- 1Natural Health Products and Metabolic Diseases Laboratory, Department of Pharmacology and Physiology, Université de Montréal, Montréal, QC, Canada
- 2Canadian Institutes of Health Research Team in Aboriginal Antidiabetic Medicines, Montréal, QC, Canada
- 3Department of Pharmacognosy, University of Beni-Suef, Beni-Suef, Egypt
- 4Nell Hodgson Woodruff School of Nursing, Emory University, Atlanta, GA, United States
- 5Department of Chemistry, Manipal Institute of Technology, Manipal University, Manipal, India
- 6Faculty of Pharmacy, Hebron University, Hebron, Palestine
- 7Department of Natural Drugs, Faculty of Pharmacy, University of Veterinary and Pharmaceutical Sciences Brno, Brno, Czechia
- 8Department of Pharmaceutical Botany, Iuliu Hatieganu University of Medicine and Pharmacy, Cluj-Napoca, Romania
- 9ICHAT and Institute for Life Sciences, University of Agricultural Sciences and Veterinary Medicine, Cluj-Napoca, Romania
- 10Applied Biotechnology Research Center, Baqiyatallah University of Medical Sciences, Tehran, Iran
- 11Dipartimento di Farmacia, University of Salerno, Fisciano, Italy
- 12Institute of Genetics and Animal Breeding, Polish Academy of Sciences, Jastrzebiec, Poland
- 13Department of Pharmacognosy, University of Vienna, Vienna, Austria
- 14Department of Vascular Biology and Thrombosis Research, Center for Physiology and Pharmacology, Medical University of Vienna, Vienna, Austria
Metabolic syndrome is a cluster of three or more metabolic disorders including insulin resistance, obesity, and hyperlipidemia. Obesity has become the epidemic of the twenty-first century with more than 1.6 billion overweight adults. Due to the strong connection between obesity and type 2 diabetes, obesity has received wide attention with subsequent coining of the term “diabesity.” Recent studies have identified unique contributions of the immensely diverse gut microbiota in the pathogenesis of obesity and diabetes. Several mechanisms have been proposed including altered glucose and fatty acid metabolism, hepatic fatty acid storage, and modulation of glucagon-like peptide (GLP)-1. Importantly, the relationship between unhealthy diet and a modified gut microbiota composition observed in diabetic or obese subjects has been recognized. Similarly, the role of diet rich in polyphenols and plant polysaccharides in modulating gut bacteria and its impact on diabetes and obesity have been the subject of investigation by several research groups. Gut microbiota are also responsible for the extensive metabolism of polyphenols thus modulating their biological activities. The aim of this review is to shed light on the composition of gut microbes, their health importance and how they can contribute to diseases as well as their modulation by polyphenols and polysaccharides to control obesity and diabetes. In addition, the role of microbiota in improving the oral bioavailability of polyphenols and hence in shaping their antidiabetic and antiobesity activities will be discussed.
Definition of Human Microbiota
The human microbiota (microflora) refers to an aggregation of a mixture of microorganisms (e.g., bacteria, fungi, archaea, and viruses) that live in human tissues such as skin, uterus, lungs, and in the gastrointestinal tract without causing pathological responses in the host (Hugon et al., 2016). Instead these organisms form a symbiotic relationship with their host. In this relationship, the host provides a shelter and nutrition for the microbes, while the microbes provide the host with essential metabolites such as vitamin K, thiamine, biotin, folic acid, and vitamin B12, digesting polysaccharides into simpler molecules, boosting the immunity to combat pathogens, and competing with the latter for survival (Santacruz et al., 2010; D'Aimmo et al., 2012; LeBlanc et al., 2013). Beyond their traditional benefits, recent studies have implicated microbes residing human gut in modulating brain development (Wang and Wang, 2016), altering metabolic functions (Martinez et al., 2016), hormones and neurochemical production (Baothman et al., 2016). Some 30 years ago, human gut microbiota was largely ignored as a causative factor for illnesses and as a target for pharmacological intervention. Scientists were focused mainly on finding a novel pathogenic bacteria residing in the gut as an underlying cause of illnesses and as a target for drug development (Eggerth and Gagnon, 1933; Dalton, 1951; Walther and Millwood, 1951). In recent years, there has been an increasing trend in this area of research, and the focus has been shifted toward identifying microbial composition of the human gut microbiota (Figure 1), the factors altering this composition, and relating it to the pathogenesis of some diseases such as diabetes (Yamaguchi et al., 2016), autism (Berding and Donovan, 2016), obesity (Valsecchi et al., 2016), and other disorders (Gondalia et al., 2012; Baothman et al., 2016). In this review, we discuss the current advances in human gut microbiota, specifically their identity and diversity within the gastrointestinal tract of healthy adults as well as their contribution to metabolic diseases. We also review current knowledge about the effects of polyphenols and polysaccharides on gut microbiota and their role in controlling obesity and diabetes.
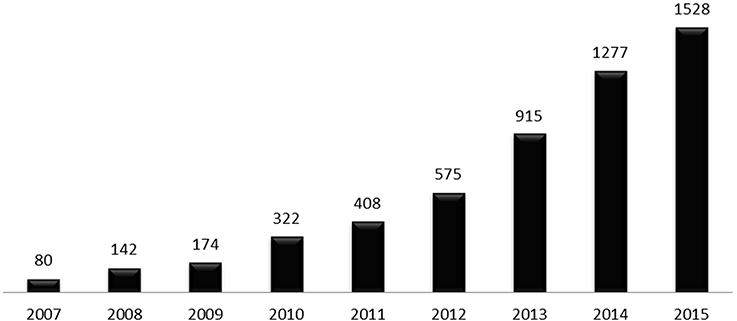
Figure 1. Number of publications related to the human gut microbiota in the last decade, per year. Data were obtained by searching Pubmed (http://www.ncbi.nlm.nih.gov/pubmed/) database using the term “human gut microbiota” on 4th November 2016.
Identity of Microbes Colonizing Human Gut
Microbes (bacteria, archaea, fungi, virus) colonizing human gut establish a complex ecosystem with current evidences confirming large diversity in their number and identity throughout the whole gastrointestinal tract (Hugon et al., 2016). It is now accepted that each individual hold a unique set of microbes (Callaway, 2015) whose composition is highly affected by many factors such as ethnicity, age, environment, and diet (Ursell et al., 2012; Yatsunenko et al., 2012). The exact composition of gut microbiota seems far from being fully identified in view of fast advances in “OMICS” techniques as well as improved culture conditions allowing for identification of new sets of bacteria, archaea, fungi, and viruses, their reclassification or renaming of already taxonomically known microbes (Hugon et al., 2016).
Major Bacteria Colonizing Healthy Human Gut
Earlier studies aiming to reveal the identity of microbiota in the gut of healthy humans have relied on culturing the microbes from fecal samples. Bacteria constitute most of the microbes colonizing human gut with species belonging to Firmicutes and Proteobacteria phyla being predominant (Eggerth and Gagnon, 1933; Zubrzycki and Spaulding, 1962). To a lesser extent, species belonging to other phyla such as Actinobacteria, Bacteroidetes, and Fusobacteria are also present (Eggerth and Gagnon, 1933; Zubrzycki and Spaulding, 1962). In later studies, several bacterial species, mostly anaerobic, have been identified in fecal samples obtained from 20 healthy individuals using improved cultural techniques (Moore and Holdeman, 1974; Holdeman et al., 1976). At least 400 bacterial species were suggested to colonize human gut and a total of 113 species have been fully recognized in the tested samples (Moore and Holdeman, 1974; Holdeman et al., 1976); major microbes identified being summarized in Tables 1–5.
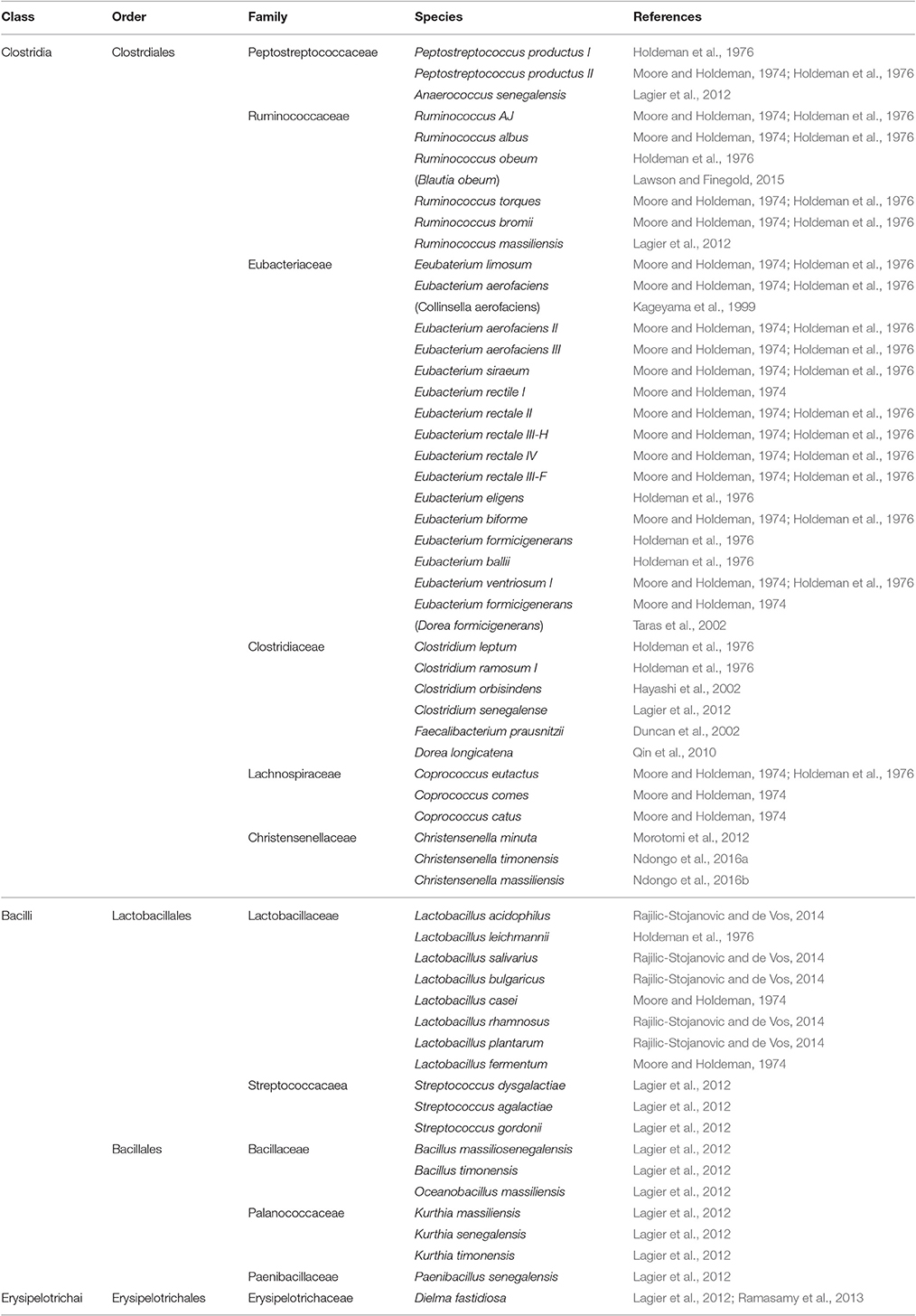
Table 1. Firmicutes phylum (class, order, family) and major species constituting the microbiota in healthy human gut.
Until recently, culture-based studies aimed to identify bacterial species in human gut were considered to be limited in their ability to fully evaluate bacterial diversity in human gut. This was based on the fact that such techniques cannot recover all species residing in the human gut (Gerritsen et al., 2011). Lau and colleagues showed that culture-enriched techniques associated with molecular profiling are alternative effective techniques to study microbial diversity in human gut (Lau et al., 2016). Fecal samples from two healthy donors have been cultured aerobically and anaerobically in 33 different culture media creating 66 different culture conditions. Cultured samples were then analyzed using 16S rRNA gene sequencing. In the previous study (Lau et al., 2016), 95% of the bacteria residing human gut has been recovered with 79 new Lachnospiraceae isolates being identified. Using a similar approach involving culturing fecal samples obtained from two healthy African donors under 212 different culture conditions, 340 species belonging to seven phyla, and 117 genera have been identified in addition to 24 “novel” microbes (Lagier et al., 2012). Advances in culture conditions have proven to be effective techniques, with currently over than 1,000 microbes reported to be effectively cultured (Rajilic-Stojanovic and de Vos, 2014). These microbes cluster in four main phyla with 450 species in Firmicutes, 214 in Proteobacteria, 164 in Actinobacteria, and 99 in Bacteroidetes (Rajilic-Stojanovic and de Vos, 2014). Collectively, these culture-based studies indicated that human gut is a complex ecological system colonized by thousands of microbes which are quite variable and diverse in-term of their quantity and identity.
Recent studies involving state-of-the-art techniques such as 16S rDNA gene, metagenomic sequencing, and molecular fingerprinting have rapidly increased our knowledge about the identity of the microbes residing in our gut (Hayashi et al., 2002; Knapp et al., 2010; Qin et al., 2010). These studies not only confirmed the identity of the microbes established by cultural techniques, but have also confirmed the incidence on “novel” microbes never detected by culture techniques. Authors characterized several “novel” microbes belonging to Clostridium genus in fecal samples obtained from three healthy Japanese individuals by using 16S rDNA libraries and culture-based techniques (Hayashi et al., 2002). In 2004, Akkermansia muciniphila a novel mucin-degrading bacterium was also identified from a healthy human volunteer using 16S rRNA-dependent approaches (Derrien et al., 2004). A. muciniphila is a Gram-negative bacterium that resides in the mucus layer and constitutes 3–5% of gut microbiota and is thus considered to be the most abundant mucus-degrading bacteria in healthy humans (Schneeberger et al., 2015). Interestingly, the abundance of A. muciniphila was found to decrease in obese and diabetic animals following high-fat diet feeding as well as in obese and diabetic human subjects (Schneeberger et al., 2015). In larger scale studies (Qin et al., 2010; Human Microbiome Project Consortium, 2012), the diversity of the microbes in human gut was examined using metagenomic analysis. Species belonging to Firmicutes, Actinobacteria, Bacteroides, and Proteobacteriaphyla were reported (Tables 1–4). However, other species belonging to Tenercutes, Spirochaetes, Cyanobacteria, and Verrucomicrobia phyla were reported to be present but at lesser densities (Human Microbiome Project Consortium, 2012).
Firmicutes Phylum
Firmicutes phylum (Table 1) represents the most diverse and the largest group of microbes making up 60–80% of the total microbes colonizing the GIT of healthy adults. Firmicutes include Gram-positive species with few exceptions such as Faecalibacterium prausnitzii (Duncan et al., 2002) previously known as Fusobacterium prausnitzii in the phylum Fusobacteria, and Christensenella minuta (Morotomi et al., 2012), C. massiliensis (Ndongo et al., 2016b), and C. timonensis (Ndongo et al., 2016a). Firmicutes phylum is classified into two main classes, Clostridia which includes the genera belonging to Christensenellaceae, Clostridiaceae, Eubacteriaceae, Lachnospiraceae, Peptococcaceae, Peptostreptococcaceae, Ruminococcaceae, and Veillonellaceae families and Bacilli which are divided into two main orders; Bacillales which includes the genera belonging to Paenibacillaceae, Planococcaceae, Bacillaceae, and Staphylococcaceae, and Lactobacillales which includes genera belonging to Aerococcaceae, Carnobacteriaceae, Lactobacillaceae, Leuconstocaceae, Lactococccaceae, and Streptococcaceae. A third class called Erysipelotrichales (e.g., Dielma fastidiosa) has been reported to present in human gut (Ramasamy et al., 2013).
Proteobacteria Phylum
Proteobacteria phylum (Table 2) comprises highly diverse Gram-negative microbes colonizing the GIT of healthy adults. It contains five major classes, α, β, γ, δ, and ε Proteobacteria with γ-class dominating other classes (Shin et al., 2015). Members of the phylum Proteobacteria, mostly belonging to γ-proteobacteria, have a low abundance in the gut of healthy humans (Shin et al., 2015). The γ-proteobacteria constitute the most diverse group of bacteria in the phylum Proteobacteria with six orders and nine families included. Several members in Aeromonadaceae and Vibreonaceae have been considered as pathogens detected in human intestine (Janda and Abbott, 1998, 2010; Hou et al., 2011). The γ-proteobacteria contain several genera which belong to Enterobacteriaceae, Pasteurellaceae Succinivibrionaceae, Pseudomonadaceae, and Moraxellaceae families. Many species of Enterobacteriaceae are a normal part of the gut flora found in the intestines of humans with their intensity increasing with age (Hopkins et al., 2001). Escherichia coli is one of the most important species colonizing the healthy human gut. The healthy human gut contains non-harmful strains of E. coli. Several other strains of E. coli have been associated with the outbreaks of diarrhea in children (Fagundes Neto et al., 1979; Bratoeva et al., 1994).
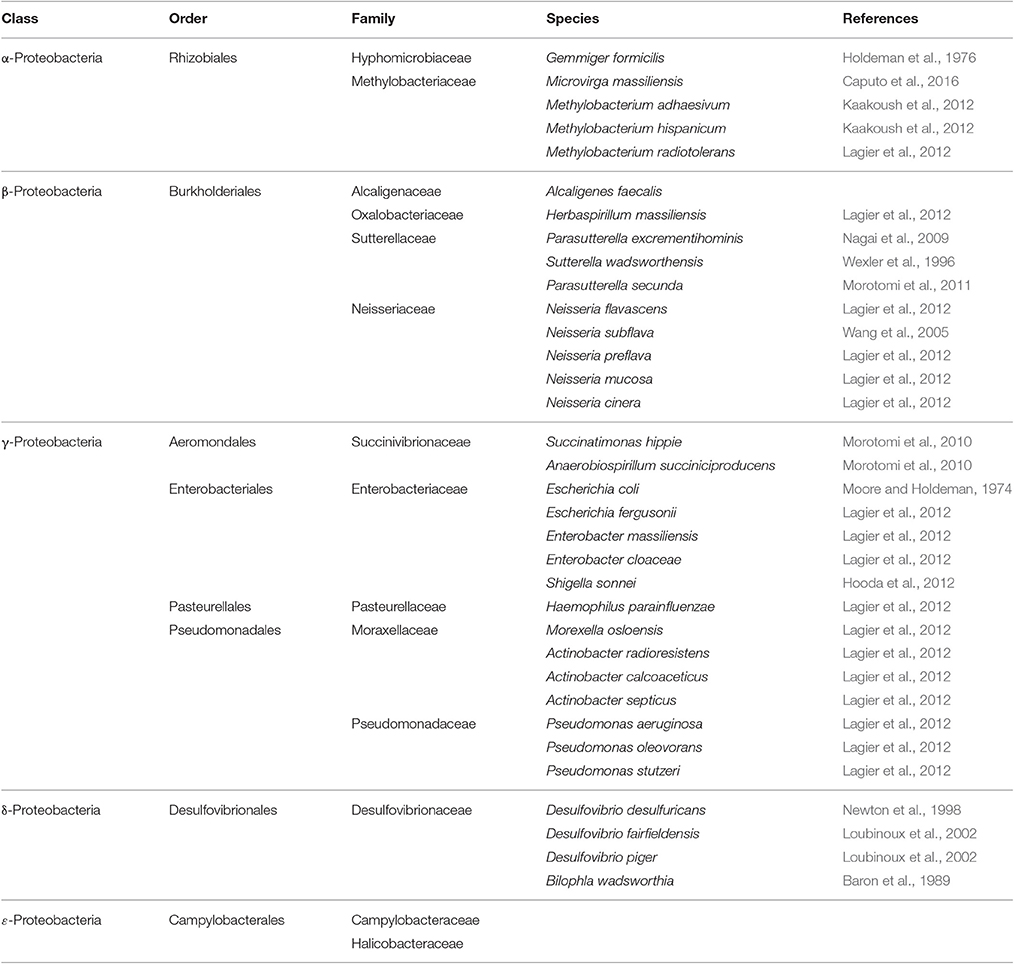
Table 2. Proteobacteria phylum (class, order, family) and major species constituting the microbiota in healthy human gut.
The class δ-proteobacteria includes a branch of predominantly aerobic sulfate-reducing bacteria that belongs to Desulfovibrionaceae family (Beaumont et al., 2016). The interest in this class was due to the ability of its species to generate the highly toxic hydrogen sulfide (H2S) molecules as part of their metabolic pathways. Overproduction of H2S has been associated with pathogenesis of ulcerative colitis and colon cancer (Beaumont et al., 2016).
The class β-Proteobacteria contains species belonging to Neisseria genera (Table 2). Neisseria meningitis and Neisseria gonorrhea are amongst the most studied species classified within this group (Corless et al., 2001; Unemo et al., 2014). Both species are Gram-negative coccoid bacteria, once thought to be restricted to humans and are part of the microflora of the upper respiratory and genito-urinary tracts but have not yet been recognized in the intestine. Despite being considered as part of the normal intestinal flora, Neisseria subflave has been linked to meningitis (Baraldes et al., 2000). Sutterellaceae species are frequently detected in human gut. They have received much attention in recent year because their density is reported to increase in inflammatory bowel diseases and in children suffering from autism and down-syndrome (De Angelis et al., 2013; Hiippala et al., 2016).
Actinobacteria Phylum
Actinobacteria phylum (Table 3) constituting healthy human gut microbiota includes diverse Gram-positive species that comprise three orders and 19 families. The Bifidobacterium species are one of the major genera of bacteria that are frequently detected exclusively in human gut and have been isolated from infant feces since 1900 (Rajilic-Stojanovic et al., 2007). Some Bifidobacterium species are critical to the health of the gut and are now considered as essential constituents of the probiotics used in the treatment of inflammatory bowel disease with no obvious side effects (Ghouri et al., 2014). In contrast to Bifidobacteriales species, the Actinomycetales species, despite being a highly diverse group of bacteria, have a relatively low abundance in healthy human gut: (102–103) cells/g of fecal sample (Hoyles et al., 2012). Amongst most detected genera are Corynebacterium and Propionibacterium (Eggerth, 1935; Moore and Holdeman, 1974; Holdeman et al., 1976). Species in Propionibacterium are capable of producing propionic acid from lactic acid and also vitamin B12 (Kiatpapan and Murooka, 2002), making them an ideal species to be included in probiotics (Kiatpapan and Murooka, 2002).

Table 3. Actinobacteria phylum (class, order, family) and major species constituting the microbiota in healthy human gut.
Bacteroidetes Phylum
Bacteroidetes phylum (Table 4) includes several large classes of Gram-negative, non-spore forming, anaerobic or aerobic, and rod-shaped bacteria that are widely distributed in the environment and in the human guts (Belizário and Napolitano, 2015). By far, species in the class Bacteroidia have been most extensively studied due to their relevance to human metabolic processes. These species play a vital role in metabolizing complex molecules such as proteins and polysaccharides such as cellulose, pectin and xylans into simpler molecules used as source of energy (Wexler, 2007; Xu et al., 2007; Sakamoto and Ohkuma, 2012). Bacteroidia colonizing human gut include species clustered in Bacteroidaceae, Porphyromonadaceae, Prevotellaceae, and Rikenellaceae families. They are symbiotic and diverse bacteria making up a substantial portion of the normal flora residing in lower GIT (1010–1011 cells per gram of human feces; Holdeman et al., 1976). Other bacterial species belonging to classes sphingobacteria and flavobacteria have occasionally been detected in healthy human gut (Lagier et al., 2012), however their significance in the gut has yet to be demonstrated.
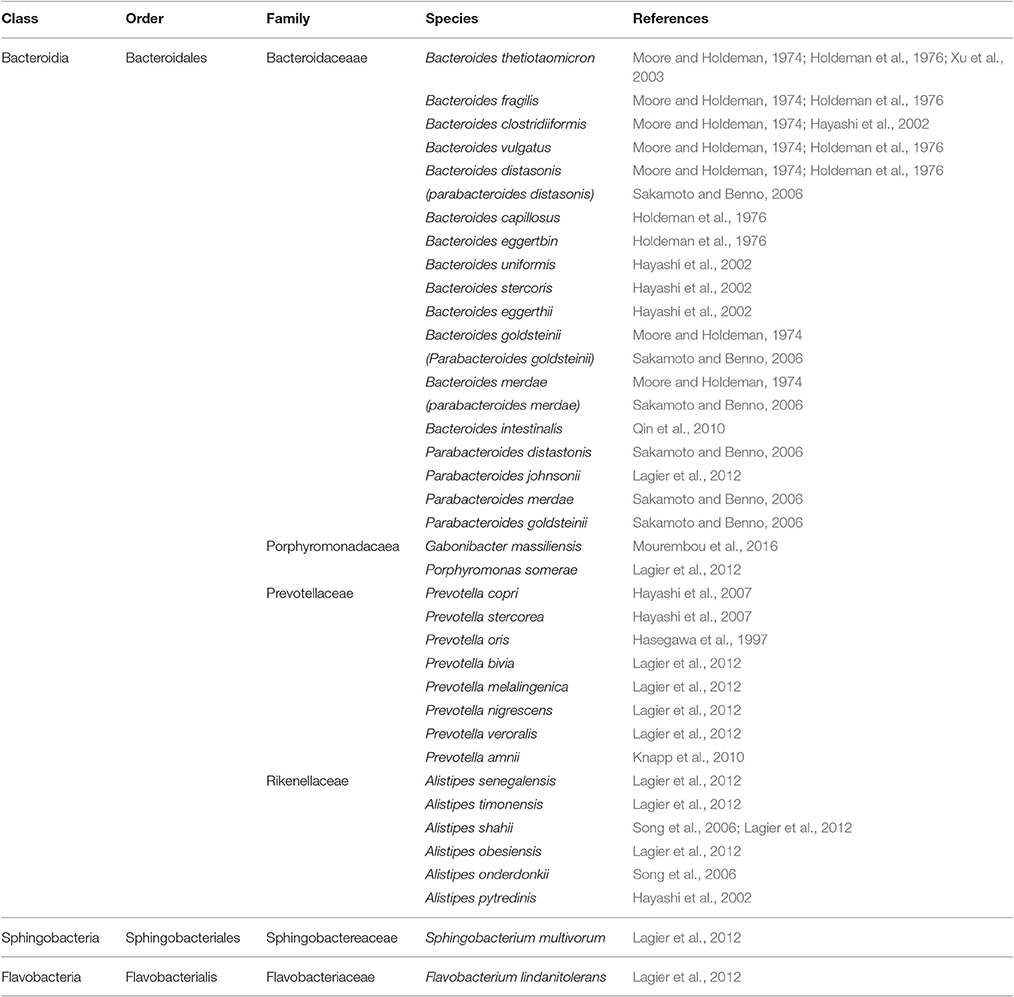
Table 4. Bacteroidetes phylum (class, order, family) and major species constituting the microbiota in healthy human gut.
Fusobacteria Phylum
Fusobactera phylum (Table 5) includes Gram-negative, non-sporeforming, anaerobic bacilli frequently detected in human gut (Walter et al., 2002). This phylum includes species belonging to Fusobacteriaceae and Leptotrichiaceae families. Species within Fusobacteriaceae were considered to be limited to the oral cavity until 1966 when it was shown that they could be detected in fecal samples (Van Houte and Gibbons, 1966). Fusobacteriaceae species appear to be directly related to the health of the gut. Their density increases in inflammatory diseases such as appendicitis (Swidsinski et al., 2012), ulcerative colitis (Rajilic-Stojanovic et al., 2013) and colon cancer (Kostic et al., 2012). Leptotrichiaceae species appears to be detected mainly in human elderly gut (Hayashi et al., 2005) and female reproductive system (Thilesen et al., 2007).
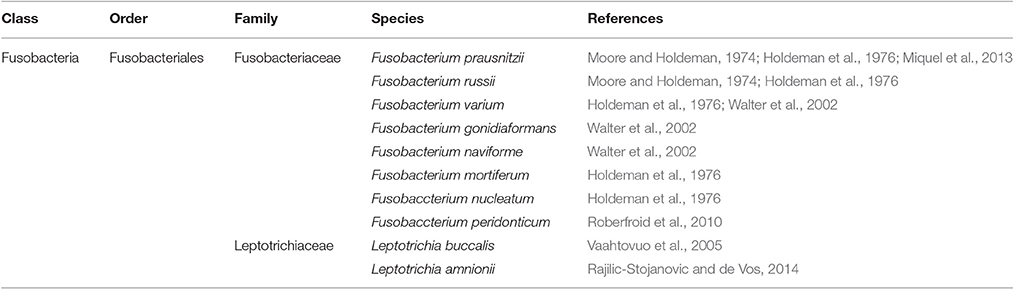
Table 5. Fusobacteria phylum (class, order, family) and major species constituting the microbiota in healthy human gut.
Archaea, Viruses, and Fungi Colonizing Healthy Human Gut
Healthy human gut microbiota includes archaea, fungi, and viruses as consistent residents (Parfrey et al., 2011; Lloyd-Price et al., 2016; Rehman et al., 2016), even though in some cases at low densities. A small number of archaeal genera have been identified in the healthy human microbiota, primarily in the gut. Historically, archaea were classified as bacteria with the name (archaebacteria) before being reclassified in a specific domain (Woese et al., 1990; Pace, 2006), since they have characteristic features unique enough to separate them from bacteria and Eukaryota domains (Cavicchioli, 2011). In total, eight archaeal species have been associated with the human GIT (Rajilic-Stojanovic and de Vos, 2014). Species of the Methanobrevibacter genus, namely; Methanobervibacter smithii, M. ruminantium, M. stadtmaniae, and M. luminyensis, are the most prevalent (Rajilic-Stojanovic and de Vos, 2014; Horz, 2015) in the healthy gut. M. smithii in particular, has been found to be well-adapted to inhabit the human gut. These species are implicated in optimizing the digestion process of dietary polysaccharides in association with other microbes (Samuel et al., 2007) and they are capable of producing methane (methanogenes) from CO2 and H2. Their abundance in infants is relatively low (102–106 cell/g of fecal sample) and increases during adulthood reaching up to 1010 cells/g fecal samples. Such changes in densities explain the absence of methane gas, as detected by a breath test, during infancy in contrast to adulthood (Gaci et al., 2014).
Archaeal science appears to be in its early stages. Unlike bacteria, archaeal species are largely ignored as a topic in microbiology, possibly due to the lack of appropriate genomic tools to reveal their existence and their diversity (Horz, 2015). Accordingly, it is quite likely that the archaeal domain represents a diverse community with several yet unknown taxa, waiting to be identified. Although, some archaeal species (methanogens) appears to be associated with gut inflammatory diseases, including constipation, the question arises as to whether it is necessary to identify all taxa. Even if they are rare and/or present only in very low abundance, their precise role in disease development remains unknown (Gaci et al., 2014; Horz, 2015).
Many viral species have been reported to colonize the healthy human gut system forming a symbiotic relationship with their bacterial (bacteriophages) and human hosts. These viruses constitute the so called “human gut virome” (Zou et al., 2016). Each individual holds a unique viral profile, highlighting the high interpersonal variability of the healthy human gut virome (Minot et al., 2011; Scarpellini et al., 2015). The human virome includes viruses from seven families, namely Herpesviridae, Polyomaviridae, Papillomaviridae, Adenoviridae, Anelloviridae, Parvoviridae, and Circoviridae. Not all families have been detected in healthy human gut (Wylie et al., 2014). In a large scale metagenomic study involving 102 healthy individuals, roseolovirus (Herpesviridae), alphapapillomavirus and gammapapillomavirus (Papillomaviridae), mastadenovirus (Adenoviridae), polyomavirus (Polyomaviridae), gyrovirus (Circoviridae), and some other unclassified viruses were generally detected in fecal samples (Wylie et al., 2014).
A better understanding of human virome composition and dynamics should confirm that it is an important factor contributing to human health (Scarpellini et al., 2015). Indeed, several recent studies have implicated gut virome in regulating/stabilizing their host bacterial species and subsequently maintaining microbial diversity in the gastrointestinal tract (Minot et al., 2011; Abeles and Pride, 2014; Scarpellini et al., 2015).
In addition, several fungal species have been reported to colonize healthy human gut, even if at low amounts (Ott et al., 2008; Sokol et al., 2016). The most prevalent fungal species detected in the healthy human gut are clustered into three phyla; Ascomycota, Basidiomycota, and Zygomycota. Candida albicans and Candida rugosa have routinely been detected in human gut (Ott et al., 2008; Sokol et al., 2016). Most Candida species under normal conditions form symbiotic/commensal relationships with the host. However, when host environmental conditions favor the outgrowth of C. albicans, excessive colonization can lead to infection and invasion of host tissues (Ott et al., 2008; Sokol et al., 2016).
Diversity Along Human Gut
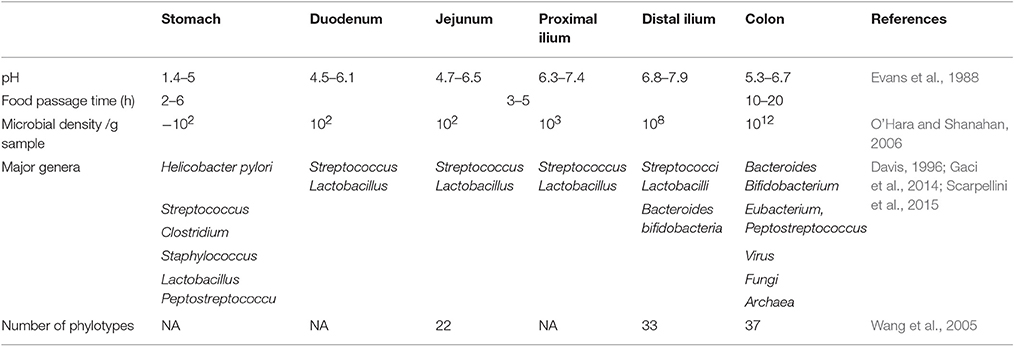
The human gut is colonized by higher numbers of bacteria and more diverse species compared to other parts of the body (Quigley, 2013). The microbial composition of the gut flora varies along the gut (Table 6), with stomach and small intestine having relatively lower microbial diversity (Guarner and Malagelada, 2003; O'Hara and Shanahan, 2006). In contrast, the colon is densely colonized by microbes reaching up to 1012 cells per gram of intestinal content (O'Hara and Shanahan, 2006). There are between 300 and 1,000 different bacterial species, but the vast majority of bacteria (99%) come from 50 to 60 species (Guarner and Malagelada, 2003; Rajilic-Stojanovic et al., 2007). Because of their abundance in the intestine, bacteria represents ~60% of fecal dry mass (Stephen and Cummings, 1980). Fungi, archaea, and viruses are also present in the gut flora, but less is known about their activities (Lozupone et al., 2012).
Stomach Flora
Healthy human stomach has no longer been considered as a “sterile organ” since the discovery of H. pylori (Marshall and Warren, 1984). It is rather now considered as a harbor for many bacterial species, dominated by five major phyla: Firmicutes, Bacteroidites, Actinobacteria, Fusobacteria, and Proteobacteria. Generally, the healthy human stomach is dominated by Prevotella, Streptococcus, Veillonella, Rothia, and Haemophilus (Bik et al., 2006; Zilberstein et al., 2007). Characterizing healthy stomach microbiota has come from studies involving gastric biopsies and stomach juices collected from individuals routinely performing upper gastrointestinal endoscopy for dyspepsia (Li et al., 2009; Engstrand and Lindberg, 2013). Using a small subunit 16S rDNA clone library approach (Bik et al., 2006), 128 phylotypes have been shown to colonize the stomach, coming from the five major phyla mentioned above. Essentially similar findings have been reported by other researchers despite differences in techniques (Delgado et al., 2013) and the ethnic background of biopsy donors (Li et al., 2009; Delgado et al., 2013; Engstrand and Lindberg, 2013), suggesting that some homogeneity may exist in the composition of stomach microbiota.
The contribution of stomach microbiota to disease pathogenesis has not yet been fully explored, although an alteration in the density of Firmicutes phylum, particularly the Streptococcus and Prevotella genera, has been reported in patients with H. pylori infection and stomach cancer (Wroblewski and Peek, 2016). It remains pivotal to know to what extent H. pylori infection or stomach cancer affects the composition of stomach microbiota and whether restoring stomach biota, either naturally or pharmacologically, can modulate the outcome of an H. pylori infection or cancer progression (Wroblewski and Peek, 2016).
Intestinal Flora
Most of the studies describing gut microbial composition in health adults have usually involved fecal samples, representing the diversity of the microbes present in the large intestine (Finegold et al., 1974; Moore and Holdeman, 1974; Holdeman et al., 1976). The small intestine, however, is more acidic, has higher levels of oxygen, less transit time, and has effective immune-cell-mediated antimicrobial factors compared with the colon (Keshav, 2006). Such physiological variations promote less microbial diversity in small intestine compared with colon. Accordingly, only fast growing facultative anaerobes, which tolerate the bactericidal effects of bile acid and effectively compete for carbohydrate, will dominate this part of the gut (Zoetendal et al., 2012). There are limited studies aimed to assess small intestinal microbial diversity (Hartman et al., 2009; Zoetendal et al., 2012). This is partly due to the technical difficulties in collecting samples for analysis.
Small intestine microbiota has been examined in three different anatomical locations, namely jejunum, ileum, and distal ileum. Despite the fact that each part is characterized by its own microbial composition, it appears that the profile of the jejunum is closely related to that of the stomach; with Bacilli, mainly of the Streptococcaceae species, dominating this section (50–70%) (Wang et al., 2005; Zoetendal et al., 2012). Even in elderly, moving along the small intestine toward the colon, the intensity of Bacilli species in the ileum and distal ileum drop remarkably to reach 20 and 5%, respectively (Hayashi et al., 2005; Zoetendal et al., 2012). Instead Clostridia species such as IX, XIVb XIVa, and IV form up to 30% of the microbiome, while Bacteroidetes species occupy a proportion of 49% at these locations (Wang et al., 2005; Zoetendal et al., 2012).
Human Gut Microbiota in Human Health and Disease
There is increasing evidence that differences in the structure, function, and diversity of the human gut microbiota are associated with states of human health and disease.
Establishment of Gut Microbes
The first 2 years of life mark a dynamic period in which the gut microbiome builds from the initial microbial repository at birth and adjusts until the composition and function are more like that of an adult (Koenig et al., 2011). The type of birthing delivery strongly influences which microbes are present upon the initial establishment of an individual's gut microbiota. Infants born vaginally develop gut microbiota that are more similar to their mothers than those born via cesarean section; this also conferring some functional differences. For example, gut microbiota in infants born vaginally tend to express a lower proportion of antibiotic resistance genes (Bäckhed et al., 2015). The developing gut microbiota is strongly influenced by the infant's diet and life events. Over time, the infant begins harboring microbes capable of digesting complex sugars and starch (Koenig et al., 2011; Bäckhed et al., 2015). As more types of foods are introduced into the infant's diet, the bacterial diversity in the gut increases. For infants who are breastfed, the discontinuation of breastfeeding appears to be the strongest factor driving the change in gut microbiota structure from the less diverse infant microbiome to the more diverse adult phenotype (Koenig et al., 2011; Bäckhed et al., 2015).
Host genetic signature has also been reported to contribute, to a certain extent, to the types of microbes present within an individual's gut. Although, studies generally suggest that genetic effects do not exert a strong global influence on which microbes colonize the gut, monozygotic twin pairs display more similar microbiota than their dizygotic counterparts (Turnbaugh et al., 2009; Smith et al., 2013; Goodrich et al., 2014). Moreover, even if few bacterial taxa exhibit strong heritability, the strongest are associated with clinically meaningful phenotypic differences (Goodrich et al., 2014). Recent evidence suggests that there is cross talk between the gut microbiome and host genetic signature that results from altered gene expression (Richards et al., 2016). In other words, differential gene expression in host cells allow for specific types of bacteria to colonize one individual over another, and the way a cell responds to bacteria (or the community of microbes present) may vary depending on the host's genetic make-up. However, environment and diet appear to be the strongest drivers of microbiota composition, with some of the observed variance attributable to host genetics (Richards et al., 2016).
Gut Microbiota and Human Health
Despite inter-individual differences in the structure and diversity of the human gut microbiome, the microbial metabolic and functional pathways remain stable among healthy individuals (Human Microbiome Project Consortium, 2012). The human gut microbiome encodes at least 10-fold more genes than the human genome and functional redundancy among some of these genes allow different microbes to create individualized communities that will carry out the same functions to maintain homeostasis and a symbiotic relationship with the human host (Ley et al., 2006; Qin et al., 2010). The functional redundancy is important for maintaining a favorable environment within the gut to ensure survival of the bacteria, while also contributing to human metabolism and health (Parekh et al., 2014). This is significant because the bacteria present in gut microbiome communities help liberate carbohydrates and other nutrients from the diet that could otherwise not be utilized by the human host (Larsbrink et al., 2014). For example, certain species of Bacteriodetes can metabolize xyloglucans, a complex carbohydrate found in dietary fiber that contributes to human's carbohydrate intake. At least one of the taxa capable of this type of metabolism is consistently reported in studies evaluating gut microbiota (Larsbrink et al., 2014). Since products of gut microbe metabolism, like that of xyloglucans, contribute to the symbiotic relationship between host and microbe, there has been increasing interest in how the microbes and their metabolic products contribute to human health and disease.
As mentioned, Firmicutes and Bacteroidetes are the most predominant phyla in the human gut and affect the production of short-chain fatty acids (Rosenbaum et al., 2015). Short-chain fatty acids (SCFA) produced from indigestible carbohydrates by gut microbes are used as energy by various human tissues (Rosenbaum et al., 2015). Some studies have observed that the proportions of bacteria present in these phyla may contribute to metabolic outcomes in the host, in part by altering the amount of short-chain fatty acids produced (Turnbaugh et al., 2006, 2009). Greater density of Bacteroidetes has been associated with increased butyrate and propionate levels that contribute to a healthy body weight by suppressing hunger and helping to maintain glucose homeostasis (Lin et al., 2012). Butyrate produced by the gut microbes also contributes to health by providing energy for colonic epithelial cells and inhibiting inflammation, particularly that induced by lipopolysaccharide (Cani et al., 2007; Belkaid and Hand, 2014).
A. muciniphila, is a Gram-negative bacterium of the order Verrucomicrobiales. Despite containing lipopolysaccharides (LPS), a well-known endotoxin, a study by Everard et al. (2013) has demonstrated the lack of direct association between Gram-negative bacteria and gut or metabolic endotoxemia (Everard et al., 2013). On the contrary, oligofructose administration which has been reported to restore A. muciniphila levels mitigated endotoxemia caused by HFD (Everard et al., 2013). Other studies also reported the seemingly counterintuitive results that A. muciniphila preserves the integrity of the intestinal mucous and intestinal barrier function and counteracts the deleterious effect of HFD on gut permeability despite having week mucin-degrading activities (Cani et al., 2009; Belzer and de Vos, 2012; Everard et al., 2013). One explanation is that this bacterium protects against inflammation through increasing the levels of the anti-inflammatory intestinal endocannabinoids which control gut barrier and gut peptide secretion (Everard et al., 2013). In addition, higher levels of A. muciniphila were associated with greater enteroendocrine L-cell activity, hence more secretion of glucagon-like peptides GLP-1 and GLP-2 (Cani et al., 2009). Gut peptides including GLPs were reported to control epithelial barrier proliferation and integrity (Belzer and de Vos, 2012). It was demonstrated that A. muciniphila controls GLP-2 secretion through increasing levels of 2-oleoylglycerol (2-OG) and reduces metabolic endotoxemia via increasing 2-arachidonoylglycerol (2-AG) levels (Everard et al., 2013). However, the link between the enhanced production of endocannabinoids and the beneficial effects of A. muciniphila needs further investigation. Other studies reported that A. muciniphila levels were reduced in diseases involving dysfunctional intestinal barrier such as irritable bowel syndrome (IBS) and chronic granulomatous colitis (Falcone et al., 2016). However, some authors reported contradictory results; for example a Chinese MGWAS study stated that A. muciniphila did not improve mucous layer thickness (Tilg and Moschen, 2014), while Ganesh et al. attributed exacerbated inflammation in Salmonella typhimurium-infected mice to the mucin-degrading effect of A. muciniphila (Ganesh et al., 2013). Such conflicting studies need further clarification.
In general, imbalances in the types of bacteria present in the gut microbiota are thought to contribute to disease in part by altering different metabolic processes and/or pathways in the host.
Gut Microbiota and Disease
Differences in the composition of the gut microbiota in humans that relate to disease have been reported for several conditions including, but not limited to, cardiovascular disease (Holmes et al., 2008; Wang et al., 2011), type 2 diabetes (Larsen et al., 2010), and obesity (Turnbaugh et al., 2009). Interest in studying the human gut microbiota related to disease has increased, in part due to the fact that it appears to be highly influenced by interventions that ameliorate symptoms or disease such as medications and diet. For example, individuals that eat red meat exhibit greater levels of the gut metabolite trimethylamine-N-oxide (TMAO) than non-meat eaters. Importantly, TMAO is associated with increased plaque formation in arteries (Koeth et al., 2013; Tang et al., 2013). The fact that products from microbial metabolism play an integral role in many metabolic pathways of the host, suggests that complex disorders with metabolic components may benefit from targeted alterations in the microbiota through dietary or supplement interventions. For this review, we will focus specifically on reviewing the microbiome literature as it relates to metabolic syndrome.
Overview of Metabolic Syndrome
Metabolic syndrome is a group of factors that collectively raise the risk for other chronic and acute disease processes. The greater the number of the risk factors an individual has, the higher the risk for other poor health outcomes and disease such as heart attacks, stroke, and type 2 diabetes. Metabolic syndrome risk factors are closely linked with obesity and sedentary lifestyles (Alberti et al., 2009; Kaur, 2014). There are several different reports that include diagnostic criteria for metabolic syndrome that are utilized in different areas of the world (Alberti et al., 2009; Kaur, 2014). Despite some minor difference in criteria, individuals presenting with metabolic syndrome exhibit 2–3 of the following characteristics: central obesity, abnormal serum lipid levels (high triglycerides and/or low high-density lipoprotein), high blood pressure, and elevated blood glucose levels. Some groups contain diagnostic metrics for insulin resistance, whereas others do not since it is difficult to uncouple from obesity (Alberti et al., 2009; Kaur, 2014).
The mechanistic pathways for obesity are complex due to a multiplicity of genetic and lifestyle factors that contribute to weight gain, although insulin is a critical regulator of adipocyte biology (Grundy et al., 2004). Insulin inhibits lipolysis and stimulates glucose transport as well as triglyceride synthesis (Grundy et al., 2004). When lipolysis is stimulated in insulin resistant individuals, large amounts of circulating fatty acids and inflammatory markers are released from expanded adipose tissue mass (Grundy et al., 2004; Afsar and Elsurer, 2014). When these processes occur consistently over time, elevations in weight and blood sugar become apparent and begin to influence other metabolic processes (Grundy et al., 2004; Afsar and Elsurer, 2014). The increase in circulating free fatty acid levels contributes to the development of triglyceride reservoirs in muscle and liver, resulting in decreased glucose uptake in muscle, increased hepatic gluconeogenesis, and increased blood cholesterol and triglyceride levels which may also contribute to rising blood pressure (Afsar and Elsurer, 2014). The resultant pro-inflammatory and pro-thrombotic states increase the risk for cardiovascular disease and type 2 diabetes (Afsar and Elsurer, 2014). Since the aforementioned components of metabolic syndrome overlap to some degree mechanistically, similar overlap in differences in microbiota have been reported based on metabolic syndrome diagnostic criteria (Rial et al., 2016).
Gut Microbiota and Obesity
Diagnostic criteria for obesity, particularly central or abdominal obesity, vary by guidelines and populations being assessed. Despite differences in diagnostic cutoffs determining obesity, several research groups have reported differences in gut microbiota related to body weight (Turnbaugh et al., 2009). Some of the first ground breaking studies investigating the gut microbiota in humans sought to evaluate the relationship with obesity. The first of which identified a decrease in diversity of gut microbiota in obese individuals, whereas this diversity was increased with dietary weight loss (Turnbaugh et al., 2009). A meta-analysis found that studies tend to report shifts in different microbes associated with obesity (Walters et al., 2014). This may be related to the functional redundancy in the gut microbiota, since multiple different taxa can contribute to the same metabolic pathway. Walters and colleagues also noted that some of the variability of results reported between studies was due to differences in laboratory protocols and analytic pipelines. For example, in the Turnbaugh and colleagues twin study (Turnbaugh et al., 2009), a de novo approach was used which assigns operational taxonomic units (OTUs) without an external reference. In the meta-analysis assessing results of obesity associated microbiome changes using a closed-reference OTU assignment process, where sequences that do not match a reference data set are excluded; there were no significant differences in the microbiota between obese and lean individuals (Turnbaugh et al., 2009). In addition to some of equivocal results observed when different laboratory and data processing pipelines are used, there is the question of the reproducibility of murine studies in humans because there are significant differences in how the different species maintain energy homeostasis as diet and metabolic demands are different (Turnbaugh et al., 2009). There are few studies evaluating processes identified as contributing to obesity in murine models and in human populations, the same applies to most of the components of metabolic syndrome (Rosenbaum et al., 2015). Studies are increasingly evaluating host genetic signatures as well as the microbiota to address these issues. Studies designed in this manner have identified features in the gut microbiome that suggest a strong relationship between certain microbes and host genomic loci that are linked to obesity in human populations (Le Chatelier et al., 2013). Despite this ambiguity, continuing studies of the human gut microbiota aim to refine result of the work that has been done thus far.
Gut Microbiota and Dyslipidemia
All diagnostic guidelines for metabolic syndrome include metrics of dyslipidemia, particularly high levels of triglycerides and low levels of high-density lipoprotein (HDL) (Kaur, 2014). All consistently categorize triglycerides as elevated when serum levels are above 150 mg/dL, and there is some variability between guidelines about what constitutes low HDL levels among men and women (Kaur, 2014). Some bacterial taxa from the human gut are present in artherosclerotic plaques, although the community of microbes within the plaque most closely resembles bacterial taxa that predominate in the oral cavity (Koren et al., 2011). Two uncharacterized taxa in the human gut, from the Erysipelotrichaceae and Lachnospiraceae families, were correlated with total cholesterol and LDL levels, but no association was observed related to serum HDL levels (Koren et al., 2011). A recent study evaluating the relationship between the human gut microbiota and blood lipid levels found that the gut microbiota has significant differences in the same metrics that are characteristics of metabolic syndrome, namely obesity, serum triglyceride, and HDL levels (Fu et al., 2015). Unlike Koren's team, Fu and colleagues did not find any variability in the gut microbiota composition to be associated with elevated levels of low-density lipoproteins (LDL) or total cholesterol levels, which are not included as diagnostic criteria for metabolic syndrome (Fu et al., 2015). Fu's team identified 34 bacterial taxa that were associated with obesity, triglyceride and HDL levels in their study group. Further, they determined that the gut microbiota composition accounted for nearly 26% of the variance observed in HDL levels. These data suggest that treatments aimed at altering the gut microbiota to increase levels of HDL cholesterol may have potential to be highly effective (Fu et al., 2015).
Gut Microbiota and Blood Sugar
Obesity and adiposity are directly related to increased risk for elevated blood glucose levels and type 2 diabetes (Alberti et al., 2009). All diagnostic guidelines contain metrics for impaired fasting glucose or impaired glucose tolerance, with or without diabetes (Kaur, 2014). Some guidelines for metabolic syndrome use glucose metrics interchangeably with insulin resistance metrics (Kaur, 2014). Differences in gut microbiota have been reported in individuals with type 2 diabetes, with elevated levels of Bacteroidetes and Proteobacteria and lower levels of Firmicutes than healthy individuals (Larsen et al., 2010). Interestingly, in Larsen's study, they did not observe the commonly reported decreased level of Bacteroidetes in individuals that were diabetics and also obese. However, others have observed that the composition of the gut microbiota in obese individuals with insulin resistance or elevated glucose levels agree with previous studies reporting lower levels of Bacteroidetes and butyrate producing species (Qin et al., 2012; Vrieze et al., 2012). As discussed previously, obesity can contribute to the development of this phenotype by increasing insulin resistance, resulting in higher levels of glucose remaining in the serum. A small intervention study (N = 18, n = 9/group) reported an increase in bacterial diversity in the gut when microbiota from lean donors was transplanted into obese recipients; with an associated increase in butyrate producing bacteria and subsequent increase in insulin sensitivity (Vrieze et al., 2012).
A. muciniphila levels were found to be inversely associated to obesity and diabetes in several animal and human studies (Tilg and Moschen, 2014). The role of A. muciniphila in reducing inflammation could provide protection against the development of type 2 diabetes (Schneeberger et al., 2015). In addition, administration of A. muciniphila reduced body weight, adipose tissue inflammation, lipidemia, and hyperglycemia in diabetic and obese animals and increased adipose tissue browning and fatty acid oxidation (Tilg and Moschen, 2014). However, two recent studies reported increased levels of A. muciniphila in animals fed with high fat and high carbohydrate diet (Carmody et al., 2015; Hamilton et al., 2015). In human studies, it was found that under certain levels of A. muciniphila, human subjects responded less efficiently to caloric restriction diet in terms of reduction of hypergycemia, insulin resistance and inflammatory markers (Hamilton et al., 2015). However, there is no simple relationship between levels of A. municiphila and inflammation, and the threshold beyond which a shift occurs between healthy to pathological conditions is still unknown (Hamilton et al., 2015).
Together, these studies suggest that the composition/diversity of gut microbiota may contribute to elevated glucose levels and insulin resistance in individuals with metabolic syndrome.
Gut Microbiota and Metabolic Syndrome
Many of the studies evaluating the gut microbiota related to the risk factors associated with metabolic syndrome report associations among multiple features of metabolic syndrome (Rial et al., 2016). Given that multiple risks occur concurrently, even in the absence of metabolic syndrome (i.e., having only 2 of the associated risk factors), it is difficult to disentangle the individual contribution to disease state without considering confounding factors. Some of the associations with multiple risk factors suggest that the gut microbiome is intimately involved with observed variations over and above those related to age, sex, and host genetics (Rial et al., 2016). Notably 12 bacterial OTUs have been associated with variability across three traits (BMI, triglycerides, and HDL levels) (Fu et al., 2015). Others have found that lower diversity associated with obesity is also associated with insulin resistance and dyslipidemia (Le Chatelier et al., 2013).
Because of confounding of characteristics associated with obesity, dyslipidemia, and elevated blood glucose levels, some scientists have evaluated the human gut microbiota composition specifically related to metabolic syndrome. Similar to what is observed in obese individuals by Turnbaugh's group (Turnbaugh et al., 2009), a lower level of diversity of gut microbes was present in individuals with metabolic syndrome; and some of the taxa associated with metabolic syndrome are linked to a genetic variant in the apolipoprotein A5 gene (APOA5) (Lim et al., 2016). Specifically, Lim and colleagues noted that specific taxa associated with metabolic syndrome were not all correlated with each characteristic of metabolic syndrome in the same way. For example, they found Lactobacillus to be correlated with central obesity and fasting blood sugar, but negatively correlated with HDL levels (Lim et al., 2016). This suggests that metabolites from gut metabolism contribute to multiple mechanistic pathways involved in metabolic syndrome and that there may be different combinations of contributing microbes depending on which characteristics of metabolic syndrome are present in an individual (Lim et al., 2016). This suggests that multiple interventions may be effective for treating and preventing metabolic syndrome by targeting mechanistic pathways associated with different characteristics of metabolic syndrome.
Therapeutic Modulation of Gut Microbiota to Restore Lipid and Glucose Homeostasis
Polyphenols
Polyphenols constitute a large group of heterogeneous secondary metabolites found almost ubiquitously in the plant kingdom. The daily intake of dietary phenols is estimated to be larger than 1 g, which is 10 times higher than vitamin C intake from diet (Scalbert et al., 2005). Their chemical structures are characterized by the presence of polyhydroxyphenyl units and range from simple monomers and oligomers to highly polymeric compounds with molecular weight reaching up to 30,000 Da, such as condensed tannins (Tsao, 2010). Polyphenols can be further classified according to their chemical structures into two subgroups: flavonoids and non-flavonoid polyphenols, including phenolic acids. They predominantly exist in combination with sugars or acylated sugars (glycosides), but may also occur in other conjugated structures (amides, esters, and methyl ethers) or in their free forms (Tsao, 2010).
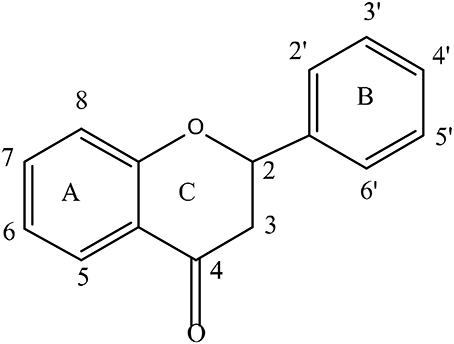
Flavonoids form the largest group of polyphenolic compounds with more than 6000 compounds identified and/or isolated from plant sources (Kumar and Pandey, 2013). For a polyphenol to be classified as a flavonoid, its structure has to contain a benzo-pyrane (chromane) moiety included in a C6-C3-C6 structural backbone (Figure 2), in which the two C6 units are benzyl rings (ring A and ring B) and C3 unit is the chromane ring (ring C). Depending on the hydroxylation and substitution patterns, as well as the degree of saturation of the chromane ring, flavonoids can be further divided into several classes such as: flavones, flavonols, flavanones, flavanonol, flavan-3-ols, anthocyanins, and isoflavones (Figure 3). Condensed tannins, also known as proanthocyanidins or polyflavonoid tannins, are polymers of flavan-3-ols or flavan-3,4-diols (Figure 3). In all flavonoid classes, the ring B is attached to the carbon-2 of ring C; with the exception of isoflavonoids in which the attachment between the two rings takes places at carbon-3 of ring C. Interestingly, this particular arrangement has conferred isoflavones the ability to interact with estrogen receptor and to act as weak phytoestrogens (Ozdal et al., 2016).
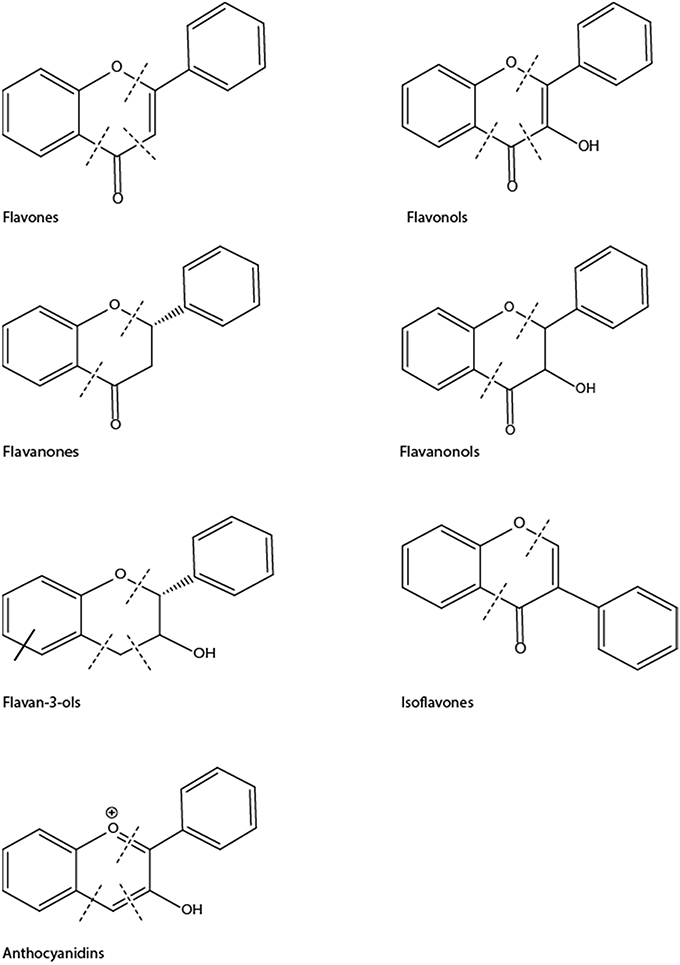
Figure 3. Chemical structures of main classes of flavonoids showing positions of potential C-ring cleavage (——) or A-ring cleavage (——).
The nonflavonoid-polyphenols have more heterogeneous structures and broadly comprises hydrolysable tannins (gallotannins and elagitannins), lignans stilbenes, and phenolic acids). Phenolic acids in food are mainly benzoic acid or cinnamic acid derivatives (Lafay and Gil-Izquierdo, 2008). Some phenolic acids such as p-coumaric and hydroxyl-cinnamic acids are considered “functional polyphenols” despite their monophenolic structures because they share many properties with polyphenols (Pereira et al., 2009).
Polyphenols are produced in plants to serve important roles such as protection against different environmental stressors and pathological aggressions, thus acting as primary defense mechanisms (phytoalexins). For this reason, polyphenols are endowed with excellent antioxidant, antifungal, antibacterial, and photo-protective properties (Li et al., 2014).
Over the last few decades, research on polyphenols has expanded exponentially and many other biological activities and health benefits have been attributed to polyphenols. These include, but are not limited to, anti-inflammatory, anticancer, and protective activities against inflammatory and oxidative stress-induced disorders such as aging, rheumatoid arthritis, diabetes mellitus, cardiovascular and neurodegenerative diseases (Li et al., 2014). As data accumulated from research studying the mode of actions of polyphenols, it soon became clear that their health benefits go beyond antioxidant potential (Scalbert et al., 2005).
Scientific interest in polyphenols has rapidly grown in the late twentieth century following many epidemiological studies linking the consumption of polyphenol-rich food such as fruits and vegetables to lower risk of developing cancer and cardiovascular diseases (Pandey and Rizvi, 2009). Of note, the hypothesis that polyphenols constitute the active ingredients in food was supported by a wealth of in vitro and animal studies using isolated pure phenolic compounds (Gao and Hu, 2010). However, bioavailability studies in animal and human subjects have shown poor absorption and extensive hepatic metabolism of phenolic compounds, thus raising concerns on the applicability of in vitro studies in which parent phenolic compounds were used (Gao and Hu, 2010). In addition, these compounds were tested at concentrations (in μM) much higher than those achieved in vivo (in nM) (Gao and Hu, 2010). Thus, the following question arises: why are polyphenols bioactive despite low plasma concentrations? In order to answer this question, it should be taken into account that the dietary intake of polyphenols is estimated to reach values up to g quantities/day which is equivalent to mM levels of polyphenols and their metabolites in the gut (Grosso et al., 2017). It has thus been proposed that gut microbiota play a crucial rule in polyphenols' metabolism and activities (Williamson, 2013).
This section focuses on polyphenols-microbiota reciprocal interactions and their pivotal role in attenuating metabolic syndrome and type 2 diabetes mellitus.
Polyphenols as Antidiabetic and Anti-Obesity Agents
The antidibetic potential of polyphenols has been extensively studied and documented. In vitro studies show that polyphenols can inhibit the enzymes of dietary carbohydrate and lipid digestion such as α-glucosidase, α-amylase, and pancreatic lipase therefore reducing glucose and fatty acid intestinal absorption (Hanhineva et al., 2010). Several polyphenols such as quercetin, tea catechins, chlorogenic caffeic, and gallic acids were also reported to inhibit glucose absorption in intestinal Caco2 cell line as well as brush-border-membrane vesicles of porcine jejunum via the inhibition of sodium-dependent SGLT1 transporters (Hanhineva et al., 2010). GLUT2 was similarly inhibited by several flavonoids including quercetin, myricetin, neohesperidin, and catechin (Johnston et al., 2005; Kwon et al., 2007).
The insulin-responsive GLUT4, another glucose transporter responsible for glucose uptake in insulin-sensitive tissues, seems to be the target of many polyphenols. Some polyphenols stimulated GLUT4 translocation in adipocytes or skeletal muscle cells by activating either the insulin-mediated phosphatidylinositide 3-kinase (PI3-K)/Akt or the AMP-activated protein kinase (AMPK) pathways (Eid et al., 2015; Hajiaghaalipour et al., 2015). Regulation of AMPK activity can also leads to the activation of a class of protein deacetylases known as sirtuins. Activation of sirtuin 1 (Sirt1) is involved in the antiaging and anti-inflammatory effects of polyphenols such as resveratrol, querectin, catechins, and piceatannol (Chung et al., 2010). Polyphenols may also enhance glucose homeostasis by modulating hepatic glucose metabolism. Flavonoids such as tea catechins, quercetin and citrus flavonoids attenuated hepatic gluconeogensis in diabetic mice and rats through the inhibition of the key gluconeogenic enzymes glucose-6-phosphatase and phosphoenolpyruvate carboxykinase (Bahadoran et al., 2013; Eid and Haddad, 2017). Hepatic glucose kinase and glycogen synthase can also be modulated by polyphenols such as ferulic and hydroxylcinnamic acid derivatives (Bahadoran et al., 2013).
The molecular mechanisms of polyphenols' antidiabetic actions also include stimulation of insulin production and protection of pancreatic β cell against hyperglycemia-induced oxidative stress and promotion of β cell proliferation and survival in both in vitro and in vivo studies (Vinayagam and Xu, 2015).
Because of their unique chemical structures, polyphenols are powerful antioxidants. Green tea had the ability to scavenge 100% of superoxide anion and 86% of other reactive oxygen species (ROS) (Umeno et al., 2016). The radical scavenging properties of polyphenols were studied in vitro and were attributed to either modulation of enzymes responsible for the production of ROS including cyclooxygenase, lipoxygenase, xanthine oxidase, microsomal monooxygenase, and NADH oxidase (Bahadoran et al., 2013) or to the enhancement of the endogenous antioxidative defense system through the modulation of antioxidant enzymes like superoxide dismutase, catalase, and glutathione reducatse (Bahadoran et al., 2013). However, it is not clear if these in vitro results could be reproduced in vivo (Halliwell et al., 2005). In addition, when biomarkers of the antioxidant activities of polyphenols in animal and human subjects, including decreased LDL and DNA oxidation and increased plasma total antioxidant potential, were evaluated using fruit juices, soy or vegetables, it was found that the observed beneficial effect might not be attributed to polyphenols (Halliwell et al., 2005). The same authors proposed that the antioxidant activities of polyphenols could actually occur in the GI tract before absorption. They argued that, at best, polyphenol plasma concentrations do not exceed 1 μM, which are much lower than the concentrations they can reach in the gut, whereby the microbial fermentation products of polyphenols could be responsible for a greater proportion of antioxidant activities (Halliwell et al., 2005).
In addition to their antidiabetic properties, the antiobesity potential of some polyphenols is well documented in cell culture, animal and clinical studies (Wang et al., 2014). Resveratrol as well as green tea extract and its catechins, particularly epigallocatechin gallate (EGCG), were reported to suppress adipocyte differentiation in 3T3-L1 cell line through the activation of AMPK and the down regulation of adipogenic factors including peroxisome proliferator activator receptor γ (PPARγ) and CCAAT/enhancer binding protein α (C/EBPα) (Wang et al., 2014). Animal studies supported the effects of the aforementioned flavonoids on obesity-related inflammation and other obesity parameters such as body weight, total lipids, cholesterol and triglyceride (Khurana et al., 2013). In addition, enhanced fat oxidation in adipose tissue and skeletal muscle was reported in two obesity models: diet-induced obesity and ob/ob mice models treated with flavonoids (Khurana et al., 2013).
Clinical and epidemiological studies further confirmed the beneficial effects of certain polyphenols. A meta-analysis of 24 randomized clinical trials (RCTs) has documented the improvement of insulin resistance after daily consumption of flavonoid-rich cocoa beverages (Shrime et al., 2011; Ellinger and Stehle, 2016). Similarly, ingestion of chocolate, cacao or flavan-3-ol has significantly reduced insulin resistance and major cardiovascular diseases risk (CVD) factors in another meta-analysis of 42 RCTs (Hooper et al., 2008). Nevertheless, some other RCTs reported inconsistent results. For example, consumption of green tea containing 456 mg catechins (Fukino et al., 2005) or isoflavonoids in a 6-month RCT in postmenopausal Chinese women (Liu et al., 2010) did not prove beneficial regarding glucose levels or insulin resistance markers. Epidemiological studies also yielded conflicting results. A meta-analysis of 6 prospective cohort studies involving 284.806 participants has reported a significant inverse association between total polyphenols consumption and risk for type 2 diabetes in the US population (Liu et al., 2014). However, some inconsistent effects were found in other subgroups (Liu et al., 2014). Similar negative results were reported from other cross-sectional studies attempting to relate protection against diabetes to the intake of total flavonols and flavones (Song et al., 2005) or anthocyanins in the Iowa women (Nettleton et al., 2006). Inconsistent findings were also reported from RCTs investigating the antiobesity effects of flavonoids such as EGCG, resveratrol, and curcumin (Wang et al., 2014). Several factors might account for such inconclusive antidiabetic and anti-obesity clinical outcomes. Those include the duration of the study, age, gender, and ethnicity of participants or the varieties of polyphenols present in the diet.
Metabolism of Polyphenols by Gut Microbiota
Initial bioavailability studies have reported poor absorption of flavonoids from food sources (Thilakarathna and Rupasinghe, 2013). Polyphenols are rarely found in nature in the free form. The majority of flavonoids monomer, except catechins, exist as β-glycosides (Marin et al., 2015). Catechins, being in the free form, are rapidly absorbed from the small intestine. On the other hand, the sugar moiety of the flavonoid glycosides determines their site of absorption. Hollman et al. (1995) detected the presence of quercetin glucoside in the circulation after ingestion of onion powder. Transport through SGLT1 was suggested as the mechanism of absorption (Hollman et al., 1995). Other studies suggested that glucosides could also be hydrolyzed by lactase phlorizin hydrolase (LPH), a β-glucosidase present in the intestinal brush border microvilli, to release the free aglycone. Absorbed agylcones mainly undergo hepatic phase II metabolism to form methyl, sulfate, and glucuronide conjugates (Marin et al., 2015). Other glycosides such as rhamnosides, arabinosides, and galactosides as well as some glucosides including cyanidin-3-glucoisde and the isoflavonoid daidzein-7-glucoside are not efficiently hydrolyzed by this enzyme and are moved to the colon where they are hydrolyzed and degraded by the colon microbiota (Marin et al., 2015).
Studies have reported that only 5–10% of dietary polyphenols can be absorbed from the small intestine. The remaining non absorbed (90–95%) polyphenols reach the colon at high concentrations (up to the mM range) where they undergo, together with the conjugates secreted in bile, deconjugation, and degradation by microbial enzymes to smaller phenolic compounds before being absorbed (Stevens and Maier, 2016). Oligomeric flavonoids are first degraded in the stomach to monomers and dimers while larger polymeric compounds such as ellagitannins are taken to the colon to be degraded by resident bacteria to their monomers (Kumar and Pandey, 2013).
Phenolic acids existing in free form, or conjugated to sugars, quinic, or shikimic acids, are absorbed from the small intestine after the hydrolysis of the conjugated forms. Importantly, the most abundant phenolic acids caffeic acid and ferulic acids are found generally bound to cell wall components such as lignins and polysaccharides and are therefore subject to colon microbial metabolism (Russell et al., 2009).
Gut microbes are not only capable of cleaving polyphenols glycosides and glucuronides, but they can also cleave the carbon-carbon bonds of heterocyclic and aromatic rings as well as decarboxylate, dehydoxylate, and hydrogenate alkene side chains (Stevens and Maier, 2016). The site of C-C cleavage seems to depend on the flavonoid subclass: in anthocyanidins, flavonols, flavones, and flavanones, the cleavage occurs in ring-C, while in flavanols, it also takes place in ring-A (Figure 3).
Phenolic acids are the major metabolites of flavonoids detected in circulation and urine (Stevens and Maier, 2016). Ring-C cleavage of anthocyanins produces many phenolic acids such protocatechuic (PCA), gallic, 3-O-methyl-gallic, syringic, p-coumaric, and vanillic acids, as well as 2,4,6-trihydroxybenzaldehyde (Duda-Chodak et al., 2015). Interestingly, many biological activities have been attributed to PCA such as antioxidant, anti-inflammatory, antihyperglycemic, antiatherogenic, hypocholeserolemic, anticancer, and neuroprotective actions (Duda-Chodak et al., 2015). In addition, anthocyanin metabolites, namely gallic acid, 3-O-methyl-gallic acid, and 2,4,6-trihydroxybenzaldehyde, were more effective as cytotoxic compounds than the parent anthocyanin (Forester and Waterhouse, 2010). Similarly, flavonols such as quercetin are metabolized to PCA and 2-(2,4,6-trihyrdoxyphenyl)-acetic acid, which can be further converted to their o-methyl conjugates. Flavanols such as catechin, epicatechin, and ECGC undergo both A-ring and C-ring cleavage to form 3-hydroxyphenylacetic acid, 3-hydroxyphenylpropionic acid, 3,4-dihydroxyphenylacetic acid, and 3-hydroxyphenyl-γ-valerolactone. Demethylation and ring-C fission of the isoflavonoid diadezein generates the major metabolite O-demethyl-langlensin (Stevens and Maier, 2016). Ellagitannins are degraded to ellagic acid, which is further metabolized by the colon microflora to the phenolic metabolites called urolithins. Phenolic acids such as caffeic acid and its ester chlorogenic acid are metabolized by the microflora to phenylpropionic, benzoic and, hippuric acids (Gonthier et al., 2003). Caffeic acid can also undergo decarboxylation and transformation by gut microbiota into 4-ethyl catechol (Ozdal et al., 2016). Lignans such as pinoresinol and syringaresinol are biotransformed to enterolactone and enterodiol, two phtoestrogens responsible for estrogenic agonistic/antagonistic activities of lignans (Ozdal et al., 2016). Finally, microbial communities also possess glucuronidases and sulphatases, thus transforming phase II metabolites into the free form and enabling their absorption into the general circulation (Cardona et al., 2013).
The biological activities of the microbial metabolites of polyphenols are still under investigation. However, because they are efficiently absorbed by the GI tract, they are believed to be responsible for the health promoting activities of polyphenols (Cardona et al., 2013). In a recent study, the two metabolites 3-hydroxyhippuric acid and 3-hydroxyphenyl acetic acid were found to reach and accumulate in the brain of rats treated with grape seed phenol extract in a dose dependent manner (Wang D. et al., 2015).
Modulation of Microbiota by Polyphenols
Because of their distinctive structures, polyphenols have shown a great potential as antimicrobial agents against human pathogens in experimental and clinical studies (Coppo and Marchese, 2014). Their activities against gastrointestinal tract pathogens (Selma et al., 2009) are of particular interest to this review. Commensal and probiotic microorganisms such as strains of Lactobacillus, Streptoccoccus, Bifidobacteria, and the yeast Saccharomyces boulardii are known to preserve epithelial integrity, protect against enteric pathogen, and serve other important function such as modulating sugar and lipid metabolism. The gut can also be inhabited by pathogenic bacteria that cause GI illness such as inflammatory bowel disease caused by Clostridium difficile infection (Bien et al., 2013). Dysbiosis is the term that describes the imbalance between the beneficial and pathogenic bacteria. It has been associated with many chronic diseases including metabolic disorders such as obesity and type 2 diabetes (Carding et al., 2015). Since Firmicutes possess fewer enzymes than Bacteriodetes capable of degrading glycans, products of polyphenol glycoside hydrolysis, it was suggested that the intake of polyphenols could influence the composition of gut microbiota in favor of beneficial bacteria with health promoting functions (Cardona et al., 2013). Thus, it is deemed necessary to investigate the effect of polyphenols on human gut bacterial growth.
The modulation of gut microbiota by polyphenols was firstly studied in vitro. One study examined the effect of some flavonoids aglycones and their glycosides on representative species of human gut microbial communities (Bacteroides galacturonicus, Lactobacillus sp., Enterococcus coccae, Bifidobacterium catenulatum, Ruminococcus gauvreauii, E. coli). The aglycones quercetin, naringenin and hesperetin but not their glycosides rutin, naringin and hesperidin inhibited the growth of certain tested bacteria (Duda-Chodak et al., 2015). In the same study, catechin had no impact on the growth of the aforementioned bacteria (Duda-Chodak et al., 2015). In another in vitro study, the flavonol galangin but not quercetin or fisetin inhibited the growth of B. adolescens (Kawabata et al., 2013). Similarly, the effects of the common dietary polyphenols on the growth and adhesion of commensal and pathogenic bacteria to enterocytes were studied in the Caco2 cell line (Parkar et al., 2008). At their physiological concentration, all polyphenols, except rutin, affected the viability of representative gut flora. However, one of the major limitations of in vitro studies is that 80% of the bacteria in the human gut are uncultured. Interestingly, incubation of anthocyanins with fecal bacteria in stirred, batch-culture fermentation vessels simulating human colon conditions resulted in an increase of the beneficial Bifidobacterium spp. and Lactobacillus-Enterococcus spp. levels (Hidalgo et al., 2012).
More interesting results have emerged from in vivo and clinical studies. Ellagitannin-rich pomegranate extract (POME) and ellagitannins main metabolite urolithin-A were reported to significantly enhance the growth of Bifidobacterium and Lactobacillus spp. in a rat model of colitis. Interestingly, urolithin-A has shown more powerful anti-inflammatory properties than the parent ellagitannin-rich extract (Larrosa et al., 2010). In a clinical study, the consumption of 1,000 mg of POME by 20 healthy volunteers for 4 weeks resulted in beneficial changes in the gut microbiota of participants who were able to metabolize ellagitannins to urolithin-A. These changes include reduction in Firmicutes levels and the induction of the growth of Enterobacter, Lactobacillus, E. coli, and Verrucomicrobia (A. muciphila). The latter two bacterial strains were reported to preserve the intestinal mucus and the glycocalyx layer and to reduce the intestinal permeability frequently associated with development of inflammation and metabolic diseases (Zhang and Zhang, 2013).
Of note, administration of both quercetin and resveratrol (Etxeberria et al., 2015), or resveratrol alone (Qiao et al., 2014), to obese rat fed high fat/high sucrose (HFHS) or high fat diets was reported to reduce F/B ratio (Qiao et al., 2014; Etxeberria et al., 2015). Moreover, it decreased the abundance of bacteria associated with diet-induced obesity, such as Bacillus spp., Eubacterium cylindroide, and Erysipelotrichaceae (Etxeberria et al., 2015) and increased the growth of Bifidobacterium and Lactobacillus (Qiao et al., 2014; Etxeberria et al., 2015).
The highly polymeric apple procyanidins administered for 20 weeks to C57BL/6J mice fed a HFHS diet attenuated weight gain and caused a reduction in inflammation and gut permeability, as well as modulation of lipid metabolizing genes (Masumoto et al., 2016). Similarly, apple peal polyphenols upregulated antioxidant enzymes and attenuated experimental inflammatory bowel disease (Denis et al., 2016). Furthermore, F/B proportion was markedly decreased and Akkermansia levels were increased eight-fold by the end of the treatment (Masumoto et al., 2016). Grape/red wine polyphenols (Roopchand et al., 2015) and procyanidin-rich cranberry extract (Anhe et al., 2015a) showed similar effects on obesity, metabolic syndrome parameters and levels of Akkermansia in mice fed a high-fat diet (Anhe et al., 2015a; Roopchand et al., 2015).
Collectively, these studies reveal that polyphenols act as prebiotic supplements that can positively modulate gut microbiota composition mainly through the enrichment of beneficial bacteria and the inhibition of pathogenic bacterial growth (Anhe et al., 2015b; Le Barz et al., 2015; Fändriks, 2017).
Polysaccharides
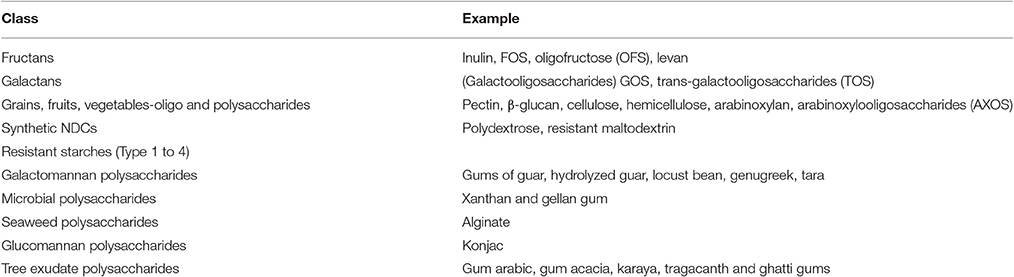
Carbohydrates are classified based on the number of linked carbohydrate units into mono-, oligo-, and poly-saccharides (Cummings and Stephen, 2007). For instance, glucose, fructose, and galactose are monosaccharides whereas lactose, sucrose, and maltose are disaccharides. Common oligosaccharides include fructo-oligosaccharides (FOS), galacto-oligosaccharides (GOS), and mannan oligosaccharides (MOS). Arabinoxylans, cellulose, chitin, and pectins are examples of polysaccharides having linear or branched structures (Cummings and Stephen, 2007).
Another classification is based on whether carbohydrates are digestible and non-digestible. Starch, dextrin, glycogen are some of the digestible polysaccharides, while non-digestible carbohydrates (NDCs) (Paeschke and Aimutis, 2010) are listed in Table 7 (Kaushik et al., 1989).
Gut Microbiota and Dietary Intake
Some of the gut's microbial species are infulenced by the composition of the diet (Kok et al., 1996; Wu et al., 2011; Pyra et al., 2012). Firmicutes and Proteobacteria (Prevotella enterotype) thrive under diets that are rich in carbohydrates and simple sugars (such as glucose and fructose), while Bacteroidetes and Actinobacteria (Bacteroides enterotype) are favored by diets rich in saturated fat and animal protein. Fat and protein decrease the microbial diversity of the intestine, whereas carbohydrates increase it (Wu et al., 2011). Oligo- and poly-saccharides enhance the concentration of the species belonging to Actinobacteria and Firmicutes phyla except for Staphylococcus (Staphylococcus aureus), Clostridium (C. difficile, Clostridium leptum, Clostridium perfringens). Proteobacteria phyla show the opposite trend, whereas Bacteroidetes and Verrucomicrobia show a mixed trend (Wu et al., 2011).
Actions of Oligo- and Poly-saccharides
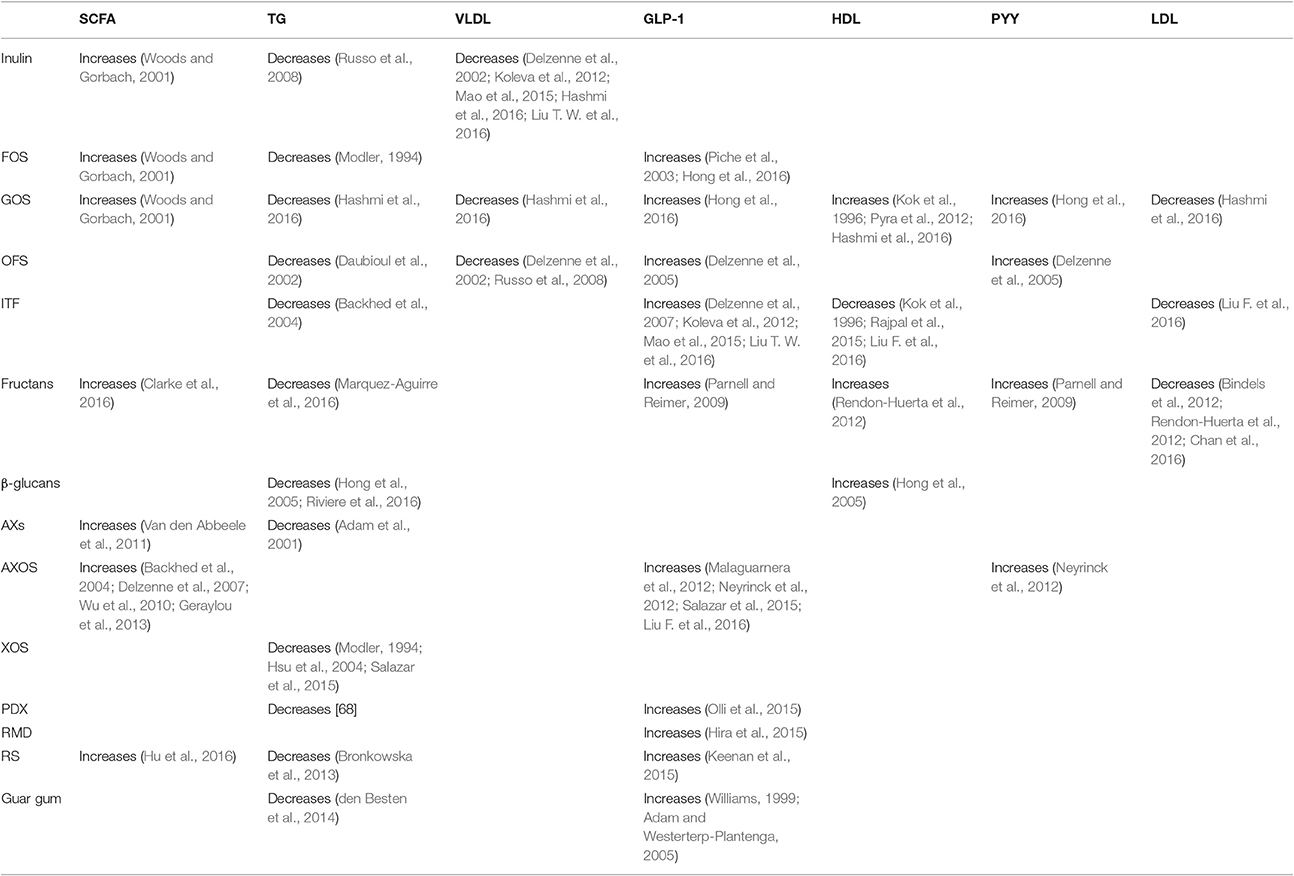
Oligo- and poly-saccharides exert a significant influence on the gut microbiota. In general, oligo- and poly-saccharides increase short-chain fatty acids (SCFA), GLP-1, and PYY, while decreasing triglycerides, VLDL, and LDL, with a mixed trend being observed for HDL (Morrison and Preston, 2016), as shown in Table 8. These changes and their metabolic consequences will be summarized below.
Firstly, the fermentation of non-digestible carbohydrates takes place in the gut and produces SCFAs, with acetate, propionate, and butyrate (Morrison and Preston, 2016) present in a ratio of 3:1:1 (Hoverstad et al., 1984). More specifically, A. municiphilla produce propionates (Derrien et al., 2004) while Ruminococcus bromii (Ze et al., 2012) F. prausnitzii, Eubacterium rectale, Eubacterium hallii, and Ruminococcus bromii, produce butyrate (Louis et al., 2010). Propionates and butyrates lower lipogenesis and serum cholesterol level while activating intestinal gluconeogenesis (IGN) which contributes to glucose homeostasis (Hosseini et al., 2011).
SCFA are ligands of the free fatty acid receptor (FFAR) 2 and 3, also known as G-protein coupled receptor (GPR) 43 and 41, respectively. FFAR2 is activated by acetate and propionate, while FFAR3 is activated by propionate and butyrate (Kaji et al., 2014). Propionate is involved in gluconeogenic activity while acetate and butyrate contribute to lipogenic activity (den Besten et al., 2013a). These three gut bacterial products (acetate, propionate, and butyrate) thus regulate gluconeogenesis and lipogenesis, thereby modulating hepatic lipid and glucose homeostasis, notably through PPAR-γ (den Besten et al., 2015). SCFAs produced from dietary fibers can also increase LPS (Blaut and Klaus, 2012), the main component of gram-negative bacteria (Delzenne and Cani, 2011). Plasma LPS participates in the development of insulin resistance, as discussed by Carvalho et al. (2012). In contrast to SCFAs, high concentrations of omega-3 fatty acids reduce LPS (Kaliannan et al., 2015), as do Gram-positive bacteria (Bifidobacterium and Lactobacillus) (Zhang et al., 2010).
On the other hand, the activity of gut microbiota influences triglycerides, which represent stored glycerol-linked fatty acids (den Besten et al., 2013b). Different concentrations of cholesterol, lipoproteins, and triglycerides determine the classification of circulating lipids into chylomicrons, very low-density lipoproteins (VLDL), intermediate-density lipoproteins (IDL), low-density lipoproteins (LDL), and high-density lipoproteins (HDL) (Cox and Garcia-Palmieri, 1990). It is well-established that diets rich in fructose can increase triglycerides (Schaefer et al., 2009). High triglyceride levels, in turn, bring out undesirable effects on lipid metabolism (Reiser, 1985) and, as detailed above, are one of the hallmarks of the metabolic syndrome. In contrast, decreased triglyceride levels reduce fatty acid synthase activity (Morand et al., 1994) and provide beneficial health effects. Several polysaccharides such as inulin, FOS, GOS, OFS, ITF, fructans, β-glucans, arabinoxylans, xylo-oligosaccharides, polydextrose, resistant starch, and guar gum decrease triglyceride levels (de Deckere et al., 1993).
Dietary carbohydrates also modulate important incretin hormones, namely glucose-dependent insulinotropic peptide (GIP) and glucagon-like peptide-1 (GLP-1) (Edholm et al., 2010). Endocrine K-cells secrete GIP in response to carbohydrates and fat ingestion, (Andersen et al., 1978) whereas, GLP-1 is secreted by L cells (Kieffer and Habener, 1999) in response to luminal sugars, amino acids and fatty acids (Holst, 2007). GLP-1 stimulates the release of insulin, suppresses the release of glucagon (Nadkarni et al., 2014), lowers the blood glucose (Nauck et al., 1993), and plays a major role in glucose homeostasis (Kieffer and Habener, 1999; Deacon, 2004). GIP exerts similar effects (Yabe and Seino, 2011). Oligo- and poly-saccharides such as fructo-oligosaccharides (FOS), Glucooligosaccharides (GOS), oligofructoses (OFS), inulin-type fructans (ITF), fructans, Arabinoxylooligosaccharides (AXOS), resistant maltodextrin, resistant starch, and guar gum increase GLP-1 (Wang X. et al., 2015).
Another important peptide hormone modulated by dietary carbohydrates is peptide YY (PYY), which is secreted by α-cells of the pancreatic islets in response to the food that is ingested (Shi et al., 2015). External administration of SCFAs releases PYY and GLP-1 into plasma (Freeland and Wolever, 2010), while generated SCFAs can increase PYY concentration only (Cherbut et al., 1998). When PYY concentration increases, serum insulin level also increases, which improves glucose tolerance (Shi et al., 2015) leading to GH regulation (Boey et al., 2007). Therefore, the higher level of GLP-1, GIP, and PYY is desirable.
Gut microbiota modulates the brown adipose tissues (BATs) (Mestdagh et al., 2012). When stimulated, FFA are used-up to clear the TGs (Bartelt et al., 2011) in turn regulating glucose homeostasis (Stanford et al., 2013) (Peirce and Vidal-Puig, 2013).
Sodium-glucose transport protein-1 (SGLT-1) and glucose transporter-2 (GLUT-2) also participate in maintaining GH (Kellett and Helliwell, 2000).
Summary
The healthy human gut represents a complex and highly variable ecological system consisting of several microbes belonging to bacteria, fungi and virus domains, in addition to host epithelial cells. Firmicutes, Bacteroidetes, Proteobacteria, Actinobacteria, and Fusobacteria are the major phyla colonizing the stomach and intestine of healthy adults. The exact role of gut microbiota is not fully elucidated but many studies implicate microbiota to perform tasks that are known to be useful for the human host such as modulating intermediate metabolism and the immune system. Diet-induced changes in the composition/diversity of gut microbes are thus believed to participate in the pathogenesis of certain diseases through modifying different metabolic processes in the host.
Less diverse intestinal microbiota have been reported in metabolic disorders. While the association between increased Firmicutes/Bacteroidetes ratio and metabolic diseases is still controversial, more recent studies associated Akkermansia and Lactobacillus species with central obesity and fasting hyperglycemia.
Polyphenols, oligo-, and poly-saccharides can influence the composition of gut microbiota by favoring beneficial bacteria and inhibiting growth and activity of pathogenic species and thus constitute a promising avenue for the prevention and treatment of metabolic disorders.
Future Challenges and Opportunities
Given all the above considerations, the perspectives for targeting the gut microbiome in the context of metabolic diseases keeps being highly relevant and timely. First and foremost, there remains a need to refine research on specific microbes that may be more specifically involved in metabolic diseases rather than considering broader categories, such as phyla. Among challenges that should be met, more studies should focus on the roles and potential mechanisms of action of non-bacterial gut microbes, since these remain poorly understood. Continued research efforts should also result in the better understanding of the modes of action of pre- and pro-biotics in metabolic diseases, notably in terms of metabolic and inflammatory mediators.
There also remains a lot to be done to further elucidate the intricate interactions between prebiotics (polyphenols and fibers) on the one hand, and probiotics (gut microbes), on the other, notably in what pertains to the metabolism of prebiotics by the latter and the influence this has on the bioactivity of the former. In this context, experimental approaches and tools have now evolved that can meet this challenge. For instance, one could think of combining bacterial metagenomics with plant metabolomics and hence study relationships through the use of powerful bioinformatics.
Overall, and in a very pragmatic sense, academics and industrial partners will need to work together to develop safe and reliable products that can help prevent and mitigate the ill effects of metabolic syndrome and related obesity and diabetes. In this context, a promising approach may be to further explore symbiotic products that can combine pre- and pro-biotics in novel and efficient ways.
Author Contributions
HE, MW, NA, AQ, AA, and PH have written the first draft of the manuscript. SH, AM, SN, and LR revised and improved the first draft. All authors have seen and agreed on the final submitted version of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
AA acknowledges the support by the Polish KNOW (Leading National Research Centre) Scientific Consortium “Healthy Animal—Safe Food,” decision of Ministry of Science and Higher Education No. 05-1/KNOW2/2015, and by the Austrian Science Fund (FWF): P25971–B23.
References
Abeles, S. R., and Pride, D. T. (2014). Molecular bases and role of viruses in the human microbiome. J. Mol. Biol. 426, 3892–3906. doi: 10.1016/j.jmb.2014.07.002
Adam, A., Levrat-Verny, M. A., Lopez, H. W., Leuillet, M., Demigne, C., and Remesy, C. (2001). Whole wheat and triticale flours with differing viscosities stimulate cecal fermentations and lower plasma and hepatic lipids in rats. J. Nutr. 131, 1770–1776.
Adam, T. C., and Westerterp-Plantenga, M. S. (2005). Nutrient-stimulated GLP-1 release in normal-weight men and women. Horm. Metab. Res. 37, 111–117. doi: 10.1055/s-2005-861160
Afsar, B., and Elsurer, R. (2014). The relationship between central hemodynamics, morning blood pressure surge, glycemic control and sodium intake in patients with type 2 diabetes and essential hypertension. Diabetes Res. Clin. Pract. 104, 420–426. doi: 10.1016/j.diabres.2014.03.011
Alberti, K. G., Eckel, R. H., Grundy, S. M., Zimmet, P. Z., Cleeman, J. I., Donato, K. A., et al. (2009). Harmonizing the metabolic syndrome. Circulation 120:1640. doi: 10.1161/CIRCULATIONAHA.109.192644
Andersen, D. K., Elahi, D., Brown, J. C., Tobin, J. D., and Andres, R. (1978). Oral glucose augmentation of insulin secretion. Interactions of gastric inhibitory polypeptide with ambient glucose and insulin levels. J. Clin. Invest. 62, 152–161. doi: 10.1172/JCI109100
Anhe, F. F., Roy, D., Pilon, G., Dudonne, S., Matamoros, S., Varin, T. V., et al. (2015a). A polyphenol-rich cranberry extract protects from diet-induced obesity, insulin resistance and intestinal inflammation in association with increased Akkermansia spp. population in the gut microbiota of mice. Gut 64, 872–883. doi: 10.1136/gutjnl-2014-307142
Anhe, F. F., Varin, T. V., Le Barz, M., Desjardins, Y., Levy, E., Roy, D., et al. (2015b). Gut microbiota dysbiosis in obesity-linked metabolic diseases and prebiotic potential of polyphenol-rich extracts. Curr. Obes. Rep. 4, 389–400. doi: 10.1007/s13679-015-0172-9
Backhed, F., Ding, H., Wang, T., Hooper, L. V., Koh, G. Y., Nagy, A., et al. (2004). The gut microbiota as an environmental factor that regulates fat storage. Proc. Natl. Acad. Sci. U.S.A. 101, 15718–15723. doi: 10.1073/pnas.0407076101
Bäckhed, F., Roswall, J., Peng, Y., Feng, Q., Jia, H., Kovatcheva-Datchary, P., et al. (2015). Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe 17, 690–703. doi: 10.1016/j.chom.2015.04.004
Bahadoran, Z., Mirmiran, P., and Azizi, F. (2013). Dietary polyphenols as potential nutraceuticals in management of diabetes: a review. J. Diabetes Metab. Disord. 12:43. doi: 10.1186/2251-6581-12-43
Baothman, O. A., Zamzami, M. A., Taher, I., Abubaker, J., and Abu-Farha, M. (2016). The role of Gut Microbiota in the development of obesity and Diabetes. Lipids Health Dis. 15:108. doi: 10.1186/s12944-016-0278-4
Baraldes, M. A., Domingo, P., Barrio, J. L., Pericas, R., Gurgui, M., and Vazquez, G. (2000). Meningitis due to Neisseria subflava: case report and review. Clin. Infect. Dis. 30, 615–617. doi: 10.1086/313700
Baron, E. J., Summanen, P., Downes, J., Roberts, M. C., Wexler, H., and Finegold, S. M. (1989). Bilophila wadsworthia, gen. nov. and sp. nov., a unique gram-negative anaerobic rod recovered from appendicitis specimens and human faeces. J. Gen. Microbiol. 135, 3405–3411. doi: 10.1099/00221287-135-12-3405
Bartelt, A., Bruns, O. T., Reimer, R., Hohenberg, H., Ittrich, H., Peldschus, K., et al. (2011). Brown adipose tissue activity controls triglyceride clearance. Nat. Med. 17, 200–205. doi: 10.1038/nm.2297
Beaumont, M., Andriamihaja, M., Lan, A., Khodorova, N., Audebert, M., Blouin, J. M., et al. (2016). Detrimental effects for colonocytes of an increased exposure to luminal hydrogen sulfide: the adaptive response. Free Radic. Biol. Med. 93, 155–164. doi: 10.1016/j.freeradbiomed.2016.01.028
Belizário, J. E., and Napolitano, M. (2015). Human microbiomes and their roles in dysbiosis, common diseases, and novel therapeutic approaches. Front. Microbiol. 6:1050. doi: 10.3389/fmicb.2015.01050
Belkaid, Y., and Hand, T. W. (2014). Role of the microbiota in immunity and inflammation. Cell 157, 121–141. doi: 10.1016/j.cell.2014.03.011
Belzer, C., and de Vos, W. M. (2012). Microbes inside–from diversity to function: the case of Akkermansia. ISME J. 6, 1449–1458. doi: 10.1038/ismej.2012.6
Berding, K., and Donovan, S. M. (2016). Microbiome and nutrition in autism spectrum disorder: current knowledge and research needs. Nutr. Rev. 74, 723–736. doi: 10.1093/nutrit/nuw048
Bien, J., Palagani, V., and Bozko, P. (2013). The intestinal microbiota dysbiosis and Clostridium difficile infection: is there a relationship with inflammatory bowel disease? Therap. Adv. Gastroenterol. 6, 53–68. doi: 10.1177/1756283X12454590
Bik, E. M., Eckburg, P. B., Gill, S. R., Nelson, K. E., Purdom, E. A., Francois, F., et al. (2006). Molecular analysis of the bacterial microbiota in the human stomach. Proc. Natl. Acad. Sci. U.S.A. 103, 732–737. doi: 10.1073/pnas.0506655103
Bindels, L. B., Porporato, P., Dewulf, E. M., Verrax, J., Neyrinck, A. M., Martin, J. C., et al. (2012). Gut microbiota-derived propionate reduces cancer cell proliferation in the liver. Br. J. Cancer 107, 1337–1344. doi: 10.1038/bjc.2012.409
Blaut, M., and Klaus, S. (2012). Intestinal microbiota and obesity. Handb. Exp. Pharmacol. 209, 251–273. doi: 10.1007/978-3-642-24716-3_11
Boey, D., Sainsbury, A., and Herzog, H. (2007). The role of peptide YY in regulating glucose homeostasis. Peptides 28, 390–395. doi: 10.1016/j.peptides.2006.07.031
Bratoeva, M. P., Wolf, M. K., Marks, J. K., and Cantey, J. R. (1994). A case of diarrhea, bacteremia, and fever caused by a novel strain of Escherichia coli. J. Clin. Microbiol. 32, 1383–1386.
Bronkowska, M., Orzel, D., Lozna, K., Styczynska, M., Biernat, J., Gryszkin, A., et al. (2013). Effect of resistant starch RS4 added to the high-fat diets on selected biochemical parameters in Wistar rats. Rocz. Panstw. Zakl. Hig. 64, 19–24.
Cani, P. D., Amar, J., Iglesias, M. A., Poggi, M., Knauf, C., Bastelica, D., et al. (2007). Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes 56, 1761–1772. doi: 10.2337/db06-1491
Cani, P. D., Possemiers, S., Van de Wiele, T., Guiot, Y., Everard, A., Rottier, O., et al. (2009). Changes in gut microbiota control inflammation in obese mice through a mechanism involving GLP-2-driven improvement of gut permeability. Gut 58, 1091–1103. doi: 10.1136/gut.2008.165886
Caputo, A., Lagier, J. C., Azza, S., Robert, C., Mouelhi, D., Fournier, P. E., et al. (2016). Microvirga massiliensis sp. nov., the human commensal with the largest genome. Microbiol.Open 5, 307–322. doi: 10.1002/mbo3.329
Carding, S., Verbeke, K., Vipond, D. T., Corfe, B. M., and Owen, L. J. (2015). Dysbiosis of the gut microbiota in disease. Microb. Ecol. Health Dis. 26:26191. doi: 10.3402/mehd.v26.26191
Cardona, F., Andres-Lacueva, C., Tulipani, S., Tinahones, F. J., and Queipo-Ortuno, M. I. (2013). Benefits of polyphenols on gut microbiota and implications in human health. J. Nutr. Biochem. 24, 1415–1422. doi: 10.1016/j.jnutbio.2013.05.001
Carmody, R. N., Gerber, G. K., Luevano, J. M. Jr., Gatti, D. M., Somes, L., Svenson, K. L., et al. (2015). Diet dominates host genotype in shaping the murine gut microbiota. Cell Host Microbe 17, 72–84. doi: 10.1016/j.chom.2014.11.010
Carvalho, B. M., Guadagnini, D., Tsukumo, D. M., Schenka, A. A., Latuf-Filho, P., Vassallo, J., et al. (2012). Modulation of gut microbiota by antibiotics improves insulin signalling in high-fat fed mice. Diabetologia 55, 2823–2834. doi: 10.1007/s00125-012-2648-4
Cavicchioli, R. (2011). Archaea–timeline of the third domain. Nat. Rev. Microbiol. 9, 51–61. doi: 10.1038/nrmicro2482
Chan, C., Hyslop, C. M., Shrivastava, V., Ochoa, A., Reimer, R. A., and Huang, C. (2016). Oligofructose as an adjunct in treatment of diabetes in NOD mice. Sci. Rep. 6:37627. doi: 10.1038/srep37627
Cherbut, C., Ferrier, L., Roze, C., Anini, Y., Blottiere, H., Lecannu, G., et al. (1998). Short-chain fatty acids modify colonic motility through nerves and polypeptide YY release in the rat. Am. J. Physiol. 275(6 Pt 1), G1415–G1422.
Chung, S., Yao, H., Caito, S., Hwang, J.-W., Arunachalam, G., and Rahman, I. (2010). Regulation of SIRT1 in cellular functions: role of polyphenols. Arch. Biochem. Biophys. 501, 79–90. doi: 10.1016/j.abb.2010.05.003
Clarke, S. T., Green-Johnson, J. M., Brooks, S. P., Ramdath, D. D., Bercik, P., Avila, C., et al. (2016). beta2-1 Fructan supplementation alters host immune responses in a manner consistent with increased exposure to microbial components: results from a double-blinded, randomised, cross-over study in healthy adults. Br. J. Nutr. 115, 1748–1759. doi: 10.1017/S0007114516000908
Coppo, E., and Marchese, A. (2014). Antibacterial activity of polyphenols. Curr. Pharm. Biotechnol. 15, 380–390. doi: 10.2174/138920101504140825121142
Corless, C. E., Guiver, M., Borrow, R., Edwards-Jones, V., Fox, A. J., and Kaczmarski, E. B. (2001). Simultaneous detection of Neisseria meningitidis, Haemophilus influenzae, and Streptococcus pneumoniae in suspected cases of meningitis and septicemia using real-time PCR. J. Clin. Microbiol. 39, 1553–1558. doi: 10.1128/JCM.39.4.1553-1558.2001
Cox, R. A., and Garcia-Palmieri, M. R. (1990). “Cholesterol, triglycerides, and associated lipoproteins,” in Clinical Methods: The History, Physical, and Laboratory Examinations, 3rd Edn., eds H. K. Walker, W. D. Hall, and J. W. Hurst (Boston, MA: Butterworths), 153–160.
Cummings, J. H., and Stephen, A. M. (2007). Carbohydrate terminology and classification. Eur. J. Clin. Nutr. 61(Suppl. 1), S5–S18. doi: 10.1038/sj.ejcn.1602936
D'Aimmo, M. R., Mattarelli, P., Biavati, B., Carlsson, N. G., and Andlid, T. (2012). The potential of bifidobacteria as a source of natural folate. J. Appl. Microbiol. 112, 975–984. doi: 10.1111/j.1365-2672.2012.05261.x
Dalton, H. W. (1951). Implantation of B. coli into the human intestine. Ir. J. Med. Sci. 308, 384–386. doi: 10.1007/BF02956866
Daubioul, C., Rousseau, N., Demeure, R., Gallez, B., Taper, H., Declerck, B., et al. (2002). Dietary fructans, but not cellulose, decrease triglyceride accumulation in the liver of obese Zucker fa/fa rats. J. Nutr. 132, 967–973.
Davis, C. P. (1996). “Normal flora,” in Medical Microbiology, 4th Edn., ed S. Baron (Galveston, TX: University of Texas Medical Branch). Available online at: https://www.ncbi.nlm.nih.gov/books/NBK7617/
De Angelis, M., Piccolo, M., Vannini, L., Siragusa, S., De Giacomo, A., Serrazzanetti, D. I., et al. (2013). Fecal microbiota and metabolome of children with autism and pervasive developmental disorder not otherwise specified. PLoS ONE 8:e76993. doi: 10.1371/journal.pone.0076993
de Deckere, E. A., Kloots, W. J., and van Amelsvoort, J. M. (1993). Resistant starch decreases serum total cholesterol and triacylglycerol concentrations in rats. J. Nutr. 123, 2142–2151.
Deacon, C. F. (2004). Therapeutic strategies based on glucagon-like peptide 1. Diabetes 53, 2181–2189. doi: 10.2337/diabetes.53.9.2181
Delgado, S., Cabrera-Rubio, R., Mira, A., Suarez, A., and Mayo, B. (2013). Microbiological survey of the human gastric ecosystem using culturing and pyrosequencing methods. Microb. Ecol. 65, 763–772. doi: 10.1007/s00248-013-0192-5
Delzenne, N. M., and Cani, P. D. (2011). Gut microbiota and the pathogenesis of insulin resistance. Curr. Diabetes Rep. 11, 154–159. doi: 10.1007/s11892-011-0191-1
Delzenne, N. M., Cani, P. D., Daubioul, C., and Neyrinck, A. M. (2005). Impact of inulin and oligofructose on gastrointestinal peptides. Br. J. Nutr. 93(Suppl. 1), S157–S161. doi: 10.1079/bjn20041342
Delzenne, N. M., Cani, P. D., Daubioul, C., and Neyrinck, A. M. (2007). Impact of inulin and oligofructose on gastrointestinal peptides. Br. J. Nutr. 93:S157. doi: 10.1079/BJN20041342
Delzenne, N. M., Daubioul, C., Neyrinck, A., Lasa, M., and Taper, H. S. (2002). Inulin and oligofructose modulate lipid metabolism in animals: review of biochemical events and future prospects. Br. J. Nutr. 87(Suppl. 2), S255–S259. doi: 10.1079/BJN/2002545
den Besten, G., Bleeker, A., Gerding, A., van Eunen, K., Havinga, R., van Dijk, T. H., et al. (2015). Short-chain fatty acids protect against high-fat diet-induced obesity via a PPARgamma-dependent switch from lipogenesis to fat oxidation. Diabetes 64, 2398–2408. doi: 10.2337/db14-1213
den Besten, G., Havinga, R., Bleeker, A., Rao, S., Gerding, A., van Eunen, K., et al. (2014). The short-chain fatty acid uptake fluxes by mice on a guar gum supplemented diet associate with amelioration of major biomarkers of the metabolic syndrome. PLoS ONE 9:e107392. doi: 10.1371/journal.pone.0107392
den Besten, G., Lange, K., Havinga, R., van Dijk, T. H., Gerding, A., van Eunen, K., et al. (2013a). Gut-derived short-chain fatty acids are vividly assimilated into host carbohydrates and lipids. Am. J. Physiol. Gastrointest. Liver Physiol. 305, G900–G910. doi: 10.1152/ajpgi.00265.2013
den Besten, G., van Eunen, K., Groen, A. K., Venema, K., Reijngoud, D.-J., and Bakker, B. M. (2013b). The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J. Lipid Res. 54, 2325–2340. doi: 10.1194/jlr.R036012
Denis, M. C., Roy, D., Yeganeh, P. R., Desjardins, Y., Varin, T., Haddad, N., et al. (2016). Apple peel polyphenols: a key player in the prevention and treatment of experimental inflammatory bowel disease. Clin. Sci. 130, 2217–2237. doi: 10.1042/CS20160524
Derrien, M., Vaughan, E. E., Plugge, C. M., and de Vos, W. M. (2004). Akkermansia muciniphila gen. nov., sp. nov., a human intestinal mucin-degrading bacterium. Int. J. Syst. Evol. Microbiol. 54(Pt 5), 1469–1476. doi: 10.1099/ijs.0.02873-0
Duda-Chodak, A., Tarko, T., Satora, P., and Sroka, P. (2015). Interaction of dietary compounds, especially polyphenols, with the intestinal microbiota: a review. Eur. J. Nutr. 54, 325–341. doi: 10.1007/s00394-015-0852-y
Duncan, S. H., Hold, G. L., Harmsen, H. J., Stewart, C. S., and Flint, H. J. (2002). Growth requirements and fermentation products of Fusobacterium prausnitzii, and a proposal to reclassify it as Faecalibacterium prausnitzii gen. nov., comb. nov. Int. J. Syst. Evol. Microbiol. 52(Pt 6), 2141–2146. doi: 10.1099/00207713-52-6-2141
Duranti, S., Milani, C., Lugli, G. A., Mancabelli, L., Turroni, F., Ferrario, C., et al. (2016). Evaluation of genetic diversity among strains of the human gut commensal Bifidobacterium adolescentis. Sci. Rep. 6:23971. doi: 10.1038/srep23971
Edholm, T., Degerblad, M., Gryback, P., Hilsted, L., Holst, J. J., Jacobsson, H., et al. (2010). Differential incretin effects of GIP and GLP-1 on gastric emptying, appetite, and insulin-glucose homeostasis. Neurogastroenterol. Motil. 22, 1191–1200. doi: 10.1111/j.1365-2982.2010.01554.x
Eggerth, A. H. (1935). The gram-positive non-spore-bearing anaerobic bacilli of human feces. J. Bacteriol. 30, 277–299.
Eggerth, A. H., and Gagnon, B. H. (1933). The bacteroides of human feces. J. Bacteriol. 25, 389–413.
Eid, H. M., and Haddad, P. S. (2017). The antidiabetic potential of quercetin: underlying mechanisms. Curr. Med. Chem. 24, 355–364. doi: 10.2174/0929867323666160909153707
Eid, H. M., Nachar, A., Thong, F., Sweeney, G., and Haddad, P. S. (2015). The molecular basis of the antidiabetic action of quercetin in cultured skeletal muscle cells and hepatocytes. Pharmacogn. Mag. 11, 74–81. doi: 10.4103/0973-1296.149708
Ellinger, S., and Stehle, P. (2016). Impact of cocoa consumption on inflammation processes—a critical review of randomized controlled trials. Nutrients 8:321. doi: 10.3390/nu8060321
Engstrand, L., and Lindberg, M. (2013). Helicobacter pylori and the gastric microbiota. Best Pract. Res. Clin. Gastroenterol. 27, 39–45. doi: 10.1016/j.bpg.2013.03.016
Etxeberria, U., Arias, N., Boque, N., Macarulla, M. T., Portillo, M. P., Martinez, J. A., et al. (2015). Reshaping faecal gut microbiota composition by the intake of trans-resveratrol and quercetin in high-fat sucrose diet-fed rats. J. Nutr. Biochem. 26, 651–660. doi: 10.1016/j.jnutbio.2015.01.002
Evans, D. F., Pye, G., Bramley, R., Clark, A. G., Dyson, T. J., and Hardcastle, J. D. (1988). Measurement of gastrointestinal pH profiles in normal ambulant human subjects. Gut 29, 1035–1041. doi: 10.1136/gut.29.8.1035
Everard, A., Belzer, C., Geurts, L., Ouwerkerk, J. P., Druart, C., Bindels, L. B., et al. (2013). Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc. Natl. Acad. Sci. U.S.A. 110, 9066–9071. doi: 10.1073/pnas.1219451110
Fagundes Neto, U., Patricio, F. R., Wehba, J., Reis, M. H., Gianotti, O. F., and Trabulsi, L. R. (1979). An Escherichia coli strain that causes diarrhea by invasion of the small intestinal mucosa and induces monosaccharide intolerance. Arq. Gastroenterol. 16, 205–208.
Falcone, E. L., Abusleme, L., Swamydas, M., Lionakis, M. S., Ding, L., Hsu, A. P., et al. (2016). Colitis susceptibility in p47(phox−/−) mice is mediated by the microbiome. Microbiome 4, 13. doi: 10.1186/s40168-016-0159-0
Fändriks, L. (2017). Roles of the gut in the metabolic syndrome: an overview. J. Intern. Med. 281, 319–336. doi: 10.1111/joim.12584
Finegold, S. M., Attebery, H. R., and Sutter, V. L. (1974). Effect of diet on human fecal flora: comparison of Japanese and American diets. Am. J. Clin. Nutr. 27, 1456–1469.
Forester, S. C., and Waterhouse, A. L. (2010). Gut metabolites of anthocyanins, gallic acid, 3-O-methylgallic acid, and 2,4,6-trihydroxybenzaldehyde, inhibit cell proliferation of Caco-2 cells. J. Agric. Food Chem. 58, 5320–5327. doi: 10.1021/jf9040172
Freeland, K. R., and Wolever, T. M. (2010). Acute effects of intravenous and rectal acetate on glucagon-like peptide-1, peptide YY, ghrelin, adiponectin and tumour necrosis factor-alpha. Br. J. Nutr. 103, 460–466. doi: 10.1017/S0007114509991863
Fu, J., Bonder, M. J., Cenit, M. C., Tigchelaar, E. F., Maatman, A., Dekens, J. A., et al. (2015). The gut microbiome contributes to a substantial proportion of the variation in blood lipids. Circ. Res. 117, 817–824. doi: 10.1161/CIRCRESAHA.115.306807
Fukino, Y., Shimbo, M., Aoki, N., Okubo, T., and Iso, H. (2005). Randomized controlled trial for an effect of green tea consumption on insulin resistance and inflammation markers. J. Nutr. Sci. Vitaminol. 51, 335–342. doi: 10.3177/jnsv.51.335
Gaci, N., Borrel, G., Tottey, W., O'Toole, P. W., and Brugere, J. F. (2014). Archaea and the human gut: new beginning of an old story. World J. Gastroenterol. 20, 16062–16078. doi: 10.3748/wjg.v20.i43.16062
Ganesh, B. P., Klopfleisch, R., Loh, G., and Blaut, M. (2013). Commensal Akkermansia muciniphila exacerbates gut inflammation in Salmonella Typhimurium-infected gnotobiotic mice. PLoS ONE 8:e74963. doi: 10.1371/journal.pone.0074963
Gao, S., and Hu, M. (2010). Bioavailability challenges associated with development of anti-cancer phenolics. Mini Rev. Med. Chem. 10, 550–567. doi: 10.2174/138955710791384081
Geraylou, Z., Souffreau, C., Rurangwa, E., Maes, G. E., Spanier, K. I., Courtin, C. M., et al. (2013). Prebiotic effects of arabinoxylan oligosaccharides on juvenile Siberian sturgeon (Acipenser baerii) with emphasis on the modulation of the gut microbiota using 454 pyrosequencing. FEMS Microbiol. Ecol. 86, 357–371. doi: 10.1111/1574-6941.12169
Gerritsen, J., Smidt, H., Rijkers, G. T., and de Vos, W. M. (2011). Intestinal microbiota in human health and disease: the impact of probiotics. Genes Nutr. 6, 209–240. doi: 10.1007/s12263-011-0229-7
Ghouri, Y. A., Richards, D. M., Rahimi, E. F., Krill, J. T., Jelinek, K. A., and DuPont, A. W. (2014). Systematic review of randomized controlled trials of probiotics, prebiotics, and synbiotics in inflammatory bowel disease. Clin. Exp. Gastroenterol. 7, 473–487. doi: 10.2147/CEG.S27530
Gondalia, S. V., Palombo, E. A., Knowles, S. R., Cox, S. B., Meyer, D., and Austin, D. W. (2012). Molecular characterisation of gastrointestinal microbiota of children with autism (with and without gastrointestinal dysfunction) and their neurotypical siblings. Autism Res. 5, 419–427. doi: 10.1002/aur.1253
Gonthier, M. P., Verny, M. A., Besson, C., Remesy, C., and Scalbert, A. (2003). Chlorogenic acid bioavailability largely depends on its metabolism by the gut microflora in rats. J. Nutr. 133, 1853–1859.
Goodrich, J. K., Waters, J. L., Poole, A. C., Sutter, J. L., Koren, O., Blekhman, R., et al. (2014). Human genetics shape the gut microbiome. Cell 159, 789–799. doi: 10.1016/j.cell.2014.09.053
Grosso, G., Stepaniak, U., Micek, A., Stefler, D., Bobak, M., and Pająk, A. (2017). Dietary polyphenols are inversely associated with metabolic syndrome in Polish adults of the HAPIEE study. Eur. J. Nutr. 56, 1409–1420. doi: 10.1007/s00394-016-1187-z
Grundy, S. M., Brewer, H. B., Cleeman, J. I., Smith, S. C., and Lenfant, C. (2004). Definition of metabolic syndrome. Circulation 109:433. doi: 10.1161/01.CIR.0000111245.75752.C6
Guarner, F., and Malagelada, J. R. (2003). Gut flora in health and disease. Lancet 361, 512–519. doi: 10.1016/S0140-6736(03)12489-0
Hajiaghaalipour, F., Khalilpourfarshbafi, M., and Arya, A. (2015). Modulation of glucose transporter protein by dietary flavonoids in type 2 diabetes mellitus. Int. J. Biol. Sci. 11, 508–524. doi: 10.7150/ijbs.11241
Halliwell, B., Rafter, J., and Jenner, A. (2005). Health promotion by flavonoids, tocopherols, tocotrienols, and other phenols: direct or indirect effects? Antioxidant or not? Am. J. Clin. Nutr. 81(1 Suppl.), 268S–276S.
Hamilton, M. K., Boudry, G., Lemay, D. G., and Raybould, H. E. (2015). Changes in intestinal barrier function and gut microbiota in high-fat diet-fed rats are dynamic and region dependent. Am. J. Physiol. Gastrointest. Liver Physiol. 308, G840–G851. doi: 10.1152/ajpgi.00029.2015
Hanhineva, K., Torronen, R., Bondia-Pons, I., Pekkinen, J., Kolehmainen, M., Mykkanen, H., et al. (2010). Impact of dietary polyphenols on carbohydrate metabolism. Int. J. Mol. Sci. 11, 1365–1402. doi: 10.3390/ijms11041365
Hartman, A. L., Lough, D. M., Barupal, D. K., Fiehn, O., Fishbein, T., Zasloff, M., et al. (2009). Human gut microbiome adopts an alternative state following small bowel transplantation. Proc. Natl. Acad. Sci. U.S.A. 106, 17187–17192. doi: 10.1073/pnas.0904847106
Hasegawa, H., Sung, J. H., and Benno, Y. (1997). Role of human intestinal Prevotella oris in hydrolyzing ginseng saponins. Planta Med. 63, 436–440. doi: 10.1055/s-2006-957729
Hashmi, A., Naeem, N., Farooq, Z., Masood, S., Iqbal, S., and Naseer, R. (2016). Effect of prebiotic galacto-oligosaccharides on serum lipid profile of hypercholesterolemics. Probiotics Antimicrob. Proteins 8, 19–30. doi: 10.1007/s12602-016-9206-1
Hayashi, H., Sakamoto, M., and Benno, Y. (2002). Phylogenetic analysis of the human gut microbiota using 16S rDNA clone libraries and strictly anaerobic culture-based methods. Microbiol. Immunol. 46, 535–548. doi: 10.1111/j.1348-0421.2002.tb02731.x
Hayashi, H., Shibata, K., Sakamoto, M., Tomita, S., and Benno, Y. (2007). Prevotella copri sp. nov. and Prevotella stercorea sp. nov., isolated from human faeces. Int. J. Syst. Evol. Microbiol. 57(Pt 5), 941–946. doi: 10.1099/ijs.0.64778-0
Hayashi, H., Takahashi, R., Nishi, T., Sakamoto, M., and Benno, Y. (2005). Molecular analysis of jejunal, ileal, caecal and recto-sigmoidal human colonic microbiota using 16S rRNA gene libraries and terminal restriction fragment length polymorphism. J. Med. Microbiol. 54(Pt 11), 1093–1101. doi: 10.1099/jmm.0.45935-0
Hidalgo, M., Oruna-Concha, M. J., Kolida, S., Walton, G. E., Kallithraka, S., Spencer, J. P., et al. (2012). Metabolism of anthocyanins by human gut microflora and their influence on gut bacterial growth. J. Agric. Food Chem. 60, 3882–3890. doi: 10.1021/jf3002153
Hiippala, K., Kainulainen, V., Kalliomaki, M., Arkkila, P., and Satokari, R. (2016). Mucosal prevalence and interactions with the epithelium indicate commensalism of Sutterella spp. Front. Microbiol. 7:1706. doi: 10.3389/fmicb.2016.01706
Hira, T., Ikee, A., Kishimoto, Y., Kanahori, S., and Hara, H. (2015). Resistant maltodextrin promotes fasting glucagon-like peptide-1 secretion and production together with glucose tolerance in rats. Br. J. Nutr. 114, 34–42. doi: 10.1017/S0007114514004322
Holdeman, L. V., Good, I. J., and Moore, W. E. (1976). Human fecal flora: variation in bacterial composition within individuals and a possible effect of emotional stress. Appl. Environ. Microbiol. 31, 359–375.
Hollman, P. C., de Vries, J. H., van Leeuwen, S. D., Mengelers, M. J., and Katan, M. B. (1995). Absorption of dietary quercetin glycosides and quercetin in healthy ileostomy volunteers. Am. J. Clin. Nutr. 62, 1276–1282.
Holmes, E., Loo, R. L., Stamler, J., Bictash, M., Yap, I. K., Chan, Q., et al. (2008). Human metabolic phenotype diversity and its association with diet and blood pressure. Nature 453, 396–400. doi: 10.1038/nature06882
Holst, J. J. (2007). The physiology of glucagon-like peptide 1. Physiol. Rev. 87, 1409–1439. doi: 10.1152/physrev.00034.2006
Hong, K. B., Kim, J. H., Kwon, H. K., Han, S. H., Park, Y., and Suh, H. J. (2016). Evaluation of prebiotic effects of high-purity galactooligosaccharides in vitro and in vivo. Food Technol. Biotechnol. 54, 156–163. doi: 10.17113/ftb.54.02.16.4292
Hong, K., Jang, K. H., Lee, J. C., Kim, S., Kim, M. K., Lee, I. Y., et al. (2005). Bacterial beta-glucan exhibits potent hypoglycemic activity via decrease of serum lipids and adiposity, and increase of UCP mRNA expression. J. Microbiol. Biotechnol. 15:8.
Hooda, S., Boler, B. M., Serao, M. C., Brulc, J. M., Staeger, M. A., Boileau, T. W., et al. (2012). 454 pyrosequencing reveals a shift in fecal microbiota of healthy adult men consuming polydextrose or soluble corn fiber. J. Nutr. 142, 1259–1265. doi: 10.3945/jn.112.158766
Hooper, L., Kroon, P. A., Rimm, E. B., Cohn, J. S., Harvey, I., Le Cornu, K. A., et al. (2008). Flavonoids, flavonoid-rich foods, and cardiovascular risk: a meta-analysis of randomized controlled trials. Am. J. Clin. Nutr. 88, 38–50.
Hopkins, M. J., Sharp, R., and Macfarlane, G. T. (2001). Age and disease related changes in intestinal bacterial populations assessed by cell culture, 16S rRNA abundance, and community cellular fatty acid profiles. Gut 48, 198–205. doi: 10.1136/gut.48.2.198
Horz, H. P. (2015). Archaeal lineages within the human microbiome: absent, rare or elusive? Life (Basel) 5, 1333–1345. doi: 10.3390/life5021333
Hosseini, E., Grootaert, C., Verstraete, W., and Van de Wiele, T. (2011). Propionate as a health-promoting microbial metabolite in the human gut. Nutr. Rev. 69, 245–258. doi: 10.1111/j.1753-4887.2011.00388.x
Hou, C. C., Lai, C. C., Liu, W. L., Chao, C. M., Chiu, Y. H., and Hsueh, P. R. (2011). Clinical manifestation and prognostic factors of non-cholerae Vibrio infections. Eur. J. Clin. Microbiol. Infect. Dis. 30, 819–824. doi: 10.1007/s10096-011-1162-9
Hoverstad, T., Fausa, O., Bjorneklett, A., and Bohmer, T. (1984). Short-chain fatty acids in the normal human feces. Scand. J. Gastroenterol. 19, 375–381.
Hoyles, L., Honda, H., Logan, N. A., Halket, G., La Ragione, R. M., and McCartney, A. L. (2012). Recognition of greater diversity of Bacillus species and related bacteria in human faeces. Res. Microbiol. 163, 3–13. doi: 10.1016/j.resmic.2011.10.004
Hsu, C. K., Liao, J. W., Chung, Y. C., Hsieh, C. P., and Chan, Y. C. (2004). Xylooligosaccharides and fructooligosaccharides affect the intestinal microbiota and precancerous colonic lesion development in rats. J. Nutr. 134, 1523–1528.
Hu, Y., Le Leu, R. K., Christophersen, C. T., Somashekar, R., Conlon, M. A., Meng, X. Q., et al. (2016). Manipulation of the gut microbiota using resistant starch is associated with protection against colitis-associated colorectal cancer in rats. Carcinogenesis 37, 366–375. doi: 10.1093/carcin/bgw019
Hugon, P., Lagier, J. C., Colson, P., Bittar, F., and Raoult, D. (2016). Repertoire of human gut microbes. Microb. Pathog. 106, 103–112. doi: 10.1016/j.micpath.2016.06.020
Human Microbiome Project Consortium (2012). Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214. doi: 10.1038/nature11234
Janda, J. M., and Abbott, S. L. (1998). Evolving concepts regarding the genus Aeromonas: an expanding Panorama of species, disease presentations, and unanswered questions. Clin. Infect. Dis. 27, 332–344. doi: 10.1086/514652
Janda, J. M., and Abbott, S. L. (2010). The genus Aeromonas: taxonomy, pathogenicity, and infection. Clin. Microbiol. Rev. 23, 35–73. doi: 10.1128/CMR.00039-09
Johnston, K., Sharp, P., Clifford, M., and Morgan, L. (2005). Dietary polyphenols decrease glucose uptake by human intestinal Caco-2 cells. FEBS Lett. 579, 1653–1657. doi: 10.1016/j.febslet.2004.12.099
Kaakoush, N. O., Day, A. S., Huinao, K. D., Leach, S. T., Lemberg, D. A., Dowd, S. E., et al. (2012). Microbial dysbiosis in pediatric patients with crohn's disease. J. Clin. Microbiol. 50, 3258–3266. doi: 10.1128/JCM.01396-12
Kageyama, A., Benno, Y., and Nakase, T. (1999). Phylogenetic and phenotypic evidence for the transfer of Eubacterium aerofaciens to the genus Collinsella as Collinsella aerofaciens gen. nov., comb. nov. Int. J. Syst. Bacteriol. 49(Pt 2), 557–565. doi: 10.1099/00207713-49-2-557
Kaji, I., Karaki, S.-I., and Kuwahara, A. (2014). Short-chain fatty acid receptor and its contribution to glucagon-like peptide-1 release. Digestion 89, 31–36. doi: 10.1159/000356211
Kaliannan, K., Wang, B., Li, X. Y., Kim, K. J., and Kang, J. X. (2015). A host-microbiome interaction mediates the opposing effects of omega-6 and omega-3 fatty acids on metabolic endotoxemia. Sci. Rep. 5:11276. doi: 10.1038/srep11276
Kaur, J. (2014). A comprehensive review on metabolic syndrome. Cardiol. Res. Pract. 2014:943162. doi: 10.1155/2014/943162
Kaushik, S. J., Medale, F., Fauconneau, B., and Blanc, D. (1989). Effect of digestible carbohydrates on protein/energy utilization and on glucose metabolism in rainbow trout (Salmo gairdneri R.). Aquaculture 79, 63–74. doi: 10.1016/0044-8486(89)90446-8
Kawabata, K., Sugiyama, Y., Sakano, T., and Ohigashi, H. (2013). Flavonols enhanced production of anti-inflammatory substance(s) by Bifidobacterium adolescentis: prebiotic actions of galangin, quercetin, and fisetin. Biofactors 39, 422–429. doi: 10.1002/biof.1081
Keenan, M. J., Zhou, J., Hegsted, M., Pelkman, C., Durham, H. A., Coulon, D. B., et al. (2015). Role of resistant starch in improving gut health, adiposity, and insulin resistance. Adv. Nutr. 6, 198–205. doi: 10.3945/an.114.007419
Kellett, G. L., and Helliwell, P. A. (2000). The diffusive component of intestinal glucose absorption is mediated by the glucose-induced recruitment of GLUT2 to the brush-border membrane. Biochem. J. 350(Pt 1), 155–162. doi: 10.1042/bj3500155
Keshav, S. (2006). Paneth cells: leukocyte-like mediators of innate immunity in the intestine. J. Leukoc. Biol. 80, 500–508. doi: 10.1189/jlb.1005556
Khurana, S., Venkataraman, K., Hollingsworth, A., Piche, M., and Tai, T. C. (2013). Polyphenols: benefits to the cardiovascular system in health and in aging. Nutrients 5, 3779–3827. doi: 10.3390/nu5103779
Kiatpapan, P., and Murooka, Y. (2002). Genetic manipulation system in propionibacteria. J. Biosci. Bioeng. 93, 1–8. doi: 10.1016/S1389-1723(02)80045-7
Kieffer, T. J., and Habener, J. F. (1999). The glucagon-like peptides. Endocr. Rev. 20, 876–913. doi: 10.1210/edrv.20.6.0385
Knapp, B. A., Seeber, J., Rief, A., Meyer, E., and Insam, H. (2010). Bacterial community composition of the gut microbiota of Cylindroiulus fulviceps (diplopoda) as revealed by molecular fingerprinting and cloning. Folia Microbiol. (Praha). 55, 489–496. doi: 10.1007/s12223-010-0081-y
Koenig, J. E., Spor, A., Scalfone, N., Fricker, A. D., Stombaugh, J., Knight, R., et al. (2011). Succession of microbial consortia in the developing infant gut microbiome. Proc. Natl. Acad. Sci. U.S.A. 108(Suppl.), 4578–4585. doi: 10.1073/pnas.1000081107
Koeth, R. A., Wang, Z., Levison, B. S., Buffa, J. A., Org, E., Sheehy, B. T., et al. (2013). Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat. Med. 19, 576–585. doi: 10.1038/nm.3145
Kok, N., Roberfroid, M., and Delzenne, N. (1996). Dietary oligofructose modifies the impact of fructose on hepatic triacylglycerol metabolism. Metab. Clin. Exp. 45, 1547–1550. doi: 10.1016/S0026-0495(96)90186-9
Koleva, P. T., Valcheva, R. S., Sun, X., Ganzle, M. G., and Dieleman, L. A. (2012). Inulin and fructo-oligosaccharides have divergent effects on colitis and commensal microbiota in HLA-B27 transgenic rats. Br. J. Nutr. 108, 1633–1643. doi: 10.1017/S0007114511007203
Koren, O., Spor, A., Felin, J., Fak, F., Stombaugh, J., Tremaroli, V., et al. (2011). Human oral, gut, and plaque microbiota in patients with atherosclerosis. Proc. Natl. Acad. Sci. U.S.A. 108(Suppl. 1), 4592–4598. doi: 10.1073/pnas.1011383107
Kostic, A. D., Gevers, D., Pedamallu, C. S., Michaud, M., Duke, F., Earl, A. M., et al. (2012). Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 22, 292–298. doi: 10.1101/gr.126573.111
Kumar, S., and Pandey, A. K. (2013). Chemistry and biological activities of flavonoids: an overview. Sci. World J. 2013, 16. doi: 10.1155/2013/162750
Kwon, O., Eck, P., Chen, S., Corpe, C. P., Lee, J. H., Kruhlak, M., et al. (2007). Inhibition of the intestinal glucose transporter GLUT2 by flavonoids. FASEB J. 21, 366–377. doi: 10.1096/fj.06-6620com
Lafay, S., and Gil-Izquierdo, A. (2008). Bioavailability of phenolic acids. Phytochem. Rev. 7:301. doi: 10.1007/s11101-007-9077-x
Lagier, J. C., Armougom, F., Million, M., Hugon, P., Pagnier, I., Robert, C., et al. (2012). Microbial culturomics: paradigm shift in the human gut microbiome study. Clin. Microbiol. Infect. 18, 1185–1193. doi: 10.1111/1469-0691.12023
Larrosa, M., Gonzalez-Sarrias, A., Yanez-Gascon, M. J., Selma, M. V., Azorin-Ortuno, M., Toti, S., et al. (2010). Anti-inflammatory properties of a pomegranate extract and its metabolite urolithin-A in a colitis rat model and the effect of colon inflammation on phenolic metabolism. J. Nutr. Biochem. 21, 717–725. doi: 10.1016/j.jnutbio.2009.04.012
Larsbrink, J., Rogers, T. E., Hemsworth, G. R., McKee, L. S., Tauzin, A. S., Spadiut, O., et al. (2014). A discrete genetic locus confers xyloglucan metabolism in select human gut Bacteroidetes. Nature 506, 498–502. doi: 10.1038/nature12907
Larsen, N., Vogensen, F. K., van den Berg, F. W., Nielsen, D. S., Andreasen, A. S., Pedersen, B. K., et al. (2010). Gut microbiota in human adults with type 2 diabetes differs from non-diabetic adults. PLoS ONE 5:e9085. doi: 10.1371/journal.pone.0009085
Lau, J. T., Whelan, F. J., Herath, I., Lee, C. H., Collins, S. M., Bercik, P., et al. (2016). Capturing the diversity of the human gut microbiota through culture-enriched molecular profiling. Genome Med. 8:72. doi: 10.1186/s13073-016-0327-7
Lawson, P. A., and Finegold, S. M. (2015). Reclassification of Ruminococcus obeum as Blautiaobeum comb. nov. Int. J. Syst. Evol. Microbiol. 65, 789–793. doi: 10.1099/ijs.0.000015
Le Barz, M., Anhe, F. F., Varin, T. V., Desjardins, Y., Levy, E., Roy, D., et al. (2015). Probiotics as complementary treatment for metabolic disorders. Diabetes Metab. J. 39, 291–303. doi: 10.4093/dmj.2015.39.4.291
Le Chatelier, E., Nielsen, T., Qin, J., Prifti, E., Hildebrand, F., Falony, G., et al. (2013). Richness of human gut microbiome correlates with metabolic markers. Nature 500, 541–546. doi: 10.1038/nature12506
LeBlanc, J. G., Milani, C., de Giori, G. S., Sesma, F., van Sinderen, D., and Ventura, M. (2013). Bacteria as vitamin suppliers to their host: a gut microbiota perspective. Curr. Opin. Biotechnol. 24, 160–168. doi: 10.1016/j.copbio.2012.08.005
Ley, R. E., Peterson, D. A., and Gordon, J. I. (2006). Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell 124, 837–848. doi: 10.1016/j.cell.2006.02.017
Li, A.-N., Li, S., Zhang, Y.-J., Xu, X.-R., Chen, Y.-M., and Li, H.-B. (2014). Resources and biological activities of natural polyphenols. Nutrients 6, 6020–6047. doi: 10.3390/nu6126020
Li, X.-X., Wong, G. L.-H., To, K.-F., Wong, V. W.-S., Lai, L. H., Chow, D. K.-L., et al. (2009). Bacterial microbiota profiling in gastritis without helicobacter pylori infection or non-steroidal anti-inflammatory drug use. PLoS ONE 4:e7985. doi: 10.1371/journal.pone.0007985
Lim, M. Y., You, H. J., Yoon, H. S., Kwon, B., Lee, J. Y., Lee, S., et al. (2016). The effect of heritability and host genetics on the gut microbiota and metabolic syndrome. Gut. 66, 1031–1038. doi: 10.1136/gutjnl-2015-311326
Lin, H. V., Frassetto, A., Kowalik, E. J. Jr., Nawrocki, A. R., Lu, M. M., Kosinski, J. R., et al. (2012). Butyrate and propionate protect against diet-induced obesity and regulate gut hormones via free fatty acid receptor 3-independent mechanisms. PLoS ONE 7:e35240. doi: 10.1371/journal.pone.0035240
Liu, F., Prabhakar, M., Ju, J., Long, H., and Zhou, H. W. (2016). Effect of inulin-type fructans on blood lipid profile and glucose level: a systematic review and meta-analysis of randomized controlled trials. Eur. J. Clin. Nutr. 71, 9–20. doi: 10.1038/ejcn.2016.156
Liu, T. W., Cephas, K. D., Holscher, H. D., Kerr, K. R., Mangian, H. F., Tappenden, K. A., et al. (2016). Nondigestible fructans alter gastrointestinal barrier function, gene expression, histomorphology, and the microbiota profiles of diet-induced obese C57BL/6J mice. J. Nutr. 146, 949–956. doi: 10.3945/jn.115.227504
Liu, Y. J., Zhan, J., Liu, X. L., Wang, Y., Ji, J., and He, Q. Q. (2014). Dietary flavonoids intake and risk of type 2 diabetes: a meta-analysis of prospective cohort studies. Clin. Nutr. 33, 59–63. doi: 10.1016/j.clnu.2013.03.011
Liu, Z. M., Chen, Y. M., Ho, S. C., Ho, Y. P., and Woo, J. (2010). Effects of soy protein and isoflavones on glycemic control and insulin sensitivity: a 6-mo double-blind, randomized, placebo-controlled trial in postmenopausal Chinese women with prediabetes or untreated early diabetes. Am. J. Clin. Nutr. 91, 1394–1401. doi: 10.3945/ajcn.2009.28813
Lloyd-Price, J., Abu-Ali, G., and Huttenhower, C. (2016). The healthy human microbiome. Genome Med. 8, 51. doi: 10.1186/s13073-016-0307-y
Loubinoux, J., Bronowicki, J. P., Pereira, I. A., Mougenel, J. L., and Faou, A. E. (2002). Sulfate-reducing bacteria in human feces and their association with inflammatory bowel diseases. FEMS Microbiol. Ecol. 40, 107–112. doi: 10.1111/j.1574-6941.2002.tb00942.x
Louis, P., Young, P., Holtrop, G., and Flint, H. J. (2010). Diversity of human colonic butyrate-producing bacteria revealed by analysis of the butyryl-CoA:acetate CoA-transferase gene. Environ. Microbiol. 12, 304–314. doi: 10.1111/j.1462-2920.2009.02066.x
Lozupone, C. A., Stombaugh, J. I., Gordon, J. I., Jansson, J. K., and Knight, R. (2012). Diversity, stability and resilience of the human gut microbiota. Nature 489, 220–230. doi: 10.1038/nature.11550
Malaguarnera, M., Vacante, M., Antic, T., Giordano, M., Chisari, G., Acquaviva, R., et al. (2012). Bifidobacterium longum with fructo-oligosaccharides in patients with non alcoholic steatohepatitis. Dig. Dis. Sci. 57, 545–553. doi: 10.1007/s10620-011-1887-4
Mao, B., Li, D., Zhao, J., Liu, X., Gu, Z., Chen, Y. Q., et al. (2015). Metagenomic insights into the effects of fructo-oligosaccharides (FOS) on the composition of fecal microbiota in mice. J. Agric. Food Chem. 63, 856–863. doi: 10.1021/jf505156h
Marin, L., Miguelez, E. M., Villar, C. J., and Lombo, F. (2015). Bioavailability of dietary polyphenols and gut microbiota metabolism: antimicrobial properties. Biomed Res. Int. 2015:905215. doi: 10.1155/2015/905215
Marquez-Aguirre, A. L., Camacho-Ruiz, R. M., Gutierrez-Mercado, Y. K., Padilla-Camberos, E., Gonzalez-Avila, M., Galvez-Gastelum, F. J., et al. (2016). Fructans from agave tequilana with a lower degree of polymerization prevent weight gain, hyperglycemia and liver steatosis in high-fat diet-induced obese mice. Plant Foods Hum. Nutr. 71, 416–421. doi: 10.1007/s11130-016-0578-x
Marshall, B. J., and Warren, J. R. (1984). Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. Lancet 1, 1311–1315. doi: 10.1016/S0140-6736(84)91816-6
Martinez, K. B., Pierre, J. F., and Chang, E. B. (2016). The gut microbiota: the gateway to improved metabolism. Gastroenterol. Clin. North Am. 45, 601–614. doi: 10.1016/j.gtc.2016.07.001
Masumoto, S., Terao, A., Yamamoto, Y., Mukai, T., Miura, T., and Shoji, T. (2016). Non-absorbable apple procyanidins prevent obesity associated with gut microbial and metabolomic changes. Sci. Rep. 6:31208. doi: 10.1038/srep31208
Mestdagh, R., Dumas, M. E., Rezzi, S., Kochhar, S., Holmes, E., Claus, S. P., et al. (2012). Gut microbiota modulate the metabolism of brown adipose tissue in mice. J. Proteome Res. 11, 620–630. doi: 10.1021/pr200938v
Minot, S., Sinha, R., Chen, J., Li, H., Keilbaugh, S. A., Wu, G. D., et al. (2011). The human gut virome: inter-individual variation and dynamic response to diet. Genome Res. 21, 1616–1625. doi: 10.1101/gr.122705.111
Miquel, S., Martin, R., Rossi, O., Bermudez-Humaran, L. G., Chatel, J. M., Sokol, H., et al. (2013). Faecalibacterium prausnitzii and human intestinal health. Curr. Opin. Microbiol. 16, 255–261. doi: 10.1016/j.mib.2013.06.003
Modler, H. W. (1994). Bifidogenic factors—sources, metabolism and applications. Int. Dairy J. 4, 383–407. doi: 10.1016/0958-6946(94)90055-8
Moore, W. E., and Holdeman, L. V. (1974). Human fecal flora: the normal flora of 20 Japanese-Hawaiians. Appl. Microbiol. 27, 961–979.
Morand, C., Levrat, M. A., Besson, C., Demigne, C., and Remesy, C. (1994). Effects of a diet rich in resistant starch on hepatic lipid metabolism in the rat. J. Nutr. Biochem. 5, 138–144. doi: 10.1016/0955-2863(94)90085-X
Morotomi, M., Nagai, F., and Watanabe, Y. (2011). Parasutterella secunda sp. nov., isolated from human faeces and proposal of Sutterellaceae fam. nov. in the order Burkholderiales. Int. J. Syst. Evol. Microbiol. 61(Pt 3), 637–643. doi: 10.1099/ijs.0.023556-0
Morotomi, M., Nagai, F., and Watanabe, Y. (2012). Description of Christensenella minuta gen. nov., sp. nov., isolated from human faeces, which forms a distinct branch in the order Clostridiales, and proposal of Christensenellaceae fam. nov. Int. J. Syst. Evol. Microbiol. 62(Pt 1), 144–149. doi: 10.1099/ijs.0.026989-0
Morotomi, M., Nagai, F., Watanabe, Y., and Tanaka, R. (2010). Succinatimonas hippei gen. nov., sp. nov., isolated from human faeces. Int. J. Syst. Evol. Microbiol. 60(Pt 8), 1788–1793. doi: 10.1099/ijs.0.015958-0
Morrison, D. J., and Preston, T. (2016). Formation of short chain fatty acids by the gut microbiota and their impact on human metabolism. Gut Microbes 7, 189–200. doi: 10.1080/19490976.2015.1134082
Mourembou, G., Rathored, J., Lekana-Douki, J. B., Ndjoyi-Mbiguino, A., Khelaifia, S., Robert, C., et al. (2016). Description of Gabonibacter massiliensis gen. nov., sp. nov., a new member of the family porphyromonadaceae isolated from the human gut microbiota. Curr. Microbiol. 73, 867–877. doi: 10.1007/s00284-016-1137-2
Nadkarni, P., Chepurny, O. G., and Holz, G. G. (2014). Regulation of glucose homeostasis by GLP-1. Prog. Mol. Biol. Transl. Sci. 121, 23–65. doi: 10.1016/B978-0-12-800101-1.00002-8
Nagai, F., Morotomi, M., Sakon, H., and Tanaka, R. (2009). Parasutterella excrementihominis gen. nov., sp. nov., a member of the family Alcaligenaceae isolated from human faeces. Int. J. Syst. Evol. Microbiol. 59(Pt 7), 1793–1797. doi: 10.1099/ijs.0.002519-0
Nauck, M. A., Heimesaat, M. M., Orskov, C., Holst, J. J., Ebert, R., and Creutzfeldt, W. (1993). Preserved incretin activity of glucagon-like peptide 1 [7-36 amide] but not of synthetic human gastric inhibitory polypeptide in patients with type-2 diabetes mellitus. J. Clin. Invest. 91, 301–307. doi: 10.1172/JCI116186
Ndongo, S., Dubourg, G., Khelaifia, S., Fournier, P. E., and Raoult, D. (2016a). Christensenella timonensis, a new bacterial species isolated from the human gut. New Microbes New Infect 13, 32–33. doi: 10.1016/j.nmni.2016.05.010
Ndongo, S., Khelaifia, S., Fournier, P. E., and Raoult, D. (2016b). Christensenella massiliensis, a new bacterial species isolated from the human gut. New Microbes New Infect. 12, 69–70. doi: 10.1016/j.nmni.2016.04.014
Nettleton, J. A., Harnack, L. J., Scrafford, C. G., Mink, P. J., Barraj, L. M., and Jacobs, D. R. Jr. (2006). Dietary flavonoids and flavonoid-rich foods are not associated with risk of type 2 diabetes in postmenopausal women. J. Nutr. 136, 3039–3045.
Newton, D. F., Cummings, J. H., Macfarlane, S., and Macfarlane, G. T. (1998). Growth of a human intestinal Desulfovibrio desulfuricans in continuous cultures containing defined populations of saccharolytic and amino acid fermenting bacteria. J. Appl. Microbiol. 85, 372–380. doi: 10.1046/j.1365-2672.1998.00522.x
Neyrinck, A. M., Van Hee, V. F., Piront, N., De Backer, F., Toussaint, O., Cani, P. D., et al. (2012). Wheat-derived arabinoxylan oligosaccharides with prebiotic effect increase satietogenic gut peptides and reduce metabolic endotoxemia in diet-induced obese mice. Nutr. Diabetes 2:e28. doi: 10.1038/nutd.2011.24
O'Hara, A. M., and Shanahan, F. (2006). The gut flora as a forgotten organ. EMBO Rep. 7, 688–693. doi: 10.1038/sj.embor.7400731
Olli, K., Salli, K., Alhoniemi, E., Saarinen, M., Ibarra, A., Vasankari, T., et al. (2015). Postprandial effects of polydextrose on satiety hormone responses and subjective feelings of appetite in obese participants. Nutr. J. 14:2. doi: 10.1186/1475-2891-14-2
Ott, S. J., Kuhbacher, T., Musfeldt, M., Rosenstiel, P., Hellmig, S., Rehman, A., et al. (2008). Fungi and inflammatory bowel diseases: alterations of composition and diversity. Scand. J. Gastroenterol. 43, 831–841. doi: 10.1080/00365520801935434
Ozdal, T., Sela, D. A., Xiao, J., Boyacioglu, D., Chen, F., and Capanoglu, E. (2016). The reciprocal interactions between polyphenols and gut microbiota and effects on bioaccessibility. Nutrients 8:78. doi: 10.3390/nu8020078
Paeschke, T. M., and Aimutis, W. R. (eds.). (2010). “Appendix nondigestible carbohydrates: structure and sources,” in Nondigestible Carbohydrates and Digestive Health (Ames, IA: Blackwell Publishing Ltd.), 321–329.
Pandey, K. B., and Rizvi, S. I. (2009). Plant polyphenols as dietary antioxidants in human health and disease. Oxid. Med. Cell. Longev. 2, 270–278. doi: 10.4161/oxim.2.5.9498
Parekh, P. J., Arusi, E., Vinik, A. I., and Johnson, D. A. (2014). The role and influence of gut microbiota in pathogenesis and management of obesity and metabolic syndrome. Front. Endocrinol. 5:47. doi: 10.3389/fendo.2014.00047
Parfrey, L. W., Walters, W. A., and Knight, R. (2011). Microbial eukaryotes in the human microbiome: ecology, evolution, and future directions. Front. Microbiol. 2:153. doi: 10.3389/fmicb.2011.00153
Parkar, S. G., Stevenson, D. E., and Skinner, M. A. (2008). The potential influence of fruit polyphenols on colonic microflora and human gut health. Int. J. Food Microbiol. 124, 295–298. doi: 10.1016/j.ijfoodmicro.2008.03.017
Parnell, J. A., and Reimer, R. A. (2009). Weight loss during oligofructose supplementation is associated with decreased ghrelin and increased peptide YY in overweight and obese adults. Am. J. Clin. Nutr. 89, 1751–1759. doi: 10.3945/ajcn.2009.27465
Peirce, V., and Vidal-Puig, A. (2013). Regulation of glucose homoeostasis by brown adipose tissue. Lancet Diabetes Endocrinol. 1, 353–360. doi: 10.1016/S2213-8587(13)70055-X
Pereira, D. M., Valentão, P., Pereira, J. A., and Andrade, P. B. (2009). Phenolics: from chemistry to biology. Molecules 14, 2202–2211. doi: 10.3390/molecules14062202
Piche, T., des Varannes, S. B., Sacher-Huvelin, S., Holst, J. J., Cuber, J. C., and Galmiche, J. P. (2003). Colonic fermentation influences lower esophageal sphincter function in gastroesophageal reflux disease. Gastroenterology 124, 894–902. doi: 10.1053/gast.2003.50159
Pyra, K. A., Saha, D. C., and Reimer, R. A. (2012). Prebiotic fiber increases hepatic acetyl CoA carboxylase phosphorylation and suppresses glucose-dependent insulinotropic polypeptide secretion more effectively when used with metformin in obese rats. J. Nutr. 142, 213–220. doi: 10.3945/jn.111.147132
Qiao, Y., Sun, J., Xia, S., Tang, X., Shi, Y., and Le, G. (2014). Effects of resveratrol on gut microbiota and fat storage in a mouse model with high-fat-induced obesity. Food Funct. 5, 1241–1249. doi: 10.1039/c3fo60630a
Qin, J., Li, R., Raes, J., Arumugam, M., Burgdorf, K. S., Manichanh, C., et al. (2010). A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464, 59–65. doi: 10.1038/nature08821
Qin, J., Li, Y., Cai, Z., Li, S., Zhu, J., Zhang, F., et al. (2012). A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 490, 55–60. doi: 10.1038/nature11450
Rajilic-Stojanovic, M., and de Vos, W. M. (2014). The first 1000 cultured species of the human gastrointestinal microbiota. FEMS Microbiol. Rev. 38, 996–1047. doi: 10.1111/1574-6976.12075
Rajilic-Stojanovic, M., Shanahan, F., Guarner, F., and de Vos, W. M. (2013). Phylogenetic analysis of dysbiosis in ulcerative colitis during remission. Inflamm. Bowel Dis. 19, 481–488. doi: 10.1097/MIB.0b013e31827fec6d
Rajilic-Stojanovic, M., Smidt, H., and de Vos, W. M. (2007). Diversity of the human gastrointestinal tract microbiota revisited. Environ. Microbiol. 9, 2125–2136. doi: 10.1111/j.1462-2920.2007.01369.x
Rajpal, D. K., Klein, J. L., Mayhew, D., Boucheron, J., Spivak, A. T., Kumar, V., et al. (2015). Selective spectrum antibiotic modulation of the gut microbiome in obesity and diabetes rodent models. PLoS ONE 10:e0145499. doi: 10.1371/journal.pone.0145499
Ramasamy, D., Lagier, J. C., Nguyen, T. T., Raoult, D., and Fournier, P. E. (2013). Non contiguous-finished genome sequence and description of Dielma fastidiosa gen. nov., sp. nov., a new member of the Family Erysipelotrichaceae. Stand. Genomic Sci. 8, 336–351. doi: 10.4056/sigs.3567059
Ramirez-Farias, C., Slezak, K., Fuller, Z., Duncan, A., Holtrop, G., and Louis, P. (2009). Effect of inulin on the human gut microbiota: stimulation of Bifidobacterium adolescentis and Faecalibacterium prausnitzii. Br. J. Nutr. 101, 541–550. doi: 10.1017/S0007114508019880
Rehman, A., Rausch, P., Wang, J., Skieceviciene, J., Kiudelis, G., Bhagalia, K., et al. (2016). Geographical patterns of the standing and active human gut microbiome in health and IBD. Gut 65, 238–248. doi: 10.1136/gutjnl-2014-308341
Reiser, S. (1985). Effect of dietary sugars on metabolic risk factors associated with heart disease. Nutr. Health 3, 203–216.
Rendon-Huerta, J. A., Juarez-Flores, B., Pinos-Rodriguez, J. M., Aguirre-Rivera, J. R., and Delgado-Portales, R. E. (2012). Effects of different sources of fructans on body weight, blood metabolites and fecal bacteria in normal and obese non-diabetic and diabetic rats. Plant Foods Hum. Nutr. 67, 64–70. doi: 10.1007/s11130-011-0266-9
Rial, S. A., Karelis, A. D., Bergeron, K.-F., and Mounier, C. (2016). Gut microbiota and metabolic health: the potential beneficial effects of a medium chain triglyceride diet in obese individuals. Nutrients 8:281. doi: 10.3390/nu8050281
Richards, A. L., Burns, M. B., Alazizi, A., Barreiro, L. B., Pique-Regi, R., Blekhman, R., et al. (2016). Genetic and transcriptional analysis of human host response to healthy gut microbiota. mSystems 1:e00067–16. doi: 10.1128/mSystems.00067-16
Riviere, A., Selak, M., Lantin, D., Leroy, F., and De Vuyst, L. (2016). Bifidobacteria and butyrate-producing colon bacteria: importance and strategies for their stimulation in the human gut. Front. Microbiol. 7:979. doi: 10.3389/fmicb.2016.00979
Roberfroid, M., Gibson, G. R., Hoyles, L., McCartney, A. L., Rastall, R., Rowland, I., et al. (2010). Prebiotic effects: metabolic and health benefits. Br. J. Nutr. 104(Suppl. 2), S1–S63. doi: 10.1017/s0007114510003363
Roopchand, D. E., Carmody, R. N., Kuhn, P., Moskal, K., Rojas-Silva, P., Turnbaugh, P. J., et al. (2015). Dietary polyphenols promote growth of the gut bacterium Akkermansia muciniphila and attenuate high-fat diet-induced metabolic syndrome. Diabetes 64, 2847–2858. doi: 10.2337/db14-1916
Rosenbaum, M., Knight, R., and Leibel, R. L. (2015). The gut microbiota in human energy homeostasis and obesity. Trends Endocrinol. Metab. 26, 493–501. doi: 10.1016/j.tem.2015.07.002
Russell, W. R., Labat, A., Scobbie, L., Duncan, G. J., and Duthie, G. G. (2009). Phenolic acid content of fruits commonly consumed and locally produced in Scotland. Food Chem. 115, 100–104. doi: 10.1016/j.foodchem.2008.11.086
Russo, F., Chimienti, G., Riezzo, G., Pepe, G., Petrosillo, G., Chiloiro, M., et al. (2008). Inulin-enriched pasta affects lipid profile and Lp(a) concentrations in Italian young healthy male volunteers. Eur. J. Nutr. 47, 453–459. doi: 10.1007/s00394-008-0748-1
Sakamoto, M., and Benno, Y. (2006). Reclassification of Bacteroides distasonis, Bacteroides goldsteinii and Bacteroides merdae as Parabacteroides distasonis gen. nov., comb. nov., Parabacteroides goldsteinii comb. nov. and Parabacteroides merdae comb. nov. Int. J. Syst. Evol. Microbiol. 56(Pt 7), 1599–1605. doi: 10.1099/ijs.0.64192-0
Sakamoto, M., and Ohkuma, M. (2012). Reclassification of Xylanibacter oryzae Ueki et al. 2006 as Prevotella oryzae comb. nov., with an emended description of the genus Prevotella. Int. J. Syst. Evol. Microbiol. 62(Pt 11), 2637–2642. doi: 10.1099/ijs.0.038638-0
Salazar, N., Dewulf, E. M., Neyrinck, A. M., Bindels, L. B., Cani, P. D., Mahillon, J., et al. (2015). Inulin-type fructans modulate intestinal Bifidobacterium species populations and decrease fecal short-chain fatty acids in obese women. Clin. Nutr. 34, 501–507. doi: 10.1016/j.clnu.2014.06.001
Samuel, B. S., Hansen, E. E., Manchester, J. K., Coutinho, P. M., Henrissat, B., Fulton, R., et al. (2007). Genomic and metabolic adaptations of Methanobrevibacter smithii to the human gut. Proc. Natl. Acad. Sci. U.S.A. 104, 10643–10648. doi: 10.1073/pnas.0704189104
Santacruz, A., Collado, M. C., Garcia-Valdes, L., Segura, M. T., Martin-Lagos, J. A., Anjos, T., et al. (2010). Gut microbiota composition is associated with body weight, weight gain and biochemical parameters in pregnant women. Br. J. Nutr. 104, 83–92. doi: 10.1017/S0007114510000176
Scalbert, A., Manach, C., Morand, C., Remesy, C., and Jimenez, L. (2005). Dietary polyphenols and the prevention of diseases. Crit. Rev. Food Sci. Nutr. 45, 287–306. doi: 10.1080/1040869059096
Scarpellini, E., Ianiro, G., Attili, F., Bassanelli, C., De Santis, A., and Gasbarrini, A. (2015). The human gut microbiota and virome: potential therapeutic implications. Digest. Liver Dis. 47, 1007–1012. doi: 10.1016/j.dld.2015.07.008
Schaefer, E. J., Gleason, J. A., and Dansinger, M. L. (2009). Dietary fructose and glucose differentially affect lipid and glucose homeostasis. J. Nutr. 139, 1257S–1262S. doi: 10.3945/jn.108.098186
Schneeberger, M., Everard, A., Gómez-Valadés, A. G., Matamoros, S., Ramírez, S., Delzenne, N. M., et al. (2015). Akkermansia muciniphila inversely correlates with the onset of inflammation, altered adipose tissue metabolism and metabolic disorders during obesity in mice. Sci. Rep. 5:16643. doi: 10.1038/srep16643
Selma, M. V., Espin, J. C., and Tomas-Barberan, F. A. (2009). Interaction between phenolics and gut microbiota: role in human health. J. Agric. Food Chem. 57, 6485–6501. doi: 10.1021/jf902107d
Shi, Y. C., Loh, K., Bensellam, M., Lee, K., Zhai, L., Lau, J., et al. (2015). Pancreatic PYY is critical in the control of insulin secretion and glucose homeostasis in female mice. Endocrinology 156, 3122–3136. doi: 10.1210/en.2015-1168
Shin, N. R., Whon, T. W., and Bae, J. W. (2015). Proteobacteria: microbial signature of dysbiosis in gut microbiota. Trends Biotechnol. 33, 496–503. doi: 10.1016/j.tibtech.2015.06.011
Shrime, M. G., Bauer, S. R., McDonald, A. C., Chowdhury, N. H., Coltart, C. E., and Ding, E. L. (2011). Flavonoid-rich cocoa consumption affects multiple cardiovascular risk factors in a meta-analysis of short-term studies. J. Nutr. 141, 1982–1988. doi: 10.3945/jn.111.145482
Smith, M. I., Yatsunenko, T., Manary, M. J., Trehan, I., Mkakosya, R., Cheng, J., et al. (2013). Gut microbiomes of Malawian twin pairs discordant for kwashiorkor. Science 339, 548–554. doi: 10.1126/science.1229000
Sokol, H., Leducq, V., Aschard, H., Pham, H. P., Jegou, S., Landman, C., et al. (2016). Fungal microbiota dysbiosis in IBD. Gut 66, 1039–1048. doi: 10.1136/gutjnl-2015-310746
Song, Y., Kononen, E., Rautio, M., Liu, C., Bryk, A., Eerola, E., et al. (2006). Alistipes onderdonkii sp. nov. and Alistipes shahii sp. nov., of human origin. Int. J. Syst. Evol. Microbiol. 56(Pt 8), 1985–1990. doi: 10.1099/ijs.0.64318-0
Song, Y., Manson, J. E., Buring, J. E., Sesso, H. D., and Liu, S. (2005). Associations of dietary flavonoids with risk of type 2 diabetes, and markers of insulin resistance and systemic inflammation in women: a prospective study and cross-sectional analysis. J. Am. Coll. Nutr. 24, 376–384. doi: 10.1080/07315724.2005.10719488
Stanford, K. I., Middelbeek, R. J., Townsend, K. L., An, D., Nygaard, E. B., Hitchcox, K. M., et al. (2013). Brown adipose tissue regulates glucose homeostasis and insulin sensitivity. J. Clin. Invest. 123, 215–223. doi: 10.1172/JCI62308
Stephen, A. M., and Cummings, J. H. (1980). The microbial contribution to human faecal mass. J. Med. Microbiol. 13, 45–56. doi: 10.1099/00222615-13-1-45
Stevens, J. F., and Maier, C. S. (2016). The chemistry of gut microbial metabolism of polyphenols. Phytochem. Rev. 15, 425–444. doi: 10.1007/s11101-016-9459-z
Swidsinski, A., Dorffel, Y., Loening-Baucke, V., Tertychnyy, A., Biche-Ool, S., Stonogin, S., et al. (2012). Mucosal invasion by fusobacteria is a common feature of acute appendicitis in Germany, Russia, and China. Saudi J. Gastroenterol. 18, 55–58. doi: 10.4103/1319-3767.91734
Tang, W. H., Wang, Z., Levison, B. S., Koeth, R. A., Britt, E. B., Fu, X., et al. (2013). Intestinal microbial metabolism of phosphatidylcholine and cardiovascular risk. N. Engl. J. Med. 368, 1575–1584. doi: 10.1056/NEJMoa1109400
Taras, D., Simmering, R., Collins, M. D., Lawson, P. A., and Blaut, M. (2002). Reclassification of Eubacterium formicigenerans Holdeman and Moore 1974 as Dorea formicigenerans gen. nov., comb. nov., and description of Dorea longicatena sp. nov., isolated from human faeces. Int. J. Syst. Evol. Microbiol. 52(Pt 2), 423–428. doi: 10.1099/00207713-52-2-423
Thilakarathna, S. H., and Rupasinghe, H. P. (2013). Flavonoid bioavailability and attempts for bioavailability enhancement. Nutrients 5, 3367–3387. doi: 10.3390/nu5093367
Thilesen, C. M., Nicolaidis, M., Lokebo, J. E., Falsen, E., Jorde, A. T., and Muller, F. (2007). Leptotrichia amnionii, an emerging pathogen of the female urogenital tract. J. Clin. Microbiol. 45, 2344–2347. doi: 10.1128/JCM.00167-07
Tilg, H., and Moschen, A. R. (2014). Microbiota and diabetes: an evolving relationship. Gut 63, 1513–1521. doi: 10.1136/gutjnl-2014-306928
Tsao, R. (2010). Chemistry and biochemistry of dietary polyphenols. Nutrients 2, 1231–1246. doi: 10.3390/nu2121231
Turnbaugh, P. J., Hamady, M., Yatsunenko, T., Cantarel, B. L., Duncan, A., Ley, R. E., et al. (2009). A core gut microbiome in obese and lean twins. Nature 457, 480–484. doi: 10.1038/nature07540
Turnbaugh, P. J., Ley, R. E., Mahowald, M. A., Magrini, V., Mardis, E. R., and Gordon, J. I. (2006). An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444, 1027–1031. doi: 10.1038/nature05414
Umeno, A., Horie, M., Murotomi, K., Nakajima, Y., and Yoshida, Y. (2016). Antioxidative and antidiabetic effects of natural polyphenols and isoflavones. Molecules 21:708. doi: 10.3390/molecules21060708
Unemo, M., Golparian, D., and Hellmark, B. (2014). First three Neisseria gonorrhoeae isolates with high-level resistance to azithromycin in Sweden: a threat to currently available dual-antimicrobial regimens for treatment of gonorrhea? Antimicrob. Agents Chemother. 58, 624–625. doi: 10.1128/AAC.02093-13
Ursell, L. K., Clemente, J. C., Rideout, J. R., Gevers, D., Caporaso, J. G., and Knight, R. (2012). The interpersonal and intrapersonal diversity of human-associated microbiota in key body sites. J. Allergy Clin. Immunol. 129, 1204–1208. doi: 10.1016/j.jaci.2012.03.010
Vaahtovuo, J., Korkeamaki, M., Munukka, E., Viljanen, M. K., and Toivanen, P. (2005). Quantification of bacteria in human feces using 16S rRNA-hybridization, DNA-staining and flow cytometry. J. Microbiol. Methods 63, 276–286. doi: 10.1016/j.mimet.2005.03.017
Valsecchi, C., Carlotta Tagliacarne, S., and Castellazzi, A. (2016). Gut microbiota and obesity. J. Clin. Gastroenterol. 50(Suppl. 2), S157–S158. doi: 10.1097/mcg.0000000000000715
Van den Abbeele, P., Gerard, P., Rabot, S., Bruneau, A., El Aidy, S., Derrien, M., et al. (2011). Arabinoxylans and inulin differentially modulate the mucosal and luminal gut microbiota and mucin-degradation in humanized rats. Environ. Microbiol. 13, 2667–2680. doi: 10.1111/j.1462-2920.2011.02533.x
Van Houte, J., and Gibbons, R. J. (1966). Studies of the cultivable flora of normal human feces. Antonie Van Leeuwenhoek 32, 212–222. doi: 10.1007/BF02097463
Vinayagam, R., and Xu, B. (2015). Antidiabetic properties of dietary flavonoids: a cellular mechanism review. Nutr. Metab. 12:60. doi: 10.1186/s12986-015-0057-7
Vrieze, A., Van Nood, E., Holleman, F., Salojärvi, J., Kootte, R. S., Bartelsman, J. F. W. M., et al. (2012). Transfer of intestinal microbiota from lean donors increases insulin sensitivity in individuals with metabolic syndrome. Gastroenterology 143, 913.e917–916.e917. doi: 10.1053/j.gastro.2012.06.031
Walter, J., Margosch, D., Hammes, P. W., and Hertel, C. (2002). Detection of fusobacterium species in human feces using genus-specific PCR primers and denaturing gradient gel electrophoresis. Microb. Ecol. Health Dis. 14, 129–132. doi: 10.1080/089106002320644294
Walters, W. A., Xu, Z., and Knight, R. (2014). Meta-analyses of human gut microbes associated with obesity and IBD. FEBS Lett. 588, 4223–4233. doi: 10.1016/j.febslet.2014.09.039
Walther, W. W., and Millwood, E. G. (1951). Presence of certain serological types of Bact. coli in the human intestine. Br. Med. J. 2, 156–157. doi: 10.1136/bmj.2.4724.156
Wang, D., Ho, L., Faith, J., Ono, K., Janle, E. M., Lachcik, P. J., et al. (2015). Role of intestinal microbiota in the generation of polyphenol derived phenolic acid mediated attenuation of Alzheimer's disease β-amyloid oligomerization. Mol. Nutr. Food Res. 59, 1025–1040. doi: 10.1002/mnfr.201400544
Wang, H. X., and Wang, Y. P. (2016). Gut microbiota-brain axis. Chin. Med. J. 129, 2373–2380. doi: 10.4103/0366-6999.190667
Wang, M., Ahrne, S., Jeppsson, B., and Molin, G. (2005). Comparison of bacterial diversity along the human intestinal tract by direct cloning and sequencing of 16S rRNA genes. FEMS Microbiol. Ecol. 54, 219–231. doi: 10.1016/j.femsec.2005.03.012
Wang, S., Moustaid-Moussa, N., Chen, L., Mo, H., Shastri, A., Su, R., et al. (2014). Novel insights of dietary polyphenols and obesity. J. Nutr. Biochem. 25, 1–18. doi: 10.1016/j.jnutbio.2013.09.001
Wang, X., Liu, H., Chen, J., Li, Y., and Qu, S. (2015). Multiple factors related to the secretion of glucagon-like peptide-1. Int. J. Endocrinol. 2015:651757. doi: 10.1155/2015/651757
Wang, Z., Klipfell, E., Bennett, B. J., Koeth, R., Levison, B. S., Dugar, B., et al. (2011). Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature 472, 57–63. doi: 10.1038/nature09922
Wexler, H. M. (2007). Bacteroides: the good, the bad, and the nitty-gritty. Clin. Microbiol. Rev. 20, 593–621. doi: 10.1128/CMR.00008-07
Wexler, H. M., Reeves, D., Summanen, P. H., Molitoris, E., McTeague, M., Duncan, J., et al. (1996). Sutterella wadsworthensis gen. nov., sp. nov., bile-resistant microaerophilic Campylobacter gracilis-like clinical isolates. Int. J. Syst. Bacteriol. 46, 252–258. doi: 10.1099/00207713-46-1-252
Williams, C. M. (1999). Effects of inulin on lipid parameters in humans. J. Nutr. 129(7 Suppl.), 1471S–1473S.
Williamson, G. (2013). Possible effects of dietary polyphenols on sugar absorption and digestion. Mol. Nutr. Food Res. 57, 48–57. doi: 10.1002/mnfr.201200511
Woese, C. R., Kandler, O., and Wheelis, M. L. (1990). Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc. Natl. Acad. Sci. U.S.A. 87, 4576–4579. doi: 10.1073/pnas.87.12.4576
Woods, M., and Gorbach, S. (2001). “Influences of fiber on the ecology of the intestinal flora,” in CRC Handbook of Dietary Fiber in Human Nutrition, 3rd Edn., ed G. A. Spiller (CRC Press), 257–270.
Wroblewski, L. E., and Peek, R. M. Jr. (2016). Helicobacter pylori, cancer, and the gastric microbiota. Adv. Exp. Med. Biol. 908, 393–408. doi: 10.1007/978-3-319-41388-4_19
Wu, G. D., Chen, J., Hoffmann, C., Bittinger, K., Chen, Y. Y., Keilbaugh, S. A., et al. (2011). Linking long-term dietary patterns with gut microbial enterotypes. Science 334, 105–108. doi: 10.1126/science.1208344
Wu, T., Yang, Y., Zhang, L., and Han, J. (2010). [Systematic review of the effects of inulin-type fructans on blood lipid profiles: a meta-analysis]. Wei Sheng Yan Jiu 39, 172–176.
Wylie, K. M., Mihindukulasuriya, K. A., Zhou, Y., Sodergren, E., Storch, G. A., and Weinstock, G. M. (2014). Metagenomic analysis of double-stranded DNA viruses in healthy adults. BMC Biol. 12:71. doi: 10.1186/s12915-014-0071-7
Xu, J., Bjursell, M. K., Himrod, J., Deng, S., Carmichael, L. K., Chiang, H. C., et al. (2003). A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science 299, 2074–2076. doi: 10.1126/science.1080029
Xu, J., Mahowald, M. A., Ley, R. E., Lozupone, C. A., Hamady, M., Martens, E. C., et al. (2007). Evolution of symbiotic bacteria in the distal human intestine. PLoS Biol. 5:e156. doi: 10.1371/journal.pbio.0050156
Yabe, D., and Seino, Y. (2011). Two incretin hormones GLP-1 and GIP: comparison of their actions in insulin secretion and beta cell preservation. Prog. Biophys. Mol. Biol. 107, 248–256. doi: 10.1016/j.pbiomolbio.2011.07.010
Yamaguchi, Y., Adachi, K., Sugiyama, T., Shimozato, A., Ebi, M., Ogasawara, N., et al. (2016). Association of intestinal microbiota with metabolic markers and dietary habits in patients with type 2 diabetes. Digestion 94, 66–72. doi: 10.1159/000447690
Yatsunenko, T., Rey, F. E., Manary, M. J., Trehan, I., Dominguez-Bello, M. G., Contreras, M., et al. (2012). Human gut microbiome viewed across age and geography. Nature 486, 222–227. doi: 10.1038/nature11053
Ze, X., Duncan, S. H., Louis, P., and Flint, H. J. (2012). Ruminococcus bromii is a keystone species for the degradation of resistant starch in the human colon. ISME J. 6, 1535–1543. doi: 10.1038/ismej.2012.4
Zhang, W., Gu, Y., Chen, Y., Deng, H., Chen, L., Chen, S., et al. (2010). Intestinal flora imbalance results in altered bacterial translocation and liver function in rats with experimental cirrhosis. Eur. J. Gastroenterol. Hepatol. 22, 1481–1486. doi: 10.1097/meg.0b013e32833eb8b0
Zhang, Y., and Zhang, H. (2013). Microbiota associated with type 2 diabetes and its related complications. Food Sci. Hum. Wellness 2, 167–172. doi: 10.1016/j.fshw.2013.09.002
Zilberstein, B., Quintanilha, A. G., Santos, M. A., Pajecki, D., Moura, E. G., Alves, P. R., et al. (2007). Digestive tract microbiota in healthy volunteers. Clinics (Sao Paulo) 62, 47–54. doi: 10.1590/S1807-59322007000100008
Zoetendal, E. G., Raes, J., van den Bogert, B., Arumugam, M., Booijink, C. C., Troost, F. J., et al. (2012). The human small intestinal microbiota is driven by rapid uptake and conversion of simple carbohydrates. ISME J. 6, 1415–1426. doi: 10.1038/ismej.2011.212
Zou, S., Caler, L., Colombini-Hatch, S., Glynn, S., and Srinivas, P. (2016). Research on the human virome: where are we and what is next. Microbiome 4:32. doi: 10.1186/s40168-016-0177-y
Keywords: microbiota, natural products, food ingredients, obesity, metabolic diseases
Citation: Eid HM, Wright ML, Anil Kumar NV, Qawasmeh A, Hassan STS, Mocan A, Nabavi SM, Rastrelli L, Atanasov AG and Haddad PS (2017) Significance of Microbiota in Obesity and Metabolic Diseases and the Modulatory Potential by Medicinal Plant and Food Ingredients. Front. Pharmacol. 8:387. doi: 10.3389/fphar.2017.00387
Received: 28 February 2017; Accepted: 02 June 2017;
Published: 30 June 2017.
Edited by:
Kalin Yanbo Zhang, University of Hong Kong, Hong KongReviewed by:
Adeyemi Oladapo Aremu, University of KwaZulu-Natal, South AfricaJelena Cvejic, University of Novi Sad, Serbia
Copyright © 2017 Eid, Wright, Anil Kumar, Qawasmeh, Hassan, Mocan, Nabavi, Rastrelli, Atanasov and Haddad. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Atanas G. Atanasov, YS5hdGFuYXNvdi5tYWlsYm94QGdtYWlsLmNvbQ==
Pierre S. Haddad, cGllcnJlLmhhZGRhZEB1bW9udHJlYWwuY2E=
 Hoda M. Eid
Hoda M. Eid Michelle L. Wright
Michelle L. Wright N. V. Anil Kumar
N. V. Anil Kumar Abdel Qawasmeh
Abdel Qawasmeh Sherif T. S. Hassan
Sherif T. S. Hassan Andrei Mocan
Andrei Mocan Seyed M. Nabavi
Seyed M. Nabavi Luca Rastrelli
Luca Rastrelli Atanas G. Atanasov
Atanas G. Atanasov Pierre S. Haddad
Pierre S. Haddad