- Pediatric Surgery Department, The First Affiliated Hospital of Zhengzhou University, Zhengzhou, China
Background: Pediatric solid tumors are a common malignant disease in children, and more and more studies have proved that there is an inseparable relationship between adult tumors and intestinal microbiome, but the changes in the intestinal microbiota of pediatric solid tumor (PST) patients have been scarcely examined. This study aims to examine the differences in the intestinal microbiota features between patients diagnosed with PST and healthy controls (HCs).
Methods: To elucidate the unique characteristics of the gut microbiota in pediatric patients with solid tumors, we recruited 23 PST patients and 20 HCs. A total of 43 stool samples were gathered, and then 16S rRNA sequencing was performed.
Results: We noticed a noticeable pattern of elevated diversity in the gut microbiota within the PST groups. The differences in microbial communities among two groups were remarkable, regarding the analysis at the class level, the abundance of Bacilli was markedly increased in PST patients compared to HCs (P < 0.05), regarding the analysis at the genus level, The presence of Enterococcus was significantly higher in PST cases compared to HCs (P < 0.01), while Lachnospiraceae unclassified, Lachnospira, Haemophilus and Colidextribacter in PST cases, the abundance was significantly reduced. (P < 0.05), 6 genera, including Bacilli, Lactobacillales, Enterococcaceae and Morganella, showed a significant enrichment compared to healthy controls, while 10 genera, including Bilophila, Colidextribacter, Pasteurellales, Haemophilus, Lachnospiraceae unclassified, Lachnospira and Fusobacteriales, were significant reduction in the PST groups.
Conclusion: Our research conducted the characterization analysis of the gut microbiota in PST patients for the first time. More importantly, there are some notable differences in the gut microbiota between PST patients and healthy controls, which we believe is an interesting finding.
Introduction
Childhood malignancies have been a gradual increase in the incidence over the past three decades, and the incidence has been rising at an annual rate of about 1% when considering all types of cancer combined (1). This translates to approximately 400,000 cases per year globally (2), and standing as the second leading cause of mortality in developed countries after accidents (3). There are numerous types of PST, with wilms tumor, osteosarcoma, and neuroblastoma being the most common types of childhood cancers (4). Neuroblastoma, constituting roughly 15% of all cancer-related fatalities, stands as the prevalent solid tumor among infants (5). Osteosarcoma stands as the predominant primary malignant bone tumor among the demographic of children and adolescents (6). Wilms tumor prevails as the primary kidney tumor in childhood, constituting 90% of occurrences (7). The occurrence of PST is linked to early developmental processes (8). Considering their emergence within actively growing tissues, childhood solid tumors can be viewed as diseases arising from dysregulated development (9). PST demonstrates high metastatic potential, poor prognosis, and substantial resistance to current therapeutic methods (10). Cancer continues to be a prominent contributor to mortality in the pediatric and adolescent populations (11). Each year, around 15,000 children and adolescents aged 0–19 receive a diagnosis of cancer in the United States (12). It is crucial for primary care physicians and parents to have a thorough understanding of the initial symptoms of childhood malignancies to enable effective early detection and treatment (13). When it comes to diagnosis, creating tests for children has proven to be more challenging (14).
The human microbiota encompasses all microorganisms, such as bacteria, viruses, fungi, archaea, and protozoa, residing within the human body (15). The primary habitat of microorganisms is the gut, but a plethora of microorganisms can also be found in various parts of the human body, including digestive system, skin, reproductive system and respiratory system (16). In human bodys, there is no denying that the largest microbial ecosystem is the gut microbiota (17). Recently, due to the progress in gut microbiome study, there has been an increasing acknowledgment of the intimate link between gut microbes and diseases, such as nonalcoholic fatty liver (18), obesity (19), heart failure (20), diabetes (21) and depression (22). Meanwhile, a growing body of research suggests an inseparable relationship between the occurrence and development of adult tumors and the gut microbiota. For instance, resident gut bacteria can influence patients' response to cancer immunotherapy (23), the gut microbiota possesses a unique ability to influence the tumor microenvironment as well as the metabolism of chemotherapy drugs or medications (24), the gut microbiota can modulate melanoma patients' response to PD-1 immunotherapy (25), the gut microbiota and its metabolites change with different stages of lung cancer development (26), the composition of the gut microbiota in patients diagnosed with colorectal cancer differs significantly from that of healthy individuals (27), based on specific changes in the gut microbiota, it is possible to screen for gut microbiota-targeted biomarkers and establish a diagnostic model, potentially serving as a non-invasive tool for distinguishing hepatocellular carcinoma (28), However, many aspects, including the mechanisms of onset, age of onset and characteristic features, differ between pediatric tumors and adult tumors. For instance, compared to adult tumors, childhood tumors generally exhibit a lower overall mutation burden (9). Similarly, genomic sequencing research has highlighted distinctions between adult and pediatric tumors. Unlike many adult tumors, which exhibit significant somatic mutations, pediatric tumors generally feature a limited number of somatic mutations in their cells (29). Therefore, we cannot indiscriminately merge the diagnosis and treatment of PST with those of adult tumors.
Over the recent decades, there has been an increasing enthusiasm for exploring the human microbiome and its implications for both well-being and illness. The primary emphasis of these investigations has revolved around understanding the changes in microbial community composition across various circumstances (30). However, research on the gut microbiome of PST patients has been limited thus far. This is indeed the purpose behind our undertaking of this research. This research explores the gut microbiome attributes in pediatric patients with solid tumors in the central region of China. We conducted analysis of fecal samples from 20 HCs and 23 PST patients using the 16S rRNA MiSeq sequencing method. The aim was to investigate the distinctive composition of gut microbiota in PST patients and observe the specific changes of intestinal microecology in children with solid tumor.
Materials and methods
Participant information
This research was rigorously planned and executed in accordance with the PRoBE guidelines (Prospective Specimen Collection and Retrospective Blinded Evaluation), in adherence to the principles set forth in the Helsinki Declaration and the Regulations of Good Clinical Practice. This research collected fecal samples for analysis and all samples from the First Affiliated Hospital of Zhengzhou University. Every participant was newly diagnosed with PST. The participants in this research were identified as individuals diagnosed with PST through pathological examination and tissue biopsy. The participants exclusively comprised patients who were newly diagnosed with PST. The exclusion criteria are as follows: (1) Simultaneous existence of additional illnesses, (2) Administration of antibiotics in the last eight weeks, and (3) medical history involves past treatments or tumor removal surgery such as chemotherapy and radiotherapy. There were 50 fecal samples were gathered in the scope of this research. These included 25 samples from patients diagnosed with PST and 25 samples from HCs. After thorough screening, 20 HCs and 23 PST patients were selected for the study, ensuring matching criteria for sex, age and BMI. In the end, 16S rRNA MiSeq sequencing was performed on 43 fecal samples were collected from both PST and HCs.
Clinicopathological and demographic information of participants was obtained through both electronic medical records and questionnaires from the hospital (Table 1).
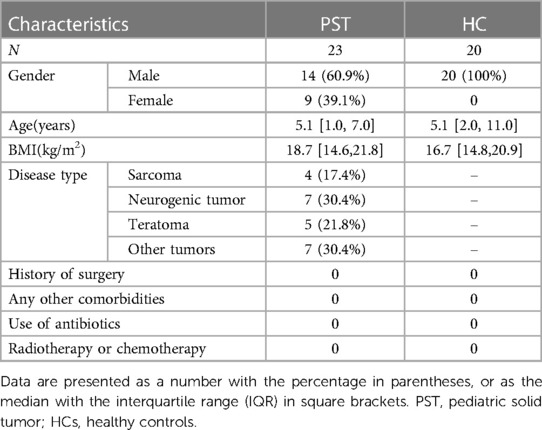
Table 1 Demographic characteristics of patients with pediatric solid tumor and the age- and gender-matched healthy controls.
Collection of human fecal samples
Participants provided a new stool specimen for the research. Standard stool examinations were conducted to evaluate the texture of the stool. The specimens were split into five portions, each weighing 200 mg, and immediately stored at −80°C.
DNA extraction and PCR amplification
Consistent techniques were employed across each sample, and the procedures were consistently executed by laboratory staff members. The specimen was blended with 790 μl of a sterile lysis buffer, this mixture was then placed in a 2 ml screw-cap tube containing 1 g glass beads(0.1 mm BioSpec Products, Inc., USA). The blend was thoroughly vortexed and subsequently subjected to incubation at 70°C for one hour. Afterward, the combination underwent bead beating at the highest speed setting for 10 min. Using The E.Z.N.A.® Stool DNA Kit (Omega Bio-tek, Inc., GA), Bacterial DNA extraction was carried out in accordance with the instructions furnished by the producer. The isolated DNA was preserved at −20°C. Amplification of the 16S rRNA genes was performed targeting the V3–V4 region using the DNA extracted from individual samples as a template. In each sample, PCR was utilized to amplify the V3–V4 region, employing primers R2 and F1 with sequences 5′-CCTACGGGNGGCWGCAG-3′ and 5′-GACTACHVGGGTATCTAATCC-3′, respectively. Following the specified protocol. PCR reactions were performed utilizing the EasyCycler 96 PCR system provided by Analytik Jena Corp., AG.
Miseq sequencing and sequence data processing
The amplified products from different samples were tagged with unique indexes and combined in equal proportions to enable sequencing. The sequencing procedure was carried out on the Miseq platform (Illumina Inc., USA) by Shanghai Mobio Biomedical Technology Co. Ltd., adhering to the manufacturer's guidelines. After arranging the chosen readings, they were categorized into specific sample groups using dedicated barcodes. Following that, the primers and barcodes were eliminated.
Utilizing FLASH v. 1.2.10 to combine paired-end sequences from individual libraries. Utilizing default parameters to enable the merging of sequences with overlapping regions (31). The merged reads obtained from the FLASH procedure underwent an evaluation of quality. Utilizing UCHIME version 4.2.40 to detect and remove potential chimera sequences (32). All nucleotide sequences from the samples were deposited into the NCBI database(National Center for Biotechnology Information) in the United States under the project accession number PRJNA1026296.
OTU clustering and taxonomic annotation
The same quantity of reads was randomly chosen from each of the samples. Following this, The UPARSE pipeline was employed for operational taxonomic unit (OTU) classification (33). We computed the total count of OTUs (operational taxonomic units) across different taxonomic levels, such as genus, family, order, class and phylum (34).
Bacterial diversity and taxonomic analysis
Utilizing the “vegan” R package, we calculated the Shannon index and Simpson index to assess bacterial community diversity. Moreover, ace estimators were employed to estimate the abundance of the bacterial community. Using Venn diagrams to illustrate the similarity and overlap of OTUs, showing common and distinct units present in different samples. Additionally, the creation of heatmaps to visually represent dominant species in the dataset was facilitated by the utilization of Heatmap Builder. We generated bar plots to illustrate the microbial community by analyzing the species composition. Utilized for performing Principal Coordinates Analysis (PCoA) and Non-Metric Multidimensional Scaling (NMDS) was the “vegan” R package, and the objective was to clarify the microbial differences among the samples. To aid in identification, used for computing both weighted and unweighted UniFrac distances was the “phyloseq” package. The evolutionary relationships among bacteria were visualized through phylogenetic trees. Bacterial taxonomic analyses were conducted at various levels, including genus, family, order, class and phylum. Afterward, we employed Wilcoxon rank-sum tests to assess microbiome differences among two groups. Furthermore, applying the LEfSe (linear discriminant analysis effect size) method, we conducted LDA (linear discriminant analysis) to identify critical microbiomes that exhibit significant distinctions. Utilizing the LEfSe online tool accessible at http://huttenhower.sph.harvard.edu/lefse/e/, the analysis was performed. An LDA score threshold of log10 = 2 was set (35). Additionally, using both the Wilcoxon rank-sum test and the nonparametric Kruskal-Wallis rank-sum test to identify significant biomarkers.
Statistical analysis
Utilizing IBM SPSS Statistics version 20.0 (IBM Corp., Armonk, NY, USA), the data analysis was conducted. Statistical significance of differences between the groups was determined. Spearman's rank test was applied for correlation analysis. To compare categorical variables, Fisher's exact test was utilized, while for continuous variables, the Wilcoxon rank-sum test was utilized.
Results
Participant profiles
The study collected 50 stool samples, including 25 PST and 25 HCs. After rigorous screening and exclusion (Figure 1), 43 stool samples remained, comprising 23 PST patients and 20 HCs. Ultimately, The fecal microbiota composition of 20 HCs and 23 PST patients was analyzed.
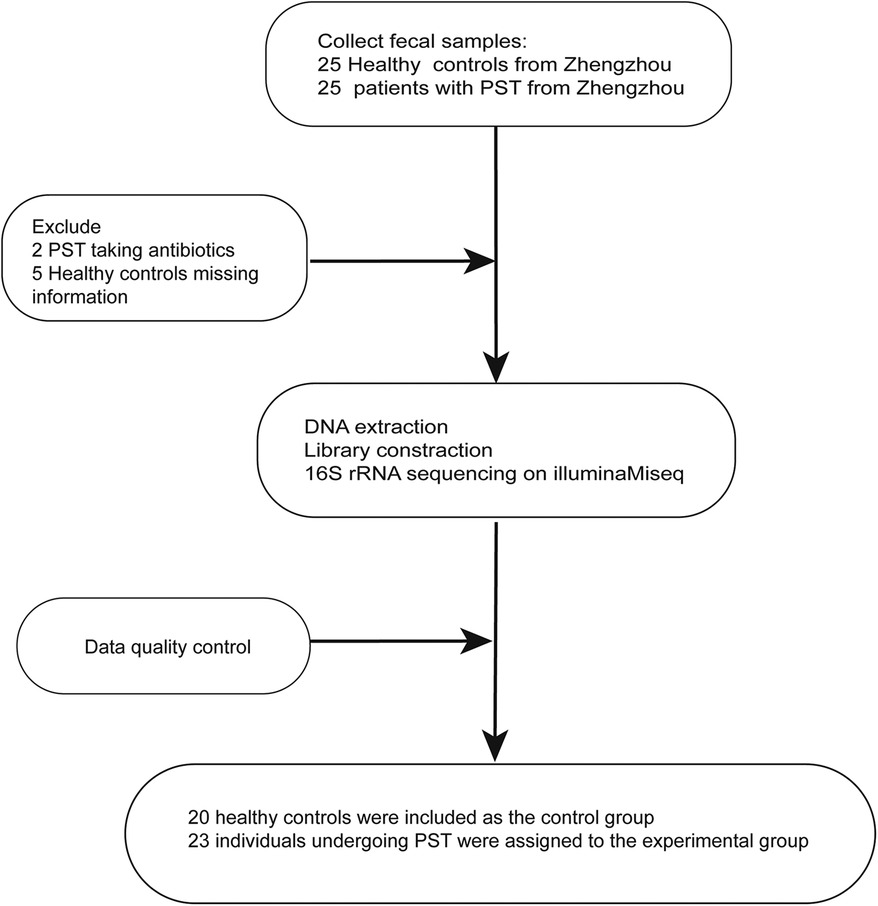
Figure 1 Study design and flow diagram. A total of 50 fecal samples from Central China were collected. After a strict pathological diagnosis and exclusion process, 23 PST patients and 20 HCs were included, 20 HCs were included as the control group, 23 individuals undergoing PST were assigned to the experimental group. PST, pediatric solid tumor; HCs, healthy controls.
The increased gut microbial diversity in PST
A marked increase in gut microbial diversity in PST patients compared to HCs. As measured by the Shannon (Figure 2A) and Simpson (Figure 2B) indices, there was a notable increase in the diversity of intestinal microbiota in PST patients. The observed and ace indices confirmed the validity of this finding (Figure 2C).
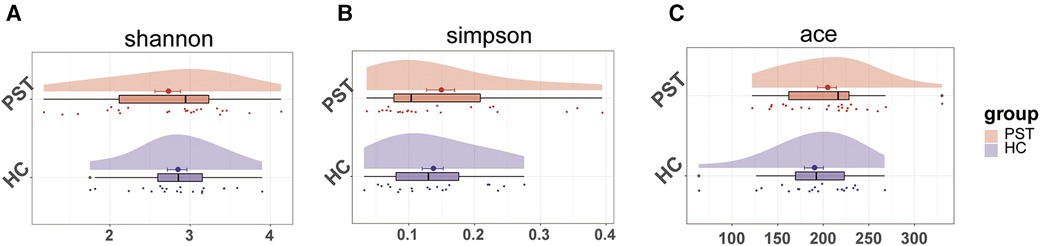
Figure 2 The microbial diversity was increased in PST patients (N = 23) vs. HCs (N = 20). As measured by the Shannon (A) and Simpson (B) index, the intestinal microbiota diversity in PST patients was significantly higher than in HCs. This finding was confirmed by the ace indices (C) PST, pediatric solid tumors; HCs, healthy controls.
Disparities in the gut microbiome between HCs and PST patients
Furthermore, beta diversity analysis was conducted to illustrate the differences in microbial community composition among individual samples. PCA (Figure 3A), PCoA (Figure 3B) and NMDS (Figure 3C) analyses estimated significant differences in OTU distribution between the two groups. Figure 3D of the Venn diagram illustrated the sharing of 431 out of 679 OTUs between the PST and HCs. Notably, among these 679 OTUs, 163 were unique to the PST group.
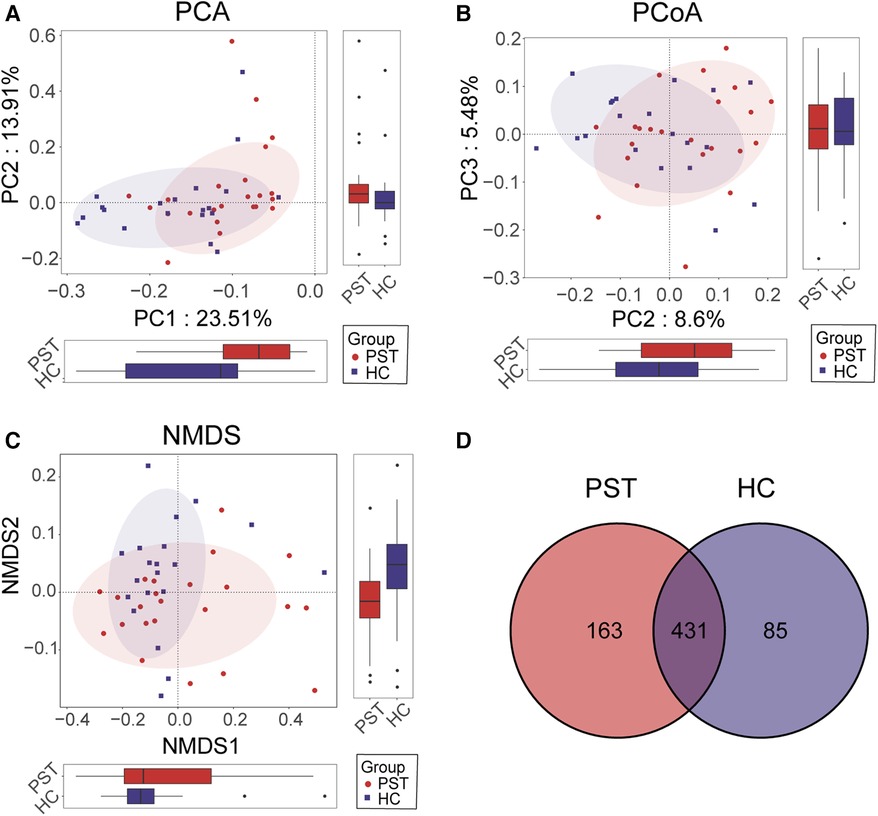
Figure 3 Comparison of beta diversity between PST patients (N = 23) and HCs (N = 20). According to PCA (A), PCOA (B) and NMDS (C) analysis, there were significant differences in OTU distribution between PST and HCs. The Venn diagram (D) revealed that out of 679 OTUs, 431 OTUs were shared between the two groups, while 163 OTUs were specific to PST. PST, pediatric solid tumor; HCs, healthy controls; OTUs, operational taxonomic units; NMDS, nonmetric multidimensional scaling; PCA, principal component analysis; PCoA, principal coordinate analysis.
Clustering of operational taxonomic units (OTUs) and taxonomic analysis
The heatmap illustrates the variation in relative abundance of OTUs between PST and HCs (Figure 4). Shades of blue are used to represent OTUs with lower relative abundances, whereas shades of red indicate higher relative abundances. A total of 8 bacterial species show an increase in PST patients, including OTU94 (Streptococcus), OTU74 (Clostridiaceae Clostridium sensu stricto), OTU279 (Bacteroides), OTU93 (Monoglobus), OTU140 (Alistipes), OTU425 (Morganella), OTU281 (Lachnospiraceae unclassified) and OTU105 (Eisenbergiella).
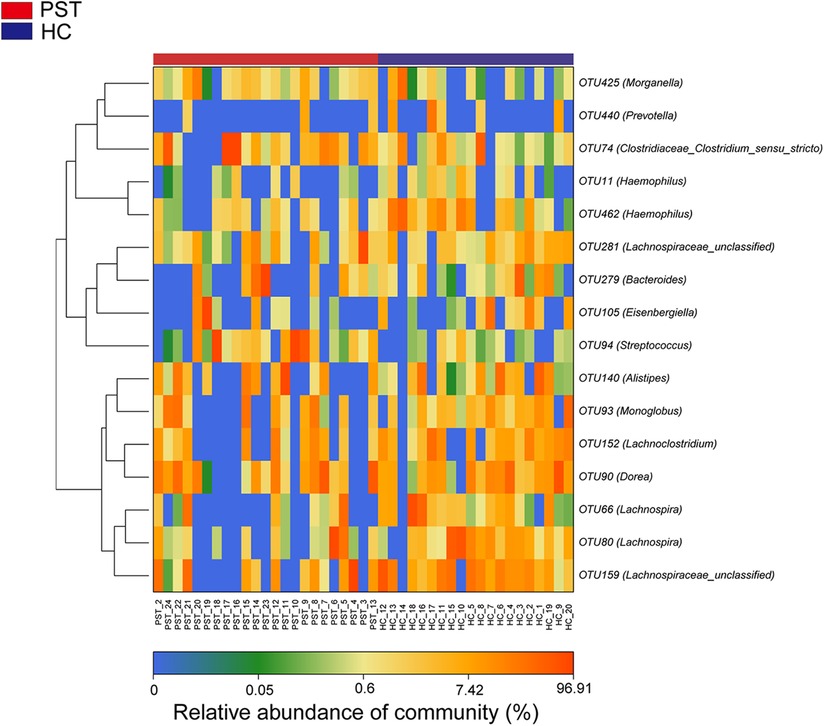
Figure 4 Heatmap displayed the relative abundance differences of OTUs between PST (N = 23) and HCs (N = 20). The relative abundance of each sample was shown, with red indicating high abundance and blue indicating low abundance. Each row represents an OTU. OTU, operational taxonomic unit; PST, pediatric solid tumors; HCs, healthy controls.
Composition and comparison of the gut microbiome in PST and HCs
The gut microbiota of PST and HCs were analyzed and compared in terms of their composition and diversity. The computed relative prevalence of individual sample was visually depicted across diverse taxonomic tiers through the utilization of OTU annotations. At the level of class, there was a predominant average proportion observed in both cohorts (nearly 99%) of Bacteroidia, Clostridia, Gammaproteobacteria, Negativicutes, Actinobacteria and Bacilli (Figure 5A). Similarly, at the genera level, including Faecalibacterium, Prevotella, Bacteroides, Bifidobacterium, Subdoligranulum, Escherichia-Shigella, Veillonella and Parabacteroides, both groups exhibited a consistent pattern where an average of 90% of the microbiota consisted of these 29 genera (Figure 5B). Regarding the classifications of class and genus, notable differences were observed between the two groups in microbial composition.
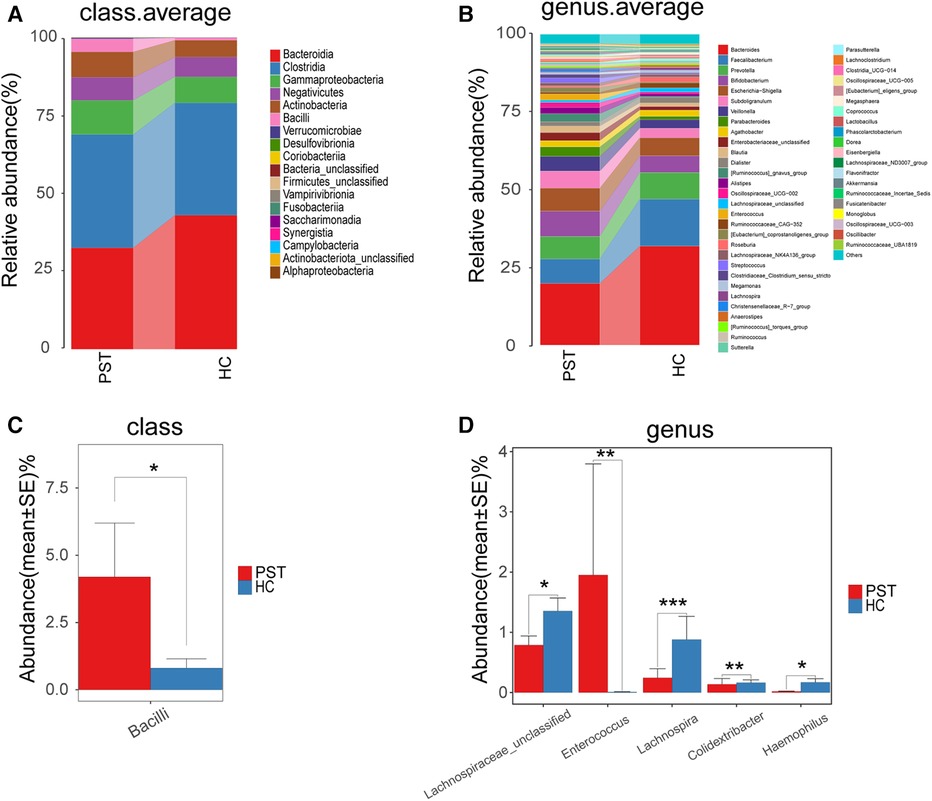
Figure 5 Composition and comparison of the gut microbiome in PST patients (N = 23) and HCs (N = 20). The class level (A) and genus level (B) composition diagrams showed the composition characteristics of the two groups of gut microbiome. The differences in the relative abundance of key bacterias in the two groups were compared at the class level (C) and the genus level (D) The relative abundance of each bacteria was represented by the mean ± SE. We used the Wilcoxon rank-sum test to evaluate whether the difference of relative abundance was significant (*P < 0.05; **P < 0.01 and ***P < 0.001). PST, pediatric solid tumor; HCs, healthy controls.
Subsequently, the gut microbiota of 23 PST and 20 HCs were compared at every taxonomy level. We utilized Wilcoxon rank-sum tests to examine substantial disparities between PST and HCs in microbial composition. In the analysis at the class level, the abundance of Bacilli was markedly elevated in PST in comparison to HCs (P < 0.05) (Figure 5C). In the analysis at the genus level, the presence of Enterococcus showed a significant elevation in PST in comparison to HCs (P < 0.01). While Lachnospiraceae unclassified, Lachnospira, Colidextribacter and Haemophilus significantly decreased (P < 0.05) (Figure 5D). In analyses conducted at both the class and genus levels, significant differences were observed.
Phylogenetic characteristics of gut microbial communities in PST
Utilizing OTU-based LDA scores and Lefse analysis, 6 genera exhibited notable enrichment in PST. Furthermore, 10 genera have been identified as predominant in the HCs, noteworthy disparities were identified among groups.
Figure 6A visually demonstrates the phylogenetic distribution of gut microbial communities between two groups. In Figure 6B, histograms illustrating LDA scores for selected taxonomic clusters are presented, indicating bacteria exhibited notable differences between two groups.
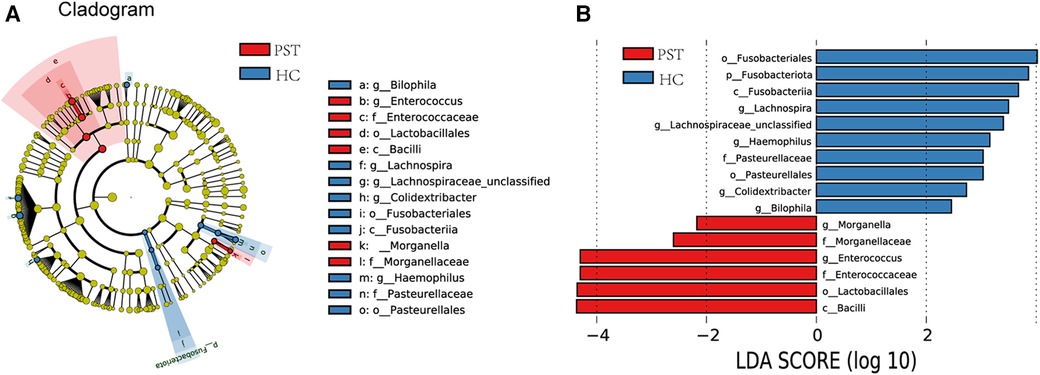
Figure 6 Lefse and LDA analysis based on OTUs characterize the microbiome between PST and HCs. The phylogenetic tree (A) diagram by the LEfSe method showed the phylogenetic distribution of the gut microbiome related to PST and HCs. The circles radiating from the inside to the outside represented the classification level from the phylum to the genus. Each circle at levels represented a classification at that level, and the diameter of the circle represents its relative abundance. The uniform coloring with no significant difference was yellow, and the biomarker with significant difference followed the grouping color for coloring. Species with an LDA score greater than 2 and a P-value less than 0.05 were considered different species. LDA score histogram (B) showed the gut microbiome with significant differences between the two groups. The higher the LDA score, the higher the importance of microbial biomarkers. The default LDA score was greater than 2, and the P-value was less than 0.05, which was considered to indicate differential species. LDA, linear discriminant analysis; LEfSe, linear discriminant analysis effect size; OTUs, operational taxonomic units; PST, pediatric solid tumor; HCs, healthy controls.
Correspondingly, 6 biomarkers, including Bacilli, Lactobacillales, Enterococcaceae, Enterococcus, Morganellaceae and Morganella, exhibited significant enrichment in PST (P < 0.05), affirming their position as predominant genera. Moreover, there was a notable decrease in the prevalence of 10 genera, including Bilophila, Colidextribacter, Pasteurellales, Haemophilus, Lachnospiraceae unclassified, Lachnospira and Fusobacteriales (P < 0.05).
Discussion
This research provides the first comprehensive analysis of gut microbiota alterations in pediatric solid tumor patients, including elements of both community composition and species diversity. Sequencing of the gut microbiota was conducted using samples collected from 23 PST and 20 HCs matched for age, sex, and BMI, all from the central region of China, using the 16S rRNA gene. Surprisingly, PST patients exhibited a substantial increase in gut microbiota diversity in comparison to the HCs. In contrast, patients diagnosed with cirrhosis and liver cancer show a decrease in microbial diversity (36). The precise factors contributing to the diversity variation among gut microbial communities remain unknown (37). Disease occurrence might be linked to changes in microbial diversity, with increases or decreases that can vary based on different diseases. For example, in individuals diagnosed with AIH (autoimmune hepatitis), there was a marked elevation in Veillonella, coupled with a concurrent reduction in Streptococcus, as opposed to HCs (38). Within the high AAC (Acute Pancreatitis) score group, there was an observed trend of decreasing microbial diversity compared to HCs (39). The group with chronic pancreatitis showed lower microbial diversity compared to the healthy controls (40). Patients diagnosed with breast cancer exhibited reduced diversity in their intestinal microbiota in comparison to HCs (41). NMDS PCA and PCoA analyses revealed notable disparities when comparing PST patients and HCs in the composition of the gut microbiota community. The utilization of LefSe analysis and the resulting LDA scores revealed significant disparities in the gut microbiota composition among two cohorts. There is undoubtedly a connection between changes in microbial diversity and the initiation and advancement of diseases, and this is precisely the purpose of conducting this research.
In this study, 6 biomarkers, including Bacilli, Lactobacillales, Enterococcaceae, Enterococcus, Morganellaceae and Morganella, exhibited significant enrichment in PST (P < 0.05), affirming their position as predominant genera. Research suggests that Bacilli bacteria may be associated with the occurrence of colorectal tumors, with bacteria such as Bacillus cereus and Bacillus subtilis found in the intestines of patients with colon cancer (42, 43). Additionally, Bacilli bacteria have potential roles in immune regulation and tumor therapy, including their influence in the tumor microenvironment and their application as immune modulators (25). Some bacteria in the Enterococcaceae family, such as Enterococcus faecalis, are commonly found in the human intestinal tract and can cause infections (44). Some studies have found an association between certain gastrointestinal diseases, such as inflammatory bowel disease, and an increase in the abundance of Enterococcaceae bacteria (45). Enterococcus and Morganella may lead to urinary tract infections and wound infections in immunocompromised or hospitalized patients (46).
As metagenomics research progresses, our comprehension of the gut microbiota has grown, elucidating their increasingly evident connection with various diseases. For instance, gut microbes influence physiology by partially breaking down dietary components and substances produced by the host, leading to the production of bioactive compounds, including toxins (47). Gut microorganisms have a central role in regulating the overall purine balance and serum uric acid (UA) levels in the host (48). The gut microbiota facilitates the anti-obesity effects of intermittent fasting by impeding the absorption of intestinal lipids (49). Differences were noted in the lipid metabolites and gut microbiome when comparing patients with Spinal Muscular Atrophy to the control subjects. The altered microbiota could potentially be associated with the disruption of lipid metabolism in individuals with Spinal Muscular Atrophy (50). Patients with hepatic encephalopathy (HE) and cirrhosis showed reduced richness and diversity of microbial species (51). Moreover, the gut microbiota holds promise for being employed as a diagnostic instrument in clinical settings. The fecal microbiome can distinguish autoimmune hepatitis patients from healthy individuals (52). The gut microbiome can distinguish epilepsy patients from healthy individuals (53). By constructing the gut metagenome, type 2 diabetes can be distinguished from the control group based on its markers. Compared to similar analyses based on human genome variations, the gut microbiome demonstrates higher specificity (54). A research has unveiled complex changes in microbial communities linked to Crohn's disease and identified microbial genes as consistent diagnostic markers applicable across diverse cohorts, including those from different cultural backgrounds and geographical (55). When it comes to the diagnosis of gastrointestinal disorders, the gut microbiome has been acknowledged as a standalone diagnostic tool. Furthermore, strategies to enhance effectiveness across diverse cohorts have been revealed by identifying factors that consistently contribute to alterations in the gut microbiome across various study groups (56). Alterations in gut microbiota composition are associated with colorectal cancer and its precancerous lesions, with increased abundance of Fusobacterium and other bacteria. These microbial characteristics can serve as biomarkers for detecting colorectal tumors (57–59). We hypothesize that detecting gut microbiota could become a new and effective method for differentiating between patients with tumors and those without. It presents benefits including minimal invasiveness, diagnostic efficiency, high acceptability and cost-effectiveness. Therefore, we consider this research to be exceptionally valuable and meaningful. Furthermore, the gut microbiota demonstrates immense potential in the treatment of diseases. For example, scientists are dedicated to addressing inflammatory bowel diseases through the restoration of intestinal microbial balance and the improvement of intestinal inflammation. Novel approaches focused on the gut microbiome, including probiotics, prebiotics, and synbiotics, are emerging as promising methods to decelerate the advancement of inflammatory bowel diseases and promote intestinal health (60). The gut microbiota serves as a potential biomarker for the diagnosis and prognosis of systemic lupus erythematosus, and it also holds promise as a potential target for the treatment of systemic lupus erythematosus (61). Modulating the gut microbiota regulates distal symmetric polyneuropathy in diabetic patients (21). Although there is limited research on the role of the gut microbiota in disease treatment currently, we firmly believe that in the near future, the gut microbiota will play a more significant role in the diagnosis and treatment of diseases, helping a larger number of people. This is the original intention behind our undertaking of this study.
Conclusion
This research offers the initial assessment of gut microbiota in PST patients. Crucially, we observed significant differences in gut microbes between the two groups, particularly, changes in bacteria such as Bacilli, Lactobacillales, Enterococcaceae, Enterococcus, Morganellaceae, and Morganella. We think this is an interesting finding. In the next step of our experiments, we will increase sample collection and begin constructing relevant diagnostic models based on their specific microbial changes.
Limitations of study
We acknowledge the limitations of this study. The sample size of the study was relatively small; it is indeed challenging to collect cases of pediatric solid tumors. We spent nearly two years gathering these cases. Many children presented with fever upon admission and were immediately treated with antibiotics, leading to their exclusion from the study. Additionally, some cases involved children who were readmitted due to relapses or post-surgery chemotherapy, and they were also excluded. Consequently, the sample size for this experiment was relatively limited.
This experiment serves as a preliminary study for the comprehensive investigation of gut microbiota changes in pediatric solid tumor patients in the next step. The aim is to observe whether there are differences in the composition of gut microbiota between pediatric solid tumor patients and the healthy control group, facilitating the subsequent research. The results are clear: there are significant differences in the microbiota composition between the two groups. This finding lays the groundwork for our next experiment.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
The studies involving humans were approved by the First Affiliated Hospital of Zhengzhou University. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study. Written informed consent was obtained from the individual(s), and minor(s)' legal guardian/next of kin, for the publication of any potentially identifiable images or data included in this article.
Author contributions
XD: Conceptualization, Data curation, Formal Analysis, Investigation, Project administration, Writing – original draft, Writing – review & editing. XC: Data curation, Formal Analysis, Investigation, Methodology, Resources, Writing – original draft, Writing – review & editing. RF: Conceptualization, Investigation, Methodology, Project administration, Resources, Writing – original draft, Writing – review & editing. JP: Conceptualization, Data curation, Formal Analysis, Investigation, Methodology, Project administration, Writing – original draft, Writing – review & editing. XC: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article.
This project was financially supported by the National Natural Science Foundation of China (Grant No. 82002959).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Steliarova-Foucher E, Fidler MM, Colombet M, Lacour B, Kaatsch P, Pineros M, et al. Changing geographical patterns and trends in cancer incidence in children and adolescents in Europe, 1991–2010 (automated childhood cancer information system): a population-based study. Lancet Oncol. (2018) 19(9):1159–69. doi: 10.1016/S1470-2045(18)30423-6
2. Ward ZJ, Yeh JM, Bhakta N, Frazier AL, Atun R. Estimating the total incidence of global childhood cancer: a simulation-based analysis. Lancet Oncol. (2019) 20(4):483–93. doi: 10.1016/S1470-2045(18)30909-4
3. Blattner-Johnson M, Jones DTW, Pfaff E. Precision medicine in pediatric solid cancers. Semin Cancer Biol. (2022) 84:214–27. doi: 10.1016/j.semcancer.2021.06.008
4. Liu F, Xiong QW, Wang JH, Peng WX. Roles of lncRNAs in childhood cancer: current landscape and future perspectives. Front Oncol. (2023) 13:1060107. doi: 10.3389/fonc.2023.1060107
5. Hagemann S, Misiak D, Bell JL, Fuchs T, Lederer MI, Bley N, et al. IGF2BP1 Induces neuroblastoma via a druggable feedforward loop with MYCN promoting 17q oncogene expression. Mol Cancer. (2023) 22(1):88. doi: 10.1186/s12943-023-01792-0
6. Eaton BR, Schwarz R, Vatner R, Yeh B, Claude L, Indelicato DJ, et al. Osteosarcoma. Pediatr Blood Cancer. (2021) 68(Suppl 2):e28352. doi: 10.1002/pbc.28352
7. Bertacca I, Pegoraro F, Tondo A, Favre C. Targeted treatment of solid tumors in pediatric precision oncology. Front Oncol. (2023) 13:1176790. doi: 10.3389/fonc.2023.1176790
8. Leichter AL, Sullivan MJ, Eccles MR, Chatterjee A. MicroRNA expression patterns and signalling pathways in the development and progression of childhood solid tumours. Mol Cancer. (2017) 16(1):15. doi: 10.1186/s12943-017-0584-0
9. Filbin M, Monje M. Developmental origins and emerging therapeutic opportunities for childhood cancer. Nat Med. (2019) 25(3):367–76. doi: 10.1038/s41591-019-0383-9
10. Aboushanab SA, Shevyrin VA, Slesarev GP, Melekhin VV, Shcheglova AV, Makeev OG, et al. Antioxidant and cytotoxic activities of kudzu roots and soy molasses against pediatric tumors and phytochemical analysis of isoflavones using HPLC-DAD-ESI-HRMS. Plants (Basel). (2022) 11(6):741. doi: 10.3390/plants11060741
11. Steliarova-Foucher E, Colombet M, Ries LAG, Moreno F, Dolya A, Bray F, et al. International incidence of childhood cancer, 2001–10: a population-based registry study. Lancet Oncol. (2017) 18(6):719–31. doi: 10.1016/S1470-2045(17)30186-9
12. Bhatia S, Tonorezos ES, Landier W. Clinical care for people who survive childhood cancer: a review. JAMA. (2023) 330(12):1175–86. doi: 10.1001/jama.2023.16875
13. Dang-Tan T, Franco EL. Diagnosis delays in childhood cancer: a review. Cancer. (2007) 110(4):703–13. doi: 10.1002/cncr.22849
14. Nierengarten MB. Liquid biopsy test could revolutionize cancer care for pediatric patients with solid tumors: although the adoption of liquid biopsy testing for solid tumors in pediatric patients with cancer has been slower than that of its adult counterpart, the development of these tests for pediatric patient diagnosis promises better monitoring and treatment: although the adoption of liquid biopsy testing for solid tumors in pediatric patients with cancer has been slower than that of its adult counterpart, the development of these tests for pediatric patient diagnosis promises better monitoring and treatment. Cancer. (2023) 129(13):1950–1. doi: 10.1002/cncr.34882
15. Belkaid Y, Naik S. Compartmentalized and systemic control of tissue immunity by commensals. Nat Immunol. (2013) 14(7):646–53. doi: 10.1038/ni.2604
16. Nenci A, Becker C, Wullaert A, Gareus R, van Loo G, Danese S, et al. Epithelial NEMO links innate immunity to chronic intestinal inflammation. Nature. (2007) 446(7135):557–61. doi: 10.1038/nature05698
17. Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. (2006) 124(4):837–48. doi: 10.1016/j.cell.2006.02.017
18. Gómez-Pérez AM, Ruiz-Limón P, Salas-Salvadó J, Vioque J, Corella D, Fitó M, et al. Gut microbiota in nonalcoholic fatty liver disease: a PREDIMED-plus trial sub analysis. Gut Microbes. (2023) 15(1):2223339. doi: 10.1080/19490976.2023.2223339
19. Lahtinen P, Juuti A, Luostarinen M, Niskanen L, Liukkonen T, Tillonen J, et al. Effectiveness of fecal Microbiota transplantation for weight loss in patients with obesity undergoing bariatric surgery: a randomized clinical trial. JAMA Netw Open. (2022) 5(12):e2247226. doi: 10.1001/jamanetworkopen.2022.47226
20. Paraskevaidis I, Xanthopoulos A, Tsougos E, Triposkiadis F. Human gut Microbiota in heart failure: trying to unmask an emerging organ. Biomedicines. (2023) 11(9):2574. doi: 10.3390/biomedicines11092574
21. Yang J, Yang X, Wu G, Huang F, Shi X, Wei W, et al. Gut microbiota modulate distal symmetric polyneuropathy in patients with diabetes. Cell Metab. (2023) 35(9):1548–62.e7. doi: 10.1016/j.cmet.2023.06.010
22. Tian P, Chen Y, Zhu H, Wang L, Qian X, Zou R, et al. Bifidobacterium breve CCFM1025 attenuates major depression disorder via regulating gut microbiome and tryptophan metabolism: a randomized clinical trial. Brain Behav Immun. (2022) 100:233–41. doi: 10.1016/j.bbi.2021.11.023
23. Routy B, Le Chatelier E, Derosa L, Duong CPM, Alou MT, Daillère R, et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science. (2018) 359(6371):91–7. doi: 10.1126/science.aan3706
24. Pandya G, Kirtonia A, Singh A, Goel A, Mohan CD, Rangappa KS, et al. A comprehensive review of the multifaceted role of the microbiota in human pancreatic carcinoma. Semin Cancer Biol. (2022) 86(Pt 3):682–92. doi: 10.1016/j.semcancer.2021.05.027
25. Gopalakrishnan V, Spencer CN, Nezi L, Reuben A, Andrews MC, Karpinets TV, et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science. (2018) 359(6371):97–103. doi: 10.1126/science.aan4236
26. Lu X, Xiong L, Zheng X, Yu Q, Xiao Y, Xie Y. Structure of gut microbiota and characteristics of fecal metabolites in patients with lung cancer. Front Cell Infect Microbiol. (2023) 13:1170326. doi: 10.3389/fcimb.2023.1170326
27. Helisz P, Dziubanek G, Krupa-Kotara K, Gwioździk W, Grajek M, Głogowska-Ligus J. Colorectal cancer and the role of the gut microbiota-do medical students know more than other young people?-cross-sectional study. Nutrients. (2022) 14(19):4185 doi: 10.3390/nu14194185
28. Ren Z, Li A, Jiang J, Zhou L, Yu Z, Lu H, et al. Gut microbiome analysis as a tool towards targeted non-invasive biomarkers for early hepatocellular carcinoma. Gut. (2019) 68(6):1014–23. doi: 10.1136/gutjnl-2017-315084
29. Sweet-Cordero EA, Biegel JA. The genomic landscape of pediatric cancers: implications for diagnosis and treatment. Science. (2019) 363(6432):1170–5. doi: 10.1126/science.aaw3535
30. Solbiati J, Frias-Lopez J. Metatranscriptome of the oral microbiome in health and disease. J Dent Res. (2018) 97(5):492–500. doi: 10.1177/0022034518761644
31. Magoč T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. (2011) 27(21):2957–63. doi: 10.1093/bioinformatics/btr507
32. Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. (2011) 27(16):2194–200. doi: 10.1093/bioinformatics/btr381
33. Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods. (2013) 10(10):996–8. doi: 10.1038/nmeth.2604
34. Kakiyama G, Pandak WM, Gillevet PM, Hylemon PB, Heuman DM, Daita K, et al. Modulation of the fecal bile acid profile by gut microbiota in cirrhosis. J Hepatol. (2013) 58(5):949–55. doi: 10.1016/j.jhep.2013.01.003
35. Lu H-F, Li A, Zhang T, Ren Z-G, He K-X, Zhang H, et al. Disordered oropharyngeal microbial communities in H7N9 patients with or without secondary bacterial lung infection. Emerg Microbes Infect. (2017) 6(12):e112. doi: 10.1038/emi.2017.101
36. Effenberger M, Waschina S, Bronowski C, Sturm G, Tassiello O, Sommer F, et al. A gut bacterial signature in blood and liver tissue characterizes cirrhosis and hepatocellular carcinoma. Hepatol Commun. (2023) 7(7):e00182. doi: 10.1097/HC9.0000000000000182
37. Rao B, Lou J, Lu H, Liang H, Li J, Zhou H, et al. Oral microbiome characteristics in patients with autoimmune hepatitis. Front Cell Infect Microbiol. (2021) 11:656674. doi: 10.3389/fcimb.2021.656674
38. Abe K, Takahashi A, Fujita M, Imaizumi H, Hayashi M, Okai K, et al. Dysbiosis of oral microbiota and its association with salivary immunological biomarkers in autoimmune liver disease. PLoS One. (2018) 13(7):e0198757. doi: 10.1371/journal.pone.0198757
39. Bao W-H, Yang W-L, Su C-Y, Lu X-H, He L, Zhang A-H. Relationship between gut microbiota and vascular calcification in hemodialysis patients. Ren Fail. (2023) 45(1):2148538. doi: 10.1080/0886022X.2022.2148538
40. Chen T, Li X, Li G, Liu Y, Huang X, Ma W, et al. Alterations of commensal microbiota are associated with pancreatic cancer. Int J Biol Markers. (2023) 38(2):89–98. doi: 10.1177/03936155231166721
41. Thu MS, Chotirosniramit K, Nopsopon T, Hirankarn N, Pongpirul K. Human gut, breast, and oral microbiome in breast cancer: a systematic review and meta-analysis. Front Oncol. (2023) 13:1144021. doi: 10.3389/fonc.2023.1144021
42. Kwong TNY, Wang X, Nakatsu G, Chow TC, Tipoe T, Dai RZW, et al. Association between bacteremia from specific microbes and subsequent diagnosis of colorectal cancer. Gastroenterology. (2018) 155(2):383–90.e8. doi: 10.1053/j.gastro.2018.04.028
43. Tomkovich S, Dejea CM, Winglee K, Drewes JL, Chung L, Housseau F, et al. Human colon mucosal biofilms from healthy or colon cancer hosts are carcinogenic. J Clin Invest. (2019) 129(4):1699–712. doi: 10.1172/JCI124196
44. Cattaneo C, Rieg S, Schwarzer G, Müller MC, Blümel B, Kern WV. Enterococcus faecalis bloodstream infection: does infectious disease specialist consultation make a difference? Infection. (2021) 49(6):1289–97. doi: 10.1007/s15010-021-01717-3
45. Lavelle A, Lennon G, O’Sullivan O, Docherty N, Balfe A, Maguire A, et al. Spatial variation of the colonic microbiota in patients with ulcerative colitis and control volunteers. Gut. (2015) 64(10):1553–61. doi: 10.1136/gutjnl-2014-307873
46. Xiang G, Lan K, Cai Y, Liao K, Zhao M, Tao J, et al. Clinical molecular and genomic epidemiology of morganella morganii in China. Front Microbiol. (2021) 12:744291. doi: 10.3389/fmicb.2021.744291
47. Cuartero MI, García-Culebras A, Nieto-Vaquero C, Fraga E, Torres-López C, Pradillo J, et al. The role of gut microbiota in cerebrovascular disease and related dementia. Br J Pharmacol. (2024) 181(6):816–39. doi: 10.1111/bph.1616737328270
48. Kasahara K, Kerby RL, Zhang Q, Pradhan M, Mehrabian M, Lusis AJ, et al. Gut bacterial metabolism contributes to host global purine homeostasis. Cell Host Microbe. (2023) 31(6):1038–53.e10. doi: 10.1016/j.chom.2023.05.011
49. Yang H, Li C, Che M, Li Y, Feng R, Sun C. Gut microbiota mediates the anti-obesity effect of intermittent fasting by inhibiting intestinal lipid absorption. J Nutr Biochem. (2023) 116:109318. doi: 10.1016/j.jnutbio.2023.109318
50. Feng Y, Cui Y, Jin J, Huang S, Wei J, Yao M, et al. The alterations of gut microbiome and lipid metabolism in patients with spinal muscular atrophy. Neurol Ther. (2023) 12(3):961–76. doi: 10.1007/s40120-023-00477-6
51. Wang Q, Chen C, Zuo S, Cao K, Li H. Integrative analysis of the gut microbiota and faecal and serum short-chain fatty acids and tryptophan metabolites in patients with cirrhosis and hepatic encephalopathy. J Transl Med. (2023) 21(1):395. doi: 10.1186/s12967-023-04262-9
52. Lou J, Jiang Y, Rao B, Li A, Ding S, Yan H, et al. Fecal microbiomes distinguish patients with autoimmune hepatitis from healthy individuals. Front Cell Infect Microbiol. (2020) 10:342. doi: 10.3389/fcimb.2020.00342
53. Cui G, Liu S, Liu Z, Chen Y, Wu T, Lou J, et al. Gut microbiome distinguishes patients with epilepsy from healthy individuals. Front Microbiol. (2021) 12:696632. doi: 10.3389/fmicb.2021.696632
54. Qin J, Li Y, Cai Z, Li S, Zhu J, Zhang F, et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. (2012) 490(7418):55–60. doi: 10.1038/nature11450
55. Gao S, Gao X, Zhu R, Wu D, Feng Z, Jiao N, et al. Microbial genes outperform species and SNVs as diagnostic markers for Crohn’s disease on multicohort fecal metagenomes empowered by artificial intelligence. Gut Microbes. (2023) 15(1):2221428. doi: 10.1080/19490976.2023.2221428
56. Li M, Liu J, Zhu J, Wang H, Sun C, Gao NL, et al. Performance of gut microbiome as an independent diagnostic tool for 20 diseases: cross-cohort validation of machine-learning classifiers. Gut Microbes. (2023) 15(1):2205386. doi: 10.1080/19490976.2023.2205386
57. Coker OO, Liu C, Wu WKK, Wong SH, Jia W, Sung JJY, et al. Altered gut metabolites and microbiota interactions are implicated in colorectal carcinogenesis and can be non-invasive diagnostic biomarkers. Microbiome. (2022) 10(1):35. doi: 10.1186/s40168-021-01208-5
58. Wong SH, Kwong TNY, Chow TC, Luk AKC, Dai RZW, Nakatsu G, et al. Quantitation of faecal Fusobacterium improves faecal immunochemical test in detecting advanced colorectal neoplasia. Gut. (2017) 66(8):1441–8. doi: 10.1136/gutjnl-2016-312766
59. Zackular JP, Rogers MA, Ruffin M, Schloss PD. The human gut microbiome as a screening tool for colorectal cancer. Cancer Prev Res (Phila). (2014) 7(11):1112–21. doi: 10.1158/1940-6207.CAPR-14-0129
60. Yuan S, Wang KS, Meng H, Hou XT, Xue JC, Liu BH, et al. The gut microbes in inflammatory bowel disease: future novel target option for pharmacotherapy. Biomed Pharmacother. (2023) 165:114893. doi: 10.1016/j.biopha.2023.114893
Keywords: gut microbiome, pediatric solid tumor, healthy controls, 16S rRNA sequencing, microbial marker
Citation: Du X, Cui X, Fan R, Pan J and Cui X (2024) Characteristics of gut microbiome in patients with pediatric solid tumor. Front. Pediatr. 12:1388673. doi: 10.3389/fped.2024.1388673
Received: 19 March 2024; Accepted: 25 June 2024;
Published: 4 July 2024.
Edited by:
Pietro Merli, Bambino Gesù Children's Hospital (IRCCS), ItalyReviewed by:
Abbas Yadegar, Shahid Beheshti University of Medical Sciences, IranSzymon Skoczen, Jagiellonian University Medical College, Poland
© 2024 Du, Cui, Fan, Pan and Cui. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xichun Cui, eGljaHVuNjM5QDEyNi5jb20=
 Xiaoran Du
Xiaoran Du Xu Cui
Xu Cui Xichun Cui
Xichun Cui