
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Pediatr. , 07 July 2023
Sec. Pediatric Infectious Diseases
Volume 11 - 2023 | https://doi.org/10.3389/fped.2023.1198278
 Julia C. Bennett1,2*
Julia C. Bennett1,2* Anne Emanuels1
Anne Emanuels1 Jessica Heimonen1
Jessica Heimonen1 Jessica O'Hanlon1
Jessica O'Hanlon1 James P. Hughes3
James P. Hughes3 Peter D. Han4,5
Peter D. Han4,5 Eric J. Chow1,2,6
Eric J. Chow1,2,6 Constance E. Ogokeh5,7
Constance E. Ogokeh5,7 Melissa A. Rolfes7
Melissa A. Rolfes7 Christine M. Lockwood4,8
Christine M. Lockwood4,8 Brian Pfau4,9
Brian Pfau4,9 Timothy M. Uyeki7
Timothy M. Uyeki7 Jay Shendure4,9
Jay Shendure4,9 Samara Hoag10
Samara Hoag10 Kairsten Fay11
Kairsten Fay11 Jover Lee11
Jover Lee11 Thomas R. Sibley11
Thomas R. Sibley11 Julia H. Rogers1,2
Julia H. Rogers1,2 Lea M. Starita4,9
Lea M. Starita4,9 Janet A. Englund12
Janet A. Englund12 Helen Y. Chu1
Helen Y. Chu1
Background: Respiratory viruses might influence Streptococcus pneumoniae nasal carriage and subsequent disease risk. We estimated the association between common respiratory viruses and semiquantitative S. pneumoniae nasal carriage density in a household setting before and during the COVID-19 pandemic.
Methods: From November 2019–June 2021, we enrolled participants in a remote household surveillance study of respiratory pathogens. Participants submitted weekly reports of acute respiratory illness (ARI) symptoms. Mid-turbinate or anterior nasal swabs were self-collected at enrollment, when ARI occurred, and, in the second year of the study only, from household contacts after SARS-CoV-2 was detected in a household member. Specimens were tested using multiplex reverse-transcription PCR for respiratory pathogens, including S. pneumoniae, rhinovirus, adenovirus, common human coronavirus, influenza A/B virus, respiratory syncytial virus (RSV) A/B, human metapneumovirus, enterovirus, and human parainfluenza virus. We estimated differences in semiquantitative S. pneumoniae nasal carriage density, estimated by the inverse of S. pneumoniae relative cycle threshold (Crt) values, with and without viral detection for any virus and for specific respiratory viruses using linear generalized estimating equations of S. pneumoniae Crt values on virus detection adjusted for age and swab type and accounting for clustering of swabs within households.
Results: We collected 346 swabs from 239 individuals in 151 households that tested positive for S. pneumoniae (n = 157 with and 189 without ≥1 viruses co-detected). Difficulty breathing, cough, and runny nose were more commonly reported among individuals with specimens with viral co-detection compared to without (15%, 80% and 93% vs. 8%, 57%, and 51%, respectively) and ear pain and headache were less commonly reported (3% and 26% vs. 16% and 41%, respectively). For specific viruses among all ages, semiquantitative S. pneumoniae nasal carriage density was greater with viral co-detection for enterovirus, RSV A/B, adenovirus, rhinovirus, and common human coronavirus (P < 0.01 for each). When stratified by age, semiquantitative S. pneumoniae nasal carriage density was significantly greater with viral co-detection among children aged <5 (P = 0.002) and 5–17 years (P = 0.005), but not among adults aged 18–64 years (P = 0.29).
Conclusion: Detection of common respiratory viruses was associated with greater concurrent S. pneumoniae semiquantitative nasal carriage density in a household setting among children, but not adults.
Streptococcus pneumoniae remains an important cause of morbidity and mortality among children and adults globally despite the availability of effective pneumococcal conjugate vaccines (PCVs) covering ≥10 S. pneumoniae serotypes (1, 2). S. pneumoniae nasopharyngeal carriage is a necessary precursor of invasive pneumococcal disease (IPD), although disease risk is influenced by a combination of host, pathogen, and environmental factors and in the majority of cases pneumococcal carriage does not result in disease (3).
Respiratory virus infection is one host factor influencing S. pneumoniae carriage and disease risk. Influenza virus infection and risk of increased pneumococcal carriage density and disease has been well-established and investigated in animal (4–6), modeling (7), ecological (8–20), and individual-level epidemiologic (21, 22) studies. Ecologic and epidemiologic studies have also associated infection with respiratory syncytial virus (RSV) and, to a lesser extent, rhinovirus, adenovirus, human metapneumovirus, and human parainfluenza virus with increased pneumococcal disease risk (10–12, 15, 17–20, 23–26).
Increased pneumococcal carriage density has been found to be associated with multiple common respiratory viruses, including rhinovirus, adenovirus, influenza virus, RSV, and common human coronavirus, although viruses and populations investigated varied across studies. Most studies were among children with acute respiratory illness (ARI) or pneumonia (21, 27–30), fewer studies included asymptomatic children (30–32), and one study to our knowledge included adults with ARI (21).
To date, studies describing the association between respiratory viruses and S. pneumoniae carriage have been conducted primarily among children and most have evaluated a small number of respiratory viruses co-detected. We aimed to estimate the association between eight common respiratory viruses and semiquantitative S. pneumoniae carriage density and to describe symptom profiles with and without viral co-detection among individuals of all ages in a community-based surveillance study among households in Seattle, Washington, USA before and during the COVID-19 pandemic.
From November 2019–June 2021, a convenience sample of participants consented and enrolled in a prospective, longitudinal household surveillance study of respiratory pathogens in the Seattle metropolitan area as part of the Seattle Flu Study (33). The study design, recruitment, eligibility, and data collection have been previously described (34). Briefly, households with ≥3 individuals and ≥1 child aged 3 months through 17 years were eligible and recruited from elementary and middle schools. Household members of all ages were eligible. One adult, designated as the household reporter, submitted weekly reports online of ARI symptoms for the entire household. ARI was defined as new or worsening acute cough or the presence of two or more other respiratory symptoms (sore throat, muscle or body aches, headache, fatigue, ear pain, sweats, fever, runny nose, chills, difficulty breathing, nausea, rash, and diarrhea).
Mid-turbinate (used during year 1 of the study from November 2019–July 2020) or anterior nasal (used during year 2 of the study from August 2020–June 2021) swabs were self-collected or collected by a parent or guardian at home. Self-swabs were collected at enrollment for all household members and once per participant when each ARI occurred. Additionally, in the second year of the study only (from August 2020–June 2021), when SARS-CoV-2 was detected among an enrolled household member, both symptomatic and asymptomatic household contacts collected self-swabs 4, 6, 8, 10, 12 and 14 days following the positive SARS-CoV-2 case. Among symptomatic individuals, clinical data were collected from individuals or a parent or guardian at swab collection and a one-week follow-up questionnaire collected data on symptoms, illness impact on school and/or work, and health care seeking. Symptom data were not collected for baseline swabs at enrollment or for household contacts of SARS-CoV-2 cases. Data were collected using Research Electronic Data Capture (REDCap) (35, 36).
Home self-collected swabs were mailed to the Northwest Genomics Center at the University of Washington at ambient temperature in universal transport media during year 1 of the study and as dry swabs in year 2 of the study. Respiratory pathogens were detected using arrayed reverse-transcription polymerase chain reaction (RT-qPCR) following total nucleic acids extraction (37). The assay has been assessed for accuracy of S. pneumoniae detection against control samples, used proprietary primer sets, and the target gene for identifying S. pneumoniae-positive specimens was hflB (Thermo Fisher Scientific assay ID Ba06439619_s1) (38). The array contained assays specific to enterovirus to avoid rhinovirus cross-reactivity but the rhinovirus assay had cross-reactivity with enterovirus and some rhinovirus detections may reflect enterovirus. Specimens with both rhinovirus and enterovirus detected were considered positive for both viruses in this analysis. Also, in year 2 of the study the rhinovirus assay was expanded to include additional rhinovirus types. S. pneumoniae relative cycle threshold (Crt) values, an alternative to cycle threshold values that is used specifically by the OpenArry platform, were based on the amplification curve only rather than all curves for a specific target (39). We used the inverse of S. pneumoniae Crt values as an estimation of semiquantitative S. pneumoniae nasal carriage density. S. pneumoniae serotyping was not performed.
The University of Washington Institutional Review Board approved this study. All participants completed informed consent (or assent and parental or guardian consent for participants <18 years of age at enrollment).
We described the proportion of all swabs collected with S. pneumoniae and/or any virus detected over the study period overall and by age group. All other analyses only included results from swabs with S. pneumoniae detected with or without the following respiratory viruses: rhinovirus, adenovirus, common human coronavirus, influenza A/B virus, RSV A/B, human metapneumovirus, enterovirus, and human parainfluenza virus. From January 1, 2020 onward, specimens were additionally tested for SARS-CoV-2. We considered individuals to be symptomatic if they reported any of the following at either sample collection or on the one-week follow-up questionnaire: difficulty breathing, chills, sweats, cough, ear pain, fatigue, fever, headache, muscle and/or body aches, runny nose, or sore throat.
We conducted a cross-sectional analysis of swabs from testing events with S. pneumoniae detected with a respiratory virus co-detected (cases) compared to S. pneumoniae-positive swabs without a respiratory virus co-detected (controls). We estimated the difference in S. pneumoniae Crt values with and without viral co-detection for any virus and for specific respiratory viruses using linear generalized estimating equations (GEE) of S. pneumoniae Crt values on virus detection adjusted for continuous age and swab type (mid-turbinate vs. anterior nasal) and accounting for clustering of persons with positive swabs within households (40). Analyses were adjusted for swab type due to the change in swabs used during the study period. For sensitivity analyses we (1) estimated differences in S. pneumoniae Crt values with and without viruses detected for symptomatic testing events and additionally adjusted for days since symptom onset, (2) stratified the analysis by pre- and post-implementation of COVID-19 restrictions (beginning in March 2020), and (3) estimated differences in S. pneumoniae Crt values with and without viruses detected excluding repeat swabs conducted by participants within 30 days. Secondary analyses included estimating differences in S. pneumoniae Crt values with and without any virus by participant age category (children aged <5 years, aged 5–17 years, and adults aged 18–64 years) and differences in S. pneumoniae Crt values with and without influenza A/B virus adjusted for current season influenza vaccination status at the time of testing. Analyses were conducted in R (R-4.1.1, R Core Team, 2021).
From November 2019–June 2021 we collected 4630 swabs from 1,700 unique individuals in 437 households. Of these, 346 (8%) were positive for S. pneumoniae and 773 (17%) were positive for any respiratory virus (excluding SARS-CoV-2). Viral detection was greatest during the 2019–early 2020 winter season, declined substantially following COVID-19 pandemic restrictions beginning in March 2020, began to increase during winter 2021, and remained high during the spring and summer months of 2021 through the end of the study in June 2021. Conversely, detection of S. pneumoniae was lower and declined to a lesser degree during the COVID-19 pandemic. Among all swabs collected (excluding swabs from household contacts of SARS-CoV-2 cases, which were collected in the second year of the study only), 30% of swabs were positive for any virus (excluding SARS-CoV-2) and 12% were positive for S. pneumoniae prior to March 2020 vs. 10% and 6%, respectively, from March–December 2020 and 21% and 5%, respectively, from January–June 2021, although these proportions are not adjusted for the change from mid-turbinate to anterior nasal swabs or the age distribution over time (Supplementary Figure S1). Trends in viral and S. pneumoniae detection over the study period were generally similar by age group and the age distribution did not change meaningfully over the study period. One exception was that S. pneumoniae detection was greatest among children and prior to the COVID-19 pandemic, the proportion of swabs with any virus only was similar to the proportion with S. pneumoniae and any virus for children aged <5 years, whereas for older children and adults a lower proportion of pre-COVID-19 pandemic swabs had S. pneumoniae with any virus co-detected relative to any virus only (Supplementary Figure S2). Although we detected SARS-CoV-2 in 35 specimens, none of these had S. pneumoniae and SARS-CoV-2 co-detected.
Subsequent analyses were restricted to 346 swabs from 239 unique individuals in 151 households that tested positive for S. pneumoniae (n = 157 swabs (45%) with and 189 (55%) without one or more viruses detected) (Table 1). The median time between swab collection for individuals with multiple swabs included in the analysis was 42 (range: 1–540 days, interquartile range: 14–103). Approximately half (49%) of participants were male, majority (79%) were White, and most were children (40% children aged <5 years, 31% children aged 5–17 years, and 29% adults aged 18–64 years). Over half (68%) of included households had a child aged <5 years and median household size was 4 individuals (range: 3–7). During the entire study period a median of one swab (range: 1–11) per participant and one swab (1–17) per household were positive for S. pneumoniae. Over half (57%) of S. pneumoniae-positive swabs without viral co-detection were baseline swabs collected at enrollment, relative to 31% of swabs with viral codetection and 19% of swabs with and 12% of swabs without viral co-detection were collected from household contacts of SARS-CoV-2 positive cases.
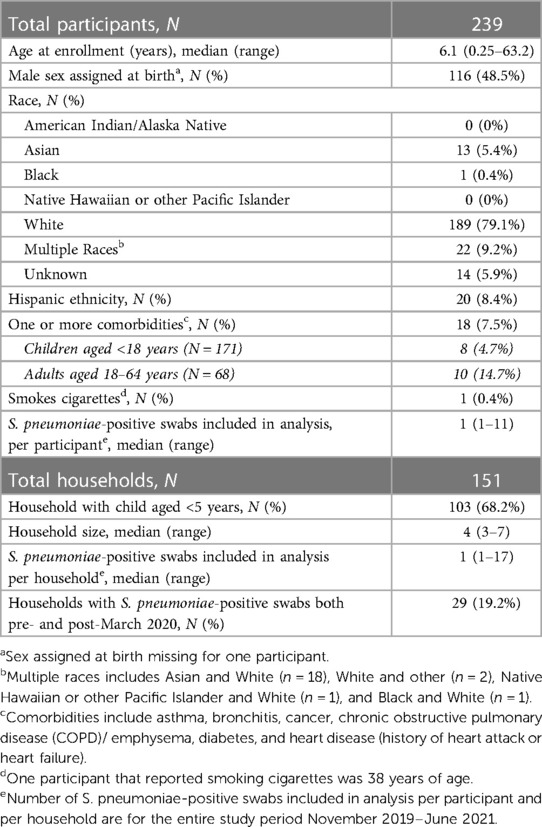
Table 1. Demographic characteristics of unique participants and households that tested positive for S. pneumoniae ≥1 times from November 2019–June 2021.
Among all S. pneumoniae-positive swabs, rhinovirus (n = 87; 25%) was the most frequently detected virus, followed by common human coronavirus [n = 30 (9%)], adenovirus [n = 23 (7%)], influenza A/B virus [n = 8 influenza A and n = 9 influenza B virus (5%)], RSV A/B [n = 9 RSV A and n = 5 RSV B (4%)], human metapneumovirus (n = 6 (2%)), human parainfluenza [n = 5 (1%)], and enterovirus [n = 4 (1%)]. Among swabs with S. pneumoniae and at least one virus, only 17% had ≥2 viruses co-detected (25 with 2 viruses co-detected and 1 with 3 and 4 viruses co-detected each) and these were mostly (69%) among children aged <5 years. The distribution of viruses co-detected with S. pneumoniae varied by age group (Supplementary Figure S3). The proportion of all S. pneumoniae-positive swabs with common human coronavirus detected was greater among adults relative to children and no swabs among adults had S. pneumoniae co-detected with human parainfluenza.
Among symptomatic individuals, days since symptom onset were similar for specimens with and without viral co-detection (median: 3 days, Table 2). Among individuals reporting any symptoms, difficulty breathing, cough, and runny rose were more commonly reported among specimens with viral co-detection (15%, 80% and 93% vs. 8%, 57%, and 51%, respectively) whereas ear pain and headache were less commonly reported among specimens with compared to without viral co-detection (3% and 26% vs. 16% and 41%, respectively). Fatigue, fever, chills or sweats, muscle and/or body aches, and sore throat were similar between specimens with and without viral co-detection (Table 2 and Supplementary Figure S4). Reported symptoms profiles differed by age group (Supplementary Table S1). Cough, fever and runny nose were more commonly reported among symptomatic children aged <5 (75%, 59%, and 90%, respectively) and 5–17 years (77%, 63%, and 71%) compared to adults aged 18–64 years (60%, 36%, and 68%) while difficulty breathing, headache, and muscle and/or body aches were more commonly reported among adults (24%, 68%, and 40%) compared to children aged <5 (12%, 6%, and 10%, respectively) and 5–17 years (6%, 40%, and 23%). Ear pain and fatigue were reported for a similar proportion of symptomatic adults (12% and 76%) and children aged 5–17 years (11% and 74%), relative to younger children (2% and 47%).
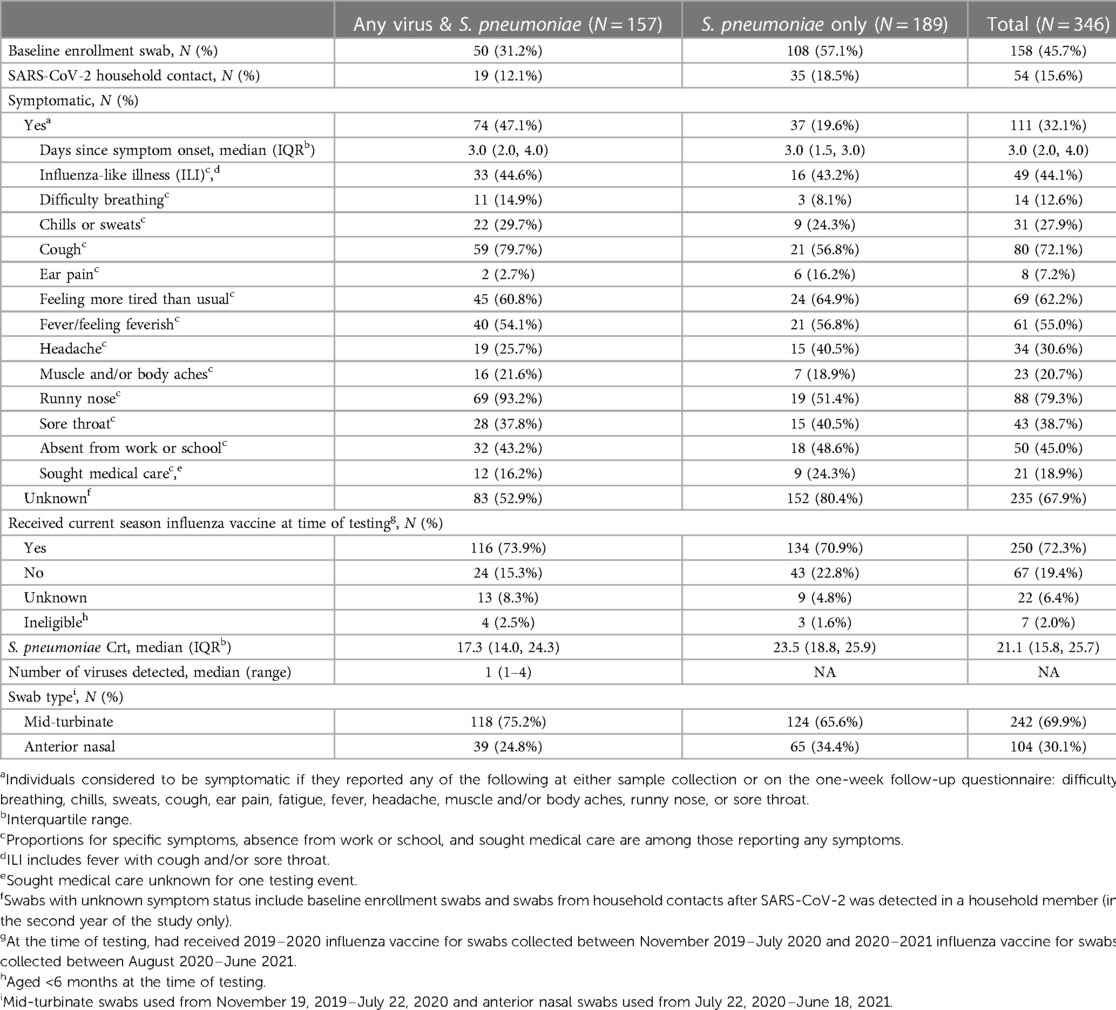
Table 2. Characteristics of participants, symptomatic events, and testing results included in analysis.
Similar proportions of testing events resulted in absence from work or school (43% vs. 49% for swabs with and without viral co-detection, respectively) and medical care seeking (16% vs. 24%) among those reporting any symptoms. Among those eligible and with known influenza vaccination status, 83% of specimens with and 76% of specimens without viral co-detection were from individuals that had received that current season's influenza vaccine at the time of sample collection. Due to the change in swab type during the study, 75% of specimens with S. pneumoniae and any virus detected were mid-turbinate swabs compared to 66% of specimens with only S. pneumoniae detected.
On average, S. pneumoniae Crt values were 2.67 units (95% CI: 1.55, 3.80, P < 0.001) lower (i.e., semiquantitative nasal carriage density was higher) for testing events with compared to without viral co-detection adjusted for age and swab type (Figure 1 and Supplementary Table S2). Of specific viruses evaluated, S. pneumoniae semiquantitative carriage density was higher with viral co-detection for enterovirus, RSV A/B, adenovirus, rhinovirus, and common human coronavirus compared to swabs without viral co-detection (P < 0.01 for each). No significant differences in S. pneumoniae semiquantitative carriage density were detected with or without influenza A/B virus, human parainfluenza and human metapneumovirus or between mid-turbinate and anterior nasal swabs. S. pneumoniae semiquantitative carriage density decreased significantly with age (P < 0.001). When analyses were stratified by age category, S. pneumoniae Crt values were significantly lower (corresponding to higher S. pneumoniae semiquantitative carriage density) with compared to without viral co-detection among children aged <5 years (2.52 units, 95% CI: 0.93, 4.11, P = 0.002) and 5–17 years (2.96 units, 0.90, 5.02, P = 0.005), but not among adults aged 18–64 years (1.08 units, −0.90, 3.05, P = 0.29) (Figure 2 and Supplementary Table S2).
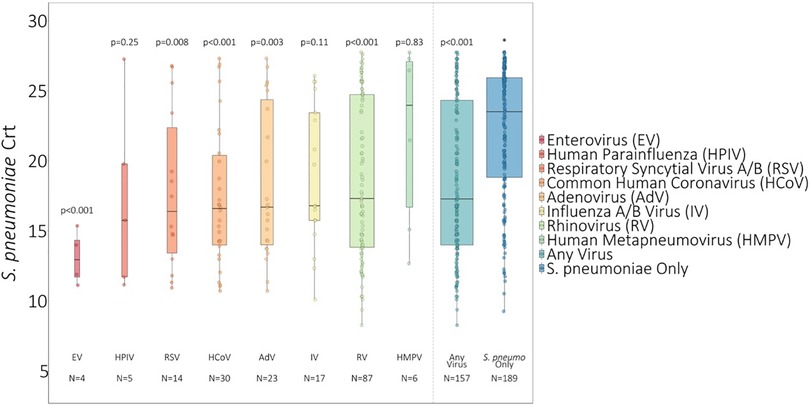
Figure 1. S. pneumoniae Crt values of swabs with and without respiratory viruses detected1. 1 Influenza A/B virus incudes n = 8 influenza A virus and n = 9 influenza B virus. RSV A/B includes n = 9 RSV A and n = 5 RSV B. * P-values comparing S. pneumoniae carriage Crt values with and without viral detection estimated using GEE of S. pneumoniae carriage Crt on virus detection adjusted for age and swab type and accounting for clustering of swabs within households. Width of boxplots corresponds to number of swabs.
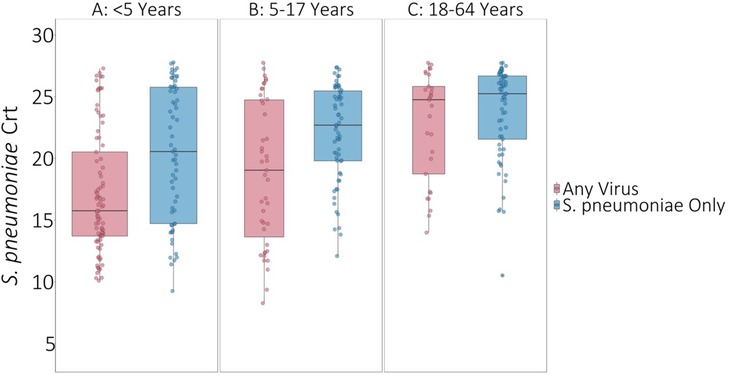
Figure 2. S. pneumoniae Crt values with and without respiratory viruses detected by age category. * P-values comparing S. pneumoniae carriage Crt values with and without viral detection estimated using GEE of S. pneumoniae carriage Crt on virus detection adjusted for age and swab type and accounting for clustering of swabs within households. Width of boxplots corresponds to number of swabs.
Results were similar when restricting to symptomatic episodes. The only exception was human parainfluenza virus, where S. pneumoniae semiquantitative carriage density was significantly higher with human parainfluenza virus detection for specimens collected from symptomatic individuals (P < 0.001) but not for all specimens (P = 0.25), although among symptomatic individuals, human parainfluenza virus was only detected in one specimen. No difference in S. pneumoniae semiquantitative carriage density was detected by days since symptom onset (Supplementary Figure S5 and Supplementary Table S2). Results were also similar between the period prior to and post-implementation of COVID-19 restrictions and when excluding repeat swabs conducted by participants within 30 days (Supplementary Figures S6–S7 and Supplementary Table S2). No difference in S. pneumoniae semiquantitative carriage density was detected comparing individuals who had vs. had not received the current season influenza vaccine at the time of testing when comparing specimens with and without influenza A/B virus (Supplementary Table S2).
In a study of remote self-collection of nasal swab specimens in families with young children in the household setting, our analysis found that multiple common respiratory viruses were associated with greater concurrent S. pneumoniae semiquantitative nasal carriage density among children but not adults. Results were similar when restricting to specimens from symptomatic individuals. We also found that symptom profiles differed between individuals with specimens with and without viral co-detection, although we are unable to ascribe the etiology of the symptoms for individuals with swabs without any viruses detected. Overall, difficulty breathing, cough, and runny rose were reported more commonly among specimens with viral co-detection whereas ear pain and headache were reported less commonly among specimens with viral co-detection compared to specimens without. Although we tested for SARS-CoV-2, none of the SARS-CoV-2-positive specimens had S. pneumoniae co-detected.
Our findings of greater S. pneumoniae semiquantitative nasal carriage density with any viral co-detection among children are consistent with another US study in healthy children (30) and our findings for specific viruses are broadly consistent with other settings, although most studies evaluated only a few respiratory viruses. In this study, lower S. pneumoniae Crt values (corresponding to higher semiquantitative nasal carriage density) were associated with enterovirus, RSV A/B, adenovirus, rhinovirus, and common human coronavirus; we did not detect a difference for influenza A/B virus, human parainfluenza, or human metapneumovirus. Our findings for rhinovirus, adenovirus, and common human coronavirus are consistent with other studies which have also associated these viruses with increased pneumococcal carriage density (21, 27–29, 31, 32). Findings have been variable for RSV, which was associated with increased pneumococcal carriage density in this study, among hospitalized children in Vietnam (28), and among children with ARI in Kenya (29), but not among healthy children attending daycare in the UK (32). Influenza virus infection is a long-established risk factor for IPD (8, 9, 14, 22) but we did not find influenza A/B virus detection to be significantly associated with lower S. pneumoniae Crt values in this study. This finding for influenza virus is not consistent with the Vietnam study among hospitalized children (28) and the South Africa study among individuals of all ages hospitalized with ARI (21), but is consistent with the UK study of healthy children attending daycare, which found no association with influenza virus (32). Variability across studies, however, may be attributable to differences in study populations (e.g., healthy individuals vs. those with ARI) and sample collection methods (e.g., nasal vs. nasopharyngeal swabs). Similar to the UK study among children attending day care, we did not detect a difference in pneumococcal carriage density for human parainfluenza virus. None of these prior studies evaluated the association with enterovirus, which we found to be associated with higher semiquantative pneumococcal carriage density, or human metapneumovirus, which we did not find to be associated with semiquantative pneumococcal carriage density.
Our study found that viral co-detection was associated with increased S. pneumoniae semiquantitative nasal carriage density among children aged <5 years and 5–17 years, but not among adults aged 18–64 years and that increasing age was associated with lower S. pneumoniae semiquantitative nasal carriage density overall. One other study investigating S. pneumoniae carriage and viral co-detections among individuals of all ages hospitalized with ARI in South Africa found that adults had less pneumococcal nasopharyngeal colonization compared to children overall, but this study did not estimate the association between viral detection and carriage by age group. In addition, carriage and IPD serotype distributions, IPD disease burden, and pneumococcal vaccine uptake differ between the US and South Africa and thus these populations may not be directly comparable (21). The age effect observed in this study may be related to lower detection of S. pneumoniae among adults compared to children as S. pneumoniae carriage density is generally lower among adults and we used nasal rather than more sensitive nasopharyngeal swabs and did not collect oropharyngeal swabs among adults, as recommended by the WHO (41, 42). Alternatively, this may reflect differences in the processes leading to increased pneumococcal carriage between children and adults, a potential impact of a different serotype distribution between children and adults, differences in prior S. pneumoniae infection and vaccination histories, or differences in the distribution of respiratory viruses co-detected. Importantly, although eligible, our analysis did not include any adults aged ≥65 years and this pattern may not be consistent among older adults at higher risk for IPD (43).
This study was conducted over two years from November 2019–June 2021. Similar to other reports, we observed a COVID-19 pandemic nonpharmaceutical interventions-associated decline in respiratory viruses in 2020 and an out-of-season resurgence of respiratory viruses during the spring and summer months of 2021 while pneumococcal carriage declined to a lesser degree over the study period (44–49). This pattern was observed in both Israel and France, where surveillance data showed declines in the incidence of both respiratory viruses, such as RSV and influenza, and IPD following COVID-19 pandemic nonpharmaceutical interventions but no significant change in pneumococcal carriage rates, suggesting that the observed decline in IPD may be attributable to the decline in respiratory viruses rather than a change in pneumococcal carriage rates (44, 45). Our finding that RSV and common human coronavirus were associated with greater concurrent S. pneumoniae semiquantitative nasal carriage density is consistent with this hypothesis that infection with common respiratory viruses may increase risk for pneumococcal disease. However, enterovirus, adenovirus, and rhinovirus, which we found to be associated with greater concurrent S. pneumoniae semiquantitative nasal carriage density in this study, all persisted during the COVID-19 pandemic. Nonpharmaceutical interventions may have had a lesser impact on enterovirus, adenovirus, and rhinovirus due to these being non-enveloped viruses that are environmentally stable, have prolonged shedding, and are often asymptomatic or only mildly symptomatic (50). Therefore, the association between respiratory virus infection and pneumococcal disease risk may be pathogen-specific.
There were several limitations to our study. First, we cannot rule out that differences in pneumococcal carriage density found in our study are due to other underlying differences between individuals, such as genetic, immune, and environmental factors, including pneumococcal vaccination. In a study of children with ARI in Kenya where samples were taken before, during, and after viral infection, RSV and rhinovirus were associated with increased pneumococcal nasopharyngeal carriage density, but this variation was small relative to the variation in carriage density observed across individuals, leading the authors to conclude that viral co-infection may play only a small role in pneumococcal carriage density (29). Our study did not routinely collect more than one specimen per illness episode and thus we are unable to similarly estimate the variation in carriage density between individuals, evaluate changes in pneumococcal carriage density over time, or assess risk for subsequent pneumococcal disease. Second, symptom data were not collected from individuals at baseline or from household contacts of SARS-CoV-2 positive cases and thus we did not collect specimens from any individuals who confirmed not having any ARI symptoms at the time of sample collection. We therefore could not compare differences in the impact of viral co-detection on pneumococcal carriage density between symptomatic, pre-symptomatic, and asymptomatic individuals or between those with viral infection vs. those with viral detection. Our self-reported symptom data may also have underestimated symptoms that are not directly observable among young children. For example, ear pain and headache were infrequently reported among symptomatic children aged <5 years relative to older children. Third, we tested self-collected mid-turbinate and anterior nasal swabs rather than nasopharyngeal swabs, which are the current gold standard for detecting pneumococcal carriage in children, and we did not collect nasopharyngeal in addition to oropharyngeal swabs among adults, the current gold standard (41). Therefore, we likely underestimated detection of S. pneumoniae, such as in low S. pneumoniae density specimens (51). Indeed, our overall positivity rate for S. pneumoniae was very low (8%). In addition, rhinorrhea induced by viruses may have contributed to greater S. pneumoniae density in these relatively superficial samples. Specimen quality may have also been variable due to self-collection, however we have previously shown this method of self-collection to result in a high rate of adequate specimen quality (37). We also relied on S. pneumoniae Crt values as a proxy of semiquantitative carriage density rather than using the more precise gene copies/mL and our assay for detecting S. pneumoniae used proprietary primer sets and therefore data regarding the accuracy of the assay, including in comparison to real world nasopharyngeal samples with culture-based detection, are limited. Fourth, we switched from mid-turbinate to anterior nasal swabs for the second year of the study, although we were able to adjust for swab type in our analyses and did not find a difference in S. pneumoniae Crt values by swab type. Fifth, we tested for ten (and analyzed eight) common respiratory viruses and therefore cannot rule out that some swabs with S. pneumoniae only detected did not also contain a respiratory virus for which we did not test and cannot determine the etiology of symptoms for individuals with swabs without respiratory viruses detected. Some rhinovirus detections may also reflect enteroviruses due to cross-reactivity in our assay. Sixth, due to relatively small sample sizes we conducted stratified analyses for all viruses combined by age group rather than testing for interactions between age and viral co-detection in the main analysis. Seventh, our study population had relatively high rates of influenza vaccination and a very low rate of smoking and may not be generalizable to other populations. Eighth, our study included the period following implementation of nonpharmaceutical interventions to limit transmission of COVID-19, when expected seasonal peaks in respiratory viruses did not occur (50) and overall detection of viruses in the study was low, particularly during this period. Despite this, we did not find a meaningful difference in results when analyses were stratified by pre- and post-implementation of COVID-19 restrictions. Finally, S. pneumoniae positive specimens were not serotyped and we did not collect data on pneumococcal vaccination. Therefore, we could not assess the potential role of pneumococcal vaccination or specific serotypes on the association between viral co-detection and pneumococcal carriage density, an important area for continued study as some S. pneumoniae serotypes have higher invasiveness potential (52–56) and virus-S. pneumoniae associations may be serotype-specific (4, 18, 28, 57).
Despite these limitations, this longitudinal, community surveillance study included both children and adults aged <65 years of age, allowing us to evaluate the association between viral co-detection and pneumococcal carriage by age, which to our knowledge has not been previously studied for most respiratory viruses included in this study. In addition, our study was conducted in a household setting whereas the majority of prior studies evaluating the relationship between S. pneumoniae carriage and viral co-detection have been among hospitalized individuals. We also included data from the pre-COVID-19 period, during the COVID-19 pandemic, and during the out-of-season resurgence of respiratory viruses following the lifting of pandemic restrictions. Our data included eight common respiratory pathogens allowing for virus-specific estimates of the association between virus co-detection and pneumococcal carriage. Finally, we collected detailed data on symptom profiles, allowing comparisons of swabs with and without viral co-detection that is not possible for many studies relying on routinely collected surveillance and laboratory data.
In conclusion, several common respiratory viruses were associated with greater concurrent S. pneumoniae semiquantitative nasal carriage density among children but not adults before and during the COVID-19 pandemic in a community-based household setting. These findings suggest that some common respiratory viruses may increase risk of subsequent pneumococcal disease in children. The potential impact of vaccines against respiratory viruses, including influenza vaccines and future RSV vaccines, for preventing pneumococcal disease should be explored.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
The studies involving human participants were reviewed and approved by The University of Washington Institutional Review Board. Written informed consent to participate in this study was provided by the participants or by the participants’ legal guardian/ next of kin for minors.
Conceived of study: JH, JH, CO, MR, JS, TU, LS, JE, HC. Designed study tools for data collection: JH, JO, AE, PH, CO, MR, TU, LS, JE, HC. Collected data or supervised data collection: JH, JO, AE, SH, JE, HC. Analyzed data or supervised analysis: JB, AE, EC, JR, JH, JE, HC. Wrote manuscript: JB, HC. Edited manuscript: JB, EC, CO, MR, JH, JR, TU, LS, JE, HC. All authors contributed to the article and approved the submitted version.
This work was funded by Gates Ventures and the Centers for Disease Control and Prevention.
This work was only possible through the contributions of study participants and the authors deeply appreciate the time and efforts of all participant volunteers as well as school partners who assisted with study recruitment. We also thank Ron Dagan, Faculty of Health Sciences, Ben-Gurion University of the Negev, Israel, for providing us with expert technical review and guidance on the interpretation of results.
HC reports consulting with Ellume, Merck, Abbvie, Pfizer, The Bill and Melinda Gates Foundation. She has received research funding from Sanofi Pasteur, and support and reagents from Ellume and Cepheid outside of the submitted work. JE reports consulting with Ark Biopharmaceuticals, Sanofi Pasteur, Moderna, Meissa Vaccines, Astra Zeneca, and Pfizer, Inc. outside of the submitted work, and has received research funding from AstraZeneca, GlaxoSmithKline, and Pfizer.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The funders were not involved in study design and do not have any ownership over the conduct of the study, data, or rights to publish. The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fped.2023.1198278/full#supplementary-material
1. Wahl B, O’Brien KL, Greenbaum A, Majumder A, Liu L, Chu Y, et al. Burden of Streptococcus pneumoniae and Haemophilus influenzae type b disease in children in the era of conjugate vaccines: global, regional, and national estimates for 2000–15. Lancet Glob Health. (2018) 6(7):e744–57. doi: 10.1016/S2214-109X(18)30247-X
2. World Health Organization. Pneumococcal Disease. Available at: https://www.who.int/teams/health-product-policy-and-standards/standards-and-specifications/vaccine-standardization/pneumococcal-disease (Accessed 12 Sep 2022).
3. Simell B, Auranen K, Käyhty H, Goldblatt D, Dagan R, O’Brien KL. The fundamental link between pneumococcal carriage and disease. Expert Rev Vaccines. (2012) 11(7):841–55. doi: 10.1586/erv.12.53
4. McCullers JA, McAuley JL, Browall S, Iverson AR, Boyd KL, Normark BH. Influenza enhances susceptibility to natural acquisition and disease from streptococcus pneumoniae in ferrets. J Infect Dis. (2010) 202(8):1287–95. doi: 10.1086/656333
5. Nakamura S, Davis KM, Weiser JN. Synergistic stimulation of type I interferons during influenza virus coinfection promotes streptococcus pneumoniae colonization in mice. J Clin Invest. (2011) 121(9):3657–65. doi: 10.1172/JCI57762
6. Diavatopoulos DA, Short KR, Price JT, Wilksch JJ, Brown LE, Briles DE, et al. Influenza A virus facilitates streptococcus pneumoniae transmission and disease. FASEB J. (2010) 24(6):1789–98. doi: 10.1096/fj.09-146779
7. Shrestha S, Foxman B, Weinberger DM, Steiner C, Viboud C, Rohani P. Identifying the interaction between influenza and pneumococcal pneumonia using incidence data. Sci Transl Med. (2013) 5(191):191ra84. doi: 10.1126/scitranslmed.3005982
8. Fleming-Dutra KE, Taylor T, Link-Gelles R, Garg S, Jhung MA, Finelli L, et al. Effect of the 2009 influenza A(H1N1) pandemic on invasive pneumococcal pneumonia. J Infect Dis. (2013) 207(7):1135–43. doi: 10.1093/infdis/jit008
9. Weinberger DM, Simonsen L, Jordan R, Steiner C, Miller M, Viboud C. Impact of the 2009 influenza pandemic on pneumococcal pneumonia hospitalizations in the United States. J Infect Dis. (2012) 205(3):458–65. doi: 10.1093/infdis/jir749
10. Chiavenna C, Presanis AM, Charlett A, de Lusignan S, Ladhani S, Pebody RG, et al. Estimating age-stratified influenza-associated invasive pneumococcal disease in England: a time-series model based on population surveillance data. PLoS Med. (2019) 16(6):e1002829. doi: 10.1371/journal.pmed.1002829
11. Nicoli EJ, Trotter CL, Turner KME, Colijn C, Waight P, Miller E. Influenza and RSV make a modest contribution to invasive pneumococcal disease incidence in the UK. J Infect. (2013) 66(6):512–20. doi: 10.1016/j.jinf.2013.02.007
12. Ciruela P, Broner S, Izquierdo C, Hernández S, Muñoz-Almagro C, Pallarés R, et al. Invasive pneumococcal disease rates linked to meteorological factors and respiratory virus circulation (catalonia, 2006–2012). BMC Public Health. (2016) 16:400. doi: 10.1186/s12889-016-3061-6
13. Walter ND, Taylor TH, Shay DK, Thompson WW, Brammer L, Dowell SF, et al. Influenza circulation and the burden of invasive pneumococcal pneumonia during a non-pandemic period in the United States. Clin Infect Dis. (2010) 50(2):175–83. doi: 10.1086/649208
14. Morens DM, Taubenberger JK, Fauci AS. Predominant role of bacterial pneumonia as a cause of death in pandemic influenza: implications for pandemic influenza preparedness. J Infect Dis. (2008) 198(7):962–70. doi: 10.1086/591708
15. Talbot TR, Poehling KA, Hartert TV, Arbogast PG, Halasa NB, Edwards KM, et al. Seasonality of invasive pneumococcal disease: temporal relation to documented influenza and respiratory syncytial viral circulation. Am J Med. (2005) 118(3):285–91. doi: 10.1016/j.amjmed.2004.09.016
16. Nelson GE, Gershman KA, Swerdlow DL, Beall BW, Moore MR. Invasive pneumococcal disease and pandemic (H1N1) 2009, Denver, Colorado, USA. Emerg Infect Dis. (2012) 18(2):208–16. doi: 10.3201/eid1802.110714
17. Jansen AG, Sanders EA, van der Ende A, van Loon AM, Hoes AW, Hak E. Invasive pneumococcal and meningococcal disease: association with influenza virus and respiratory syncytial virus activity? Epidemiol Infect. (2008) 136(11):1448–54. doi: 10.1017/S0950268807000271
18. Ampofo K, Bender J, Sheng X, Korgenski K, Daly J, Pavia AT, et al. Seasonal invasive pneumococcal disease in children: role of preceding respiratory viral infection. Pediatrics. (2008) 122(2):229–37. doi: 10.1542/peds.2007-3192
19. Zhou H, Haber M, Ray S, Farley MM, Panozzo CA, Klugman KP. Invasive pneumococcal pneumonia and respiratory virus co-infections. Emerg Infect Dis. (2012) 18(2):294–7. doi: 10.3201/eid1802.102025
20. Murdoch DR, Jennings LC. Association of respiratory virus activity and environmental factors with the incidence of invasive pneumococcal disease. J Infect. (2009) 58(1):37–46. doi: 10.1016/j.jinf.2008.10.011
21. Wolter N, Tempia S, Cohen C, Madhi SA, Venter M, Moyes J, et al. High nasopharyngeal pneumococcal density, increased by viral coinfection, is associated with invasive pneumococcal pneumonia. J Infect Dis. (2014) 210(10):1649–57. doi: 10.1093/infdis/jiu326
22. O’Brien KL, Walters MI, Sellman J, Quinlisk P, Regnery H, Schwartz B, et al. Severe pneumococcal pneumonia in previously healthy children: the role of preceding influenza infection. Clin Infect Dis. (2000) 30(5):784–9. doi: 10.1086/313772
23. Stensballe LG, Hjuler T, Andersen A, Kaltoft M, Ravn H, Aaby P, et al. Hospitalization for respiratory syncytial virus infection and invasive pneumococcal disease in Danish children aged <2 years: a population-based cohort study. Clin Infect Dis. (2008) 46(8):1165–71. doi: 10.1086/529438
24. Weinberger DM, Grant LR, Steiner CA, Weatherholtz R, Santosham M, Viboud C, et al. Seasonal drivers of pneumococcal disease incidence: impact of bacterial carriage and viral activity. Clin Infect Dis. (2014) 58(2):188–94. doi: 10.1093/cid/cit721
25. Peltola V, Heikkinen T, Ruuskanen O, Jartti T, Hovi T, Kilpi T, et al. Temporal association between rhinovirus circulation in the community and invasive pneumococcal disease in children. Pediatr Infect Dis J. (2011) 30(6):456–61. doi: 10.1097/INF.0b013e318208ee82
26. Weinberger DM, Klugman KP, Steiner CA, Simonsen L, Viboud C. Association between respiratory syncytial virus activity and pneumococcal disease in infants: a time series analysis of US hospitalization data. PLoS Med. (2015) 12(1):e1001776. doi: 10.1371/journal.pmed.1001776
27. Fan RR, Howard LM, Griffin MR, Edwards KM, Zhu Y, Williams JV, et al. Nasopharyngeal pneumococcal density and evolution of acute respiratory illnesses in young children, Peru, 2009–2011. Emerg Infect Dis. (2016) 22(11):1996–9. doi: 10.3201/eid2211.160902
28. Vu HTT, Yoshida LM, Suzuki M, Nguyen HAT, Nguyen CDL, Nguyen ATT, et al. Association between nasopharyngeal load of streptococcus pneumoniae, viral coinfection, and radiologically confirmed pneumonia in Vietnamese children. Pediatr Infect Dis J. (2011) 30(1):11–8. doi: 10.1097/INF.0b013e3181f111a2
29. Morpeth SC, Munywoki P, Hammitt LL, Bett A, Bottomley C, Onyango CO, et al. Impact of viral upper respiratory tract infection on the concentration of nasopharyngeal pneumococcal carriage among Kenyan children. Sci Rep. (2018) 8:11030. doi: 10.1038/s41598-018-29119-w
30. DeMuri GP, Gern JE, Eickhoff JC, Lynch SV, Wald ER. Dynamics of bacterial colonization with streptococcus pneumoniae, Haemophilus influenzae, and Moraxella catarrhalis during symptomatic and asymptomatic viral upper respiratory tract infection. Clin Infect Dis. (2018) 66(7):1045–53. doi: 10.1093/cid/cix941
31. Howard LM, Zhu Y, Griffin MR, Edwards KM, Williams JV, Gil AI, et al. Nasopharyngeal pneumococcal density during asymptomatic respiratory virus infection and risk for subsequent acute respiratory illness. Emerg Infect Dis. (2019) 25(11):2040–7. doi: 10.3201/eid2511.190157
32. Thors V, Christensen H, Morales-Aza B, Oliver E, Sikora P, Vipond I, et al. High-density bacterial nasal carriage in children is transient and associated with respiratory viral infections—implications for transmission dynamics. Pediatr Infect Dis J. (2019) 38(5):533–8. doi: 10.1097/INF.0000000000002256
33. Chu HY, Chu HY, Boeckh M, Boeckh M, Englund JA, Englund JA, et al. LB21. The seattle flu study: a community-based study of influenza. Open Forum Infect Dis. (2019) 6(Suppl 2):S1002. doi: 10.1093/ofid/ofz415.2504
34. Emanuels A, Heimonen J, O’Hanlon J, Kim AE, Wilcox N, McCulloch DJ, et al. Remote household observation for noninfluenza respiratory viral illness. Clin Infect Dis. (2020) 73(11):e4411–8. doi: 10.1093/cid/ciaa1719
35. Harris PA, Taylor R, Minor BL, Elliott V, Fernandez M, O’Neal L, et al. The REDCap consortium: building an international community of software platform partners. J Biomed Inform. (2019) 95:103208. doi: 10.1016/j.jbi.2019.103208
36. Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG. Research electronic data capture (REDCap)—a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform. (2009) 42(2):377–81. doi: 10.1016/j.jbi.2008.08.010
37. Kim AE, Brandstetter E, Wilcox N, Heimonen J, Graham C, Han PD, et al. Evaluating specimen quality and results from a community-wide, home-based respiratory surveillance study. J Clin Microbiol. (2021) 59(5):e02934–20. doi: 10.1128/JCM.02934-20
38. A complete workflow solution for detecting respiratory tract microbiota using OpenArray technology.
39. Thermo Fisher Scientific Inc. Crt, a relative threshold method for qPCR data analysis on the QuantStudio 12 K Flex system with OpenArray technology (2016).
40. Liang KY, Zeger SL. Longitudinal data analysis using generalized linear models. Biometrika. (1986) 73(1):13–22. doi: 10.1093/biomet/73.1.13
41. Satzke C, Turner P, Virolainen-Julkunen A, Adrian PV, Antonio M, Hare KM, et al. Standard method for detecting upper respiratory carriage of streptococcus pneumoniae: updated recommendations from the world health organization pneumococcal carriage working group. Vaccine. (2013) 32(1):165–79. doi: 10.1016/j.vaccine.2013.08.062
42. Roca A, Bottomley C, Hill PC, Bojang A, Egere U, Antonio M, et al. Effect of age and vaccination with a pneumococcal conjugate vaccine on the density of pneumococcal nasopharyngeal carriage. Clin Infect Dis. (2012) 55(6):816–24. doi: 10.1093/cid/cis554
43. Pilishvili T, Bennett NM. Pneumococcal disease prevention among adults: strategies for the use of pneumococcal vaccines. Vaccine. (2015) 33:D60–5. doi: 10.1016/j.vaccine.2015.05.102
44. Danino D, Ben-Shimol S, Van Der Beek BA, Givon-Lavi N, Avni YS, Greenberg D, et al. Decline in pneumococcal disease in young children during the COVID-19 pandemic in Israel associated with suppression of seasonal respiratory viruses, despite persistent pneumococcal carriage: a prospective cohort study. Clin Infect Dis. (2021) 75(1):e1154–e1164. doi: 10.1093/cid/ciab1014
45. Rybak A, Levy C, Angoulvant F, Auvrignon A, Gembara P, Danis K, et al. Association of nonpharmaceutical interventions during the COVID-19 pandemic with invasive pneumococcal disease, pneumococcal carriage, and respiratory viral infections among children in France. JAMA Netw Open. (2022) 5(6):e2218959. doi: 10.1001/jamanetworkopen.2022.18959
46. Olsen SJ, Winn AK, Budd AP, Prill MM, Steel J, Midgley CM, et al. Changes in influenza and other respiratory virus activity during the COVID-19 pandemic — United States, 2020–2021. MMWR Morb Mortal Wkly Rep. (2021) 70:1013–9. doi: 10.15585/mmwr.mm7029a1
47. Haddadin Z, Schuster JE, Spieker AJ, Rahman H, Blozinski A, Stewart L, et al. Acute respiratory illnesses in children in the SARS-CoV-2 pandemic: prospective multicenter study. Pediatrics. (2021) 148(2):e2021051462. doi: 10.1542/peds.2021-051462
48. Huang QS, Wood T, Jelley L, Jennings T, Jefferies S, Daniells K, et al. Impact of the COVID-19 nonpharmaceutical interventions on influenza and other respiratory viral infections in New Zealand. Nat Commun. (2021) 12(1):1001. doi: 10.1038/s41467-021-21157-9
49. Foley DA, Yeoh DK, Minney-Smith CA, Martin AC, Mace AO, Sikazwe CT, et al. The interseasonal resurgence of respiratory syncytial virus in Australian children following the reduction of coronavirus disease 2019–related public health measures. Clin Infect Dis. (2021) 73(9):e2829–30. doi: 10.1093/cid/ciaa1906
50. Chow EJ, Uyeki TM, Chu HY. The effects of the COVID-19 pandemic on community respiratory virus activity. Nat Rev Microbiol. (2022) 21(3):195–201. doi: 10.1038/s41579-022-00807-9
51. Grijalva CG, Griffin MR, Edwards KM, Johnson M, Gil AI, Verastegui H, et al. Concordance between RT-PCR-based detection of respiratory viruses from nasal swabs collected for viral testing and nasopharyngeal swabs collected for bacterial testing. J Clin Virol. (2014) 60(3):309–12. doi: 10.1016/j.jcv.2014.04.011
52. Varon E, Cohen R, Béchet S, Doit C, Levy C. Invasive disease potential of pneumococci before and after the 13-valent pneumococcal conjugate vaccine implementation in children. Vaccine. (2015) 33(46):6178–85. doi: 10.1016/j.vaccine.2015.10.015
53. Brueggemann AB, Peto TEA, Crook DW, Butler JC, Kristinsson KG, Spratt BG. Temporal and geographic stability of the serogroup-specific invasive disease potential of Streptococcus pneumoniae in children. J Infect Dis. (2004) 190(7):1203–11. doi: 10.1086/423820
54. Hanage WP, Kaijalainen TH, Syrjänen RK, Auranen K, Leinonen M, Mäkelä PH, et al. Invasiveness of serotypes and clones of Streptococcus pneumoniae among children in Finland. Infect Immun. (2005) 73(1):431–5. doi: 10.1128/IAI.73.1.431-435.2005
55. Shouval DS, Greenberg D, Givon-Lavi N, Porat N, Dagan R. Site-specific disease potential of individual Streptococcus pneumoniae serotypes in pediatric invasive disease, acute otitis media and acute conjunctivitis. Pediatr Infect Dis J. (2006) 25(7):602–7. doi: 10.1097/01.inf.0000220231.79968.f6
56. Cohen R, Levy C, Ouldali N, Goldrey M, Béchet S, Bonacorsi S, et al. Invasive disease potential of pneumococcal serotypes in children after PCV13 implementation. Clin Infect Dis. (2021) 72(8):1453–6. doi: 10.1093/cid/ciaa917
57. Greenberg D, Givon-Lavi N, Faingelernt Y, Ben-Shimol S, Avni YS, Bar-Ziv J, et al. Nasopharyngeal pneumococcal carriage during childhood community-acquired alveolar pneumonia: relationship between specific serotypes and coinfecting viruses. J Infect Dis. (2017) 215(7):1111–6. doi: 10.1093/infdis/jiw613
Keywords: streptococcus pneumoniae, pneumococcal, nasal carriage, respiratory, viruses, COVID-19 pandemic
Citation: Bennett JC, Emanuels A, Heimonen J, O'Hanlon J, Hughes JP, Han PD, Chow EJ, Ogokeh CE, Rolfes MA, Lockwood CM, Pfau B, Uyeki TM, Shendure J, Hoag S, Fay K, Lee J, Sibley TR, Rogers JH, Starita LM, Englund JA and Chu HY (2023) Streptococcus pneumoniae nasal carriage patterns with and without common respiratory virus detections in households in Seattle, WA, USA before and during the COVID-19 pandemic. Front. Pediatr. 11:1198278. doi: 10.3389/fped.2023.1198278
Received: 31 March 2023; Accepted: 23 June 2023;
Published: 7 July 2023.
Edited by:
Josette Raymond, Hôpital de Bicêtre, FranceReviewed by:
Robert Cohen, Activ, Association Clinique et Thérapeutique Infantile du Val-de-Marne, France© 2023 Bennett, Emanuels, Heimonen, O'Hanlon, Hughes, Han, Chow, Ogokeh, Rolfes, Lockwood, Pfau, Uyeki, Shendure, Hoag, Fay, Lee, Sibley, Rogers, Starita, Englund and Chu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Julia C. Bennett jubenn94@uw.edu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.