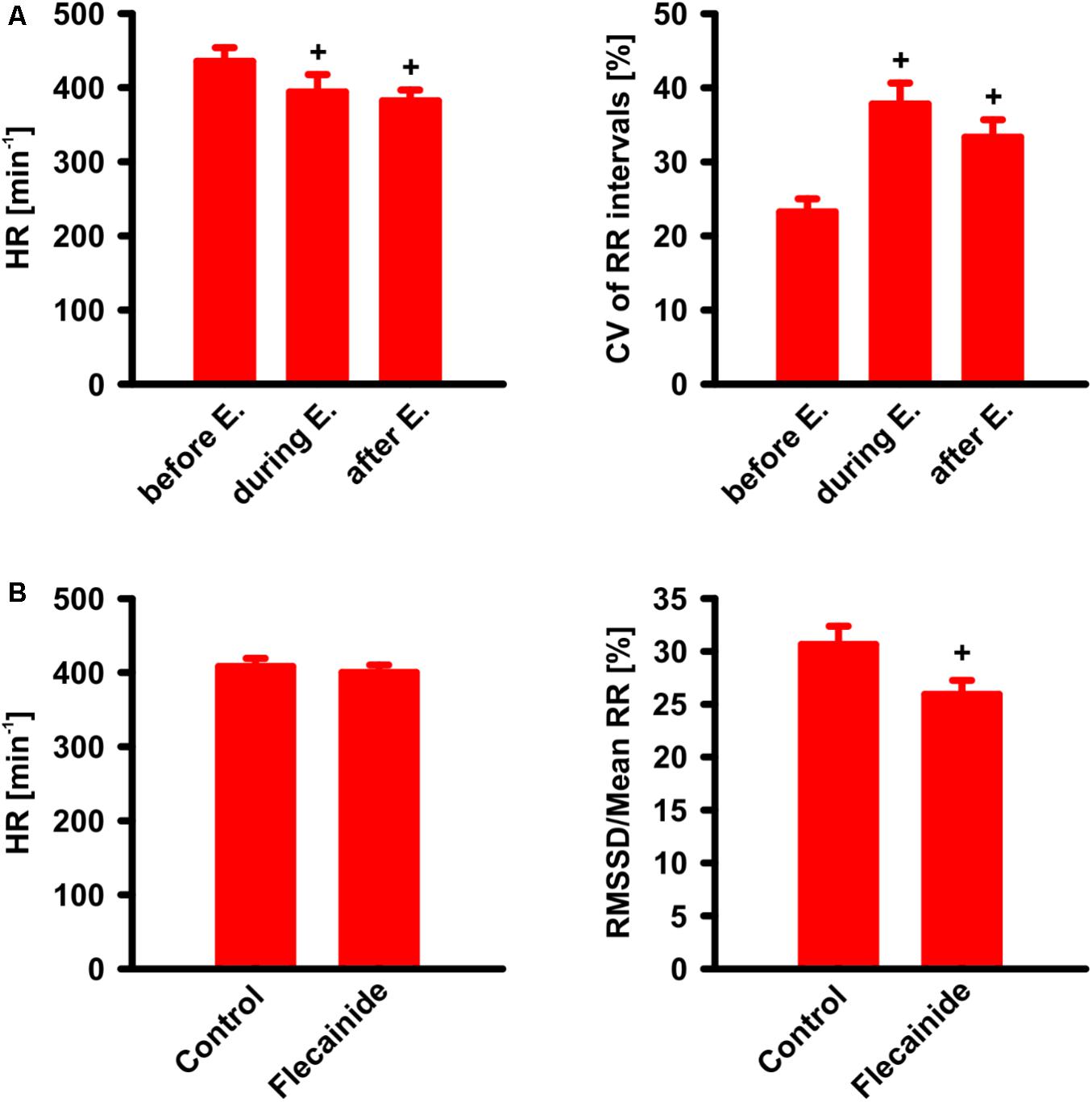
FIGURE 6. Drug effects on CREMT/T animals with AF. (A) Effect of esmolol (E.) i.v. infusion (10 mg/kg/min during 1 min) on heart rate (HR) and coefficient of variation (CV) of RR intervals. Note that, as known and expected from humans, HR is found decreased during and shortly (2 min) after infusion, whereas CV of RR intervals was increased. +p < 0.05 vs. before esmolol (n = 8). (B) Effect of flecainide. ECG was recorded during 90 min after i.v.-administration of flecainide (5 mg/kg) and compared to a previous control ECG (90 min) without flecainide administration. Note that beat-to-beat variability (measured as the root mean square of successive differences, relative to mean RR) was decreased after flecainide +p < 0.05 vs. control (n = 15). All data given as mean ± SEM.
In this study we describe the establishment of a breeding line homozygous for the insertion of human CREM-IbΔC-X cDNA. Creation of the original transgenic mouse model was described previously (Müller et al., 2005). During this original study, two independent founder lines were obtained, both of which showed the key phenotypic change of dilated atria going along with cardiac-specific expression of the CREM repressor isoform HIbII. As it is improbable that DNA insertion events occurred at the same loci in both founder lines it was thus reasonable to conclude that the atrial phenotype was indeed a consequence of transgene expression, and not due to unspecific gene insertion effects, such as disruption of other genes at insertion sites. However, one of the founder lines showed a markedly increased early mortality, which led to breeding failure. Whether this was caused by the many times stronger expression of CREM repressor isoforms or by other effects remained to be speculated. It was proposed (Davis et al., 2012) that transgenic models driven by the αMHC promoter should only be used with care to evaluate atrial phenotypes, as in the atria this promoter is already active throughout embryonic development, and massive overexpression of wild-type proteins alone could already lead to atrial hypertrophy, as was the case in a model overexpressing MYL3 (James et al., 1999). However, in this case, only one founder line grossly overexpressing the transgene showed this phenotype. Several other models overexpressing negative regulators of CRE-mediated transcription consistently showed atrial dilation (Fentzke et al., 1998; Okamoto et al., 2001; Kehat et al., 2006b), and downregulation of CREB target genes was connected to AF susceptibility in humans (Deshmukh et al., 2015). It therefore seems reasonable to suggest specific effects by which CREM-IbΔC-X overexpression contributes to the development of the atrial phenotype. We sought to generate animals homozygous for the insertion site of the transgene by mating hemizygous parents. Indeed, a Southern blot, which could also clearly identify wild-types, showed two different levels of transgenic DNA content in the genome of the progeny. As it is highly unlikely that more than one insertion site (or more specifically, more than one chromosome containing insertion sites) was left after several generations of breeding, this strongly hinted to a homozygous presence of this insertion site. This assumption could later be verified in offspring from breeding pairs only consisting of animals deemed homozygous, which showed doubled CREM-IbΔC-X cDNA content in a quantitative PCR when compared to transgenic offspring from the hemizygous breeding colony. Moreover, immunological detection of protein products translated from the transgene revealed that a product of the same size as in hearts from CREMT/W animals could be identified in hearts from CREMT/T animals, but in a roughly doubled quantity, albeit with greater variation between animals.
Going along with, and probably as a consequence of this increased expression of a CREM repressor isoform, the atrial phenotype developed more quickly and into a more pronounced state in CREMT/T animals. While 7 weeks old CREMT/W animals did not yet show signs of atrial hypertrophy, an increase in relative atrial weights was already apparent in CREMT/T animals of the same age. At 14 and 26 weeks of age both hemizygous and homozygous animals had developed atrial hypertrophy, but also at this age hypertrophy was more distinct in CREMT/T, with the difference rather increasing with older age. Simultaneously, development of spontaneous atrial fibrillation was accelerated in CREMT/T. While slightly more than half of the hemizygous animals had shown episodes of spontaneous AF by the age of 22 weeks, all homozygous animals had developed spontaneous AF already by the age of 10 weeks. We performed a first comparative assessment of the levels of mRNAs encoding ion channels or gap junction proteins (Cacna1c, Kcnd2, Scn5a, Kcne1, Kcnv2, Gja5), ion channel regulators (Fxyd1), and proteins involved in ion channel localization and stabilization (Ank2, Cav3), structural and functional organization of intercalated disks (Cxadr), signal transduction (Akap9), gene transcription (Mef2c), and myosin function (Myl3) to study the expression of genes linked to AF development and discussed in the context of the hemizygous model (Seidl et al., 2017). While many genes were regulated similarly in 14 weeks old CREMT/W and CREMT/T animals (vs. WT), some of the genes were only differently expressed in CREMT/T. Although the time course of gene expression changes and its interconnection with development of the AF phenotype was out of the scope of this study, it can be speculated that this was a consequence of the more advanced phenotype in the homozygous model. Thus, the homozygous model may well become a valuable addition to the hemizygous model also in future studies addressing molecular mechanisms of AF development.
In ventricular cardiomyocytes isolated from CREMT/W hearts arrhythmogenic alterations in Ca2+-handling and K+-currents were identified, leading to an increased propensity to ventricular extrasystoles especially during β-adrenergic stimulation (Schulte et al., 2016). It is supposable that this might also be aggravated compared to CREMT/W. Nevertheless, overall survival of CREMT/T and CREMT/W animals was comparable up to the age of 26 weeks (and not different from WT), allowing the conclusion that there was no increased prevalence of life-threatening ventricular arrhythmias up to this age. In our present study, a mild ventricular hypertrophy (vs. WT) was found in CREMT/T animals while being absent from CREMT/W. In the original study describing the hemizygous model (Müller et al., 2005), both newly established lines showed ventricular hypertrophy (Tg2 strongly, Tg1 mildly), going along with an increased ventricular function in Tg1 (Tg2 could not be measured due to early mortality and breeding failure, as mentioned above). In CREMT/W (descending from Tg1), this hypertrophy probably regressed with decreasing gene dose due to breeding. Assessment of ventricular fibrosis in 14 weeks old CREMT/T and CREMT/W revealed no significant difference of interstitial and perivascular collagen content, both being well in the range reported for WT from the same background in another study (Seeland et al., 2007). In short, given these findings and the unchanged survival of CREMT/T vs. CREMT/W, we have no reason to suspect overt ventricular pathology in CREMT/T, while we currently cannot formally rule it out.
In summary, this study indicates that in homozygous CREM-IbΔC-X transgenic animals a doubled gene dosage leads to an increased expression of gene products, which in turn accelerates and aggravates the atrial phenotype, yet without causing increased early mortality.
Drugs used as antiarrhythmic agents can be roughly categorized into those used for rhythm control and those used for rate control (Kirchhof et al., 2016). We evaluated one substance out of each group to verify that CREMT/T animals adequately reproduce effects typically found in human AF. Esmolol is a short-acting β1-adrenoceptor antagonist, whose indications include quick achievement of rate control in AF patients with hemodynamically relevant tachycardia. Although our model, with a mean heart rate of about 400 min-1 under isoflurane anesthesia, is not as such mimicking this situation, a significant reduction of heart rate was observed during and shortly after esmolol infusion. Concurrently, heart rate variability was increased. Comparable results were obtained in a study whose authors treated human AF patients with a β-adrenoceptor antagonist: heart rates decreased, while heart rate variability increased, the latter effect being attributed to relative changes in vagal tone (van den Berg et al., 1997). Thus, to this extent, our model quite adequately reflects conditions in humans.
Flecainide is a Na+-channel blocker belonging to the class Ic antiarrhythmics, being effective in ventricular as well as supraventricular arrhythmias. In patients with non-permanent AF, but without concomitant structural ventricular disease, it can typically be used to attempt pharmacologic cardioversion. In this study, we applied flecainide to relatively young mice to ensure that the animals had not been in AF for a prolonged time. Nevertheless, in none of the experiments a definite conversion to sinus rhythm with visible P waves could be observed during 90 min of follow-up. Comparing ECGs recorded immediately after flecainide application to control ECGs recorded from the same animals few days previously, we found a decrease in beat-to-beat RR variability. At the same time, the dependence of RR interval duration on randomness was reduced by flecainide treatment. Both parameters, an increase in beat-to-beat RR variability as well as more randomly distributed RR interval durations, are used in an algorithm to detect the occurrence of AF in ECG recordings (Dash et al., 2009). The reduction of one of these parameters as well as the trend to reduction of the other might hint at a measurable atrial effect of flecainide in our AF model, yet not enough to induce conversion to sinus rhythm. However, flecainide was attributed vagolytic properties in a study on human subjects (Fauchier et al., 1998). Although this study does not allow differentiation between effects of onetime and prolonged flecainide application and does not assess AF patients, it still cannot be excluded that the effect of flecainide on beat-to-beat RR variability in our model might, at least in part, be attributable to vegetative mechanisms. Ultimately, it must be speculated that CREMT/T animals very rapidly progress into permanent AF.
In this study we could show that, by establishing a breeding colony homozygous for the insertion of the CREM-IbΔC-X transgene, one specific phenotype of this animal model, i.e., atrial hypertrophy and spontaneous atrial fibrillation, could be intensified and its development accelerated, with 100% of homozygous mice presenting AF within 10 weeks. At the same time, except for a mild ventricular hypertrophy, no unspecific side effects became obvious, and no increased early mortality up to an age of 6 months appeared. While further studies may help to elucidate effects of the increased gene dose on the arrhythmogenic substrate underlying AF development in the hemizygous model, this study clearly suggests the homozygous model as a feasible and economic animal disease model of AF, with rapid onset of the phenotype and low concomitant mortality.
This study was carried out in accordance with the requirements of relevant German laws and regulations (“Tierschutzgesetz [TSchG]” and “Tierschutz-Versuchstierverordnung [TierSchVersV]”) as well as European Union Directive 2010/63/EU. The protocol was approved by the animal protection commission of the “Landesamt für Natur, Umwelt und Verbraucherschutz (LANUV) Nordrhein-Westfalen.”
FM, FS, and JS conceived of the study. KH, BS, BVS, JS, FS, and MS performed the experiments and analyzed and interpreted their data. FS drafted the manuscript, with parts provided by BVS and MS. All authors thoroughly reviewed the manuscript, revised it for important intellectual content, and approved the final version.
This work was supported by the European Union as part of the EUTRAF project and by Deutsche Forschungsgemeinschaft (DFG) grant DFG Mu1376/11-3.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors wish to thank Melanie Voß for her excellent technical assistance and Florentina Pluteanu for providing primers for the qRT-PCR experiments.
Altarejos, J. Y., and Montminy, M. (2011). CREB and the CRTC co-activators: sensors for hormonal and metabolic signals. Nat. Rev. Mol. Cell Biol. 12, 141–151. doi: 10.1038/nrm3072
Bukowska, A., Felgendreher, M., Scholz, B., Wolke, C., Schulte, J. S., Fehrmann, E., et al. (2018). CREM-transgene mice: an animal model of atrial fibrillation and thrombogenesis. Thromb. Res. 163, 172–179. doi: 10.1016/j.thromres.2017.07.033
Chelu, M. G., Sarma, S., Sood, S., Wang, S., Oort, R. J., van, Skapura, D. G., et al. (2009). Calmodulin kinase II–mediated sarcoplasmic reticulum Ca2+ leak promotes atrial fibrillation in mice. J. Clin. Invest. 119, 1940–1951. doi: 10.1172/JCI37059
Chugh, S. S., Havmoeller, R., Narayanan, K., Singh, D., Rienstra, M., Benjamin, E. J., et al. (2014). Worldwide epidemiology of atrial fibrillation. Circulation 129, 837–847. doi: 10.1161/CIRCULATIONAHA.113.005119
Dash, S., Chon, K. H., Lu, S., and Raeder, E. A. (2009). Automatic real time detection of atrial fibrillation. Ann. Biomed. Eng. 37, 1701–1709. doi: 10.1007/s10439-009-9740-z
Davis, J., Maillet, M., Miano, J. M., and Molkentin, J. D. (2012). Lost in transgenesis: a user’s guide for genetically manipulating the mouse in cardiac research. Circ. Res. 111, 761–777. doi: 10.1161/CIRCRESAHA.111.262717
Deshmukh, A., Barnard, J., Sun, H., Newton, D., Castel, L., Pettersson, G., et al. (2015). Left atrial transcriptional changes associated with atrial fibrillation susceptibility and persistence. Circ. Arrhythm. Electrophysiol. 8, 32–41. doi: 10.1161/CIRCEP.114.001632
Fauchier, L., Babuty, D., Autret, M. L., Poret, P., Cosnay, P., and Fauchier, J. P. (1998). Effect of flecainide on heart rate variability in subjects without coronary artery disease or congestive heart failure. Cardiovasc. Drugs Ther. 12,483–486. doi: 10.1023/A:1007710301259
Fentzke, R. C., Korcarz, C. E., Lang, R. M., Lin, H., and Leiden, J. M. (1998). Dilated cardiomyopathy in transgenic mice expressing a dominant-negative CREB transcription factor in the heart. J. Clin. Invest. 101, 2415–2426. doi: 10.1172/JCI2950
Gallagher, A. M., Staa, T. P., van Murray-Thomas, T., Schoof, N., Clemens, A., Ackermann, D., et al. (2014). Population-based cohort study of warfarin-treated patients with atrial fibrillation: incidence of cardiovascular and bleeding outcomes. BMJ Open 4:e003839. doi: 10.1136/bmjopen-2013-003839
James, J., Osinska, H., Hewett, T. E., Kimball, T., Klevitsky, R., Witt, S., et al. (1999). Transgenic over-expression of a motor protein at high levels results in severe cardiac pathology. Transgenic Res. 8, 9–22. doi: 10.1023/A:1008894507995
Kehat, I., Hasin, T., and Aronheim, A. (2006a). The role of basic leucine zipper protein-mediated transcription in physiological and pathological myocardial hypertrophy. Ann. N. Y. Acad. Sci. 1080, 97–109. doi: 10.1196/annals.1380.009
Kehat, I., Heinrich, R., Ben-Izhak, O., Miyazaki, H., Gutkind, J. S., and Aronheim, A. (2006b). Inhibition of basic leucine zipper transcription is a major mediator of atrial dilatation. Cardiovasc. Res. 70, 543–554. doi: 10.1016/j.cardiores.2006.02.018
Kirchhof, P., Benussi, S., Kotecha, D., Ahlsson, A., Atar, D., Casadei, B., et al. (2016). 2016 ESC Guidelines for the management of atrial fibrillation developed in collaboration with EACTS. Eur. J. Cardiothorac. Surg. 37, 2893–2962.
Kirchhof, P., Marijon, E., Fabritz, L., Li, N., Wang, W., Wang, T., et al. (2013). Overexpression of cAMP-response element modulator causes abnormal growth and development of the atrial myocardium resulting in a substrate for sustained atrial fibrillation in mice. Int. J. Cardiol. 166, 366–374. doi: 10.1016/j.ijcard.2011.10.057
Li, N., Chiang, D. Y., Wang, S., Wang, Q., Sun, L., Voigt, N., et al. (2014). Ryanodine receptor–mediated calcium leak drives progressive development of an atrial fibrillation substrate in a transgenic mouse model. Circulation 129, 1276–1285. doi: 10.1161/CIRCULATIONAHA.113.006611
Müller, F. U., Bokník, P., Knapp, J., Neumann, J., Vahlensieck, U., Oetjen, E., et al. (1998). Identification and expression of a novel isoform of cAMP response element modulator in the human heart. FASEB J. 12, 1191–1199. doi: 10.1096/fasebj.12.12.1191
Müller, F. U., Lewin, G., Baba, H. A., Bokník, P., Fabritz, L., Kirchhefer, U., et al. (2005). Heart-directed expression of a human cardiac isoform of camp-response element modulator in transgenic mice. J. Biol. Chem. 280, 6906–6914. doi: 10.1074/jbc.M407864200
Müller, F. U., Neumann, J., and Schmitz, W. (2000). Transcriptional regulation by cAMP in the heart. Mol. Cell. Biochem. 212, 11–17. doi: 10.1023/A:1007176030884
Nishida, K., Michael, G., Dobrev, D., and Nattel, S. (2010). Animal models for atrial fibrillation: clinical insights and scientific opportunities. Europace 12, 160–172. doi: 10.1093/europace/eup328
Okamoto, Y., Chaves, A., Chen, J., Kelley, R., Jones, K., Weed, H. G., et al. (2001). Transgenic mice with cardiac-specific expression of activating transcription factor 3, a stress-inducible gene, have conduction abnormalities and contractile dysfunction. Am. J. Pathol. 159, 639–650. doi: 10.1016/S0002-9440(10)61735-X
Riley, G., Syeda, F., Kirchhof, P., and Fabritz, L. (2012). An introduction to murine models of atrial fibrillation. Front. Physiol. 3:296. doi: 10.3389/fphys.2012.00296
Rockman, H. A., Hamilton, R. A., Jones, L. R., Milano, C. A., Mao, L., and Lefkowitz, R. J. (1996). Enhanced myocardial relaxation in vivo in transgenic mice overexpressing the beta2-adrenergic receptor is associated with reduced phospholamban protein. J. Clin. Invest. 97, 1618–1623. doi: 10.1172/JCI118587
Schulte, J. S., Fehrmann, E., Tekook, M. A., Kranick, D., Fels, B., Li, N., et al. (2016). Cardiac expression of the CREM repressor isoform CREM-IbΔC-X in mice leads to arrhythmogenic alterations in ventricular cardiomyocytes. Basic Res. Cardiol. 111:15. doi: 10.1007/s00395-016-0532-y
Seeland, U., Selejan, S., Engelhardt, S., Müller, P., Lohse, M. J., and Böhm, M. (2007). Interstitial remodeling in β1-adrenergic receptor transgenic mice. Basic Res. Cardiol. 102, 183–193. doi: 10.1007/s00395-006-0635-y
Seidl, M. D., Stein, J., Hamer, S., Pluteanu, F., Scholz, B., Wardelmann, E., et al. (2017). Characterization of the genetic program linked to the development of atrial fibrillation in CREM-IbΔC-X mice. Circ. Arrhythm. Electrophysiol. 10:e005075. doi: 10.1161/CIRCEP.117.005075
van den Berg, M. P., Haaksma, J., Brouwer, J., Tieleman, G., Mulder, G., and Crijns, J. G. M. (1997). Heart rate variability in patients with atrial fibrillation is related to vagal tone. Circulation 96, 1209–1216. doi: 10.1161/01.CIR.96.4.1209
Keywords: atrial fibrillation, atrial hypertrophy, animal models of disease, antiarrhythmics, cAMP response element modulator
Citation: Stümpel FT, Stein J, Himmler K, Scholz B, Seidl MD, Skryabin BV and Müller FU (2018) Homozygous CREM-IbΔC-X Overexpressing Mice Are a Reliable and Effective Disease Model for Atrial Fibrillation. Front. Pharmacol. 9:706. doi: 10.3389/fphar.2018.00706
Received: 16 February 2018; Accepted: 11 June 2018;
Published: 05 July 2018.
Edited by:
Ajay Sharma, Chapman University, United StatesReviewed by:
Scott M. MacDonnell, Regeneron Pharmaceuticals, Inc., United StatesCopyright © 2018 Stümpel, Stein, Himmler, Scholz, Seidl, Skryabin and Müller. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Frank T. Stümpel, ZnN0dWVAdW5pLW11ZW5zdGVyLmRl
†These authors have contributed equally to this work.
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.