
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Pharmacol. , 26 September 2016
Sec. Cancer Molecular Targets and Therapeutics
Volume 7 - 2016 | https://doi.org/10.3389/fphar.2016.00327
This article is part of the Research Topic Targeting the ErbB/Her receptor family for cancer therapy: connecting oncogenes with immunity View all 8 articles
Advances in molecular pathology have changed the landscape of oncology. The ability to interrogate tissue samples for oncogene amplification, driver mutations, and other molecular alterations provides clinicians with an enormous level of detail about their patient’s cancer. In some cases, this information informs treatment decisions, especially those related to targeted anti-cancer therapies. However, in terms of immune-based therapies, it is less clear how to use such information. Likewise, despite studies demonstrating the pivotal role of neoantigens in predicting responsiveness to immune checkpoint blockade, it is not known if the expression of neoantigens impacts the response to targeted therapies despite a growing recognition of their diverse effects on immunity. To realize the promise of ‘personalized medicine’, it will be important to develop a more integrated understanding of the relationships between oncogenic events and processes governing anti-tumor immunity. One area of investigation to explore such relationships centers on defining how ErbB/HER activation and signal transduction influences antigen processing and presentation.
Stated simply, oncogenic events are not immunologically null (Seliger, 2014). Defining the immunologic impact of oncogenic events will be a critical area of research as advances in molecular diagnostics parallel the expansion and availability of cancer immunotherapy (Whiteside et al., 2016). Alterations in DNA sequence, copy number variation (CNV), transcriptional profile, and epigenetics can all influence the expression of genes of the immune system, immunologic processes, and ultimately anti-tumor immunity. The importance of antigen processing and presentation in this regard is underscored by recent studies that have defined the impact of mutation-derived neoantigens on the response to immune checkpoint blockade (Sorensen et al., 2009; van Rooij et al., 2013; Schumacher et al., 2014; Snyder et al., 2014; Kelderman et al., 2015; Le et al., 2015; Van Allen et al., 2015). These recent findings, further highlight the importance of defining interactions between oncogenes, oncogenic signal transduction, and the antigen processing machinery (APM) (Seliger, 2014). Moreover, because the APM plays a critical role in cancer immunoediting, understanding how anti-cancer therapies (of all types) influence the APM will be equally important (Galluzzi et al., 2012; Wargo et al., 2014; Okwan-Duodu et al., 2015; Ward et al., 2016). The goal of this article is to provide a framework for understanding interactions between oncogenic ErbB/HER signal transduction and the APM, using the expression of major histocompatibility complex (MHC) molecules as a model.
Recent studies confirming the relevance of neoantigens, also referred to as tumor specific antigens (TSA), provides enormous rationale to understand how oncogenic events impact the ability of tumor cells to process and present antigenic peptides. Moreover, being able to pharmacologically manipulate the expression of proteins that make up the APM takes on additional relevance since defects within the APM are common in human cancer and such alterations will likely influence the peptide-MHC (pMHC) repertoires presented by tumors cells (Seliger et al., 2000; Garrido et al., 2016). The dual ability of ErbB/HER signaling to modulate large transcriptional programs and influence MHC expression suggests that alterations in the ErbB/HER family of ligands and/or receptors will change the pMHC repertoire present at the surface of tumor cells. Alterations of ErbB/HER receptors and ligands occurring in human cancer are shown in Tables 1 and 2. This concept was supported by the fact that ErbB/HER ligands and receptors can greatly influence the cellular transcriptome, which in turn influences the pMHC repertoire (Choi et al., 2007; Fortier et al., 2008; Nagashima et al., 2008). In addition, the complex nature of ErbB/HER signaling that results from having numerous ligands, receptors, and mechanistically distinct activating events, supports the idea that there will be differential effects on MHC expression. While it will likely be cell-context dependent, defining ErbB/HER oncogenic signaling in this manner provides an experimental approach that can further our understanding of interactions between oncogenes and immunity. Moreover, this approach lends itself to validation using cell-based systems, genetically engineered mouse models, and human pathology samples. Some of these concepts are shown in Figure 1.
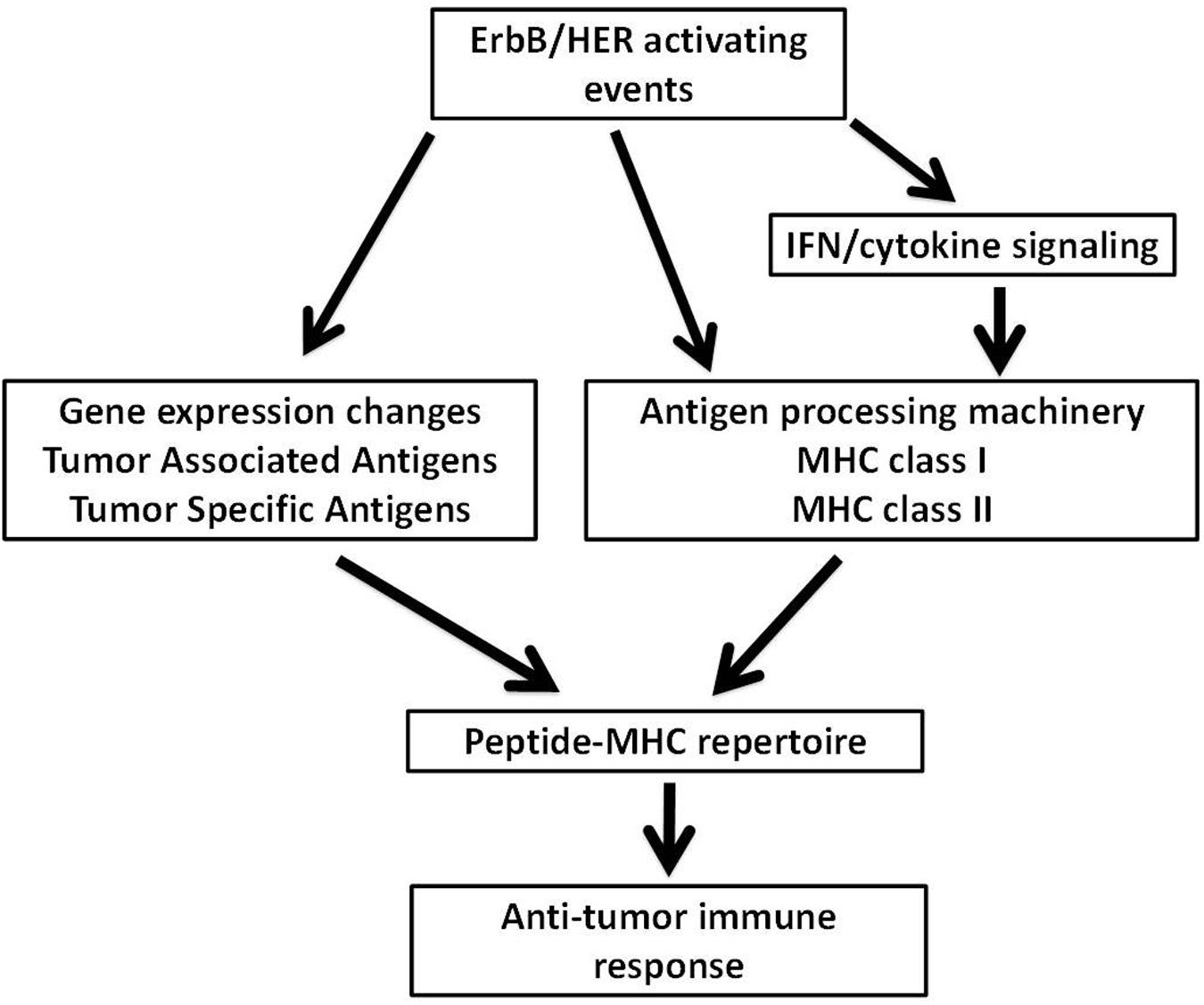
FIGURE 1. Possible mechanisms through which ErbB/HER oncogenic activating events influence the peptide-MHC (pMHC) repertoire. ErbB/HER oncogenic events may influence the peptide-MHC repertoire via several possible mechanisms. ErbB/HER oncogenic activation alters the transcriptome of tumor cells which may include the expression of tumor associated antigens and tumor specific antigens (TSA) such as neoantigens. By altering the transcriptome of tumor cells, ErbB/HER oncogenic events influence the pool of available proteins available for antigen processing and presentation. In addition, ErbB/HER oncogenic signaling may influence the expression of components of the antigen processing machinery (APM) including MHCI and MHCII molecules directly or indirectly by altering the cellular response to cytokines. This will alter the nature and density of the pMHC repertoire. Qualitatively and quantitatively, the nature of the pMHC repertoire may influence T cell-mediated anti-tumor immune responses.
Major histocompatibility complex molecules are cell surface glycoproteins that function to present self- and antigen-derived peptides to cells of the immune system. In humans, MHC molecules are encoded by human leukocyte antigen (HLA) genes and these terms are often used interchangeably. MHC class I (MHCI) molecules consist of two polypeptide chains, the alpha chain (HLA-A, B, C) and ß2-microglobulin which are non-covalently associated on the cell surface. The alpha-1 and alpha-2 domains of the MHC class I molecule form the peptide binding cleft which binds and presents cytosol-derived peptides that are 8-11 amino acids in length to CD8 T lymphocytes (Comber and Philip, 2014). As MHCI molecules function in concert with CD8 T cells for immune surveillance against infections and malignancies, these molecules are expressed on the surface of virtually all nucleated cells and MHCI expression can be further increased (induced) by type I and type II interferons (IFNs) as well as other cytokines. Underscoring their important role in controlling malignancies and infections is the fact that tumor cells and microbes alike such as the human immunodeficiency virus (HIV) and Mycobacterium tuberculosis (Mtb) harbor mechanisms that down-regulate MHCI expression to facilitate immune escape (Ferris et al., 2006; Pennini et al., 2006; Choma et al., 2015; Concha-Benavente et al., 2016).
MHC class II (MHCII) molecules function to present antigen to CD4 T lymphocytes generating helper T cell responses that are critical for effective adaptive immune responses against infection and cancer (DeSandro et al., 1999; Accolla et al., 2014). Each MHCII molecule (HLA-DR, DP, DQ) is composed of an alpha and beta polypeptide chain that non-covalently associate at the cell surface with one subunit from each chain forming the peptide binding cleft. MHCII molecules bind peptides of 13-17 amino acids in length that are generated by proteolysis in lysosomes and endosomes and are constitutively expressed on the surface of antigen presenting cells (APCs) such as B cells, macrophages, and dendritic cells (DCs; Roche and Furuta, 2015).
In order to understand how oncogenic signal transduction might influence the expression of MHC molecules, it is important to review some aspects of MHC expression regulation. In general, though not exclusively, MHC molecules are regulated transcriptionally and epigenetically (van den Elsen et al., 2004; Choi et al., 2011; Kobayashi and van den Elsen, 2012). This regulation is orchestrated at several levels involving complex interactions between regulatory DNA sequences, within the MHC locus (such as promoters and enhancers), DNA-binding transcription factors (TFs) that bind these sequences, transcriptional co-activators (NLRC5/CITA for MHCI and CIITA for MHCII) and the formation of complex looping structures that involve interactions with epigenetic enzymes and chromatin (Gobin et al., 2001; Meissner et al., 2012). There are many excellent detailed reviews available on the regulation of MHCI and MHCII molecules (van den Elsen et al., 1998; van den Elsen, 2011; Devaiah and Singer, 2013; Neerincx et al., 2013).
When considering interactions between oncogenic signaling and MHCI expression, two types of expression need to be considered. Constitutive expression refers to the level of MHCI molecules expressed under physiologic conditions and varies between different tissues due in part to differences in epigenetic marks (Kotekar et al., 2008). In addition to constitutive expression, increases that occur in response to cytokines are referred to as inducible expression. Defects in both types of MHCI expression occur in human cancer (Garrido et al., 2010). Mechanistically, constitutive MHCI expression is regulated by distinct regions within MHCI promoters that are binding sites for TFs such as NFkB, IRF-1, and CREB. The inducible expression of MHCI molecules occurs in response to cytokines such as type I and type II interferons (IFNs) and tumor necrosis factor-alpha (TNF-α) and is mediated through changes in TFs, co-activators, and other proteins that occur in response to the inducing cytokine (van den Elsen, 2011). Thus, when considering interactions between ErbB/HER signaling and MHC expression, the status of both constitutive and inducible MHCI expression warrant attention.
In contrast to MHCI, MHCII is expressed constitutively only on specialized cells of the immune system such as DCs and B cells. The expression of MHCII is regulated largely via the activity of the MHCII co-activator (CIITA) which itself is regulated by distinct promoters that are active in DCs and B cells (named pI and pIII, respectively). Interestingly, aberrant constitutive MHCII expression occurs on some tumor cells such as melanoma (Martins et al., 2007; Degenhardt et al., 2010). Despite this restricted constitutive expression pattern, MHCII molecules are inducible by IFN-γ in most cell types; a unique feature of IFN-γ. The fact that MHCI and MHCII molecules are inducible by cytokines is highly relevant to ErbB/HER signaling because as outlined in more detail below, there are functional links between cytokine signaling and ErbB/HER signaling. Moreover, ErbB/HER signaling can also influence the expression of the TFs that bind to the promoters of HLA class I genes such as NFκB and IRF-1 (Andersen et al., 2008; Shostak and Chariot, 2015). Thus, ErbB/HER signaling pathways are well poised to alter MHC expression (positively or negatively) via mechanisms that are not entirely understood.
In the setting of cancer, MHC molecules play the pivotal role of presenting processed tumor antigens to CD4 and CD8 lymphocytes in order to generate a tumor-specific cytotoxic response (Seliger, 2008b; Hastings, 2013). As tumor antigens are ultimately derived from self, barriers to the activation of an antitumor T cell response exist intrinsically as T cells with affinity for self-antigens are deleted during T cell development. Further, tumor cells can create an immunosuppressive microenvironment to suppress anti-tumor T cell responses (Kim et al., 2006). As mentioned earlier, one mechanism through which tumor cells evade detection by the immune system is the down regulation of the surface expression of MHCI molecules. This can occur via many mechanisms including the genetic loss of the MHC locus, epigenetic silencing, and many others (Marincola et al., 2000; Seliger, 2008a,b, 2014; van der Burg et al., 2016). Importantly, while some of these defects are irreversible, others are not and in these cases the expression of MHCI molecules can be corrected or ‘rescued’ by cytokines (such as IFNs) and medications such as metformin, inhibitors of DNA methylation, histone deacetylase (HDAC) inhibitors, and other approaches including targeted therapies as outlined later in this review (Seliger and Pfizenmaier, 1989; Komatsu and Hayashi, 1998; Magner et al., 2000; Lopez-Albaitero et al., 2006; Khan et al., 2008; Han et al., 2011; Lampen and van Hall, 2011; Oliveras-Ferraros et al., 2012). Many immune-based therapies hinge on the generation of effector CD4 and/or CD8 T cells. As such, being able to increase MHCI and/or MHCII levels pharmacologically on tumor cells would be an attractive complement to CD4/CD8-based immunotherapy because CD4- and CD8 T cell responses are functionally influenced by the pMHC density present at the cell surface. For MHCI, as pMHCI density is increased, T cell activation becomes more efficient and the inhibition seen in the setting of an excessively long TCR-pMHCI interaction half-life is attenuated (Gonzalez et al., 2005). For MHCII, higher pMHCII density can elicit a more robust CD4 T cell response than the same pMHCII at a lower density (Vanguri et al., 2013). While it is less clear if increases in pMHC density can functionally alter an anti-tumor immune response, increasing pMHC density via the aforementioned approaches offers another possible avenue to manipulate interactions between tumor cells and therapeutic T cells.
In addition to their level of expression, the peptides bound to MHC molecules play a crucial role in anti-tumor immunity. As mentioned previously, the presentation of strongly antigenic peptides (such as neoantigens derived from mutations) via MHCI or MHCII can have an enormous impact on clinical responses to immunotherapies such as those targeting immune checkpoints (mediated by CTLA-4 signaling and the PD-1/PD-L1 axis) (Lu and Robbins, 2016; Ward et al., 2016). With this in mind, it will be crucial to understand how oncogenic events and anti-cancer therapies influence antigen processing, antigen presentation, and the pool of antigenic peptides that can be displayed on the cell surface of tumors via MHC molecules and detected by anti-tumor CD4- and/or CD8 T lymphocytes (Chang and Ferrone, 2007; Hastings, 2013).
Given their canonical roles in cancer biology and immunology, respectively, it is not surprising that attempts to identify interactions between the ErbB/HER and MHC molecules goes back three decades (Schreiber et al., 1984). These initial papers examined the ability of EGFR ligands to influence MHC expression. For example, it was found that epidermal growth factor (EGF) treatment could reduce the binding of KE-2 antibodies (which recognize an epitope on the heavy chain of HLA class I molecules) to A431 cells; a finding that pre-dates the seminal paper by Sporn and Roberts (1985) regarding the autocrine secretion of growth factors such as TGF-α by cancer cells (Schreiber et al., 1984; Sporn and Roberts, 1985). In regards to MHCII, links between MHCII expression and ErbB/HER ligands dates back over two decades where it was demonstrated that the induction of HLA-DR molecules by IFN could be attenuated by ErbB/HER ligands EGF and/or transforming growth factor-α (TGFA) in thyroid epithelial cells and keratinocytes (Lahat et al., 1992; Mitra and Nickoloff, 1992). These early studies provided evidence that there were signals initiated by EGFR activation that had a repressive effect on the expression of MHCI and MHCII molecules at least in some cellular contexts.
When considering ErbB/HER-MHC interactions, it is important to consider the crosstalk that exists between ErbB/HER receptors and members of the cytokine receptor superfamily (Kaczmarski and Mufti, 1991; Prenzel et al., 2000). For example, in response to cytokines such as IFN-γ that induce MHC expression, ErbB/HER activation can occur via the protease-dependent release of ErbB/HER ligands (Burova et al., 2007). As a result, the cellular response to cytokines that induce MHC molecules involves the activation of ErbB/HER receptors. This places the EGFR and other members of the ErbB/HER family, as well as those pathways downstream, in a unique position to influence the cellular response to cytokines and thus MHC induction. This becomes even more important when one considers the dual nature of MHCI regulation described earlier since malignant cells can harbor defects in constitutive MHCI expression yet retain their responsiveness to cytokines. In such a scenario, it is possible that oncogenic ErbB/MHC signaling would be particularly impactful since MHCI expression on tumor cells would be effectively dependent on cytokines present in the tumor microenvironment for MHCI expression yet rendered less responsive to cytokines due to aberrant ErbB/HER signaling.
The ligands for ErbB/HER family members are EGF-family growth factors that include EGF, TGFA, amphiregulin (AREG), heparin-binding EGF-like growth factor (HBEGF), betacellulin (BTC), epigen (EPGN), epiregulin (EREG), and four neuregulins (NRG1-4; are also called heregulins) (Normanno et al., 2003a; Schneider and Wolf, 2009). These ligands bind ErbB/HER receptors with varying specificities and induce the formation of homo- and heterodimers creating a large number of possible receptor-ligand combinations (Table 3) (Roskoski, 2014). Through these interactions, ErbB/HER ligands initiate signal transduction via complex signaling networks that lead to large-scale changes in gene expression (Normanno et al., 2006). ErbB/HER ligands are widely expressed in epithelial tissues including those that give rise to the most common forms of human cancer such as the skin, gut, lungs and the aerodigestive tract. These ligands exert distinct effects based upon the receptors they engage and the nature of the downstream signals they transmit (O-Charoenrat et al., 2002; Zaiss et al., 2015). For example, by activating different receptors, ligands induce signals with distinct intensities and durations that in turn contribute to ligand-specific effects on gene expression (Nagashima et al., 2007). Moreover, ligands activating the same ErbB/HER receptor bind with different affinities causing distinct biological outcomes (Macdonald-Obermann and Pike, 2014). Transgenic and knockout mouse models have demonstrated these differences underscoring the fact that these ligands exert distinct effects even within the same tissue context (Table 4) (Normanno et al., 2006). In the skin, forced expression of TGF-α causes papillomas at sites of mechanical irritation, forced expression of AREG induces psoriatic-like lesions, and forced expression of betacellulin causes altered hair development (Vassar and Fuchs, 1991; Dominey et al., 1993; Cook et al., 1997; Schneider et al., 2008). Knockout studies have also illustrated the different roles for ErbB/HER ligands within the same tissue as mice deficient in both EGF and TGF-α expression show impaired prostate development while mice deficient only in TGF-α show increased proliferation of prostate tissue compared to wild-type (Abbott et al., 2003). This suggested that intact TGF-α signaling is required to control EGF-stimulated proliferation of prostate tissue. Similar observations were made in the central and peripheral nervous systems where EGF, TGF-α, and HBEGF are all expressed. Experiments in mice deficient in TGF-α demonstrated normal development and functioning of the peripheral nervous system suggesting EGF and HBEGF signaling are sufficient and compensate for a lack of TGF-α deficient signaling (Xian and Zhou, 2004). Despite the above phenotypes, the impact of ErbB/HER ligands on adaptive immune responses in these models are incompletely understood in part because these ligands were initially characterized for their ability to influence processes such as cell growth and survival. However, recently there has been a growing appreciation for the immune functions of ErbB/HER ligands.
One important role ErbB/HER ligands in the context of adaptive immunity was recently uncovered when it was demonstrated that regulatory T cells (T regs) express the EGFR and respond to EGFR ligands such as amphiregulin (AREG) (Zaiss et al., 2013). In addition, a role for AREG in ultraviolet radiation (UVR)-mediated immunosuppression has recently been reported (Meulenbroeks et al., 2015). The ability of immune cells including CD4 and CD8 T cells to produce AREG further underscores the expanding appreciation for the role of ErbB/HER ligands in adaptive immune responses including those against tumors (Zaiss et al., 2006; Kwong et al., 2010; Qi et al., 2012; Burzyn et al., 2013). Despite these examples, the functional role of ErbB/HER ligands in shaping anti-tumor immune responses in vivo is poorly defined though the ability of these ligands to influence the APM and MHC expression places them in a central position to do so.
As mentioned above, in the 1980s and 1990s, several groups found that treatment of cells with ErbB/HER ligands such as EGF and TGFA repress the expression/induction of MHCI and MHCII molecules (Schreiber et al., 1984; Lahat et al., 1992; Mitra and Nickoloff, 1992). More recently, we have also shown that EGF can repress the induction of MHCII molecules by IFN-γ (Pollack et al., 2011). Likewise, others have shown similar effects of ErbB/HER ligands. For example, in their studies on esophageal and gastric cancer, Mimura et al. demonstrated that MHCI was repressed by the neuregulin NRG-1-β1 (Mimura et al., 2013). Likewise, in their studies on prostate cancer, Chen et al. (2015) showed that EGF could repress MHCI expression. However, in these and other studies, not all cells respond to ErbB/HER ligands in the same manner; cells responding to one ligand may fail to respond to another, and in some cells, ErbB/HER ligands have no effect on MHC expression (Mimura et al., 2013; Vantourout et al., 2014; Chen et al., 2015). Such differences in the responses to ErbB/HER ligands are likely due to the complex nature of ErbB/HER signaling and MHC expression. For example, the cellular response to ErbB/HER ligands depends on ErbB/HER expression patterns and is likely influenced by the (activation) status of downstream enzymes (such as RAS and RAF) and other proteins that impact the response to ErbB/HER inhibition (Gazdar, 2009). Moreover, as mentioned earlier, when considering the impact of ErbB/HER ligands on MHC expression both constitutive and inducible MHCI expression warrant investigation as some effects may require cytokine treatment(s) to be manifest. Table 5 provides a summary of studies demonstrating repression of MHC molecules by ErbB/HER ligands (in at least some contexts).
Further complicating the picture, especially as it relates to ErbB/HER-MHCI interactions, is the fact that ErbB/HER signaling activates pathways known to have opposing effects on MHC expression. For example, ErbB/HER signaling can activate NFκB, IRF-1, and Stat1, known positive regulators of MHC expression, while at the same time activate the MAPK pathway which can negatively regulate MHC expression (Figure 2) (Pedersen et al., 2005; Andersen et al., 2008; van den Elsen, 2011; Mimura et al., 2013; Sapkota et al., 2013; Chen et al., 2015; Shostak and Chariot, 2015). Therefore, the balance of these positive and negative regulators of MHC expression induced in response to ErbB/HER activation may ultimately dictate their impact on MHC expression. The study by Mimura et al. highlights this point. In their model, a clear hierarchy existed as EGF failed to influence MHCI expression, whereas NRG-1β1 repressed it and both the MAPK and PI3K pathways were implicated in modulating MHCI expression (Mimura et al., 2013). Thus, while many questions remain regarding the impact of ErbB/HER ligands on the APM there is growing evidence that in some contexts these ligands influence MHC expression. Moreover, based upon the expression pattern of ErbB/HER receptors present on a given tumor cell, some ligands may influence MHC expression whereas others will not. Figure 2 illustrates some of the above concepts.
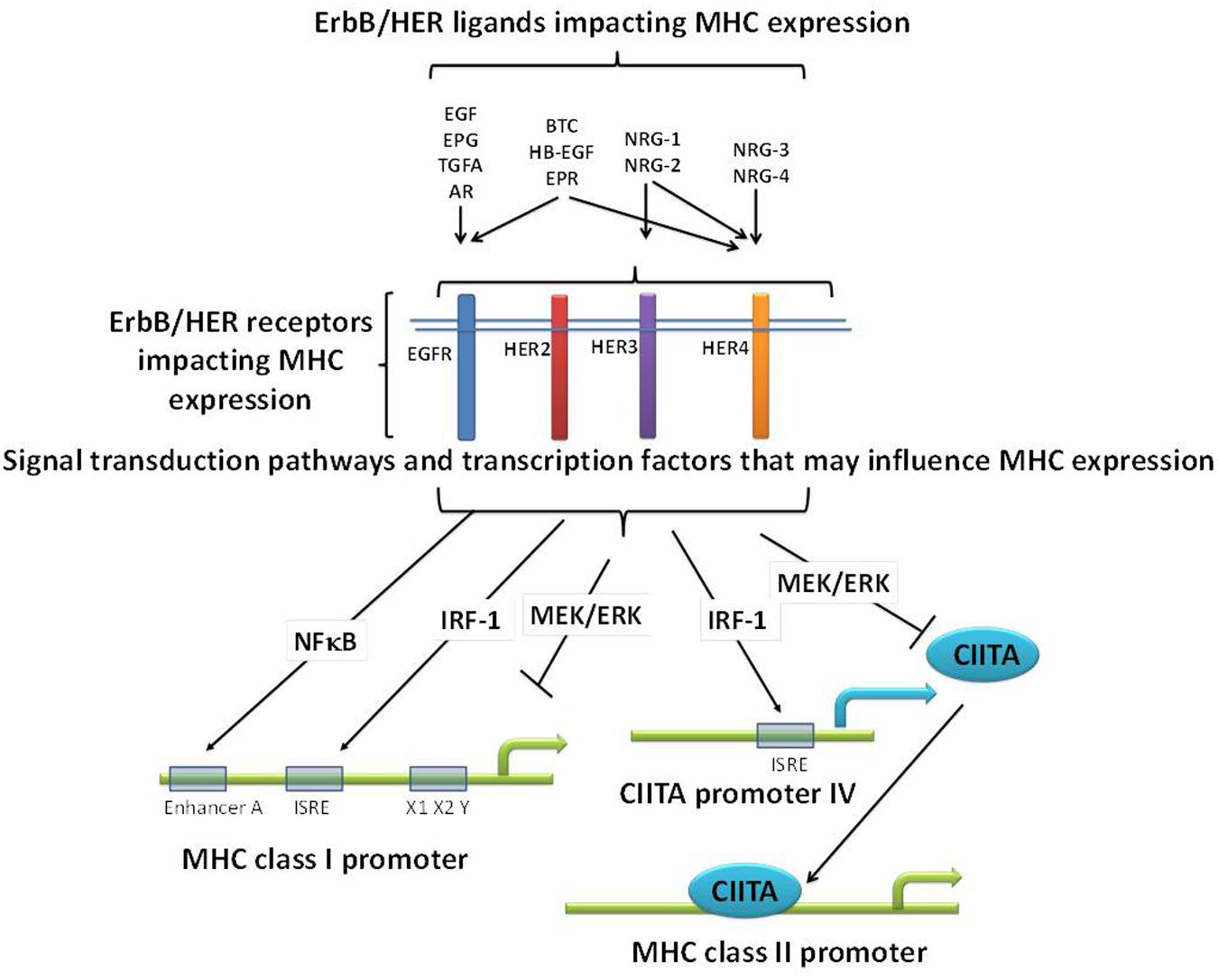
FIGURE 2. Potential mechanisms relating ErbB/HER ligands, receptors, and MHC expression. ErbB/HER ligands bind and activate different ErbB/HER receptors initiating distinct signal transduction pathways and cellular effects. As a result, there may be differences across ligand groups based upon the receptors and downstream signaling pathways they activate. In addition, even ErbB/HER ligands that bind the same receptors can have distinct impacts on downstream signaling and as such may have distinct effects on MHC expression. ErbB/HER ligands and receptors activate signaling pathways known to negatively (such as the MAPK pathway), and positively (such as NFκB and IRF-1), regulate MHC expression via the binding of transcription factors (TFs) to MHC promoters or the promoters of co-activators such as CIITA. As a result, in any given cellular context, the sum of these opposing effects will likely determine the impact of ErbB/HER activation and inhibition on MHC expression.
The four members of the ErbB/HER family include the EGFR (also known as ErbB1 and HER1), ErbB2 [also known as HER2 and Neu, ErbB3 (HER3), and ErbB4 (HER4)] (Roskoski, 2014). Structurally, these receptors contain an extracellular ligand-binding domain, a transmembrane domain, and a cytoplasmic domain that contains tyrosine kinase activity (Lemmon et al., 2014). There are some noteworthy exceptions to the preceding description. Namely, ErbB2/HER2/neu has no known ligand, and ErbB3/HER3 has impaired tyrosine kinase activity. ErbB/HER receptors can signal as homo- or heterodimers creating an array of combinations that have distinct signaling outputs (Roskoski, 2014). Moreover, hierarchies exist such that the ErbB2/ErbB3 heterodimer is felt to be the most potent ErbB pairing signaling combination (Pinkas-Kramarski et al., 1996; Tzahar et al., 1996; Baselga and Swain, 2009). ErbB/HER receptors are widely expressed on normal and malignant cell types (Salomon et al., 1995). Genetic knockout of ErbB/HER receptors in mice leads to alterations in numerous tissues (Table 6). In addition to their classical roles in regulating cellular proliferation, survival, angiogenesis, apoptosis, and the cell cycle, these receptors have recently been implicated in the control of epithelial-to-mesenchymal transition (EMT) (Chen et al., 2015). Thus, ErbB/HER family members can regulate a variety of complex processes including immune responses as outlined below.
Before reviewing the immune effects of ErbB/HER proteins, it is worth highlighting the fact that these proteins have been examined from markedly different yet valid perspectives (Seliger and Kiessling, 2013). From the cancer biology perspective, ErbB/HER proteins are canonical oncogenic enzymes that drive tumor cell proliferation/survival/angiogenesis and metastasis (Hynes and Lane, 2005). This perspective considered little the immune effects of ErbB/HER proteins yet fueled enormous efforts to inhibit ErbB/HER expression and/or activity (Mendelsohn and Baselga, 2000; Normanno et al., 2003a). Contrasting this view is the tumor immunology perspective that considers ErbB/HER proteins as bonafide tumor antigens (Zaks and Rosenberg, 1998; Nistico et al., 1999; Disis et al., 2000). From this perspective, ErbB/HER-derived peptides including those derived from activating mutations, such as the EGFRvIII variant with a mutation in the extracellular domain of the receptor, have the potential to stimulate an anti-tumor immune response innately or via vaccination (Li and Wong, 2008; Nedergaard et al., 2012; Paff et al., 2014; Schneble et al., 2014). Vaccine studies using a 14-amino acid peptide spanning the mutated extracellular domain of EGFRvIII (CDX-110) fused to adjuvant have shown durable oncogene-specific antibody and CD8 T cell responses as well as improved survival in glioblastoma multiforme patients (Sampson et al., 2008). In this case, the expression of ErbB/HER proteins on tumor cells would be required for successful anti-cancer therapy and by targeting mutations specific to cancer cells these therapies result in minimal bystander damage of healthy somatic cells expressing normal ErbB/HER proteins. Other studies incorporate both perspectives by investigating interactions between oncogenic ErbB/HER proteins and proteins central to immune responses; namely MHC molecules.
Attempts to establish links between the expression of ErbB/HER family members and MHC molecules have been the subject of study for many years (Nouri et al., 1995; Nistico et al., 1997; Lollini et al., 1998). Using a forced-expression approach, Herrmann et al. (2004) demonstrated that the expression of ErbB2/HER2 was associated with decreases in MHCI and other components of the APM. Complementing these findings using siRNA-based approaches, Choudhury et al. (2004) demonstrated that the loss of ErbB2/HER2 was associated with increases in MHCI molecules. These studies provided links between ErbB/HER proteins and MHC expression. The development of pharmacologic ErbB/HER inhibitors has provided additional evidence supporting interactions between ErbB/HER proteins and MHC molecules.
Because MHC molecules govern interactions between tumor cells and CD4 and CD8 T cells, early studies examined the effects of ErbB/HER inhibition on MHC expression. In one early study, no effect of the anti-HER monoclonal antibody (mAb) trastuzumab on MHC expression was seen though the treatment did enhance tumor lysis by MHCI-restricted cytotoxic T lymphocytes (CTLs) (Kono et al., 2004). Our group explored the impact of EGFR inhibitors on MHCI and MHCII expression and found that small molecule tyrosine kinase inhibitors (TKIs) (such as PD168393 and AG1478) as well as the EGFR blocking antibody cetuximab (alone or with the ErbB2/HER2/neu blocking antibody trastuzumab) could enhance the induction of MHCI and MCHII molecules by IFN-γ (Pollack et al., 2011). In some experimental models, the addition of either PD168393 or cetuximab alone augmented MHCI even in the absence of IFN-γ. Subsequent studies have confirmed that at least under some circumstances ErbB/HER inhibitors can enhance MHCI and/or MHCII expression. For example, erlotinib, cetuximab and the pan ErbB/HER inhibitor dacomitinib increased the induction of MHCII molecules by IFN-γ in head and neck cancer cell lines (Kumai et al., 2013, 2015). In their study on non-small cell lung cancer, Okita et al. (2015) found that gefitinib upregulated MHCI expression in some cellular contexts. Moreover, anti-ErbB2/HER2/Neu therapy increased MHCII levels in vivo (Mortenson et al., 2013). Further illustrating relevant functional interactions between ErbB/HER inhibition and MHC molecules was a study demonstrating that EGFRI resistance is associated with defects in MHCI expression (Garrido et al., 2014). The above studies illustrate that under some contexts, ErbB/HER proteins and inhibitors thereof can influence the expression of MHC molecules. Table 7 lists studies reporting increases in MHCI and/or MHCII by ErbB/HER inhibition.
While the above provide evidence that ErbB/HER activity impacts MHC expression, it is important to note that just as was the case for ErbB/HER ligands, not all cells respond to ErbB/HER inhibition with increases in MHC expression. We and others have shown that treatment of some cells with inhibitors of ErbB/HER proteins does not alter MHC expression (Pollack et al., 2011; Mimura et al., 2013; Okita et al., 2015). Thus, in some cells, ErbB/HER activity is coupled to MHC expression, whereas in other cells this is not the case. While the mechanisms underpinning these interactions are still being explored, some cells that are unresponsive to ErbB/HER inhibition respond to inhibitors of downstream enzymes particularly those inhibiting enzymes in the mitogen-activated protein kinase (MAPK) pathway as outlined below and shown in Figure 3.
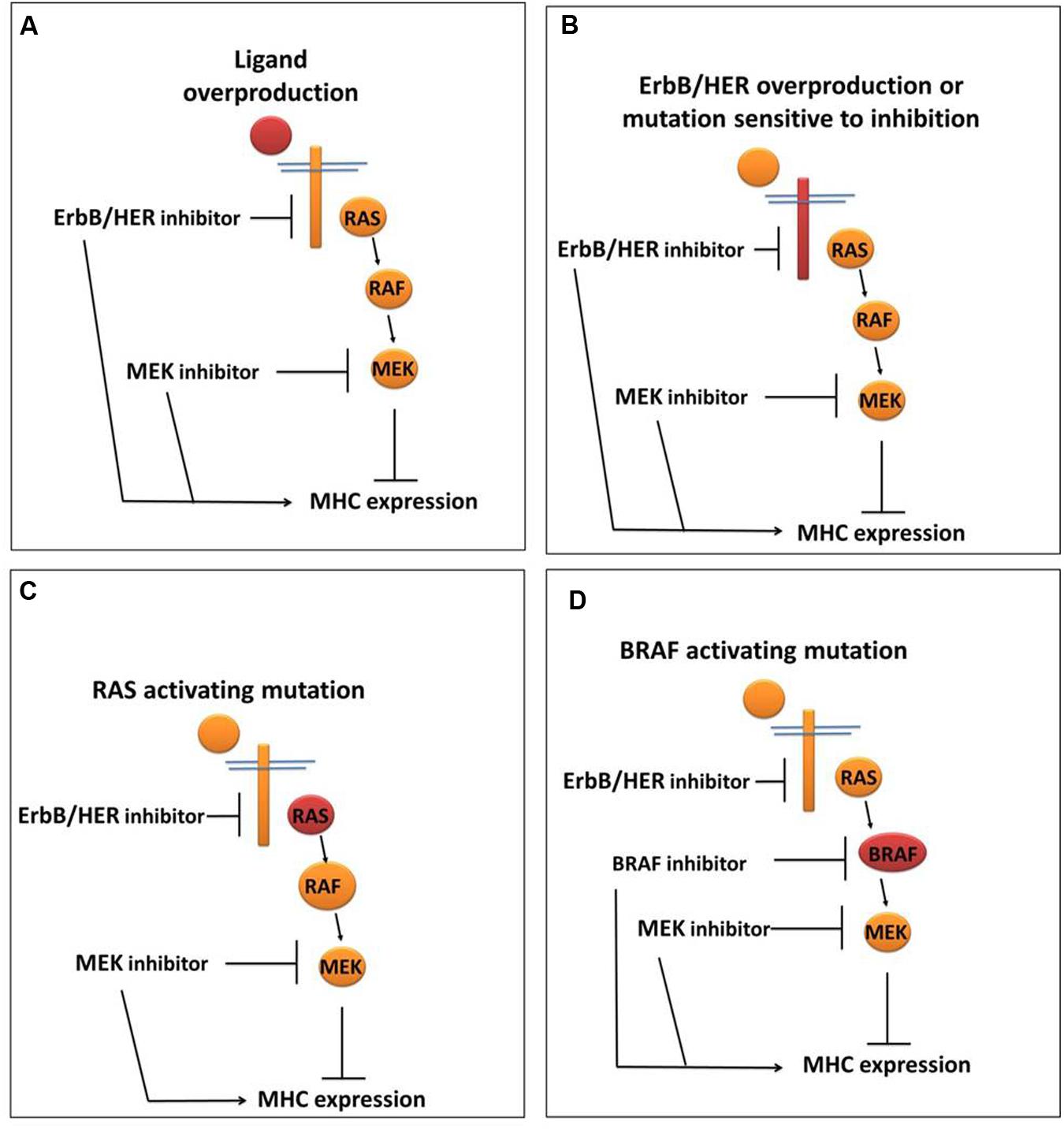
FIGURE 3. ErbB/HER downstream signaling pathways may influence the ability of ErbB/HER and other kinase inhibitors to modulate MHC expression. In settings where the MAPK pathway is actively repressing MHC expression, the location of the activating/oncogenic event (indicated in red) will likely influence the response to ErbB/HER and/or other kinase inhibitors. (A) In the setting of ErbB/HER ligand overproduction, inhibition of ErbB/HER activity (via an ErbB/HER inhibitor) and/or enzymes in the MAPK signaling pathway may increase MHC expression. (B) In the setting of ErbB/HER activating mutations and/or overproduction, an ErbB/HER inhibitor and/or MAPK inhibitor may increase MHC expression. (C) In the setting of an activating RAS mutation, the effect of an ErbB/HER inhibitor may be null/minimal because MAPK signaling is being driven by activated RAS, whereas a downstream MAPK inhibitor may increase MHC expression. (D) In the setting of an activating mutation in BRAF, as with an activating RAS mutation, an ErbB/HER inhibitor would not likely change MHC expression whereas a BRAF inhibitor and/or a MEK inhibitor may increase MHC expression.
ErbB/HER proteins are signaling hubs that can initiate signal transduction via a variety of downstream pathways that include the RAS/RAF/MAPK pathway, the phosphoinositide-3-kinase (PI3K) pathway, the phospholipase C-γ (PLC-γ) pathway and signal transducers and activators of transcription (STATS) (Pines et al., 2010). Evidence for direct links between enzymes downstream of ErbB/HER proteins, such as RAS, and the APM have existed for over twenty years (Seliger et al., 1988, 1996, 1998; Seliger and Pfizenmaier, 1989; Lohmann et al., 1996). In addition, links between RAS mutations and MHC expression have been reported in colorectal carcinoma (Atkins et al., 2004). More recently, inhibitors of the aforementioned pathways have been exploited to examine interactions between these pathways and MHC expression. Akin to the development of ErbB/HER inhibitors, the main paradigm fueling the development these inhibitors (such as those targeting BRAFV600E and MEK to inhibit the MAPK pathway and those targeting the PI3K pathway) centered on their potential to block proliferative and survival signals rather than their immune effects (Rodon et al., 2013; Zhao and Adjei, 2014; Sidaway, 2015; Thorpe et al., 2015). Despite this, there is clear evidence that inhibition of enzymes downstream of ErbB/HER receptors, especially the MAPK pathway, alters MHC expression in some contexts.
Early studies using pre-clinical MEK inhibitors (such as PD98059) revealed that MEK inhibition augments MHCI and MHCII expression (as well as CD86, CD80, and CD40) on and allostimulation by growth factor-dependent DC in the presence of TNF-α (Yanagawa et al., 2002). Likewise, another group reported that PD98059 could increase MHCII expression on monocyte-derived DCs (moDCs) (Aiba et al., 2003). These studies provided early links between the MAPK pathway and MHC expression in the context of professional APCs. As mentioned earlier, similar to tumor cells, microbes such as Mtb can down-regulate MHC expression to avoid detection by the immune system. Mechanistically, the MAPK pathway has been implicated since the down regulation of the MHCII master regulator CIITA in macrophages by Mtb was mediated by MAPK signaling and reversed by the MEK inhibitor U0126 (Pennini et al., 2006). These observations support subsequent studies demonstrating that MAPK inhibitors influence MHC expression in the context of cancer.
Kono and colleagues reported that the expression of melanoma differentiation antigens (MDAs) increased in the presence of U0126 and PD98059 (Kono et al., 2006). While they found no effect on MHC expression, a subsequent study by Sers et al. (2009) demonstrated that treatment of cells with the MEK inhibitor U0126 led to increases in MHCI expression. The FDA approval and clinical use of BRAFV600E and MEK inhibitors (such as vemurafenib, dabrafenib, and trametinib) have further underscored the immune effects of MAPK inhibition in the context of cancer. For example, vemurafenib induced changes in the expression of melanoma differentiation antigens (MDA), and the tumor microenvironment in patients including increases in CD8+ T cell infiltration (Boni et al., 2010; Frederick et al., 2013). Again, in these studies, no changes in MHC expression were seen, yet they underscored the potent immunologic effects induced by disruption of oncogenic MAPK signaling.
Our group looked at the ability of vemurafenib to influence the induction of MHCI and MHCII molecules by IFN-γ and IFN-γ2b. We found that in BRAFV600E mutant melanoma cell lines lacking wild type BRAF the induction of MHCI and MHCII was enhanced in the presence of vemurafenib at nanomolar concentrations and that basal levels of MHCI decreased with the forced expression of BRAFV600E (Sapkota et al., 2013). Further support identifying the MAPK pathway (and phospho-ERK in particular) as a dominant regulator of MHCI expression (in esophageal and gastric cancer) comes from the study by Mimura and colleagues (Mimura et al., 2013). More recently, others have reported increases in MHC expression when oncogenic MAPK signaling is disrupted especially in the context of cytokine treatment or adoptive cell therapy (Mimura et al., 2013; Hu-Lieskovan et al., 2015; Sabbatino et al., 2016; Whipple et al., 2016). Thus, inhibitors of MAPK signaling can influence the expression and induction of MHC molecules in some settings through several possible mechanisms as outlined below. Table 8 lists examples where inhibitors of enzymes in the MAPK pathway increase MHC expression.
One mechanism implicated in mediating MAPK-MHCII interactions is through the activity of the MHCII co-activator (and ‘master regulator’) CIITA. CIITA is critical to the generation of immune responses as it is responsible for regulating the expression of MHC class II molecules (Choi et al., 2011). CIITA does not directly bind DNA, but instead regulates transcription by interacting with TFs and elements of the enhanceosome complex affecting chromatin remodeling and transcription initiation (Reith and Boss, 2008; Devaiah and Singer, 2013). The activity of CIITA is highly regulated through numerous post-translational modifications including ubiquitination, acetylation, and phosphorylation (Wu et al., 2009; Morgan et al., 2015). Early work demonstrated that post-translational phosphorylation of various serine residues of CIITA, were crucial to its ability to localize to the nucleus and increase MHC II expression (Greer et al., 2004). In this study, Greer and colleagues demonstrated that the loss of these phosphorylation sites was associated with an increase in the activation of endogenous MHCII genes. This finding was subsequently confirmed using dominant negative proteins and the ERK inhibitor PD98059 both of which increased MHC II induction by CIITA by attenuating ERK activity (Voong et al., 2008). Thus, while many questions remain, the MAPK pathway likely impacts MHCII expression in part via changes in CIITA post-translational modifications.
Compared to MHCII, the mechanisms through which MHCI is regulated by MAPK signaling is less well defined. Analogous to CIITA for MHCII, NLRC5/CITA has been shown to be a key regulator of MHCI expression that has been implicated in cancer (Meissner et al., 2010; Kobayashi and van den Elsen, 2012; Yoshihama et al., 2016). We have found that vemurafenib increases in NLRC5/CITA mRNA induction by IFN-γ (Sapkota et al., 2013). In addition to possible effects on NLRC5/CITA, others have identified a novel gain-of-function activity of BRAFV600E that directly targets MHCI protein for degradation (Bradley et al., 2015). This study underscores the intimate links that exist between oncogenes and MHC expression.
Given the complex nature of oncogenic signaling and the regulation of immune responses, many complementary approaches need to be used in order to fully understand how ErbB/HER oncogenic signal transduction influences the expression of MHC molecules and the APM. These will likely include biochemical studies, cell-based studies, murine models, and a detailed analysis of human samples. While bioinformatics approaches are integral to all of the aforementioned models, they are particularly useful in the setting of large tumor tissue databases (Cooper et al., 2015; Li et al., 2015). Having databases containing genomic, transcriptomic, and proteomic information provides an invaluable opportunity to characterize relationships between genomic events and those relevant to immunity in the context of human cancer (Rutledge et al., 2013; Saba et al., 2015). An example relevant to MHCI expression and regulation in human cancer is outlined below.
NLRC5/CITA was known to be an important regulator of MHCI molecules, yet until recently, its role in the setting of human cancer was poorly defined. In a recently published study by Yoshihama and colleagues, our understanding of the role of NLRC5 in human cancer was greatly expanded (Yoshihama et al., 2016). The authors in this study included bioinformatics-based approaches to assess the expression pattern of NLRC5/CITA and MHCI in human cancer thereby providing a much better understanding of how NLRC5/CITA expression correlates with other immune parameters in the setting of human tumors. Moreover, these authors used this approach to determine that expression of NLRC5/CITA correlated with survival in several cancer types including melanoma, rectal cancer, bladder cancer, uterine cancer, cervical cancer and head/neck cancer. This study illustrates how genetic information from human tumor databases can be combined with bioinformatics to generate clinically relevant information. A brief description of some of these resources and approaches are reviewed in the next paragraph.
A number of public bioinformatics resources exist to investigate the relationship between immune parameters, oncogenic driver mutations and signaling network activation. The Cancer Genome Atlas1 is a public data resource of the National Cancer Institute that has produced comprehensive genomic, epigenomic, transcriptomic, and proteomic profiles of over 11,000 patients with 33 distinct cancer types. The data generated by TCGA is readily accessible via a number of stand-alone public resources that facilitate exploration of mutational spectra and copy number events, gene expression and their relationship to clinical outcomes including the cBioPortal2, the Broad Institute Tumor Portal3, and the Cancer Regulome Explorer4(Cerami et al., 2012; Gao et al., 2013; Lawrence et al., 2014). The use of these resources and others will provide important avenues to examine interactions between ErbB/HER ligands, receptors, and the APM to understand interactions between these important oncogenes and adaptive immune responses in a more comprehensive manner.
In summary, functional links between ErbB/HER ligands, receptors, and MHC molecules demonstrates the intimate connections that can exist between oncogenes and genes that regulate antigen processing and presentation. The clinical use of ErbB/HER inhibitors provides a unique opportunity to define these relationships in more depth and to gain insight into how signaling pathways defined for their role in cancer influence fundamental immunologic processes. This information will enhance our ability to use targeted therapies with more insight and more rationally combine them with immune-based therapies.
AK, drafting and conception of the review; MS, critical feedback for the review; LC, drafting and conception of the review; HK, critical feedback for the review; BP, conception, drafting, and final approval of the review.
BP receives funding from the Winship Cancer Institute Skin Cancer and Melanoma Fund, a Career Development Award from the Melanoma Research Foundation, and a Merit Review Award from the Department of Veterans Affairs.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors’ appreciated the comments and feedback by Michael Rossi.
Abbott, B. D., Lin, T. M., Rasmussen, N. T., Albrecht, R. M., Schmid, J. E., and Peterson, R. E. (2003). Lack of expression of EGF and TGF-alpha in the fetal mouse alters formation of prostatic epithelial buds and influences the response to TCDD. Toxicol. Sci. 76, 427–436. doi: 10.1093/toxsci/kfg238
Accolla, R. S., Lombardo, L., Abdallah, R., Raval, G., Forlani, G., and Tosi, G. (2014). Boosting the MHC class II-restricted tumor antigen presentation to CD4+ T helper cells: a critical issue for triggering protective immunity and Re-orienting the tumor microenvironment toward an anti-tumor state. Front. Oncol. 4:32. doi: 10.3389/fonc.2014.00032
Aiba, S., Manome, H., Nakagawa, S., Mollah, Z. U., Mizuashi, M., Ohtani, T., et al. (2003). p38 Mitogen-activated protein kinase and extracellular signal-regulated kinases play distinct roles in the activation of dendritic cells by two representative haptens, NiCl2 and 2,4-dinitrochlorobenzene. J. Invest. Dermatol. 120, 390–399. doi: 10.1046/j.1523-1747.2003.12065.x
Al-Kasspooles, M., Moore, J. H., Orringer, M. B., and Beer, D. G. (1993). Amplification and over-expression of the EGFR and erbB-2 genes in human esophageal adenocarcinomas. Int. J. Cancer 54, 213–219. doi: 10.1002/ijc.2910540209
Andersen, P., Pedersen, M. W., Woetmann, A., Villingshoj, M., Stockhausen, M. T., Odum, N., et al. (2008). EGFR induces expression of IRF-1 via STAT1 and STAT3 activation leading to growth arrest of human cancer cells. Int. J. Cancer 122, 342–349. doi: 10.1002/ijc.23109
Atkins, D., Breuckmann, A., Schmahl, G. E., Binner, P., Ferrone, S., Krummenauer, F., et al. (2004). MHC class I antigen processing pathway defects, ras mutations and disease stage in colorectal carcinoma. Int. J. Cancer 109, 265–273. doi: 10.1002/ijc.11681
Baselga, J., and Swain, S. M. (2009). Novel anticancer targets: revisiting ERBB2 and discovering ERBB3. Nat. Rev. Cancer 9, 463–475. doi: 10.1038/nrc2656
Beckhardt, R. N., Kiyokawa, N., Xi, L., Liu, T. J., Hung, M. C., El-Naggar, A. K., et al. (1995). HER-2/neu oncogene characterization in head and neck squamous cell carcinoma. Arch. Otolaryngol. Head Neck Surg. 121, 1265–1270. doi: 10.1001/archotol.1995.01890110041008
Blair, J. A., Rauh, D., Kung, C., Yun, C. H., Fan, Q. W., Rode, H., et al. (2007). Structure-guided development of affinity probes for tyrosine kinases using chemical genetics. Nat. Chem. Biol. 3, 229–238. doi: 10.1038/nchembio866
Boni, A., Cogdill, A. P., Dang, P., Udayakumar, D., Njauw, C. N., Sloss, C. M., et al. (2010). Selective BRAFV600E inhibition enhances T-cell recognition of melanoma without affecting lymphocyte function. Cancer Res. 70, 5213–5219. doi: 10.1158/0008-5472.CAN-10-0118
Bradley, S. D., Chen, Z., Melendez, B., Talukder, A., Khalili, J. S., Rodriguez-Cruz, T., et al. (2015). BRAFV600E Co-opts a conserved MHC class I internalization pathway to diminish antigen presentation and CD8+ T-cell recognition of Melanoma. Cancer Immunol. Res. 3, 602–609. doi: 10.1158/2326-6066.CIR-15-0030
Burova, E., Vassilenko, K., Dorosh, V., Gonchar, I., and Nikolsky, N. (2007). Interferon gamma-dependent transactivation of epidermal growth factor receptor. FEBS Lett. 581, 1475–1480. doi: 10.1016/j.febslet.2007.03.002
Burzyn, D., Kuswanto, W., Kolodin, D., Shadrach, J. L., Cerletti, M., Jang, Y., et al. (2013). A special population of regulatory T cells potentiates muscle repair. Cell 155, 1282–1295. doi: 10.1016/j.cell.2013.10.054
Canueto, J., Cardenoso, E., Garcia, J. L., Santos-Briz, A., Castellanos-Martin, A., Fernandez-Lopez, E., et al. (2016). EGFR expression is associated with poor outcome in cutaneous squamous cell carcinoma. Br. J. Dermatol. doi: 10.1111/bjd.14936 [Epub ahead of print].
Carey, L. A., Perou, C. M., Livasy, C. A., Dressler, L. G., Cowan, D., Conway, K., et al. (2006). Race, breast cancer subtypes, and survival in the Carolina Breast Cancer Study. JAMA 295, 2492–2502. doi: 10.1001/jama.295.21.2492
Cerami, E., Gao, J., Dogrusoz, U., Gross, B. E., Sumer, S. O., Aksoy, B. A., et al. (2012). The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401–404. doi: 10.1158/2159-8290.CD-12-0095
Chan, S. Y., and Wong, R. W. (2000). Expression of epidermal growth factor in transgenic mice causes growth retardation. J. Biol. Chem. 275, 38693–38698. doi: 10.1074/jbc.M004189200
Chang, C. C., and Ferrone, S. (2007). Immune selective pressure and HLA class I antigen defects in malignant lesions. Cancer Immunol. Immunother. 56, 227–236. doi: 10.1007/s00262-006-0183-1
Chen, X. H., Liu, Z. C., Zhang, G., Wei, W., Wang, X. X., Wang, H., et al. (2015). TGF-beta and EGF induced HLA-I downregulation is associated with epithelial-mesenchymal transition (EMT) through upregulation of snail in prostate cancer cells. Mol. Immunol. 65, 34–42. doi: 10.1016/j.molimm.2014.12.017
Choi, K., Creighton, C. J., Stivers, D., Fujimoto, N., and Kurie, J. M. (2007). Transcriptional profiling of non-small cell lung cancer cells with activating EGFR somatic mutations. PLoS ONE 2:e1226. doi: 10.1371/journal.pone.0001226
Choi, N. M., Majumder, P., and Boss, J. M. (2011). Regulation of major histocompatibility complex class II genes. Curr. Opin. Immunol. 23, 81–87. doi: 10.1016/j.coi.2010.09.007
Choma, M. K., Lumb, J., Kozik, P., and Robinson, M. S. (2015). A genome-wide screen for machinery involved in downregulation of MHC class I by HIV-1 Nef. PLoS ONE 10:e0140404. doi: 10.1371/journal.pone.0140404
Choudhury, A., Charo, J., Parapuram, S. K., Hunt, R. C., Hunt, D. M., Seliger, B., et al. (2004). Small interfering RNA (siRNA) inhibits the expression of the Her2/neu gene, upregulates HLA class I and induces apoptosis of Her2/neu positive tumor cell lines. Int. J. Cancer 108, 71–77. doi: 10.1002/ijc.11497
Comber, J. D., and Philip, R. (2014). MHC class I antigen presentation and implications for developing a new generation of therapeutic vaccines. Ther. Adv. Vaccines 2, 77–89. doi: 10.1177/2051013614525375
Concha-Benavente, F., Srivastava, R., Ferrone, S., and Ferris, R. L. (2016). Immunological and clinical significance of HLA class I antigen processing machinery component defects in malignant cells. Oral Oncol. 58, 52–58. doi: 10.1016/j.oraloncology.2016.05.008
Cook, P. W., Piepkorn, M., Clegg, C. H., Plowman, G. D., Demay, J. M., Brown, J. R., et al. (1997). Transgenic expression of the human amphiregulin gene induces a psoriasis-like phenotype. J. Clin. Invest. 100, 2286–2294. doi: 10.1172/JCI119766
Cooper, L. A., Kong, J., Gutman, D. A., Dunn, W. D., Nalisnik, M., and Brat, D. J. (2015). Novel genotype-phenotype associations in human cancers enabled by advanced molecular platforms and computational analysis of whole slide images. Lab. Invest. 95, 366–376. doi: 10.1038/labinvest.2014.153
Crone, S. A., Zhao, Y. Y., Fan, L., Gu, Y., Minamisawa, S., Liu, Y., et al. (2002). ErbB2 is essential in the prevention of dilated cardiomyopathy. Nat. Med. 8, 459–465. doi: 10.1038/nm0502-459
Degenhardt, Y., Huang, J., Greshock, J., Horiates, G., Nathanson, K., Yang, X., et al. (2010). Distinct MHC gene expression patterns during progression of melanoma. Genes Chromosomes Cancer 49, 144–154. doi: 10.1002/gcc.20728
DeSandro, A., Nagarajan, U. M., and Boss, J. M. (1999). The bare lymphocyte syndrome: molecular clues to the transcriptional regulation of major histocompatibility complex class II genes. Am. J. Hum. Genet. 65, 279–286. doi: 10.1086/302519
Devaiah, B. N., and Singer, D. S. (2013). CIITA and its dual roles in MHC gene transcription. Front. Immunol. 4:476. doi: 10.3389/fimmu.2013.00476
Disis, M. L., Knutson, K. L., Schiffman, K., Rinn, K., and Mcneel, D. G. (2000). Pre-existent immunity to the HER-2/neu oncogenic protein in patients with HER-2/neu overexpressing breast and ovarian cancer. Breast Cancer Res. Treat. 62, 245–252. doi: 10.1023/A:1006438507898
Dominey, A. M., Wang, X. J., King, L. E. Jr., Nanney, L. B., Gagne, T. A., Sellheyer, K., et al. (1993). Targeted overexpression of transforming growth factor alpha in the epidermis of transgenic mice elicits hyperplasia, hyperkeratosis, and spontaneous, squamous papillomas. Cell Growth Differ. 4, 1071–1082.
Erickson, S. L., O’shea, K. S., Ghaboosi, N., Loverro, L., Frantz, G., Bauer, M., et al. (1997). ErbB3 is required for normal cerebellar and cardiac development: a comparison with ErbB2-and heregulin-deficient mice. Development 124, 4999–5011.
Erwin, C. R., Helmrath, M. A., Shin, C. E., Falcone, RA Jr, Stern, L. E., and Warner, B. W. (1999). Intestinal overexpression of EGF in transgenic mice enhances adaptation after small bowel resection. Am. J. Physiol. 277, G533–G540.
Falck, V. G., and Gullick, W. J. (1989). c-erbB-2 oncogene product staining in gastric adenocarcinoma. An immunohistochemical study. J. Pathol, 159, 107–111.
Ferris, R. L., Whiteside, T. L., and Ferrone, S. (2006). Immune escape associated with functional defects in antigen-processing machinery in head and neck cancer. Clin. Cancer Res. 12, 3890–3895. doi: 10.1158/1078-0432.CCR-05-2750
Fortier, M. H., Caron, E., Hardy, M. P., Voisin, G., Lemieux, S., Perreault, C., et al. (2008). The MHC class I peptide repertoire is molded by the transcriptome. J. Exp. Med. 205, 595–610. doi: 10.1084/jem.20071985
Frederick, D. T., Piris, A., Cogdill, A. P., Cooper, Z. A., Lezcano, C., Ferrone, C. R., et al. (2013). BRAF inhibition is associated with enhanced melanoma antigen expression and a more favorable tumor microenvironment in patients with metastatic melanoma. Clin. Cancer Res. 19, 1225–1231. doi: 10.1158/1078-0432.CCR-12-1630
Frederick, L., Wang, X. Y., Eley, G., and James, C. D. (2000). Diversity and frequency of epidermal growth factor receptor mutations in human glioblastomas. Cancer Res. 60, 1383–1387.
Galluzzi, L., Senovilla, L., Zitvogel, L., and Kroemer, G. (2012). The secret ally: immunostimulation by anticancer drugs. Nat. Rev. Drug Discov. 11, 215–233. doi: 10.1038/nrd3626
Gao, J., Aksoy, B. A., Dogrusoz, U., Dresdner, G., Gross, B., Sumer, S. O., et al. (2013). Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 6, pl1. doi: 10.1126/scisignal.2004088
Garrido, F., Aptsiauri, N., Doorduijn, E. M., Garcia Lora, A. M., and Van Hall, T. (2016). The urgent need to recover MHC class I in cancers for effective immunotherapy. Curr. Opin. Immunol. 39, 44–51. doi: 10.1016/j.coi.2015.12.007
Garrido, F., Cabrera, T., and Aptsiauri, N. (2010). “Hard” and “soft” lesions underlying the HLA class I alterations in cancer cells: implications for immunotherapy. Int. J. Cancer 127, 249–256. doi: 10.1002/ijc.25270
Garrido, G., Rabasa, A., Garrido, C., Lopez, A., Chao, L., Garcia-Lora, A. M., et al. (2014). Preclinical modeling of EGFR-specific antibody resistance: oncogenic and immune-associated escape mechanisms. Oncogene 33, 3129–3139. doi: 10.1038/onc.2013.288
Gassmann, M., Casagranda, F., Orioli, D., Simon, H., Lai, C., Klein, R., et al. (1995). Aberrant neural and cardiac development in mice lacking the ErbB4 neuregulin receptor. Nature 378, 390–394. doi: 10.1038/378390a0
Gazdar, A. F. (2009). Activating and resistance mutations of EGFR in non-small-cell lung cancer: role in clinical response to EGFR tyrosine kinase inhibitors. Oncogene 28(Suppl. 1), S24–S31. doi: 10.1038/onc.2009.198
Gobin, S. J., Van Zutphen, M., Westerheide, S. D., Boss, J. M., and Van Den Elsen, P. J. (2001). The MHC-specific enhanceosome and its role in MHC class I and beta(2)-microglobulin gene transactivation. J. Immunol. 167, 5175–5184. doi: 10.4049/jimmunol.167.9.5175
Golub, M. S., Germann, S. L., and Lloyd, K. C. (2004). Behavioral characteristics of a nervous system-specific erbB4 knock-out mouse. Behav. Brain Res. 153, 159–170. doi: 10.1016/j.bbr.2003.11.010
Gonzalez, P. A., Carreno, L. J., Coombs, D., Mora, J. E., Palmieri, E., Goldstein, B., et al. (2005). T cell receptor binding kinetics required for T cell activation depend on the density of cognate ligand on the antigen-presenting cell. Proc. Natl. Acad. Sci. U.S.A. 102, 4824–4829. doi: 10.1073/pnas.0500922102
Grandis, J. R., and Tweardy, D. J. (1993). Elevated levels of transforming growth factor alpha and epidermal growth factor receptor messenger RNA are early markers of carcinogenesis in head and neck cancer. Cancer Res. 53, 3579–3584.
Greer, S. F., Harton, J. A., Linhoff, M. W., Janczak, C. A., Ting, J. P., and Cressman, D. E. (2004). Serine residues 286, 288, and 293 within the CIITA: a mechanism for down-regulating CIITA activity through phosphorylation. J. Immunol. 173, 376–383. doi: 10.4049/jimmunol.173.1.376
Han, W., Pan, H., Jiang, L., Wei, K., Zou, D., and Zhang, Z. (2011). A novel approach to rescue immune escape in oral squamous cell carcinoma: Combined use of interferon-gamma and LY294002. Oncol. Rep. 25, 181–187.
Hastings, K. T. (2013). GILT: shaping the MHC class II-restricted peptidome and CD4(+) T cell-mediated immunity. Front. Immunol. 4:429. doi: 10.3389/fimmu.2013.00429
Herrmann, F., Lehr, H. A., Drexler, I., Sutter, G., Hengstler, J., Wollscheid, U., et al. (2004). HER-2/neu-mediated regulation of components of the MHC class I antigen-processing pathway. Cancer Res. 64, 215–220. doi: 10.1158/0008-5472.CAN-2522-2
Hirata, A., Hosoi, F., Miyagawa, M., Ueda, S., Naito, S., Fujii, T., et al. (2005). HER2 overexpression increases sensitivity to gefitinib, an epidermal growth factor receptor tyrosine kinase inhibitor, through inhibition of HER2/HER3 heterodimer formation in lung cancer cells. Cancer Res. 65, 4253–4260. doi: 10.1158/0008-5472.CAN-04-2748
Hofmann, M., Stoss, O., Shi, D., Buttner, R., Van De Vijver, M., Kim, W., et al. (2008). Assessment of a HER2 scoring system for gastric cancer: results from a validation study. Histopathology 52, 797–805. doi: 10.1111/j.1365-2559.2008.03028.x
Howlader, N., Altekruse, S. F., Li, C. I., Chen, V. W., Clarke, C. A., Ries, L. A., et al. (2014). US incidence of breast cancer subtypes defined by joint hormone receptor and HER2 status. J. Natl. Cancer Inst. 106, dju055. doi: 10.1093/jnci/dju055
Huang, H. E., Chin, S. F., Ginestier, C., Bardou, V. J., Adelaide, J., Iyer, N. G., et al. (2004). A recurrent chromosome breakpoint in breast cancer at the NRG1/neuregulin 1/heregulin gene. Cancer Res. 64, 6840–6844. doi: 10.1158/0008-5472.CAN-04-1762
Hu-Lieskovan, S., Mok, S., Homet Moreno, B., Tsoi, J., Robert, L., Goedert, L., et al. (2015). Improved antitumor activity of immunotherapy with BRAF and MEK inhibitors in BRAF(V600E) melanoma. Sci. Transl. Med. 7, 279ra241. doi: 10.1126/scitranslmed.aaa4691
Hynes, N. E., and Lane, H. A. (2005). ERBB receptors and cancer: the complexity of targeted inhibitors. Nat. Rev. Cancer 5, 341–354. doi: 10.1038/nrc1609
Jhappan, C., Stahle, C., Harkins, R. N., Fausto, N., Smith, G. H., and Merlino, G. T. (1990). TGF alpha overexpression in transgenic mice induces liver neoplasia and abnormal development of the mammary gland and pancreas. Cell 61, 1137–1146. doi: 10.1016/0092-8674(90)90076-Q
Jones, F. E., and Stern, D. F. (1999). Expression of dominant-negative ErbB2 in the mammary gland of transgenic mice reveals a role in lobuloalveolar development and lactation. Oncogene 18, 3481–3490. doi: 10.1038/sj.onc.1202698
Jones, F. E., Welte, T., Fu, X. Y., and Stern, D. F. (1999). ErbB4 signaling in the mammary gland is required for lobuloalveolar development and Stat5 activation during lactation. J. Cell Biol. 147, 77–88. doi: 10.1083/jcb.147.1.77
Kaczmarski, R. S., and Mufti, G. J. (1991). The cytokine receptor superfamily. Blood Rev. 5, 193–203. doi: 10.1016/0268-960X(91)90036-C
Kelderman, S., Schumacher, T. N., and Kvistborg, P. (2015). Mismatch repair-deficient cancers are targets for anti-PD-1 therapy. Cancer Cell 28, 11–13. doi: 10.1016/j.ccell.2015.06.012
Khan, A. N., Gregorie, C. J., and Tomasi, T. B. (2008). Histone deacetylase inhibitors induce TAP, LMP, Tapasin genes and MHC class I antigen presentation by melanoma cells. Cancer Immunol. Immunother. 57, 647–654.
Kim, R., Emi, M., Tanabe, K., and Arihiro, K. (2006). Tumor-driven evolution of immunosuppressive networks during malignant progression. Cancer Res. 66, 5527–5536. doi: 10.1158/0008-5472.CAN-05-4128
Kobayashi, K. S., and van den Elsen, P. J. (2012). NLRC5: a key regulator of MHC class I-dependent immune responses. Nat. Rev. Immunol. 12, 813–820. doi: 10.1038/nri3339
Komatsu, Y., and Hayashi, H. (1998). Histone deacetylase inhibitors up-regulate the expression of cell surface MHC class-I molecules in B16/BL6 cells. J Antibiot. (Tokyo) 51, 89–91. doi: 10.7164/antibiotics.51.89
Kono, K., Sato, E., Naganuma, H., Takahashi, A., Mimura, K., Nukui, H., et al. (2004). Trastuzumab (Herceptin) enhances class I-restricted antigen presentation recognized by HER-2/neu-specific T cytotoxic lymphocytes. Clin. Cancer Res. 10, 2538–2544. doi: 10.1158/1078-0432.CCR-03-0424
Kono, M., Dunn, I. S., Durda, P. J., Butera, D., Rose, L. B., Haggerty, T. J., et al. (2006). Role of the mitogen-activated protein kinase signaling pathway in the regulation of human melanocytic antigen expression. Mol. Cancer Res. 4, 779–792. doi: 10.1158/1541-7786.MCR-06-0077
Kotekar, A. S., Weissman, J. D., Gegonne, A., Cohen, H., and Singer, D. S. (2008). Histone modifications, but not nucleosomal positioning, correlate with major histocompatibility complex class I promoter activity in different tissues in vivo. Mol. Cell. Biol. 28, 7323–7336. doi: 10.1128/MCB.00889-08
Kumai, T., Matsuda, Y., Oikawa, K., Aoki, N., Kimura, S., Harabuchi, Y., et al. (2013). EGFR inhibitors augment antitumour helper T-cell responses of HER family-specific immunotherapy. Br. J. Cancer 109, 2155–2166. doi: 10.1038/bjc.2013.577
Kumai, T., Ohkuri, T., Nagato, T., Matsuda, Y., Oikawa, K., Aoki, N., et al. (2015). Targeting HER-3 to elicit antitumor helper T cells against head and neck squamous cell carcinoma. Sci. Rep. 5:16280. doi: 10.1038/srep16280
Kwong, B. Y., Roberts, S. J., Silberzahn, T., Filler, R. B., Neustadter, J. H., Galan, A., et al. (2010). Molecular analysis of tumor-promoting CD8+ T cells in two-stage cutaneous chemical carcinogenesis. J. Invest. Dermatol. 130, 1726–1736. doi: 10.1038/jid.2009.362
Lahat, N., Sheinfeld, M., Sobel, E., Kinarty, A., and Kraiem, Z. (1992). Divergent effects of cytokines on human leukocyte antigen-DR antigen expression of neoplastic and non-neoplastic human thyroid cells. Cancer 69, 1799–1807. doi: 10.1002/1097-0142(19920401)69:7<1799::AID-CNCR2820690723>3.0.CO;2-8
Lampen, M. H., and van Hall, T. (2011). Strategies to counteract MHC-I defects in tumors. Curr. Opin. Immunol. 23, 293–298. doi: 10.1016/j.coi.2010.12.005
Lawrence, M. S., Stojanov, P., Mermel, C. H., Robinson, J. T., Garraway, L. A., Golub, T. R., et al. (2014). Discovery and saturation analysis of cancer genes across 21 tumour types. Nature 505, 495–501. doi: 10.1038/nature12912
Le, D. T., Uram, J. N., Wang, H., Bartlett, B. R., Kemberling, H., Eyring, A. D., et al. (2015). PD-1 blockade in tumors with mismatch-repair deficiency. N. Engl. J. Med. 372, 2509–2520. doi: 10.1056/NEJMoa1500596
Lee, K. F., Simon, H., Chen, H., Bates, B., Hung, M. C., and Hauser, C. (1995). Requirement for neuregulin receptor erbB2 in neural and cardiac development. Nature 378, 394–398. doi: 10.1038/378394a0
Lei, S., Appert, H. E., Nakata, B., Domenico, D. R., Kim, K., and Howard, J. M. (1995). Overexpression of HER2/neu oncogene in pancreatic cancer correlates with shortened survival. Int. J. Pancreatol. 17, 15–21.
Lemmon, M. A., Schlessinger, J., and Ferguson, K. M. (2014). The EGFR family: not so prototypical receptor tyrosine kinases. Cold Spring Harb. Perspect. Biol. 6, a020768. doi: 10.1101/cshperspect.a020768
Leu, M., Bellmunt, E., Schwander, M., Farinas, I., Brenner, H. R., and Muller, U. (2003). Erbb2 regulates neuromuscular synapse formation and is essential for muscle spindle development. Development 130, 2291–2301. doi: 10.1242/dev.00447
Li, G., and Wong, A. J. (2008). EGF receptor variant III as a target antigen for tumor immunotherapy. Expert Rev. Vaccines 7, 977–985. doi: 10.1586/14760584.7.7.977
Li, M. M., Monzon, F. A., Biegel, J. A., Jobanputra, V., Laffin, J. J., Levy, B., et al. (2015). A multicenter, cross-platform clinical validation study of cancer cytogenomic arrays. Cancer Genet. 208, 525–536. doi: 10.1016/j.cancergen.2015.08.002
Lohmann, S., Wollscheid, U., Huber, C., and Seliger, B. (1996). Multiple levels of MHC class I down-regulation by ras oncogenes. Scand. J. Immunol. 43, 537–544. doi: 10.1046/j.1365-3083.1996.d01-73.x
Lollini, P. L., Nicoletti, G., Landuzzi, L., De Giovanni, C., Rossi, I., Di Carlo, E., et al. (1998). Down regulation of major histocompatibility complex class I expression in mammary carcinoma of HER-2/neu transgenic mice. Int. J. Cancer 77, 937–941. doi: 10.1002/(SICI)1097-0215(19980911)77:6<937::AID-IJC24>3.3.CO;2-O
Lopez-Albaitero, A., Nayak, J. V., Ogino, T., Machandia, A., Gooding, W., Deleo, A. B., et al. (2006). Role of antigen-processing machinery in the in vitro resistance of squamous cell carcinoma of the head and neck cells to recognition by CTL. J. Immunol. 176, 3402–3409. doi: 10.4049/jimmunol.176.6.3402
Lu, Y. C., and Robbins, P. F. (2016). Targeting neoantigens for cancer immunotherapy. Int. Immunol. 28, 365–370. doi: 10.1093/intimm/dxw026
Luetteke, N. C., Qiu, T. H., Fenton, S. E., Troyer, K. L., Riedel, R. F., Chang, A., et al. (1999). Targeted inactivation of the EGF and amphiregulin genes reveals distinct roles for EGF receptor ligands in mouse mammary gland development. Development 126, 2739–2750.
Lynch, T. J., Bell, D. W., Sordella, R., Gurubhagavatula, S., Okimoto, R. A., Brannigan, B. W., et al. (2004). Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N. Engl. J. Med. 350, 2129–2139. doi: 10.1056/NEJMoa040938
Macdonald-Obermann, J. L., and Pike, L. J. (2014). Different epidermal growth factor (EGF) receptor ligands show distinct kinetics and biased or partial agonism for homodimer and heterodimer formation. J. Biol. Chem. 289, 26178–26188. doi: 10.1074/jbc.M114.586826
Magner, W. J., Kazim, A. L., Stewart, C., Romano, M. A., Catalano, G., Grande, C., et al. (2000). Activation of MHC class I, II, and CD40 gene expression by histone deacetylase inhibitors. J. Immunol. 165, 7017–7024.
Marincola, F. M., Jaffee, E. M., Hicklin, D. J., and Ferrone, S. (2000). Escape of human solid tumors from T-cell recognition: molecular mechanisms and functional significance. Adv. Immunol. 74, 181–273. doi: 10.1016/S0065-2776(08)60911-6
Martins, I., Deshayes, F., Baton, F., Forget, A., Ciechomska, I., Sylla, K., et al. (2007). Pathologic expression of MHC class II is driven by mitogen-activated protein kinases. Eur. J. Immunol. 37, 788–797. doi: 10.1002/eji.200636620
Meden, H., and Kuhn, W. (1997). Overexpression of the oncogene c-erbB-2 (HER2/neu) in ovarian cancer: a new prognostic factor. Eur. J. Obstet. Gynecol. Reprod. Biol. 71, 173–179. doi: 10.1016/S0301-2115(96)02630-9
Meissner, T. B., Li, A., Biswas, A., Lee, K. H., Liu, Y. J., Bayir, E., et al. (2010). NLR family member NLRC5 is a transcriptional regulator of MHC class I genes. Proc. Natl. Acad. Sci. U.S.A. 107, 13794–13799. doi: 10.1073/pnas.1008684107
Meissner, T. B., Liu, Y. J., Lee, K. H., Li, A., Biswas, A., Van Eggermond, M. C., et al. (2012). NLRC5 cooperates with the RFX transcription factor complex to induce MHC class I gene expression. J. Immunol. 188, 4951–4958. doi: 10.4049/jimmunol.1103160
Mendelsohn, J., and Baselga, J. (2000). The EGF receptor family as targets for cancer therapy. Oncogene 19, 6550–6565. doi: 10.1038/sj.onc.1204082
Meulenbroeks, C., Van Weelden, H., Schwartz, C., Voehringer, D., Redegeld, F. A., Rutten, V. P., et al. (2015). Basophil-derived amphiregulin is essential for UVB irradiation-induced immune suppression. J. Invest. Dermatol. 135, 222–228. doi: 10.1038/jid.2014.329
Miettinen, P. J., Berger, J. E., Meneses, J., Phung, Y., Pedersen, R. A., Werb, Z., et al. (1995). Epithelial immaturity and multiorgan failure in mice lacking epidermal growth factor receptor. Nature 376, 337–341. doi: 10.1038/376337a0
Mimura, K., Shiraishi, K., Mueller, A., Izawa, S., Kua, L. F., So, J., et al. (2013). The MAPK pathway is a predominant regulator of HLA-A expression in esophageal and gastric cancer. J. Immunol. 191, 6261–6272. doi: 10.4049/jimmunol.1301597
Mitra, R. S., and Nickoloff, B. J. (1992). Epidermal growth factor and transforming growth factor-alpha decrease gamma interferon receptors and induction of intercellular adhesion molecule (ICAM-1) on cultured keratinocytes. J. Cell. Physiol. 150, 264–268. doi: 10.1002/jcp.1041500207
Morgan, J. E., Shanderson, R. L., Boyd, N. H., Cacan, E., and Greer, S. F. (2015). The class II transactivator (CIITA) is regulated by post-translational modification cross-talk between ERK1/2 phosphorylation, mono-ubiquitination and Lys63 ubiquitination. Biosci. Rep. 35:e00233. doi: 10.1042/BSR20150091
Moroni, M., Veronese, S., Benvenuti, S., Marrapese, G., Sartore-Bianchi, A., Di Nicolantonio, F., et al. (2005). Gene copy number for epidermal growth factor receptor (EGFR) and clinical response to antiEGFR treatment in colorectal cancer: a cohort study. Lancet Oncol. 6, 279–286. doi: 10.1016/S1470-2045(05)70102-9
Mortenson, E. D., Park, S., Jiang, Z., Wang, S., and Fu, Y. X. (2013). Effective anti-neu-initiated antitumor responses require the complex role of CD4+ T cells. Clin. Cancer Res. 19, 1476–1486. doi: 10.1158/1078-0432.CCR-12-2522
Nagashima, T., Oyama, M., Kozuka-Hata, H., Yumoto, N., Sakaki, Y., and Hatakeyama, M. (2008). Phosphoproteome and transcriptome analyses of ErbB ligand-stimulated MCF-7 cells. Cancer Genomics Proteomics 5, 161–168.
Nagashima, T., Shimodaira, H., Ide, K., Nakakuki, T., Tani, Y., Takahashi, K., et al. (2007). Quantitative transcriptional control of ErbB receptor signaling undergoes graded to biphasic response for cell differentiation. J. Biol. Chem. 282, 4045–4056. doi: 10.1074/jbc.M608653200
Neal, D. E., Marsh, C., Bennett, M. K., Abel, P. D., Hall, R. R., Sainsbury, J. R., et al. (1985). Epidermal-growth-factor receptors in human bladder cancer: comparison of invasive and superficial tumours. Lancet 1, 366–368. doi: 10.1016/S0140-6736(85)91386-8
Nedergaard, M. K., Hedegaard, C. J., and Poulsen, H. S. (2012). Targeting the epidermal growth factor receptor in solid tumor malignancies. Biodrugs 26, 83–99. doi: 10.2165/11599760-000000000-00000
Neerincx, A., Castro, W., Guarda, G., and Kufer, T. A. (2013). NLRC5, at the heart of antigen presentation. Front. Immunol. 4:397. doi: 10.3389/fimmu.2013.00397
Niikura, H., Sasano, H., Sato, S., and Yajima, A. (1997). Expression of epidermal growth factor-related proteins and epidermal growth factor receptor in common epithelial ovarian tumors. Int. J. Gynecol. Pathol. 16, 60–68. doi: 10.1097/00004347-199701000-00010
Nistico, P., Mottolese, M., Cascioli, S., Benevolo, M., Del Bello, D., Di Modugno, F., et al. (1999). Host immunosurveillance contributes to the control of erbB-2 overexpression in HLA-A2-breast-cancer patients. Int. J. Cancer 84, 598–603. doi: 10.1002/(SICI)1097-0215(19991222)84:6<598::AID-IJC10>3.0.CO;2-7
Nistico, P., Mottolese, M., Mammi, C., Benevolo, M., Del Bello, D., Rubiu, O., et al. (1997). Low frequency of ErbB-2 proto-oncogene overexpression in human leukocyte antigen-A2-positive breast cancer patients. J. Natl. Cancer Inst. 89, 319–321. doi: 10.1093/jnci/89.4.319
Normanno, N., Bianco, C., De Luca, A., Maiello, M. R., and Salomon, D. S. (2003a). Target-based agents against ErbB receptors and their ligands: a novel approach to cancer treatment. Endocr. Relat. Cancer 10, 1–21. doi: 10.1677/erc.0.0100001
Normanno, N., De Luca, A., Bianco, C., Strizzi, L., Mancino, M., Maiello, M. R., et al. (2006). Epidermal growth factor receptor (EGFR) signaling in cancer. Gene 366, 2–16. doi: 10.1016/j.gene.2005.10.018
Normanno, N., Maiello, M. R., and De Luca, A. (2003b). Epidermal growth factor receptor tyrosine kinase inhibitors (EGFR-TKIs): simple drugs with a complex mechanism of action? J. Cell. Physiol. 194, 13–19. doi: 10.1002/jcp.10194
Nouri, A. M., Hussain, R. F., and Oliver, R. T. (1995). Epidermal growth factor-induced protection of tumour cell susceptibility to cytolysis. Eur. J. Cancer 31a, 963–969. doi: 10.1016/0959-8049(95)00120-4
O-Charoenrat, P., Rhys-Evans, P. H., Modjtahedi, H., and Eccles, S. A. (2002). The role of c-erbB receptors and ligands in head and neck squamous cell carcinoma. Oral Oncol. 38, 627–640. doi: 10.1016/S1368-8375(02)00029-5
Okita, R., Wolf, D., Yasuda, K., Maeda, A., Yukawa, T., Saisho, S., et al. (2015). Contrasting effects of the cytotoxic anticancer drug gemcitabine and the EGFR tyrosine kinase inhibitor gefitinib on NK cell-mediated cytotoxicity via regulation of NKG2D ligand in non-small-cell lung cancer cells. PLoS ONE 10:e0139809. doi: 10.1371/journal.pone.0139809
Okwan-Duodu, D., Pollack, B. P., Lawson, D., and Khan, M. K. (2015). Role of radiation therapy as immune activator in the era of modern immunotherapy for metastatic malignant melanoma. Am. J. Clin. Oncol. 38, 119–125. doi: 10.1097/COC.0b013e3182940dc3
Oliveras-Ferraros, C., Cufi, S., Vazquez-Martin, A., Menendez, O. J., Bosch-Barrera, J., Martin-Castillo, B., et al. (2012). Metformin rescues cell surface major histocompatibility complex class I (MHC-I) deficiency caused by oncogenic transformation. Cell Cycle 11, 865–870. doi: 10.4161/cc.11.5.19252
Oxnard, G. R., Binder, A., and Janne, P. A. (2013). New targetable oncogenes in non-small-cell lung cancer. J. Clin. Oncol. 31, 1097–1104. doi: 10.1200/JCO.2012.42.9829
Paez, J. G., Janne, P. A., Lee, J. C., Tracy, S., Greulich, H., Gabriel, S., et al. (2004). EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science 304, 1497–1500. doi: 10.1126/science.1099314
Paff, M., Alexandru-Abrams, D., Hsu, F. P., and Bota, D. A. (2014). The evolution of the EGFRvIII (rindopepimut) immunotherapy for glioblastoma multiforme patients. Hum. Vaccin. Immunother. 10, 3322–3331. doi: 10.4161/21645515.2014.983002
Park, S. K., Miller, R., Krane, I., and Vartanian, T. (2001). The erbB2 gene is required for the development of terminally differentiated spinal cord oligodendrocytes. J. Cell Biol. 154, 1245–1258. doi: 10.1083/jcb.200104025
Pedersen, M. W., Pedersen, N., Damstrup, L., Villingshoj, M., Sonder, S. U., Rieneck, K., et al. (2005). Analysis of the epidermal growth factor receptor specific transcriptome: effect of receptor expression level and an activating mutation. J. Cell. Biochem. 96, 412–427. doi: 10.1002/jcb.20554
Pennini, M. E., Pai, R. K., Schultz, D. C., Boom, W. H., and Harding, C. V. (2006). Mycobacterium tuberculosis 19-kDa lipoprotein inhibits IFN-gamma-induced chromatin remodeling of MHC2TA by TLR2 and MAPK signaling. J. Immunol. 176, 4323–4330. doi: 10.4049/jimmunol.176.7.4323
Pines, G., Kostler, W. J., and Yarden, Y. (2010). Oncogenic mutant forms of EGFR: lessons in signal transduction and targets for cancer therapy. FEBS Lett. 584, 2699–2706. doi: 10.1016/j.febslet.2010.04.019
Pinkas-Kramarski, R., Soussan, L., Waterman, H., Levkowitz, G., Alroy, I., Klapper, L., et al. (1996). Diversification of Neu differentiation factor and epidermal growth factor signaling by combinatorial receptor interactions. EMBO J. 15, 2452–2467.
Pollack, B. P., Sapkota, B., and Cartee, T. V. (2011). Epidermal growth factor receptor inhibition augments the expression of MHC class I and II genes. Clin. Cancer Res. 17, 4400–4413. doi: 10.1158/1078-0432.CCR-10-3283
Prenzel, N., Zwick, E., Leserer, M., and Ullrich, A. (2000). Tyrosine kinase signalling in breast cancer. Epidermal growth factor receptor: convergence point for signal integration and diversification. Breast Cancer Res. 2, 184–190.
Prickett, T. D., Agrawal, N. S., Wei, X., Yates, K. E., Lin, J. C., Wunderlich, J. R., et al. (2009). Analysis of the tyrosine kinome in melanoma reveals recurrent mutations in ERBB4. Nat. Genet. 41, 1127–1132. doi: 10.1038/ng.438
Qi, Y., Operario, D. J., Georas, S. N., and Mosmann, T. R. (2012). The acute environment, rather than T cell subset pre-commitment, regulates expression of the human T cell cytokine amphiregulin. PLoS ONE 7:e39072. doi: 10.1371/journal.pone.0039072
Raj, E. H., Skinner, A., Mahji, U., Nirmala, K. N., Ravichandran, K., Shanta, V., et al. (2001). Neuregulin 1-alpha expression in locally advanced breast cancer. Breast 10, 41–45. doi: 10.1054/brst.2000.0182
Reith, W., and Boss, J. M. (2008). New dimensions of CIITA. Nat. Immunol. 9, 713–714. doi: 10.1038/ni0708-713
Roche, P. A., and Furuta, K. (2015). The ins and outs of MHC class II-mediated antigen processing and presentation. Nat. Rev. Immunol. 15, 203–216. doi: 10.1038/nri3818
Rodon, J., Dienstmann, R., Serra, V., and Tabernero, J. (2013). Development of PI3K inhibitors: lessons learned from early clinical trials. Nat. Rev. Clin. Oncol. 10, 143–153. doi: 10.1038/nrclinonc.2013.10
Rolitsky, C. D., Theil, K. S., Mcgaughy, V. R., Copeland, L. J., and Niemann, T. H. (1999). HER-2/neu amplification and overexpression in endometrial carcinoma. Int. J. Gynecol. Pathol. 18, 138–143. doi: 10.1097/00004347-199904000-00007
Roskoski, R. Jr. (2014). The ErbB/HER family of protein-tyrosine kinases and cancer. Pharmacol. Res. 79, 34–74. doi: 10.1016/j.phrs.2013.11.002
Rusch, V., Klimstra, D., Venkatraman, E., Pisters, P. W., Langenfeld, J., and Dmitrovsky, E. (1997). Overexpression of the epidermal growth factor receptor and its ligand transforming growth factor alpha is frequent in resectable non-small cell lung cancer but does not predict tumor progression. Clin. Cancer Res. 3, 515–522.
Rutledge, W. C., Kong, J., Gao, J., Gutman, D. A., Cooper, L. A., Appin, C., et al. (2013). Tumor-infiltrating lymphocytes in glioblastoma are associated with specific genomic alterations and related to transcriptional class. Clin. Cancer Res. 19, 4951–4960. doi: 10.1158/1078-0432.CCR-13-0551
Saba, N. F., Wilson, M., Doho, G., Dasilva, J., Benjamin Isett, R., Newman, S., et al. (2015). Mutation and transcriptional profiling of formalin-fixed paraffin embedded specimens as companion methods to immunohistochemistry for determining therapeutic targets in Oropharyngeal Squamous Cell Carcinoma (OPSCC): a pilot of proof of principle. Head Neck Pathol. 9, 223–235. doi: 10.1007/s12105-014-0566-0
Sabbatino, F., Wang, Y., Scognamiglio, G., Favoino, E., Feldman, S. A., Villani, V., et al. (2016). Antitumor activity of BRAF inhibitor and IFNalpha combination in BRAF-mutant melanoma. J. Natl. Cancer Inst. 108, djv435. doi: 10.1093/jnci/djv435
Saffari, B., Jones, L. A., El-Naggar, A., Felix, J. C., George, J., and Press, M. F. (1995). Amplification and overexpression of HER-2/neu (c-erbB2) in endometrial cancers: correlation with overall survival. Cancer Res. 55, 5693–5698.
Salomon, D. S., Brandt, R., Ciardiello, F., and Normanno, N. (1995). Epidermal growth factor-related peptides and their receptors in human malignancies. Crit. Rev. Oncol. Hematol. 19, 183–232. doi: 10.1016/1040-8428(94)00144-I
Sampson, J. H., Archer, G. E., Mitchell, D. A., Heimberger, A. B., and Bigner, D. D. (2008). Tumor-specific immunotherapy targeting the EGFRvIII mutation in patients with malignant glioma. Semin. Immunol. 20, 267–275. doi: 10.1016/j.smim.2008.04.001
Sandgren, E. P., Luetteke, N. C., Palmiter, R. D., Brinster, R. L., and Lee, D. C. (1990). Overexpression of TGF alpha in transgenic mice: induction of epithelial hyperplasia, pancreatic metaplasia, and carcinoma of the breast. Cell 61, 1121–1135. doi: 10.1016/0092-8674(90)90075-P
Sapkota, B., Hill, C. E., and Pollack, B. P. (2013). Vemurafenib enhances MHC induction in BRAFV600E homozygous melanoma cells. Oncoimmunology 2, e22890. doi: 10.4161/onci.22890
Sato, K., Moriyama, M., Mori, S., Saito, M., Watanuki, T., Terada, K., et al. (1992). An immunohistologic evaluation of C-erbB-2 gene product in patients with urinary bladder carcinoma. Cancer 70, 2493–2498. doi: 10.1002/1097-0142(19921115)70:10<2493::AID-CNCR2820701017>3.0.CO;2-K
Sauter, G., Moch, H., Moore, D., Carroll, P., Kerschmann, R., Chew, K., et al. (1993). Heterogeneity of erbB-2 gene amplification in bladder cancer. Cancer Res. 53, 2199–2203.
Schaefer, G., Shao, L., Totpal, K., and Akita, R. W. (2007). Erlotinib directly inhibits HER2 kinase activation and downstream signaling events in intact cells lacking epidermal growth factor receptor expression. Cancer Res. 67, 1228–1238. doi: 10.1158/0008-5472.CAN-06-3493
Scher, H. I., Sarkis, A., Reuter, V., Cohen, D., Netto, G., Petrylak, D., et al. (1995). Changing pattern of expression of the epidermal growth factor receptor and transforming growth factor alpha in the progression of prostatic neoplasms. Clin. Cancer Res. 1, 545–550.
Schneble, E. J., Berry, J. S., Trappey, F. A., Clifton, G. T., Ponniah, S., Mittendorf, E., et al. (2014). The HER2 peptide nelipepimut-S (E75) vaccine (NeuVax) in breast cancer patients at risk for recurrence: correlation of immunologic data with clinical response. Immunotherapy 6, 519–531. doi: 10.2217/imt.14.22
Schneider, M. R., Antsiferova, M., Feldmeyer, L., Dahlhoff, M., Bugnon, P., Hasse, S., et al. (2008). Betacellulin regulates hair follicle development and hair cycle induction and enhances angiogenesis in wounded skin. J. Invest. Dermatol. 128, 1256–1265. doi: 10.1038/sj.jid.5701135
Schneider, M. R., and Wolf, E. (2009). The epidermal growth factor receptor ligands at a glance. J. Cell. Physiol. 218, 460–466. doi: 10.1002/jcp.21635
Schreiber, A. B., Schlessinger, J., and Edidin, M. (1984). Interaction between major histocompatibility complex antigens and epidermal growth factor receptors on human cells. J. Cell Biol. 98, 725–731. doi: 10.1083/jcb.98.2.725
Schumacher, T., Bunse, L., Pusch, S., Sahm, F., Wiestler, B., Quandt, J., et al. (2014). A vaccine targeting mutant IDH1 induces antitumour immunity. Nature 512, 324–327. doi: 10.1038/nature13387
Seliger, B. (2008a). Different regulation of MHC class I antigen processing components in human tumors. J. Immunotoxicol. 5, 361–367. doi: 10.1080/15476910802482870
Seliger, B. (2008b). Molecular mechanisms of MHC class I abnormalities and APM components in human tumors. Cancer Immunol. Immunother. 57, 1719–1726. doi: 10.1007/s00262-008-0515-4
Seliger, B. (2014). The link between MHC class I abnormalities of tumors, oncogenes, tumor suppressor genes, and transcription factors. J. Immunotoxicol. 11, 308–310. doi: 10.3109/1547691X.2013.875084
Seliger, B., Harders, C., Lohmann, S., Momburg, F., Urlinger, S., Tampe, R., et al. (1998). Down-regulation of the MHC class I antigen-processing machinery after oncogenic transformation of murine fibroblasts. Eur. J. Immunol. 28, 122–133. doi: 10.1002/(SICI)1521-4141(199801)28:01<122::AID-IMMU122>3.0.CO;2-F
Seliger, B., Harders, C., Wollscheid, U., Staege, M. S., Reske-Kunz, A. B., and Huber, C. (1996). Suppression of MHC class I antigens in oncogenic transformants: association with decreased recognition by cytotoxic T lymphocytes. Exp. Hematol. 24, 1275–1279.
Seliger, B., and Kiessling, R. (2013). The two sides of HER2/neu: immune escape versus surveillance. Trends Mol. Med. 19, 677–684. doi: 10.1016/j.molmed.2013.08.003
Seliger, B., Killian, M., and Pfizenmaier, K. (1988). Distinct mechanisms of interferon-gamma and tumor necrosis factor-alpha action in oncogene-transformed mouse fibroblasts. J. Cell. Biochem. 38, 205–212. doi: 10.1002/jcb.240380308
Seliger, B., Maeurer, M. J., and Ferrone, S. (2000). Antigen-processing machinery breakdown and tumor growth. Immunol. Today 21, 455–464. doi: 10.1016/S0167-5699(00)01692-3
Seliger, B., and Pfizenmaier, K. (1989). Post-transcriptional downregulation of MHC class I expression in oncogene-transformed cells is reverted by IFN-gamma and TNF-alpha. J. Immunogenet. 16, 315–320. doi: 10.1111/j.1744-313X.1989.tb00477.x
Sers, C., Kuner, R., Falk, C. S., Lund, P., Sueltmann, H., Braun, M., et al. (2009). Down-regulation of HLA Class I and NKG2D ligands through a concerted action of MAPK and DNA methyltransferases in colorectal cancer cells. Int. J. Cancer 125, 1626–1639. doi: 10.1002/ijc.24557
Shintani, S., Funayama, T., Yoshihama, Y., Alcalde, R. E., and Matsumura, T. (1995). Prognostic significance of ERBB3 overexpression in oral squamous cell carcinoma. Cancer Lett. 95, 79–83. doi: 10.1016/0304-3835(95)03866-U
Shostak, K., and Chariot, A. (2015). EGFR and NF-kappaB: partners in cancer. Trends Mol. Med. 21, 385–393. doi: 10.1016/j.molmed.2015.04.001
Sibilia, M., Steinbach, J. P., Stingl, L., Aguzzi, A., and Wagner, E. F. (1998). A strain-independent postnatal neurodegeneration in mice lacking the EGF receptor. EMBO J. 17, 719–731. doi: 10.1093/emboj/17.3.719
Sibilia, M., and Wagner, E. F. (1995). Strain-dependent epithelial defects in mice lacking the EGF receptor. Science 269, 234–238. doi: 10.1126/science.7618085
Sidaway, P. (2015). Targeted therapies: drug addiction revealed in BRAF and MEK inhibitor-resistant melanoma cells. Nat. Rev. Clin. Oncol. 12:189. doi: 10.1038/nrclinonc.2015.28
Slattery, M. L., John, E. M., Stern, M. C., Herrick, J., Lundgreen, A., Giuliano, A. R., et al. (2013). Associations with growth factor genes (FGF1, FGF2, PDGFB, FGFR2, NRG2, EGF, ERBB2) with breast cancer risk and survival: the breast cancer health disparities study. Breast Cancer Res. Treat. 140, 587–601. doi: 10.1007/s10549-013-2644-5
Snyder, A., Makarov, V., Merghoub, T., Yuan, J., Zaretsky, J. M., Desrichard, A., et al. (2014). Genetic basis for clinical response to CTLA-4 blockade in melanoma. N. Engl. J. Med. 371, 2189–2199. doi: 10.1056/NEJMoa1406498
Sorensen, M. R., Holst, P. J., Pircher, H., Christensen, J. P., and Thomsen, A. R. (2009). Vaccination with an adenoviral vector encoding the tumor antigen directly linked to invariant chain induces potent CD4(+) T-cell-independent CD8(+) T-cell-mediated tumor control. Eur. J. Immunol. 39, 2725–2736. doi: 10.1002/eji.200939543
Soung, Y. H., Lee, J. W., Kim, S. Y., Wang, Y. P., Jo, K. H., Moon, S. W., et al. (2006). Somatic mutations of the ERBB4 kinase domain in human cancers. Int. J. Cancer 118, 1426–1429. doi: 10.1002/ijc.21507
Sporn, M. B., and Roberts, A. B. (1985). Autocrine growth factors and cancer. Nature 313, 745–747. doi: 10.1038/313745a0
Tanner, M., Hollmen, M., Junttila, T. T., Kapanen, A. I., Tommola, S., Soini, Y., et al. (2005). Amplification of HER-2 in gastric carcinoma: association with Topoisomerase IIalpha gene amplification, intestinal type, poor prognosis and sensitivity to trastuzumab. Ann. Oncol. 16, 273–278. doi: 10.1093/annonc/mdi064
Thorpe, L. M., Yuzugullu, H., and Zhao, J. J. (2015). PI3K in cancer: divergent roles of isoforms, modes of activation and therapeutic targeting. Nat. Rev. Cancer 15, 7–24. doi: 10.1038/nrc3860
Threadgill, D. W., Dlugosz, A. A., Hansen, L. A., Tennenbaum, T., Lichti, U., Yee, D., et al. (1995). Targeted disruption of mouse EGF receptor: effect of genetic background on mutant phenotype. Science 269, 230–234. doi: 10.1126/science.7618084
Troyer, K. L., and Lee, D. C. (2001). Regulation of mouse mammary gland development and tumorigenesis by the ERBB signaling network. J. Mammary Gland Biol. Neoplasia 6, 7–21. doi: 10.1023/A:1009560330359
Tzahar, E., Waterman, H., Chen, X., Levkowitz, G., Karunagaran, D., Lavi, S., et al. (1996). A hierarchical network of interreceptor interactions determines signal transduction by Neu differentiation factor/neuregulin and epidermal growth factor. Mol. Cell. Biol. 16, 5276–5287. doi: 10.1128/MCB.16.10.5276
Umelo, I., Noeparast, A., Chen, G., Renard, M., Geers, C., Vansteenkiste, J., et al. (2016). Identification of a novel HER3 activating mutation homologous to EGFR-L858R in lung cancer. Oncotarget 7, 3068–3083. doi: 10.18632/oncotarget.6585
Van Allen, E. M., Miao, D., Schilling, B., Shukla, S. A., Blank, C., Zimmer, L., et al. (2015). Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science 350, 207–211. doi: 10.1126/science.aad0095
van den Elsen, P. J. (2011). Expression regulation of major histocompatibility complex class I and class II encoding genes. Front. Immunol. 2:48. doi: 10.3389/fimmu.2011.00048
van den Elsen, P. J., Gobin, S. J., Van Eggermond, M. C., and Peijnenburg, A. (1998). Regulation of MHC class I and II gene transcription: differences and similarities. Immunogenetics 48, 208–221. doi: 10.1007/s002510050425
van den Elsen, P. J., Holling, T. M., Kuipers, H. F., and Van Der Stoep, N. (2004). Transcriptional regulation of antigen presentation. Curr. Opin. Immunol. 16, 67–75. doi: 10.1016/j.coi.2003.11.015
van der Burg, S. H., Arens, R., Ossendorp, F., Van Hall, T., and Melief, C. J. (2016). Vaccines for established cancer: overcoming the challenges posed by immune evasion. Nat. Rev. Cancer 16, 219–233. doi: 10.1038/nrc.2016.16
van Rooij, N., Van Buuren, M. M., Philips, D., Velds, A., Toebes, M., Heemskerk, B., et al. (2013). Tumor exome analysis reveals neoantigen-specific T-cell reactivity in an ipilimumab-responsive melanoma. J. Clin. Oncol. 31, e439–e442. doi: 10.1200/JCO.2012.47.7521
Vanguri, V., Govern, C. C., Smith, R., and Huseby, E. S. (2013). Viral antigen density and confinement time regulate the reactivity pattern of CD4 T-cell responses to vaccinia virus infection. Proc. Natl. Acad. Sci. U.S.A. 110, 288–293. doi: 10.1073/pnas.1208328110
Vantourout, P., Willcox, C., Turner, A., Swanson, C. M., Haque, Y., Sobolev, O., et al. (2014). Immunological visibility: posttranscriptional regulation of human NKG2D ligands by the EGF receptor pathway. Sci. Transl. Med. 6, 231ra249. doi: 10.1126/scitranslmed.3007579
Vassar, R., and Fuchs, E. (1991). Transgenic mice provide new insights into the role of TGF-alpha during epidermal development and differentiation. Genes Dev. 5, 714–727. doi: 10.1101/gad.5.5.714
Voong, L. N., Slater, A. R., Kratovac, S., and Cressman, D. E. (2008). Mitogen-activated protein kinase ERK1/2 regulates the class II transactivator. J. Biol. Chem. 283, 9031–9039. doi: 10.1074/jbc.M706487200
Ward, J. P., Gubin, M. M., and Schreiber, R. D. (2016). The role of neoantigens in naturally occurring and therapeutically induced immune responses to cancer. Adv. Immunol. 130, 25–74. doi: 10.1016/bs.ai.2016.01.001
Wargo, J. A., Cooper, Z. A., and Flaherty, K. T. (2014). Universes collide: combining immunotherapy with targeted therapy for cancer. Cancer Discov. 4, 1377–1386. doi: 10.1158/2159-8290.CD-14-0477
Watanabe, K., Tachibana, O., Sata, K., Yonekawa, Y., Kleihues, P., and Ohgaki, H. (1996). Overexpression of the EGF receptor and p53 mutations are mutually exclusive in the evolution of primary and secondary glioblastomas. Brain Pathol. 6, 217–223; discussion223–214. doi: 10.1111/j.1750-3639.1996.tb00848.x
Weiner, D. B., Nordberg, J., Robinson, R., Nowell, P. C., Gazdar, A., Greene, M. I., et al. (1990). Expression of the neu gene-encoded protein (P185neu) in human non-small cell carcinomas of the lung. Cancer Res. 50, 421–425.
Whipple, C. A., Boni, A., Fisher, J. L., Hampton, T. H., Tsongalis, G. J., Mellinger, D. L., et al. (2016). The mitogen-activated protein kinase pathway plays a critical role in regulating immunological properties of BRAF mutant cutaneous melanoma cells. Melanoma Res. 26, 223–235. doi: 10.1097/CMR.0000000000000244
Whiteside, T. L., Demaria, S., Rodriguez-Ruiz, M. E., Zarour, H. M., and Melero, I. (2016). Emerging opportunities and challenges in cancer immunotherapy. Clin. Cancer Res. 22, 1845–1855. doi: 10.1158/1078-0432.CCR-16-0049
Wiesen, J. F., Young, P., Werb, Z., and Cunha, G. R. (1999). Signaling through the stromal epidermal growth factor receptor is necessary for mammary ductal development. Development 126, 335–344.
Wong, R. W. (2003). Transgenic and knock-out mice for deciphering the roles of EGFR ligands. Cell. Mol. Life Sci. 60, 113–118. doi: 10.1007/s000180300007
Wong, R. W., Kwan, R. W., Mak, P. H., Mak, K. K., Sham, M. H., and Chan, S. Y. (2000). Overexpression of epidermal growth factor induced hypospermatogenesis in transgenic mice. J. Biol. Chem. 275, 18297–18301. doi: 10.1074/jbc.M001965200
Wu, S. J., Jiang, S. Y., Wu, J., and Xiong, G. L. (2015). Association between EGF +61 A > G polymorphism and gastric cancer risk: a meta-analysis. J. Huazhong Univ. Sci. Technolog. Med. Sci. 35, 327–332. doi: 10.1007/s11596-015-1432-3
Wu, X., Kong, X., Luchsinger, L., Smith, B. D., and Xu, Y. (2009). Regulating the activity of class II transactivator by posttranslational modifications: exploring the possibilities. Mol. Cell. Biol. 29, 5639–5644. doi: 10.1128/MCB.00661-09
Xian, C. J., and Zhou, X. F. (2004). EGF family of growth factors: essential roles and functional redundancy in the nerve system. Front. Biosci. 9:85–92.
Xie, W., Paterson, A. J., Chin, E., Nabell, L. M., and Kudlow, J. E. (1997). Targeted expression of a dominant negative epidermal growth factor receptor in the mammary gland of transgenic mice inhibits pubertal mammary duct development. Mol. Endocrinol. 11, 1766–1781. doi: 10.1210/mend.11.12.0019
Yamanaka, Y., Friess, H., Kobrin, M. S., Buchler, M., Beger, H. G., and Korc, M. (1993a). Coexpression of epidermal growth factor receptor and ligands in human pancreatic cancer is associated with enhanced tumor aggressiveness. Anticancer. Res. 13, 565–569.
Yamanaka, Y., Friess, H., Kobrin, M. S., Buchler, M., Kunz, J., Beger, H. G., et al. (1993b). Overexpression of HER2/neu oncogene in human pancreatic carcinoma. Hum. Pathol. 24, 1127–1134. doi: 10.1016/0046-8177(93)90194-L
Yanagawa, Y., Iijima, N., Iwabuchi, K., and Onoe, K. (2002). Activation of extracellular signal-related kinase by TNF-alpha controls the maturation and function of murine dendritic cells. J. Leukoc. Biol. 71, 125–132.
Yano, T., Doi, T., Ohtsu, A., Boku, N., Hashizume, K., Nakanishi, M., et al. (2006). Comparison of HER2 gene amplification assessed by fluorescence in situ hybridization and HER2 protein expression assessed by immunohistochemistry in gastric cancer. Oncol. Rep. 15, 65–71.
Yoshihama, S., Roszik, J., Downs, I., Meissner, T. B., Vijayan, S., Chapuy, B., et al. (2016). NLRC5/MHC class I transactivator is a target for immune evasion in cancer. Proc. Natl. Acad. Sci. U.S.A. 113, 5999–6004. doi: 10.1073/pnas.1602069113
Zaiss, D. M., Gause, W. C., Osborne, L. C., and Artis, D. (2015). Emerging functions of amphiregulin in orchestrating immunity, inflammation, and tissue repair. Immunity 42, 216–226. doi: 10.1016/j.immuni.2015.01.020
Zaiss, D. M., Van Loosdregt, J., Gorlani, A., Bekker, C. P., Grone, A., Sibilia, M., et al. (2013). Amphiregulin enhances regulatory T cell-suppressive function via the epidermal growth factor receptor. Immunity 38, 275–284. doi: 10.1016/j.immuni.2012.09.023
Zaiss, D. M., Yang, L., Shah, P. R., Kobie, J. J., Urban, J. F., and Mosmann, T. R. (2006). Amphiregulin, a TH2 cytokine enhancing resistance to nematodes. Science 314:1746. doi: 10.1126/science.1133715
Zaks, T. Z., and Rosenberg, S. A. (1998). Immunization with a peptide epitope (p369-377) from HER-2/neu leads to peptide-specific cytotoxic T lymphocytes that fail to recognize HER-2/neu+ tumors. Cancer Res. 58, 4902–4908.
Zhao, Y., and Adjei, A. A. (2014). The clinical development of MEK inhibitors. Nat. Rev. Clin. Oncol. 11, 385–400. doi: 10.1038/nrclinonc.2014.83
Keywords: MHC class I, MHC class II, EGFR, ErbB receptors, immunology, signal transduction, gene expression regulation
Citation: Kersh AE, Sasaki M, Cooper LA, Kissick HT and Pollack BP (2016) Understanding the Impact of ErbB Activating Events and Signal Transduction on Antigen Processing and Presentation: MHC Expression as a Model. Front. Pharmacol. 7:327. doi: 10.3389/fphar.2016.00327
Received: 15 July 2016; Accepted: 06 September 2016;
Published: 26 September 2016.
Edited by:
Rolando Pérez Rodríguez, Centre of Molecular Immunology, CubaReviewed by:
Anna Rita Migliaccio, Icahn School of Medicine at Mount Sinai, USACopyright © 2016 Kersh, Sasaki, Cooper, Kissick and Pollack. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Brian P. Pollack, YnJpYW4ucG9sbGFja0BlbW9yeS5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.