- 1School of Perfume and Aroma Technology, Shanghai Institute of Technology, Shanghai, China
- 2School of Food and Biological Engineering, Shaanxi University of Science and Technology, Xi'an, China
The metabolite profiles and distributions of procyanidin B2 were qualitatively described using UPLC-DAD-ESI-IT-TOF-MSn without help of reference standards, and a possible metabolic pathway was proposed in the present study. Summarily, 53 metabolites (24 new metabolites) were detected as metabolites of procyanidin B2, and 45 of them were tentatively identified. Twenty seven metabolites were assigned as similar metabolites of (−)-epicatechin by scission of the flavanol interflavanic bond C4–C8, including 16 aromatic metabolites, 5 conjugated metabolites, 3 ring-cleavage metabolites, and 2 phenylvalerolactone metabolites. Additionally, 14 metabolites were conjugates of free procyanidin B2, comprising 9 methylation metabolites, 8 sulfation metabolites, 5 hydration metabolites, 2 hydroxylation metabolites, 1 hydrogenation metabolites, and 1 glucuronidation metabolites. The results of metabolite distributions in organs indicated that the conjugated reaction of free procyanidin B2 mainly occurred in liver and diversified metabolites forms were observed in small intestine. The metabolic components of procyanidin B2 identified in mice provided useful information for further study of the bioactivity and mechanism of its action.
Introduction
Proanthocyanidins are one subclass of polyphenolic compounds that can be detectable in a wide variety of natural sources, such as nuts, beans, apples, cocoa, and tea (Prior and Gu, 2005; Xu et al., 2010; Li et al., 2013). Proanthocyanidins have been known as one kind of strong antioxidant compounds to prevent or ameliorate metabolic syndrome and to reduce the incidence of cardiovascular diseases and cancer (Veluri et al., 2006; Terra et al., 2011; Guerrero et al., 2013; Margalef et al., 2015).
The most ubiquitous proanthocyanidin present in nature source is the procyanidin B2, where the flavan-3-ols are linked via an interflavan bond between the benzylic C-4 carbon of the heterocyclic ring of the upper unit and the C-8 carbon of the flavan-3-ol A-ring of the lower unit (4→8) (Figure 1; Stoupi et al., 2010a,c). There has been a great increase in knowledge about the bioactivities of procyanidins extraction from natural sources rich in procyanidin B2. However, the absorption and metabolism progress of procyanidin B2 was complex. Procyanidin B2 has been reported to be slightly absorbed without conjugation or methylation (Clifford, 2004; Appeldoorn et al., 2009b; Stoupi et al., 2010a). Interestingly, the beneficial effects of procyanidin B2 may in part be due to the metabolites formed in the tissue rather than the parent compounds (Del Rio et al., 2013; Guerrero et al., 2013). Furthermore, Spencer et al. (2001) found that (−)-epicatechin was the major metabolites of purified procyanidin B2 after transfer across the rat small intestine via LC-MS technology. Shoji et al. (2006) detected methylated B-type dimmer in rat plasma after oral administration apple procyanidins using the Porter method and LC-MS. However, the metabolites of procyanidin B2 have not been still completely clear, and only a few metabolites forms of procyanidin B2 were found in vivo, such as methylation, phenolic acids, and monomer forms.
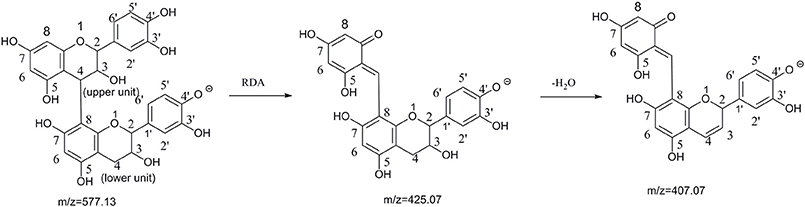
Figure 1. The pathway of retro-Diels-Alder(RDA) cleavage of procyanidin B2 and binding site label (Sun and Miller, 2003; Stoupi et al., 2010b).
To gain a more comprehensive understanding of the metabolism of procyanidin B2, it was necessary to profile metabolites and reveal the distribution of these metabolites in vivo, but the metabolites profiling of procyanidin B2 was difficult because of little structural information reported previously. Therefore, one powerful analysis technology is essential. In the past few years, triple quadrupole mass spectrometry coupled with chromatographic separation has become a powerful and frequently used technique for metabolite identification, whereas they are challenged for complex biological samples containing many compounds because of low sensitivity in full scan mode. Notably, limited reports using one novel strategy demonstrated that LC combined with hybrid ion trap/time-of flight mass spectrometry (LC-IT-TOF) could be effectively applied to discover the metabolites of polyphenolic compounds, such as (+)-catechin, (−)-epicatechin, and Paeoniae Radix Rubra extract (Liang et al., 2013, 2014). Undoubtedly, LC-IT-TOF technology characterized with a wide mass range, high sensitivity in full scan mode, high resolving power, and multistage fragmentations enables to the powerful identification of functional compounds undergoing multiple and unpredictable metabolites from biological matrix in lack of authentic compounds for verification (Ferrer and Thurman, 2007; Wang et al., 2009; Tian et al., 2013).
Therefore, the aims of this study were to explore more information of the in vivo metabolites of procyanidin B2 (Epicatechin-(4β → 8)-epicatechin), profile the metabolites in mice tissues, and analyze the possible metabolic pathways of procyanidin B2 via the UPLC-DAD-IT-TOF-MSn technique without the help of metabolites reference standards.
Materials and Methods
Chemicals and Materials
Acetonitrile (HPLC-grade), methanol (HPLC-grade), and formic acid (HPLC-grade) were purchased from Aladdin Industrial Corporation. Ultrapure water was prepared by Milli-Q water purification system (Madrid, Spain). Sodium carboxymethyl cellulose (CMC-Na) was purchased from Aladdin Industrial Corporation. Standards of procyanidin B2 and (−)-epicatechin were purchased from Tauto Biotech Corporation. (Shanghai, China). The purities of all standards were over 99% based on HPLC analysis.
Animal Experiments
Eighteen male 10-week C57BL/6 mice (23 ± 2 g) were obtained from SLAC Laboratory Animal center (Shanghai, China). All animals were housed in an environmentally controlled breeding room (24 ± 2°C, 60 ± 10% humidity) for 7 days with unrestricted chow and water. Then, the mice were randomly divided into 2 groups with nine mice per group (Group A: the procyanidin B2 group and Group B: control group) before the beginning of experiments. The procyanidin B2 was dissolved in 0.5% CMC-Na water solution and orally administered to group A at a dose of 800 mg/kg body weight, and the mice in group B were administered 0.5% CMC-Na water solution in the same way. The mice were administered for 3 days at 9:00 am. All experiments were performed in compliance with the Chinese legislation on the use and care of laboratory animals and were approved by the Shanghai Institute of Technology Committee on Animal Care and Use.
Sample Collection and Pretreatment
Urine sample was collected within 2 days after the first administration. Finally, all urine samples from one group were merged into one sample. Whereafter, the urine samples were dried by Boyikang FD-80 freezer dryer (Beijing, China). Next, 1.00 g of dried sample was redissolved by 2 mL methanol. Then, the solution was centrifuged at 2,795 g for 15 min (Sigma 3k-18, Sigma, Germany) after ultrasonic bath for 30 min. Afterward, the supernatant was stored in −80°C(MDF-1156, SANYO, Japan) before the next step of analysis (Liang et al., 2013).
The blood samples were collected in heparinized tubes at 1, 1.5, and 2 h by extracting eye bowl after the first administration of the last day under anesthesia state, and then centrifuged for 15 min at 1,789 g for obtaining the plasma samples. All plasma samples from one group were merged into one sample. Finally, each plasma sample (3 ml) was deproteinized with ice-cold acetonitrile (9 ml) in an ultrasonic bath for 20 min, and then was centrifuged at 2,795 g for 15 min to remove precipitated protein, and the solid residue was washed for three times using ice-cold acetonitrile (1 ml). Then, the merged supernatant was dried with a gentle flow of nitrogen in ice bath. The residue was redissolved in 3 ml methanol and was centrifuged at 2,795 g for 15 min after vortex oscillation. The supernatant was reconstituted to dryness, and was quickly redissolved in 200 μL methanol with vortex oscillation. The treated samples were centrifuged at 11,180 g for 10 min before injecting the system of high-performance liquid chromatography.
The pretreatment of heart, liver, brain, spleen, lungs, kidneys, or small intestine was homogenized by a homogenizer (PRO 200, Oxford, USA), which was suspended in methanol (1/10, g/ml). Then, the homogenates were in the ice-cold ultrasonic bath for 30 min and centrifuged at 2,795 g for 15 min. Afterward, the supernatant was evaporated to dryness in vacuum at 45°C, and then quickly redissolved in 2 mL methanol. The pretreated samples were stored at −80°C before analysis. All the samples were filtered through 0.45 μm nylon membrane before injecting the system (Liang et al., 2013).
Instruments and Conditions
The UPLC-DAD-ESI-IT-TOF analysis of the metabolites was performed using Shimadzu LC-30AD instrument equipped with two LC-30AD pumps, an SIL-30AC autosampler, SPD-M20A, CTO-20AC column oven, and CBM-20A system controller, and coupled to an IT-TOF mass spectrometer with an ESI interface.
The chromatography separations were performed on a Shim-pack HR-ODS column (150 mm × 2.1 mm, 3 μm). The column was eluted with a gradient mobile phase A of water-formic acid (100: 0.1, v/v) and B of acetonitrile, The elution gradient was 0–2 min, 5–10% B, 2–4 min, 10–12% B, 4–7 min, 12–18% B, 7–13 min, 18–27% B, 13–32 min, 27–56% B, 32–37 min, 56–100% B, and 37–47 min 100% B. The flow rate was 0.2 mL/min; the injection volume was 5 μL and the column oven temperature was kept at 30°C.
The tandem mass spectrometry analyses were carried out on an IT-TOF (Shimadzu, Japan) with the full scan over m/z 100–900 (MS1) and m/z 50–900 (MS2 and MS3) in the ion model of negative(NI) and positive(PI). The parameters were as follows: heat block and curved desolvation line temperature, 200°C; nebulizing nitrogen gas flow, 1.5 L/min; interface voltage: (+), 4.5 kv; (−), 3.5 kv; detector voltage, 1.56 kv; relative collision-induced dissociation energy, 50%.
Methods of Analyzing the Metabolites
To find more information of metabolites, in this study, we adopted a specially pattern for metabolites screening and identification. The procedures included three steps, (1) collecting the information of metabolites of procyanidin B2 by searching the literature databases and bioinformatic databases on line (Chemspider: http://www.chemspider.com, http://metlin.scripps.edu/); (2) searching the possible mass-to-charge ratio of metabolites by extracted ion chromatograms (EICs) in the total ion chromatograms (TICs) comparing with the TICs of control group using Profiling Solution version 3.60 and metabolites with the aid of MetlID Solution 1.2 software; (3) the known metabolites were confirmed by the databases described above. The unknown metabolites were tentatively identified in several steps: firstly, the type of substitution was identified based on the characteristic fragment ions and neutral losses in MSn spectrum; second, the skeleton structure was speculated in comparison with the mass fragmentation patterns of reference standard procyanidin B2; third, the proposed isomeric metabolites were further confirmed by characteristic fragment ions in MSn spectrum, n-octanol/water partition coefficient (ClogP value) and predominant conjugation site reported by other literature (Liang et al., 2013, 2014). All the data was processed by Shimadzu software of Formula Predictor version 1.20.
Results and Discussion
Fragmentation Behaviors of the Reference Compounds in Negative Ion Mode
Since it has been well proven that the mass fragmentation patterns of metabolites are always similar to the parent compounds, the fragmentation patterns analysis of parent compound is very helpful for the metabolite characterizations. Therefore, this study conducted a detailed fragmentation of procyanidin B2 and (−)-epicatechin authentic standard in UPLC-DAD-ESI-IT-TOF (shown in Figure 2), which provided guidance in the subsequent structure of metabolites. Procyanidin B2 showed the [M-H]− at m/z 577.1260 (C30H25O12), which was broken into characteristic fragment ions at m/z 425.0845 (C22H17O9), 407.0764 (C22H15O8), 289.0715 (C15H14O6), and m/z 245.0796 (C14H14O4) in the MS2 spectrum. The (−)-epicatechin showed the characteristic fragment ions at m/z 245.0799 (C14H14O4), 205.0476 (C11H10O4), 137.0258 (C7H6O3), and m/z 125.0258 (C6H6O3).
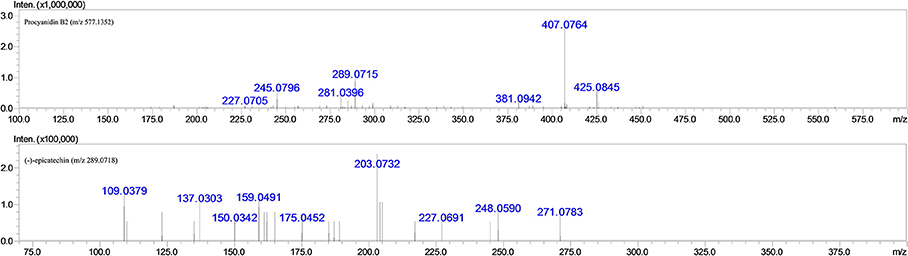
Figure 2. MS2 fragmentation data obtained in negative ion detection mode of procyanidin B2 and (−)-epicatechin.
The MS full-scan might reveal all the ion chromatograms of the expected or unexpected metabolites. The peaks detected in the ion chromatograms of sample group correspond to the mass-to-charge ratio (m/z) of the possible metabolites compared with the ion chromatograms of control group (shown in Figures 3 and 4). According to predicted gains and neutral losses in molecular masses of the metabolites compared with the molecular mass of the parent compound, the multiple metabolites from biological matrix could be effectively revealed based on the advantage of LC-IT-TOF technology in absence of authentic standards for calibration compounds.
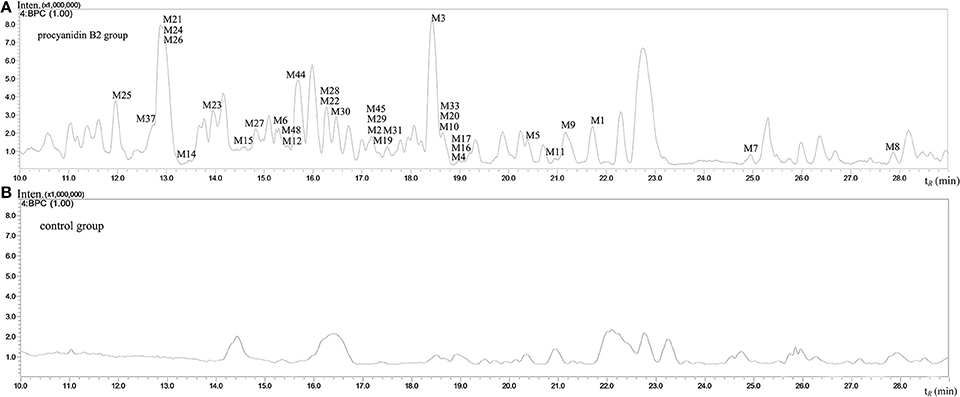
Figure 3. The negative ion base peak chromatograms of mice urine. (A) The BPC of urine of Procyanidin B2 group; (B) The BPC of urine of control group.
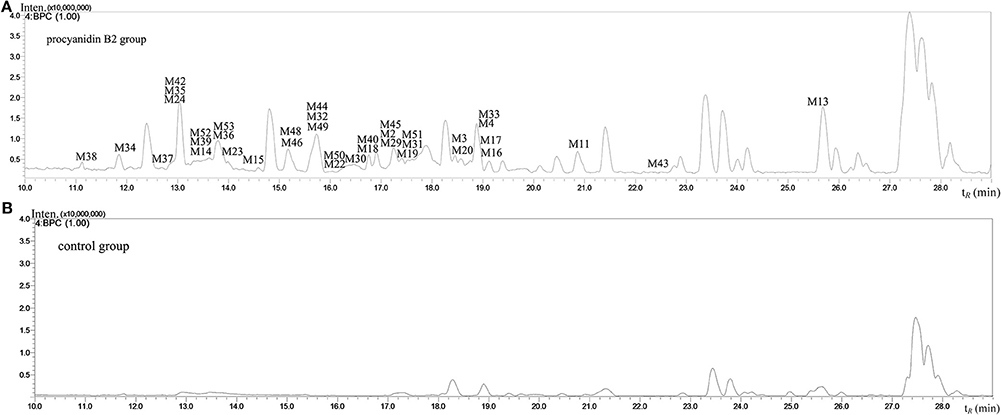
Figure 4. The negative ion base peak chromatograms of mice plasma. (A) The BPC of plasma of Procyanidin B2 group; (B) The BPC of plasma of control group.
Characterization of Metabolites of Procyanidin B2
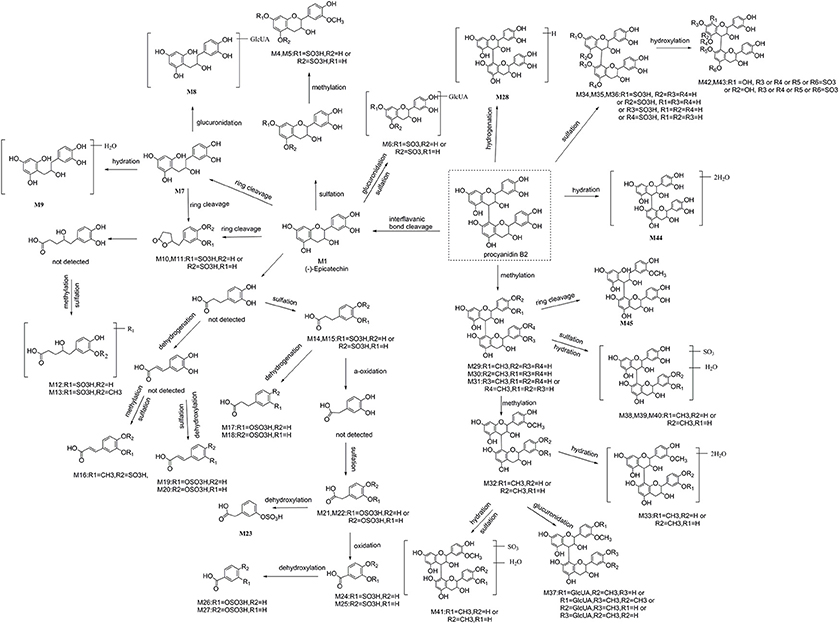
A total of 53 compounds were detected as metabolites of procyanidin B2 (45 of them were identified). As for 45 preliminarily identified metabolites, 27 metabolites (Mr < 577.13) were degraded from procyanidin B2 by cleavage of the flavanol interflavanic bond C4–C8. Additionally, 14 conjugates were formed, including 9 methylation metabolites, 8 sulfation conjugates, 5 hydration metabolites, 2 hydroxylation metabolites, 1 hydrogenation metabolite, and 1 glucuronidation metabolite. Interestingly, 24 compounds as metabolites of procyanidin B2 were newly found, including 8 methylation metabolites, 9 sulfation conjugates, 2 hydroxylation metabolites, 7 hydration metabolites, 1 hydrogenation metabolite, 1 glucuronidation metabolite, and 8 unidentified metabolites. The data was summarized in Tables 1, 2.
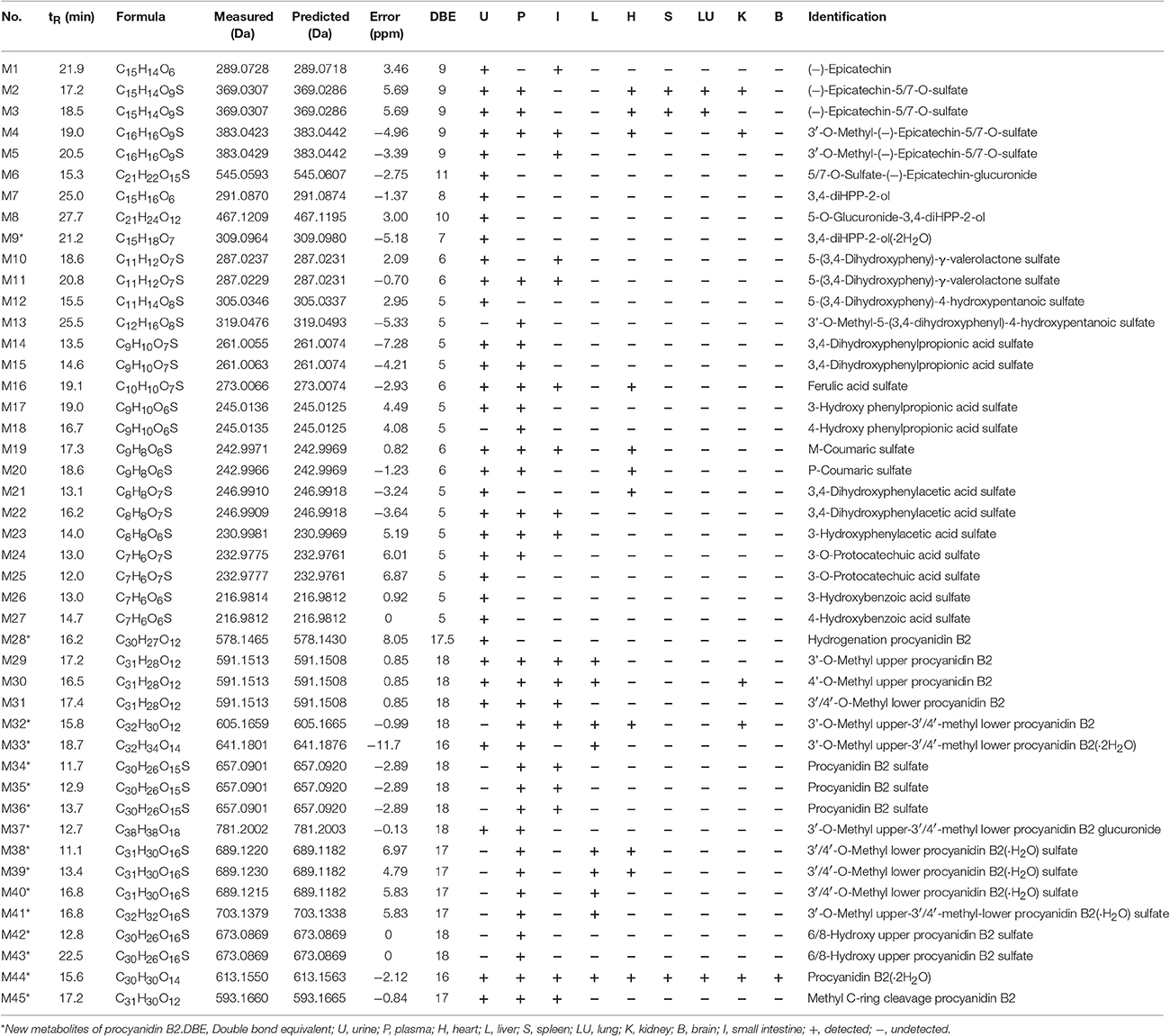
Table 1. MS data obtained in negative ion detection mode of 45 identification metabolites in urine, plasma, and organ distributions of mice.
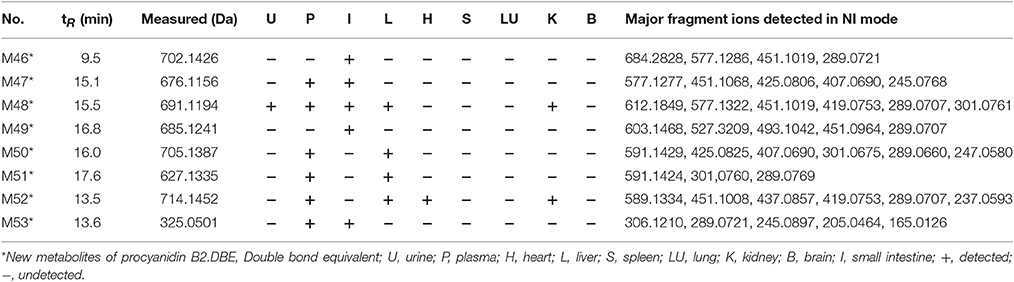
Table 2. MSn data obtained in negative ion detection mode of 8 unidentified metabolites in urine, plasma, and organ distributions of mice.
Identification of the Monomer Metabolites: M1
M1 was calculated to be C15H14O6 on the basis of high-resolution mass spectrometry (HRMS) data. Since the typical fragment ions at m/z 245.07, 205.04, and 137.02 were noticed, which were in high accordance with the reference (−)-epicatechin, as generated by Retro-Diels-Alder (RDA) cleavage in accordingly with the previous reports (Liang et al., 2014), the M1 was confirmed as (−)-epicatechin formed by cleavage of interflavanic bond of procyanidin B2.
Conjugated Metabolites of (−)-Epicatechin: M2–M6
In the MSn spectra of M2-M6, the same fragmentation ions at m/z 289.06, 245.07, and 137.02 were found, which were similar with the fragment behaviors of (−)-epicatechin. Therefore, M2-M6 were the metabolic compounds of (−)-epicatechin. The molecular formula of M2 and M3 showed the same [M-H]− ion at m/z 369.03, which could fragment into [M-SO3-H]− at m/z 289.07 by neutral loss of 79.95 Da. Since the typical A-ring fragment ions at m/z 216.98 and 137.02 of M2 and M3 were noticed, which were produced by RDA cleavage, we could deduce that the sulfonic group should be conjugated at A-ring. Hence, it was concluded that M2 and M3 were (−)-epicatechin-5/7-O-sulfate. M4 and M5 manifested the same [M-H]− at m/z 383.04 in their MSn spectra, and the same [M-CH2-H]− at m/z 369.03 was detected by neutral loss of 14.01 Da (elemental composition: CH2). Accordingly, M4 and M5 were presumed to be methylation M2/M3 with the preferable site of C-3′-OH of 3′-O-methyl-5/7-O-sulfate (−)-epicatechin (Selma et al., 2009; Liang et al., 2014). Because the molecular formula of M6 was determined to be C21H22O15S with the [M-C6H8O6-H]− at m/z 369.03 by a neutral loss of 176.03 Da (elemental composition: C6H8O6) in the NI MS2 spectra, M6 was supposed to be one metabolite of (−)-epicatechin, which was formed through glucuronidation and sulfation. But we could not determine its exact binding site, so it was tentatively inferred to be 5/7-O-sulfate-epicatechin-O-glucuronide (Liang et al., 2014).
Metabolites from C-Ring Cleavage of (−)-Epicatechin Monomer: M7–M9
The molecular formulas of M7-M9 were calculated to be C15H16O6, C21H24O12, and C15H18O7, respectively, with the [M-H]− at m/z 291.08, 467.12, and 309.09 based on the HRMS data. In the NI MSn spectra, M7, M8, and M9 had the same characteristic fragment ions at m/z 247.09 (elemental composition: C14H15O4), 167.03 (elemental composition: C8H7O4), and 123.04 (elemental composition: C7H7O2), showing the similar fragmentation pathways of 1-(3′,4′-dihydroxyphenyl)-3-(2′,4′, 6′-trihydroxyphenyl)-propan-2-ol (3, 4-diHPP-2-ol) (Liang et al., 2014; Margalef et al., 2014). As a result, M7 was assigned as 3,4-diHPP-2-ol. M8 showed the [M-C6H8O6-H]− at m/z 291.08 by neutral loss 176.03 Da (elemental composition: C6H8O6), so it was preliminarily identified as glucuronic acid conjugates of 3,4-diHPP-2-ol. According to the glucuronidation of (−)-epicatechin preferably occurred at the C-5-OH of A-ring (Shali et al., 1991), M8 was tentatively identified as 5-O-glucuronide-3,4-diHPP-2-ol. As for M9, the neutral loss of 18.01 Da (elemental composition: H2O) was observed. Therefore, M9 was identified as hydration products of 3,4-diHPP-2-ol.
Phenylvalerolactone Metabolites: M10–M11
The molecular formula of M10-M11 were calculated to be C11H12O7S with the same [M-SO3-H]− at m/z 207.06 by the neutral loss of 79.95 Da (elemental composition: SO3), and the precursor ion [M-SO3-H]− at m/z 207.06 can break into characteristic fragment ions at m/z 163.07, and m/z 122.04. Moreover, the fragmentation pathways of the aglucon were in good accordance with that of 5-(3,4-dihydroxyphenyl)-γ-valerolactone (Appeldoorn et al., 2009a; Urpi-Sarda et al., 2009). Thus, they were confirmed to be 5-(3,4-dihydroxyphenyl)-γ-valerolactone sulfate.
Aromatic Acid Metabolites: M12–M27
Summarily, 16 aromatic acid metabolites are tentatively identified, including 2 phenylvalenic acid metabolites, 7 phenylpropionic acid metabolites, 3 phenylacetic acids metabolites, and 4 benzoic acid metabolites. Their structures are shown in Figure 5.
Phenylvalenic Acid Metabolites: M12–M13
The molecular formula of M12 was C11H14O8S with [M-H]− at m/z 305.03, yielding fragmentation ions at m/z 225.07, 207.06, and 163.07, which were same with the characteristic fragment ions of 5-(3,4-dihydroxyphenyl)-γ-valerolactone. Since the neutral loss of 79.95 Da (elemental composition: SO3) was observed in their HRMS spectra, M12 was tentatively identified as the ring open product of 5-(3,4-dihydroxyphenyl)-γ-valerolactone sulfate with molecular formula of C11H12O7S of 5-(3,4-dihydroxyphenyl)-4-hydroxypentanoic acid sulfate (Liang et al., 2013). As the molecular weight of M13 was 14.01 Da (elemental composition: CH2) larger than M12, M13 was determined to be methyl M12, Furthermore, according to the major methylation site of (−)-epicatechin confirmed by the previous reports, the methyl conjugates site was on the C-3 position of M12 with the similarly conjugation rule (De Pascual-Teresa et al., 2000; Ou and Gu, 2014).
Phenylpropionic Acid Metabolites: M14–M20
In the NI MSn spectra of M14 and M15, [M-SO3-H]− at m/z 181.05 formed by the neutral loss of 79.95Da (elemental composition: SO3) was detected, the characteristic ions yielded by aglucon at m/z 137.06, and 122.04 in the MSn spectra were similar with the fragment behaviors of the 2-(3-hydroxy-4-methoxyphenyl) acetic acid (Liang et al., 2014). Hence, M14 and M15 were provisionally identified as 3,4-dihyroxyphenylpropionic acid sulfate. M16 showed molecular ion [M-H]− at m/z 273.00, indicating that it was calculated to be C10H10O7S. Furthermore, the characteristic fragmentation ions at m/z 193.05 and 134.04 were observed, which were in high accordance with those of the ferulic acid according to the previous literature (Liang et al., 2014). Accordingly, M16 was identified to be ferulic acid sulfate. The molecular formula of M17 and M18 were determined to be C9H10O6S based on their HRMS data. In their NI MSn spectra, the [M-SO3-H]− at m/z 165.05 was detected, implying the neutral loss of 79.95 Da (elemental composition: SO3). Moreover, the fragment of [M-SO3-H]− was identical to be hydroxy phenylpropionic acid because of previously reported (Urpi-Sarda et al., 2009). Therefore, M17 and M18 were identified as hydroxy phenylpropionic acid sulfate. Furthermore, Gonthier et al. reported that 3-hydroxy phenylpropionic acid was a preferred metabolite of (−)-epicatechin (Gonthier et al., 2003). Meanwhile, the peak area of M17 (peak area: 381, 733) was much larger than M18 (peak area: 268, 056). Accordingly, we assigned M17 as 3-hydroxy phenylpropionic acid sulfate and M18 as 4-hydroxyphenylpropionic acid sulfate. The molecular formula of M19 and M20 were identified as C9H8O6S based on the HRMS data. In their NI MSn spectra, the [M-SO3-H]− at m/z 163.04 was observed, which was formed by a neutral loss of 79.95 Da (elemental composition: SO3). The fragmentation behaviors of [M-SO3-H]− were the same as coumaric acid. Moreover, the m-coumaric acid was preferred metabolite in mice in comparison with p-coumaric acid (Touriño et al., 2011). Hence, M19 with a bigger peak area (peak area: 110, 777) was found to be m-coumaric sulfate, and the M20 (peak area: 69, 181) was identified as p-coumaric sulfate.
Phenylacetic Acids Metabolites: M21–M23
M21 and M22 showed the same [M-H]− ion at m/z 246.99, and yielded fragment ions at m/z 167.03 formed by the neutral loss of 79.95 Da (elemental composition: SO3). In addition, the characteristic fragmentation ions at m/z 123.04 of 3,4-dihydroxyphenylacetic acid for only characteristic catabolism of procyanidin B2 via á-oxidation of 3,4-dihydroxyphenylpropionic acid was also observed (Spencer et al., 2000; Gu et al., 2004). Hence, M21 and M22 were 3,4-dihydroxyphenylacetic acid sulfate. The molecular ion [M-H]− of M23 at m/z 230.99 was observed in the MSn spectra, which yielded the fragment ions at m/z 151.04, generated by a neutral loss of 79.95 Da (elemental composition: SO3). Moreover, the characteristic fragmentation at m/z 107.05 of 3-hydroxyphenylacetic acid was also detected in the MSn spectra of M23 (Urpi-Sarda et al., 2009). As results, M23 was determined to be 3-hydroxyphenylacetic acid sulfate.
Benzoic Acid Metabolites: M24–M27
M24 and M25 showed the same molecular ion at m/z 232.97, and yielded [M-SO3-H]− at m/z 153.02 in their NI MSn spectra, which was formed by a neutral loss of 79.95 Da (elemental composition: SO3), suggesting that they were sulfate conjugates. Moreover, the [M-SO3-H]− was identified to be protocatechuic acid comparing with the previous studies. Therefore, M24 (peak area: 1, 692, 371) was identified as 3-O-protocatechuic acid sulfate, while M25 (peak area: 624, 480) was assigned as 4-O-protocatechuic acid sulfate according to the previous reports (Vasudevan and Mahadevan, 1990; Urpi-Sarda et al., 2009). M26 and M27 showed the same molecular ion [M-H]− at m/z 216.98 with the molecular formula of C7H6O6S, and yielded fragment ions at m/z 137.02 and m/z 93.03, which were the characteristic fragment ions of 3/4-hydroxybenzoic acid (Gonthier et al., 2003). Accordingly, they were identified as 3/4-hydroxybenzoic acid sulfate. Moreover, 3-hydroxybenzoic acid sulfate had a smaller ClogP (−1.409) than 4-hydroxybenzoic acid sulfate (ClogP = 0.345), which suggested that the former should have a shorter retention time. Accordingly, M26 (tR = 13.0 min) was tentatively identified as 3-hydroxybenzoic acid sulfate, and M27 (tR = 17.7 min) was 4-hydroxybenzoic acid sulfate.
Conjugated Metabolites of Procyanidin B2 (Mr > 577): M28–M44
A total of 17 conjugated metabolites were identified, which had the same characteristic fragmentation behaviors at m/z 425.08 and 407.07, as generated from RDA cleavage (Shown in Figure 1) of procyanidin B2.
Hydrogenation Reaction of Procyanidin B2: M28
M28 showed the molecular ion at 578.14, and yielded the [M-2H]− at m/z 577.13 (elemental composition: C30H26O12), which could produce characteristic fragmentations of m/z 425.08, 407.07, and 289.07. And the fragmentation behaviors of the aglucon were identical to that of procyanidin B2 by comparison with the reference compounds. Moreover, M28 increased the metabolite mass by 1.01 Da in the structure of procyanidin B2. As a result, M28 was inferred to be product of procyanidin B2 hydrogenation reaction.
Metabolites Formed through Methylation: M29–M33
M29, M30, and M31 had the same molecular ion at m/z 591.15, and the [M-CH2-H]− at m/z 577.13 was observed by a neutral loss of 14.01 (elemental composition: CH2) in their NI MSn spectra. In the NI MSn spectra of M29 and M30, the fragment ions of m/z 303.08, 407.07, and 289.06 were also detected. Therefore, the methylation site was likely on the B-ring of upper (−)-epicatechin unit of procyanidin B2. According to the previously confirmed major methylation site of (−)-epicatechin of 3′-OH, similarly, M29 (peak area: 252, 351) was tentatively identified as 3′-O-methyl upper (−)-epicatechin unit of procyanidin B2 because of its massive quantity, and M30 (peak area: 172, 287) was determined to be 4′-O-methyl upper (−)-epicatechin unit (Shali et al., 1991; Stoupi et al., 2010a). Based on the MSn data of M31, the characteristic fragment ions at m/z 407.07 and 421.04 were observed, which were formed by RDA cleavage (shown in Figure 1). Accordingly, M31 was tentatively identified to be 3′/4′-O-methyl lower (−)-epicatechin unit. Based on the HRMS data of M32, the molecular formula of the metabolites was determined to be C32H30O12, with two additional CH2 units in comparison with procyanidin B2. In the NI MSn spectra of M32, the characteristic fragment ions at m/z 407.08 and 421.09 were also observed. Hence, it could be concluded that a CH2 unit should be in the 3′-OH or 4′-OH of lower (−)-epicatechin unit. According to the confirmed major methylation site of M29, M32 was temporarily identified as 3′-O-methyl upper (−)-epicatechin-3′/4′-methyl lower (−)-epicatechin.
The molecular formula of M33 was C32H34O14, as calculated from its HRMS data with m/z at 641.18, which yielded [M-2H2O-H]− at m/z 605.16 formed by a neutral loss of 36.02 Da (elemental composition: H4O2) in the NI MSn spectrum. Therefore, M33 was tentatively identified as double hydration M32.
Metabolites Formed through Sulfation: M34–M36
In the NI MSn spectra of M34, M35, and M36, the same [M-SO3-H]− at m/z 577.13 was detected, which was formed by a neutral loss of 79.95 Da (elemental composition: SO3). In addition, the [M-SO3-H]− could yield the fragment ions at m/z 407.07 and 425.07, which were the RDA characteristic fragments of procyanidin B2 (shown in Figure 1). As a result, they were identified as procyanidin B2 sulfate. Furthermore, the [M-SO3-H]− at m/z 369.02, 216.98, and 137.02 were observed, and sulfate (−)-epicatechin preferably occurred at C-5-OH or C-7-OH based on several previous studies. Hence, sulfonic group was concluded to conjugate to the 5/7-OH of A-ring of upper or lower (−)-epicatechin unit, however the accurate site of M34, M35, and M36 was unclear (Liang et al., 2013, 2014).
Metabolites Formed through Methylation and Glucuronidation: M37
M37 had the molecular formula of C38H38O18, as calculated from their HRMS data. In their NI MSn spectra, the [M-C6H8O6-H]− at m/z 605.16 was observed, which was formed by a neutral loss of 176.03 Da (elemental composition: C6H8O6). Further, the characteristic fragmentation ions at m/z 245.08, m/z 421.10 were noticed, which were formed by the RDA cleavage of (−)-epicatechin unit, suggested that the B-ring was substituted by a glucuronic group (Liang et al., 2014). Therefore, the glucuronidation group of M37 was supposed to be in 4′-O-upper (−)-epicatechin unit of M32 or 3′/4′-O-lower (−)-epicatechin unit of M32.
Metabolites Formed through Methylation and Sulfation: M38–M41
In the MSn spectra of M38, M39, and M40, the same molecular ion [M-H]− at m/z 689.12 was found, which yielded the fragmentation ions at m/z 591.15, 407.07, and 289.06. Thus, they were identified to be the metabolites of procyanidin B2. Moreover, the molecular weight were 97.97 Da (elemental composition: H2O and SO3) larger than the [M-H]− at m/z 591.15 of M31, and the characteristic fragment ions at m/z 451.09, 465.11 were detected in the NI MS2 spectra. Hence, the methyl group was lined to the C-3′/4′-OH of lower (−)-epicatechin unit, but the exact site could not be determined, and the binding sites of hydration and sulfation were also unclear. In the MSn spectra of M41, the [M-H]− at m/z 703.13 was observed, which yielded the fragment ions at m/z 605.16 and m/z 289.06. Additionally, the molecular weight were 97.97 Da (elemental composition: H2O and SO3) larger than the [M-H]− at m/z 605.16 of M32. As results, it was presumably identified to be the metabolites of procyanidin B2 of hydration sulfate M32.
Metabolites Formed through Hydroxylation and Sulfation: M42–M43
The M42 and M43 showed the same molecular ion at m/z 673.08, which were 16.01 Da (elemental composition: O) larger than molecular ion at m/z 657.09 of M34, suggesting that they contained one additional oxygen atom. M42 and M43 produced an associated RDA fragment at m/z 423.07, 407.07, and a MS2 fragmentation ion at m/z 289.06 possibly upon cleavage of the interflavan bond and the release of an unchanged (−)-epicatechin unit (Stoupi et al., 2010a). This observation indicated that the oxygen was not inserted in the lower flavanol unit. M42 and M43 were assigned as tentatively as either 8-hydroxy M34 or 6-hydroxy upper (−)-epicatechin unit of M34.
Metabolites Formed through Hydration: M44
The molecular formula of M44 were calculated to be C30H30O14 with the [M-H]− at m/z 613.15 based on the HRMS data. The molecular weight of M44 was 36.02 Da (elemental composition: H4O2) larger than 577.13, which yielded the same characteristic ions as those of the reference standard procyanidin B2. Therefore, M44 was tentatively identified as double hydration metabolite of procyanidin B2.
Metabolites Formed by C-Ring Cleavage in Upper(−)-Epicatechin Unit of Procyanidin B2: M45
M45 had the molecular formula C31H30O12 with the [M-CH2-H]− at m/z 579.15 and m/z 289.07 in the NI MSn spectra, which was confirmed as methyl C-ring cleavage of procyanidin B2 based on previous reports. Although there should be two potential sites for reductive cleavage of the C-ring (upper unit and lower unit), the site of upper (−)-epicatechin unit was more accessible to reaction (Stoupi et al., 2010b). As results, M45 could be defined as upper C-ring cleavage of M29.
Unidentified Metabolites of Procyanidin B2: M46–M53
In NI MSn spectra of all compounds, the [M-H]− at m/z 702.1426, 676.1156 691.1194, 685.1241, 705.1387, 627.1335, 714.1452, and 325.0501 were detected, respectively, which were identified as the metabolites of procyanidin B2 in comparison with the fragmentation behaviors (shown in Table 2). In addition, m/z 591.14 was observed in MSn spectra of M50 and M51, respectively. Hence, M50 and M51 should be the methyl procyanidin B2, but the structure of all compounds consisting of complicated aglucon and misty conjugated sites were not fully identified with pure high-resolution mass spectrometry data in the absence of applicable standard and guidance from previous literature.
Possible Metabolic Pathway of Procyanidin B2 in Mice
The MSn data with high mass accuracy provided much information to tentatively analyze the structure of metabolites under negative ion mode. The primary objective was to characterize the relatively comprehensive metabolites of the procyanidin B2 in mice in the present study. Therefore, we analyzed and compared the metabolites in urine, plasma, and organ tissue samples following oral routes of administration. The base peak chromatograms of the mice urine and plasma samples detected in negative ion mode are shown in Figures 3 and 4, respectively. 53 metabolites of procyanidin B2 (24 new metabolites) showed that the procyanidin B2 was metabolized diversely (shown in Tables 1, 2). Although the previous study reported that glucuronide or sulfate metabolites of dimeric procyanidins had not been detected in biological fluids, we identified the intact procyanidin B2 conjugates of 10 sulfate metabolites and 1 glucuronide metabolites in this work (Appeldoorn et al., 2009a,b). Meanwhile, aromatic acid metabolites were very common in the metabolic pathways of procyanidins after cleavage of the interflavan bond.
Furthermore, a possible metabolic pathway was proposed in Figure 5. The metabolites of procyanidin B2 could be categorized into two major groups: (a) cleavage of interflavanic bond (C4–C8) of procyanidin B2 (Mr < 577) and formation of small molecule material, including (−)-epicatechin, C-ring cleavage of epicatechin metabolites, phenylvalerolactone metabolites, and aromatic acid metabolites; (b) metabolites with intact interflavanic bond C4–C8, including procyanidin B2 conjugates, partial heterocyclic ring cleavage products, and their conjugates.
In general, the conjugation process (glucuronidation, sulfation, and methylation) affected the bioactive properties of the parent compound. Sulfation and glucuronidation had been reported to improve a considerable attenuation of biological activity. Additionally, the case of methylation seemed to be more complex because the incorporation of methyl groups reduced the number of available OH group, but increased the lipophilic nature of the compound, which could be advantageous for cellular uptake by passive diffusion (Donovan et al., 2007; Monagas et al., 2010). In particular, we found 32 sulfation metabolites, 3 glucuronidation metabolites, and 14 methylation metabolites, respectively. Besides, M4 and M5 exhibited the potential inhibition of lipid oxidation and inhibition of apoptosis induced by oxidized LDL. M1 was identified as beneficial effects in vascular disaster (Schroeter et al., 2001; Su et al., 2004). In the case of M10, M11, M21, and M22, they have been reported to be a slight increase in antioxidant activity (Grimm et al., 2004). In addition, the biological activities of metabolites of procyanidin B2 were still largely unknown, thus the bioactivities of metabolites were also worthy to further research.
Distribution of the Procyanidin B2 in Mice
In this work, the metabolic profile of procyanidin B2 in mice urine, plasma and organs were qualitatively described by UPLC-DAD-ESI-IT-TOF method. The numbers of all detected metabolites in plasma, urine, small intestine, liver, heart, kidney, lung, brain and spleen were 37, 34, 23, 13, 12, 7, 3, 1, and 3, respectively.
According to previous reports, liver had more importance in the metabolization process of polyphenols showing a higher metabolite concentration (Appeldoorn et al., 2009a). In this work, all metabolites found in the liver was Mr > 577, which indicated that cleavage of interflavanic bond could not occur in the liver, and conjugation reaction (glucuronidated, sulfated, and methylated) was the major reaction formation with intact procyanidin B2. 34 metabolites in urine and 5 in the kidney showed that the urinary path was the main procyanidin B2 excretion pathway, which was consistent with the conclusion of previous literature (Serra et al., 2011). There were 12 metabolites observed in the heart, which would enhance their described beneficial effects. The presence of products in such tissues as the heart may be related to the potential health benefits of procyanidin B2, especially in the context of cardiovascular health effect (Fraga et al., 2011). 23 metabolites were detected in small intestine, including M1, M4–M5, M10–M11, M16, M19, M22–M23, M29–M32, M34–M36, M44–M45, M46–M49, and M53. Furthermore, 14 metabolites were the conjugation metabolites with intact procyanidin B2, suggesting that the procyanidin B2 could pass intact through the gastrointestinal tract, reaching the small intestine where it was transformed by the intestinal microbiota before absorption, and these metabolites could reach the systemic circulation to be transported to other tissue where they could be further biotransformed according to the previous reports (Aura, 2008; Crozier et al., 2009; Monagas et al., 2010; Margalef et al., 2015).
Conclusions
It was the first time that the absorption and metabolism of oral administration of pure procyanidin B2 in mice was explored using UPLC-DAD-ESI-IT-TOF in the present study. In total, 45 metabolites of procyanidin B2 were preliminarily identified, and 24 of them were firstly reported. The results showed that the small intestine represented the largest proportion of metabolites in comparison with other tissues, and liver was the major conjugated reaction organ tissue of procyanidin B2. Meanwhile, aromatic acid metabolites were very common in the metabolic pathways of procyanidin B2 after cleavage of the interflavan bond. In conclusion, we profiled the metabolites of procyanidin B2 in mice and revealed the distribution of metabolites. Our findings improve understanding of the metabolism and bioactive action mechanism of procyanidin B2. As for 24 new metabolites, the conjugation types and structure skeletons could be preliminarily determined using the IT-TOF technique. To ascertain their exact structures, the characterization of metabolites of procyanidin B2 in vivo should be further confirmed in the future work.
Author Contributions
YX designed the experiments and wrote the paper. ZH and ZY performed the experiments and analyzed data. YZ analyzed data and contributed analysis tools. TL performed the experiments. XZ conceived the experiments and revised the paper. DC analyzed data and revised the paper.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was financially supported by Program of National Natural science foundation of China (grant no. 31401504).
References
Appeldoorn, M. M., Vincken, J.-P., Aura, A.-M., Hollman, P. C. H., and Gruppen, H. (2009a). Procyanidin dimers are metabolized by human microbiota with 2-(3,4-dihydroxyphenyl)acetic acid and 5-(3,4-dihydroxyphenyl)-γ-valerolactone as the major metabolites. J. Agric. Food Chem. 57, 1084–1092. doi: 10.1021/jf803059z
Appeldoorn, M. M., Vincken, J. P., Gruppen, H., and Hollman, P. C. H. (2009b). Procyanidin dimers A1, A2, and B2 are absorbed without conjugation or methylation from the small intestine of mice. J. Nutr. 139, 1469–1473. doi: 10.3945/jn.109.106765
Aura, A. M. (2008). Microbial metabolism of dietary phenolic compounds in the colon. Phytochem. Rev. 7, 407–429. doi: 10.1007/s11101-008-9095-3
Clifford, M. N. (2004). Diet-derived phenols in plasma and tissues and their implications for health. Planta Med. 70, 1103–l114. doi: 10.1055/s-2004-835835
Crozier, A., Jaganath, I. B., and Clifford, M. N. (2009). Cheminform abstract: dietary phenolics: chemistry, bioavailability and effects on health. Nat. Prod. Rep. 26, 1001–1043. doi: 10.1039/b802662a
Del Rio, D., Rodriguez-Mateos, A., Spencer, J. P. E., Tognolini, M., Borges, G., and Crozier, A. (2013). Dietary (poly)phenolics in human health: structures, bioavailability, and evidence of protective effects against chronic diseases. Antioxid. Redox Signal. 18, 1818–1892. doi: 10.1089/ars.2012.4581
De Pascual-Teresa, S., Santos-Buelga, C., Rivas-Gonzalo, J. C., and Agric, J. (2000). Analysis of flavanols in beverages by high-performance liquid chromatography with chemical reaction detection. Food. Chem. 48, 5331–5337. doi: 10.1021/jf000549h
Donovan, J. L., Manach, C., Faulks, R. M., and Kroon, P. A. (2007). “Absorption and metabolism of dietary plant secondary metabolites,” in Plant Secondary Metabolites: Occurrence, Structure and Role in the Human Diet, eds A. Crozier, M. N. Clifford, and H. Ashihara (Blackwell Publishing Ltd.), 303–351. doi: 10.1002/9780470988558.ch8
Ferrer, I., and Thurman, E. M. (2007). Multi-residue method for the analysis of 101 pesticides and their degradates in food and water samples by liquid chromatography/time-of-flight mass spectrometry. J. Chromatogr. A 1175, 24–37. doi: 10.1016/j.chroma.2007.09.092
Fraga, C. G., Litterio, M. C., Prince, P. D., Calabró, V., Piotrkowski, B., and Galleano, M. (2011). Cocoa flavanols: effects on vascular nitric oxide and blood pressure. J. Clin. Biochem. Nutr. 48, 63–67. doi: 10.3164/jcbn.11-010FR
Gonthier, M.-P., Donovan, J. L., Texier, O., Felgines, C., Remesy, C., and Scalbert, A. (2003). Metabolism of dietary procyanidins in rats. Free Radic. Biol. Med. 35, 837–844. doi: 10.1016/S0891-5849(03)00394-0
Grimm, T., Schäfer, A., and Högger, P. (2004). Antioxidant activity and inhibition of matrix metalloproteinases by metabolites of maritime pine bark extract (pycnogenol). Free Radic. Biol. Med. 36, 811–822. doi: 10.1016/j.freeradbiomed.2003.12.017
Guerrero, L., Margalef, M., Pons, Z., Quiñones, M., Arola, L., and Arola-Arnal, A. (2013). Serum metabolites of proanthocyanidin-administered rats decrease lipid synthesis in hepg2 cells. J. Nutr. Biochem. 24, 2092–2099. doi: 10.1016/j.jnutbio.2013.08.001
Gu, L., Kelm, M. A., Hammerstone, J. F., Beecher, G., Holden, J., Haytowitz, D., et al. (2004). Concentrations of proanthocyanidins in common foods and estimations of normal consumption. J. Nutr. 134, 613–617.
Li, S., Sui, Y., Xiao, J., Wu, Q., Hu, B., Xie, B., et al. (2013). Absorption and urinary excretion of a-type procyanidin oligomers from Litchi chinensis, pericarp in rats by selected ion monitoring liquid chromatography–mass spectrometry. Food Chem. 138, 1536–1542. doi: 10.1016/j.foodchem.2012.09.120
Liang, J., Xu, F., Zhang, Y.-Z., Huang, S., Zang, X.-Y., Zhao, X., et al. (2013). The profiling and identification of the absorbed constituents and metabolites of Paeoniae Radix Rubra decoction in rat plasma and urine by the HPLC-DAD-ESI-IT-TOF-MSn technique: a novel strategy for the systematic screening and identification of absorbed constituents and metabolites from traditional Chinese medicines. J. Pharm. Biomed. Anal. 83, 108–121. doi: 10.1016/j.jpba.2013.04.029
Liang, J., Xu, F., Zhang, Y.-Z., Zang, X.-Y., Wang, D., Shang, M.-Y., et al. (2014). The profiling and identification of the metabolites of (+)-catechin and study on their distribution in mice by HPLC-DAD- ESI-IT-TOF- MSn technique. Biomed. Chromatogr. 28, 401–411. doi: 10.1002/bmc.3034
Margalef, M., Pons, Z., Bravo, F. I., Muguerza, B., and Arola-Arnal, A. (2015). Plasma kinetics and microbial biotransformation of grape seed flavanols in rats. J. Funct. Foods 12, 478–488. doi: 10.1016/j.jff.2014.12.007
Margalef, M., Pons, Z., Muguerza, B., and Arola-Arnal, A. (2014). A rapid method to determine colonic microbial metabolites derived from grape flavanols in rat plasma by liquid chromatography-tandem mass spectrometry. J. Agric. Food Chem. 62, 7698–7706. doi: 10.1021/jf5019752
Monagas, M., Urpi-Sarda, M., Sánchez-Patán, F., Llorach, R., Garrido, I., Gómez-Cordovés, C., et al. (2010). Insights into the metabolism and microbial biotransformation of dietary flavan-3-ols and the bioactivity of their metabolites. Food Funct. 1, 233–253. doi: 10.1039/c0fo00132e
Ou, K. Q., and Gu, L. W. (2014). Absorption and metabolism of proanthocyanidins. J. Funct. Foods 7, 43–53. doi: 10.1016/j.jff.2013.08.004
Prior, R. L., and Gu, L. (2005). Occurrence and biological significance of proanthocyanidins in the American diet. Phytochemistry 66, 2264–2280. doi: 10.1016/j.phytochem.2005.03.025
Schroeter, H., Spencer, J. P. E., Rice-Evans, C., and Williams, R. J. (2001). Flavonoids protect neurons from oxidized low-density-lipoprotein-induced apoptosis involving c-jun n-terminal kinase (jnk), c-jun and caspase-3. Biochem. J. 358(Pt 3), 547–557. doi: 10.1042/bj3580547
Selma, M. V., Espín, J. C., and Tomás-Barberán, F. A. (2009). Interaction between phenolics and gut microbiota: role in human health. J. Agric. Food Chem. 57, 6485–6501. doi: 10.1021/jf902107d
Serra, A., Macià, A., Romero, M.-P., Anglès, N., Morelló, J. R., and Motilva, M. J. (2011). Distribution of procyanidins and their metabolites in rat plasma and tissues after an acute intake of hazelnut extract. Food Funct. 2, 562–568. doi: 10.1039/c1fo10083a
Shali, N. A., Curtis, C. G., Powell, G. M., and Roy, A. B. (1991). Sulfation of the flavonoids quercetin and catechin by rat liver. Xenobiotca. 21, 881–893.
Shoji, T., Masumoto, S., Moriichi, N., Akiyama, H., Kanda, T., Ohtake, Y., et al. (2006). Apple procyanidin oligomers absorption in mice after oral administration: analysis of procyanidins in plasma using the porter method and high-performance liquid chromatography/tandem mass spectrometry. J. Agric. Food Chem. 54, 884–892. doi: 10.1021/jf052260b
Spencer, J. P. E., Chaudry, F., Pannala, A. S., Srai, S. K., Debnam, E., and Rice-Evans, C. (2000). Decomposition of cocoa procyanidins in the gastric milieu. Biochem. Biophys. Res. Commun. 272, 236–241. doi: 10.1006/bbrc.2000.2749
Spencer, J. P. E., Schroeter, H., Shenoy, B., Srai, S. K., Debnam, E. S., and Rice-Evans, C. (2001). Epicatechin is the primary bioavailable form of the procyanidin dimers B2 and B5 after transfer across the small intestine. Biochem. Biophys. Res. Commun, 285, 588–593. doi: 10.1006/bbrc.2001.5211
Stoupi, S., Williamson, G., Drynan, J. W., Barron, D., and Clifford, M. N. (2010b). Procyanidin B2 catabolism by human fecal microflora: partial characterizationof ‘dimeric’ intermediates. Arch. Biochem. Biophys. 501, 73–78. doi: 10.1016/j.abb.2010.02.009
Stoupi, S., Williamson, G., Drynan, J. W., Barron, D., and Clifford, M. N. (2010c). A comparison of the in vitro biotransformation of (−)-epicatechin and procyanidin B2 by human faecal microbiota. Mol. Nutr. Food Res. 54, 747–759. doi: 10.1002/mnfr.200900123
Stoupi, S., Williamson, G., Viton, F., Barron, D., King, L. J., Brown, J. E., et al. (2010a). In vivo bioavailability, absorption, excretion, and pharmacokinetics of [14C]proscyanidin B2 in male mice. Drug Metab. Dispos. 38, 287–291. doi: 10.1124/dmd.109.030304
Su, Y. L., Xu, J. Z., Ng, C. H., Leung, L. K., Huang, Y., and Chen, Z. Y. (2004). Antioxidant activity of tea theaflavins and methylated catechins in canola oil. J. Oil Fat Indust. 81, 269–274. doi: 10.1007/s11746-004-0894-7
Sun, W. X., and Miller, J. M. (2003). Tandem mass spectrometry of the b-type procyanidins in wine and b-type dehydrodicatechins in an autoxidation mixture of (+)-catechin and (−)-epicatechin. J. Mass Spectrom. 38, 438–446. doi: 10.1002/jms.456
Terra, X., Pallarés, V., Ardèvol, A., Bladé, C., Fernández-Larrea, J., Pujadas, G., et al. (2011). Modulatory effect of grape seed procyanidins on local systemic inflammation in diet induced obesity rats. J. Nutr. Biochem. 22, 380–387. doi: 10.1016/j.jnutbio.2010.03.006
Tian, J. X., Peng, C., Xu, L., Tian, Y., and Zhang, Z. J. (2013). In vitro, metabolism study of strychnos, alkaloids using high-performance liquid chromatography combined with hybrid ion trap/time-of-flight mass spectrometry. Biomed. Chromatogr. 27, 775–783. doi: 10.1002/bmc.2860
Touriño, S., Pérez-Jiménez, J., Mateos-Martín, M. L., Fuguet, E., Vinardell, M. P., Cascante, M., et al. (2011). Metabolites in contact with the rat digestive tract after ingestion of a phenolic-rich dietary fiber matrix. J. Agric. Food Chem. 59, 5955–5963. doi: 10.1021/jf200159f
Urpi-Sarda, M., Garrido, I., Monagas, M., Gómez-Cordovés, C., Medina-Remón, A., Andres-Lacueva, C., et al. (2009). Profile of plasma and urine metabolites after the intake of almond[Prunus dulcis (Mill) D.A.Webb] polyphenols in humans. J. Agric. Food Chem. 57, 10134–10142. doi: 10.1021/jf901450z
Vasudevan, N., and Mahadevan, A. (1990). Utilization of catechin by micro organisms. Curr. Sci. 59, 1323–1325.
Veluri, R., Singh, R. P., Liu, Z. J., Thompson, J. A., Agarwal, R., and Agarwal, C. (2006). Fractionation of grape seed extract and identification of gallicacid as one of the major active constitution causing growth inhibition and apoptotic death of DU145 human prostate carcinoma cells. Carcinogenesis 27, 1445–1453. doi: 10.1093/carcin/bgi347
Wang, Y., Hao, H., Wang, G., Tu, P., Jiang, Y., Liang, Y., et al. (2009). An approach to identifying sequential metabolites of a typical phenylethanoid glycoside, echinacoside, based on liquid chromatography-ion trap-time of flight mass spectrometry analysis. Talanta 80, 572–580. doi: 10.1016/j.talanta.2009.07.027
Keywords: procyanidin B2, metabolic profile, identification, distribution, UPLC-DAD-ESI -IT-TOF-MSn
Citation: Xiao Y, Hu Z, Yin Z, Zhou Y, Liu T, Zhou X and Chang D (2017) Profiling and Distribution of Metabolites of Procyanidin B2 in Mice by UPLC-DAD-ESI-IT-TOF-MSn Technique. Front. Pharmacol. 8:231. doi: 10.3389/fphar.2017.00231
Received: 09 February 2017; Accepted: 12 April 2017;
Published: 04 May 2017.
Edited by:
Jianbo Xiao, University of Macau, ChinaReviewed by:
Zhimin Xu, Louisiana State University, USARuili Yang, South China Agricultural University, China
Copyright © 2017 Xiao, Hu, Yin, Zhou, Liu, Zhou and Chang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaoli Zhou, emhvdXhsc2l0QDE2My5jb20=
Ying Xiao, eGlhb3lpbmdAc2l0LmVkdS5jbg==
 Ying Xiao
Ying Xiao Zhongzhi Hu1
Zhongzhi Hu1