- 1Department of General Biology, Institute of Biological Sciences, Federal University of Minas Gerais (UFMG), Belo Horizonte, Brazil
- 2UFMG Graduate Program in Genetics, Institute of Biological Sciences, Federal University of Minas Gerais, Belo Horizonte, Brazil
- 3Department of Pharmacology, Institute of Biosciences of Botucatu, Universidade Estadual Paulista, Botucatu, Brazil
Hypertension is a leading cause of cardiovascular mortality, but only about half of patients on antihypertensive therapy achieve blood pressure control. Preeclampsia is defined as pregnancy-induced hypertension and proteinuria, and is associated with increased maternal and perinatal mortality and morbidity. Similarly, a large number of patients with preeclampsia are non-responsive to antihypertensive therapy. Pharmacogenomics may help to guide the personalized treatment for non-responsive hypertensive patients. There is evidence for the association of genetic variants with variable response to the most commonly used antihypertensive drugs. However, further replication is needed to confirm these associations in different populations. The failure to replicate findings from single-locus association studies has prompted the search for novel statistical methods for data analysis, which are required to detect the complex effects from multiple genes to drug response phenotypes. Notably, gene–gene interaction analyses have been applied to pharmacogenetic studies, including antihypertensive drug response. In this perspective article, we present advances of considering the interactions among genetic polymorphisms of different candidate genes within pathways relevant to antihypertensive drug response, and we highlight recent findings related to gene–gene interactions on pharmacogenetics of hypertension and preeclampsia. Finally, we discuss the future directions that are needed to unravel additional genes and variants involved in the responsiveness to antihypertensive drugs.
Introduction
Pharmacogenomics aims to elucidate the contribution of genetic variants to interindividual variability in drug responses, such as efficacy, dose requirements and adverse reactions. Candidate gene association studies have been performed to assess the effects of single polymorphisms or haplotypes, the combination of alleles at multiple polymorphisms, on drug response phenotypes. In the past decade, genome-wide association studies (GWAS) in pharmacogenomics have led to the identification of variants associated with efficacy and adverse drug reactions. However, much of the genetic heritability to drug response phenotypes appears to be hidden in multigenic and multifactorial complex traits (Zanger, 2010). It remains uncertain how many genes and genetic variants contribute to pharmacological traits, how common and rare variants affect response and whether gene–gene interactions play a role (Chhibber et al., 2014).
Gene–gene interactions among common regulatory variants can have strong effects in drug response phenotypes (Sadee et al., 2014). Notably, findings from 108 GWAS in pharmacogenomics revealed that 96.4% of the 928 associated single nucleotide polymorphisms (SNPs) are located in non-coding regions (Luizon and Ahituv, 2015). These findings suggest that most of common variants associated with variability in drug response are regulatory. However, most of the SNPs used in GWAS arrays were selected as informative genetic markers with higher allele frequencies and more likely to have greater power to detect association. In addition, the associated SNPs in GWAS are not necessarily the causal variant responsible for the phenotype, but may be in high linkage disequilibrium with the causal variant (Luizon and Ahituv, 2015). Notably, the failure to replicate findings from pharmacogenetic studies, particularly in the analysis of single-locus, has prompted the search for novel statistical methods to analyze the data. These methods take into account the biological complexity underlying drug response and examine potential epistatic interactions that may predict drug response, as reviewed elsewhere (Motsinger et al., 2007; Pander et al., 2010).
The multifactorial nature of drug response phenotypes require novel approaches to detect complex genetic effects from multiple minor impact genes, entire pathways, gene–gene and gene-environment interactions (Zanger, 2010). Therefore, research in pharmacogenomics should take into account the interactions among polymorphisms from different genes within drug response pathways. In this perspective article, we present current advances focused on gene–gene interactions associated with response to antihypertensive drugs, including non-coding SNPs found to be associated in GWAS and candidate gene studies, which are relevant to pharmacogenomics of hypertension and preeclampsia (Figure 1).
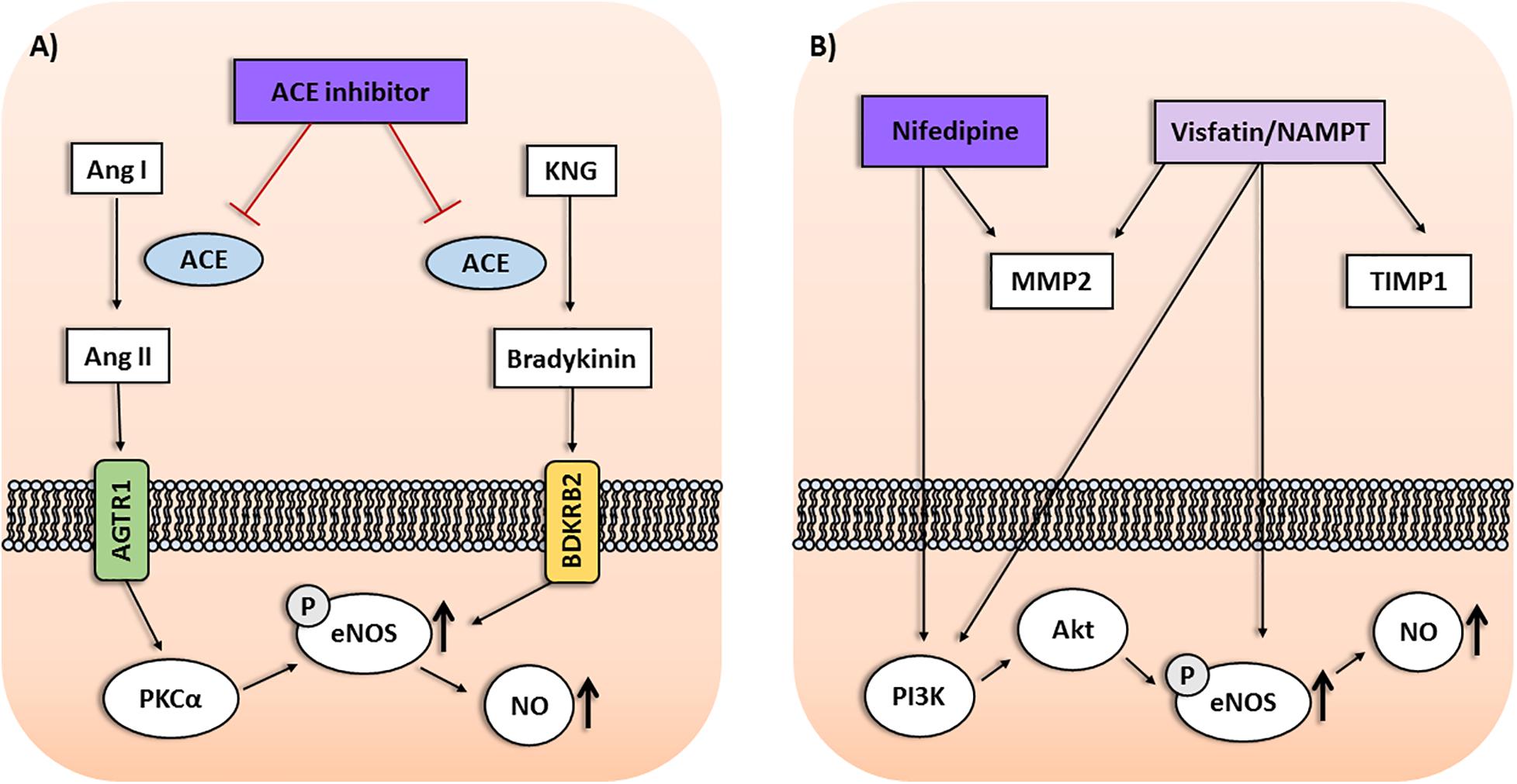
FIGURE 1. (A) Interactions among Protein Kinase C Alpha (PRKCA), NOS3, and BDKRB2 gene products within the ACE inhibitor pathway may affect the antihypertensive response to enalapril. Protein kinase C alpha (PKCα) upregulates the endothelial nitric oxide synthase (eNOS) activity, leading to enhanced nitric oxide (NO) production and vasodilation. Moreover, stimulating bradykinin receptors on endothelial cells may result in eNOS upregulation mediated by PKCα. (B) Interactions among genes of the visfatin/NAMPT pathway and response to nifedipine in preeclampsia. NAMPT was associated with antihypertensive response in preeclampsia depending on TIMP1 and MMP2 genotypes. Nifedipine may affect circulating MMP levels in hypertension. Nifedipine and visfatin/NAMPT increase PI3K/Akt pathway and eNOS activity, resulting in enhanced NO production. Ang I, Angiotensin I; Ang II, Angiotensin II; AGTR1, Angiotensin II Receptor Type 1; KNG, Kininogen.
Gene–Gene Interactions in Pharmacogenomics of Hypertension
Hypertension is the most prevalent modifiable risk factor for stroke and coronary heart disease, affects approximately 1 billion individuals worldwide and is a leading cause of cardiovascular mortality. GWAS have also improved the understanding on hypertension genetics, and provide evidence to suggest that much of the genetic influence on hypertension reside within non-coding elements and operate through gene–gene interactions (Padmanabhan and Joe, 2017). The polygenic nature of hypertension indicates that single loci may not be considered as relevant clinical target for all individuals (Padmanabhan and Joe, 2017). These findings highlight the importance of assessing the interactions among multiple loci when studying complex traits, including drug response phenotypes.
Antihypertensive therapy is associated with a significant reduction in mortality, and blood pressure (BP) is monitored in response to treatment and dosage adjusted on the basis of BP response and adverse effects. However, only about half of patients on antihypertensive therapy achieve BP control (Cooper-DeHoff and Johnson, 2016). Genetic variation may partially account for the interindividual variability in response to antihypertensive drugs. Candidate gene studies and GWAS have identified common genetic polymorphisms associated with response to antihypertensive drugs, as reviewed elsewhere (Fontana et al., 2015; Cooper-DeHoff and Johnson, 2016; Manunta et al., 2016). Although no examples have been sufficiently validated for clinical use, the available findings suggest promise and pharmacogenomics holds the potential to guide the personalized treatment for patients who are non-responsive to hypertensive drugs (Cooper-DeHoff and Johnson, 2016).
Hydrochlorothiazide is among the most commonly prescribed antihypertensive drugs. A GWAS found an intronic SNP (rs16960228, A/G) of the PRKCA gene encoding protein kinase C alpha (PKCα) to be associated with BP response to hydrochlorothiazide in two cohorts and replicated in other cohorts of European Americans (Turner et al., 2013). Systolic and diastolic BP responses were consistently greater in carriers of the GA+AA genotypes than in homozygote GG carriers across all study samples. The rs16960228 SNP was also associated with diastolic BP response to the β-blocker atenolol, with the opposite direction of effect, which may be explained by the different pharmacologic mechanisms compared to thiazide diuretics (Turner et al., 2013; Cooper-DeHoff and Johnson, 2016).
Angiotensin-converting enzyme inhibitors (ACEi) are also widely used to treat hypertension, including enalapril. The mechanism of action involves the reduction of angiotensin II formation, but a secondary mechanism is related to vasodilation produced by nitric oxide (NO) as a result of endothelial nitric oxide synthase (eNOS) activation. Both mechanisms seem to be affected by PKC signaling, as reviewed elsewhere (Oliveira-Paula et al., 2017) (Figure 1A). Recently, the rs16960228 SNP of PRKCA gene was also associated with BP responses in hypertensive patients classified as poor or good responders to enalapril. The GA+AA genotypes, and the A allele, were associated with worse responses to enalapril (Oliveira-Paula et al., 2017). Previous expression studies have shown that PRKCA expression was significantly higher among carriers of the GA+AA genotypes than among those with the GG genotype (Turner et al., 2013). It is possible that the GG genotype can enhance the responses to ACEi, which increase PKCα. Conversely, it seems reasonable to suggest that PKCα signaling could be less affected by ACEi in patients carrying the GA+AA genotypes, resulting in worse responses to these drugs. However, the molecular mechanisms supporting these suggestions remain to be proved (Oliveira-Paula et al., 2017).
Interactions among genes within the ACEi pathway might also influence the responses to enalapril. Polymorphisms of the bradykinin receptor B2 (BDKRB2) and nitric oxide synthase 3 (NOS3) genes were associated with the antihypertensive responses to enalapril (Silva et al., 2013; Oliveira-Paula et al., 2016). However, the CC genotype for the rs1799722 SNP (C-58T) in the promoter region of BDKRB2 gene was not associated with the response to enalapril in single-locus analysis. Interestingly, the gene–gene interaction analysis showed that the CC genotype for the rs1799722 SNP was associated with response to enalapril depending on genotypes for the rs2070744 SNP (T-786C) in the promoter region of NOS3 gene. The combination with the TT genotype was more frequent in poor responders, while the combination with TC genotype was more frequent in good responders (Silva et al., 2013). These findings are obscured when single BDKRB2 genotypes alone are considered, and were found to be associated with drug response only when performing gene–gene interaction analysis.
Furthermore, interactions among PRKCA, BDKRB2 and NOS3 genes were also shown to affect the response to enalapril (Oliveira-Paula et al., 2017). In single-locus analysis, the GG genotype for the rs16960228 GWAS SNP of PRKCA gene was associated with better responses to enalapril. However, the combinations between BDKRB2 and NOS3 genes found to be associated with either poor responders or good responders to enalapril described above (Silva et al., 2013) were significant only when combined with the GG genotype for the rs16960228 SNP of PRKCA gene (Oliveira-Paula et al., 2017). PKCα increases the transcription of NOS3 gene and upregulates eNOS activity, leading to enhanced NO production and vasodilation. Moreover, ACEi increases tissue bradykinin levels, stimulating bradykinin receptors on endothelial cells, which may result in NOS3 upregulation mediated by PKCα, as reviewed elsewhere (Oliveira-Paula et al., 2017). These gene–gene interactions could affect eNOS activity and NO bioavailability (Figure 1A). Thus, to evaluate circulating nitrite levels (a marker of endogenous NO formation) and plasma eNOS activity may be a possible approach to clarify the underlying molecular mechanisms. In this regard, NOS3 polymorphisms were shown to modify plasma and blood nitrite concentrations (Metzger et al., 2013; Oliveira-Paula et al., 2016). However, further studies are needed to examine the effects of gene interactions within this pathway on circulating nitrite levels and eNOS activity in patients treated with ACEi (Oliveira-Paula et al., 2017). Taken together, these findings highlight the relevance of take into consideration gene–gene interaction analysis in pharmacogenomics of hypertension.
Gene–Gene Interactions in Pharmacogenomics of Preeclampsia
Preeclampsia is a major cause of maternal and perinatal morbidity and mortality, defined as pregnancy-induced hypertension and proteinuria after 20 weeks of gestation. It is a multisystem disorder that has implications for future maternal health, in particular an increased risk of cardiovascular disease. Preeclampsia is a complex genetic disorder, and it is probable that no single gene or variant will be identified as responsible for all cases of preeclampsia (Williams and Morgan, 2012). This scenario highlights the need to focus on more than one candidate gene, and on interactions among polymorphisms of different genes rather than on the effect of single polymorphisms. Indeed, interactions among SNPs in candidate genes within pathways relevant to disease pathophysiology were associated with the susceptibility to preeclampsia (Luizon et al., 2012, 2014, 2017b).
Antihypertensive therapy in preeclampsia includes methyldopa, nifedipine, hydralazine, and labetalol. These antihypertensive drugs allow the prolongation of gestation, thereby decreasing fetal and maternal adverse outcomes. However, a large subgroup of pregnant with preeclampsia is non-responsive to antihypertensive therapy and is associated with the worst clinical parameters (Luizon et al., 2017a). The use of pharmacogenomics may maximize the treatment outcome by reducing maternal and fetal morbidity and mortality associated with preeclampsia, as well as reducing adverse events associated with pharmacological therapy (Williams and Morgan, 2012). However, pharmacogenetics of antihypertensive therapy in preeclampsia was the scope of just a few studies, which examined the association of genetic polymorphisms with subgroups of patients who were responsive or non-responsive to antihypertensive therapy (Luizon et al., 2017a). Patients with preeclampsia were classified according to response to the initial drug of choice, methyldopa, and to the total antihypertensive therapy, including nifedipine and hydralazine, the latter used in cases of hypertensive crisis, according to the criteria of responsiveness reviewed elsewhere (Luizon et al., 2017a).
These pharmacogenetic studies included polymorphisms that modify the expression of candidate genes related to the pathophysiology of preeclampsia (Luizon et al., 2017a). Notably, endothelial dysfunction is associated with both hypertension and proteinuria. NO plays an important role in regulating endothelial function, and the eNOS enzyme is responsible for NO synthesis in the cardiovascular system. Reduced expression of eNOS consequently results in reduced NO bioavailability which plays a significant role in the endothelial dysfunction associated with preeclampsia (Sandrim et al., 2008). Therefore, eNOS is considered as a potential target for therapy for preeclampsia (Williams and Morgan, 2012). Calcium channel blockers (including nifedipine) may improve endothelial function and restore NO bioavailability, which could produce its effects by enhancing NO formation, thus counteracting the impaired NO bioavailability in preeclampsia (Sandrim et al., 2008) (Figure 1B). Nifedipine was shown to increase PI3K/Akt pathway and to increase eNOS activity and expression, as reviewed elsewhere (Luizon et al., 2016). Therefore, NOS3 polymorphisms that modulate NO synthesis could also interfere with the response to nifedipine. Notably, polymorphisms and haplotypes of NOS3 gene were associated with the disease (Muniz et al., 2012) or with the responsive and non-responsive subgroups of patients with preeclampsia (Sandrim et al., 2010).
Drugs that target the systemic vascular dysfunction were proposed as potential therapies for preeclampsia. Visfatin, an adipokine also known as nicotinamide phosphorybosil transferase (NAMPT), is considered a potential biomarker for vascular endothelial dysfunction. Notably, visfatin/NAMPT was shown to increase the expression and activity of eNOS in human endothelial cells, leading to an enhanced production of NO, as reviewed elsewhere (Peiro et al., 2010). Therefore, NAMPT polymorphisms that modulate visfatin/NAMPT levels could be evaluated as biomarkers for this pathway. Notably, the rs1319501 SNP (T-423C) in the promoter region and the rs3801266 SNP (A/G) in the intron 1 of NAMPT gene were shown to be associated with the susceptibility to preeclampsia (Luizon et al., 2015), and to affect plasma visfatin/NAMPT levels in patients who were non-responsive to total antihypertensive therapy (Luizon et al., 2017b). The TC+CC genotypes of the rs1319501 SNP and the AG+GG genotypes of the rs3801266 SNP were associated with lower and higher visfatin/NAMPT levels, respectively (Luizon et al., 2017b). However, further research on the interplay among visfatin/NAMPT levels, NO bioavailability and the response to antihypertensive therapy in preeclampsia are needed (Luizon et al., 2017b).
Visfatin/NAMPT induces endothelial cell proliferation and migration mediated by signaling pathways relevant to preeclampsia. It enhances the levels and activation of matrix metalloproteinases (MMP)-2/9 while decreasing the levels of tissue inhibitor of metalloproteinases (TIMP)-1/2 (Peiro et al., 2010). MMPs are endopeptidases that may interact with vasoactive peptides and contribute to the endothelial dysfunction in preeclampsia. Notably, the GG genotype, and the G allele, of the rs2070584 (T/G) SNP of TIMP1 gene were associated with both the disease and with the patients with preeclampsia who were non-responsive to antihypertensive therapy (Luizon et al., 2014). Conversely, SNPs of MMP2 and NAMPT genes were not found to be associated with the responsiveness to antihypertensive therapy in preeclampsia, as reviewed elsewhere (Luizon et al., 2017a). Interestingly, gene–gene interaction analysis showed that the GG genotype for the rs2070584 SNP of TIMP1 was associated with preeclampsia and with responsiveness to total antihypertensive therapy in preeclampsia depending on genotypes for the rs3801266 SNP (A/G) of NAMPT gene. The combination with the AA was more frequent in the responsive patients, while the combination with the AG genotype was more frequent in the non-responsive patients (Luizon et al., 2017b). These combinations were significant only when combined with the CC genotype of the rs2285053 SNP (C-735C) of MMP2 gene. These findings are obscured when single MMP2 and NAMPT genotypes alone are considered, as they were not associated with the responsiveness in the single-locus analysis. Therefore, they further highlight the importance of gene–gene interactions in the genetics (Luizon et al., 2012) and pharmacogenetics of preeclampsia (Luizon et al., 2017a,b). Notably, calcium channel blockers (like nifedipine) were shown to affect circulating MMP levels in essential hypertensive patients, as reviewed elsewhere (Luizon et al., 2017a). However, further research into the molecular mechanisms underlying the interactions among NAMPT, TIMP1 and MMP2 and their relationship with the response to antihypertensive therapy in preeclampsia is needed (Figure 1B).
Conclusion and Perspectives
Here, we present current findings related to gene–gene interactions on pharmacogenomics of hypertension (Silva et al., 2013; Oliveira-Paula et al., 2017) and preeclampsia (Luizon et al., 2014, 2017b). Of note, the reported association findings should be replicated in different populations. Moreover, the molecular mechanisms supporting these gene–gene interactions remain to be proved. Regarding preeclampsia, plasma from non-responsive and responsive patients was shown to evoke different gene expression profiles in human umbilical vein endothelial cells (HUVEC). Notably, nifedipine and hydralazine may act by upregulate or downregulate genes found to be downregulated or upregulated in HUVEC incubated with plasma from non-responsive patients (Luizon et al., 2016). The use of transcriptomics data holds promise for potential insights and implications to the pharmacogenomics of hypertension (Cooper-DeHoff and Johnson, 2016).
We expect that epigenomics research focused on the regulatory role of GWAS associated SNPs located in non-coding regions might provide a deeper understanding of pathways involved in the response to antihypertensive drugs. Notably, more than 96% of the associated SNPs from GWAS in pharmacogenomics reside in non-coding regions, which suggest these common variants are regulatory (Luizon and Ahituv, 2015). In addition, gene–gene interactions among common regulatory variants can have strong effects in drug response phenotypes (Sadee et al., 2014). Therefore, we further expect that gene–gene interaction analysis are more likely to be used for pharmacogenomics in the future.
Taken together, these efforts will guide further studies focused on gene–gene interactions within pathways relevant to antihypertensive drug response, which may enable the identification of novel pharmacogenomic biomarkers to guide personalized medicine for hypertension and preeclampsia.
Author Contributions
MRL and DAP drafted the perspective article. All authors revised and approved it for publication.
Funding
Research was funded by grants from the National Council for Scientific and Technological Development (CNPq-Brazil, Grant#2014-5/305587) and the Fundação de Amparo a Pesquisa do Estado de São Paulo (FAPESP-Brazil, Grant#2015/20461-8). MRL was supported by the Young Talent Attraction Fellowship (BJT) from CNPq-Brazil (Grant#400295/2014-8). DAP is supported by a fellowship of the Coordination of Improvement of Higher Level Personnel (CAPES-Brazil).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Chhibber, A., Kroetz, D. L., Tantisira, K. G., Mcgeachie, M., Cheng, C., Plenge, R., et al. (2014). Genomic architecture of pharmacological efficacy and adverse events. Pharmacogenomics 15, 2025–2048. doi: 10.2217/pgs.14.144
Cooper-DeHoff, R. M., and Johnson, J. A. (2016). Hypertension pharmacogenomics: in search of personalized treatment approaches. Nat. Rev. Nephrol. 12, 110–122. doi: 10.1038/nrneph.2015.176
Fontana, V., Luizon, M. R., and Sandrim, V. C. (2015). An update on the pharmacogenetics of treating hypertension. J. Hum. Hypertens. 29, 283–291. doi: 10.1038/jhh.2014.76
Luizon, M. R., and Ahituv, N. (2015). Uncovering drug-responsive regulatory elements. Pharmacogenomics 16, 1829–1841. doi: 10.2217/pgs.15.121
Luizon, M. R., Belo, V. A., Palei, A. C., Amaral, L. M., Lacchini, R., Sandrim, V. C., et al. (2015). Effects of NAMPT polymorphisms and haplotypes on circulating visfatin/NAMPT levels in hypertensive disorders of pregnancy. Hypertens. Res. 38, 361–366. doi: 10.1038/hr.2015.15
Luizon, M. R., Caldeira-Dias, M., Deffune, E., Fernandes, K. S., Cavalli, R. C., Tanus-Santos, J. E., et al. (2016). Antihypertensive therapy in pre-eclampsia: effects of plasma from nonresponsive patients on endothelial gene expression. Pharmacogenomics 17, 1121–1127. doi: 10.2217/pgs-2016-0033
Luizon, M. R., Palei, A. C., Cavalli, R. C., and Sandrim, V. C. (2017a). Pharmacogenetics in the treatment of pre-eclampsia: current findings, challenges and perspectives. Pharmacogenomics 18, 571–583. doi: 10.2217/pgs-2016-0198
Luizon, M. R., Palei, A. C., Sandrim, V. C., Amaral, L. M., Machado, J. S., Lacchini, R., et al. (2014). Tissue inhibitor of matrix metalloproteinase-1 polymorphism, plasma TIMP-1 levels, and antihypertensive therapy responsiveness in hypertensive disorders of pregnancy. Pharmacogenomics J. 14, 535–541. doi: 10.1038/tpj.2014.26
Luizon, M. R., Palei, A. C. T., Belo, V. A., Amaral, L. M., Lacchini, R., Duarte, G., et al. (2017b). Gene-gene interactions in the NAMPT pathway, plasma visfatin/NAMPT levels, and antihypertensive therapy responsiveness in hypertensive disorders of pregnancy. Pharmacogenomics J. 17, 427–434. doi: 10.1038/tpj.2016.35
Luizon, M. R., Sandrim, V. C., Palei, A. C., Lacchini, R., Cavalli, R. C., Duarte, G., et al. (2012). Epistasis among eNOS, MMP-9 and VEGF maternal genotypes in hypertensive disorders of pregnancy. Hypertens. Res. 35, 917–921. doi: 10.1038/hr.2012.60
Manunta, P., Ferrandi, M., Cusi, D., Ferrari, P., Staessen, J., and Bianchi, G. (2016). Personalized therapy of hypertension: the past and the future. Curr. Hypertens. Rep. 18:24. doi: 10.1007/s11906-016-0632-y
Metzger, I. F., Luizon, M. R., Lacchini, R., Ishizawa, M. H., and Tanus-Santos, J. E. (2013). Effects of endothelial nitric oxide synthase tagSNPs haplotypes on nitrite levels in black subjects. Nitric Oxide 28, 33–38. doi: 10.1016/j.niox.2012.10.002
Motsinger, A. A., Ritchie, M. D., and Reif, D. M. (2007). Novel methods for detecting epistasis in pharmacogenomics studies. Pharmacogenomics 8, 1229–1241. doi: 10.2217/14622416.8.9.1229
Muniz, L., Luizon, M. R., Palei, A. C., Lacchini, R., Duarte, G., Cavalli, R. C., et al. (2012). eNOS tag SNP haplotypes in hypertensive disorders of pregnancy. DNA Cell Biol. 31, 1665–1670. doi: 10.1089/dna.2012.1768
Oliveira-Paula, G. H., Lacchini, R., Luizon, M. R., Fontana, V., Silva, P. S., Biagi, C., et al. (2016). Endothelial nitric oxide synthase tagSNPs influence the effects of enalapril in essential hypertension. Nitric Oxide 55–56, 62–69. doi: 10.1016/j.niox.2016.03.006
Oliveira-Paula, G. H., Luizon, M. R., Lacchini, R., Fontana, V., Silva, P. S., Biagi, C., et al. (2017). Gene-gene interactions among PRKCA, NOS3 and BDKRB2 polymorphisms affect the antihypertensive effects of enalapril. Basic Clin. Pharmacol. Toxicol. 120, 284–291. doi: 10.1111/bcpt.12682
Padmanabhan, S., and Joe, B. (2017). Towards precision medicine for hypertension: a review of genomic, epigenomic, and microbiomic effects on blood pressure in experimental rat models and humans. Physiol. Rev. 97, 1469–1528. doi: 10.1152/physrev.00035.2016
Pander, J., Wessels, J. A., Mathijssen, R. H., Gelderblom, H., and Guchelaar, H. J. (2010). Pharmacogenetics of tomorrow: the 1 + 1 = 3 principle. Pharmacogenomics 11, 1011–1017. doi: 10.2217/pgs.10.87
Peiro, C., Romacho, T., Carraro, R., and Sanchez-Ferrer, C. F. (2010). Visfatin/PBEF/Nampt: a new cardiovascular target? Front. Pharmacol. 1:135. doi: 10.3389/fphar.2010.00135
Sadee, W., Hartmann, K., Seweryn, M., Pietrzak, M., Handelman, S. K., and Rempala, G. A. (2014). Missing heritability of common diseases and treatments outside the protein-coding exome. Hum. Genet. 133, 1199–1215. doi: 10.1007/s00439-014-1476-7
Sandrim, V. C., Palei, A. C., Luizon, M. R., Izidoro-Toledo, T. C., Cavalli, R. C., and Tanus-Santos, J. E. (2010). eNOS haplotypes affect the responsiveness to antihypertensive therapy in preeclampsia but not in gestational hypertension. Pharmacogenomics J. 10, 40–45. doi: 10.1038/tpj.2009.38
Sandrim, V. C., Palei, A. C., Metzger, I. F., Gomes, V. A., Cavalli, R. C., and Tanus-Santos, J. E. (2008). Nitric oxide formation is inversely related to serum levels of antiangiogenic factors soluble fms-like tyrosine kinase-1 and soluble endogline in preeclampsia. Hypertension 52, 402–407. doi: 10.1161/HYPERTENSIONAHA.108.115006
Silva, P. S., Fontana, V., Luizon, M. R., Lacchini, R., Silva, WA Jr, Biagi, C., et al. (2013). eNOS and BDKRB2 genotypes affect the antihypertensive responses to enalapril. Eur. J. Clin. Pharmacol. 69, 167–177. doi: 10.1007/s00228-012-1326-2
Turner, S. T., Boerwinkle, E., O’connell, J. R., Bailey, K. R., Gong, Y., Chapman, A. B., et al. (2013). Genomic association analysis of common variants influencing antihypertensive response to hydrochlorothiazide. Hypertension 62, 391–397. doi: 10.1161/HYPERTENSIONAHA.111.00436
Williams, P. J., and Morgan, L. (2012). The role of genetics in pre-eclampsia and potential pharmacogenomic interventions. Pharmgenomics Pers. Med. 5, 37–51. doi: 10.2147/PGPM.S23141
Keywords: antihypertensive therapy, gene–gene interactions, epistasis, hypertension, pathways, pharmacogenomics, preeclampsia
Citation: Luizon MR, Pereira DA and Sandrim VC (2018) Pharmacogenomics of Hypertension and Preeclampsia: Focus on Gene–Gene Interactions. Front. Pharmacol. 9:168. doi: 10.3389/fphar.2018.00168
Received: 01 December 2017; Accepted: 15 February 2018;
Published: 28 February 2018.
Edited by:
Wanqing Liu, Wayne State University, United StatesReviewed by:
Reginald F. Frye, University of Florida, United StatesVijay Suppiah, University of South Australia, Australia
Copyright © 2018 Luizon, Pereira and Sandrim. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Marcelo R. Luizon, bHVpem9ubXJAZ21haWwuY29t
 Marcelo R. Luizon
Marcelo R. Luizon Daniela A. Pereira
Daniela A. Pereira Valeria C. Sandrim
Valeria C. Sandrim