- 1Department of Public Health Laboratory Sciences, School of Public Health, Hengyang Medical School, University of South China, Hengyang, China
- 2Shenzhen Center for Disease Control and Prevention, Shenzhen, China
- 3Shenzhen Research Center for Communicable Disease Control and Prevention, Chinese Academy of Medical Sciences, Shenzhen, China
- 4School of Public Health, Shanxi Medical University, Taiyuan, China
- 5Pediatric Department, Shenzhen Maternity and Child Healthcare Hospital, Shenzhen, China
Purpose: To explore the clinical characteristics of Micrococcus luteus bloodstream infection in an infant and characterize the phenotype and genotype of the isolated strains, as well as seek suitable infection models for assessing virulence.
Methods: Clinical data was collected from an infant patient diagnosed with M. luteus bloodstream infection. Metagenomic sequencing was performed on the isolated blood sample. The strain was isolated and underwent mass spectrometry, biochemical tests, antibiotic susceptibility assays, and whole-genome sequencing. The Galleria mellonella infection model was used to assess M. luteus virulence.
Results: Patient responded poorly to cephalosporins, but recovered after Linezolid treatment. Metagenomic sequencing identified M. luteus as the predominant species in the sample, confirming infection. They were identified as M. luteus with a high confidence level of 98.99% using mass spectrometry. The strain showed positive results for Catalase, Oxidase, and Urea tests, and negative results for Mannose, Xylose, Lactose, Mannitol, Arginine, and Galactose tests, consistent with the biochemical profile of M. luteus reference standards. M. luteus susceptibility to 15 antibiotics was demonstrated and no resistance genes were detected. Predicted virulence genes, including clpB, were associated with metabolic pathways and the type VI secretion system. The infection model demonstrated dose-dependent survival rates.
Conclusion: The infant likely developed a bloodstream infection with M. luteus due to compromised immunity. Although the isolated strain is sensitive to cephalosporin antibiotics and has low pathogenicity in infection models, clinical treatment with cephalosporins was ineffective. Linezolid proved to be effective, providing valuable guidance for future clinical management of such rare infections.
1. Introduction
Micrococcus luteus, a Gram-positive coccus of the genus Micrococcaceae, is widely distributed in the environment, including in soil, air, water and on animals (1, 2). M. luteus is rarely reported in clinical cases (3) though, as an opportunistic pathogen (4), it has been reported to cause disease in susceptible patients, such as those with malnutrition and poor immunity (5, 6). For example, Albertson et al. (7) reported septic shock caused by M. luteus in a patient with valveless heart disease and an implanted prosthesis. Buonsenso et al. (8) reported natural valvular complications of pulmonary infarction in a pediatric patient resulting from M. luteus infection. Although reports of human infection with M. luteus are rare, determining potential antibiotic susceptibility and virulence of M. luteus may ensure effective treatment of people susceptible to infection.
Galleria mellonella has no ethical constraints and a short life cycle (9). The G. mellonella immune system exhibits both humoral and cellular components, and in some aspects the immune response is similar to the innate immune response of mammals (10). As such, G. mellonella larvae have been successfully utilized as a model organism in a variety of bacterial infection experiments (11). In addition, there is no reports indicating animal infection model of M. luteus.
In this study we identified a case of bloodstream infection, analyzed the patient's clinical treatment, identified the pathogen, and explored its pathogenicity to provide information about this rare infection. The virulence of the pathogen was evaluated by using a model organism of M. luteus infecting G. mellonella larvae, providing a basis for studying the pathogenesis of M. luteus.
2. Materials and methods
2.1. Collecting case information
This study involved the collection of patient information from an individual who exhibited recurrent fever symptoms and had two consecutive positive blood cultures for M. luteus. The gathered data encompassed age, gender, results of laboratory tests, administered treatments, and clinical outcome.
2.2. Pathogen identification
2.2.1. Isolation culture and staining
A sample of the patient's blood was inoculated onto a plate containing Columbia blood agar medium (Guangzhou Detgerm Microbiogical Science Ltd, Guangdong, China) and kept at a constant temperature of 37°C for a 48 h incubation. After incubation, individual colonies with a consistent appearance were picked for culturing. A Gram staining kit (Huankai Microbial, Guangdong, China) was used to stain bacteria for microscopic observation using a Zeiss fully automatic positive fluorescence microscope Axio Imager M2 (ZEISS, Oberkochen, Germany) (12).
2.2.2. Mass spectrometry identification
Using the direct coating method, Escherichia coli ATCC8739 was used as the calibration strain, and a small amount of pure, cultured individual colonies were evenly coated with 1μl inoculation loop to target plate spots. Immediately, 1 ul of alpha-cyano-4-hydroxy-cinnamic acid (CHCA) substrate was added and, when the substrate was completely dry, strain identification occurred using the standard operating procedure of Matrix-Assisted Laser Desorption/Ionization Time of Flight Mass Spectrometry (MALDI-TOF/MS; BioMerieux, Lyon, France).
2.2.3. Physiological and biochemical identification
A homogenous bacterial suspension, equivalent to a turbidity of 0.50–0.63 McFarland, was prepared by calibrating the turbidimeter (BioMerieux, Lyon, France) with a sterile swab and inoculating identical colonies into prepared sterile saline tubes. The bacterial suspension tubes and Gram-positive bacterial identification cards (Biomerieux, Lyon, France) were placed in the card rack of the VITEK 2 Compact Automatic Microbiological Identification and Drug Sensitivity Analyzer and operated according to the guidelines. The Gram-positive bacterial identification cards include multiple biochemical details such as peroxidase, oxidase, mannose, xylose, lactose, mannitol, urea, galactose, and others. The bacterial identification results were referred to the Bergey’s Manual of Determinative Bacteriology (13).
2.2.4. Antibiotic susceptibility test
The broth microdilution method (Fosun Diagnostics, Shanghai, China) was used to test the antibiotic sensitivity of M. luteus strains. Penicillin, Vancomycin, Erythromycin, Clindamycin, Moxifloxacin, Cefotaxime, Tetracycline, Linezolid, Cotrimoxazole, Meropenem, Chloramphenicol, Amoxicillin, Levofloxacin, Teicoplanin, and Cefepime were tested. Breakpoints for sensitive (S), intermediate (I), and resistant (R) were defined by the Clinical and Laboratory Standards Institute (CLSI) document M45 (http://www.clsi.org), and Staphylococcus aureus ATCC 29213 was used as the control.
2.2.5. Bacterial genomic DNA extraction and whole genome sequencing
The total DNA of Gram-positive bacteria was extracted using the Ezup column-based bacterial DNA extraction kit (Sangon Biotech, Shanghai, China), and the quality of nucleic acids was confirmed using a NanoDrop one ultra-micro spectrophotometer (Thermo Fisher Scientific, Massachusetts, America). Small fragment libraries, with an average insert size of 350 bp, were prepared by Beijing Novozymes Technology Co. using the Illumina NovaSeq 6000 platform. Sequencing evaluation and screening were then performed. Trimmomatic software (v0.39) (14) was used to quality-control filter the raw data and obtain valid data, and SPAdes gene assembly software (V3.9.1) (15) was used to splice and assemble the valid sequences. Kraken2 software (16) was used to identify the possible pathogens for the valid sequences after successful splicing. Investigation of carriage of antibiotic resistance and virulence genes was assessed comprehensively using the ResFinder7 database (17), the CARD database (18) and the VFDB6 database (19). Chiplot (https://www.chiplot.online/) was used to visualize the gene density, gene function annotation, and CG content of the whole bacterial genome.
2.2.6. Nucleic acid extraction of blood pathogens and sequencing of microbial genomes
After blood cultures were established as positive for Gram-positive cocci, pathogenic DNA was extracted from blood samples using the QIAamp DNA Blood Mini Kit by (QIAGEN, Guangdong, China). Metagenomic sequencing was performed by Novogene (Beijing, China). Using Readfq (v8) (http://github.com/lh3/readfq), preprocess raw data was obtained from the Illumina HiSeq sequencing platform for subsequent analysis (Clean Data). Bowtie2 software (v2.2.4) (20) was used to delete host readings and MEGAHIT software (v1.0.4 beta) (21) was used to assemble and analyze Clean Data. DIAMOND (v0.9.9.110) (22) was used to integrate unigenes with the National Center for Biotechnology Information (NCBI) non-redundant (NR) database (https://www.ncbi.nlm.nih.gov/), compare bacteria, fungi, archaea, and viruses, and annotate species information using Lowest Common Ancestor (LCA) algorithm on January 18, 2018. Short-read sequencing data from isolated samples have been deposited in the NCBI Sequence Read Archive under the BioProject PRJNA973819.
2.2.7. Galleria mellonella infection model
The isolated strain of M. luteus was inoculated into a blood plate, and incubated at 37°C for 48 h. Single bacterial colonies were then placed into 2 ml of nutrient broth (Huankai Microbial, Guangdong, China) for 12 h then centrifuged at 8,000 r/min for 3 min and washed three times with physiological saline. Bacteria were resuspended in 1 ml of physiological saline and the following bacterial suspension doses were prepared: 5 × 102 CFU/ml (low dose), 5 × 104 CFU/ml (medium dose) and 5 × 106 CFU/ml (high dose). G. mellonella larvae (250–350 mg/piece) were fasted for 24 h then 10 larvae per dose group (high, medium, and low) were injected with 10 μl of the corresponding bacterial suspension using a 10 μl micro syringe. Three forms of control were set: a blank control group of 10 larvae (without any treatment), a puncture group of 10 larvae (with a micro syringe puncture without injection of physiological saline), and a physiological saline group of 10 larvae (injected with 10 μl of physiological saline). G. mellonella larvae were incubated at 37°C for 72 h after which surface color changes, response to stimuli, and survival of the larvae were observed every 24 h. G. mellonella were considered dead when unresponsive to external stimuli (23–26).
2.3. Statistical analyses
Mean ± standard deviation (SD) was used to describe the survival data of G. mellonella. Between-group comparisons were conducted using the log-rank test in SPSS v20.0 software. The survival curves were plotted using GraphPad Prism v9.3 software. For all tests, P ≤ 0.001 indicates a statistically significant difference.
3. Results
3.1. Patient clinical data
In July 2022, the patient, a 7-month-and-26-day-old infant male, presented at Shenzhen Maternal and Child Health Hospital with a fever peak of 38.3°C and two instances of non-jet vomiting with no obvious cause. The first and second days of admission, the patient received Cephalosporin antibiotics, along with fluid replacement, antipyretics, and other symptomatic treatments. However, recurrent fever persisted, and routine blood examinations showed an increase in C-reactive protein (CRP) to 110.51 mg/l, an elevation in procalcitonin from 0.46 ng/ml to 0.74 ng/ml, and an erythrocyte sedimentation rate of 90 mm/h. When a patient is suspected of having Bloodstream Infection upon admission to the pediatric ward, the standard procedure for sampling involves collecting blood samples from both sides of the patient's limbs at different time points. The blood culture reports indicated the presence of Gram-positive cocci, with the blood culture isolate identified as M. luteus. Cerebrospinal fluid analysis revealed no bacteria or fungi, excluding the possibility of intracranial infection. The patient's immunoglobulin levels are within the normal range, and there is no history of recurrent infections. After initiating treatment with Linezolid, the patient's body temperature returned to normal, and CRP levels decreased. The patient was diagnosed with: (i) bloodstream infection; (ii) acute gastroenteritis; (iii) acute upper respiratory tract infection; (iv) acute otitis media (bilateral); (v) mild anemia.
3.2. Pathogen identification
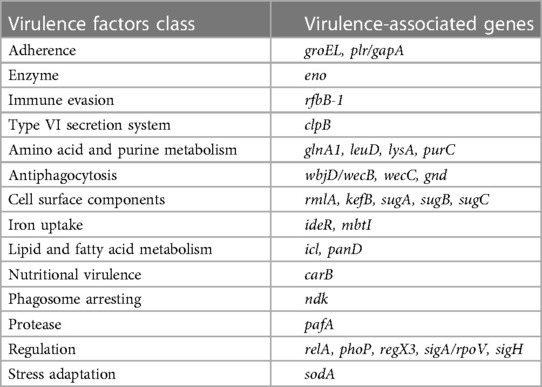
Bacterial growth was relatively slow at 37°C over 24 h, and colony morphology was relatively small. After 48 h of incubation, colony size had increased to 1–2 mm and presented with a pale yellow, round and raised smooth surface with a neat edge (Figure 1A). Microscopic examination after Gram staining of bacterial smears showed Gram-positive cocci, most of which were arranged in pairs, fours, or clusters (Figure 1B). Mass spectrometry analysis identified the cocci to be M. luteus with a confidence percentage of 98.90%. The results of physicochemical characterization of the strains showed that the isolates of M. luteus were consistent with the results of reference in terms of colony color, growth temperature, motility, peroxidase, oxidase, mannose, xylose, lactose, mannitol, urea, galactose, and other physiological and biochemical characteristics (Table 1). The antibiotic susceptibility test showed that the isolated M. luteus strain was sensitive to 15 antibiotics, including Penicillin, Vancomycin, Erythromycin, Clindamycin, Moxifloxacin, Cefotaxime, Tetracycline, Linezolid, Cotrimoxazole, Meropenem, Chloramphenicol, Amoxicillin, Levofloxacin, Teicoplanin and Cefepime.
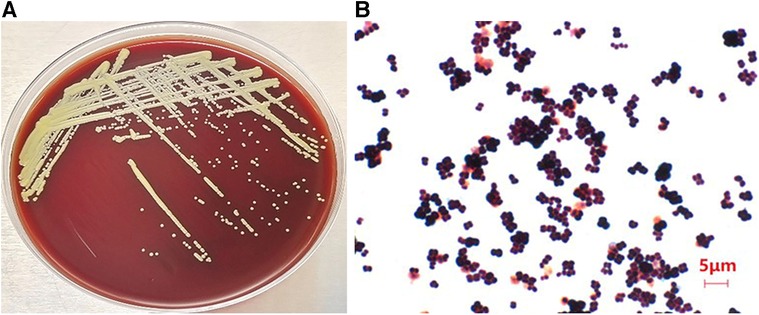
Figure 1. Images of isolated bacteria to depict morphology of (A) the colony on a blood plate and (B) gram positive cocci indicated by gram staining of a bacterial smear of picked colonies (×100 magnification).
3.3. Genome analysis of the isolated strain
The genome was reassembled from scratch to obtain 57 sized contigs, with a total genome length of 2494966 bp and a guanine-cytosine (guanine-cytosine content, GC) content of 72.92%. Among the 2,291 predicted genes, 2,234 encode proteins, and 53 tRNAs, 1 tmRNA, and 3 rRNAs are also predicted (Figure 2). The results of species identification confirmed the bacterium to be M. luteus. Analysis of M. luteus virulence genes demonstrated a dominance of regulating amino acids and purine metabolism genes, as well as those involved in lipid and fatty acid metabolism, iron absorption, nutritional virulence, cell surface composition and type VI secretion system (T6SS) (Table 2). The commonly used ResFinder7 and CARD databases did not predict resistance genes.
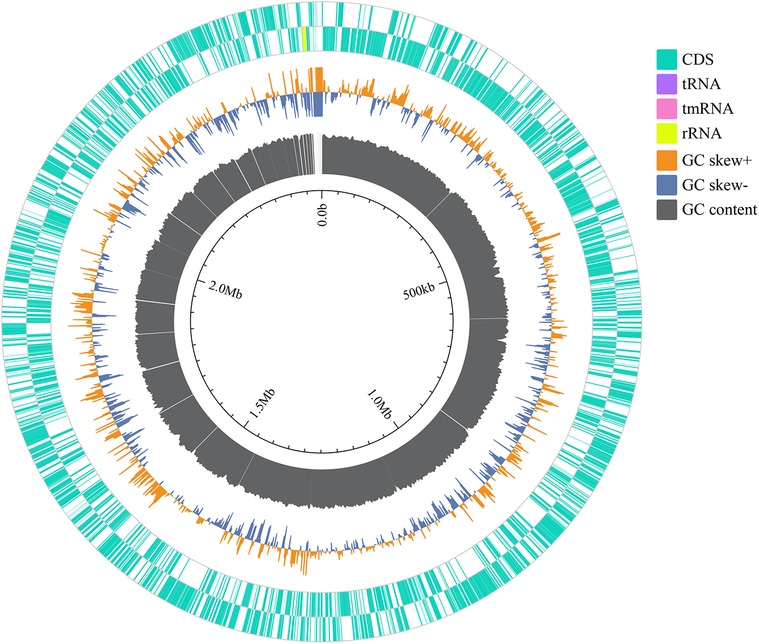
Figure 2. The genomic circular representation of the Micrococcus luteus isolate. From outside to inside, the various circles represent genes on coding DNA sequence (CDS green), RNA genes (tRNAs purple, tmRNAs pink, rRNAs yellow), GC skew, and GC content.
3.4. Genomic analysis of blood pathogens
The relative abundance of species at different taxonomic levels, based on metagenomic data of blood pathogens, indicates that M. luteus contributed the highest proportion at 18.14%. Janibacter hoylei, Streptococcus pneumoniae and other Micrococcus spp., were also annotated by the LCA algorithm aside from the ‘Other pathogens’ category (Figure 3). ‘Others’ in the species richness pie chart indicates that the program could not predict the classification level according to the prescribed rules and information in the database.
Figure 3. The relative abundance of the top 10 species of blood pathogens at the species level is reflected in the patient.
3.5. Infection and survival of Galleria mellonella larvae
After 24 h inoculation with M. luteus, the G. mellonella larvae of the high dose group began to blacken and die. There was no larval mortality in the low and medium dose groups. The blank control group, puncture group, and physiological saline group showed no mortality of G. mellonella larvae, with a light-yellow appearance and sensitive response to external stimuli.
At 24 h post-inoculation, G. mellonella larvae survival rates in low, medium, and high dose groups of M. luteus isolate were 100% (10/10), 100% (10/10) and 70% (7/10) respectively. After 48 h of vaccination, the survival rates of low, medium, and high dose groups were 100% (10/10), 100% (10/10), and 60% (6/10), respectively; After 72 h of vaccination, the survival rates of the three groups were 100% (10/10), 100% (10/10), and 50% (5/10), respectively. There was a statistically significant difference in survival rates between the control group and the experimental group (P < 0.001), and there was a statistically significant difference in survival rates between low, medium, and high doses (P < 0.001) (Figure 4).

Figure 4. Survival of Galleria mellonella following infection by Micrococcus luteus strains. The data shown are means ± SD from three independent experiments recorded for 72 h. Differences in survival were calculated using the log-rank test for multiple comparisons. Differences were considered statistically significant at P < 0.001.
4. Discussion
Micrococcus luteus is an opportunistic Gram-positive coccus widely distributed in water, air, soil and other environments (1), which seldom was reported as human pathogenesis, but it could arose infection in some specific situation (3, 27). Currently, it is still challenging to distinguish between M. luteus infection and contamination. Clinical judgment, positive blood culture bottles, short time to growth period (time from bacterial inoculation to detection), and new microbiologic technologies are considered possible methods to avoid blood culture contamination (28). In this research, we were able to isolate M. luteus from positive blood cultures obtained from bilateral limbs at two different time points and identify a high concentration of M. luteus in the blood using metagenomics. Compare to blood culture contamination, the above method could further indicates that we found a blood infection caused by M. luteus. Considering the clinical information, the patient lacks a history of recurrent infections, and the immunoglobulin levels fall within the normal range, eliminating the possibility of hereditary immunocompromised disease. The presence of inflammation in the upper respiratory and gastrointestinal tracts, coupled with mild anemia and the young age of only 7 months and 26 days, renders the patient more susceptible to infections. Consequently, there is a likelihood of a lower immune level, creating an opportunity for opportunistic infection by M. luteus.
Effective treatment of M. luteus is important for people at increased risk of infection. There have been reports (28) indicating that Cephalosporins and Quinolones are effective empirical antibiotics for treating M. luteus. Vancomycin and Teicoplanin should be considered for potential extensively drug-resistant M. luteus strains. In this case, although antibiotic susceptibility testing showed that M. luteus was susceptible to Cephalosporins and other antibiotics, the patient continued to experience a fever and elevated inflammatory markers, such as C-reactive protein and procalcitonin, even after receiving cephalosporin treatment. Due to the possibility of rash in infant clinical treatment with Vancomycin, Linezolid, with a similar antibacterial spectrum to vancomycin, was eventually used and resulted in a decrease in body temperature. In summary, the empirically effective antibiotic, Cephalosporins, demonstrated inconsistency between its in antibiotic susceptibility test and actual clinical treatment outcomes. On the other hand, it's crucial to exercise caution regarding the adverse effects of Vancomycin in infant use. Ultimately, Linezolid proved to be an effective treatment for the infant. However, the research has its limitations. The incidence of bloodstream infections caused by M. luteus is low, and as of November 2023, only four articles related to M. luteus bloodstream infections were found on PubMed. There is limited research attention on M. luteus bloodstream infections, and consensus on empirical treatment for M. luteus infections has yet to be established. While the patient's clinical treatment implies the necessity of contemplating the potential for Cephalosporin resistance and the successful application of Linezolid in treating this infection, the limitation of having only one case emphasizes the need for additional case data on M. luteus bloodstream infections to support our conclusions. Furthermore, it is crucial to continue exploring the optimal treatment for M. luteus bloodstream infections.
M. luteus, a group of actinomycetes widely used in biotechnology, is considered as a new hospital pathogen (6, 29). Although the use of bioinformatics analysis could predict functionality, there is usually lack of validated pathogenicity to establish animal infection models that could help increase understanding of disease mechanisms (30, 31). At present, there are few reports on animal models of M. luteus. The results of the infection model of M. luteus larvae constructed in this study show that the larvae of G. mellonella can withstand low infection doses, generate immune responses against M. luteus, and kill the larvae of G. mellonella in a short time at high dose concentrations. The mortality and dose concentration are dependent, suggesting that G. mellonella larvae could be used as a model for the construction of M. luteus infection. The survival rate of bacteria could be used to judge the virulence of the strain, providing an important internal model selection for the infection of M. luteus. Based on the comprehensive clinical manifestations, although the patient experiences recurrent fever and elevated inflammatory markers, there are no clinical signs of sepsis. Moreover, M. luteus exhibits low toxicity when isolated from the blood of pediatric patients, as it can only kill larvae at high concentrations.
The virulence genes of the M. luteus are isolated in this study, including genes that regulate T6SS. T6SS is a nanomoleular complex, regulated by clpB gene, that can release virulence factors to target host cells (32) and exists widely in gram-negative bacteria. T6SS is involved in bacterial colonization, enhanced survival, adhesive modification, internalization and escape from the immune system (33) and can cause the destruction of lipid membrane and cytoskeleton (32). While we found the clpB gene of T6SS in the isolated strain of Gram positive coccus M. luteus, the potential role in bacterial virulence and human pathogenicity remains to be further explored.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/sra/PRJNA973819, PRJNA973819.
Ethics statement
The studies involving human participants and Galleria mellonella were reviewed and approved by Ethics Committee of the Shenzhen Center for Disease Control and Prevention (QS2023040067). Written informed consent from the participants’ legal guardian/next of kin was not required to participate in this study in accordance with the national legislation and the institutional requirements. The manuscript presents research on animals that do not require ethical approval for their study.
Author contributions
XS: Formal Analysis, Funding acquisition, Methodology, Resources, Supervision, Writing – review & editing. SQ: Conceptualization, Data curation, Formal Analysis, Investigation, Methodology, Project administration, Software, Validation, Visualization, Writing – original draft, Writing – review & editing. LJ: Visualization, Writing – review & editing. HL: Software, Visualization, Writing – review & editing. SW: Writing – review & editing. QC: Writing – review & editing. XZ: Writing – review & editing. QH: Writing – review & editing. TF: Writing – review & editing. SC: Writing – review & editing. WC: Writing – review & editing. SX: Writing – review & editing. MJ: Writing – review & editing. RC: Writing – review & editing. YG: Writing – review & editing. QB: Writing – review & editing. DH: Writing – review & editing. PL: Investigation, Methodology, Project administration, Resources, Supervision, Validation, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article.
This research was supported by the Doctoral Fund of Shenzhen Maternity and Child Healthcare Hospital (Fund Number: FYA2018028), the National Natural Science Foundation of China (No. 81773436), the Science and Technology Planning Project of Guangdong Province of China (grant 2021 B1212030009), the Research Foundation of Shenzhen Science and Technology Emergency Key Technology Program (JSGG20220301090007009), the Key Scientific and Technological Project of Shenzhen Science and Technology Innovation Committee (KCXFZ202002011006190), the Shenzhen Key Medical Discipline Construction Fund (SZXK064), the Non-profit Central Research Institute Fund of Chinese Academy of Medical Sciences (2020-PT330-006), the Sanming Project of Medicine in Shenzhen (No. SZSM202211023), and the Sanming Project of Medicine in Shenzhen (NO. SZSM201811071).
Acknowledgments
We would like to thank Qin Gao from Xinhua Harvard International Healthcare Innovation Collaboration Initiatives for helpful discussions and for reviewing our manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Young M, Artsatbanov V, Beller HR, Chandra G, Chater KF, Dover LG, et al. Genome sequence of the fleming strain of Micrococcus luteus, a simple free-living actinobacterium. J Bacteriol. (2010) 192(3):841–60. doi: 10.1128/JB.01254-09
2. Becerra SC, Roy DC, Sanchez CJ, Christy RJ, Burmeister DM. An optimized staining technique for the detection of gram positive and gram negative bacteria within tissue. BMC Res Notes. (2016) 9(1):216. doi: 10.1186/s13104-016-1902-0
3. Erbasan F. Brain abscess caused by Micrococcus luteus in a patient with systemic lupus erythematosus: case-based review. Rheumatol Int. (2018) 38(12):2323–8. doi: 10.1007/s00296-018-4182-2
4. Magee JT, Burnett IA, Hindmarch JM, Spencer RC. Micrococcus and Stomatococcus spp. from human infections. J Hosp Infect. (1990) 16(1):67–73. doi: 10.1016/0195-6701(90)90050-X
5. Eiff C, Kuhn N, Herrmann M, Weber S, Peters G. Micrococcus luteus as a cause of recurrent bacteremia. Pediatr Infect Dis J. (1996) 15(8):711–3. doi: 10.1097/00006454-199608000-00019
6. Khan A, Aung TT, Chaudhuri D. The first case of native mitral valve endocarditis due to Micrococcus luteus and review of the literature. Case Rep Cardiol. (2019) 2019:5907319. doi: 10.1155/2019/5907319
7. Albertson D, Natsios GA, Gleckman R. Septic shock with Micrococcus luteus. Arch Intern Med. (1978) 138(3):487–8. doi: 10.1001/archinte.1978.03630270093032
8. Buonsenso D, Lombardo A, Fregola A, Ferrari V, Piastra M, Calvani M, et al. First report of Micrococcus luteus native valve endocarditis complicated with pulmonary infarction in a pediatric patient: case report and literature review. Pediatr Infect Dis J. (2021) 40(7):e284–e6. doi: 10.1097/INF.0000000000003133
9. Tsai CJ-Y, Loh JMS, Proft T. Galleria mellonella infection models for the study of bacterial diseases and for antimicrobial drug testing. Virulence. (2016) 7(3):214–29. doi: 10.1080/21505594.2015.1135289
10. Velikova N, Kavanagh K, Wells JM. Evaluation of Galleria mellonella larvae for studying the virulence of Streptococcus suis. BMC Microbiol. (2016) 16(1):291. doi: 10.1186/s12866-016-0905-2
11. Durieux M-F, Melloul É, Jemel S, Roisin L, Dardé M-L, Guillot J, et al. Galleria mellonella as a screening tool to study virulence factors of Aspergillus fumigatus. Virulence. (2021) 12(1):818–34. doi: 10.1080/21505594.2021.1893945
12. Froböse NJ, Bjedov S, Schuler F, Kahl BC, Kampmeier S, Schaumburg F. Gram staining: a comparison of two automated systems and manual staining. J Clin Microbiol. (2020) 58(12):e01914–20. doi: 10.1128/JCM.01914-20
13. Breed RS, Murray EG, Hitchens AP. The outline classification used in the bergey manual of determinative bacteriology. Bacteriol Rev. (1944) 8(4):255–60. doi: 10.1128/br.8.4.255-260.1944
14. Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics. (2014) 30(15):2114–20. doi: 10.1093/bioinformatics/btu170
15. Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, et al. Spades: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. (2012) 19(5):455–77. doi: 10.1089/cmb.2012.0021
16. Wood DE, Salzberg SL. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol. (2014) 15(3):R46. doi: 10.1186/gb-2014-15-3-r46
17. Bortolaia V, Kaas RS, Ruppe E, Roberts MC, Schwarz S, Cattoir V, et al. Resfinder 4.0 for predictions of phenotypes from genotypes. J Antimicrob Chemother. (2020) 75(12):3491–500. doi: 10.1093/jac/dkaa345
18. Alcock BP, Raphenya AR, Lau TTY, Tsang KK, Bouchard M, Edalatmand A, et al. Card 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. (2020) 48(D1):D517–D25. doi: 10.1093/nar/gkz935
19. Liu B, Zheng D, Jin Q, Chen L, Yang J. Vfdb 2019: a comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res. (2019) 47(D1):D687–D92. doi: 10.1093/nar/gky1080
20. Karlsson FH, Fåk F, Nookaew I, Tremaroli V, Fagerberg B, Petranovic D, et al. Symptomatic atherosclerosis is associated with an altered gut metagenome. Nat Commun. (2012) 3:1245. doi: 10.1038/ncomms2266
21. Feng Q, Liang S, Jia H, Stadlmayr A, Tang L, Lan Z, et al. Gut microbiome development along the colorectal adenoma-carcinoma sequence. Nat Commun. (2015) 6:6528. doi: 10.1038/ncomms7528
22. Huson DH, Mitra S, Ruscheweyh H-J, Weber N, Schuster SC. Integrative analysis of environmental sequences using Megan4. Genome Res. (2011) 21(9):1552–60. doi: 10.1101/gr.120618.111
23. Ménard G, Rouillon A, Cattoir V, Donnio P-Y. Galleria mellonella as a suitable model of bacterial infection: past, present and future. Front Cell Infect Microbiol. (2021) 11:782733. doi: 10.3389/fcimb.2021.782733
24. Tao Y, Duma L, Rossez Y. Galleria mellonella as a good model to study Acinetobacter baumannii pathogenesis. Pathogens. (2021) 10(11):1483. doi: 10.3390/pathogens10111483
25. Asai M, Li Y, Newton SM, Robertson BD, Langford PR. Galleria mellonella-intracellular bacteria pathogen infection models: the ins and outs. FEMS Microbiol Rev. (2023) 47(2):fuad011. doi: 10.1093/femsre/fuad011
26. Piatek M, Sheehan G, Kavanagh K. Galleria mellonella: the versatile host for drug discovery, in vivo toxicity testing and characterising host-pathogen interactions. Antibiotics(Basel). (2021) 10(12):1545. doi: 10.3390/antibiotics10121545
27. Ianniello NM, Andrade DC, Ivancic S, Eckardt PA, Ramirez JCL. Native valve infective endocarditis due to Micrococcus luteus in a non-hodgkin’s lymphoma patient. ID Cases. (2019) 18:e00657. doi: 10.1016/j.idcr.2019.e00657
28. Zhu M, Zhu Q, Yang Z, Liang Z. Clinical characteristics of patients with Micrococcus luteus bloodstream infection in a Chinese tertiary-care hospital. Pol J Microbiol. (2021) 70(3):321–6. doi: 10.33073/pjm-2021-030
29. Li Y, Sun Z-Z, Rong J-C, Xie B-B. Comparative genomics reveals broad genetic diversity, extensive recombination and nascent ecological adaptation in Micrococcus luteus. BMC Genomics. (2021) 22(1):124. doi: 10.1186/s12864-021-07432-5
30. Mikulak E, Gliniewicz A, Przygodzka M, Solecka J. Galleria mellonella L. as model organism used in biomedical and other studies. Przegl Epidemiol. (2018) 72(1):57–73. PMID: 2966738129667381
31. Prakoso D, Zhu X, Rajeev S. Galleria mellonella infection model to evaluate pathogenic and nonpathogenic Leptospira strains. Vet Microbiol. (2022) 264:109295. doi: 10.1016/j.vetmic.2021.109295
32. Prakash SR, Kiran K. Bacterial type VI secretion system (T6ss): an evolved molecular weapon with diverse functionality. Biotechnol Lett. (2023) 45(3):309–31. doi: 10.1007/s10529-023-03354-2
Keywords: Micrococcus luteus, bloodstream infection, pathogenesis, genomics, virulence
Citation: Shi X, Qiu S, Ji L, Lu H, Wu S, Chen Q, Zou X, Hu Q, Feng T, Chen S, Cui W, Xu S, Jiang M, Cai R, Geng Y, Bai Q, Huang D and Liu P (2023) Pathogenetic characterization of a Micrococcus luteus strain isolated from an infant. Front. Pediatr. 11:1303040. doi: 10.3389/fped.2023.1303040
Received: 27 September 2023; Accepted: 4 December 2023;
Published: 22 December 2023.
Edited by:
Eugenia Bezirtzoglou, Democritus University of Thrace, GreeceReviewed by:
Kondapalli Kasturi, Acharya Nagarjuna University, IndiaMohsen Norouzinia, Shahid Beheshti University of Medical Sciences, Iran
© 2023 Shi, Qiu, Ji, Lu, Wu, Chen, Zou, Hu, Feng, Chen, Cui, Xu, Jiang, Cai, Geng, Bai, Huang and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Peihui Liu bHBoNTIxNTIxQDEyNi5jb20=
†These authors have contributed equally to this work and share first authorship
 Xiaolu Shi
Xiaolu Shi Shuxiang Qiu
Shuxiang Qiu Liyin Ji
Liyin Ji Huiqun Lu
Huiqun Lu Shuang Wu2
Shuang Wu2 Qinghua Hu
Qinghua Hu Peihui Liu
Peihui Liu