
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Pharmacol. , 07 March 2018
Sec. Experimental Pharmacology and Drug Discovery
Volume 9 - 2018 | https://doi.org/10.3389/fphar.2018.00203
This article is part of the Research Topic Advances in Biopharmaceutics View all 25 articles
Numerous bacteria utilize molecular communication systems referred to as quorum sensing (QS) to synchronize the expression of certain genes regulating, among other aspects, the expression of virulence factors and the synthesis of biofilm. To achieve this process, bacteria use signaling molecules, known as autoinducers (AIs), as chemical messengers to share information. Naturally occurring strategies that interfere with bacterial signaling have been extensively studied in recent years, examining their potential to control bacteria. To interfere with QS, bacteria use quorum sensing inhibitors (QSIs) to block the action of AIs and quorum quenching (QQ) enzymes to degrade signaling molecules. Recent studies have shown that these strategies are promising routes to decrease bacterial pathogenicity and decrease biofilms, potentially enhancing bacterial susceptibility to antimicrobial agents including antibiotics and bacteriophages. The efficacy of QSIs and QQ enzymes has been demonstrated in various animal models and are now considered in the development of new medical devices against bacterial infections, including dressings, and catheters for enlarging the therapeutic arsenal against bacteria.
Quorum sensing (QS) is a molecular mechanism by which bacteria communicate to collectively adapt their behavior according to cell density and the surrounding environment (Figure 1). This communication system enables bacteria to undertake processes that are costly and non-effective at low cell density but that become useful for the whole community at high cell density such as virulence factor synthesis, biofilm formation, and protease and siderophore production (Heilmann et al., 2015). QS consists in the production and sensing of small extracellular molecules, known as autoinducers (AIs), that are released in proportion to cell density (Papenfort and Bassler, 2016). In Gram-positive bacteria, autoinducing peptides (AIPs) were widely studied and reported to induce QS. AIPs are specific to species and strains and have been described in Staphylococcus spp., Clostridium spp., or Enterococcus spp., among others, AIPs (Figure 2; Monnet et al., 2016). Many Gram-negative bacteria, including Pseudomonas spp., Acinetobacter spp., or Burkholderia spp., were reported to use a different class of autoinducers: the acyl-homoserine lactones (AHLs) (Schuster et al., 2013). AHLs are composed of a lactone ring and an aliphatic acyl chain varying in length and modifications (Schuster et al., 2013). A wide variety of other signaling molecules was also identified (Hawver et al., 2016), including fatty acids used by Xanthomonas spp., Burkholderia spp., Xylella spp. (Zhou et al., 2017), ketones (Vibrio spp. and Legionella spp.; Tiaden and Hilbi, 2012), epinephrine, norepinephrine and AI-3 (enterohemorrhagic bacteria; Kendall and Sperandio, 2007) or quinolones (Pseudomonas aeruginosa; Heeb et al., 2011). Finally, AI-2, a furanosyl borate diester, is used by both Gram-negative and Gram-positive bacteria (Chen et al., 2002; Figure 2). Most Gram-negative bacteria combine several QS systems to integrate different signals either hierarchically, as P. aeruginosa in which four QS systems (las, rhl, iqs, and pqs) act in a network (Lee and Zhang, 2015), or in parallel, as in Vibrio harveyi in which three systems are integrated into one regulatory cascade (Plener et al., 2015).
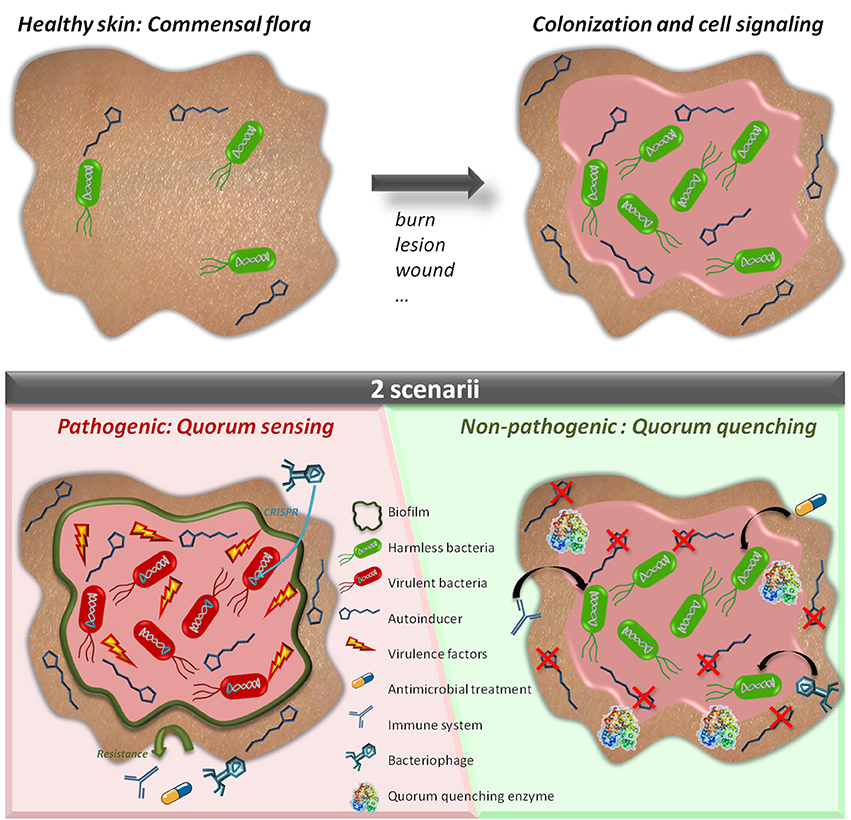
Figure 1. Quorum sensing and quorum quenching in a wounded tissue. The skin usually harbors a natural and commensal flora which is not pathogenic (Upper Left). When a wound or a lesion occurs, bacteria colonize the wounded tissue and further develop being in a favorable environment (Upper Right). While growing, bacteria produce communication molecules (autoinducers). If the molecules are not degraded (Bottom Left), bacteria can synchronize their behavior to secrete virulence factors and produce biofilms which may prevent efficiency of antibiotic or phage therapy. The wound is infected. If the autoinducers are degraded (Bottom Right) bacteria do not synchronize their behavior and remain harmless and defenseless. The wound remains colonized but no infection occurs.
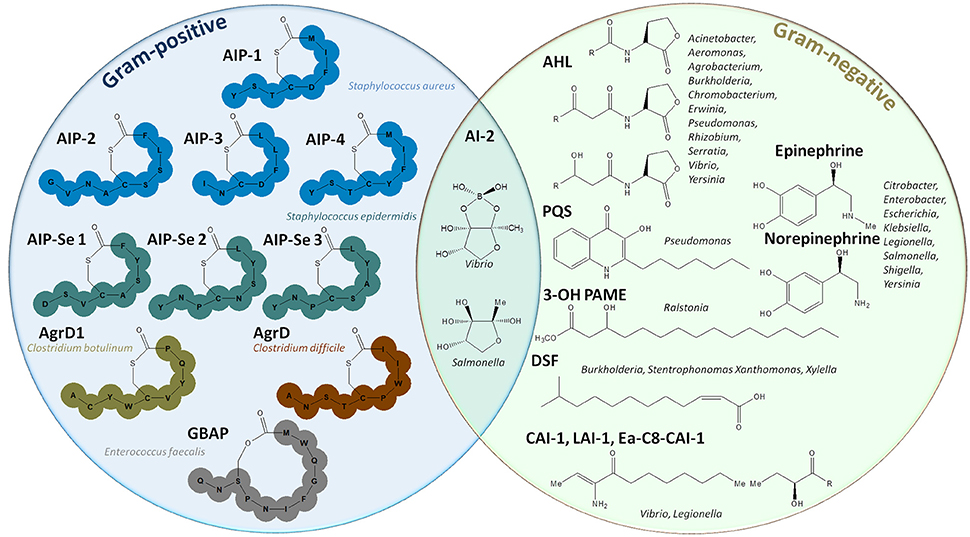
Figure 2. Representation of autoinducer molecules. The left circle represents autoinducing peptides used by Gram-positive bacteria such as Staphylococcus spp., Clostridium spp., Enterococcus faecalis (Monnet et al., 2016). The right circle gives an overview of the different molecules used in Gram-negative quorum sensing: acyl homoserine lactones (AHLs) (Schuster et al., 2013), quinolones (PQS), 4-hydroxypalmitate methyl ester (3-OH-PAME) (Flavier et al., 1997), fatty acids (DSF) (Zhou et al., 2017), epinephrine, and norepinephrine (Kendall and Sperandio, 2007). In the middle, the different forms of AI-2, a furanosyl diester, used by both Gram-positive and Gram-negative bacteria are depicted (Chen et al., 2002).
Interferences with QS are termed quorum quenching (QQ) (Figure 1). QQ was discovered as a naturally occurring phenomenon first described in 2000 with the identification of a QQ enzyme able to degrade AHL signals from Erwinia carotovora (Dong et al., 2000). The enzymatic hydrolysis of AHL led to the disruption of the QS signal. The disruption of bacterial communication can be achieved by several processes: (i) interfering with the production or perception of AIs via small molecules referred to as quorum sensing inhibitors (QSIs) (Tang and Zhang, 2014), (ii) scavenging of AIs by quorum quenching antibodies (Park et al., 2007), and macromolecules such as cyclodextrins (Kato et al., 2006, 2007; Morohoshi et al., 2013), or (iii) by extracellular hydrolysis of the AIs using QQ enzymes (Fetzner, 2015; Figure 3). Several antagonist peptides have been identified among natural compounds or designed to quench Gram-positive bacteria and many QSIs, mainly targeting Gram-negative QS and AI-2 mediated QS, have also been reported (Tang and Zhang, 2014; Singh et al., 2016). Such compounds can be natural products, like polyphenols isolated from tea or honey, ajoene from garlic, eugenol from clove or many others produced by marine organisms and fungi (Tang and Zhang, 2014; Delago et al., 2016), or they can be synthetic, such as 5-fluorouracil (5-FU) or azithromycin (Ueda et al., 2009; Swatton et al., 2016). Many QQ enzymes and macromolecules (Amara et al., 2011; Fetzner, 2015) as well as natural or synthetic QSIs (Dembitsky et al., 2011; Galloway et al., 2011; Stevens et al., 2011; Kalia, 2013; Delago et al., 2016) have been reported to date and exhaustively reviewed. Patents associated with these compounds (Pan and Ren, 2009; Romero et al., 2012; Jiang and Li, 2013) as well as routes to access novel molecules (Scutera et al., 2014) have also been discussed. The mechanisms used by the different QSIs are not always known and most probably differ from one QSI to another (Defoirdt et al., 2013). Some molecules inhibiting QS such as azithromycin are also considered as antibiotics as they can inhibit bacterial growth above a certain concentration (Nalca et al., 2006).
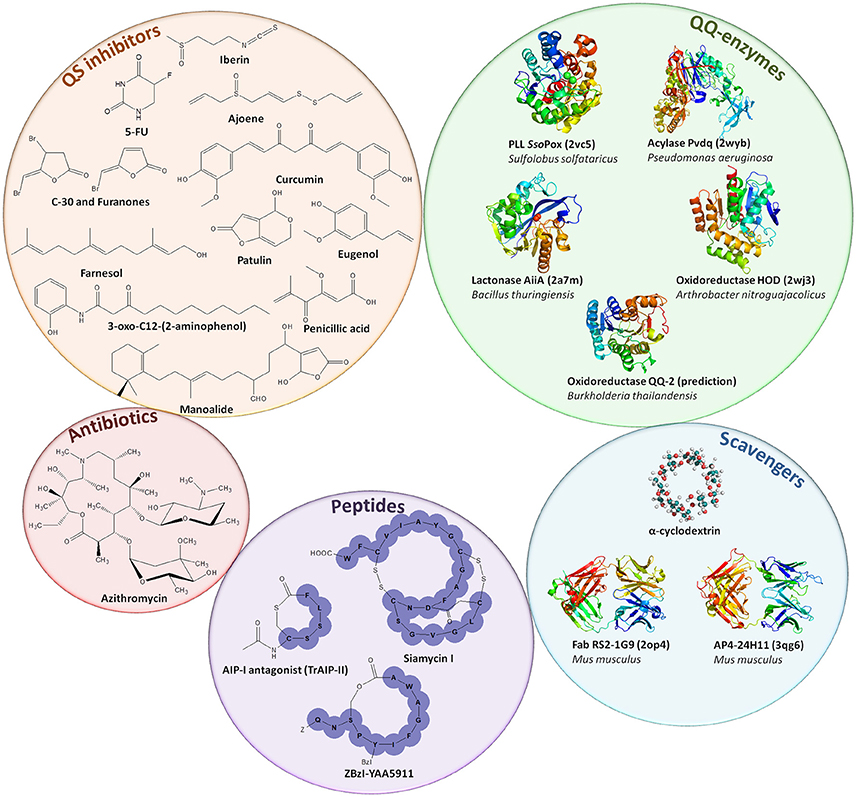
Figure 3. Representation of quorum quenching agents. Quorum sensing inhibitors, mainly acting against AHL or AI-2-based QS, are depicted in the orange circle (Tang and Zhang, 2014). Antibiotics such as azithromycin can be used as QSI at sub-inhibitory concentrations (Swatton et al., 2016). Purple circle represents the QQ peptides used to inhibit Gram-positive QS (Singh et al., 2016). Blue circle represents molecules used to scavenge AIs such as cyclodextrins or derivatives (Morohoshi et al., 2013) and antibodies scavenging AHL (Fab RS2-1G9) or AIP (AP4-24H11) (Park et al., 2007). Green circle depicts QQ enzymes that disrupt AHLs (SsoPox, Pvdq, and AiiA), the quinolone PQS (HOD) and AI-2 signals (QQ-2) (Fetzner, 2015).
Currently identified QQ enzymes mainly target AHLs and AI-2 mediated QS: phosphotriesterase-like lactonases (PLLs), lactonases, acylases, and oxidoreductases degrade AHL signals (Fetzner, 2015) and oxidoreductases target AI-2 (Dong et al., 2000; Weiland-Bräuer et al., 2016). As QS induces noxious traits such as biofilm formation or virulence, the disruption of bacterial communication appears as a promising strategy to prevent bacteria from synchronizing their virulent behavior. Therefore, QQ approaches may have applications in numerous fields such as agronomy, water engineering, and the marine industry and is particularly relevant in health care (Bzdrenga et al., 2017).
In the current context of the rise of antibiotic tolerance and resistance, novel therapeutic approaches are needed (Kaye and Pogue, 2015). The ability of QQ approaches to inhibit bacterial virulence and biofilm is appealing as this latter is associated with increased antibiotic tolerance (Stewart and William Costerton, 2001). Biofilm formation is triggered via QS and consists in a heterogeneous multi-cellular structure attached to a solid surface, embedded in an extracellular matrix (de la Fuente-Núñez et al., 2013). The extracellular matrix, made of polysaccharides, proteins and extracellular DNA, may prevent some antibiotics from successfully penetrating the cells, inducing antibiotic tolerance (Otto, 2006). The bacterial cells embedded in the matrix also have a slower growth rate and an altered metabolism which further reduces antibiotic efficiency (Olsen, 2015). In addition, biofilm environments combine high cell density and high selection pressure increasing the rate with which resistant cells appear through mutations or gene transfer (Driffield et al., 2008). Biofilms also shelter persistent cells, a non-inheritable trait denoting a subpart of cells present in any bacterial population that will survive antibiotic treatment as a result of being in a different physiological state at the time of the treatment (Brauner et al., 2016). Bacteria in biofilms are considered to be 100–1,000 times more tolerant to antimicrobial compounds as compared to their planktonic lifestyle (Olsen, 2015). Biofilms can also be at the source of infections: it is considered that between 65 and 80% of infections are biofilm associated infections, either by directly infecting the tissue such as lung infection in the case of cystic fibrosis or via a contaminated device such as a catheter (Lebeaux et al., 2013). Hospital-acquired infections (HAIs) affect between 6 and 10% of health care patients in developed countries, the most frequent type of infections being urinary tract infections (Klevens et al., 2007; Lobdell et al., 2012). Eliminating biofilms in health care devices and environment is a challenge to limit and treat HAIs. It is, therefore, essential to develop alternative or complementary treatments to conventional antimicrobial and antibiotic products. To this end, QQ and phage therapy are increasingly studied (Kostakioti et al., 2013; Rémy et al., 2016a).
This review highlights the latest findings and biopharmaceutical perspectives of QQ as well as its potential complementarity with antimicrobial agents, antibiotics and bacteriophages. The eukaryotic models used to prove the efficiency of QQ as a successful anti-virulence and anti-biofilm strategy and the medical applications with QQ devices are also summarized.
As QS involves a global change in bacterial gene expression and cell physiology, the relationship between QS and antibiotic tolerance is multi-faceted. For example, the addition of AHLs to a logarithmic culture of P. aeruginosa was shown to increase the number of persister cells in the population after treatment with carbenicillin and ciprofloxacin (Möker et al., 2010). Furthermore, transcriptomic analysis with the QS transcription regulator MvfR (PqsR) in P. aeruginosa PA14 revealed that QS induces the expression of peroxidases which provide protection against reactive oxygen species (H2O2) and β-lactam antibiotics (Maura et al., 2016). In another study using P. aeruginosa PAO1, VqsM, a global regulator that induces QS, was shown to mediate antibiotic tolerance by inducing the expression of nfxB, an antibiotic resistance regulator, providing increased tolerance to quinolones, tetracycline, and kanamycin via regulation of mexC-mexD-oprJ operon expression (Poole et al., 1996; Liang et al., 2014).
Although some physiological aspects may be involved in QS-mediated tolerance to antibiotics, many reports focus on the importance of biofilm in antibiotic tolerance of bacteria (Høiby et al., 2010), causing many difficulties for treatments of clinical infections (Høiby et al., 2011). Those effects have been frequently observed with P. aeruginosa (Strateva and Yordanov, 2009) in both model and clinical strains (Hill et al., 2005) as well as in other species such as Klebsiella pneumoniae (Vuotto et al., 2014, 2017) and Staphylococcus aureus (Savage et al., 2013). The particular conditions that the biofilm mode of growth provides to bacteria favors the development of different defense mechanisms and phenotypes: physical barrier, modification of gene expression, and cellular physiological states (e.g., persister cell) (de la Fuente-Núñez et al., 2013). In P. aeruginosa, the rhl, and las QS systems are essential for biofilm formation and their disruption is correlated with a higher sensitivity to the host immune system and antimicrobial compounds (Davies et al., 1998; Shih and Huang, 2002; Bjarnsholt, 2005b). Moreover in P. aeruginosa, another QS system, the pqs system, has been demonstrated to mediate a programmed cell death inducing extracellular DNA release which promotes biofilm formation and antibiotic tolerance, benefitting the rest of the cell population (Hazan et al., 2016). In clinical isolates of Acinetobacter baumannii, the presence of levofloxacin or meropenem antibiotics was reported to induce the overexpression of an efflux pump which stimulates the release of AHL and thus enhances the formation of QS-mediated biofilm, increasing antibiotic tolerance (He et al., 2015).
Regarding the important role of QS in biofilm formation and antibiotic tolerance, combination therapy with QQ was investigated. In P. aeruginosa, the use of a pharmacological compound, benzamide-benzimidazole, inhibiting the QS regulator MvfR (PqsR) decreased biofilm formation and restored antibiotic susceptibility (Starkey et al., 2014; Maura and Rahme, 2017). Baicalin hydrate and hamamelitannin, an AHL-targeting QSI and a peptide-based QSI, enhanced biofilm disruption in both Gram-negative (P. aeruginosa and Burkholderia cepacia complex) and Gram-positive (S. aureus) bacteria and showed synergistic effects in cotreatment with tobramycin and clindamycin or vancomycin respectively both in vitro and in vivo (Brackman et al., 2011b). From aminoglycosides (Jakobsen et al., 2012; Stenvang et al., 2016) to quinolones (Guo et al., 2016), polypeptides (Furiga et al., 2016; Bulman et al., 2017), cephalosporins (Maura and Rahme, 2017), and glycopeptides (Das et al., 2016), the efficiency of a large range of antibiotics is enhanced by the addition of QSIs.
Taken together these results suggest that using QSIs is a potential way of increasing antibiotic sensitivity and thereby lower antibiotic active doses. Additionally, similar trend and efficiency have also been observed with a lactonase QQ enzyme and the antibiotic ciprofloxacin in a mice model (Gupta et al., 2015). Combining antimicrobial agents and QQ was showed to have encouraging synergistic effects, highlighting that QQ is a good strategy to decrease antibiotics use and fight against the increasing problem of antibiotic resistance. Nevertheless, if QQ can help to prevent and treat infections, it cannot be used on its own to treat acute infections by antibiotic resistant strains.
Recently, interest has considerably increased in phage therapy as a way of treating infections caused by multi-drug resistant bacteria (Pires et al., 2017). Bacteriophages are the most abundant bacterial predators on the planet and they are still used to treat bacterial infections in Eastern Europe (Brüssow and Hendrix, 2002). Although bacteriophages represent an interesting solution to circumvent antibiotic resistance, bacteria have also developed resistance mechanisms to counteract phage actions (Labrie et al., 2010). Firstly, phage entry can be decreased by extracellular matrix production, or by modifying the phage receptor structure or expression (Chapman-McQuiston and Wu, 2008; Labrie et al., 2010). Once inside the cell, phage DNA can be recognized and degraded by restriction enzymes or the adaptive inducible CRISPR-Cas (clustered regularly interspaced short palindromic repeat and CRISPR associated proteins) system (Barrangou et al., 2007; Labrie et al., 2010). Other potential metabolism adaptations could also provide bacteriophage resistance (Qin et al., 2017). As the risk of phage infection rises with cell density (Abedon, 2012), QS-mediated resistance would provide protection during high risk conditions while limiting the overall fitness cost of those mechanisms (Hall et al., 2011).
The relationship between QS and bacteriophage sensitivity was originally observed in P. aeruginosa (Glessner et al., 1999), but the ability of QS to regulate phage defense mechanisms in Escherichia coli was only demonstrated years later (Høyland-Kroghsbo et al., 2013). The authors showed that AHLs induced a reduction in the phages lambda and chi adsorption rate by reducing the number of receptors at the cell surface. In Vibrio cholerae, a deficiency in AI synthase genes, and thereby in QS induction, reduced phage resistance which could be restored by the addition of exogenous AIs, AI-2, and CAI-1 (Hoque et al., 2016). This increased phage resistance upon QS activation was explained by the downregulation of O-antigen synthesis which decreased phage adsorption and by an increase in hemagglutinin protease production which was shown to inactivate phages (Hoque et al., 2016). Similarly, the addition of synthetic AHLs to a Vibrio anguillarum QS deficient strain improved phage resistance (Tan et al., 2015). Indeed, AHL production was negatively correlated with phage receptor ompK expression. Recently, it was shown in Serratia marcescens that the CRISPR-Cas immune system was also under QS regulation (Patterson et al., 2016). Both the acquisition of immunity and DNA degradation mechanism coordinated by this system were negatively impacted by the absence of QS signal in a synthase mutant. Besides, by analyzing former datasets (Bowden et al., 2013; Gao et al., 2015), the authors suggested that this type of regulation might occur in Pectobacterium atrosepticum as well as in Burkholderia glumae. Similar QS control of CRISPR-Cas system was demonstrated in P. aeruginosa PA14 in which the expression of the CRISPR-Cas genes is down-regulated in a strain deleted for both AI synthase genes lasI and rhlI (Høyland-Kroghsbo et al., 2017). To evaluate whether QQ would increase phage sensitivity, QSI effects were investigated on resistance mechanisms and on phage susceptibility phenotypes. The use of penicillic acid increased P. aeruginosa sensitivity to phages by increasing the proportion of sensitive viable cells in the population (Qin et al., 2017). Finally, the QSI baicalin was shown to inhibit the QS stimulation of CRISPR-Cas system in P. aeruginosa which could prevent the use of this adaptive system by bacteria in case of phage infection (Høyland-Kroghsbo et al., 2017).
In light of these findings, the use of QQ compounds is a highly promising way to develop new therapeutic applications. Indeed, their use in combination with phage therapy treatments could increase bacterial sensitivity to phages by synergistic effects. In addition, disturbing the QS of one species was demonstrated to induce a reduction in total biomass in multimicrobial cultures under phage infection, leading to the consideration that QQ combined with phage therapy could as well be efficient against polymicrobial infections (Mumford and Friman, 2017). To prove the efficiency of QQ as antivirulent agent proper in vivo assays and proof of concepts on animal models should be performed.
In order to evaluate the role of QS in pathogenicity several models have been developed over the past few years. Three models, from a simple unicellular model to complex models, commonly used to assess the benefits of QQ and studies conducted on humans are summarized below.
Free-living amoebae are eukaryotic organisms found either in a resting (cyst) or a vegetative (trophozoite) form feeding on bacteria among other organisms (algae or fungi). In this way, they use phagocytosis coupled with lysosomal digestion which is close to macrophage bacterial elimination pathway (Greub and Raoult, 2004; Matz and Kjelleberg, 2005; Hilbi et al., 2007). Considering these close interactions, amoeba were considered to test bacterial production of virulence factors (Cosson et al., 2002; Clamens et al., 2017), biofilm (Matz et al., 2004, 2005), and secretion systems (Pukatzki et al., 2002, 2006; Matz et al., 2008). Classically, the evaluation of bacterial virulence in amoebae relies on the capacity of amoebae to grow or not to grow in the presence of pathogenic bacteria (Cosson et al., 2002; Pukatzki et al., 2002). The link between virulence factors and QS in different bacterial species was evaluated using this approach and was extensively described for P. aeruginosa. QS-deficient mutants of P. aeruginosa had a decreased virulence toward the amoeba Dictyostelium discoideum (Cosson et al., 2002; Pukatzki et al., 2002). Although this model is fast and convenient for large scale experiments such as screening assays, the use of this model is limited, as both culture conditions and amoeba species may strongly affect the results (Weitere et al., 2005). Despite these limitations the amoeba model was recently used to test a QQ enzyme based on the well-characterized assay with P. aeruginosa and D. discoideum (Clamens et al., 2017). The over-production of P. aeruginosa PA14 aliphatic amidase AmiE resulted in a disruption of QS and reduction of virulence in a D. discoideum plate killing assay (Clamens et al., 2017).
The roundworm Caenorhabditis elegans is a widely used multicellular organism model to study microbial virulence (Tan et al., 1999b; Garsin et al., 2001; Alegado et al., 2003; Ermolaeva and Schumacher, 2014). Like amoebae, C. elegans is a convenient model for high throughput evaluation of QS impact on bacterial virulence (Tan et al., 1999b; Garsin et al., 2001; Rasmussen et al., 2005; O'Loughlin et al., 2013). However, in contrast to amoebae, C. elegans has an innate immune system which results in a closer comparison with the human immune response (Ermolaeva and Schumacher, 2014) and is particularly relevant for studying pathogenicity. Classically, C. elegans is fed using the bacteria of interest and the survival rate is followed. Two types of assays can be performed: (i) a fast killing assay which leads to worm death in a few hours to assess the presence of toxins, and (ii) a slow killing assay with death occurring after several days to evaluate bacterial colonization (Tan et al., 1999a; Köthe et al., 2003; Park et al., 2017).
In order to decipher the importance of QS in virulence, many experiments were dedicated to study the pathogenicity of bacterial mutants impaired in AI synthesis or perception. QS inactivation in different P. aeruginosa strains resulted in a decrease in worm mortality (Darby et al., 1999; Tan et al., 1999b; O'Loughlin et al., 2013; Mukherjee et al., 2017). The C. elegans model was also used to show the link between QS and virulence of other various Gram-negative bacteria including Chromobacterium violaceum (Swem et al., 2009), E. coli (Lee et al., 2011), Yersinia pseudotuberculosis (Atkinson et al., 2011), B. cepacia (Köthe et al., 2003), Burkholderia cenocepacia (Deng et al., 2012), or Burkholderia pseudomallei (Song et al., 2005). Moreover, the link between QS and pathogenicity was also shown for Gram-positive bacteria such as Enterococcus faecalis (Garsin et al., 2001; Sifri et al., 2002) and S. aureus (Sifri et al., 2003). Considered as a whole, these studies highlight that QS triggers virulence in many bacteria.
In addition to genetic mutations, the roundworm model was used, alongside traditional in vitro tests, to prove the efficiency of QSIs as well as QQ enzymes or bacteria (Table 1). Though the impact on survival may vary according to the assay used and the culture conditions, all the QQ agents tested were shown to efficiently decrease virulence in both Gram-positive and negative bacteria and thus enhancing C. elegans survival up to 100% notably with the QQ enzyme BpiB09 targeting AHLs (Bijtenhoorn et al., 2011). The QSI having the more drastic effect on C. elegans survival after infection with P. aeruginosa PAO1 is 4-nitro-pyridine-N-oxide, a non-toxic chemical compound, which almost fully restored worm survival (Rasmussen et al., 2005). The most efficient natural QSIs are extracts from Conocarpus, Callistemon vinimalis, or Bucida buceras with a restoration of survival up to 87% (Adonizio et al., 2008; Table 1). Moreover, a synergistic effect with antibiotics was reported for the QSIs baicalin and hamamelitannin (Brackman et al., 2011b).
Caenorhabditis elegans is a highly valuable invertebrate model enabling high throughput screening (for bacterial mutants or QQ compounds) and gives a very deep insight into virulence regulation, modulation by QQ agents and, in general, by anti-infective molecules (Kong et al., 2016). In most cases, QQ with either QSI or enzymes seems to be able to reduce mortality due to a wide range of bacteria in C. elegans and thus gives a relevant proof of concept of QQ as antivirulent agent in a multicellular organism. However, it also has some limitations, such as the living parameters of the worm which differ from bacterial ones (e.g., growth temperature around 20°C), and the physiopathology of the roundworm which is very different from the human one. Furthermore, as for amoebae, the influence of assay conditions on the outcome of the assay have been highlighted by several studies (Mahajan-Miklos et al., 1999; Tan et al., 1999a; Gallagher and Manoil, 2001).
Mammalian models, such as rats or mice, are commonly used to decipher the impact of QS in bacterial infections. Indeed, mutations or deletions of QS related genes were shown to reduce the mortality or severity of the infections in the lungs (Pearson et al., 2000; Wu et al., 2001; Lesprit et al., 2003; Sokol, 2003), wound burns (Rumbaugh et al., 1999; Tan et al., 1999b), peritonitis (Sifri et al., 2002), the prostate (Nelson et al., 2009), and the intraperitoneal foreign body model (Christensen et al., 2007), with the exception of Staphylococcus epidermidis (Xu et al., 2006). In the vast majority of cases, QQ approaches result in a decrease in mortality, accelerate recovery and reduce bacterial colonization.
In lung infection models, P. aeruginosa colonization or related mortality was reduced by furanones (Hentzer et al., 2003; Wu, 2004), sub-minimal inhibitory concentrations of azithromycin (Hoffmann et al., 2007), garlic extract (Bjarnsholt, 2005a), and also by the inhalation of the lactonase SsoPox (Hraiech et al., 2014). In skin wound models, a wide range of QSIs reduced S. aureus pathogenicity (Cirioni et al., 2013; Simonetti et al., 2016; Muhs et al., 2017; Todd et al., 2017). Similar inhibition of pathogenicity was observed with an AIP-targeting antibody (Park et al., 2007). Treatment with QS inhibiting peptide strongly reduced S. epidermidis colonization in a graft associated infection (Balaban et al., 2003). The efficiency of QQ was also demonstrated in a burn wound infection model with P. aeruginosa and an AHL degrading enzyme (Gupta et al., 2015) or PqsR (MvfR) inhibitors (Lesic et al., 2007) and, in an excision injury model, with the use of tea polyphenols as QSIs (Yin et al., 2015). Moreover, the combination effect of QQ molecules and antibiotics in vivo was demonstrated against both Gram-positive and negative bacteria. Indeed, for B. cenocepacia, the combination of baicalin and tobramycin allowed to reduce lung colonization by 2 and 1 log of CFU compare to control and antibiotic alone respectively (Brackman et al., 2011a). The use of ciprofloxacin and a lactonase to treat P. aeruginosa wound burn infection enabled to reduce mortality and global bacterial dissemination to internal mice organs (Gupta et al., 2015). Furthermore, the cotreatment with a QSI and an antibiotic also drastically reduced colonization of artificial foreign body (e.g., catheter or implants) by S. aureus (Cirioni et al., 2013; Simonetti et al., 2016), S. epidermidis (Balaban et al., 2003), and P. aeruginosa (Christensen et al., 2012; Das et al., 2016). Those examples increased the interest of reducing antibiotic tolerance by QQ either in infected organs or medical device associated infections.
Murine models are useful and common tools to investigate the QQ impact on bacterial infections thanks to their adaptive and innate immune systems together with a physiology closely related to human beings. Furthermore, they are usually necessary and required as preclinical tests before starting human trials. At this stage of drug development, QQ seems to demonstrate great efficiency to reduce either morbidity or deleterious impacts for a wide variety of infections. However, murine models are less prone to screening steps because of practical and ethical problems unlike C. elegans or amoeba (van der Worp et al., 2010). Furthermore, some physiological aspects of a pathology are not fully mimicked in murine models like wound healing or inflammation (van der Worp et al., 2010; Seok et al., 2013; Abdullahi et al., 2014).
So far, only previously approved or commercialized QSIs were used in clinical trials, even if their primary use and approved biological activity did not relate to bacterial QS at all, but rather to their bactericidal, antimicrobial activities (antibiotics) or their cytotoxicity (anti-cancer molecules) (Walz et al., 2010; van Delden et al., 2012).
In the early 2000s, azithromycin (Figure 3) was used in clinical trials to treat cystic fibrosis (Wolter et al., 2002; Saiman et al., 2003) and pulmonary transplanted patients (Gerhardt et al., 2003). This macrolide antibiotic improved patient's quality of life but did not lead to a decrease of bacterial load (Saiman et al., 2003). At the same period, the ability of azithromycin at non bactericidal concentrations to disrupt bacterial signaling in P. aeruginosa was demonstrated in vitro (Tateda et al., 2001). Later, the impact of azithromycin on P. aeruginosa QS in ventilator-associated pneumonia patients was evaluated (van Delden et al., 2012). The authors described beneficial anti-virulence effects of azithromycin in a high-risk group of patients, yet results were not significant enough.
Garlic is also known for its QQ properties (Rasmussen et al., 2005) and was used in a trial to treat cystic fibrosis patients, although, no clear evidence has emerged of the curative effect of garlic extract on patient health (Smyth et al., 2010).
Finally, the anti-cancer drug (Longley et al., 2003), 5-FU, a pyrimidine analog (Figure 3), was demonstrated to inhibit QS-regulated virulence in P. aeruginosa in vitro (Ueda et al., 2009) and was further used for the coating of functionalized catheters, which were shown to be efficient during clinical trials (Jacobsen et al., 2008; Walz et al., 2010).
In the end, very few QQ molecules reached human clinical trials but they tend to demonstrate some beneficial effects of QSIs. Although many proofs of concept were performed in animal models, further efforts have to be dedicated to the validation of this approach in clinical phases to confirm its therapeutic relevance.
Medical devices are involved in numerous HAIs (Neoh et al., 2017). Multi-drug resistant and/or biofilm forming bacteria are mainly responsible for HAIs causing severe medical complications, high morbidity and risk of mortality. Considering the ability of QQ to prevent bacterial virulence (Grandclément et al., 2016), the development of novel medical devices using QQ agents is of outmost interest. New generations of catheters (Mandakhalikar et al., 2016), dressings (Rémy et al., 2016a; Bzdrenga et al., 2017), aerosols (Hraiech et al., 2014), contact lenses (Jain et al., 2016), implantable devices (Francolini et al., 2017), or orthopedic and trauma devices (Moriarty et al., 2016) are currently being developed (Table 2).
QSIs were first considered for the functionalization of catheters. Covalently-attached furanones were shown to decrease biofilm formation by S. epidermidis ATCC 35984 and to control infection for 65 days in an in vivo sheep model (Hume et al., 2004). 5-FU was used to coat a central venous catheter and was demonstrated to be efficient and comparable to classically used chlorhexidine/silver sulfadiazine coated catheters in a clinical study involving 960 adult patients in 25 US intensive care units (Jacobsen et al., 2008; Walz et al., 2010). Although the link to QS was not made by the authors, the 5-FU coated catheters showed reduced contamination levels, by Gram-negative bacteria, as compared to the traditional coating which could be a clue as to interference with AHL dependent QS in this study. Poly(ethylene glycol)-based coating containing the QSI DHP (5-methylene-1-(prop-2-enoyl)-4-(2-fluorophenyl)-dihydropyrrol-2-one) was recently shown to reduce S. aureus strain 38 and P. aeruginosa MH602 colonization (Ozcelik et al., 2017). Combinations of DHP and furanone derivatives were also covalently attached onto glass surfaces and significantly reduced the adhesion of S. aureus SA38 and P. aeruginosa PAO1 (Taunk et al., 2016). A delivery system based on varnishes releasing the QSI thiazolidinedione-8 (TZD-8) was used on catheters and were active against Candida albicans biofilms (Shenderovich et al., 2015). Notably, honey polyphenols were introduced into a scaffold of selenium nanovectors for quenching P. aeruginosa PAO1 in vitro and in vivo (Prateeksha et al., 2017).
For agr-based QS in S. aureus, inhibiting peptides were also successfully incorporated into biomaterials. Macrocyclic peptides were loaded into non-woven polymer nanofibers by electrospinning and showed to retain biological activity against S. aureus after releasing over a 3 week period (Kratochvil et al., 2017). Click chemistry was also considered for covalently coating surfaces with pro- and anti-QS peptides, AIP-I and TrAIP-II respectively and showed efficacy against S. aureus strains (Kim et al., 2017). The synergy of QS inhibiting peptide FS3 with antibiotics was also demonstrated, with daptomycin being highly effective against staphylococcal infections when combined with a FS3-coated prosthesis (Cirioni et al., 2013). Similarly, for several strains of S. epidermidis, the RNAIII-Inhibiting peptide (RIP) was efficient in reducing infection when incorporated into a Dacron graft (Balaban et al., 2003).
Although QS inhibiting materials were obtained after covalent immobilization of QSIs or peptides, QQ enzymes were also thoroughly investigated as these compounds, acting on secreted autoinducers, do not need direct contact with the cells to disrupt communication. Acylase from Aspergillus melleus was successfully incorporated into polyurethane coatings and silicon catheters reducing biofilm formation of P. aeruginosa ATCC 10145 and PAO1 respectively (Ivanova et al., 2015b; Grover et al., 2016). Combination of the acylase with α-amylase from Bacillus amyloliquefaciens delayed biofilm development of both P. aeruginosa ATCC 10145 and E. coli ATCC 25922 for up to 7 days in an in vivo rabbit model (Ivanova et al., 2015a). Acylase from porcine kidney was also immobilized on carboxylated polyaniline nanofibers for the development of nanobiocatalysts limiting biofilm formation of P. aeruginosa PAO1. Topical treatment involving lactonase from Bacillus sp. ZA12 was also investigated in a burn infection model on mice using P. aeruginosa PAO1 (Gupta et al., 2015). Application of a lactonase-containing gel after 106 bacteria burn infection prevented systemic spread, decreased mortality, and showed synergistic effect with ciprofloxacin.
Because enzyme stability is a major bottleneck in the development of bio-based materials, catalysts from extremophile environments were considered. Particularly, PLL SsoPox from Sulfolobus solfataricus was found to be a highly attractive way of quenching bacterial virulence (Rémy et al., 2016a; Bzdrenga et al., 2017). This highly robust enzyme (Hiblot et al., 2012; Rémy et al., 2016b), was first immobilized onto nanoalumina membranes while retaining strong efficacy for reducing virulence factor secretions, pyocyanin and elastase of P. aeruginosa PAO1 (Ng et al., 2011). The variant enzyme SsoPox-W263I was further shown to significantly reduce the virulence of 51 clinical isolates of P. aeruginosa from diabetic foot ulcerations and kept its efficiency toward PAO1 after immobilization into polyurethane coating via glutaraldehyde crosslinking (Guendouze et al., 2017). The in vivo use of this variant was also reported through intratracheal administration and drastically enhanced the survival rate in a rat pneumonia model infected by P. aeruginosa PAO1 (Hraiech et al., 2014).
In addition to the studies using AHL-based QS quenchers, a recent report described the use of the AI-2 processing kinase LsrK. This enzyme was attached to a capsule of biological polymers chitosan and alginate supplemented with ATP substrate and reduced AI-2 mediated QS (Rhoads et al., 2017).
QQ-based devices have raised special attention considering that they could prevent HAIs by limiting bacterial virulence and biofilm formation. However, further efforts have to be dedicated to validate the proof of concepts in vivo and in clinical phases. The efficacy of these devices has to be demonstrated not only in model bacterial strains but also on genetically and phenotypically diverse clinical isolates. Although the development of medical devices is less constrained than for drugs, further regulatory concerns have to be considered to confirm the potential of the techniques for therapeutic applications. Nevertheless, the wide spectrum of both QSI and QQE as well as the numerous examples of their medical relevance would pave the way to the emergence of innovative devices.
Over the past 15 years, many studies have demonstrated that QQ molecules and QQ approaches have great potential as anti-infective agents against a broad range of bacteria. This is evidenced by the numerous studies demonstrating the benefit of these approaches in functionalizing medical devices. To date, little is known about potential resistance mechanisms that bacteria could develop to overcome QQ (Defoirdt et al., 2010; García-Contreras et al., 2013). The apparition of resistance phenomenon results from the natural process of evolution in a context of selection pressure which favors the growth of resistant strains. This is the case for antibiotics that apply high selection pressure, through growth inhibition, for sensitive strains (Davies and Davies, 2010). If some QSIs such as azithromycin lead to severe growth inhibition, others have only moderate or no effect on growth rate (García-Contreras et al., 2015). Apparition of QQ resistant bacteria is possible but its rate might be slower as compared to antibiotic resistance and will depend on the type of QQ (QSIs or QQEs) and its impact on bacterial growth (García-Contreras et al., 2016). QQ resistant strains have already been reported either from laboratory experiments or from clinical samples, notably strains with lower uptake or higher efflux of QSIs (Maeda et al., 2012). Nevertheless, little is known on how QQ resistant strains would over grow QQ sensitive strains nor on how the population would evolve. Most studies performed to address this question were performed in vitro and using QS mutants or QSIs, the results obtained so far were not consistent (Griffin et al., 2004; Gerdt and Blackwell, 2014; García-Contreras et al., 2016). To limit QQ resistance, QQ agents should be carefully chosen to keep growth deleterious effects minimal. Many QSIs have toxic activities and need to enter the cells in order to be active. QQ enzymes may represent ideal candidates combining a minimal, if any, selection pressure and potent inhibitory effects on biofilm formation and virulence (Guendouze et al., 2017). Further studies involving QQ enzymes should be performed to evaluate potential QQ resistance mechanisms using QQ enzymes. Nonetheless, QQ is a promising strategy to extend the therapeutic arsenal available to treat bacterial infections in complement to classical antimicrobial agents and antibiotics or reemerging bacteriophages.
The broad effect of QS on the physiology of bacteria shows that QQ would be an appropriate strategy not only to reducing bacterial virulence but also in terms of restoring antibiotic tolerance by decreasing biofilm formation and in terms of decreasing bacterial phage resistance, paving the way for future combination therapies.
Remarkably, the disruption of bacterial signaling, a communication system central to microbial communities (McFall-Ngai et al., 2013), has implications that go beyond the single bacteria physiology. Indeed, the gut microbiota of fishes fed with probiotic bacteria, producing QQ enzymes, was modified and the population of pathogenic Aeromonas hydrophila was reduced (Zhou et al., 2016). In another approach, a recent study showed the ability of E. coli overproducing AI-2 to counter the impact of streptomycin-induced gut dysbiosis potentially underlining the role of quorum sensing in the context of complex microbiota (Thompson et al., 2015). More studies are needed to delineate the effects induced by QQ strategies at both the single bacterial species level and in the context of communities. Future investigations will determine the breadth of the action of QQ molecules and their potential in being used as therapy, combination therapy and as coating agents in medical devices.
BR, SM, LP, ME, EC, and DD: conceived and designed the work; BR, SM, LP, and DD: performed survey and drafted the paper; BR, SM, LP, ME, EC, and DD: critically revised the manuscript. All authors read and approved the final manuscript.
ME and EC have a patent WO2014167140 A1 licensed to Gene&GreenTK.
LP, DD, and EC report personal fees from Gene&GreenTK during the conduct of the study.
The other authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The handling editor declared a past co-authorship with the authors ME, EC, and DD.
This work is granted by a project RAPID (LACTO-TEX) from Direction Générale de l'Armement (DGA, France). This work was also supported by Investissements d'avenir program (Méditerranée Infection 10-IAHU-03) of the French Agence Nationale de la Recherche (ANR). BR is a Ph.D. student granted by the Emplois Jeunes Doctorants program of Région Provence-Alpes-Côte d'Azur (PACA, France). SM is Ph.D. student granted by Direction Générale de l'Armement (DGA).
Abdullahi, A., Amini-Nik, S., and Jeschke, M. G. (2014). Animal models in burn research. Cell. Mol. Life Sci. 71, 3241–3255. doi: 10.1007/s00018-014-1612-5
Abedon, S. T. (2012). Spatial vulnerability: bacterial arrangements, microcolonies, and biofilms as responses to low rather than high phage densities. Viruses 4, 663–687. doi: 10.3390/v4050663
Adonizio, A., Leal, S. M., Ausubel, F. M., Mathee, K. (2008). Attenuation of Pseudomonas aeruginosa virulence by medicinal plants in a Caenorhabditis elegans model system. J. Med. Microbiol. 57, 809–813. doi: 10.1099/jmm.0.47802-0
Alegado, R. A., Campbell, M. C., Chen, W. C., Slutz, S. S., and Tan, M.-W. (2003). Characterization of mediators of microbial virulence and innate immunity using the Caenorhabditis elegans host–pathogen model. Cell. Microbiol. 5, 435–444. doi: 10.1046/j.1462-5822.2003.00287.x
Amara, N., Krom, B. P., Kaufmann, G. F., and Meijler, M. M. (2011). Macromolecular inhibition of quorum sensing: enzymes, antibodies, and beyond. Chem. Rev. 111, 195–208. doi: 10.1021/cr100101c
Atkinson, S., Goldstone, R. J., Joshua, G. W., Chang, C.-Y., Patrick, H. L., Cámara, M., et al. (2011). Biofilm development on Caenorhabditis elegans by Yersinia is facilitated by quorum sensing-dependent repression of type III secretion. PLoS Pathog. 7:e1001250. doi: 10.1371/journal.ppat.1001250
Balaban, N., Giacometti, A., Cirioni, O., Gov, Y., Ghiselli, R., Mocchegiani, F., et al. (2003). Use of the quorum-sensing inhibitor RNAIII-inhibiting peptide to prevent biofilm formation in vivo by drug-resistant Staphylococcus epidermidis. J. Infect. Dis. 187, 625–630. doi: 10.1086/345879
Barrangou, R., Fremaux, C., Deveau, H., Richards, M., Boyaval, P., Moineau, S., et al. (2007). CRISPR provides acquired resistance against viruses in prokaryotes. Science 315, 1709–1712. doi: 10.1126/science.1138140
Bijtenhoorn, P., Mayerhofer, H., Müller-Dieckmann, J., Utpatel, C., Schipper, C., Hornung, C., et al. (2011). A novel metagenomic short-chain dehydrogenase/reductase attenuates pseudomonas aeruginosa biofilm formation and virulence on Caenorhabditis elegans. PLoS ONE 6:e26278. doi: 10.1371/journal.pone.0026278
Bjarnsholt, T. (2005a). Garlic blocks quorum sensing and promotes rapid clearing of pulmonary Pseudomonas aeruginosa infections. Microbiology 151, 3873–3880. doi: 10.1099/mic.0.27955-0
Bjarnsholt, T. (2005b). Pseudomonas aeruginosa tolerance to tobramycin, hydrogen peroxide and polymorphonuclear leukocytes is quorum-sensing dependent. Microbiology 151, 373–383. doi: 10.1099/mic.0.27463-0
Bowden, S. D., Eyres, A., Chung, J. C., Monson, R. E., Thompson, A., Salmond, G. P., et al. (2013). Virulence in Pectobacterium atrosepticum is regulated by a coincidence circuit involving quorum sensing and the stress alarmone, (p)ppGpp. Mol. Microbiol. 90, 457–471. doi: 10.1111/mmi.12369
Brackman, G., Celen, S., Hillaert, U., Van Calenbergh, S., Cos, P., Maes, L., et al. (2011a). Structure-activity relationship of cinnamaldehyde analogs as inhibitors of AI-2 based quorum sensing and their effect on virulence of Vibrio spp. PLoS ONE 6:e16084. doi: 10.1371/journal.pone.0016084
Brackman, G., Cos, P., Maes, L., Nelis, H. J., and Coenye, T. (2011b). Quorum sensing inhibitors increase the susceptibility of bacterial biofilms to antibiotics in vitro and in vivo. Antimicrob. Agents Chemother. 55, 2655–2661. doi: 10.1128/AAC.00045-11
Brauner, A., Fridman, O., Gefen, O., and Balaban, N. Q. (2016). Distinguishing between resistance, tolerance and persistence to antibiotic treatment. Nat. Rev. Microbiol. 14, 320–330. doi: 10.1038/nrmicro.2016.34
Brüssow, H., and Hendrix, R. W. (2002). Phage genomics: small is beautiful. Cell 108, 13–16. doi: 10.1016/S0092-8674(01)00637-7
Bulman, Z. P., Ly, N. S., Lenhard, J. R., Holden, P. N., Bulitta, J. B., and Tsuji, B. T. (2017). Influence of rhlR and lasR on polymyxin pharmacodynamics in Pseudomonas aeruginosa and implications for quorum sensing inhibition with azithromycin. Antimicrob. Agents Chemother. 61:e00096-16. doi: 10.1128/AAC.00096-16
Bzdrenga, J., Daudé, D., Rémy, B., Jacquet, P., Plener, L., Elias, M., et al. (2017). Biotechnological applications of quorum quenching enzymes. Chem. Biol. Interact. 267, 104–115. doi: 10.1016/j.cbi.2016.05.028
Chapman-McQuiston, E., and Wu, X. L. (2008). Stochastic receptor expression allows sensitive bacteria to evade phage attack. part i: experiments. Biophys. J. 94, 4525–4536. doi: 10.1529/biophysj.107.120212
Chen, X., Schauder, S., Potier, N., Van Dorsselaer, A., Pelczer, I., Bassler, B. L., et al. (2002). Structural identification of a bacterial quorum-sensing signal containing boron. Nature 415, 545–549. doi: 10.1038/415545a
Christensen, L. D., Moser, C., Jensen, P. Ø., Rasmussen, T. B., Christophersen, L., Kjelleberg, S., et al. (2007). Impact of Pseudomonas aeruginosa quorum sensing on biofilm persistence in an in vivo intraperitoneal foreign-body infection model. Microbiology 153, 2312–2320. doi: 10.1099/mic.0.2007/006122-0
Christensen, L. D., van Gennip, M., Jakobsen, T. H., Alhede, M., Hougen, H. P., Høiby, N., et al. (2012). Synergistic antibacterial efficacy of early combination treatment with tobramycin and quorum-sensing inhibitors against Pseudomonas aeruginosa in an intraperitoneal foreign-body infection mouse model. J. Antimicrob. Chemother. 67, 1198–1206. doi: 10.1093/jac/dks002
Christiaen, S. E., Matthijs, N., Zhang, X.-H., Nelis, H. J., Bossier, P., and Coenye, T. (2014). Bacteria that inhibit quorum sensing decrease biofilm formation and virulence in Pseudomonas aeruginosa PAO1. Pathog. Dis. 70, 271–279. doi: 10.1111/2049-632X.12124
Cirioni, O., Mocchegiani, F., Cacciatore, I., Vecchiet, J., Silvestri, C., Baldassarre, L., et al. (2013). Quorum sensing inhibitor FS3-coated vascular graft enhances daptomycin efficacy in a rat model of staphylococcal infection. Peptides 40, 77–81. doi: 10.1016/j.peptides.2012.12.002
Clamens, T., Rosay, T., Crépin, A., Grandjean, T., Kentache, T., Hardouin, J., et al. (2017). The aliphatic amidase AmiE is involved in regulation of Pseudomonas aeruginosa virulence. Sci. Rep. 7:41178. doi: 10.1038/srep41178
Cosson, P., Zulianello, L., Join-Lambert, O., Faurisson, F., Gebbie, L., Benghezal, M., et al. (2002). Pseudomonas aeruginosa virulence analyzed in a dictyostelium discoideum host system. J. Bacteriol. 184, 3027–3033. doi: 10.1128/JB.184.11.3027-3033.2002
Darby, C., Cosma, C. L., Thomas, J. H., and Manoil, C. (1999). Lethal paralysis of Caenorhabditis elegans by Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 96, 15202–15207. doi: 10.1073/pnas.96.26.15202
Das, M. C., Sandhu, P., Gupta, P., Rudrapaul, P., De, U. C., Tribedi, P., et al. (2016). Attenuation of Pseudomonas aeruginosa biofilm formation by Vitexin: A combinatorial study with azithromycin and gentamicin. Sci. Rep. 6:23347. doi: 10.1038/srep23347
Davies, D. G., Parsek, M. R., Pearson, J. P., Iglewski, B. H., Costerton, J. W., and Greenberg, E. P. (1998). The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 280, 295–298. doi: 10.1126/science.280.5361.295
Davies, J., and Davies, D. (2010). Origins and evolution of antibiotic resistance. Microbiol. Mol. Biol. Rev. 74, 417–433. doi: 10.1128/MMBR.00016-10
Defoirdt, T., Boon, N., and Bossier, P. (2010). Can bacteria evolve resistance to quorum sensing disruption? PLoS Pathog. 6:e1000989. doi: 10.1371/journal.ppat.1000989
Defoirdt, T., Brackman, G., and Coenye, T. (2013). Quorum sensing inhibitors: how strong is the evidence? Trends Microbiol. 21, 619–624. doi: 10.1016/j.tim.2013.09.006
de la Fuente-Núñez, C., Reffuveille, F., Fernández, L., and Hancock, R. E. (2013). Bacterial biofilm development as a multicellular adaptation: antibiotic resistance and new therapeutic strategies. Curr. Opin. Microbiol. 16, 580–589. doi: 10.1016/j.mib.2013.06.013
Delago, A., Mandabi, A., and Meijler, M. M. (2016). Natural quorum sensing inhibitors–small molecules, big messages. Isr. J. Chem. 56, 310–320. doi: 10.1002/ijch.201500052
Dembitsky, V. M., Al Quntar, A. A., and Srebnik, M. (2011). Natural and synthetic small boron-containing molecules as potential inhibitors of bacterial and fungal quorum sensing. Chem. Rev. 111, 209–237. doi: 10.1021/cr100093b
Deng, Y., Schmid, N., Wang, C., Wang, J., Pessi, G., Wu, D., et al. (2012). Cis-2-dodecenoic acid receptor RpfR links quorum-sensing signal perception with regulation of virulence through cyclic dimeric guanosine monophosphate turnover. Proc. Natl. Acad. Sci. U.S.A. 109, 15479–15484. doi: 10.1073/pnas.1205037109
Dong, Y.-H., Xu, J.-L., Li, X.-Z., and Zhang, L.-H. (2000). AiiA, an enzyme that inactivates the acylhomoserine lactone quorum-sensing signal and attenuates the virulence of Erwinia carotovora. Proc. Natl. Acad. Sci. U.S.A. 97, 3526–3531. doi: 10.1073/pnas.97.7.3526
Driffield, K., Miller, K., Bostock, J. M., O'Neill, A. J., and Chopra, I. (2008). Increased mutability of Pseudomonas aeruginosa in biofilms. J. Antimicrob. Chemother. 61, 1053–1056. doi: 10.1093/jac/dkn044
Ermolaeva, M. A., and Schumacher, B. (2014). Insights from the worm: the, C. elegans model for innate immunity. Semin. Immunol. 26, 303–309. doi: 10.1016/j.smim.2014.04.005
Fetzner, S. (2015). Quorum quenching enzymes. J. Biotechnol. 201, 2–14. doi: 10.1016/j.jbiotec.2014.09.001
Flavier, A. B., Clough, S. J., Schell, M. A., and Denny, T. P. (1997). Identification of 3-hydroxypalmitic acid methyl ester as a novel autoregulator controlling virulence in Ralstonia solanacearum. Mol. Microbiol. 26, 251–259. doi: 10.1046/j.1365-2958.1997.5661945.x
Francolini, I., Vuotto, C., Piozzi, A., and Donelli, G. (2017). Antifouling and antimicrobial biomaterials: an overview. APMIS 125, 392–417. doi: 10.1111/apm.12675
Furiga, A., Lajoie, B., El Hage, S., Baziard, G., and Roques, C. (2016). Impairment of Pseudomonas aeruginosa biofilm resistance to antibiotics by combining the drugs with a new quorum-sensing inhibitor. Antimicrob. Agents Chemother. 60, 1676–1686. doi: 10.1128/AAC.02533-15
Gallagher, L. A., and Manoil, C. (2001). Pseudomonas aeruginosa PAO1 Kills Caenorhabditis elegans by cyanide poisoning. J. Bacteriol. 183, 6207–6214. doi: 10.1128/JB.183.21.6207-6214.2001
Galloway, W. R., Hodgkinson, J. T., Bowden, S. D., Welch, M., and Spring, D. R. (2011). Quorum sensing in gram-negative bacteria: small-molecule modulation of AHL and AI-2 quorum sensing pathways. Chem. Rev. 111, 28–67. doi: 10.1021/cr100109t
Gao, R., Krysciak, D., Petersen, K., Utpatel, C., Knapp, A., Schmeisser, C., et al. (2015). Genome-wide RNA sequencing analysis of quorum sensing-controlled regulons in the plant-associated burkholderia glumae PG1 strain. Appl. Environ. Microbiol. 81, 7993–8007. doi: 10.1128/AEM.01043-15
García-Contreras, R., Maeda, T., and Wood, T. K. (2013). Resistance to quorum-quenching compounds. Appl. Environ. Microbiol. 79, 6840–6846. doi: 10.1128/AEM.02378-13
García-Contreras, R., Maeda, T., and Wood, T. K. (2016). Can resistance against quorum-sensing interference be selected? ISME J. 10, 4–10. doi: 10.1038/ismej.2015.84
García-Contreras, R., Peréz-Eretza, B., Jasso-Chávez, R., Lira-Silva, E., Roldán-Sánchez, J. A., González-Valdez, A., et al. (2015). High variability in quorum quenching and growth inhibition by furanone C-30 in Pseudomonas aeruginosa clinical isolates from cystic fibrosis patients. Pathog. Dis. 73:ftv040. doi: 10.1093/femspd/ftv040
Garsin, D. A., Sifri, C. D., Mylonakis, E., Qin, X., Singh, K. V., Murray, B. E., et al. (2001). A simple model host for identifying Gram-positive virulence factors. Proc. Natl. Acad. Sci. U.S.A. 98, 10892–10897. doi: 10.1073/pnas.191378698
Gerdt, J. P., and Blackwell, H. E. (2014). Competition studies confirm two major barriers that can preclude the spread of resistance to quorum-sensing inhibitors in bacteria. ACS Chem. Biol. 9, 2291–2299. doi: 10.1021/cb5004288
Gerhardt, S. G., McDyer, J. F., Girgis, R. E., Conte, J. V., Yang, S. C., and Orens, J. B. (2003). Maintenance azithromycin therapy for bronchiolitis obliterans syndrome: results of a pilot study. Am. J. Respir. Crit. Care Med. 168, 121–125. doi: 10.1164/rccm.200212-1424BC
Glessner, A., Smith, R. S., Iglewski, B. H., and Robinson, J. B. (1999). Roles of Pseudomonas aeruginosa las andrhl quorum-sensing systems in control of twitching motility. J. Bacteriol. 181, 1623–1629.
Grandclément, C., Tannières, M., Moréra, S., Dessaux, Y., and Faure, D. (2016). Quorum quenching: role in nature and applied developments. FEMS Microbiol. Rev. 40, 86–116. doi: 10.1093/femsre/fuv038
Greub, G., and Raoult, D. (2004). Microorganisms resistant to free-living amoebae. Clin. Microbiol. Rev. 17, 413–433. doi: 10.1128/CMR.17.2.413-433.2004
Griffin, A. S., West, S. A., and Buckling, A. (2004). Cooperation and competition in pathogenic bacteria. Nature 430, 1024–1027. doi: 10.1038/nature02744
Grover, N., Plaks, J. G., Summers, S. R., Chado, G. R., Schurr, M. J., and Kaar, J. L. (2016). Acylase-containing polyurethane coatings with anti-biofilm activity. Biotechnol. Bioeng. 113, 2535–2543. doi: 10.1002/bit.26019
Guendouze, A., Plener, L., Bzdrenga, J., Jacquet, P., Rémy, B., Elias, M., et al. (2017). Effect of quorum quenching lactonase in clinical isolates of Pseudomonas aeruginosa and comparison with quorum sensing inhibitors. Front. Microbiol. 8:227. doi: 10.3389/fmicb.2017.00227
Guo, Q., Wei, Y., Xia, B., Jin, Y., Liu, C., Pan, X., et al. (2016). Identification of a small molecule that simultaneously suppresses virulence and antibiotic resistance of Pseudomonas aeruginosa. Sci. Rep. 6:19141. doi: 10.1038/srep19141
Gupta, P., Chhibber, S., and Harjai, K. (2015). Efficacy of purified lactonase and ciprofloxacin in preventing systemic spread of Pseudomonas aeruginosa in murine burn wound model. Burns 41, 153–162. doi: 10.1016/j.burns.2014.06.009
Hall, A. R., Scanlan, P. D., Morgan, A. D., and Buckling, A. (2011). Host–parasite coevolutionary arms races give way to fluctuating selection. Ecol. Lett. 14, 635–642. doi: 10.1111/j.1461-0248.2011.01624.x
Hawver, L. A., Jung, S. A., and Ng, W.-L. (2016). Specificity and complexity in bacterial quorum-sensing systems. FEMS Microbiol. Rev. 40, 738–752. doi: 10.1093/femsre/fuw014
Hazan, R., Que, Y. A., Maura, D., Strobel, B., Majcherczyk, P. A., Hopper, L. R., et al. (2016). Auto poisoning of the respiratory chain by a quorum-sensing-regulated molecule favors biofilm formation and antibiotic tolerance. Curr. Biol. 26, 195–206. doi: 10.1016/j.cub.2015.11.056
He, X., Lu, F., Yuan, F., Jiang, D., Zhao, P., Zhu, J., et al. (2015). Biofilm Formation caused by clinical Acinetobacter baumannii isolates is associated with overexpression of the AdeFGH efflux pump. Antimicrob. Agents Chemother. 59, 4817–4825. doi: 10.1128/AAC.00877-15
Heeb, S., Fletcher, M. P., Chhabra, S. R., Diggle, S. P., Williams, P., and Cámara, M. (2011). Quinolones: from antibiotics to autoinducers. FEMS Microbiol. Rev. 35, 247–274. doi: 10.1111/j.1574-6976.2010.00247.x
Heilmann, S., Krishna, S., and Kerr, B. (2015). Why do bacteria regulate public goods by quorum sensing?—how the shapes of cost and benefit functions determine the form of optimal regulation. Front. Microbiol. 6:767. doi: 10.3389/fmicb.2015.00767
Hentzer, M., Wu, H., Andersen, J. B., Riedel, K., Rasmussen, T. B., Bagge, N., et al. (2003). Attenuation of Pseudomonas aeruginosa virulence by quorum sensing inhibitors. EMBO J. 22, 3803–3815. doi: 10.1093/emboj/cdg366
Hiblot, J., Gotthard, G., Chabriere, E., and Elias, M. (2012). Characterisation of the organophosphate hydrolase catalytic activity of SsoPox. Sci. Rep. 2:779. doi: 10.1038/srep00779
Hilbi, H., Weber, S. S., Ragaz, C., Nyfeler, Y., and Urwyler, S. (2007). Environmental predators as models for bacterial pathogenesis. Environ. Microbiol. 9, 563–575. doi: 10.1111/j.1462-2920.2007.01238.x
Hill, D., Rose, B., Pajkos, A., Robinson, M., Bye, P., Bell, S., et al. (2005). Antibiotic susceptibilities of Pseudomonas aeruginosa isolates derived from patients with cystic fibrosis under aerobic, anaerobic, and biofilm conditions. J. Clin. Microbiol. 43, 5085–5090. doi: 10.1128/JCM.43.10.5085-5090.2005
Hoffmann, N., Lee, B., Hentzer, M., Rasmussen, T. B., Song, Z., Johansen, H. K., et al. (2007). Azithromycin blocks quorum sensing and alginate polymer formation and increases the sensitivity to serum and stationary-growth-phase killing of Pseudomonas aeruginosa and attenuates chronic, P. aeruginosa lung infection in Cftr/Mice. Antimicrob. Agents Chemother. 51, 3677–3687. doi: 10.1128/AAC.01011-06
Høiby, N., Bjarnsholt, T., Givskov, M., Molin, S., and Ciofu, O. (2010). Antibiotic resistance of bacterial biofilms. Int. J. Antimicrob. Agents 35, 322–332. doi: 10.1016/j.ijantimicag.2009.12.011
Høiby, N., Ciofu, O., Johansen, H. K., Song, Z., Moser, C., Jensen, P. Ø., et al. (2011). The clinical impact of bacterial biofilms. Int. J. Oral Sci. 3, 55–65. doi: 10.4248/IJOS11026
Hoque, M. M., Naser, I. B., Bari, S. M., Zhu, J., Mekalanos, J. J., and Faruque, S. M. (2016). Quorum regulated resistance of Vibrio cholerae against environmental bacteriophages. Sci. Rep. 6:37956. doi: 10.1038/srep37956
Høyland-Kroghsbo, N. M., Maerkedahl, R. B., and Svenningsen, S. L. (2013). A quorum-sensing-induced bacteriophage defense mechanism. MBio 4:e00362-12. doi: 10.1128/mBio.00362-12
Høyland-Kroghsbo, N. M., Paczkowski, J., Mukherjee, S., Broniewski, J., Westra, E., Bondy-Denomy, J., et al. (2017). Quorum sensing controls the Pseudomonas aeruginosa CRISPR-Cas adaptive immune system. Proc. Natl. Acad. Sci. U.A.S. 114, 131–135. doi: 10.1073/pnas.1617415113
Hraiech, S., Hiblot, J., Lafleur, J., Lepidi, H., Papazian, L., Rolain, J.-M., et al. (2014). Inhaled lactonase reduces Pseudomonas aeruginosa quorum sensing and mortality in rat Pneumonia. PLoS ONE 9:e107125. doi: 10.1371/journal.pone.0107125
Hume, E. B., Baveja, J., Muir, B., Schubert, T. L., Kumar, N., Kjelleberg, S., et al. (2004). The control of Staphylococcus epidermidis biofilm formation and in vivo infection rates by covalently bound furanones. Biomaterials 25, 5023–5030. doi: 10.1016/j.biomaterials.2004.01.048
Husain, F. M., Ahmad, I., Al-thubiani, A. S., Abulreesh, H. H., AlHazza, I. M., and Aqil, F. (2017). Leaf extracts of Mangifera indica L. Inhibit quorum sensing – regulated production of virulence factors and biofilm in test bacteria. Front. Microbiol. 8:727. doi: 10.3389/fmicb.2017.00727
Husain, F. M., Ahmad, I., Asif, M., and Tahseen, Q. (2013). Influence of clove oil on certain quorum-sensing-regulated functions and biofilm of Pseudomonas aeruginosa and Aeromonas hydrophila. J. Biosci. 38, 835–844. doi: 10.1007/s12038-013-9385-9
Husain, F. M., Ahmad, I., Khan, M. S., Ahmad, E., Tahseen, Q., Khan, M. S., et al. (2015a). Sub-MICs of Mentha piperita essential oil and menthol inhibits AHL mediated quorum sensing and biofilm of Gram-negative bacteria. Front. Microbiol. 6:420. doi: 10.3389/fmicb.2015.00420
Husain, F. M., Ahmad, I., Khan, M. S., and Al-Shabib, N. A. (2015b). Trigonella foenum-graceum (Seed) extract interferes with quorum sensing regulated traits and biofilm formation in the strains of Pseudomonas aeruginosa and Aeromonas hydrophila. Evid. Based Complement. Alternat. Med. 2015:879540. doi: 10.1155/2015/879540
Ivanova, K., Fernandes, M. M., Francesko, A., Mendoza, E., Guezguez, J., Burnet, M., et al. (2015a). Quorum-quenching and matrix-degrading enzymes in multilayer coatings synergistically prevent bacterial biofilm formation on urinary catheters. ACS Appl. Mater. Interfaces. 7, 27066–27077. doi: 10.1021/acsami.5b09489
Ivanova, K., Fernandes, M. M., Mendoza, E., and Tzanov, T. (2015b). Enzyme multilayer coatings inhibit Pseudomonas aeruginosa biofilm formation on urinary catheters. Appl. Microbiol. Biotechnol. 99, 4373–4385. doi: 10.1007/s00253-015-6378-7
Jacobsen, S. M., Stickler, D. J., Mobley, H. L., and Shirtliff, M. E. (2008). Complicated catheter-associated urinary tract infections due to Escherichia coli and Proteus mirabilis. Clin. Microbiol. Rev. 21, 26–59. doi: 10.1128/CMR.00019-07
Jain, N., Bhosale, P., and Tale, V. (2016). Biofilm formation on contact lenses by bacterial pathogens. J. Pharm. Res. 10, 50–53.
Jakobsen, T. H., Gennip, M., van, Phipps, R. K., Shanmugham, M. S., Christensen, L. D., Alhede, M., et al. (2012). Ajoene, a sulfur-rich molecule from garlic, inhibits genes controlled by quorum sensing. Antimicrob. Agents Chemother. 56, 2314–2325. doi: 10.1128/AAC.05919-11
Jiang, T., and Li, M. (2013). Quorum sensing inhibitors: a patent review. Expert Opin. Ther. Pat. 23, 867–894. doi: 10.1517/13543776.2013.779674
Kalia, V. C. (2013). Quorum sensing inhibitors: an overview. Biotechnol. Adv. 31, 224–245. doi: 10.1016/j.biotechadv.2012.10.004
Kato, N., Morohoshi, T., Nozawa, T., Matsumoto, H., and Ikeda, T. (2006). Control of gram-negative bacterial quorum sensing with cyclodextrin immobilized cellulose ether gel. J. Incl. Phenom. Macrocycl. Chem. 56, 55–59. doi: 10.1007/s10847-006-9060-y
Kato, N., Tanaka, T., Nakagawa, S., Morohoshi, T., Hiratani, K., and Ikeda, T. (2007). Control of virulence factor expression in opportunistic pathogens using cyclodextrin immobilized gel. J. Incl. Phenom. Macrocycl. Chem. 57, 419–423. doi: 10.1007/s10847-006-9228-5
Kaye, K. S., and Pogue, J. M. (2015). Infections caused by resistant gram-negative bacteria: epidemiology and management. Pharmacotherapy 35, 949–962. doi: 10.1002/phar.1636
Kendall, M. M., and Sperandio, V. (2007). Quorum sensing by enteric pathogens. Curr. Opin. Gastroenterol. 23, 10–15. doi: 10.1097/MOG.0b013e3280118289
Kim, M. K., Zhao, A., Wang, A., Brown, Z. Z., Muir, T. W., Stone, H. A., et al. (2017). Surface-attached molecules control Staphylococcus aureus quorum sensing and biofilm development. Nat. Microbiol. 2:17080. doi: 10.1038/nmicrobiol.2017.80
Klevens, R. M., Edwards, J. R., Richards, C. L., Horan, T. C., Gaynes, R. P., Pollock, D. A., et al. (2007). Estimating health care-associated infections and deaths in U.S. hospitals, (2002). Public Health Rep. 122, 160–166. doi: 10.1177/003335490712200205
Kong, C., Eng, S.-A., Lim, M.-P., and Nathan, S. (2016). Beyond traditional antimicrobials: a Caenorhabditis elegans model for discovery of novel anti-infectives. Front. Microbiol. 7:1956. doi: 10.3389/fmicb.2016.01956
Kostakioti, M., Hadjifrangiskou, M., and Hultgren, S. J. (2013). Bacterial biofilms: development, dispersal, and therapeutic strategies in the dawn of the postantibiotic era. Cold Spring Harb. Perspect. Med. 3:a010306. doi: 10.1101/cshperspect.a010306
Köthe, M., Antl, M., Huber, B., Stoecker, K., Ebrecht, D., Steinmetz, I., et al. (2003). Killing of Caenorhabditis elegans by Burkholderia cepacia is controlled by the cep quorum-sensing system. Cell. Microbiol. 5, 343–351. doi: 10.1046/j.1462-5822.2003.00280.x
Kratochvil, M. J., Yang, T., Blackwell, H. E., and Lynn, D. M. (2017). Nonwoven polymer nanofiber coatings that inhibit quorum sensing in Staphylococcus aureus: toward new nonbactericidal approaches to infection control. ACS Infect. Dis. 3, 271–280. doi: 10.1021/acsinfecdis.6b00173
Labrie, S. J., Samson, J. E., and Moineau, S. (2010). Bacteriophage resistance mechanisms. Nat. Rev. Microbiol. 8, 317–327. doi: 10.1038/nrmicro2315
Lebeaux, D., Chauhan, A., Rendueles, O., and Beloin, C. (2013). From in vitro to in vivo models of bacterial biofilm-related infections. Pathogens 2, 288–356. doi: 10.3390/pathogens2020288
Lee, J., Lee, I., Nam, J., Hwang, D. S., Yeon, K.-M., and Kim, J. (2017). Immobilization and stabilization of acylase on carboxylated polyaniline nanofibers for highly effective antifouling application via quorum quenching. ACS Appl. Mater. Interfaces 9, 15424–15432. doi: 10.1021/acsami.7b01528
Lee, J., and Zhang, L. (2015). The hierarchy quorum sensing network in Pseudomonas aeruginosa. Protein Cell 6, 26–41. doi: 10.1007/s13238-014-0100-x
Lee, K.-M., Lim, J., Nam, S., Yoon, M. Y., Kwon, Y.-K., Jung, B. Y., et al. (2011). Inhibitory effects of broccoli extract on Escherichia coli O157:H7 quorum sensing and in vivo virulence: effects of broccoli extract on bacterial virulence. FEMS Microbiol. Lett. 321, 67–74. doi: 10.1111/j.1574-6968.2011.02311.x
Lesic, B., Lépine, F., Déziel, E., Zhang, J., Zhang, Q., Padfield, K., et al. (2007). Inhibitors of pathogen intercellular signals as selective anti-infective compounds. PLoS Pathog. 3:e126. doi: 10.1371/journal.ppat.0030126
Lesprit, P., Faurisson, F., Join-Lambert, O., Roudot-Thoraval, F., Foglino, M., Vissuzaine, C., et al. (2003). Role of the quorum-sensing system in experimental Pneumonia due to Pseudomonas aeruginosa in rats. Am. J. Respir. Crit. Care Med. 167, 1478–1482. doi: 10.1164/rccm.200207-736BC
Liang, H., Deng, X., Li, X., Ye, Y., and Wu, M. (2014). Molecular mechanisms of master regulator VqsM mediating quorum-sensing and antibiotic resistance in Pseudomonas aeruginosa. Nucleic Acids Res. 42, 10307–10320. doi: 10.1093/nar/gku586
Lin, Y.-H., Xu, J.-L., Hu, J., Wang, L.-H., Ong, S. L., Leadbetter, J. R., et al. (2003). Acyl-homoserine lactone acylase from Ralstonia strain XJ12B represents a novel and potent class of quorum-quenching enzymes. Mol. Microbiol. 47, 849–860. doi: 10.1046/j.1365-2958.2003.03351.x
Lobdell, K. W., Stamou, S., and Sanchez, J. A. (2012). Hospital-acquired infections. Surg. Clin. North Am. 92, 65–77. doi: 10.1016/j.suc.2011.11.003
Longley, D. B., Harkin, D. P., and Johnston, P. G. (2003). 5-Fluorouracil: mechanisms of action and clinical strategies. Nat. Rev. Cancer 3, 330–338. doi: 10.1038/nrc1074
Maeda, T., García-Contreras, R., Pu, M., Sheng, L., Garcia, L. R., Tomás, M., et al. (2012). Quorum quenching quandary: resistance to antivirulence compounds. ISME J. 6, 493–501. doi: 10.1038/ismej.2011.122
Mahajan-Miklos, S., Tan, M.-W., Rahme, L. G., and Ausubel, F. M. (1999). Molecular mechanisms of bacterial virulence elucidated using a Pseudomonas aeruginosa–Caenorhabditis elegans pathogenesis model. Cell 96, 47–56. doi: 10.1016/S0092-8674(00)80958-7
Mandakhalikar, K. D., Chua, R. R., and Tambyah, P. A. (2016). New technologies for prevention of catheter associated urinary tract infection. Curr. Treat. Options Infect. Dis. 8, 24–41. doi: 10.1007/s40506-016-0069-5
Matz, C., Bergfeld, T., Rice, S. A., and Kjelleberg, S. (2004). Microcolonies, quorum sensing and cytotoxicity determine the survival of Pseudomonas aeruginosa biofilms exposed to protozoan grazing. Environ. Microbiol. 6, 218–226. doi: 10.1111/j.1462-2920.2004.00556.x
Matz, C., and Kjelleberg, S. (2005). Off the hook – how bacteria survive protozoan grazing. Trends Microbiol. 13, 302–307. doi: 10.1016/j.tim.2005.05.009
Matz, C., McDougald, D., Moreno, A. M., Yung, P. Y., Yildiz, F. H., and Kjelleberg, S. (2005). Biofilm formation and phenotypic variation enhance predation-driven persistence of Vibrio cholerae. Proc. Natl. Acad. Sci. U.S.A. 102, 16819–16824. doi: 10.1073/pnas.0505350102
Matz, C., Moreno, A. M., Alhede, M., Manefield, M., Hauser, A. R., Givskov, M., et al. (2008). Pseudomonas aeruginosa uses type III secretion system to kill biofilm-associated amoebae. ISME J. 2, 843–852. doi: 10.1038/ismej.2008.47
Maura, D., Hazan, R., Kitao, T., Ballok, A. E., and Rahme, L. G. (2016). Evidence for direct control of virulence and defense gene circuits by the Pseudomonas aeruginosa quorum sensing regulator, MvfR. Sci. Rep. 6:34083. doi: 10.1038/srep34083
Maura, D., and Rahme, L. G. (2017). Pharmacological inhibition of the Pseudomonas aeruginosa MvfR quorum sensing system interferes with biofilm formation and potentiates antibiotic-mediated biofilm disruption. Antimicrob. Agents Chemother. 61:e01362-17. doi: 10.1128/AAC.01362-17
McFall-Ngai, M., Hadfield, M. G., Bosch, T. C., Carey, H. V., Domazet-Lošo, T., Douglas, A. E., et al. (2013). Animals in a bacterial world, a new imperative for the life sciences. Proc. Natl. Acad. Sci. U.S.A. 110, 3229–3236. doi: 10.1073/pnas.1218525110
Möker, N., Dean, C. R., and Tao, J. (2010). Pseudomonas aeruginosa increases formation of multidrug-tolerant persister cells in response to quorum-sensing signaling molecules. J. Bacteriol. 192, 1946–1955. doi: 10.1128/JB.01231-09
Monnet, V., Juillard, V., and Gardan, R. (2016). Peptide conversations in Gram-positive bacteria. Crit. Rev. Microbiol. 42, 339–351. doi: 10.3109/1040841X.2014.948804
Moriarty, T. F., Kuehl, R., Coenye, T., Metsemakers, W.-J., Morgenstern, M., Schwarz, E. M., et al. (2016). Orthopaedic device-related infection: current and future interventions for improved prevention and treatment. EFORT Open Rev. 1, 89–99. doi: 10.1302/2058-5241.1.000037
Morohoshi, T., Tokita, K., Ito, S., Saito, Y., Maeda, S., Kato, N., et al. (2013). Inhibition of quorum sensing in gram-negative bacteria by alkylamine-modified cyclodextrins. J. Biosci. Bioeng. 116, 175–179. doi: 10.1016/j.jbiosc.2013.01.022
Muhs, A., Lyles, J. T., Parlet, C. P., Nelson, K., Kavanaugh, J. S., Horswill, A. R., et al. (2017). Virulence inhibitors from Brazilian peppertree block quorum sensing and abate dermonecrosis in skin infection models. Sci. Rep. 7:42275. doi: 10.1038/srep42275
Mukherjee, S., Moustafa, D., Smith, C. D., Goldberg, J. B., and Bassler, B. L. (2017). The RhlR quorum-sensing receptor controls Pseudomonas aeruginosa pathogenesis and biofilm development independently of its canonical homoserine lactone autoinducer. PLOS Pathog. 13:e1006504. doi: 10.1371/journal.ppat.1006504
Mumford, R., and Friman, V.-P. (2017). Bacterial competition and quorum-sensing signalling shape the eco-evolutionary outcomes of model in vitro phage therapy. Evol. Appl. 10, 161–169. doi: 10.1111/eva.12435
Musthafa, K. S., Balamurugan, K., Pandian, S. K., and Ravi, A. V. (2012a). 2,5-Piperazinedione inhibits quorum sensing-dependent factor production in Pseudomonas aeruginosa PAO1. J. Basic Microbiol. 52, 679–686. doi: 10.1002/jobm.201100292
Musthafa, K. S., Sivamaruthi, B. S., Pandian, S. K., and Ravi, A. V. (2012b). Quorum sensing inhibition in Pseudomonas aeruginosa PAO1 by antagonistic compound phenylacetic acid. Curr. Microbiol. 65, 475–480. doi: 10.1007/s00284-012-0181-9
Nalca, Y., Jänsch, L., Bredenbruch, F., Geffers, R., Buer, J., and Häussler, S. (2006). Quorum-sensing antagonistic activities of azithromycin in Pseudomonas aeruginosa PAO1: a global approach. Antimicrob. Agents Chemother. 50, 1680–1688. doi: 10.1128/AAC.50.5.1680-1688.2006
Nelson, L. K., D'Amours, G. H., Sproule-Willoughby, K. M., Morck, D. W., and Ceri, H. (2009). Pseudomonas aeruginosa las and rhl quorum-sensing systems are important for infection and inflammation in a rat prostatitis model. Microbiology 155, 2612–2619. doi: 10.1099/mic.0.028464-0
Neoh, K. G., Li, M., Kang, E.-T., Chiong, E., and Tambyah, P. A. (2017). Surface modification strategies for combating catheter-related complications: recent advances and challenges. J. Mater. Chem. B 5, 2045–2067. doi: 10.1039/C6TB03280J
Ng, F. S., Wright, D. M., and Seah, S. Y. (2011). Characterization of a phosphotriesterase-like lactonase from Sulfolobus solfataricus and its immobilization for disruption of quorum sensing. Appl. Environ. Microbiol. 77, 1181–1186. doi: 10.1128/AEM.01642-10
O'Loughlin, C. T., Miller, L. C., Siryaporn, A., Drescher, K., Semmelhack, M. F., and Bassler, B. L. (2013). A quorum-sensing inhibitor blocks Pseudomonas aeruginosa virulence and biofilm formation. Proc. Natl. Acad. Sci. U.S.A. 110, 17981–17986. doi: 10.1073/pnas.1316981110
Olsen, I. (2015). Biofilm-specific antibiotic tolerance and resistance. Eur. J. Clin. Microbiol. Infect. Dis. 34, 877–886. doi: 10.1007/s10096-015-2323-z
Otto, M. (2006). Bacterial evasion of antimicrobial peptides by biofilm formation. Curr. Top. Microbiol. Immunol. 306, 251–258. doi: 10.1007/3-540-29916-5_10
Ozcelik, B., Ho, K. K. K., Glattauer, V., Willcox, M., Kumar, N., and Thissen, H. (2017). Poly(ethylene glycol)-based coatings combining low-biofouling and quorum-sensing inhibiting properties to reduce bacterial colonization. ACS Biomater. Sci. Eng. 3, 78–87. doi: 10.1021/acsbiomaterials.6b00579
Pan, J., and Ren, D. (2009). Quorum sensing inhibitors: a patent overview. Expert Opin. Ther. Pat. 19, 1581–1601. doi: 10.1517/13543770903222293
Papaioannou, E., Wahjudi, M., Nadal-Jimenez, P., Koch, G., Setroikromo, R., and Quax, W. J. (2009). Quorum-quenching acylase reduces the virulence of Pseudomonas aeruginosa in a Caenorhabditis elegans infection model. Antimicrob. Agents Chemother. 53, 4891–4897. doi: 10.1128/AAC.00380-09
Papenfort, K., and Bassler, B. (2016). Quorum-sensing signal-response systems in gram-negative bacteria. Nat. Rev. Microbiol. 14, 576–588. doi: 10.1038/nrmicro.2016.89
Park, H.-E., Jung, Y., and Lee, S. V. (2017). Survival assays using Caenorhabditis elegans. Mol. Cells 40, 90–99. doi: 10.14348/molcells.2017.0017
Park, J., Jagasia, R., Kaufmann, G. F., Mathison, J. C., Ruiz, D. I., Moss, J. A., et al. (2007). Infection control by antibody disruption of bacterial quorum sensing signaling. Chem. Biol. 14, 1119–1127. doi: 10.1016/j.chembiol.2007.08.013
Patterson, A. G., Jackson, S. A., Taylor, C., Evans, G. B., Salmond, G. P. C., Przybilski, R., et al. (2016). Quorum sensing controls adaptive immunity through the regulation of multiple CRISPR-Cas systems. Mol. Cell 64, 1102–1108. doi: 10.1016/j.molcel.2016.11.012
Pearson, J. P., Feldman, M., Iglewski, B. H., and Prince, A. (2000). Pseudomonas aeruginosa cell-to-cell signaling is required for virulence in a model of acute pulmonary infection. Infect. Immun. 68, 4331–4334. doi: 10.1128/IAI.68.7.4331-4334.2000
Pires, D. P., Melo, L., Vilas Boas, D., Sillankorva, S., and Azeredo, J. (2017). Phage therapy as an alternative or complementary strategy to prevent and control biofilm-related infections. Curr. Opin. Microbiol. 39, 48–56. doi: 10.1016/j.mib.2017.09.004
Plener, L., Lorenz, N., Reiger, M., Ramalho, T., Gerland, U., and Jung, K. (2015). The phosphorylation flow of the Vibrio harveyi quorum-sensing cascade determines levels of phenotypic heterogeneity in the population. J. Bacteriol. 197, 1747–1756. doi: 10.1128/JB.02544-14
Poole, K., Gotoh, N., Tsujimoto, H., Zhao, Q., Wada, A., Yamasaki, T., et al. (1996). Overexpression of the mexC-mexD-oprJ efflux operon in nfxB-type multidrug-resistant strains of Pseudomonas aeruginosa. Mol. Microbiol. 21, 713–724. doi: 10.1046/j.1365-2958.1996.281397.x
Prateeksha, Singh, B. R., Shoeb, M., Sharma, S., Naqvi, A. H., Gupta, V. K., et al. (2017). Scaffold of selenium nanovectors and honey phytochemicals for inhibition of Pseudomonas aeruginosa quorum sensing and biofilm formation. Front. Cell. Infect. Microbiol. 7:93. doi: 10.3389/fcimb.2017.00093
Pukatzki, S., Kessin, R. H., and Mekalanos, J. J. (2002). The human pathogen Pseudomonas aeruginosa utilizes conserved virulence pathways to infect the social amoeba Dictyostelium discoideum. Proc. Natl. Acad. Sci. U.S.A. 99, 3159–3164. doi: 10.1073/pnas.052704399
Pukatzki, S., Ma, A. T., Sturtevant, D., Krastins, B., Sarracino, D., Nelson, W. C., et al. (2006). Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc. Natl. Acad. Sci. U.S.A. 103, 1528–1533. doi: 10.1073/pnas.0510322103
Qin, X., Sun, Q., Yang, B., Pan, X., He, Y., and Yang, H. (2017). Quorum sensing influences phage infection efficiency via affecting cell population and physiological state. J. Basic Microbiol. 57, 162–170. doi: 10.1002/jobm.201600510
Rasamiravaka, T., Vandeputte, O. M., Pottier, L., Huet, J., Rabemanantsoa, C., Kiendrebeogo, M., et al. (2015). Pseudomonas aeruginosa biofilm formation and persistence, along with the production of quorum sensing-dependent virulence factors, are disrupted by a triterpenoid coumarate ester isolated from Dalbergia trichocarpa, a tropical legume. PLoS ONE 10:e0132791. doi: 10.1371/journal.pone.0132791
Rasmussen, T. B., Bjarnsholt, T., Skindersoe, M. E., Hentzer, M., Kristoffersen, P., Köte, M., et al. (2005). Screening for Quorum-Sensing Inhibitors (QSI) by use of a novel genetic system, the QSI selector. J. Bacteriol. 187, 1799–1814. doi: 10.1128/JB.187.5.1799-1814.2005
Rémy, B., Plener, L., Elias, M., Daudé, D., and Chabrière, E. (2016a). [Enzymes for disrupting bacterial communication, an alternative to antibiotics?]. Ann. Pharm. Fr. 74, 413–420. doi: 10.1016/j.pharma.2016.06.005
Rémy, B., Plener, L., Poirier, L., Elias, M., Daudé, D., and Chabrière, E. (2016b). Harnessing hyperthermostable lactonase from Sulfolobus solfataricus for biotechnological applications. Sci. Rep. 6:37780. doi: 10.1038/srep37780
Rhoads, M. K., Hauk, P., Terrell, J., Tsao, C.-Y., Oh, H., Raghavan, S. R., et al. (2017). Incorporating LsrK AI-2 quorum quenching capability in a functionalized biopolymer capsule. Biotechnol. Bioeng. 115, 278–289. doi: 10.1002/bit.26397
Romero, M., Acuña, L., and Otero, A. (2012). Patents on quorum quenching: interfering with bacterial communication as a strategy to fight infections. Recent Pat. Biotechnol. 6, 2–12. doi: 10.2174/187220812799789208
Rudrappa, T., and Bais, H. P. (2008). Curcumin, a known phenolic from Curcuma longa, attenuates the virulence of Pseudomonas aeruginosa PAO1 in whole plant and animal pathogenicity models. J. Agric. Food Chem. 56, 1955–1962. doi: 10.1021/jf072591j
Rumbaugh, K. P., Griswold, J. A., Iglewski, B. H., and Hamood, A. N. (1999). Contribution of quorum sensing to the virulence ofpseudomonas aeruginosa in burn wound infections. Infect. Immun. 67, 5854–5862.
Saiman, L., Marshall, B. C., Mayer-Hamblett, N., Burns, J. L., Cibene, D. A., Coquillette, S., et al. (2003). Azithromycin in patients with cystic fibrosis chronically infected with Pseudomonas aeruginosa a randomized controlled trial. JAMA 290, 1749–1756. doi: 10.1001/jama.290.13.1749
Sarabhai, S., Sharma, P., and Capalash, N. (2013). Ellagic acid derivatives from terminalia chebula retz. downregulate the expression of quorum sensing genes to attenuate Pseudomonas aeruginosa PAO1 virulence. PLoS ONE 8:e53441. doi: 10.1371/journal.pone.0053441
Savage, V. J., Chopra, I., and O'Neill, A. J. (2013). Staphylococcus aureus biofilms promote horizontal transfer of antibiotic resistance. Antimicrob. Agents Chemother. 57, 1968–1970. doi: 10.1128/AAC.02008-12
Schuster, M., Sexton, D. J., Diggle, S. P., and Greenberg, E. P. (2013). Acyl-homoserine lactone quorum sensing: from evolution to application. Annu. Rev. Microbiol. 67, 43–63. doi: 10.1146/annurev-micro-092412-155635
Scutera, S., Zucca, M., and Savoia, D. (2014). Novel approaches for the design and discovery of quorum-sensing inhibitors. Expert Opin. Drug Discov. 9, 353–366. doi: 10.1517/17460441.2014.894974
Seok, J., Warren, H. S., Cuenca, A. G., Mindrinos, M. N., Baker, H. V., Xu, W., et al. (2013). Genomic responses in mouse models poorly mimic human inflammatory diseases. Proc. Natl. Acad. Sci. U.S.A. 110, 3507–3512. doi: 10.1073/pnas.1222878110
Shenderovich, J., Feldman, M., Kirmayer, D., Al-Quntar, A., Steinberg, D., Lavy, E., et al. (2015). Local sustained-release delivery systems of the antibiofilm agent thiazolidinedione-8 for prevention of catheter-associated urinary tract infections. Int. J. Pharm. 485, 164–170. doi: 10.1016/j.ijpharm.2015.02.067
Shih, P.-C., and Huang, C.-T. (2002). Effects of quorum-sensing deficiency on Pseudomonas aeruginosa biofilm formation and antibiotic resistance. J. Antimicrob. Chemother. 49, 309–314. doi: 10.1093/jac/49.2.309
Sifri, C. D., Begun, J., Ausubel, F. M., and Calderwood, S. B. (2003). Caenorhabditis elegans as a model host for Staphylococcus aureus pathogenesis. Infect. Immun. 71, 2208–2217. doi: 10.1128/IAI.71.4.2208-2217.2003
Sifri, C. D., Mylonakis, E., Singh, K. V., Qin, X., Garsin, D. A., Murray, B. E., et al. (2002). Virulence effect of Enterococcus faecalis protease genes and the quorum-sensing locus fsr in Caenorhabditis elegans and mice. Infect. Immun. 70, 5647–5650. doi: 10.1128/IAI.70.10.5647-5650.2002
Simonetti, O., Cirioni, O., Cacciatore, I., Baldassarre, L., Orlando, F., Pierpaoli, E., et al. (2016). Efficacy of the quorum sensing inhibitor FS10 alone and in combination with tigecycline in an animal model of Staphylococcal infected wound. PLOS ONE 11:e0151956. doi: 10.1371/journal.pone.0151956
Singh, R. P., Desouky, S. E., and Nakayama, J. (2016). Quorum quenching strategy targeting gram-positive pathogenic bacteria. Adv. Exp. Med. Biol. 901, 109–130. doi: 10.1007/5584_2016_1
Smyth, A. R., Cifelli, P. M., Ortori, C. A., Righetti, K., Lewis, S., Erskine, P., et al. (2010). Garlic as an inhibitor of Pseudomonas aeruginosa quorum sensing in cystic fibrosis-a pilot randomized controlled trial. Pediatr. Pulmonol. 45, 356–362. doi: 10.1002/ppul.21193
Sokol, P. A. (2003). The CepIR quorum-sensing system contributes to the virulence of Burkholderia cenocepacia respiratory infections. Microbiology 149, 3649–3658. doi: 10.1099/mic.0.26540-0
Song, Y., Xie, C., Ong, Y.-M., Gan, Y.-H., and Chua, K.-L. (2005). The BpsIR quorum-sensing system of Burkholderia pseudomallei. J. Bacteriol. 187, 785–790. doi: 10.1128/JB.187.2.785-790.2005
Starkey, M., Lepine, F., Maura, D., Bandyopadhaya, A., Lesic, B., He, J., et al. (2014). Identification of anti-virulence compounds that disrupt quorum-sensing regulated acute and persistent pathogenicity. PLoS Pathog. 10:e1004321. doi: 10.1371/journal.ppat.1004321
Stenvang, M., Dueholm, M. S., Vad, B. S., Seviour, T., Zeng, G., Geifman-Shochat, S., et al. (2016). Epigallocatechin gallate remodels overexpressed functional amyloids in Pseudomonas aeruginosa and increases biofilm susceptibility to antibiotic treatment. J. Biol. Chem. 291, 26540–26553. doi: 10.1074/jbc.M116.739953
Stevens, A. M., Queneau, Y., Soulère, L., von Bodman, S., and Doutheau, A. (2011). Mechanisms and synthetic modulators of AHL-dependent gene regulation. Chem. Rev. 111, 4–27. doi: 10.1021/cr100064s
Stewart, P. S., and William Costerton, J. (2001). Antibiotic resistance of bacteria in biofilms. Lancet 358, 135–138. doi: 10.1016/S0140-6736(01)05321-1
Strateva, T., and Yordanov, D. (2009). Pseudomonas aeruginosa – a phenomenon of bacterial resistance. J. Med. Microbiol. 58, 1133–1148. doi: 10.1099/jmm.0.009142-0
Swatton, J. E., Davenport, P. W., Maunders, E. A., Griffin, J. L., Lilley, K. S., and Welch, M. (2016). Impact of azithromycin on the quorum sensing-controlled proteome of Pseudomonas aeruginosa. PLoS ONE 11:e0147698. doi: 10.1371/journal.pone.0147698
Swem, L. R., Swem, D. L., O'Loughlin, C. T., Gatmaitan, R., Zhao, B., Ulrich, S. M., et al. (2009). A quorum-sensing antagonist targets both membrane-bound and cytoplasmic receptors and controls bacterial pathogenicity. Mol. Cell 35, 143–153. doi: 10.1016/j.molcel.2009.05.029
Tan, D., Svenningsen, S. L., and Middelboe, M. (2015). Quorum sensing determines the choice of antiphage defense strategy in Vibrio anguillarum. MBio 6:e00627-15. doi: 10.1128/mBio.00627-15
Tan, M.-W., Mahajan-Miklos, S., and Ausubel, F. M. (1999a). Killing of Caenorhabditis elegans by Pseudomonas aeruginosa used to model mammalian bacterial pathogenesis. Proc. Natl. Acad. Sci. U.S.A. 96, 715–720.
Tan, M.-W., Rahme, L. G., Sternberg, J. A., Tompkins, R. G., and Ausubel, F. M. (1999b). Pseudomonas aeruginosa killing of Caenorhabditis elegans used to identify P. aeruginosa virulence factors. Proc. Natl. Acad. Sci. U.S.A. 96, 2408–2413.
Tang, K., Su, Y., Brackman, G., Cui, F., Zhang, Y., Shi, X., et al. (2015). MomL, a novel marine-derived N-Acyl homoserine lactonase from Muricauda olearia. Appl. Environ. Microbiol. 81, 774–782. doi: 10.1128/AEM.02805-14
Tang, K., and Zhang, X.-H. (2014). Quorum quenching agents: resources for antivirulence therapy. Mar. Drugs 12, 3245–3282. doi: 10.3390/md12063245
Tateda, K., Comte, R., Pechere, J.-C., Köhler, T., Yamaguchi, K., and Van Delden, C. (2001). Azithromycin inhibits quorum sensing in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 45, 1930–1933. doi: 10.1128/AAC.45.6.1930-1933.2001
Taunk, A., Ho, K. K. K., Iskander, G., Willcox, M. D., and Kumar, N. (2016). Surface immobilization of antibacterial quorum sensing inhibitors by photochemical activation. J. Biotechnol. Biomater. 6:238. doi: 10.4172/2155-952X.1000238
Thompson, J. A., Oliveira, R. A., Djukovic, A., Ubeda, C., and Xavier, K. B. (2015). Manipulation of the quorum sensing signal AI-2 affects the antibiotic-treated gut microbiota. Cell Rep. 10, 1861–1871. doi: 10.1016/j.celrep.2015.02.049
Tiaden, A., and Hilbi, H. (2012). α-Hydroxyketone synthesis and sensing by Legionella and Vibrio. Sensors 12, 2899–2919. doi: 10.3390/s120302899
Todd, D. A., Parlet, C. P., Crosby, H. A., Malone, C. L., Heilmann, K. P., Horswill, A. R., et al. (2017). Signal biosynthesis inhibition with ambuic acid as a strategy to target antibiotic-resistant infections. Antimicrob. Agents Chemother. 61:e00263-17. doi: 10.1128/AAC.00263-17
Ueda, A., Attila, C., Whiteley, M., and Wood, T. K. (2009). Uracil influences quorum sensing and biofilm formation in Pseudomonas aeruginosa and fluorouracil is an antagonist. Microb. Biotechnol. 2, 62–74. doi: 10.1111/j.1751-7915.2008.00060.x
van Delden, C., Köhler, T., Brunner-Ferber, F., François, B., Carlet, J., and Pechère, J.-C. (2012). Azithromycin to prevent Pseudomonas aeruginosa ventilator-associated pneumonia by inhibition of quorum sensing: a randomized controlled trial. Intensive Care Med. 38, 1118–1125. doi: 10.1007/s00134-012-2559-3
van der Worp, H. B., Howells, D. W., Sena, E. S., Porritt, M. J., Rewell, S., O'Collins, V., et al. (2010). Can animal models of disease reliably inform human studies? PLoS Med. 7:e1000245. doi: 10.1371/journal.pmed.1000245
Vuotto, C., Longo, F., Balice, M., Donelli, G., and Varaldo, P. (2014). Antibiotic resistance related to biofilm formation in Klebsiella pneumoniae. Pathogens 3, 743–758. doi: 10.3390/pathogens3030743
Vuotto, C., Longo, F., Pascolini, C., Donelli, G., Balice, M. P., Libori, M. F., et al. (2017). Biofilm formation and antibiotic resistance in Klebsiella pneumoniae urinary strains. J. Appl. Microbiol. 123, 1003–1018. doi: 10.1111/jam.13533
Walz, J. M., Avelar, R. L., Longtine, K. J., Carter, K. L., Mermel, L. A., and Heard, S. O. (2010). Anti-infective external coating of central venous catheters: a randomized, noninferiority trial comparing 5-fluorouracil with chlorhexidine/silver sulfadiazine in preventing catheter colonization. Crit. Care Med. 38, 2095–2102. doi: 10.1097/CCM.0b013e3181f265ba
Weiland-Bräuer, N., Kisch, M. J., Pinnow, N., Liese, A., and Schmitz, R. A. (2016). Highly effective inhibition of biofilm formation by the first metagenome-derived AI-2 quenching enzyme. Front. Microbiol. 7:1098. doi: 10.3389/fmicb.2016.01098
Weitere, M., Bergfeld, T., Rice, S. A., Matz, C., and Kjelleberg, S. (2005). Grazing resistance of Pseudomonas aeruginosa biofilms depends on type of protective mechanism, developmental stage and protozoan feeding mode. Environ. Microbiol. 7, 1593–1601. doi: 10.1111/j.1462-2920.2005.00851.x
Wolter, J., Seeney, S., Bell, S., Bowler, S., Masel, P., and McCormack, J. (2002). Effect of long term treatment with azithromycin on disease parameters in cystic fibrosis: a randomised trial. Thorax 57, 212–216. doi: 10.1136/thorax.57.3.212
Wopperer, J., Cardona, S. T., Huber, B., Jacobi, C. A., Valvano, M. A., and Eberl, L. (2006). A quorum-quenching approach to investigate the conservation of quorum-sensing-regulated functions within the Burkholderia cepacia complex. Appl. Environ. Microbiol. 72, 1579–1587. doi: 10.1128/AEM.72.2.1579-1587.2006
Wu, H. (2004). Synthetic furanones inhibit quorum-sensing and enhance bacterial clearance in Pseudomonas aeruginosa lung infection in mice. J. Antimicrob. Chemother. 53, 1054–1061. doi: 10.1093/jac/dkh223
Wu, H., Song, Z., Givskov, M., Doring, G., Worlitzsch, D., Mathee, K., et al. (2001). Pseudomonas aeruginosa mutations in lasI and rhlI quorum sensing systems result in milder chronic lung infection. Microbiology 147, 1105–1113. doi: 10.1099/00221287-147-5-1105
Xu, L., Li, H., Vuong, C., Vadyvaloo, V., Wang, J., Yao, Y., et al. (2006). Role of the luxS quorum-sensing system in biofilm formation and virulence of Staphylococcus epidermidis. Infect. Immun. 74, 488–496. doi: 10.1128/IAI.74.1.488-496.2006
Yin, H., Deng, Y., Wang, H., Liu, W., Zhuang, X., and Chu, W. (2015). Tea polyphenols as an antivirulence compound disrupt quorum-sensing regulated pathogenicity of Pseudomonas aeruginosa. Sci. Rep. 5:16158. doi: 10.1038/srep16158
Zhou, L., Zhang, L.-H., Cámara, M., and He, Y.-W. (2017). The DSF family of quorum sensing signals: diversity, biosynthesis, and turnover. Trends Microbiol. 25, 293–303. doi: 10.1016/j.tim.2016.11.013
Keywords: quorum sensing (QS), bacterial virulence, biofilm, quorum sensing inhibitors, quorum quenching enzymes, antibioresistance, phage resistance, medical devices
Citation: Rémy B, Mion S, Plener L, Elias M, Chabrière E and Daudé D (2018) Interference in Bacterial Quorum Sensing: A Biopharmaceutical Perspective. Front. Pharmacol. 9:203. doi: 10.3389/fphar.2018.00203
Received: 03 November 2017; Accepted: 22 February 2018;
Published: 07 March 2018.
Edited by:
Patrick Masson, Kazan Federal University, RussiaCopyright © 2018 Rémy, Mion, Plener, Elias, Chabrière and Daudé. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: David Daudé, ZGF2aWQuZGF1ZGVAZ2VuZS1ncmVlbnRrLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.