- 1Department of Clinical Laboratory, the Children’ s Hospital, Zhejiang University School of Medicine, National Clinical Research Center for Child Health, Hangzhou, China
- 2Department of Dermatology, The Children' s Hospital, Zhejiang University School of Medicine, Hangzhou, China
Background: Atopic dermatitis (AD) is a common chronic inflammatory skin disease, adversely affecting nearly 20% of the pediatric population worldwide. Interleukin-4 (IL-4) and interleukin-18 (IL-18) are considered to be involved in the pathogenesis and development of AD. The aim of this study was to investigate the association of IL-4 and IL-18 gene polymorphisms with the susceptibility and severity of AD in Chinese children.
Methods: Six candidate single nucleotide polymorphisms (SNPs) in IL-4 and IL-18 genes were genotyped through multi-PCR combined with next-generation sequencing in 132 AD children and 100 healthy controls, and all the analyses were performed on blood genome DNA.
Results: The frequencies of G allele, CG genotype and CG + GG genotype of IL-4 rs2243283, as well as the haplotype IL-4/GTT (rs2243283-rs2243250-rs2243248) were all significantly decreased in AD patients compared with the controls [G vs. C: P = 0.033, OR = 0.59; CG vs. CC: P = 0.024, OR = 0.47; CG + GG vs. CC: P = 0.012, OR = 0.49; GTT vs. CCT: P = 0.011, OR = 0.65]. Moreover, the frequencies of A allele, AA genotype and AG + AA genotype of IL-18 rs7106524, along with the haplotype IL-18/CAA (rs187238-rs360718-rs7106524) were statistically increased in the severe AD patients (A vs. G: P < 0.001, OR = 2.79; AA vs. GG: P = 0.003, OR = 5.51; AG + AA vs. GG: P = 0.036, OR = 2.93; CAA vs. CAG: P = 0.001, OR = 2.86).
Conclusions: Our findings suggested that genetic variation in IL-4 rs2243283 such as G allele, CG genotype and CG + GG genotype might confer the reduced susceptibility to AD in Chinese children. Furthermore, A allele, AA genotype and AG + AA genotype of IL-18 rs7106524 explored the strong association with severity in Chinese AD children.
1. Introduction
Atopic dermatitis (AD), also known as eczema, is a recurrent inflammatory skin disease that adversely affects a significant proportion of the pediatric population (10%–20%) (1). The quality of life in patients with AD seriously decreased due to the characteristics of intense pruritus, diffuse xerosis and sleep disturbance (2). Although the pathogenesis of AD has not been completely understood, immune imbalance and skin barrier dysfunction led by the interaction of genetic predisposition, immune and microbiome are considered to be the critical causes of AD (1, 3).
It has been confirmed that the immune imbalance between T helper (Th) lymphocytes 1 and 2 can result in abnormal cytokine production, which plays a crucial role in the occurrence and development of immune-mediated and allergic disorders, such as AD, asthma, and allergic rhinitis (4, 5). Interleukin-4 (IL-4), mainly produced by mast cells and activated T cells including Th2 cells, is an important driver of type 2 immunity in these diseases. IL-4 is also considered central to the pathogenesis of AD and key drug target (6). It has been shown that IL-4 acts on both immune cells and histiocytoes, mediating multiple steps of the AD pathogenicity cascade. On the one hand, IL-4 could cause differentiation of naïve CD4+ T cells to Th2 cells and induce T cytotoxic 2 (Tc2) cells and group-2 innate lymphoid cells (ILC2) development (6, 7). Through downregulating filaggrin (FLG) expression, IL-4 also amplifies the effects of histamine and stimulates immunoglobulin E (IgE) production by B cells, which could inhibit skin barrier function and induce pruritus or anaphylaxis in skin lesions of acute AD (8, 9). On the other hand, IL-4 regulates the activity of dendritic cells, decreasing their expression of IL-12 and major histocompatibility complex (MHC) class II and increasing IL-10 production, thereby amplifying type 2 inflammation (10). Moreover, mouse model studies have established the centrality of IL-4 in the pathogenesis of AD, highlighting its capability to induce all the histopathological features of AD (6). In summary, the role of IL-4 in the pathogenesis of AD and in mediating a variety of clinical features, including skin inflammation and pruritus, is well established.
Although type 2 inflammation is the dominant immune mechanism, there is growing evidence that AD involves multiple immune pathways. Interleukin-18 (IL-18), identified as an IFN-γ inducing factor, is a pro-inflammatory cytokine that plays a critical role in Th1 lymphocytes activation. Moreover, IL-18 synergistically stimulates the production of IgE and Th2 cytokines with IL-12 (11, 12). It has been presented that IL-18 is overexpressed in both AD patients and animal model, suggesting that IL-18 might play a certain role in the pathogenesis and development of dermatitis (13). Recent evidence also demonstrated that IL-18 deficiency alleviated AD-induced lesions by reducing corticotropin-releasing hormone receptor (CRHR) 2 expression in a mouse model (14). Furthermore, Zedan et al. reported that serum IL-18 levels were significantly increased in AD patients and strongly correlated with AD activity, suggesting that IL-18 may help to assess AD activity and therefore would be useful in predicting disease progression (12).
In addition, it has been proposed that the gene polymorphisms of several cytokines are related to immune imbalance in allergic diseases such as bronchial asthma (15). Previous surveys have also indicated that genetic predisposition profoundly affected the onset and severity of AD (16, 17). To our knowledge, several single nucleotide polymorphisms (SNPs) of IL-4 and IL-18 genes have been identified their relevance to AD in Macedonian children, Saudi Arabian population, and Egyptians (18–20). Nevertheless, there have been few studies to date on the linkage between these two cytokines polymorphisms and AD in Chinese population. Therefore, in this study, three SNPs of IL-4 and three SNPs of IL-18 associated with AD or other allergic diseases in previous researches (19–23) were selected to explore their correlation with the susceptibility and severity of AD in Chinese Han children, so as to assess the population-specific prediction of IL-4 and IL-18 genetic variation for AD. This will provide researchers and clinicians with more baseline information including the identifiable risk factors, which may help predict the occurrence and prevent the progression of allergic dermatitis.
2. Materials and methods
2.1. Subjects
From February 2021 to August 2021, the children with AD attending the dermatology clinic of the Children's Hospital of Zhejiang University School of Medicine were used as the study subjects. Inclusion criteria were determined as follows: newly diagnosed AD patients in accordance with the AD diagnostic criteria of Hanifin and Rajka (1); Age less than 18 years; all subjects and their parents consented to participate in the study. Children with any other systemic inflammatory and autoimmune diseases or a history of medication for skin dysfunctions were excluded. The severity of AD was assessed by the Severity Scoring of Atopic Dermatitis (SCORAD) index (1). In order to observe the association of IL-4 and IL-18 SNPs with the severity of AD, AD cases were separated into two subgroups, namely, mild-to-moderate (SCORAD: 0–50) and severe subgroup (SCORAideD: > 50) (1, 24). Meanwhile, one hundred unrelated healthy subjects without a history of atopic or autoimmune disorders were enrolled from the physical examination department in our hospital as controls. All AD cases and controls were of Chinese Han ethnicity. The present study was approved by the Ethics Committee of the Children's Hospital of Zhejiang University School of Medicine in accordance with the principles of the Declaration of Helsinki (2021-IRB-009).
2.2. Genotype assessment
One milliliter peripheral blood samples from each subjects were collected in ethylene diamine tetraacetic acid (EDTA) anticoagulant tubes and 200 μl of each specimen was taken for DNA extraction and genotyping. DNA was extracted by Biospin genomic DNA Extraction Kit (BIOER technology, #BSC06S1) and its purity and concentration were detected by Nanodrop1000c spectrophotometer of ThermoScience Company in the United States. The integrity of DNA was observed by 1% agarose gel electrophoresis. The qualified DNA samples were kept at −20 °C for the subsequent genetic analysis.
Three IL-4 SNPs rs2243283, rs2243250, rs2243248 and three IL-18 SNPs rs187238, rs360718, rs7106524 were genotyped by multiplex PCR combined with next-generation sequencing technique on X-10 Illumina, a high-throughput genotyping platform of Shanghai BioWing Applied Biotechnology Company (http://www.biowing.com.cn/) (25). The first round PCR was carried out on Gene Amp PCR System 9600 (Norwalk, USA) in the following conditions: 95 °C for 15 min; 4 cycles of 94°C for 30 s, 60°C for 10 min and 72°C for 30 s; 20 cycles of 94°C for 30 s, 60°C for 1 min and 72°C for 30 s. 1 μl of the first round product could be used as a sample for the second round of PCR under these conditions: 95°C for 15 min; 5 cycles of 94°C for 30 s, 60°C for 4 min and 72°C for 30 s; 10 cycles of 94°C for 30 s, 65°C for 1 min and 72°C for 30 s. The detailed primers used for genotyping were shown in Table 1. Then the products of the second round PCR were purified and sequenced on the X-10 platform. All the sequencing data were analyzed by Illumabcl2fastq software.
2.3. Statistical analysis
Statistical analysis was performed by the software of Statistical Package for Social Science (SPSS) (version 22.0) (IBM, USA). Continuous variables were shown as mean ± standard deviation (SD), and comparisons between groups were made by t-test or Anova variance analysis. Categorical variables were expressed as percentages or ratios. Comparisons between groups were performed using Fisher's exact test or Pearson's chi-square test. Genotypic frequencies of all participants were examined for Hardy-Weinberg equilibrium (HWE) before analysis. HWE, linkage-disequilibrium and haplotypes were achieved via Haploview version 4.2 program (D′ > 0.5 and r2 > 0.33 indicate strong linkage disequilibrium). All P values were bidirectional, and P < 0.05 was considered statistically significant. Odds ratio (OR) and 95% confidence intervals (CIs) of different groups or subgroups were calculated using the unconditional logistic regression analysis.
3. Results
3.1. Demographics
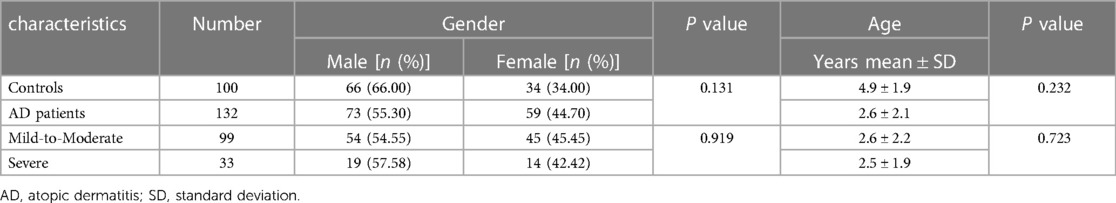
The detailed demographics of 132 AD patients and 100 controls were provided in Table 2. There were 73 males and 59 females with an average age of 2.6 ± 2.1 years in the case group, including 99 (75.00%) mild-to-moderate AD and 33 (25.00%) severe AD. The healthy controls were composed of 66 boys and 34 girls with an average age of 4.9 ± 1.9 years. No significant differences were found in term of gender and age either between the cases and controls, or between the mild-to-moderate subgroup and the severe subgroup.
3.2. Association between genetic polymorphisms and AD susceptibility
The successful genotyping rates of six SNPs were 94.4%–100%. The genotype distributions of all six SNPs in different groups or subgroups were all in agreement with HWE.
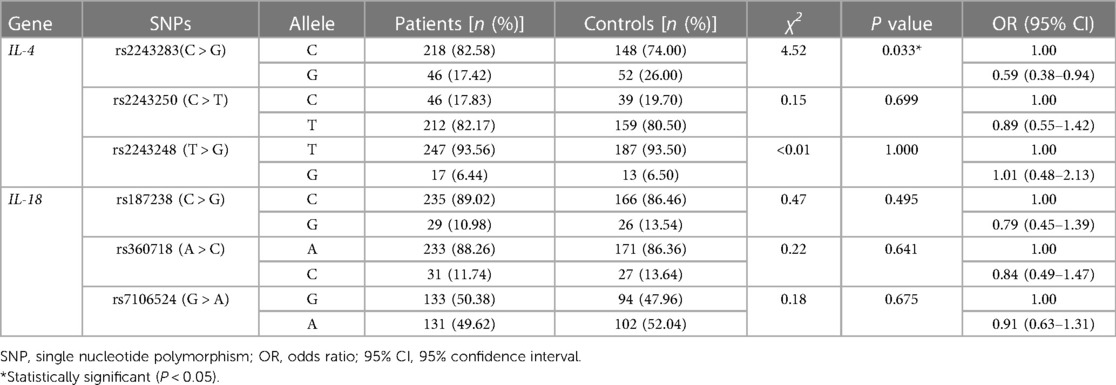
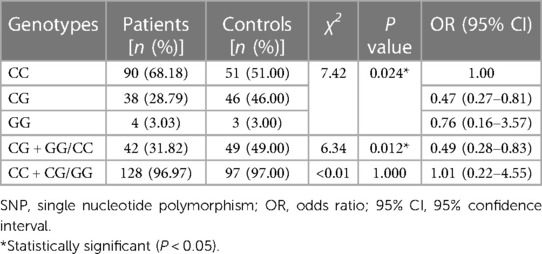
As shown in Table 3, the frequency of the mutant G allele of IL-4 rs2243283 was significantly lower in AD patients comparing to controls (G vs. C: P = 0.033, OR = 0.59, 95% CI = 0.38–0.94). Accordingly, we found that carriers with CG genotype of IL-4 rs2243283 had a statistically decreased risk of AD than carriers with CC genotype (P = 0.024; OR = 0.47, 95% CI = 0.27–0.81). Furthermore, the risk of AD susceptibility also reduced when CG and GG genotypes were analyzed together (CG + GG vs. CC: P = 0.012, OR = 0.49, 95% CI = 0.28–0.83) (Table 4). No significant differences in allele or genotype frequencies of the other five SNPs were found between AD patients and controls.
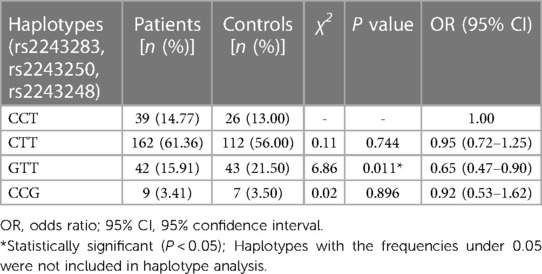
In haplotype analysis, the three SNPs of IL-4 were located in the same haplotype block with a state of linkage disequilibrium (D′ > 0.5, r2 > 0.33). Comparing to the haplotype CCT composed of IL-4 rs2243283, rs2243250 and rs2243248, the frequency of haplotype GTT in AD patients was significantly lower than that in controls (GTT vs. CCT: P = 0.011, OR = 0.65, 95% CI = 0.47–0.90) (Table 5). The frequencies of the other two haplotypes CTT and CCG did not differ between AD cases and controls.
3.3. Association between genetic polymorphisms and AD severity
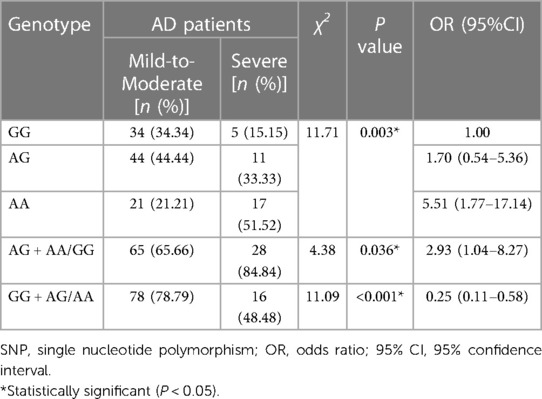
As shown in Table 6, the mutant A allele of IL-18 rs7106524 was associated with a higher risk of severe AD compared with the wild G allele (P < 0.001; OR = 2.79, 95% CI = 1.55–5.03). Consistent with the results of allele analysis, the carriers with AA genotype of IL-18 rs7106524 had an increased risk of severe AD as compared to those with GG genotype (P = 0.003; OR = 5.51, 95% CI = 1.77–17.14). Similar risk trends were observed when AA and AG genotypes were analyzed together (AG + AA vs. GG: P = 0.036, OR = 2.93, 95% CI = 1.04–8.27) (Table 7). Additionally, the frequency of rs7106524 GG + AG genotype was significantly lower than that in AA genotype (P < 0.001; OR = 0.25, 95% CI: 0.11–0.58). No correlation was found between the other five SNPs and the severity of AD.
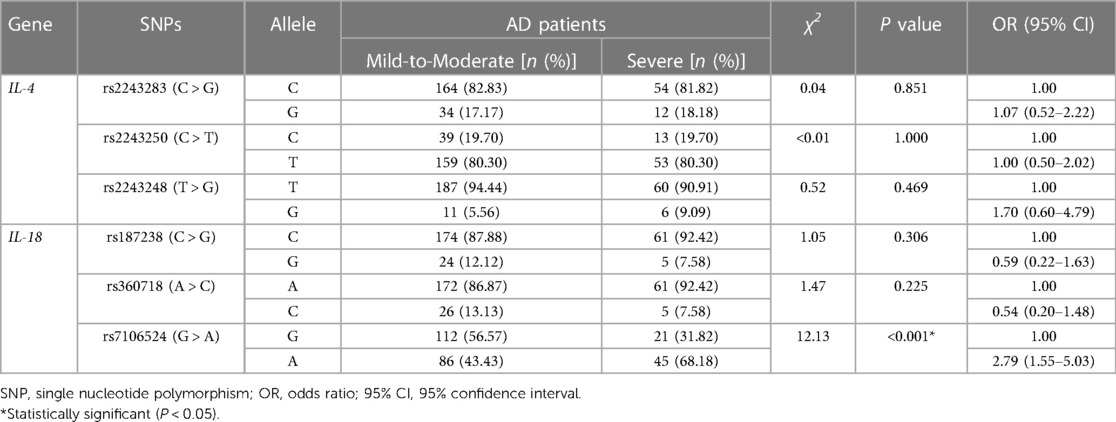
Table 6. Allele frequencies of six SNPs of IL-4 and IL-18 in patients with mild-to-moderate and severe AD.
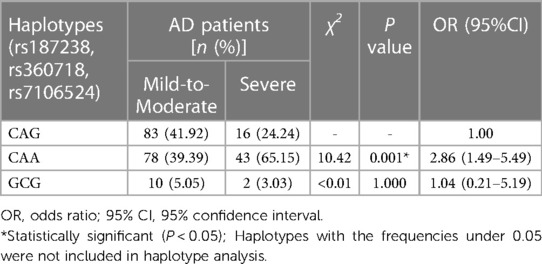
Furthermore, the haplotypes consisting of IL-18 rs187238, rs360718 and rs7106524 were analyzed for their association with AD severity. The three SNPs of IL-18 were also located in the same haplotype block with a state of linkage disequilibrium (D′ > 0.5, r2 > 0.33). As shown in Table 8, the haplotype CAA was related to an increased risk of severe AD contrasted with the wild haplotype CAG (P = 0.001; OR = 2.86, 95% CI = 1.49–5.49). Another haplotype GCG did not present a correlation to AD severity.
4. Discussion
AD has always been a hot research topic owing to its high prevalence, recurrence, disability and cost of care (26). Over the past few decades, knowledge of AD has evolved tremendously with insights into pathogenesis, epidemiology, impact of disease, and new therapies (27). Genetic predisposition, epidermal dysfunction, and T-cell driven inflammation are thought to be involved in the complex pathophysiology of AD (1). Among these, genetic predisposition has been reported as the strongest identifiable risk factor and an important contributor influencing the disease incidences and development of AD (1). The current literature has provided evidence for the relationship between genetic polymorphisms of relevant mediators and AD, and thus far, more than 34 AD-related genic causes have been identified, including FLG, IL-13, RAD50, LRRC32, IL2/IL21, PRR5l, CLEC16A/DEXI, ZNF652, RUNX3, ERBB3, IL6R, CD207, PPP2R3C, IL-7R, STAT3, ZBTB10 and TLR4, etc (1, 16, 17).
In this case-control study, the results demonstrated the association of IL-4 gene polymorphisms with the susceptibility to AD in Chinese Han children. The data showed that the mutant G allele, CG genotype and CG + GG genotype of IL-4 rs2243283 were associated with a significantly reduced risk of AD. We also found a correlation between IL-4/GTT haplotype composed of rs2243283, rs2243250, rs2243248 and lower AD risk. As reported, rs2243283 gene variants have been confirmed to correlate to some chronic inflammatory and immune diseases, such as asthma, chronic periodontitis, and autoimmune hepatitis (22). However, as we know, rare studies focused on the linkage of this SNP with AD. Therefore, our data might provide a new risk predictor for estimating AD predisposition. Except for rs2243283, the other two SNPs of IL-4 had no relationship with AD in the present study. IL-4 rs2243250 (−590C/T) is a common locus in researches on the association of IL-4 gene polymorphisms with AD. But the findings regarding the correlation between IL-4 rs2243250 and AD susceptibility are controversial. Consistent with our results were the investigations conducted in the Saudi and Macedonian pediatric population (18, 19). Inconsistent with our data were the surveys carried out among Czech children and another subset of Chinese pediatric participants (28, 29). Chinese academic Shang H reported that the 590 T and 589 T alleles may be related to the increased risk of AD, based on a case-control study that enrolled 82 AD patients and 100 healthy controls (29). Whether the discrepancy between our data and the above result is due to different sample sizes remains to be clarified by further investigations with larger numbers of participants. The contradictions in these findings also might be attributed to different genetic backgrounds and environments (30). The larger studies in populations with the same ethnic background are needed to confirm or refute these results. Additionally, no link of IL-4 genetic polymorphisms to AD severity was found in this study. With the exception of one report published in 2002 that indicated the IL-4 590 T allele was associated with the severity of AD (31), several researches in recent years showed no relevance between IL-4 rs2243250 and AD severity (29, 30).
Besides the etiology diagnosis of AD, studies on prediction of the AD severity remains challenging. Several inflammation-related gene polymorphisms are attached with the severity of AD and could be used to predict potential severity (16, 32, 33). IL-18 is a glycoprotein encoded by the IL-18 gene, which has been mapped to chromosome 11q22.2-q22.3 (34). And its length is approximately 21.61 kb (34). Although serum level of IL-18 was regarded as a possible biomarker for the severity of AD (35), there was few publications so far on the association between IL-18 genetic variations and AD severity. The present study was conducted for the first time in Chinese Han children and revealed the relation of IL-18 polymorphisms to AD severity. Our data explored that A allele, AA genotype and AG + AA genotype of IL-18 rs7106524, as well as IL-18/CAA haplotype consisting of rs187238, rs360718 and rs7106524 were associated with a high risk of severe AD, suggesting the genetic polymorphisms of IL-18 rs7106524 might be used as potential predictors of AD severity. Consequently, the results may be useful in making appropriate therapy recommendations and preventing dermatitis development. Nevertheless, the functions of the investigated polymorphisms are still not well elucidated, so further studies are still needed to explore the potential molecular mechanisms underlying the association between IL-18 rs7106524 polymorphism and AD severity.
In addition, no linkage between IL-18 genetic polymorphisms and susceptibility to AD was found, inconsistent with several previous investigations. For example, the merged quantitative analysis in a recent meta-analysis study displayed that IL-18 rs187238 polymorphisms might affect the risk of AD in overall population (36). This discrepancy between studies might still be due to the sample size and ethnicity. Future multicenter studies with a large sample size will be carried out to further explore the relationship between genetic variation in cytokines and AD. Besides, whether nucleotide polymorphisms in IL-4 and IL-18 genes affect alterations in amino acid sequences and protein secondary structure, and whether they subsequently influence expression levels and protein functions, deserve deeper consideration and require prospective work to elucidate. Perhaps, in-depth cell-specific transcriptomic investigation will guide better understanding of cellular impact and underlying mechanisms of the IL-4 and IL-18 signaling pathways, and then lead to better drug targets for AD treatment.
5. Conclusion
In summary, the results of the current study revealed that the CG genotype of IL-4 rs2243283 and the IL-4/GTT haplotype confer the decreased susceptibility to AD in Chinese pediatric population. And IL-18 rs7106524 AA genotype and the IL-18/CAA haplotype might serve as predictors of high risk for severe AD in Chinese children.
Data availability statement
The data presented in the study are deposited in the figshare repository, accession number https://doi.org/10.6084/m9.figshare.22776002.v1.
Ethics statement
The studies involving human participants were reviewed and approved by Institutional Ethics Committee of the Children's Hospital, Zhejiang University School of Medicine. Written informed consent to participate in this study was provided by the participants’ legal guardian/next of kin.
Author contributions
JS and LH completed the experimental part of the study, performed data analysis and interpretation, drafted the manuscript. HZ, WL, and RT contributed to the study conception and design. HZ and YL collected patients’ information. RT supervised the entire study and provided academic guidance throughout the study process. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by Zhejiang Provincial Natural Science Foundation of China (LY21H040002) and Zhejiang Province Research Project of Public Welfare Technology Application (LGF22H110001).
Acknowledgments
We thank all the participants and staffs who help us in the process of this study.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Langan SM, Irvine AD, Weidinger S. Atopic dermatitis. Lancet. (2020) 396(10247):345–60. doi: 10.1016/S0140-6736(20)31286-1
2. Ramirez FD, Chen S, Langan SM, Prather AA, McCulloch CE, Kidd SA, et al. Assessment of sleep disturbances and exhaustion in mothers of children with atopic dermatitis. JAMA Dermatol. (2019) 155(5):556–63. doi: 10.1001/jamadermatol.2018.5641
3. Wang HN, Ji K, Zhang LN, Xie CC, Li WY, Zhao ZF, et al. Inhibition of c-fos expression attenuates IgE-mediated mast cell activation and allergic inflammation by counteracting an inhibitory AP1/Egr1/IL-4 axis. J Transl Med. (2021) 19(1):261. doi: 10.1186/s12967-021-02932-0
4. Liang Y, Chang C, Lu Q. The genetics and epigenetics of atopic dermatitis-filaggrin and other polymorphisms. Clin Rev Allergy Immunol. (2016) 51(3):315–28. doi: 10.1007/s12016-015-8508-5
5. Piazza S, Fumagalli M, Martinelli G, Pozzoli C, Maranta N, Angarano M, et al. Hydrolyzable tannins in the management of Th1, Th2 and Th17 inflammatory-related diseases. Molecules. (2022) 27(21):7593. doi: 10.3390/molecules27217593
6. Chiricozzi A, Maurelli M, Peris K, Girolomoni G. Targeting IL-4 for the treatment of atopic dermatitis. Immunotargets Ther. (2020) 9:151–6. doi: 10.2147/ITT.S260370
7. Pelly VS, Kannan Y, Coomes SM, Entwistle LJ, Rückerl D, Seddon B, et al. IL-4-producing ILC2s are required for the differentiation of TH2 cells following heligmosomoides polygyrus infection. Mucosal Immunol. (2016) 9(6):1407–17. doi: 10.1038/mi.2016.4
8. Tsuji G, Hashimoto-Hachiya A, Kiyomatsu-Oda M, Takemura M, Ohno F, Ito T, et al. Aryl hydrocarbon receptor activation restores filaggrin expression via OVOL1 in atopic dermatitis. Cell Death Dis. (2017) 8(7):e2931. doi: 10.1038/cddis.2017.322
9. Oetjen LK, Mack MR, Feng J, Whelan TM, Niu H, Guo CJ, et al. Sensory neurons co-opt classical immune signaling pathways to mediate chronic itch. Cell. (2017) 171(1):217–28. doi: 10.1016/j.cell.2017.08.006
10. Teraki Y, Hotta T, Shiohara T. Increased circulating skin-homing cutaneous lymphocyte-associated antigen (CLA)+ type 2 cytokine-producing cells, and decreased CLA+ type 1 cytokine-producing cells in atopic dermatitis. Br J Dermatol. (2000) 143(2):373–8. doi: 10.1046/j.1365-2133.2000.03665.x
11. Lee JH, Cho DH, Park HJ. IL-18 and cutaneous inflammatory diseases. Int J Mol Sci. (2015) 16(12):29357–69. doi: 10.3390/ijms161226172
12. Zedan K, Rasheed Z, Farouk Y, Alzolibani AA, Bin Saif G, Ismail HA, et al. Immunoglobulin e, interleukin-18 and interleukin-12 in patients with atopic dermatitis: correlation with disease activity. J Clin Diagn Res. (2015) 9(4):WC01-5. doi: 10.7860/JCDR/2015/12261.5742
13. Tanaka T, Tsutsui H, Yoshimoto T, Kotani M, Matsumoto M, Fujita A, et al. Interleukin-18 is elevated in the sera from patients with atopic dermatitis and from atopic dermatitis model mice, NC/Nga. Int Arch Allergy Immunol. (2001) 125(3):236–40. doi: 10.1159/000053821
14. Chen JL, Niu XL, Gao YL, Ma L, Gao XH, Chen HD, et al. IL-18 knockout alleviates atopic dermatitis-like skin lesions induced by MC903 in a mouse model. Int J Mol Med. (2020) 46(2):880–8. doi: 10.3892/ijmm.2020.4630
15. Babusikova E, Jurecekova J, Jesenak M, Evinova A. Association of gene polymorphisms in interleukin 6 in infantile bronchial asthma. Arch Bronconeumol. (2017) 53(7):381–6. doi: 10.1016/j.arbres.2016.09.012
16. Shi J, He L, Tao R, Zheng H, Li W, Huang S, et al. TLR4 Polymorphisms as potential predictors of atopic dermatitis in Chinese Han children. J Clin Lab Anal. (2022) 36:e24385. doi: 10.1002/jcla.24385
17. Bin L, Leung DY. Genetic and epigenetic studies of atopic dermatitis. Allergy Asthma Clin Immunol. (2016) 12:52. doi: 10.1186/s13223-016-0158-5
18. Stavric K, Peova S, Trajkov D, Spiroski M. Gene polymorphisms of 22 cytokines in Macedonian children with atopic dermatitis. Iran J Allergy Asthma Immunol. (2012) 11(1):37–50.22427475
19. Hussein YM, Alzahrani SS, Alharthi AA, Alhazmi AS, Ghonaim MM, Alghamdy AA, et al. Gene polymorphism of interleukin-4, interleukin-4 receptor and STAT6 in children with atopic dermatitis in taif, Saudi Arabia. Immunol Invest. (2016) 45(3):223–34. doi: 10.3109/08820139.2015.1135943
20. Ibrahim GH, ElTabbakh MT, Gomaa AH, Mohamed EA. Interleukin-18 gene polymorphisms in Egyptian patients with allergic diseases. Am J Rhinol Allergy. (2012) 26(5):385–9. doi: 10.2500/ajra.2012.26.3806
21. Gao SJ, Zhang L, Lu W, Wang L, Chen L, Zhu Z, et al. Interleukin-18 genetic polymorphisms contribute differentially to the susceptibility to crohn’s disease. World J Gastroenterol. (2015) 21(28):8711–22. doi: 10.3748/wjg.v21.i28.8711
22. Jin T, Zhang Y, Sun Y, Wu J, Xiong Z, Yang Z. IL-4 gene polymorphisms and their relation to steroid-induced osteonecrosis of the femoral head in Chinese population. Mol Genet Genomic Med. (2019) 7(3):e563. doi: 10.1002/mgg3.563
23. Al-Naseri MA, Salman ED, Ad'Hiah AH. Association between interleukin-4 and interleukin-10 single nucleotide polymorphisms and multiple sclerosis among Iraqi patients. Neurol Sci. (2019) 40(11):2383–9. doi: 10.1007/s10072-019-04000-4
24. Yao X, Song ZQ, Li W, Liang YS, Zhao Y, Cao H, et al. Guidelines for diagnosis and treatment of atopic dermatitis in China (2020). Int J Dermatol Venereol. (2021) 4(1):1–9. doi: 10.1097/JD9.0000000000000143
25. Chen K, Zhou YX, Li K, Qi LX, Zhang QF, Wang MC, et al. A novel three-round multiplex PCR for SNP genotyping with next generation sequencing. Anal Bioanal Chem. (2016) 408(16):4371–7. doi: 10.1007/s00216-016-9536-6
26. Silverberg JI. Public health burden and epidemiology of atopic dermatitis. Dermatol Clin. (2017) 35(3):283–9. doi: 10.1016/j.det.2017.02.002
27. Tracy A, Bhatti S, Eichenfield LF. Update on pediatric atopic dermatitis. Cutis. (2020) 106(3):143–6. doi: 10.12788/cutis.0077
28. Kayserova J, Sismova K, Zentsova-Jaresova I, Katina S, Vernerova E, Polouckova A, et al. A prospective study in children with a severe form of atopic dermatitis: clinical outcome in relation to cytokine gene polymorphisms. J Investig Allergol Clin Immunol. (2012) 22(2):92–101.22533231
29. Shang H, Cao XL, Wan YJ, Meng J, Guo LH. IL-4 Gene polymorphism may contribute to an increased risk of atopic dermatitis in children. Dis Markers. (2016) 2016:1021942. doi: 10.1155/2016/1021942
30. Liang J, Liu Y, Xue R, Chen L, Chen H, Shao L, et al. Interleukin 4 -590C/T (rs2243250) polymorphism is associated with increased risk of atopic dermatitis: meta-analysis of case-control studies. Dermatitis. (2017) 28(2):144–51. doi: 10.1097/DER.0000000000000265
31. Söderhäll C, Bradley M, Kockum I, Luthman H, Wahlgren CF, Nordenskjöld M. Analysis of association and linkage for the interleukin-4 and interleukin-4 receptor b;alpha; regions in Swedish atopic dermatitis families. Clin Exp Allergy. (2002) 32(8):1199–202. doi: 10.1046/j.1365-2745.2002.01452.x
32. Gleń J, Trzeciak M, Sobjanek M, Bandurski T, Wilkowska A, Nedoszytko B, et al. Interleukin-13 promoter gene polymorphism -1112 C/T is associated with atopic dermatitis in Polish patients. Acta Dermatovenerol Croat. (2012) 20(4):231–8.
33. Wilkowska A, Gleń J, Zabłotna M, Trzeciak M, Ryduchowska M, Sobjanek M, et al. The association of GM-CSF -677A/C promoter gene polymorphism with the occurrence and severity of atopic dermatitis in a Polish population. Int J Dermatol. (2014) 53(3):e172–4. doi: 10.1111/ijd.12245
34. Al-Shobaili HA, Ahmed AA, Alnomair N, Alobead ZA, Rasheed Z. Molecular genetic of atopic dermatitis: an update. Int J Health Sci (Qassim). (2016) 10(1):96–120.27004062
35. Thijs J, Krastev T, Weidinger S, Buckens CF, de Bruin-Weller M, Bruijnzeel-Koomen C, et al. Biomarkers for atopic dermatitis: a systematic review and meta-analysis. Curr Opin Allergy Clin Immunol. (2015) 15(5):453–60. doi: 10.1097/ACI.0000000000000198
Keywords: interleukin-4, interleukin-18, atopic dermatitis, polymorphisms, susceptibility, inflammation
Citation: Shi J, He L, Zheng H, Li W, Huang S, Li Y and Tao R (2023) Association of IL-4 and IL-18 genetic polymorphisms with atopic dermatitis in Chinese children. Front. Pediatr. 11:1202100. doi: 10.3389/fped.2023.1202100
Received: 7 April 2023; Accepted: 10 May 2023;
Published: 30 May 2023.
Edited by:
Jordi Pérez-Tur, Spanish National Research Council (CSIC), SpainReviewed by:
Dorota Pastuszak-Lewandoska, Medical University of Lodz, PolandSulev Kõks, Murdoch University, Australia
© 2023 Shi, He, Zheng, Li, Huang, Li and Tao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ran Tao Y2hydGFvQHpqdS5lZHUuY24=
†These authors have contributed equally to this work and share first authorship
 Jianrong Shi1,†
Jianrong Shi1,† Lin He
Lin He Huiwen Zheng
Huiwen Zheng Wei Li
Wei Li Ran Tao
Ran Tao