- 1Department of Pharmacology, University of Washington, Seattle, WA, USA
- 2Howard Hughes Medical Institute, Chevy Chase, MD, USA
Phosphorylation events that occur in response to the second messenger cAMP are controlled spatially and temporally by protein kinase A (PKA) interacting with A-kinase anchoring proteins (AKAPs). Recent advances in understanding the structural basis for this interaction have reinforced the hypothesis that AKAPs create spatially constrained signaling microdomains. This has led to the realization that the PKA/AKAP interface is a potential drug target for modulating a plethora of cell-signaling events. Pharmacological disruption of kinase–AKAP interactions has previously been explored for disease treatment and remains an interesting area of research. However, disrupting or enhancing the association of phosphatases with AKAPs is a therapeutic concept of equal promise, particularly since they oppose the actions of many anchored kinases. Accordingly, numerous AKAPs bind phosphatases such as protein phosphatase 1 (PP1), calcineurin (PP2B), and PP2A. These multimodal signaling hubs are equally able to control the addition of phosphate groups onto target substrates, as well as the removal of these phosphate groups. In this review, we describe recent advances in structural analysis of kinase and phosphatase interactions with AKAPs, and suggest future possibilities for targeting these interactions for therapeutic benefit.
Characterizing Protein Kinase A (PKA) Anchoring
The cAMP-dependent protein kinase A (PKA) was first identified and described by Edwin G. Krebs in 1968 as catalyzing the transfer of phosphate from ATP to a target serine or threonine residue in substrate proteins (Walsh et al., 1968). Since the initial identification of this ubiquitous kinase, many studies have defined its regulation by regulatory subunits (R-subunits), of which there are four isoforms (RIα, RIβ, RIIα, RIIβ; Taylor et al., 2012). PKA regulatory subunits inhibit the activity of the PKA catalytic subunit (C-subunit) by occupying the substrate binding site of the C-subunit and preventing the phosphorylation of substrate proteins (Corbin et al., 1978). When cAMP binds to the R-subunits and inhibition is released, the C-subunit is able to assume its catalytic activity and phosphorylate nearby targets. In addition, each R-subunit isotype contains an N-terminal docking and dimerization domain (D/D domain) that is the basis for the formation of a heterotetramer composed of two R-subunits, each of which bind one C-subunit (2:2 stoichiometry; Corbin et al., 1975; Newlon et al., 1999). In addition to the formation of R-subunit dimers, this D/D domain is responsible for docking to a genetically diverse but functionally related family of proteins called A-kinase anchoring proteins (AKAPs; Scott et al., 1990; Newlon et al., 2001).
The first AKAP to be identified was microtubule-associated protein 2 (MAP2) by analysis of associated cAMP-dependent kinase activity (Theurkauf and Vallee, 1982). The number of AKAPs identified since has vastly increased due to use of a far-western technique known as the RII overlay (Carr et al., 1991), as well as through more recent development of computational algorithms designed to predict R-subunit binding regions (Burgers et al., 2015). Some of the most characterized AKAPs include AKAP79/150, gravin, AKAP15/18, and mAKAP (Wong and Scott, 2004). In addition, some AKAPs have been shown to bind RI subunit isoforms, either with dual-specificity for RI and RII, or preference for the RI types (Huang et al., 1997a,b; Lacana et al., 2002; Kovanich et al., 2010; Means et al., 2011). However, the majority of AKAPs interact primarily with RII isoforms.
A-kinase anchoring proteins tether pools of readily stimulated PKA holoenzymes to subcellular compartments and organelles through a variety of mechanisms (Langeberg and Scott, 2015). Importantly, AKAPs also bind other signaling enzymes such as phosphodiesterases (PDEs), G-protein coupled receptors (GPCRs), ion channels, and protein phosphatases to form complexes that are able to integrate and modulate multiple second messenger signaling pathways and fine-tune cellular signaling responses. Many excellent reviews have described the range of binding partners these AKAPs associate with (Wong and Scott, 2004; Carnegie et al., 2009; Welch et al., 2010; Diviani et al., 2011; Sanderson and Dell’Acqua, 2011). In this review, we focus on the structural basis for anchoring of PKA as well as the protein phosphatases that oppose cAMP-mediated signaling.
Structural Basis for PKA Anchoring
Though AKAPs are not typically related to one another on a sequence level, a common unifying feature is their ability to bind the D/D domain of R-subunit dimers through a short (14–18 residues) amphipathic helix, which appears to have arisen relatively early in evolution (Peng et al., 2015). This helix is often one of the few ordered regions, as most AKAPs are intrinsically disordered (Gold et al., 2008). Therefore, this helix serves as a short linear motif (SLiM), which is an emerging concept in cellular signaling that has important implications for protein–protein interactions and drug development (Van Roey et al., 2014). For example, a recent study examining the scaffolding properties of the yeast deubiquitinating enzyme Ubp10 showed that the interplay of SLiMs and intrinsic disorder is essential for facilitating interactions with diverse substrates and binding partners (Reed et al., 2015). SLiMs are often isolated within intrinsically disordered proteins and can serve to facilitate transient interactions which allows a single anchoring protein to interact with a dynamic range of signaling partners (Ren et al., 2008). The first atomic model of an AKAP helix was solved using peptides derived from AKAP79 and Ht31 (AKAP-Lbc) (Newlon et al., 1999). It was obtained using NMR techniques and was solved in complex with the D/D domain (residues 1–44) of RIIα. Subsequently, other structures of the D/D domain in complex with various AKAP-derived helices have been solved using X-ray crystallography (Figure 1A; Gold et al., 2006; Kinderman et al., 2006; Sarma et al., 2010). The D/D domain has been shown to adopt an anti-parallel four-helix X-type bundle that forms a platform with a hydrophobic groove. This groove is the basis for a high affinity interaction with the hydrophobic face of amphipathic AKAP helices. The D/D domain is then connected via a flexible linker to two cAMP-binding cassettes per protomer that display cooperative binding of cAMP (Vigil et al., 2004; Zawadzki and Taylor, 2004). Upon binding of cAMP, a conformational change occurs that relieves inhibition of the PKA C-subunit and allows it to phosphorylate nearby substrates. Crystal structures have been solved for the cAMP-binding cassettes in complex with C-subunit or with cAMP (Figure 1B; Su et al., 1995; Diller et al., 2001; Wu et al., 2007; Zhang et al., 2012). Together with the known structure of the D/D in complex with AKAP helices, these structures have provided insights at the atomic level about the intricate topology and organization of the different functional elements of PKA holoenzyme.
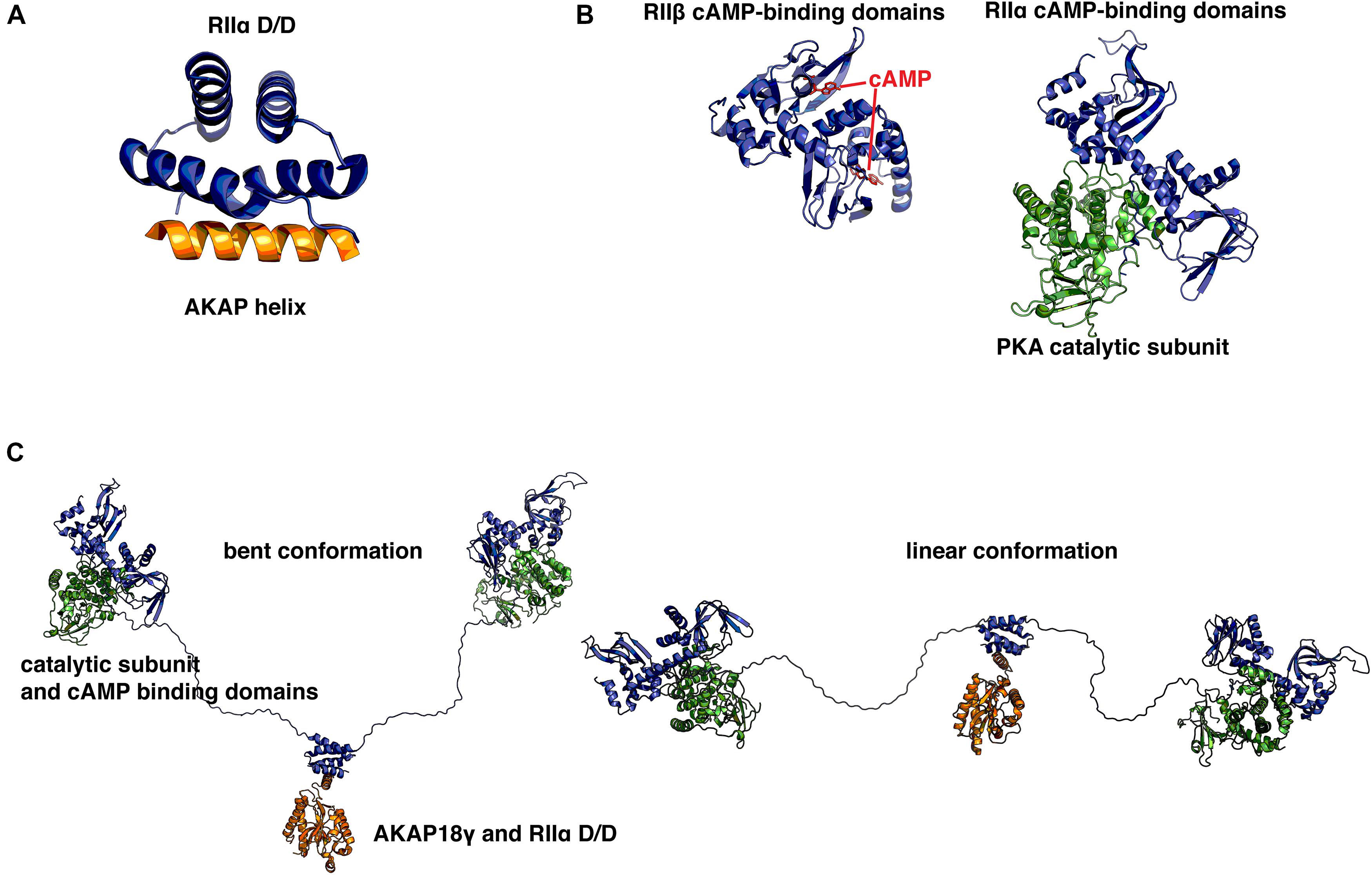
FIGURE 1. Structural basis for protein kinase A (PKA) holoenzyme formation and anchoring. (A) Crystal structure of the synthetic A-kinase anchoring protein (AKAP) helix AKAP’s (orange) in complex with the RIIα docking/dimerization (D/D) domain, residues 1–44 (blue). The AKAP amphipathic helix binds to a hydrophobic groove created by the antiparallel X-type helix bundle of the RII D/D domain. PDB ID: 2IZX. (B) Left: RIIβ cAMP-binding cassettes (blue) in complex with cAMP (red). Right: RIIα cAMP binding cassettes in complex with PKA catalytic subunit (green). With cAMP bound at each of two sites, RII releases inhibition of the catalytic subunit. When cAMP is not present, RII presents an inhibitory sequence to the active site, preventing phosphorylation of PKA substrates. PDB IDs: 1CX4, 2WVS. (C) A pseudo-atomic model of the PKA holoenzyme in complex with AKAP18γ derived from low-resolution EM data. This illustrates that the PKA holoenzyme has a constrained range of flexibility (∼300 Å) provided by AKAPs, allowing the catalytic subunits to be poised near potential substrates. PDB IDs: 3J4Q, 3J4R. Models were prepared using PyMol (Schrödinger).
Yet, there is currently no high-resolution structural information available for the 46 (in mammals) amino acid flexible linker that connects the D/D domain to the pseudosubstrate region that binds the C-subunit and to the tandem cAMP binding cassettes. Therefore, a recent study used single particle electron microscopy studies to examine the structure of an AKAP18γ-PKA holoenzyme complex (Smith et al., 2013). This study revealed that although many crystal structures of RII and C-subunits showed surface contact between each heterodimer of RII and C, the complexes likely occupy a much broader conformational space that is constrained by the length of the linker, yet facilitated by the intrinsic disorder of the linker (Figure 1C). This linker-guided conformation sampling may be a mechanism by which PKA preferentially phosphorylates substrates within the same macromolecular complexes upon elevation of cAMP levels. cAMP PDEs have been suggested to form a ‘fence’ around subcellular pools of elevated cAMP (Baillie et al., 2005). AKAP18γ has been shown to form a complex with PDE4D3 and regulate its activity via PKA phosphorylation (Stefan et al., 2007). In combination with local restraint of PKA conformations by the RII flexible linker, these local PDE fences represent an intriguing scheme by which spatiotemporal specificity may be regulated by macromolecular signaling complexes.
Targeting the PKA/AKAP Interaction for Therapeutics
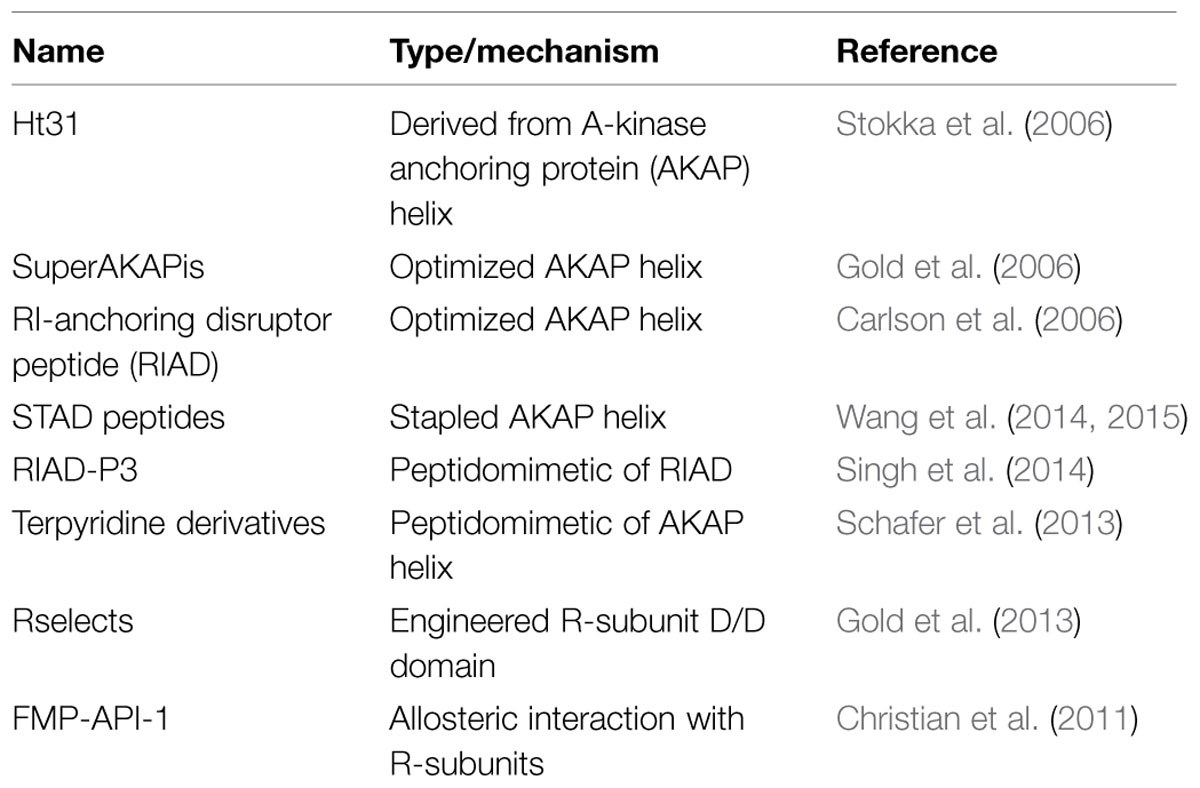
Since PKA activity modulates a variety of physiological events, such as cardiac remodeling, disrupting the PKA/AKAP interface has been a long-standing area of interest for therapeutics (Troger et al., 2012; Table 1). One of the first disruptors of the AKAP/RII interaction is the 24 amino acid peptide Ht31, named after human thyroid clone 31, which was later realized to represent a biologically active segment of the multifunctional scaffolding protein AKAP-Lbc (Carr et al., 1992). The Ht31 peptide has since been lipid modified with a stearol group to increase its membrane permeability for treatment of cell lines and elucidation of anchored PKA signaling events (Vijayaraghavan et al., 1997; Gold et al., 2012). In silico approaches have resulted in optimized peptides that mimic the AKAP amphipathic helix and bind to RII or RI with high affinity (Alto et al., 2003). Added to this, structure-based approaches have further increased the specificity of the peptide superAKAPis for RII to the low nanomolar affinity range with a 12,000-fold preference for RII over RI (Gold et al., 2006). Conversely, the RI-anchoring disruptor peptide (RIAD) has been engineered to specifically disrupt RI/AKAP interactions (Carlson et al., 2006). Peptidomimetics have been developed by several groups that mimic amphipathic helix structures and are able to disrupt RI/RII interactions with AKAPs (Schafer et al., 2013; Singh et al., 2014). Recent work has centered on developing stapled AKAP-mimetic peptides that are cell-permeable and have increased stability (Wang et al., 2014, 2015; Kennedy and Scott, 2015). This would increase the utility of these peptides for therapeutic purposes as well as for teasing apart the molecular mechanisms by which AKAPs influence local PKA signaling pathways.
Since there are numerous AKAPs and only four R-subunit isoforms, any disruptor that relies on an interaction with an R-subunit is by definition non-selective. In order to disrupt a specific AKAP’s ability to bind PKA, anchoring disruptors must bind to an AKAP helix with high affinity and recognize the unique structural features of one AKAP helix preferentially. Therefore, a phage-display screening approach, which used immobilized AKAP helices to enrich for phage variants that displayed mutant RII D/D domains, selected variants that exhibit preferential binding to specific AKAPs. These mutant D/D domains are termed Rselects, and have been shown in preliminary work to bind and label AKAPs in a cellular context as well as in to purified proteins (Gold et al., 2013). Further development of these Rselects could lead to high affinity binding variants that could disrupt individual pools of anchored PKA while allowing other anchored PKA signaling events to proceed unperturbed. The potential to isolate spatially constrained post-translational modifications is an important step forward for targeted therapeutics. However, utility of this approach as a cell based means of selectively interrupting particular PKA–AKAP interfaces has yet to be rigorously established.
Small molecule disruptors are another attractive means to pharmacologically target the PKA–AKAP interface. Although these studies are still in their formative stages there have been a few successful attempts at moderate-throughput screening for small molecule AKAP disruptors (Schachterle et al., 2015). Perhaps the most notable example is the development of 3,3′-diamino-4,4′-dihydroxydiphenylmethane (FMP-API-1), a small molecule antagonist that appears to allosterically inhibit the RII–AKAP interaction and activate anchored PKA C-subunit (Christian et al., 2011). Yet, despite extensive characterization of this compound the mechanism of action of FMP-API-1 has yet to be defined. Nonetheless, the future is bright for the discovery and development of cell soluble chemical entities that target PKA–AKAP interfaces.
Protein Phosphatase Anchoring
Classically, protein phosphatases are considered to be responsible for the opposing action to kinases, namely the removal of phosphate groups from serine, threonine, or tyrosine residues. In addition a burgeoning family of pseudokinases and pseudophosphatases are emerging a key players in cell signaling (Reiterer et al., 2014). Protein phosphatases fall into two main classes – serine–threonine phosphatases, and tyrosine phosphatases. While there are 428 serine/threonine kinases, there are only ∼40 serine/threonine phosphatases (Moorhead et al., 2007). This disparity in gene number infers that additional mechanisms come into play as a means to modulate and vary the substrate specificity of these critical regulatory enzymes. Philip and Tricia Cohen were the first to recognize that regulation of protein phosphatases by association with regulatory and targeting subunits is a crucial mechanism to allosterically modulate substrate specificity (Stralfors et al., 1985; Cohen and Cohen, 1989). Subsequently others have shown that most of the three classes of serine/threonine phosphatases are modulated by targeting subunits (Langeberg and Scott, 2015). In this article we focus exclusively on protein phosphatase 1 (PP1) and protein phosphatase 2B (PP2B, or calcineurin), since these ubiquitous phosphatases often oppose the action of PKA and are especially reliant on anchoring for their regulation.
Protein Phosphatase 1 Regulation by Auxiliary Proteins
Protein phosphatase 1 has an important role in a number of physiological processes, notably regulation of glycogen synthesis (Hubbard et al., 1990), nuclear events (Helps et al., 1998), and synaptic long term potentiation (LTP) and long term depression (LTD), (Morishita et al., 2001; Malinow and Malenka, 2002). The latter two events occur through phosphatase opposition of CaMKII and PKA phosphorylation of glutamate receptors at the post-synaptic density. The PP1 catalytic subunit (PP1c) associates with over 200 regulatory subunits, many of which bind via a conserved short linear peptide motif called the RVxF motif (Cohen, 2002; Roy and Cyert, 2009).
Some of these subunits serve primarily to inhibit the catalytic activity, such as the protein Inhibitor 1 (I-1) and dopamine and cAMP-regulated phosphoprotein 32 (DARPP32), (Williams et al., 1986). Notably some of these inhibitors are activated by PKA phosphorylation. Other regulatory subunits contain localization signatures that target PP1 to specific subcellular regions and may or may not also inhibit the enzymatic activity of PP1c. The most recognized examples of these targeting subunits are the myosin phosphatase targeting subunit MYPT1, the GM regulatory subunit, p53-binding protein 2 (53BP2), and PP1 nuclear targeting subunit (PNUTS). Recent investigation of PNUTS has highlighted several properties shared by many PP1-binding proteins (Choy et al., 2014). First, the RVxF motif serves as a short linear interaction motif (SLiM) and is responsible for the primary interaction. Second, intrinsic disorder in PNUTS facilitates extended contact with PP1 on additional surfaces to fine-tune the phosphatase. Third, binding to these surfaces inhibits activity toward some substrates without physically blocking the active site of the phosphatase (Figure 2A).
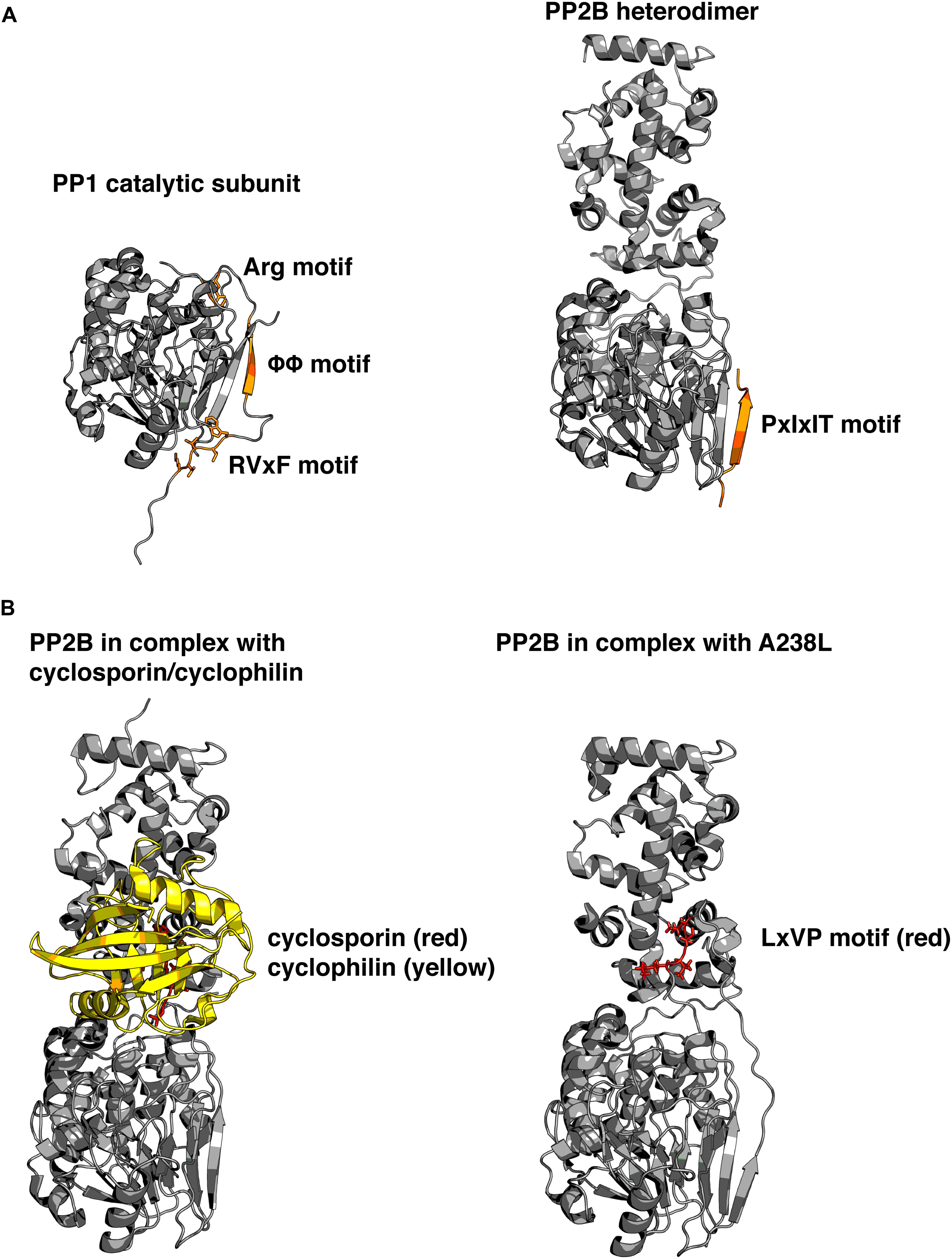
FIGURE 2. Structural basis for phosphatase regulation and anchoring. (A) Left: PP1 catalytic subunit (gray) in complex with RVxF and auxiliary anchoring motifs from protein phosphatase 1 (PP1) nuclear targeting subunit (PNUTS; orange). Right: PP2B (gray) in complex with PIAIIIT sequence from AKAP79 (orange). Comparison reveals that similar surfaces are used for anchoring, and that multiple motifs can simultaneously interact with varied portions of the molecule. PDB IDs: 4MOY, 3LL8. (B) Left: PP2B in complex with cyclosporin (red)/cyclophilin (yellow) complex. Right: PP2B in complex with a viral peptide A238L, containing a PxIxIT motif, as well as an LxVP motif (red). Cyclosporin and LxVP peptides bind to overlapping surfaces on PP2B, formed by both the catalytic and regulatory subunits of PP2B. This surface does not occlude the active site of the phosphatase, yet immunosuppressants are able to allosterically inhibit PP2B activity toward substrates. PDB IDs: 1MF8, 4F0Z. Models were prepared using PyMol (Schrödinger).
A number of AKAPs have been shown to interact with PP1c, including AKAP220 (Schillace and Scott, 1999), D-AKAP1 (Steen et al., 2000), and yotiao (Westphal et al., 1999). Likewise, some isoforms of AKAP18 are thought to sequester PP1, although it would appear that this occurs via indirect mechanisms (Singh et al., 2011). All direct PP1–AKAP interfaces utilize some version of the degenerate RVxF motif. D-AKAP1 was suggested to be involved in anchoring PP1 for efficient nuclear envelope reassembly after mitosis (Collas et al., 1999; Steen et al., 2000). AKAP220 has been shown to anchor PP1 through a modified KVxF motif, and this has been proposed to play a role in regulating the activity of glycogen synthase kinase 3β (GSK3β), through modulation of the phosphorylation state of serine 9. Phosphorylation of this residue results in suppression of GSK3β activity (Schillace et al., 2001; Tanji et al., 2002; Whiting et al., 2015). Yotiao, a product of the AKAP9 gene, also contains an RVxF motif, and has been shown to regulate the phosphorylation state of NMDA receptors through localization of PP1 (Lin et al., 1998; Westphal et al., 1999). AKAP18 does not appear to interact directly with PP1, but some reports indicate that it binds Inhibitor-1 to promote its phosphorylation by PKA (Singh et al., 2011). The net effect of this later phosphorylation event is to promote local inhibition of PP1c.
PP2B Regulation by Auxiliary Proteins
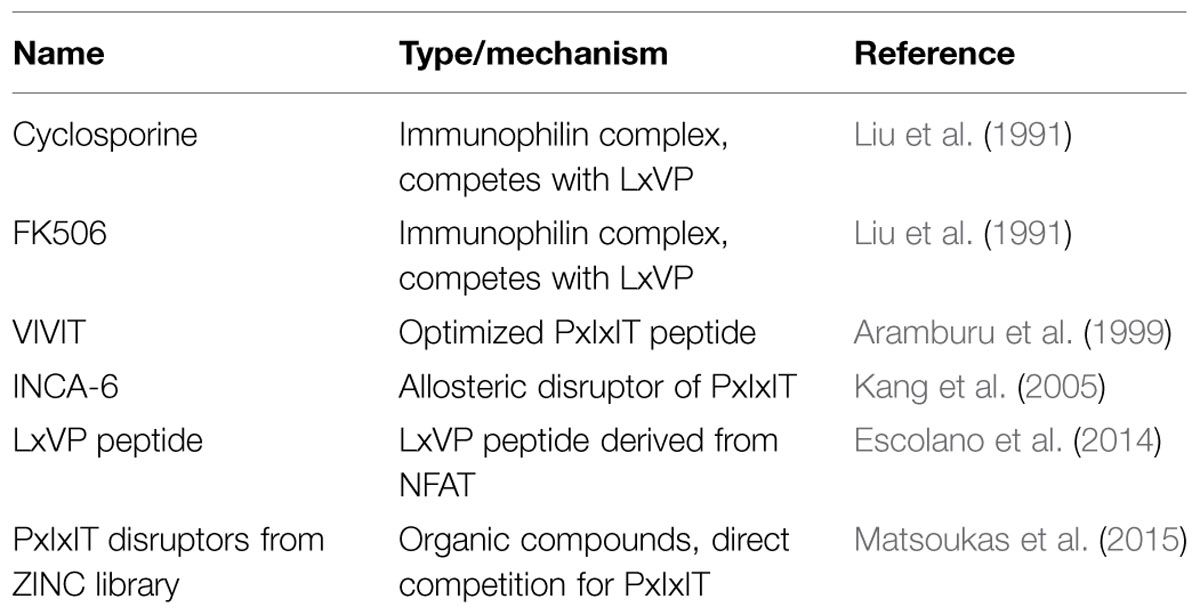
PP2B, also known as calcineurin, is a broadly expressed obligate heterodimeric protein phosphatase that is activated by calcium and calmodulin (Stewart et al., 1982). Like PP1, PP2B is involved in diverse processes such as synaptic plasticity (Mulkey et al., 1994), glucose metabolism (Hinke et al., 2012), cardiac signaling (Tandan et al., 2009), and immune responses (Clipstone and Crabtree, 1992). In addition, activation of PP2B can mobilize phosphatase cascade, through dephosphorylation of PP1 regulatory subunits (Mulkey et al., 1994). The catalytic A subunit of PP2B contains an autoinhibitory region that occludes the active site in the absence of calcium. Upon elevation of calcium levels, calcium ions bind directly to the regulatory B subunit, and to calmodulin, which in turn interacts with the autoinhibitory region and allows PP2B to resume catalytic activity toward phosphosubstrates (Li et al., 2011). Because calcium transients often envelop the whole cell rather than occurring locally, regulation of PP2B’s activity toward substrates is accomplished primarily through protein–protein interactions. The best-known PP2B substrate is the nuclear factor of activated T-cells (NFAT) family. These transcription factors contain phosphoserine-rich regions, and when dephosphorylated, dimerize and translocate to the nucleus, where they are responsible for controlling a range of transcriptional responses such as inflammation in response to immune system signaling (Li et al., 2012). The common immunosuppressants FK506 and cyclosporine target PP2B and have their primary effect through inhibition of NFAT signaling (Liu et al., 1991).
Not only is NFAT a typical PP2B substrate, it also contains two SLiMs, which are typical of PP2B interacting proteins – the PxIxIT motif, and the LxVP motif (Roy et al., 2007; Rodriguez et al., 2009). The PxIxIT motif forms a beta strand that binds to a hydrophobic groove formed by a beta sheet on the PP2B A subunit (Li et al., 2007). This surface of the PP2B A subunit is analogous to the region of PP1 which interacts with the RVxF motif (Figure 2A). Proteins that contain PxIxIT motifs include NFAT, regulator of calcineurin 1 (RCAN1; Mehta et al., 2009), TWIK-related spinal cord potassium channel (TRESK; Roy and Cyert, 2009), and notably, AKAP79/150 (Dell’Acqua et al., 2002). The LxVP motif is a degenerate sequence that binds to the interface of the A and B subunits of PP2B, and only binds to activated calcineurin (Rodriguez et al., 2009). It has been challenging to describe a consensus LxVP sequence. Therefore, many LxVP motifs have been identified without originally being aware of their identity. The first LxVP motif to be described was that of the RII subunit by Blumenthal et al. (1986), although it was not recognized as a conserved binding mode until it was found in NFAT. Many substrates of PP2B contain an LxVP motif, and it has been suggested that all efficient substrates contain some type of sequence that interacts with the LxVP binding region on PP2B (Grigoriu et al., 2013). The characterization of multiple SLiMs that interact with various surfaces of PP2B parallels that of PP1, and suggests that other mechanisms may also be in common such as fine-tuning the location and activity of PP2B through a combination of disorder and SLiMs. Recently, a structure of PP2B in complex with a viral inhibitor peptide from African swine fever revealed the binding site for the LxVP motif in atomic level detail (Grigoriu et al., 2013). This crystal structure reveals that the leucine residue occupies a pocket formed by two aromatic residues, and when these are mutated to alanine residues they no longer interact with the LxVP motif. In addition, this binding site overlaps with the binding sites for cyclosporine and FK506 complexes (Figure 2B). However, no structure has been solved of the PP2B heterodimer bound to calmodulin in the fully active state, so the question of how LxVP motifs are able to impact PP2B activity remains unclear.
Some AKAPs have been shown to bind PP2B, such as the aforementioned AKAP79, and mAKAP (Li et al., 2010). In addition, the AKAP gravin has been suggested to be in the same complex as PP2B and beta-adrenergic receptors, however, evidence for a direct interaction is not immediately apparent (Shih et al., 1999). The mAKAP interaction has been mapped to the residues 1286–1345 in the mAKAPα splice variant. However, this region does not contain an easily identifiable PxIxIT or LxVP sequence. Loss of mAKAP-PP2B binding was shown to result in reduced cardiac myocyte hypertrophy in response to norepinephrine, as well decreased atrial natriuretic factor expression. Interestingly, the pool of PP2B bound to mAKAP appeared to be active, and required to dephosphorylate NFAT efficiently in response to phenylephrine treatment. Formation of the PP2B/mAKAP complex was enhanced in vitro by calcium/calmodulin, suggesting that the interaction may occur via a similar mechanism to the LxVP motif (Li et al., 2010).
A-kinase anchoring protein 79 is perhaps the best characterized AKAP, and its interaction with PP2B has been extensively investigated. Although original studies suggested an interaction site was restricted to the N-terminal third of AKAP79 (Coghlan et al., 1995), later work described the primary site of interaction as being a PxIxIT motif from residues 337-343 (Dell’Acqua et al., 2002; Oliveria et al., 2007). Use of a transgenic mouse model in which AKAP79 lacks this region, known as the AKAP79ΔPIX mouse, has revealed that AKAP79-anchored PP2B is required for NMDA-dependent hippocampal long-term depression and NFAT signaling in neurons (Oliveria et al., 2007; Sanderson et al., 2012). Intriguingly, the AKAP79ΔPIX mouse shows improved insulin sensitivity, indicating that this interaction may be a possible therapeutic target for Type II diabetes (Hinke et al., 2012).
Because of the importance of the AKAP79–PP2B interaction, much emphasis has been placed on understanding the structural basis of this interaction. Native mass spectrometry and biochemical approaches have suggested that there is an additional interaction site for PP2B between residues 1–153 of AKAP79 that is dependent on calcium/calmodulin (Gold et al., 2011). Although a crystal structure of PP2B in complex with a synthetic PxIxIT motif was solved in 2007 (Li et al., 2007), the first structure of PP2B bound to a natural PxIxIT motif was that of AKAP79 (Li et al., 2012). This structure matched closely with the previously solved structure, in that crystal packing is such that each PIAIIIT sequence contacts two PP2B A subunits along the PxIxIT binding region. This, along with native mass spectrometry approaches, raises the question of whether AKAP79 is capable of binding two PP2B molecules simultaneously.
Because of PP2B’s importance in a range of physiological contexts, there is great interest in developing disruptors that target specific PP2B anchoring proteins (Table 2). One of the first targeted approaches resulted in an optimized high-affinity PxIxIT motif called the VIVIT peptide (Aramburu et al., 1999), which is the aforementioned synthetic peptide that was co-crystallized with PP2B. In addition, fluorescence polarization screens for small molecules that disrupt binding to the PxIxIT motif have yielded a potential candidate known as INCA-6 that is able to inhibit PP2B-NFAT signaling with similar potency to cyclosporine and FK506, but through an alternate mechanism (Kang et al., 2005). Recently, an approach disrupting the LxVP interaction in macrophages through lentiviral expression of an LxVP peptide was shown to reduce inflammation and confer resistance to arthritis and contact hypersensitivity (Escolano et al., 2014). Understanding the molecular basis for PP2B anchoring has led to potential for therapeutics and the realization that primary and secondary binding sites may both be targeted for diverse physiological effect. Because the AKAP79–PP2B interaction is important in many processes, specifically targeting this interaction may be of great promise.
Conclusion
Protein kinase A phosphorylation events and the phosphatases that oppose them are tightly regulated by anchoring proteins. Recently, the use of new and sophisticated biochemical, biophysical, and structural techniques have forged two important concepts. First, the combination of SLiMs and intrinsic disorder allow anchoring proteins to allosterically and spatially control the range and specificity of phospho-signaling. Second, AKAPs are not just static anchors, but are conformationally and compositionally flexible. This allows them to adapt to a varied and continually changing cellular signaling environment. A recent paper characterizing binding partners of the AKAP ezrin by quantitative mass spectrometry revealed that conformational switches in ezrin are accompanied by changes in the complement of enzymes present in the complex (Viswanatha et al., 2013). This may well prove to be the case for many AKAPs allowing them to perform cell type specific roles. Moreover, the concept of AKAPs as conformational switches could account for how the same anchoring protein can simultaneously perform distinct functions at multiple locations within a single cell.
These new biological insights have been demonstrated by using hybrid structural techniques such as x-ray crystallography, NMR, hydrogen/deuterium exchange experiments and crosslinking/mass spectrometry (Burns-Hamuro et al., 2005; Gold et al., 2011; Choy et al., 2014). The advent of direct electron detectors for cryo-electron microscopy has increased attainable resolutions (Campbell et al., 2012), and will likely contribute to increased structural understanding of these flexible multi-protein complexes. In addition, computational advances in understanding heterogeneous cryo-EM samples will also advance our knowledge of multiple conformational states (Behrmann et al., 2015). Already, negative-stain approaches such as random conical tilt (RCT) experiments are allowing researchers to understand structural heterogeneity in protein complexes (Veesler et al., 2014). Combining these approaches with biosensors for enzymatic activity (Mehta and Zhang, 2014; Mehta et al., 2014) will provide a more comprehensive picture of how the structural properties of anchored kinase and phosphatase complexes are able to influence local signaling in a cellular context. Finally, as exemplified by a recent structure-guided pharmacophore screen for inhibitors of PP2B anchoring, atomic resolution structural insights will guide design of small molecules that target anchoring protein interactions in the context of SLiMs and intrinsic disorder (Matsoukas et al., 2015).
Author Contributions
PN, JS: Wrote, edited, and approved final version of the manuscript and figures.
Funding
Supported by NIH grants DK054441 and DK105542.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgment
We thank members of the Scott lab for fruitful discussions, and apologize to colleagues whose deserving work we were unable to cite due to space limitations.
References
Alto, N. M., Soderling, S. H., Hoshi, N., Langeberg, L. K., Fayos, R., Jennings, P. A., et al. (2003). Bioinformatic design of A-kinase anchoring protein-in silico: a potent and selective peptide antagonist of type II protein kinase A anchoring. Proc. Natl. Acad. Sci. U.S.A. 100, 4445–4450. doi: 10.1073/pnas.0330734100
Aramburu, J., Yaffe, M. B., Lopez-Rodriguez, C., Cantley, L. C., Hogan, P. G., and Rao, A. (1999). Affinity-driven peptide selection of an NFAT inhibitor more selective than cyclosporin A. Science 285, 2129–2133. doi: 10.1126/science.285.5436.2129
Baillie, G. S., Scott, J. D., and Houslay, M. D. (2005). Compartmentalisation of phosphodiesterases and protein kinase A: opposites attract. FEBS Lett. 579, 3264–3270. doi: 10.1016/j.febslet.2005.03.089
Behrmann, E., Loerke, J., Budkevich, T. V., Yamamoto, K., Schmidt, A., Penczek, P. A., et al. (2015). Structural snapshots of actively translating human ribosomes. Cell 161, 845–857. doi: 10.1016/j.cell.2015.03.052
Blumenthal, D. K., Takio, K., Hansen, R. S., and Krebs, E. G. (1986). Dephosphorylation of cAMP-dependent protein kinase regulatory subunit (type II) by calmodulin-dependent protein phosphatase. J. Biol. Chem. 261, 8140–8145.
Burgers, P. P., Van Der Heyden, M. A., Kok, B., Heck, A. J., and Scholten, A. (2015). A systematic evaluation of protein kinase A-A-kinase anchoring protein interaction motifs. Biochemistry 54, 11–21. doi: 10.1021/bi500721a
Burns-Hamuro, L. L., Hamuro, Y., Kim, J. S., Sigala, P., Fayos, R., Stranz, D. D., et al. (2005). Distinct interaction modes of an AKAP bound to two regulatory subunit isoforms of protein kinase A revealed by amide hydrogen/deuterium exchange. Protein Sci. 14, 2982–2992. doi: 10.1110/ps.051687305
Campbell, M. G., Cheng, A., Brilot, A. F., Moeller, A., Lyumkis, D., Veesler, D., et al. (2012). Movies of ice-embedded particles enhance resolution in electron cryo-microscopy. Structure 20, 1823–1828. doi: 10.1016/j.str.2012.08.026
Carlson, C. R., Lygren, B., Berge, T., Hoshi, N., Wong, W., Tasken, K., et al. (2006). Delineation of type I protein kinase A-selective signaling events using an RI anchoring disruptor. J. Biol. Chem. 281, 21535–21545. doi: 10.1074/jbc.M603223200
Carnegie, G. K., Means, C. K., and Scott, J. D. (2009). A-kinase anchoring proteins: from protein complexes to physiology and disease. IUBMB Life 61, 394–406. doi: 10.1002/iub.168
Carr, D. W., Hausken, Z. E., Fraser, I. D., Stofko-Hahn, R. E., and Scott, J. D. (1992). Association of the type II cAMP-dependent protein kinase with a human thyroid RII-anchoring protein. Cloning and characterization of the RII-binding domain. J. Biol. Chem. 267, 13376–13382.
Carr, D. W., Stofko-Hahn, R. E., Fraser, I. D., Bishop, S. M., Acott, T. S., Brennan, R. G., et al. (1991). Interaction of the regulatory subunit (RII) of cAMP-dependent protein kinase with RII-anchoring proteins occurs through an amphipathic helix binding motif. J. Biol. Chem. 266, 14188–14192.
Choy, M. S., Hieke, M., Kumar, G. S., Lewis, G. R., Gonzalez-Dewhitt, K. R., Kessler, R. P., et al. (2014). Understanding the antagonism of retinoblastoma protein dephosphorylation by PNUTS provides insights into the PP1 regulatory code. Proc. Natl. Acad. Sci. U.S.A. 111, 4097–4102. doi: 10.1073/pnas.1317395111
Christian, F., Szaszak, M., Friedl, S., Drewianka, S., Lorenz, D., Goncalves, A., et al. (2011). Small molecule AKAP-protein kinase A (PKA) interaction disruptors that activate PKA interfere with compartmentalized cAMP signaling in cardiac myocytes. J. Biol. Chem. 286, 9079–9096. doi: 10.1074/jbc.M110.160614
Clipstone, N. A., and Crabtree, G. R. (1992). Identification of calcineurin as a key signalling enzyme in T-lymphocyte activation. Nature 357, 695–697. doi: 10.1038/357695a0
Coghlan, V. M., Perrino, B. A., Howard, M., Langeberg, L. K., Hicks, J. B., Gallatin, W. M., et al. (1995). Association of protein kinase A and protein phosphatase 2B with a common anchoring protein. Science 267, 108–112. doi: 10.1126/science.7528941
Cohen, P., and Cohen, T. W. (1989). Protein phosphatases come of age. J. Biol. Chem. 264, 21435–21438.
Collas, P., Le Guellec, K., and Tasken, K. (1999). The A-kinase-anchoring protein AKAP95 is a multivalent protein with a key role in chromatin condensation at mitosis. J. Cell Biol. 147, 1167–1180. doi: 10.1083/jcb.147.6.1167
Corbin, J. D., Keely, S. L., Soderling, T. R., and Park, C. R. (1975). Hormonal regulation of adenosine 3’,5’-monophosphate-dependent protein kinase. Adv. Cyclic Nucleotide Res. 5, 265–279.
Corbin, J. D., Sugden, P. H., West, L., Flockhart, D. A., Lincoln, T. M., and Mccarthy, D. (1978). Studies on the properties and mode of action of the purified regulatory subunit of bovine heart adenosine 3’:5’-monophosphate-dependent protein kinase. J. Biol. Chem. 253, 3997–4003.
Dell’Acqua, M. L., Dodge, K. L., Tavalin, S. J., and Scott, J. D. (2002). Mapping the protein phosphatase-2B anchoring site on AKAP79. Binding and inhibition of phosphatase activity are mediated by residues 315-360. J. Biol. Chem. 277, 48796–48802. doi: 10.1074/jbc.M207833200
Diller, T. C., Madhusudan, Xuong, N. H., and Taylor, S. S. (2001). Molecular basis for regulatory subunit diversity in cAMP-dependent protein kinase: crystal structure of the type II beta regulatory subunit. Structure 9, 73–82. doi: 10.1016/S0969-2126(00)00556-6
Diviani, D., Dodge-Kafka, K. L., Li, J., and Kapiloff, M. S. (2011). A-kinase anchoring proteins: scaffolding proteins in the heart. Am. J. Physiol. Heart Circ. Physiol. 301, H1742–H1753. doi: 10.1152/ajpheart.00569.2011
Escolano, A., Martinez-Martinez, S., Alfranca, A., Urso, K., Izquierdo, H. M., Delgado, M., et al. (2014). Specific calcineurin targeting in macrophages confers resistance to inflammation via MKP-1 and p38. EMBO J. 33, 1117–1133. doi: 10.1002/embj.201386369
Gold, M. G., Fowler, D. M., Means, C. K., Pawson, C. T., Stephany, J. J., Langeberg, L. K., et al. (2013). Engineering A-kinase anchoring protein (AKAP)-selective regulatory subunits of protein kinase A (PKA) through structure-based phage selection. J. Biol. Chem. 288, 17111–17121. doi: 10.1074/jbc.M112.447326
Gold, M. G., Lygren, B., Dokurno, P., Hoshi, N., Mcconnachie, G., Tasken, K., et al. (2006). Molecular basis of AKAP specificity for PKA regulatory subunits. Mol. Cell 24, 383–395. doi: 10.1016/j.molcel.2006.09.006
Gold, M. G., Reichow, S. L., O’neill, S. E., Weisbrod, C. R., Langeberg, L. K., Bruce, J. E., et al. (2012). AKAP2 anchors PKA with aquaporin-0 to support ocular lens transparency. EMBO Mol. Med. 4, 15–26. doi: 10.1002/emmm.201100184
Gold, M. G., Smith, F. D., Scott, J. D., and Barford, D. (2008). AKAP18 contains a phosphoesterase domain that binds AMP. J. Mol. Biol. 375, 1329–1343. doi: 10.1016/j.jmb.2007.11.037
Gold, M. G., Stengel, F., Nygren, P. J., Weisbrod, C. R., Bruce, J. E., Robinson, C. V., et al. (2011). Architecture and dynamics of an A-kinase anchoring protein 79 (AKAP79) signaling complex. Proc. Natl. Acad. Sci. U.S.A. 108, 6426–6431. doi: 10.1073/pnas.1014400108
Grigoriu, S., Bond, R., Cossio, P., Chen, J. A., Ly, N., Hummer, G., et al. (2013). The molecular mechanism of substrate engagement and immunosuppressant inhibition of calcineurin. PLoS Biol. 11:e1001492. doi: 10.1371/journal.pbio.1001492
Helps, N. R., Brewis, N. D., Lineruth, K., Davis, T., Kaiser, K., and Cohen, P. T. (1998). Protein phosphatase 4 is an essential enzyme required for organisation of microtubules at centrosomes in Drosophila embryos. J. Cell Sci. 111, 1331–1340.
Hinke, S. A., Navedo, M. F., Ulman, A., Whiting, J. L., Nygren, P. J., Tian, G., et al. (2012). Anchored phosphatases modulate glucose homeostasis. EMBO J. 31, 3991–4004. doi: 10.1038/emboj.2012.244
Huang, L. J., Durick, K., Weiner, J. A., Chun, J., and Taylor, S. S. (1997a). D-AKAP2, a novel protein kinase A anchoring protein with a putative RGS domain. Proc. Natl. Acad. Sci. U.S.A. 94, 11184–11189. doi: 10.1073/pnas.94.21.11184
Huang, L. J., Durick, K., Weiner, J. A., Chun, J., and Taylor, S. S. (1997b). Identification of a novel dual specificity protein kinase A anchoring protein, D-AKAP1. J. Biol. Chem. 272, 8057–8064. doi: 10.1074/jbc.272.12.8057
Hubbard, M. J., Dent, P., Smythe, C., and Cohen, P. (1990). Targeting of protein phosphatase 1 to the sarcoplasmic reticulum of rabbit skeletal muscle by a protein that is very similar or identical to the G subunit that directs the enzyme to glycogen. Eur. J. Biochem. 189, 243–249. doi: 10.1111/j.1432-1033.1990.tb15483.x
Kang, S., Li, H., Rao, A., and Hogan, P. G. (2005). Inhibition of the calcineurin-NFAT interaction by small organic molecules reflects binding at an allosteric site. J. Biol. Chem. 280, 37698–37706. doi: 10.1074/jbc.M502247200
Kennedy, E. J., and Scott, J. D. (2015). Selective disruption of the AKAP signaling complexes. Methods Mol. Biol. 1294, 137–150. doi: 10.1007/978-1-4939-2537-7_11
Kinderman, F. S., Kim, C., Von Daake, S., Ma, Y., Pham, B. Q., Spraggon, G., et al. (2006). A dynamic mechanism for AKAP binding to RII isoforms of cAMP-dependent protein kinase. Mol. Cell 24, 397–408. doi: 10.1016/j.molcel.2006.09.015
Kovanich, D., Van Der Heyden, M. A., Aye, T. T., Van Veen, T. A., Heck, A. J., and Scholten, A. (2010). Sphingosine kinase interacting protein is an A-kinase anchoring protein specific for type I cAMP-dependent protein kinase. Chembiochem 11, 963–971. doi: 10.1002/cbic.201000058
Lacana, E., Maceyka, M., Milstien, S., and Spiegel, S. (2002). Cloning and characterization of a protein kinase A anchoring protein (AKAP)-related protein that interacts with and regulates sphingosine kinase 1 activity. J. Biol. Chem. 277, 32947–32953. doi: 10.1074/jbc.M202841200M202841200
Langeberg, L. K., and Scott, J. D. (2015). Signalling scaffolds and local organization of cellular behaviour. Nat. Rev. Mol. Cell Biol. 16, 232–244. doi: 10.1038/nrm3966
Li, H., Pink, M. D., Murphy, J. G., Stein, A., Dell’Acqua, M. L., and Hogan, P. G. (2012). Balanced interactions of calcineurin with AKAP79 regulate Ca(2+)-calcineurin-NFAT signaling. Nat. Struct. Mol. Biol. 19, 337–345. doi: 10.1038/nsmb.2238
Li, H., Rao, A., and Hogan, P. G. (2011). Interaction of calcineurin with substrates and targeting proteins. Trends Cell Biol. 21, 91–103. doi: 10.1016/j.tcb.2010.09.011
Li, H., Zhang, L., Rao, A., Harrison, S. C., and Hogan, P. G. (2007). Structure of calcineurin in complex with PVIVIT peptide: portrait of a low-affinity signalling interaction. J. Mol. Biol. 369, 1296–1306. doi: 10.1016/j.jmb.2007.04.032
Li, J., Negro, A., Lopez, J., Bauman, A. L., Henson, E., Dodge-Kafka, K., et al. (2010). The mAKAPbeta scaffold regulates cardiac myocyte hypertrophy via recruitment of activated calcineurin. J. Mol. Cell. Cardiol. 48, 387–394. doi: 10.1016/j.yjmcc.2009.10.023
Lin, J. W., Wyszynski, M., Madhavan, R., Sealock, R., Kim, J. U., and Sheng, M. (1998). Yotiao, a novel protein of neuromuscular junction and brain that interacts with specific splice variants of NMDA receptor subunit NR1. J. Neurosci. 18, 2017–2027.
Liu, J., Farmer, J. D. J., Lane, W. L., Friedman, J., Weissman, I., and Schreiber, S. L. (1991). Calcineurin is a common target of cyclophilin-cyclosporin A and FKBP-FK506 complexes. Cell 66, 807–815. doi: 10.1016/0092-8674(91)90124-H
Malinow, R., and Malenka, R. C. (2002). AMPA receptor trafficking and synaptic plasticity. Annu. Rev. Neurosci. 25, 103–126. doi: 10.1146/annurev.neuro.25.112701.142758
Matsoukas, M. T., Aranguren-Ibanez, A., Lozano, T., Nunes, V., Lasarte, J. J., Pardo, L., et al. (2015). Identification of small-molecule inhibitors of calcineurin-NFATc signaling that mimic the PxIxIT motif of calcineurin binding partners. Sci. Signal. 8, ra63. doi: 10.1126/scisignal.2005918
Means, C. K., Lygren, B., Langeberg, L. K., Jain, A., Dixon, R. E., Vega, A. L., et al. (2011). An entirely specific type I A-kinase anchoring protein that can sequester two molecules of protein kinase A at mitochondria. Proc. Natl. Acad. Sci. U.S.A. 108, E1227–E1235. doi: 10.1073/pnas.1107182108
Mehta, S., Aye-Han, N. N., Ganesan, A., Oldach, L., Gorshkov, K., and Zhang, J. (2014). Calmodulin-controlled spatial decoding of oscillatory Ca2+ signals by calcineurin. Elife 3, e03765. doi: 10.7554/eLife.03765
Mehta, S., Li, H., Hogan, P. G., and Cunningham, K. W. (2009). Domain architecture of the regulators of calcineurin (RCANs) and identification of a divergent RCAN in yeast. Mol. Cell. Biol. 29, 2777–2793. doi: 10.1128/MCB.01197-08
Mehta, S., and Zhang, J. (2014). Using a genetically encoded FRET-based reporter to visualize calcineurin phosphatase activity in living cells. Methods Mol. Biol. 1071, 139–149. doi: 10.1007/978-1-62703-622-1_11
Moorhead, G. B., Trinkle-Mulcahy, L., and Ulke-Lemee, A. (2007). Emerging roles of nuclear protein phosphatases. Nat. Rev. Mol. Cell Biol. 8, 234–244. doi: 10.1038/nrm2126
Morishita, W., Connor, J. H., Xia, H., Quinlan, E. M., Shenolikar, S., and Malenka, R. C. (2001). Regulation of synaptic strength by protein phosphatase 1. Neuron 32, 1133–1148. doi: 10.1016/S0896-6273(01)00554-2
Mulkey, R. M., Endo, S., Shenolikar, S., and Malenka, R. C. (1994). Involvement of a calcineurin/inhibitor-1 phosphatase cascade in hippocampal long-term depression. Nature 369, 486–488. doi: 10.1038/369486a0
Newlon, M. G., Roy, M., Morikis, D., Carr, D. W., Westphal, R., Scott, J. D., et al. (2001). A novel mechanism of PKA anchoring revealed by solution structures of anchoring complexes. EMBO J. 20, 1651–1662. doi: 10.1093/emboj/20.7.1651
Newlon, M. G., Roy, M., Morikis, D., Hausken, Z. E., Coghlan, V., Scott, J. D., et al. (1999). The molecular basis for protein kinase A anchoring revealed by solution NMR. Nat. Struct. Biol. 6, 222–227. doi: 10.1038/6663
Oliveria, S. F., Dell’Acqua, M. L., and Sather, W. A. (2007). AKAP79/150 anchoring of calcineurin controls neuronal L-type Ca2+ channel activity and nuclear signaling. Neuron 55, 261–275. doi: 10.1016/j.neuron.2007.06.032
Peng, M., Aye, T. T., Snel, B., Van Breukelen, B., Scholten, A., and Heck, A. J. (2015). Spatial organization in protein kinase A signaling emerged at the base of animal evolution. J. Proteome Res. 14, 2976–2987. doi: 10.1021/acs.jproteome.5b00370
Reed, B. J., Locke, M. N., and Gardner, R. G. (2015). A conserved deubiquitinating enzyme uses intrinsically disordered regions to scaffold multiple protein-interaction sites. J. Biol. Chem. doi: 10.1074/jbc.M115.650952 [Epub ahead of print].
Reiterer, V., Eyers, P. A., and Farhan, H. (2014). Day of the dead: pseudokinases and pseudophosphatases in physiology and disease. Trends Cell Biol. 24, 489–505. doi: 10.1016/j.tcb.2014.03.008
Ren, S., Uversky, V. N., Chen, Z., Dunker, A. K., and Obradovic, Z. (2008). Short Linear Motifs recognized by SH2, SH3 and Ser/Thr Kinase domains are conserved in disordered protein regions. BMC Genomics 9(Suppl. 2):S26. doi: 10.1186/1471-2164-9-S2-S26
Rodriguez, A., Roy, J., Martinez-Martinez, S., Lopez-Maderuelo, M. D., Nino-Moreno, P., Orti, L., et al. (2009). A conserved docking surface on calcineurin mediates interaction with substrates and immunosuppressants. Mol. Cell 33, 616–626. doi: 10.1016/j.molcel.2009.01.030
Roy, J., and Cyert, M. S. (2009). Cracking the phosphatase code: docking interactions determine substrate specificity. Sci. Signal. 2, re9. doi: 10.1126/scisignal.2100re9
Roy, J., Li, H., Hogan, P. G., and Cyert, M. S. (2007). A conserved docking site modulates substrate affinity for calcineurin, signaling output, and in vivo function. Mol. Cell 25, 889–901. doi: 10.1016/j.molcel.2007.02.014
Sanderson, J. L., and Dell’Acqua, M. L. (2011). AKAP signaling complexes in regulation of excitatory synaptic plasticity. Neuroscientist 17, 321–336. doi: 10.1177/1073858410384740
Sanderson, J. L., Gorski, J. A., Gibson, E. S., Lam, P., Freund, R. K., Chick, W. S., et al. (2012). AKAP150-anchored calcineurin regulates synaptic plasticity by limiting synaptic incorporation of Ca2+-permeable AMPA receptors. J. Neurosci. 32, 15036–15052. doi: 10.1523/JNEUROSCI.3326-12.2012
Sarma, G. N., Kinderman, F. S., Kim, C., Von Daake, S., Chen, L., Wang, B. C., et al. (2010). Structure of D-AKAP2:PKA RI complex: insights into AKAP specificity and selectivity. Structure 18, 155–166. doi: 10.1016/j.str.2009.12.012
Schachterle, C., Christian, F., Fernandes, J. M., and Klussmann, E. (2015). Screening for small molecule disruptors of AKAP-PKA interactions. Methods Mol. Biol. 1294, 151–166. doi: 10.1007/978-1-4939-2537-7_12
Schafer, G., Milic, J., Eldahshan, A., Gotz, F., Zuhlke, K., Schillinger, C., et al. (2013). Highly functionalized terpyridines as competitive inhibitors of AKAP-PKA interactions. Angew. Chem. Int. Ed. Engl. 52, 12187–12191. doi: 10.1002/anie.201304686
Schillace, R. V., and Scott, J. D. (1999). Association of the type 1 protein phosphatase PP1 with the A-kinase anchoring protein AKAP220. Curr. Biol. 9, 321–324. doi: 10.1016/S0960-9822(99)80141-9
Schillace, R. V., Voltz, J. W., Sim, A. T., Shenolikar, S., and Scott, J. D. (2001). Multiple interactions within the AKAP220 signaling complex contribute to protein phosphatase 1 regulation. J. Biol. Chem. 276, 12128–12134. doi: 10.1074/jbc.M010398200
Scott, J. D., Stofko, R. E., Mcdonald, J. R., Comer, J. D., Vitalis, E. A., and Mangeli, J. (1990). Type II regulatory subunit dimerization determines the subcellular localization of the cAMP-dependent protein kinase. J. Biol. Chem. 265, 21561–21566.
Shih, M., Lin, F., Scott, J. D., Wang, H. Y., and Malbon, C. C. (1999). Dynamic complexes of beta2-adrenergic receptors with protein kinases and phosphatases and the role of gravin. J. Biol. Chem. 274, 1588–1595. doi: 10.1074/jbc.274.3.1588
Singh, A., Redden, J. M., Kapiloff, M. S., and Dodge-Kafka, K. L. (2011). The large isoforms of A-kinase anchoring protein 18 mediate the phosphorylation of inhibitor-1 by protein kinase A and the inhibition of protein phosphatase 1 activity. Mol. Pharmacol. 79, 533–540. doi: 10.1124/mol.110.065425
Singh, M., Singh, P., Vaira, D., Torheim, E. A., Rahmouni, S., Tasken, K., et al. (2014). The RIAD peptidomimetic inhibits HIV-1 replication in humanized NSG mice. Eur. J. Clin. Invest. 44, 146–152. doi: 10.1111/eci.12200
Smith, F. D., Reichow, S. L., Esseltine, J. L., Shi, D., Langeberg, L. K., Scott, J. D., et al. (2013). Intrinsic disorder within an AKAP-protein kinase A complex guides local substrate phosphorylation. Elife 2, e01319. doi: 10.7554/eLife.01319
Steen, R. L., Martins, S. B., Tasken, K., and Collas, P. (2000). Recruitment of protein phosphatase 1 to the nuclear envelope by A- kinase anchoring protein AKAP149 is a prerequisite for nuclear lamina assembly. J. Cell Biol. 150, 1251–1262. doi: 10.1083/jcb.150.6.1251
Stefan, E., Wiesner, B., Baillie, G. S., Mollajew, R., Henn, V., Lorenz, D., et al. (2007). Compartmentalization of cAMP-dependent signaling by phosphodiesterase-4D is involved in the regulation of vasopressin-mediated water reabsorption in renal principal cells. J. Am. Soc. Nephrol. 18, 199–212. doi: 10.1681/ASN.2006020132
Stewart, A. A., Ingebritsen, T. S., Manalan, A., Klee, C. B., and Cohen, P. (1982). Discovery of a Ca2+- and calmodulin-dependent protein phosphatase: probable identity with calcineurin (CaM-BP80). FEBS Lett. 137, 80–84. doi: 10.1016/0014-5793(82)80319-0
Stokka, A. J., Gesellchen, F., Carlson, C. R., Scott, J. D., Herberg, F. W., and Tasken, K. (2006). Characterization of A-kinase-anchoring disruptors using a solution-based assay. Biochem. J. 400, 493–499. doi: 10.1042/BJ20060962
Stralfors, P., Hiraga, A., and Cohen, P. (1985). The protein phosphatases involved in cellular regulation. Purification and characterisation of the glycogen-bound form of protein phosphatase- 1 from rabbit skeletal muscle. Eur. J. Biochem. 149, 295–303. doi: 10.1111/j.1432-1033.1985.tb08926.x
Su, Y., Dostmann, W. R. G., Herberg, F. W., Durick, K., Xuong, N.-H., Ten Eyck, L., et al. (1995). Regulatory subunit of protein kinase A: structure of deletion mutant with cAMP binding proteins. Science 269, 807–813. doi: 10.1126/science.7638597
Tandan, S., Wang, Y., Wang, T. T., Jiang, N., Hall, D. D., Hell, J. W., et al. (2009). Physical and functional interaction between calcineurin and the cardiac L-type Ca2+ channel. Circ. Res. 105, 51–60. doi: 10.1161/CIRCRESAHA.109.199828
Tanji, C., Yamamoto, H., Yorioka, N., Kohno, N., Kikuchi, K., and Kikuchi, A. (2002). A-kinase anchoring protein AKAP220 binds to glycogen synthase kinase-3beta (GSK-3beta) and mediates protein kinase A-dependent inhibition of GSK-3beta. J. Biol. Chem. 277, 36955–36961. doi: 10.1074/jbc.M206210200
Taylor, S. S., Ilouz, R., Zhang, P., and Kornev, A. P. (2012). Assembly of allosteric macromolecular switches: lessons from PKA. Nat. Rev. Mol. Cell Biol. 13, 646–658. doi: 10.1038/nrm3432
Theurkauf, W. E., and Vallee, R. B. (1982). Molecular characterization of the cAMP-dependent protein kinase bound to microtubule-associated protein 2. J. Biol. Chem. 257, 3284–3290.
Troger, J., Moutty, M. C., Skroblin, P., and Klussmann, E. (2012). A-kinase anchoring proteins as potential drug targets. Br. J. Pharmacol. 166, 420–433. doi: 10.1111/j.1476-5381.2011.01796.x
Van Roey, K., Uyar, B., Weatheritt, R. J., Dinkel, H., Seiler, M., Budd, A., et al. (2014). Short linear motifs: ubiquitous and functionally diverse protein interaction modules directing cell regulation. Chem. Rev. 114, 6733–6778. doi: 10.1021/cr400585q
Veesler, D., Cupelli, K., Burger, M., Graber, P., Stehle, T., and Johnson, J. E. (2014). Single-particle EM reveals plasticity of interactions between the adenovirus penton base and integrin alphaVbeta3. Proc. Natl. Acad. Sci. U.S.A. 111, 8815–8819. doi: 10.1073/pnas.1404575111
Vigil, D., Blumenthal, D. K., Heller, W. T., Brown, S., Canaves, J. M., Taylor, S. S., et al. (2004). Conformational differences among solution structures of the type Ialpha, IIalpha and IIbeta protein kinase A regulatory subunit homodimers: role of the linker regions. J. Mol. Biol. 337, 1183–1194. doi: 10.1016/j.jmb.2004.02.028
Vijayaraghavan, S., Goueli, S. A., Davey, M. P., and Carr, D. W. (1997). Protein kinase A-anchoring inhibitor peptides arrest mammalian sperm motility. J. Biol. Chem. 272, 4747–4752. doi: 10.1074/jbc.272.8.4747
Viswanatha, R., Wayt, J., Ohouo, P. Y., Smolka, M. B., and Bretscher, A. (2013). Interactome analysis reveals ezrin can adopt multiple conformational states. J. Biol. Chem. 288, 35437–35451. doi: 10.1074/jbc.M113.505669
Walsh, D. A., Perkins, J. P., and Krebs, E. G. (1968). An adenosine 3’,5’-monophosphate-dependent protein kinase from rabbit skeletal muscle. J. Biol. Chem. 243, 3763–3765.
Wang, Y., Ho, T. G., Bertinetti, D., Neddermann, M., Franz, E., Mo, G. C., et al. (2014). Isoform-selective disruption of AKAP-localized PKA using hydrocarbon stapled peptides. ACS Chem. Biol. 9, 635–642. doi: 10.1021/cb400900r
Wang, Y., Ho, T. G., Franz, E., Hermann, J. S., Smith, F. D., Hehnly, H., et al. (2015). PKA-type I selective constrained peptide disruptors of AKAP complexes. ACS Chem. Biol. 10, 1502–1510. doi: 10.1021/acschembio.5b00009
Welch, E. J., Jones, B. W., and Scott, J. D. (2010). Networking with AKAPs: context-dependent regulation of anchored enzymes. Mol. Interv. 10, 86–97. doi: 10.1124/mi.10.2.6
Westphal, R. S., Tavalin, S. J., Lin, J. W., Alto, N. M., Fraser, I. D., Langeberg, L. K., et al. (1999). Regulation of NMDA receptors by an associated phosphatase-kinase signaling complex. Science 285, 93–96. doi: 10.1126/science.285.5424.93
Whiting, J. L., Nygren, P. J., Tunquist, B. J., Langeberg, L. K., Seternes, O. M., and Scott, J. D. (2015). Protein kinase A opposes the phosphorylation-dependent recruitment of glycogen synthase kinase 3β to A-kinase anchoring protein 220. J. Biol. Chem. doi: 10.1074/jbc.M115.654822 [Epub ahead of print].
Williams, K. R., Hemmings, H. C. Jr., Lopresti, M. B., Konigsberg, W. H., and Greengard, P. (1986). DARPP-32, a dopamine- and cyclic AMP-regulated neuronal phosphoprotein. Primary structure and homology with protein phosphatase inhibitor-1. J. Biol. Chem. 261, 1890–1903.
Wong, W., and Scott, J. D. (2004). AKAP Signalling complexes: focal points in space and time. Nat. Rev. Mol. Cell Biol. 5, 959–971. doi: 10.1038/nrm1527
Wu, J., Brown, S. H., Von Daake, S., and Taylor, S. S. (2007). PKA type IIalpha holoenzyme reveals a combinatorial strategy for isoform diversity. Science 318, 274–279. doi: 10.1126/science.1146447
Zawadzki, K. M., and Taylor, S. S. (2004). cAMP-dependent protein kinase regulatory subunit type IIbeta: active site mutations define an isoform-specific network for allosteric signaling by cAMP. J. Biol. Chem. 279, 7029–7036. doi: 10.1074/jbc.M310804200
Keywords: protein kinase A (PKA), protein phosphatase 2B (PP2B), calcineurin, A-kinase anchoring protein (AKAP), intrinsic disorder, cAMP signaling, short linear interaction motifs (SLiMs)
Citation: Nygren PJ and Scott JD (2015) Therapeutic strategies for anchored kinases and phosphatases: exploiting short linear motifs and intrinsic disorder. Front. Pharmacol. 6:158. doi: 10.3389/fphar.2015.00158
Received: 16 June 2015; Accepted: 16 July 2015;
Published: 28 July 2015.
Edited by:
Frank Christian, University of Glasgow, UKReviewed by:
Doriano Lamba, Consiglio Nazionale delle Ricerche, ItalyEnno Klussmann, Max Delbrück Center for Molecular Medicine, Germany
Copyright © 2015 Nygren and Scott. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: John D. Scott, Department of Pharmacology, University of Washington, 1959 NE Pacific Street, Seattle, WA 98195, USA,c2NvdHRqZHdAdXcuZWR1
 Patrick J. Nygren
Patrick J. Nygren John D. Scott
John D. Scott