
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Pediatr. , 16 August 2023
Sec. Pediatric Hematology and Hematological Malignancies
Volume 11 - 2023 | https://doi.org/10.3389/fped.2023.1224966
This article is part of the Research Topic Diagnosis and Therapy Pediatric Hematological Malignancies - Recent Progress View all 5 articles
 Qinglin Liu1,†
Qinglin Liu1,† Xiang Yu2,3,4,†
Xiang Yu2,3,4,† Jinquan Wen5
Jinquan Wen5 Nange Yin2,3,6
Nange Yin2,3,6 Xin Liao2,7
Xin Liao2,7 Pinli Zou2,7
Pinli Zou2,7 Yuxia Guo2,3,7
Yuxia Guo2,3,7 Lin Song2,6
Lin Song2,6 Jianwen Xiao4,7,8*
Jianwen Xiao4,7,8*
Objective: To investigate the genomic signatures and prognosis of advanced-stage T cell lymphoblastic lymphoma (T-LBL) and to examine the relationship between T-LBL and T cell acute lymphoblastic leukemia (T-ALL).
Methods: 35 Chinese T-LBL children with stage III or IV disease were recruited for this study. They were treated with combination chemotherapy and whole exome sequencing. The relationship of the clinical features, prognosis and specific gene mutations was researched. Gene chips of T-LBL and T-ALL were downloaded from a database, and differential gene expression was analyzed.
Results: Germline causal gene mutations (CARS or MAP2K2) were detected in 2 patients; 3.06 ± 2.21 somatic causal gene mutations were identified in the 35 patients, and somatic mutations were observed in the NOTCH1, FBXW7, PHF6 and JAK3 genes. NOTCH1 mutations were significantly associated with FBXW7 mutations, and the age at diagnosis of patients with NOTCH1-FBXW7 mutations was less than that of patients without such mutations (P < 0.05). 32 patients achieved complete remission (CR), and 14 and 18 patients were classified into the intermediate risk (IR) group and high risk (HR) group. During a median follow-up of 44 months, 3 patients relapsed. Three-year prospective event free survival (pEFS) was 82.286%, and no significant differences of pEFS were found for different sexes, ages, or statuses of NOTCH1-FBXW7 mutations, (P > 0.05); however, the mean survival time of the IR group was longer than that of the HR group (P < 0.05). Differential expression of genes in the T-LBL and/or T-ALL datasets was analyzed using the R package limma, and 1/3 of the differentially expressed genes were found in both the T-ALL and T-LBL datasets. High expression of PI3K-Akt signal pathway genes and the USP34 gene was found in the T-LBL dataset.
Conclusion: Although T-ALL and T-LBL both originate from precursor T-cells and are considered different manifestations of the same disease and the outcome of T-LBL is favorable when using T-ALL-based chemotherapy, there are differences in the gene distribution between T-LBL and T-ALL. It seems that the PI3K-Akt signaling pathway and the USP34 gene play important roles in T-LBL, but medicines targeting the USP34 gene or the PI3K-Akt pathway may be invalid.
Lymphoblastic lymphoma (LBL) is an aggressive non-Hodgkin's lymphoma (NHL) that accounts for 20%–30% of the pediatric NHL population (1). T cell LBL (T-LBL) involves the precursor T-cell immunophenotype and represents 70%–80% of LBL cases in children (1, 2). The prognosis of T-LBL in children has remained poor historically. Acute lymphoblastic leukemia (ALL)-based chemotherapy and allogenic hematopoietic stem cell transplantation have demonstrated effective results, and event-free survival (EFS) exceeds 80%–90% even for advanced stage (stage III and IV) patients (2, 3), but the survival of relapsed and/or refractory cases remains poor (<10%–30%).
Gene mutations, such as FBXW7, NOTCH1, and PTEN mutations, play important roles in T-LBL development and are potential prognostic biomarkers for T-LBL (2, 4). Pathogenetic and molecular biological studies of T-LBL are limited due to the limitations of detection methods and sufficient available materials.
Next-generation sequencing (NGS) technologies have become the method of choice for gene mutation analysis in cancer (4, 5), but NGS data from Chinese pediatric T-LBL patients have not yet been reported. In the current study, whole exome sequencing (WES) by NGS was used to analyze T-LBL patients with the aim of gaining detailed genome-wide insight into the signature and mechanisms of tumorigenesis in pediatric T-LBL. The relationship of prognosis and specific gene mutations in children with T-LBL was also researched. Gene chips were downloaded from a database, and differential gene expression was analyzed.
Patients with newly diagnosed T-LBL admitted to Children's Hospital of Chongqing Medical University (CHCMU) and Children's Hospital of Xianyang (CHX) between January 2013 and September 2020 were enrolled in the study. Diagnosis of T-LBL was in accordance with the World Health Organization (WHO) criteria of 2008 or 2016 (6, 7), and patients were staged with the revised international pediatric NHL staging system (IPNHLSS) (8). Patients who were ≥18 years at diagnosis, diagnosed with mixed-phenotype LBL or secondary lymphoma, or had human immunodeficiency virus infection were excluded; patients who were classified with a local stage (stage I or II) and who had received chemotherapy before hospitalization were also excluded from the study.
Pathologic diagnosis of T-LBL patients was confirmed by lymph node biopsy. Immunohistochemical staining was detected as described in the literature (7); fluorescence in situ hybridization (FISH) of MYC, MLL and SIL-TAL was also performed (9, 10). Blast cells from bone marrow (BM) samples <25% was defined as the cutoff value between T-LBL (stage IV) and T-ALL (1, 6).
Patients were treated with the modified LBL-1995 Berlin-Frankfurt-Münster protocol (BFM-LBL-95) (4, 11). An intrathecal injection was administered as the protocol required, and cranial radiotherapy was carried out for patients with central nervous system (CNS) involvement. Treatment response was evaluated at three time points (TP): TP1 or TP2 (days 15 or 33 of remission induction) and TP3 (prior to consolidation). Minimal disseminated disease (MDD) levels were detected in BM samples using computed tomography (CT) or positron emission tomography computed tomography (PET-CT) screening as described in previous literature reports, and disease status was evaluated as partial remission (PR), complete remission (CR), progressive disease (PD) or refractory disease (RD) (12–14). All patients with advanced stage were regarded as the intermediate risk (IR) group at diagnosis; patients who presented with PD at TP1, PR at TP1 and CR at TP2 were considered as the high risk (HR) group; patients who did not achieve CR at TP3 were considered as having RD.
The details of the risk group classification, course of treatment and drug dosage for the modified BFM-LBL-95 protocol are listed in the Supplementary Material. Clinical data, laboratory findings and prognosis data of enrolled patients were collected and analyzed retrospectively.
Tumor DNA samples of T-LBL patients were obtained at diagnosis from formalin-fixed specimens; germline samples were collected from the oral mucosa of patients and their parents' peripheral blood (PB). Genomic DNA was extracted using a QIAmp DNA Minikit (QIAGEN, China). Genomic DNA was enriched, and sequencing was carried out (Agilent SureSelect Human All Exon V6). PCR products of the whole exome were sequenced (Illumina HiSeq PE 150 bp).
The original WES sequencing data were read using Illumina Pipeline software (version 1.3.4), and data were obtained from databases (dbSNP, 1,000 Genomes Project, ClinVar, ESP6500, ExAc, Ensembl, HGMD, UCSC, etc.). Mutated genotypes were determined using GATK, LRT, Mutation Taster and SamTools software.
The identified variants were divided into the following four categories according to previous literature reports (15) and software analysis: (1) pathogenic genotypes that were confirmed by literature reports; (2) likely pathogenic genotypes that were reported in literature reports and/or affected proteins by function prediction; (3) indefinite variants and (4) single nucleotide polymorphisms (SNPs) or single nucleotide variants (SNPs). Pathogenic genotypes and likely pathogenic genotypes were recorded as causal gene mutations, and causal gene mutations of tumor samples were confirmed by Sanger sequencing. Germline samples were cross-checked and detected by Sanger sequencing, and causal somatic or germline gene mutations were identified.
The gene chip dataset GSE29986 was downloaded from the GEO database (https://www.ncbi.nlm.nih.gov/geo/). These datasets included 20 T-LBL samples, 10 T-ALL samples and 6 ALL samples. Differential expression analysis was performed using the R package limma. First, we performed differential analysis of the T-LBL and T-ALL datasets, and we obtained upregulated and downregulated genes in the datasets (adj.P. Val <0.05 and |logFC|≥1). Next, we performed differential analysis of the T-LBL and ALL datasets, and we identified upregulated and downregulated genes in these two datasets using the same filtering threshold. Volcano plot showing differentially expressed genes (DEGs) was generated using R language ggplot2. We determined the overlapping genes from the previous two rounds of differential analysis. The overlapping upregulated and downregulated genes related to T-LBL were selected for downstream analysis.
We used R package weighted gene coexpression network analysis (WGCNA) to construct coexpression modules. Thirty-six samples were used to calculate Pearson's correlation coefficients. A power of 6 was selected. An unsigned hybrid coexpression network was then generated using the standard settings. We selected 5,000 genes to construct a topological heatmap. We performed Pearson correlation analysis between the module eigengenes and the trait data to identify module-trait relationships. Finally, we selected turquoise module (related to T-LBL) genes to construct a gene regulatory network and performed Gene Ontology (GO) enrichment analysis.
The R package clusterProfiler was used to analyze the GO enrichment of the upregulated gene and downregulated genes related to T-LBL. Biological process (BP) analysis, cellular component (CC) analysis, molecular function (MF) analysis, and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis of the selected genes were carried out. After performing WGCNA, the hub genes related to T-LBL were selected as input genes for BiNGO. We used Cytoscape software to visualize the results.
To verify the hub genes (USP34, C3 and MGP) identified in WGCNA, we explored the expression level of the hub genes in TCGA (tumor datasets). We checked the expression level of a hub gene (USP34) in different cancers in the CCLE database (https://portals.broadinstitute.org/ccle/home).
Events were defined as each of the following situations (2, 4): RD at TP3, relapsed disease, death or diagnosis of a secondary malignancy, or loss to follow-up. With follow-up to December 2020, data on the clinical features, laboratory findings, WES sequencing, treatment responses, CR rate, treatment-related mortality (TRM) and prospective event-free survival (pEFS) of the patients were collected and analyzed.
EFS was calculated from the date of diagnosis to the last follow-up, loss of follow-up or first event. SPSS 19.0 (IBM Corp., Armonk, NY) software was applied for statistical analysis. Survival curves were calculated according to the Kaplan-Meier test. Proportional differences between patient groups were analyzed by Pearson chi-squared (χ2) tests or Fisher's exact tests. A P value <0.05 was regarded as a significant difference.
Sixty patients with newly diagnosed T-LBL were admitted in the study period; 2 patients were classified as stage I or stage II, and chemotherapy was refused by 4 patients because of family choice. Twelve patients received chemotherapy, but WES sequencing failed because of insufficient available tumor samples. WES sequencing was refused by 7 families. Thirty-five patients received chemotherapy, and WES sequencing was also performed.
The 35 enrolled patients included 25 patients with stage III disease and 10 patients with stage IV disease; 26 males and 9 females were included; family members with gastric cancer were found for 2 (5.71%) patients. Age at diagnosis was 15–168 m (median value: 86 m; average value: 92.83 ± 45.09 m); serum level of lactate dehydrogenase (LDH) was 180–1,947.1 U/L (median value: 339.6 U/L, average value: 522.14 ± 462.40 U/L, normal value <220 U/L), ≥twofold normal level of LDH was detectable in 16 (45.71%) patients; mediastinal, BM or testicular involvement was found in 32 (91.43%), 10 (28.57%) or 1 (2.86%) patients, respectively; and CNS involvement was undetectable in the cohort (Table 1).
WES sequencing of the 35 patients was performed, and germline causal gene mutations (CARS or MAP2K2) were detected in 2 patients without a family history of cancer; 1–13 (average 3.06 ± 2.21; median 2) somatic causal gene mutations were identified in the 35 patients, and the somatic mutations were observed in the NOTCH1, FBXW7, PHF6 and JAK3 genes (Figure 1A). The relationship among these gene mutations was demonstrated by corplot (Figure 1B), and NOTCH1 mutations were significantly associated with FBXW7 mutations (6/35, 17.14%). The associations between NOTCH1-FBXW7 mutational status and clinical characteristics are listed in Table 2, which revealed that the age at diagnosis of patients with NOTCH1-FBXW7 mutations was less than that of patients without such mutations (P < 0.05), whereas significant differences were not found for sex distribution, LDH level or disease staging distribution (P > 0.05). These results were similar to those for pediatric T-ALL in previous literature reports (16, 17).
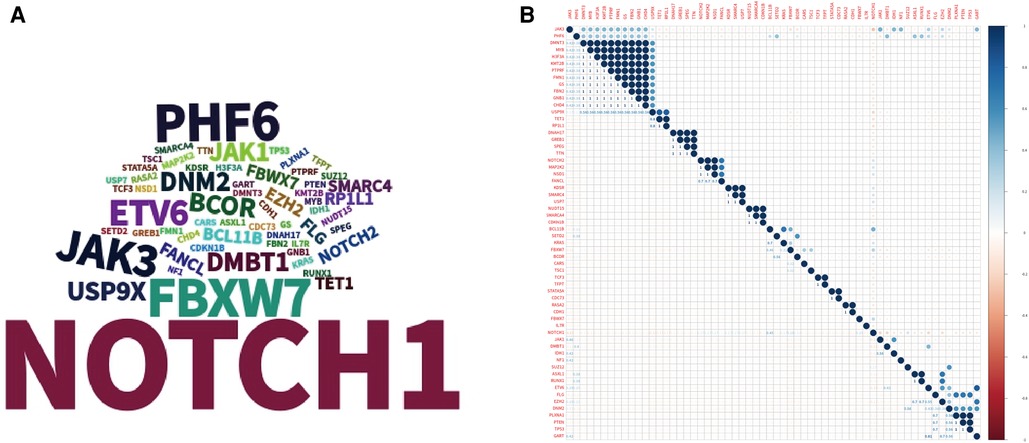
Figure 1. Landscape of gene mutations in the cohort. (A) Tagcloud show candidate genes identified by WES analysis word size according to the frequency of the gene. (B) Heatmap shows the correlations of mutated genes identified by WES.
Treatment effects were evaluated at different TP levels as the protocol required. Twelve, 21 or 2 patients were classified as CR, PR or PD at TP1 and TP2, respectively; at TP3, 1 PR patient died of sepsis, 32 achieved CR, and 2 patients were classified as PD, and the CR rate was 91.43% after they finished the course of remission induction. The 2 PD patients quit the study, and 14 and 18 patients were classified into the intermediate risk (IR) group or high risk (HR) group; chemotherapy was continued with the different risk group strategy (Table 3). With follow-up to December 2020, 3 patients relapsed. Among the 35 patients in the cohort, TRM was 2.86%, the relapse rate was 8.57%, and 3-year pEFS was 82.286% (95% CI 64.71–91.64%, Figures 2A,B). pEFS was compared among different sexes, ages, LDH levels and disease stages, but no significant differences were found (P > 0.05, Figures 2C–E).
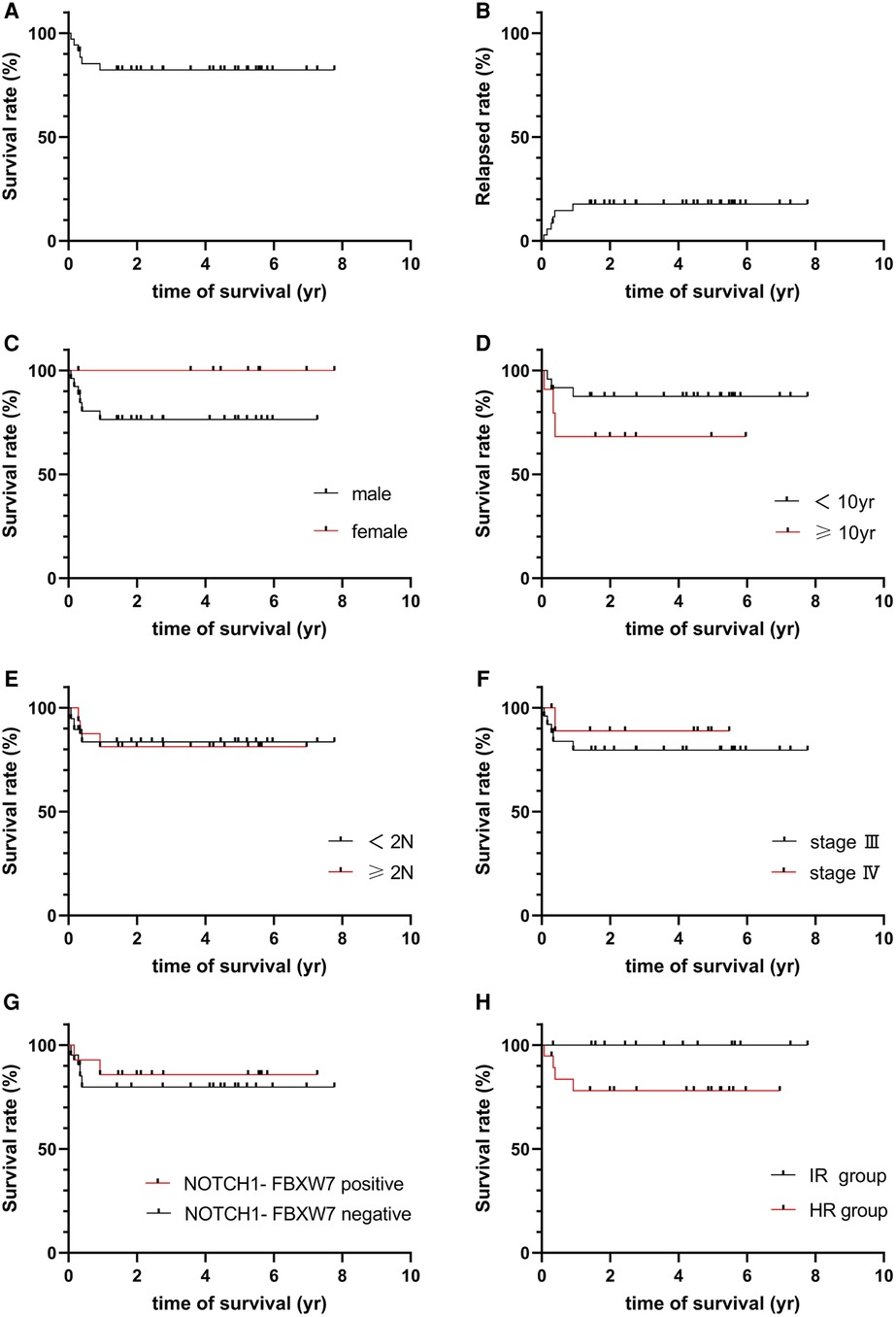
Figure 2. (A) Survival of total group. (B) Relapsed rate of total group. (C) Survival for gender groups. (D) Survival for age groups. (E) Survival for LDH level. (F) Survival for different stage. (G) Survival for NOTCH-FBXW7. (H) Survival for risk group.
The 35 patients were classified into 2 groups (group A: NOTCH1-FBXW7 mutation; group B: without the NOTCH1-FBXW7 mutation), and the treatment responses and outcomes were also calculated with different mutational statuses. There was no significant difference in the CR rate between the two groups at TP1, TP2 or TP3 (P > 0.05, Table 4). One patient in each group remained PD at TP2; the remaining patients were treated as different risk groups, and a significant difference was not found between different mutational statuses or risk groups (P > 0.05, Table 4, Figures 2F–H), but the mean survival time of the IR group was longer than that of the HR group (66.14 ± 7.88 vs. 42.07 ± 8.46 m, P < 0.05). Literature reports have shown that the NOTCH1-FBXW7 mutation is related to favorable outcomes (18–20), but our study showed that the diversity of responses might be overcome by intensive chemotherapy; however, a larger sample and multiple centers are needed to verify this hypothesis.
To further understand the pathophysiolopoiesis, differences and similarities of T-LBL and T-ALL, differential expression analysis was performed, and the results were visualized. A volcano plot was generated to show the results of the differential expression analysis; a Venn plot was generated to show the overlapping upregulated and downregulated genes from the differential expression analysis (T-LBL versus T-ALL/ALL, Figures 3A,B). Although T-ALL and T-LBL are considered different manifestations of the same disease and T-LBL is regarded as a nonleukemic phase of T-LBL, it seemed that the gene expression results were not the same between the T-LBL and T-ALL datasets and according to a literature review (21, 22); 1/3 of the differentially expressed genes were found in both the T-ALL and T-LBL datasets.
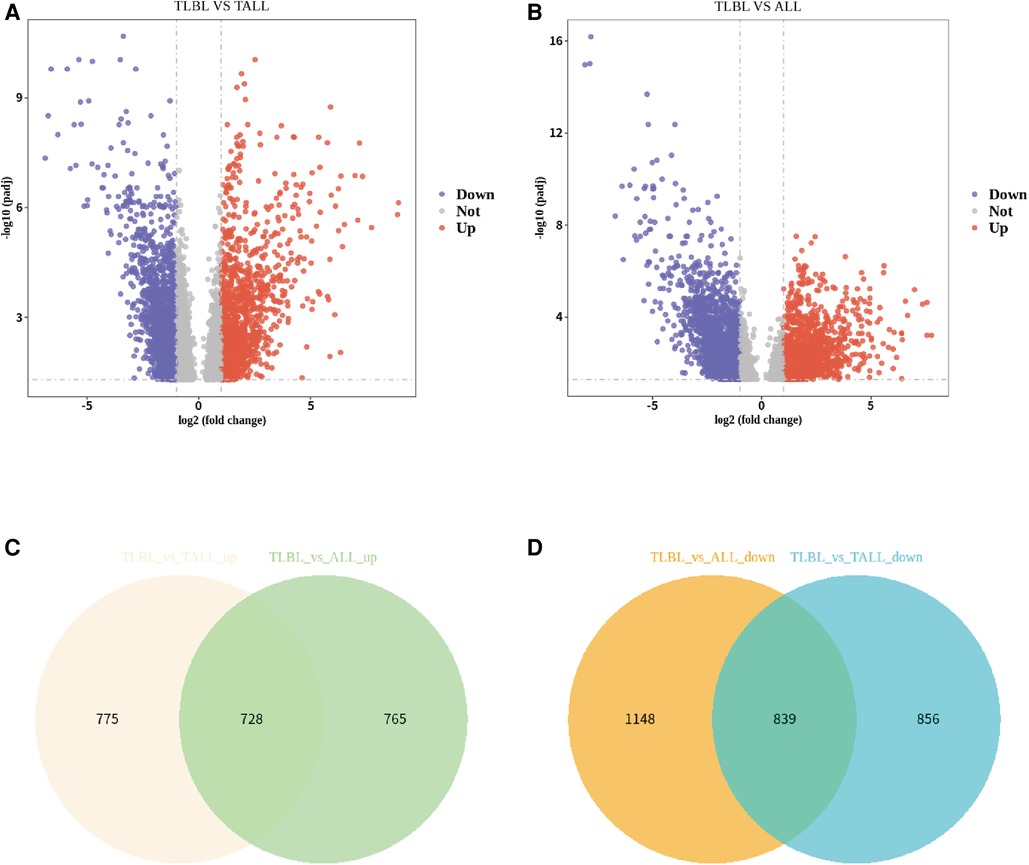
Figure 3. Differential expression analysis of T-LBL versus T-ALL/ALL. (A) Volcano plot showed differential expression analysis for TLBL and TALL microarray data. (B) Volcano plot showed differential expression analysis for TLBL and ALL microarray data. (C) Venn plot showed the overlapped up-regulated genes from differential expression analysis (TLBL versus TALL/ALL). (D) Venn plot showed the overlapped down-regulated genes from differential expression analysis (TLBL versus TALL/ALL).
Significance was determined as a P value of <0.05, and 728 overlapping upregulated genes and 839 overlapping downregulated genes were obtained (T-LBL vs. T-ALL/ALL, Figures 3C,D). The overlapping upregulated genes or downregulated genes related to T-LBL were assessed by GO and KEGG pathway enrichment analyses (BP, CC and MF), and the interactions between the gene sets and GO terms were analyzed and visualized; it appeared that the PI3K-Akt signal pathway, focal adhesion and ECM-receptor interaction play roles in the pathophysiology of T-LBL (Figure 4). However, further studies are required to confirm this result.
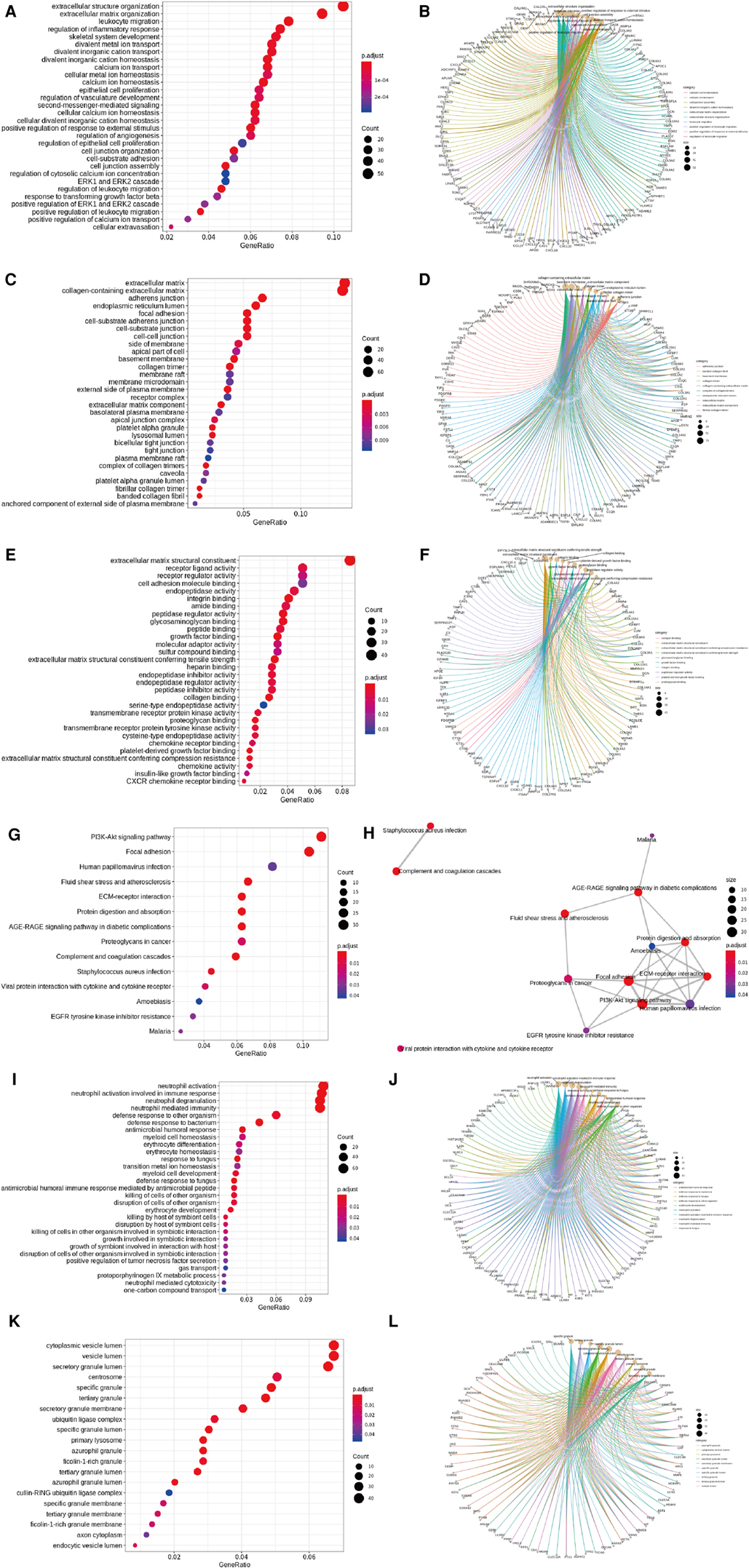
Figure 4. GO and KEGG pathway analyses. (A, B) Enriched GO terms (BP) in overlapped TLBL up-regulated genes and the interactions between the gene sets and GO terms. (C, D) Enriched GO terms (CC) in overlapped TLBL up-regulated genes and the interactions between the gene sets and GO terms. (E, F) Enriched GO terms (MF) in overlapped TLBL up-regulated genes and the interactions between the gene sets and GO terms. (G, H) Enriched KEGG pathway in overlapped TLBL up-regulated genes and interaction network between these pathways. (I, J) Enriched GO terms (BP) in overlapped TLBL down-regulated genes and the interactions between the gene sets and GO terms. (K, L) Enriched GO terms (CC) in overlapped TLBL down-regulated genes and the interactions between the gene sets and GO terms.
Sample hierarchical clustering plots and trait heatmaps from the GSE29986 dataset were generated, and the topological overlap matrix (TOM) of coexpressed genes in different modules of the top 5,000 genes is shown as a heatmap (Figure 5). Module and trait relationships were determined, and the turquoise module was the most related module to TLBL; the coexpression network of significant genes in the turquoise module (related to T-LBL) was generated (Figure 5). The mRNA expression levels of three key genes (USP34, C3 and MGP) based on TCGA tumor data were also determined, and the USP34 gene was highly expressed in the cancer cell lines included in the T-LBL and T-ALL datasets according to CCLE analysis (Figure 6).
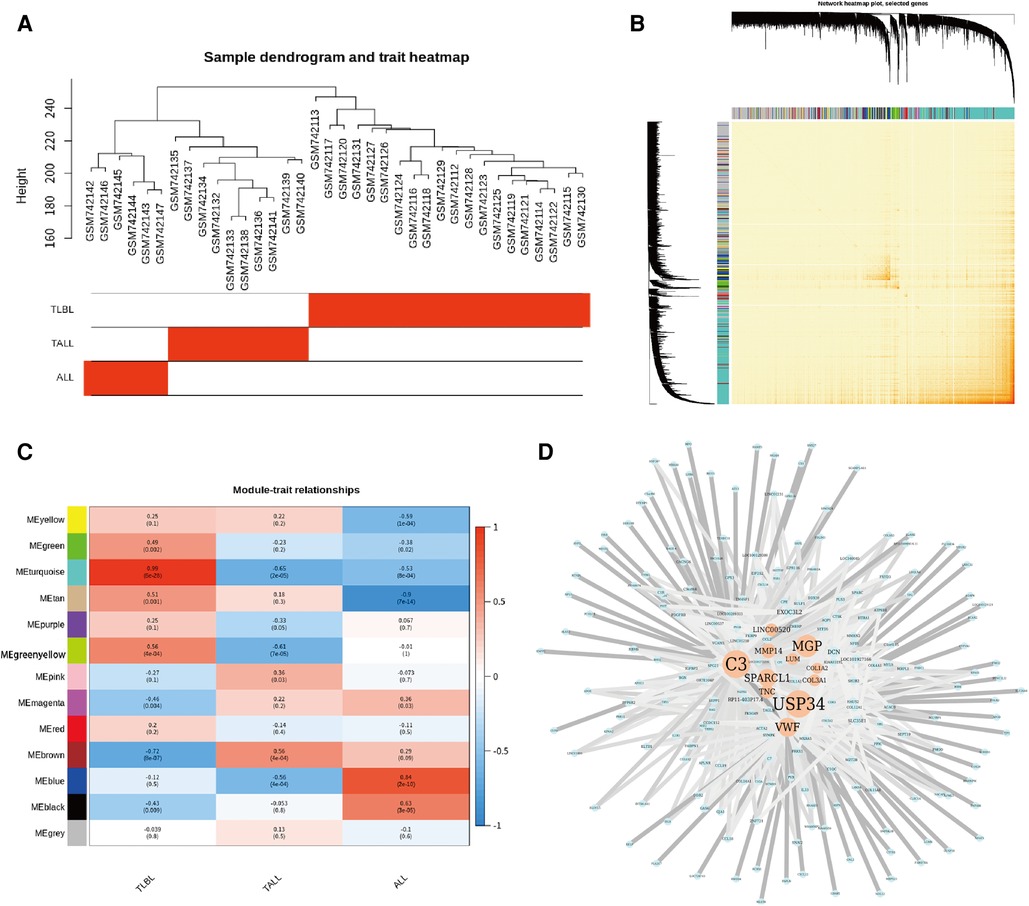
Figure 5. Coexpressed genes in different modules in the top 5000 genes. (A) Sample hierarchical clustering plot and trait heatmap from GSE29986 datasets. (B) Heatmap showed the Topological Overlap Matrix (TOM) of co-expressed genes in different modules using the top 5000 genes. (C) Module and trait relationships. The darker the module color, the more significant their relationship. The turquoise module is most related to TLBL. (D) The co-expression network of the significant genes in the turquoise Module (related to TLBL) the color depth of the edge refers to the weight, node size refers to Degree.
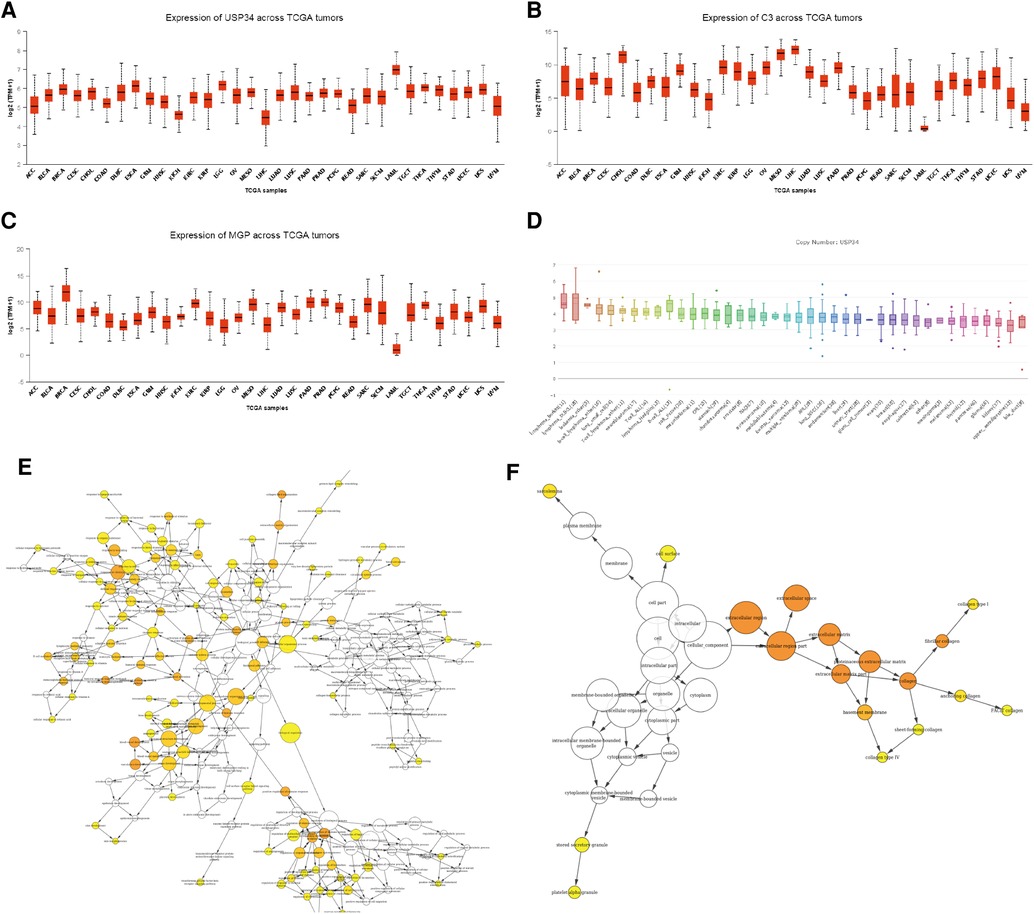
Figure 6. mRNA expression levels of three key genes. (A-C) The mRNA expression level of three key genes (USP34, C3 and MGP) based on TCGA tumors data. (D) usp34 were high expressed in cancer cell lines from CCLE analysis. (E) The gene ontology enrichment (BP) of the significant genes in the turquoise module (related to TLBL). The color depth of the nodes refers to the corrected p values of the ontologies, node size refers to enriched gene numbers. (F) The gene ontology enrichment (CC) of the significant genes in the turquoise module (related to TLBL). The color depth of the nodes refers to the corrected p values of the ontologies, node size refers to enriched gene numbers.
Ubiquitination is a common posttranslational modification of proteins and is involved in many physiological activities, such as cell division and differentiation, growth and development, transcriptional regulation, injury stress, and immune response (23). Deubiquitination enzymes have diverse structures and functions; they can remove ubiquitination modifications, affect protein function and regulate physiological activities. The ubiquitin-specific peptidase (USP) superfamily is one of the most widely known deubiquitinase families with the most diverse structures, and the USP34 gene encodes a member of the USP family that can remove ubiquitin molecules from large protein molecules and eliminate the biological functions of ubiquitinated proteins (24, 25). This USP plays a key regulatory role in the process of DNA damage repair and tumor occurrence and development (24–27).
T-LBL is an aggressive form of non-Hodgkin's lymphoma derived from precursor T cells and makes up approximately 80% of the LBL population. T-ALL is regarded as the leukemic phase of T-LBL (1, 2). Several studies have demonstrated that leukemia-based therapy (e.g., the BFM protocol series) and allo-HSCT are effective and lead to a good prognosis for T-LBL patients, and EFS exceeds 80%–90% for advanced stage patients in developed or developing countries (2, 3, 20, 28). Traditional prognostic factors have been challenging because age, sex, and multiple organ involvement have not been found to be prognostically significant by the literature (2, 3) or our study. Germline mutations (CARS or MAP2K2 mutation) were detected in 2 patients without cancer history and the relationship between tumor development and hereditary should be considered, but data were limited, further data were needed to investigate it.
Multiple gene mutations play different roles in the development, progression and carcinogenesis of T-LBL and T-ALL. Our data and the literature suggest that the common somatic mutations are NOTCH1, FBXW7 and PHF6 mutations and that NOTCH1 mutations are significantly associated with FBXW7 mutations (5, 16, 20, 29). The associations between NOTCH1-FBXW7 mutational status and clinical characteristics are listed in Table 2, which revealed that the age at diagnosis in patients with NOTCH1-FBXW7 mutations was less than that in patients without such mutations (P < 0.05), whereas significant differences were not found for sex distribution, LDH level or disease stage distribution (P > 0.05). The results were similar to those for pediatric T-ALL in previous literature reports (16, 17).
T-LBL and T-ALL both descend from precursor T-cells, and T-ALL is regarded as the leukemic phase of T-LBL. The outcomes of T-LBL are improved by ALL-based chemotherapy, but EFS of T-LBL and T-ALL in our study were different. Three-year pEFS exceeded 80% for advanced-stage T-LBL in our study, whereas 3-year pEFS was 54.1 ± 11.2% in the pediatric T-ALL population in our department (30). This difference was partly due to the different chemotherapy regimens used for the two patient groups, but the different gene distributions of T-ALL and T-LBL also played important roles in the prognosis in our opinion. Data were collected from T-LBL and T-ALL datasets and analyzed. Only one-third of upregulated genes or downregulated genes overlapped in the two datasets, while the other two-thirds did not overlap (Figure 3), revealing that T-ALL and T-LBL are similar but not the same disease. The different gene distributions of T-ALL and T-LBL may partially due to the sample sources in which DNA samples of T-LBL were acquired from formalin-fixed specimens; in In future investigations, DNA samples obtained from fresh biopsy samples were needed to investigate it; in the other hand, different gene distributions of T-ALL and T-LBL might also existed and these differences could be associated with the carcinogenesis and development of the disease. Therefore, treatment strategies need to be adjusted, and further data and research are needed to confirm this hypothesis.
Although FBXW7, NOTCH1 and PHF6 mutations were the most common somatic mutations in T-LBL patients, other gene mutations and related pathways were also detectable. The T-LBL database was evaluated by GO and KEGG pathway enrichment analyses, and it appeared that the PI3K-Akt signaling pathway and USP34 gene played important roles in T-LBL. Tumor development, proliferation and metastasis are regulated by abnormal cellular signaling pathways, and genes in the phosphoinositide 3-kinase (PI3K)/Akt pathway are the most frequently altered in human cancers. Aberrant activation of this pathway is associated with cellular transformation, tumorigenesis, cancer progression, and drug resistance (31, 32). The ubiquitin-specific protease 34 (USP34) gene and its protein are closely related to development and progression of human cancers. Research has shown that USP34 overexpression can be detected in diffuse large B cell lymphoma instead of follicular lymphoma, but a significant association has not been identified between the USP34 protein level and patient survival (33, 34). Pancreatic cancer cell proliferation and migration are promoted by USP34 overexpression via upregulation of the Akt protein, and pancreatic cancer cell apoptosis induced by an Akt inhibitor is reversed by USP34 overexpression (35). Thus, we assume that USP34 overexpression and the PI3K-Akt signaling pathway are associated with T-LBL development and progression, but anticancer therapies targeting the USP34 gene or the PI3K-Akt signaling pathway may be invalid for refractory relapsed T-LBL patients. Further laboratory studies should be performed.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
The studies involving humans were approved by Ethics Administration Office of Children Hospital of Chongqing Medical University. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation in this study was provided by the participants’ legal guardians/next of kin.
JX conceived and designed the study. QL, XY and NY prepared the figures and tables. QL, XY, JW, XL, PZ and YG analyzed and interpreted the data. LS and JX drafted the manuscript. JX revised the manuscript. All authors contributed to the article and approved the submitted version.
The study was supported by the National Natural Science Foundation of China (Project No. 81900162) and the Chongqing Science and Technology Commission of the PR China (Project No. cstc2018jsyj-jsyjX0015); the funding sources had no roles in the design and conduct of the study collection; management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and the decision to submit the manuscript for publication. The study was registered at the Chinese Clinical Trail Registry and the clinical study registration (ChiCTR1900025690 and ChiCTR-IPR-14005706).
We thank the patients and their families who participated in this study. The authors declare that there are no competing interests associated with the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fped.2023.1224966/full#supplementary-material
1. Burkhardt B, Hermiston ML. Lymphoblastic lymphoma in children and adolescents: review of current challenges and future opportunities. Br J Haematol. (2019) 185:1158–70. doi: 10.1111/bjh.15793
2. Landmann E, Burkhardt B, Zimmermann M, Meyer U, Woessmann W, Klapper W, et al. Results and conclusions of the European intergroup EURO-LB02 trial in children and adolescents with lymphoblastic lymphoma. Haematologica. (2017) 102:2086–96. doi: 10.3324/haematol.2015.139162
3. Brammer JE, Khouri I, Marin D, Ledesma C, Rondon G, Ciurea SO, et al. Stem cell transplantation outcomes in lymphoblastic lymphoma. Leuk Lymphoma. (2017) 58:366–71. doi: 10.1080/10428194.2016.1193860
4. Balbach ST, Makarova O, Bonn BR, Zimmermann M, Rohde M, Oschlies I, et al. Proposal of a genetic classifier for risk group stratification in pediatric T-cell lymphoblastic lymphoma reveals differences from adult T-cell lymphoblastic leukemia. Leukemia. (2016) 30:970–73. doi: 10.1038/leu.2015.203
5. Bonn BR, Huge A, Rohde M, Oschlies I, Klapper W, Voss R, et al. Whole exome sequencing hints at a unique mutational profile of paediatric T-cell lymphoblastic lymphoma. Br J Haematol. (2015) 168:308–13. doi: 10.1111/bjh.13105
6. Jiang M, Bennani NN, Feldman AL. Lymphoma classification update: t-cell lymphomas, Hodgkin lymphomas, and histiocytic/dendritic cell neoplasms. Expert Rev Hematol. (2017) 10:239–49. doi: 10.1080/17474086.2017.1281122
7. Swerdlow SH, Campo E, Pileri SA, Harris NL, Stein H, Siebert R, et al. The 2016 revision of the world health organization classification of lymphoid neoplasms. Blood. (2016) 127:2375–90. doi: 10.1182/blood-2016-01-643569
8. Rosolen A, Perkins SL, Pinkerton CR, Guillerman RP, Sandlund JT, Patte C, et al. Revised international pediatric non-Hodgkin lymphoma staging system. J Clin Oncol. (2015) 33:2112–28. doi: 10.1200/JCO.2014.59.7203
9. La Starza R, Pierini V, Pierini T, Nofrini V, Matteucci C, Arniani S, et al. Design of a comprehensive fluorescence in situ hybridization assay for genetic classification of T-cell acute lymphoblastic leukemia. J Mol Diagn. (2020) 22:629–39. doi: 10.1016/j.jmoldx.2020.02.004
10. La Starza R, Lettieri A, Pierini V, Nofrini V, Gorello P, Songia S, et al. Linking genomic lesions with minimal residual disease improves prognostic stratification in children with T-cell acute lymphoblastic leukaemia. Leuk Res. (2013) 37:928–35. doi: 10.1016/j.leukres.2013.04.005
11. Burkhardt B, Reiter A, Landmann E, Lang P, Lassay L, Dickerhoff R, et al. Poor outcome for children and adolescents with progressive disease or relapse of lymphoblastic lymphoma: a report from the Berlin-Frankfurt-muenster group. J Clin Oncol. (2009) 27:3363–69. doi: 10.1200/JCO.2008.19.3367
12. Becker S, Vermeulin T, Cottereau AS, Boissel N, Vera P, Lepretre S. Predictive value of 18F-FDG PET/CT in adults with T-cell lymphoblastic lymphoma: post hoc analysis of results from the GRAALL-LYSA LLO3 trial. Eur J Nucl Med Mol Imaging. (2017) 44:2034–41. doi: 10.1007/s00259-017-3776-3
13. Shi X, Zhou W, Xu M, Hua T, Guan Y. Successful treatment for adult precursor B-cell lymphoblastic lymphoma involving central nervous system monitored by 18F-fluorode oxyglucose PET/CT imaging. Front Oncol. (2020) 24:334. doi: 10.3389/fonc.2020.00334
14. Mussolin L, Buldini B, Lovisa F, Carraro E, Disarò S, Lo Nigro L, et al. Detection and role of minimal disseminated disease in children with lymphoblastic lymphoma: the AIEOP experience. Pediatr Blood Cancer. (2015) 62:1906–13. doi: 10.1002/pbc.25607
15. Li MM, Datto M, Duncavage EJ, Kulkarni S, Lindeman NI, Roy S, et al. Standards and guidelines for validating next-generation sequencing bioinformatics pipelines: a joint recommendation of the association for molecular pathology and the college of American pathologists. J Mol Diagn. (2017) 19:4–23. doi: 10.1016/j.jmoldx.2017.11.003
16. Valliyammai N, Nancy NK, Sagar TG, Rajkumar T. Study of NOTCH1 and FBXW7 mutations and its prognostic significance in south Indian T-cell acute lymphoblastic leukemia. J Pediatr Hematol Oncol. (2018) 40:e1–8. doi: 10.1097/MPH.0000000000001006
17. Kimura S, Seki M, Yoshida K, Shiraishi Y, Akiyama M, Koh K, et al. NOTCH1 pathway activating mutations and clonal evolution in pediatric T-cell acute lymphoblastic leukemia. Cancer Sci. (2019) 110:784–94. doi: 10.1111/cas.13859
18. Zuurbier L, Homminga I, Calvert V, te Winkel ML, Buijs-Gladdines JG, Kooi C, et al. NOTCH1 and/or FBXW7 mutations predict for initial good prednisone response but not for improved outcome in pediatric T-cell acute lymphoblastic leukemia patients treated on DCOG or COALL protocols. Leukemia. (2010) 24:2014–22. doi: 10.1038/leu.2010.204
19. Natarajan V, Bandapalli OR, Rajkumar T, Sagar TG, Karunakaran N. NOTCH1 and FBXW7 mutations favor better outcome in pediatric south Indian T-cell acute lymphoblastic leukemia. J Pediatr Hematol Oncol. (2015) 37:e23–30. doi: 10.1097/MPH.0000000000000290
20. Park MJ, Taki T, Oda M, Watanabe T, Yumura-Yagi K, Kobayashi R, et al. FBXW7 and NOTCH1 mutations in childhood T cell acute lymphoblastic leukaemia and T cell non-Hodgkin lymphoma. Br J Haematol. (2010) 145:198. doi: 10.1111/j.1365-2141.2009.07607.x
21. Basso K, Mussolin L, Lettieri A, Brahmachary M, Lim WK, Califano A, et al. T-cell lymphoblastic lymphoma shows differences and similarities with T-cell acute lymphoblastic leukemia by genomic and gene expression analyses. Genes Chromosomes Cancer. (2011) 50:1063–75. doi: 10.1002/gcc.20924
22. Kroeze E, Loeffen JLC, Poort VM, Meijerink JPP. T-cell lymphoblastic lymphoma and leukemia: different diseases from a common premalignant progenitor? Blood Adv. (2020) 4:3466–73. doi: 10.1182/bloodadvances.2020001822
23. Khadivi F, Razavi S, Hashemi F. Protective effects of zinc on rat sperm chromatin integrity involvement: DNA methylation, DNA fragmentation, ubiquitination and protamination after bleomycin etoposide and cis-platin treatment. Theriogenology. (2020) 142:177–83. doi: 10.1016/j.theriogenology.2019.09.039
24. Mitxitorena I, Somma D, Mitchell JP, Lepistö M, Tyrchan C, Smith EL, et al. The deubiquitinase USP7 uses a distinct ubiquitin-like domain to deubiquitinate NF-κB subunits. J Biol Chem. (2020) 295(33):11754–63. doi: 10.1074/jbc.RA120.014113
25. Sakai K, Ito C, Wakabayashi M, Kanzaki S, Ito T, Takada S, et al. Usp26 mutation in mice leads to defective spermatogenesis depending on genetic background. Sci Rep. (2019) 9:1–12. doi: 10.1038/s41598-018-37186-2
26. Gu Z, Lin C, Hu J, Xia J, Wei S, Gao D. USP34 regulated human pancreatic cancer cell survival via AKT and PKC pathways. Biol Pharm Bull. (2019) 42(4):573–9. 10.1248/bpb.b18-0064630686807
27. Lui TTH, Lacroix C, Ahmed SM, Goldenberg SJ, Leach CA, Daulat AM, et al. The ubiquitin-specific protease USP34 regulates axin stability and wnt/β-catenin signaling. Mol Cell Biol. (2011) 31:2053–65. doi: 10.1128/MCB.01094-10
28. Chenyu Z, Jianwen X, Jie Y, Yonggang C. Prognosis and side effects of T-NHL-2009 protocol on treatment of lymphoblastic lymphoma in children. Lab Med Clin. (2017) 14:1883–87. doi: 10.3969/j.issn.1672-9455.2017.13.013
29. Wang Q, Qiu H, Jiang H, Wu L, Dong S, Pan J, et al. Mutations of PHF6 are associated with mutations of NOTCH1, JAK1 and rearrangement of SET-NUP214 in T-cell acute lymphoblastic leukemia. Haematologica. (2011) 96:1808–14. doi: 10.3324/haematol.2011.043083
30. Wang M, Wen J, Guo Y, Shen Y, An X, Hu Y, et al. Clinical characterization and prognosis of T cell acute lymphoblastic leukemia with high CRLF2 gene expression in children. PLoS One. (2019) 14:e0224652. doi: 10.1371/journal.pone.0224652
31. Mayer IA, Arteaga CL. The PI3K/AKT pathway as a target for cancer treatment. Annu Rev Med. (2016) 67:11–28. doi: 10.1146/annurev-med-062913-051343
32. Burris HA. Overcoming acquired resistance to anticancer therapy: focus on the PI3K/AKT/mTOR pathway. Cancer Chemother Pharmacol. (2013) 71:829–42. doi: 10.1007/s00280-012-2043-3
33. Li C, Huang L, Lu H, Wang W, Chen G, Gu Y, et al. Expression and clinical significance of ubiquitin specific processing protease 34 in diffuse large B cell lymphoma. Mol Med Rep. (2018) 18:4543–54. doi: 10.3892/mmr.2018.9447
34. Kurşun D, Küçük C. Systematic analysis of the frequently amplified 2p15-p16.1 locus reveals PAPOLG as a potential proto-oncogene in follicular and transformed follicular lymphoma. Turk J Biol. (2019) 43:124–32. doi: 10.3906/biy-1810-2
Keywords: T cell lymphoblastic lymphoma, children, whole exome sequencing, PI3K-Akt pathway, USP34 gene
Citation: Liu Q, Yu X, Wen J, Yin N, Liao X, Zou P, Guo Y, Song L and Xiao J (2023) Genomic signatures and prognosis of advanced stage Chinese pediatric T cell lymphoblastic lymphoma by whole exome sequencing. Front. Pediatr. 11:1224966. doi: 10.3389/fped.2023.1224966
Received: 18 May 2023; Accepted: 31 July 2023;
Published: 16 August 2023.
Edited by:
Victor Aquino, University of Texas Southwestern Medical Center, United StatesReviewed by:
Concetta Micalizzi, Giannina Gaslini Institute (IRCCS), Italy© 2023 Liu, Yu, Wen, Yin, Liao, Zou, Guo, Song and Xiao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jianwen Xiao dG9tYWhhd2s2NTAyQHNvaHUuY29t
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.