- 1The First Affiliated Hospital of Henan University of Chinese Medicine, Zhengzhou, China
- 2School of Pediatrics, Henan University of Chinese Medicine, Zhengzhou, China
Background: The etiology of idiopathic scoliosis (IS) remains unclear. Gene-based studies on genetic etiology and molecular mechanisms have improved our understanding of IS and guided treatment and diagnosis. Therefore, it is imperative to explicate and demarcate the preponderant areas of inquiry, key scholars, and their aggregate scholarly output, in addition to the collaborative associations amongst publications or researchers.
Methods: Documents were retrieved from the Web of Science Core Collection (WoSCC) with the following criteria: TS = (“idiopathic scoliosis” AND gene) refined by search operators (genomic OR “hereditary substance” OR “germ plasm” OR Cistrons OR genetics OR genetic OR genes OR Polygenic OR genotype OR genome OR allele OR polygenes OR Polygene) AND DOCUMENT TYPES (ARTICLE OR REVIEW), and the timespan of 2002-01-01 to 2022-11-26. The online bibliometric analysis platform (bibliometric), bibliographic item co-occurrence matrix builder (BICOMB), CiteSpace 6.1. R6 and VOS viewer were used to evaluate articles for publications, nations, institutions, journals, references, knowledge bases, keywords, and research hotspots.
Results: A total of 479 documents were retrieved from WoSCC. Fourty-four countries published relevant articles. The country with the most significant number of articles was China, and the institution with the most significant number of articles was Nanjing University. Citation analysis formed eight meaningful clusters and 16 high-frequency keywords. (2) The citation knowledge map included single nucleotide polymorphisms, whole exome sequencing, axonal dynamin, drug development, mesenchymal stem cells, dietary intake, curve progression, zebrafish development model, extracellular matrix, and rare variants were the current research hotspots and frontiers.
Conclusions: Recent research has focused on IS-related genes, whereas the extracellular matrix and unusual variants are research frontiers and hotspots. Functional analysis of susceptibility genes will prove to be valuable for identifying this disease.
1. Introduction
Idiopathic scoliosis (IS) is a three-dimensional spinal deformity in the coronal, sagittal, and horizontal planes that occurs without underlying vertebral abnormalities or significant physiological defects. The pathogenesis of IS remains unknown and poses a considerable challenge to its prevention and treatment (1–3). Human genetic studies have revealed the existence of complex polygenic diseases with genetics and phenotypes (4–6). The consistency of IS estimates ranged from 0.13 to 0.73 with monozygotic twins and from 0.00 to 0.36 with dizygotic twins (7). Autosomal dominant inheritance, x-linked dominant inheritance, and autosomal recessive inheritance models have been reported in the literature (8). IS is significantly associated with Single Nucleotide Polymorphism (SNPs) in several genes in multiple study populations, such as Melatonin receptor 1B (MTNR1B), Estrogen receptor1 (ESR1), and Matrilin 1 (MATN1) (9). Despite the vast amount of research conducted on IS-related genes, the specific mechanism underlying altered spine anatomy remains elusive (10). Several types of pathogenic gene mutations have been identified in idiopathic scoliosis through genetic studies, including a group of the TGFB1 gene and the MiR4300HG gene that are related to the severity of the curve; a category of LBX1, NTF3 and SOCS3 that can predict progress curves; and a cohort of susceptibility loci in SOX9, MATN1, AJAP1, MMP9 and TIMP2 associated with cartilage and extracellular matrix (11, 12).
Based on these findings, idiopathic scoliosis has been extensively studied genetically, particularly regarding potential pathogenic mechanisms and susceptibility genes. However, it is unclear how the current research situation has developed. Bibliometric analysis is a method that employs objective statistical analysis to quantify the data characteristics and temporal trends of scientific literature pertinent to a given research project (13). Compared with traditional expert-written systematic reviews, bibliometric analyses provide a timely, intuitive, and unbiased way to track developments and explore specific areas of knowledge. In this study, we investigated Science Citation Index Expanded (SCIE) papers on IS-related gene studies and established a visual knowledge map of the area based on the cited references. Furthermore, we identified research hotspots using burst keywords. The above investigations provide researchers with a macro- and micro-level understanding of the entire field of knowledge.
2. Methods
2.1. Database
To ensure the stability of the search results, all data were downloaded from the Web of Science Core Collection (WoSCC) on Nov 26, 2022.
2.2. Search strategy
Based on the keyword search strategy, the search was constructed using keywords such as "idiopathic scoliosis" and "gene", and the types of filtered articles included "articles" and "reviews'. By limiting the search period to Nov 26, 2022, two researchers (LR and WL) independently confirmed the results. Detailed search strategies are presented in Table 1.
2.3. Data collection
After completing the literature search, we exported the results to a complete recording format for analysis. We collected the following fundamental data for each article: title, abstract, authors, organization, nation, journal, keywords, and references. Two researchers (LR and WL) manually screened the imported data source files using EndNote. The inclusion criteria for the literature are as follows: (1) The literature must be a study on genes or gene mutations related to human idiopathic scoliosis; (2) The publication date must be between January 1, 2002 and November 26, 2022; (3) The article must be published in the SCIE database and have a certain degree of academic authority; (4) The literature must be of high-quality research types, such as original research, review, meta-analysis, clinical trial, systematic evaluation, etc.; (5) The literature must have reliability and reproducibility, including clear research objectives, methods, results, conclusions, and sufficient data support. Exclusion criteria include: (1) Articles that are not related to genes or gene mutations related to human idiopathic scoliosis; (2) Conference abstracts, book chapters, data papers, editorial materials, and reproductions that are unrelated to the topic; (3) Publications that were published earlier than 2002 or after November 26, 2022, or unpublished documents that cannot obtain full-text or do not have enough information for further analysis. Finally, 27 articles were excluded (Figure 1).
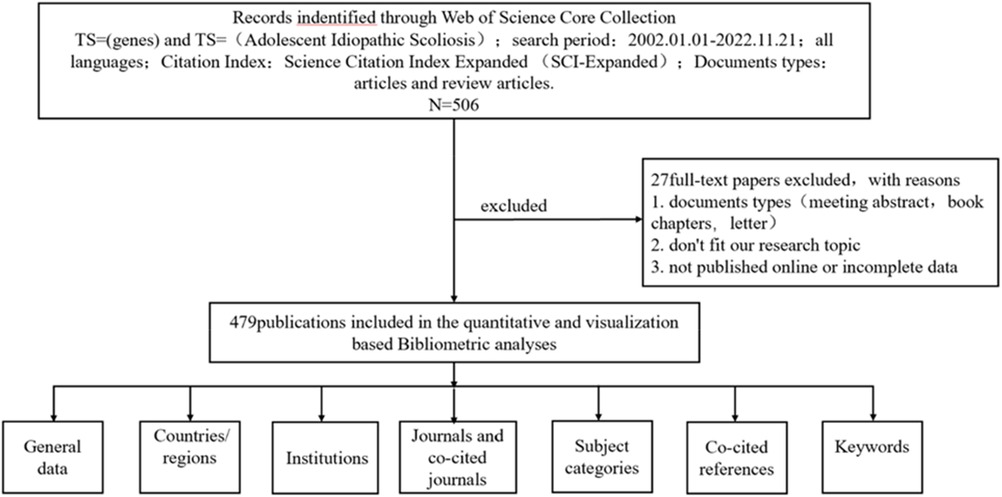
Figure 1. A frame flow diagram. The diagram displayed details selection criteria for Idiopathic Scoliosis gene publications from the WoSCC database and the steps of bibliometric analysis.
2.4. Data analysis
All aspects of publications were analyzed, including countries, institutions, journals, and keywords. An online bibliometric analysis platform (http://bibliometric.com/) and VOSviewer (University Leiden Netherlands) were used for co-occurrence analysis, and the results were displayed using collaborative network maps. The basic version of Citespace 6.1. R6 software (14) was used to analyze journals, references, and keywords. We set the operating parameters to # Years Per Slice = 1, TopN = 100, Top N% = 5%, and K = 10 by using "T" button to cluster and label the reference, after which the centrality of the nodes was calculated. The most-cited articles are then displayed.
3. Results
3.1. Distribution of literature by year of publication
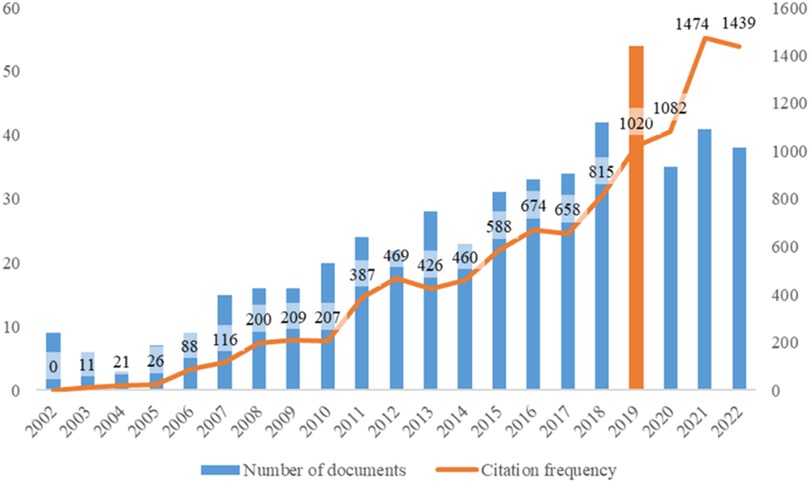
A total of 479 items were included in this topic from 2002 to Nov 26, 2022. Idiopathic scoliosis gene studies peaked in 2019, and the frequency peaked in 2021 (Figure 2).
3.2. Countries/regions and institutions
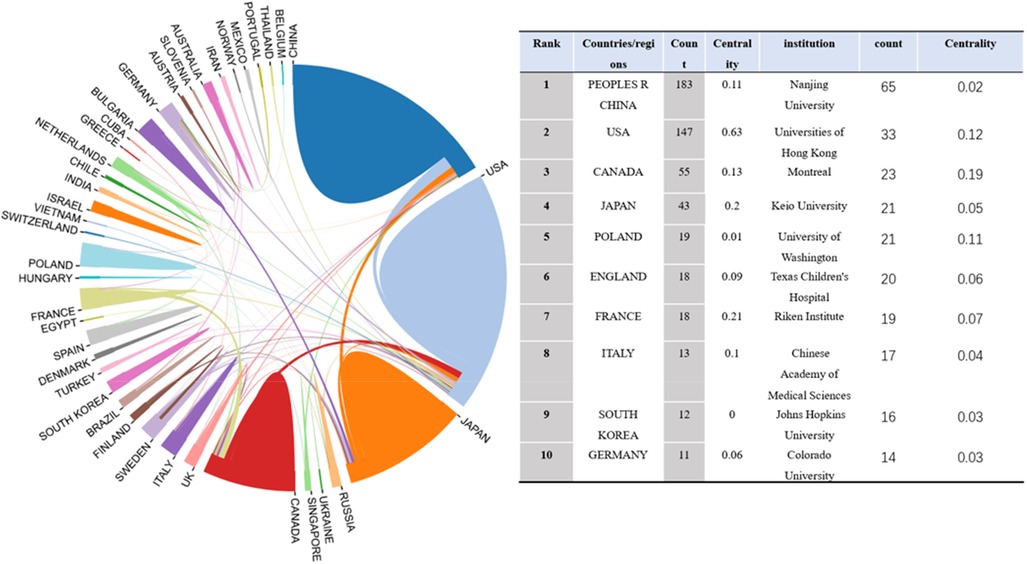
Between 2002 and Nov 26, 2022, studies on idiopathic scoliosis gene research were published by 44 nations/regions, and these nations/regions cooperated. China published the most articles (183, 38.2%), followed by the United States (147, 30.6%), Canada (55), and Japan (43) Figure 3). Most publications were published by Nanjing University (65), followed by Universities of Hong Kong (33), Montreal (23), and Keio University (21) (Figure 4).

Figure 4. Network map of institutions contributed to publications on idiopathic scoliosis-genes from 2002 to 2022.
3.3. Analysis of journals
The citation relationship in an academic journal represents the flow of knowledge in this research field. The data of the cited journals display the number of citations received by the journal in the JCR year, which reflects academic status. Table 2 summarizes the details of the top 10 journals cited. The collaborative networks of these journals are revealed in Figure 5. The most cited journal was SPINE (432), followed by BONE JOINT SURG AM (292). The bispectrum overlays of the journals are displayed in Figure 6. The left side shows cited journals, and the right side shows cited journals. The colored paths indicate citation relationships. Molecular/biology/genetics are often cited by molecular/biology/immunology and neurology/sports/ophthalmology (Figure 6).
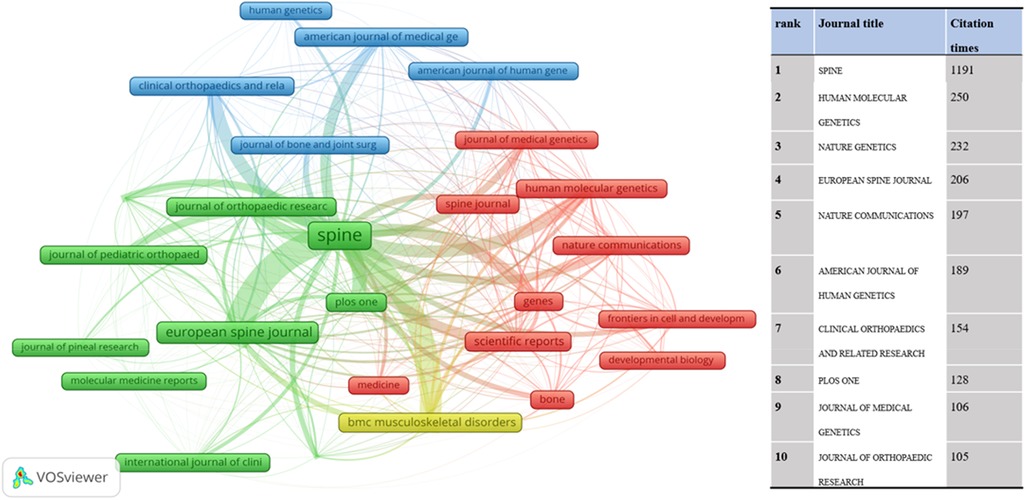
Figure 5. The network map of the journals cited contributed to publications on the idiopathic scoliosis genes from 2002 to 2022.
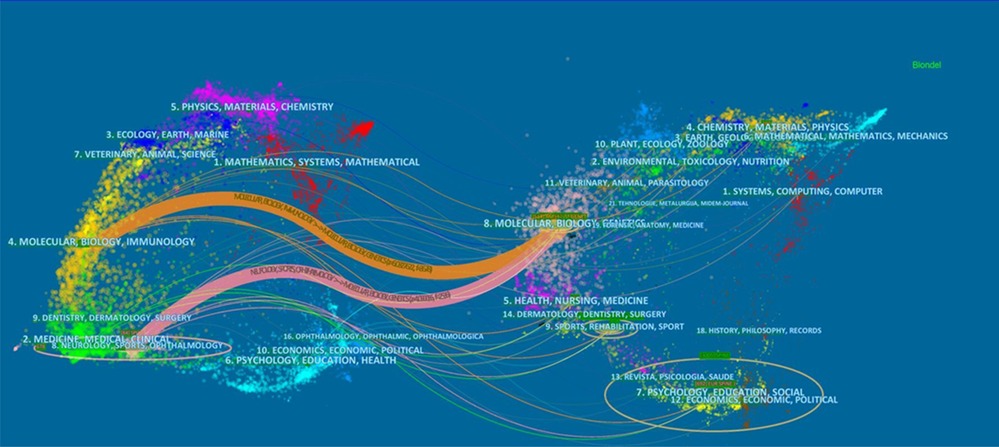
Figure 6. The dual-map overlay of journals contributed to publications on idiopathic scoliosis-genes from 2002 to 2022.
3.4. Analysis of references
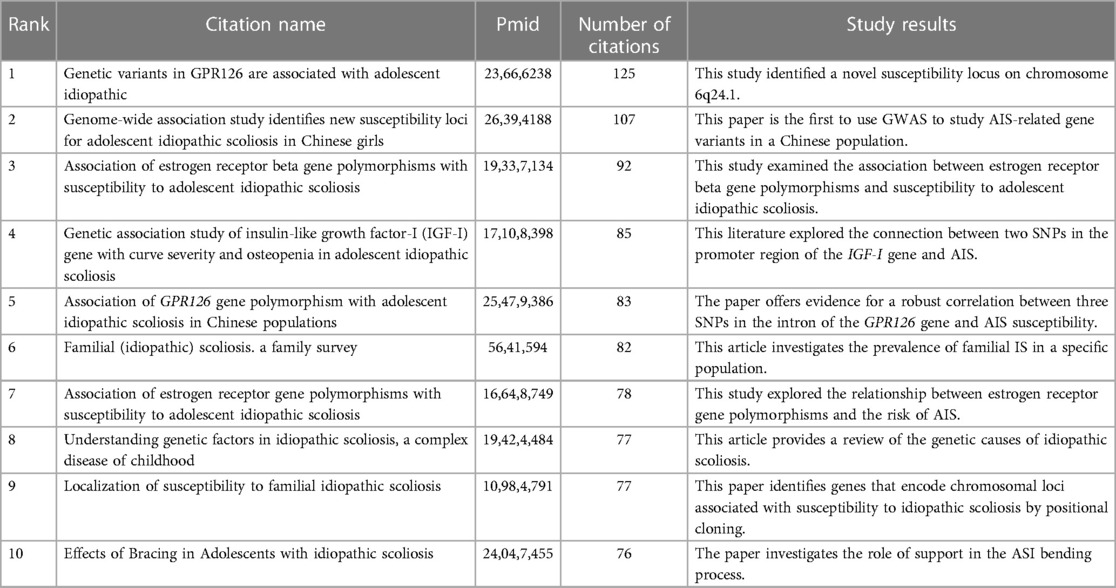
Citing papers constitute the frontiers of knowledge, and cited papers constitute the basis of knowledge. Numerous highly cited articles have been published in top journals on human idiopathic scoliosis genetic studies, including those in Nature and Science. The topics of these articles include genome-wide associations, protein-coupled receptors, biological pathways, and myelination. Reference co-citation refers to the phenomenon in which more than two articles cite the same reference. By analyzing the clusters and critical nodes in the co-citation network, the knowledge structure of the IS gene research field can be revealed (14), as well as the relationship between research themes and subfields (15). The Modularity Q score was 0.573 (> 0.5), indicating that the network was reasonably divided into loosely coupled clusters. The Weighted Mean Silhouette was 0.8489 (> 0.5), and the Harmonic Mean (Q, S) was 0.6842, indicating that the homogeneity of these clusters was acceptable (Figure 7). The largest cluster, # 0, was labeled as "single nucleotide polymorphism"; cluster # 1 was "whole exome sequencing" (9, 16–18), and cluster #2 was "axonemal dynein" (19–21). Cluster # 3 is "drug discovery" (3, 22, 23). Cluster #4 is "mesenchymal stem cell" (24–26). Cluster # 5 was "dietary intake" (27–29). Cluster # 6 was the "curve progression" (30–32), and cluster # 7 was the "zebrafish developmental model" (33, 34). The most cited references in each cluster were color-coded and shown, including those with the strongest mediation centrality (Figure 7).
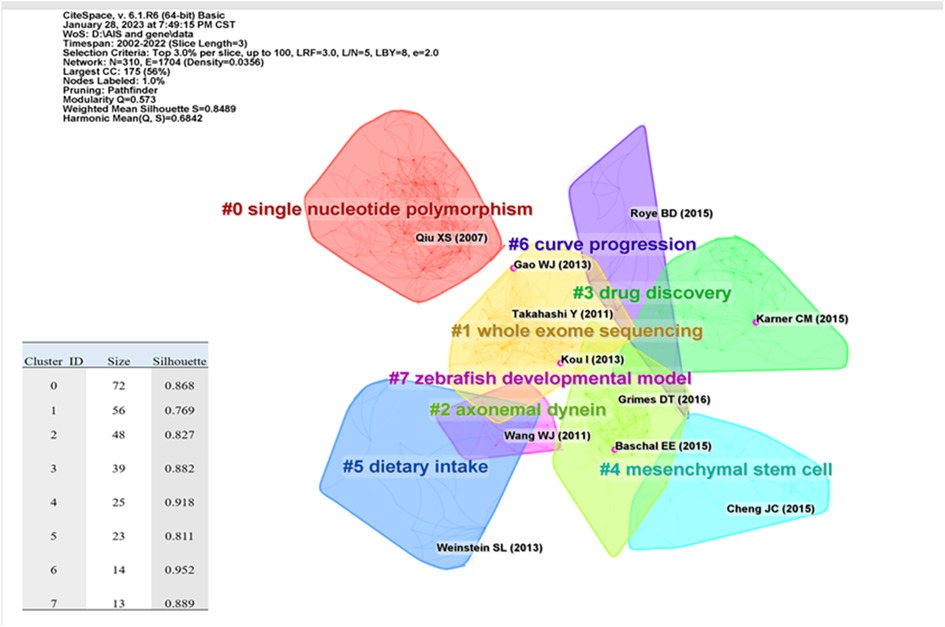
Figure 7. Citation clustering results are shown: the literature in purple circles are articles with high mediated centrality, with mediated centrality > 0.1, representing that the literature plays an essential role in the network and development.
3.5. Keywords
We extracted and analyzed keywords from the relevant literature. The top 16 keywords with the highest number of occurrences were listed (Figure 8). In addition to idiopathic scoliosis, susceptibility and genome-wide associations appeared more than 80 times. We performed an emergent analysis of keywords and analyzed the temporal trends of hotspot shifts based on the top 16 emergent keywords, such as rare variant (2018–2022) and extracellular matrix (2020–2022) (Figure 9).
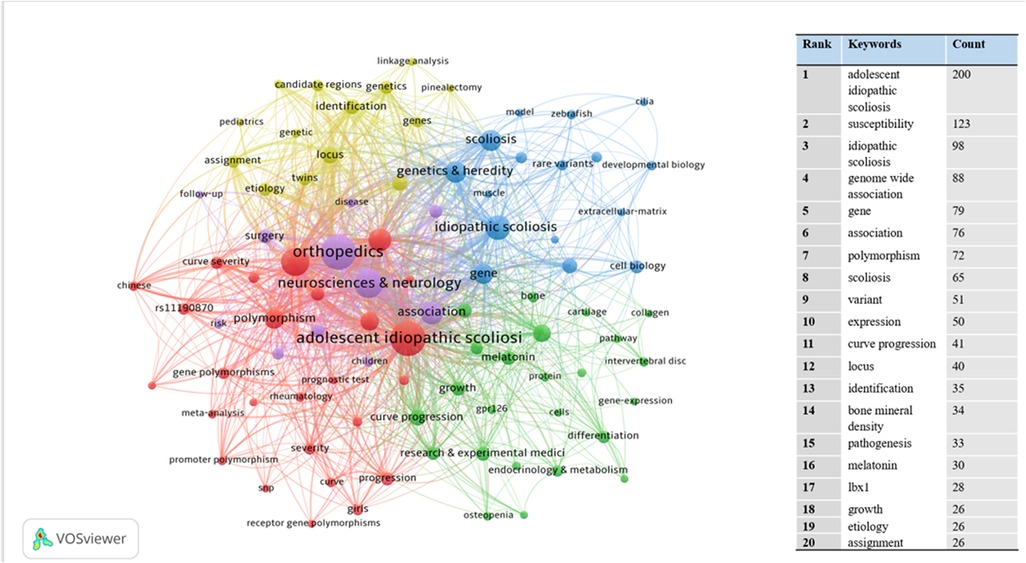
Figure 8. The network map of key worlds contributed to publications on idiopathic scoliosis-genes from 2002 to 2022. The co-occurrence of high-frequency keywords represents the focus and research trend.
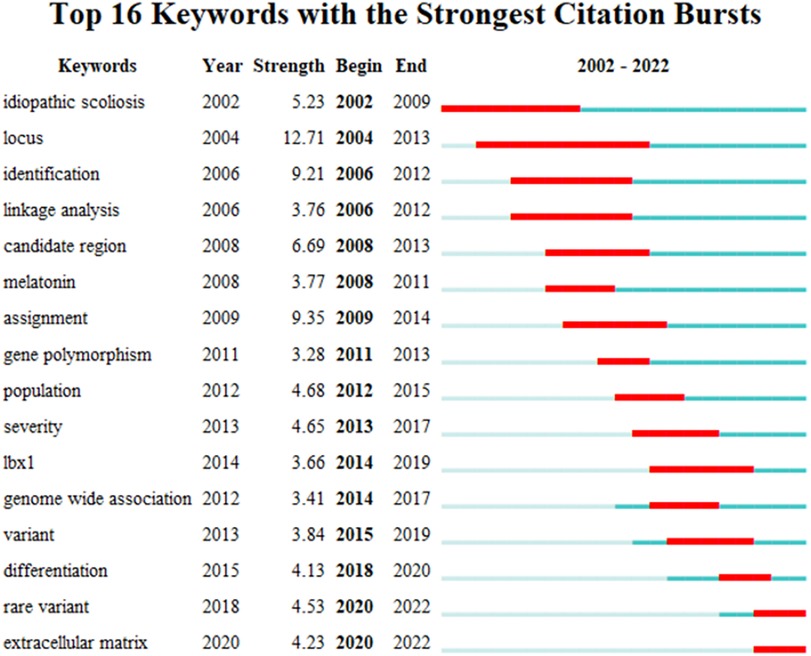
Figure 9. The left side displays the burgeoning keywords, while the right side displays the hot spot transfer trend. The red bars reflect that researchers placed high importance on this keyword at this time.
4. Discussion
4.1. Knowledge map of references
Previous research has led to a better organizational map of genetic studies on idiopathic scoliosis (Figure 7). Based on the analysis of references, we analyzed the knowledge base, study objectives, and potential research directions in this research field.
In cluster 0, studies have shown that single nucleotide polymorphisms (SNPs) in the gene promoter region may be necessary to identify IS susceptibility genes. Multiple AIS susceptibility genes, including LBX1, GPR126, BNC2, PAX1, LBX1-as1, BCL2, AJAP1, PAX3, TNIK, MEIS1, MAGI1, TGFB1, MIR4300HG, and TPH1 genes, have already been identified in Caucasian, Japanese, and Chinese populations (35, 36). However, most of these findings are difficult to replicate in multi-ethnic populations. Recent advances in IS etiology research have focused on bone morphogenetic protein (BMP4), interleukin (IL6), leptin, matrix metalloproteinase 3 (MMP3), chromosome domain caspase DNA DNA-binding protein 7 (CHD7) (8), and promoter polymorphisms of the melatonin 1 B receptor (MTNR1B) promoter (35). Furthermore, these genes do not have the same impact on the etiology of IS. There is a great deal of interaction between them, and they are closely coupled (37). Qiu. et al. (29) revealed that MTNR1B gene polymorphism is linked to adolescent idiopathic scoliosis susceptibility (AIS). Subsequent research (36, 38) has revealed that MTNR1A gene promoter polymorphism may not be related to the occurrence of AIS or the severity of the curve. Nikolova ST et al. (39) reported that combining IL-6 and MMP3 genotypes might be associated with IS susceptibility. The biological effects of IS susceptibility gene polymorphisms could explain the above occurrence, highlighting the importance of verifying gene interactions in IS gene research (40).
Currently, there is no conclusive evidence whether AIS is a single disease or a collection of multiple diseases. Research suggests that AIS may be influenced by genetic factors, environmental factors, and spinal growth and development (41–44). Additionally, a significant body of research indicates the presence of diverse morphological features, clinical presentations, and prognoses among AIS patients (45). Complexity and heterogeneity are key characteristics of AIS etiology and phenotype, suggesting that AIS can be considered as a relatively complex group of diseases.
In cluster 1, "whole exome sequencing," which is limited by the low efficiency of genetic marker definition, gene linkage analysis can only identify chromosomal regions that may contain disease-related genes (46). Methods for obtaining more genetic evidence for the IS gene through high-throughput sequencing have been widely used. Genome-wide association studies (GWAS) have identified potential common risk alleles, especially those located in or near LBX1 (20, 22) and GPR126/ADGRG6 (3, 47, 48). Whole exome sequencing (WES) studies have found rare variations in extracellular matrix genes that can lead to AIS phenotypes in families and unrelated individuals (28, 49, 50).
Among cluster 2, "axonemal dynein," kinesin family 6 (KIF6) mutants, and PTK7 were reported to be IS candidate genes. Grimes et al. (45) defined the limited development window of motor cilia by investigating the flow characteristics of cerebrospinal fluid (CSF) in a zebrafish model with a PTK7 gene mutation, thus revealing the regulatory mechanism of cilia activity recovery during scoliosis progression. Baschal et al. (20) revealed predictive variants in IS-related genes, static cilia, and other actin-based cell projections using GO enrichment analysis. Functional studies of these variants confirmed the relationship between IS and these variations. However, the current research in this area is limited, which calls for further investigation and verification of the causes of the disease to establish a foundation for its prevention and treatment.
In cluster 3,"drug discovery,"Karner et al. (51) demonstrated a GPR126 mechanism of action in axial cartilage, indicating that GPR126cAMP agonists could still repair the decrease in GPR126 signaling in cartilage cells, which might be a valuable resource for drug exposure in IS. Currently, this type of medication has demonstrated significant potential in the treatment of idiopathic scoliosis. Therefore, it may be considered as a promising candidate in the development of drugs for both preventing and treating idiopathic scoliosis.
In cluster 4, "mesenchymal stem cells," irregular bone development and decreased bone mineral density were initially discovered as bone metabolism problems in patients with AIS. Bone marrow mesenchymal stem cells (BMMSCs) have been studied for their ability to promote osteogenic differentiation and proliferation (52) to improve bone mass homeostasis (53).
BMMSCs use miRNAs for several biological activities, including transmembrane transport, cytokinesis, DNA-dependent transcription, GTPase-mediated signal transduction, and response to hypoxia. Hui SYet al. (54) discovered 54 previously unknown differentially expressed genes in AIS patients with BMMSCs, and identified the differential expression profiles of miRNAs and related pathways, providing new insights into the pathophysiology of AIS and the mechanism of bone loss.
In Cluster 5, "dietary intake". The dietary mechanisms underlying IS have been postulated to involve several hypotheses, including DNA methylation (55, 56), lipid peroxidation (57), and neurodevelopmental processes (58). As an influential environmental factor (59), diet exerts a certain influence on the patterns of methylated cytosines ("methylome") during childhood growth and development, ultimately impacting gene expression patterns (60). Dietary habits exert regulatory control over the genetic phenotype plasticity of idiopathic scoliosis during the developmental plasticity window through intricate gene regulatory mechanisms (62). Keiko Asakura et al. (63) investigated the relationship between dietary habits and IS in 2,431 subjects, 47.8% of whom had AIS, and found no significant association between dietary habits and AIS. In contrast, research (64) has discovered a correlation between dietary intake and IS in children. Several studies have reported that adolescents diagnosed with idiopathic scoliosis display notable variations in anthropometric measurements, such as increased height, reduced body mass index, and decreased bone density, compared to their age-matched counterparts (65). Daily supplementation of calcium and vitamin D has been shown to enhance the trabecular bone area, density, and strain index at the distal end of the tibia and radius. Additionally, it has been found to increase the cortical area of the tibial shaft (66). The cumulative effect of dietary imbalances across multiple generations in both parents has resulted in variations in 1C genes, related epigenetic regulatory factors, and methylation patterns of target DNA sequences (57). The metabolic adaptations induced by traditional dietary habits within a family can give rise to significant transgenerational effects (67). It is worth noting that the asymmetry of paraspinal muscles in AIS patients involves alterations in both chronomics and the genome (68, 69). These alterations are potentially associated with lipid peroxidation. Additionally, research suggests that dietary antioxidants may have the potential to mitigate the process of lipid peroxidation (58). Moreover, scientific investigations have revealed a noteworthy correlation (55) between enhanced dietary quality and the predictive factors pertaining to the accelerated aging epigenetic clock. For example, it has been observed that elevated fructose consumption induces DNA methylation in the promoter region of CPT1A, while a high-fat diet exerts an inhibitory effect on this methylation process, thereby modulating the organism's energy metabolism levels (70). While the precise molecular mechanisms underlying the impact of diet on the genetic phenotype of IS have yet to be fully elucidated, it is evident that diet, as a specific environmental factor, plays a conspicuous role in the epigenetic alterations associated with IS (59). Maintaining good dietary habits and paying attention to what we eat may be helpful in reducing the incidence of idiopathic scoliosis.
In cluster 6, which focuses on "curve progression," accurate early diagnosis and precise prediction of curve progression are crucial in mitigating the long-term adverse effects associated with AIS treatment in the management of AIS (71). Age, gender, skeletal maturity, Tanner stage, and menarche were frequently used clinical variables to identify people at risk of curve progression (72). Simultaneously, identifying the non-genetic basis of the risk of curve progression has emerged as a research hotspot. Roye BD (73) et al. Identified epigenetic differences in IS twins and genomic variations between individuals with distinct curves, clarifying that non-genetic factors must be involved in curve progression in AIS patients. Meng YC et al. (74) affirmed that epigenetic and genetic markers could be used as potential predictors of curve progression. However, the majority of gene tests associated with it have proven to be unsuccessful. The potential reasons for such outcomes may be attributed to the complex and dynamic nature of epigenetics. The emergence of epigenetics has provided new insights into AIS research (55). Epigenetic mechanisms allow organisms to respond to environmental changes through alterations in gene expression, maintaining gene silencing or activation through chemical modifications of DNA regions and protein modifications during cell development (75). As a disease with unclear pathogenesis, despite the failures of most gene tests associated with IS, the reasons behind these results may be attributed to the complex and dynamic nature of epigenetics, which is influenced by multiple factors (61). These factors include, but are not limited to, the growth and development window (41), lifestyle (59), temperature (76), geographical latitude (77), which may explain the test failures.
Despite genetic variability and a lack of suitable developmental models, little is known about the genesis and mechanism of idiopathic scoliosis in cluster 7, "zebrafish developmental model." Numerous genes with similar expression features exist in both humans and zebrafish. Influenced by genomic perturbation, bio-imaging, and high-throughput small-molecule screening, zebrafish are naturally sensitive to spinal curvature and can reasonably accept vigorous positive gene screening, hence providing strong support for the pathogenicity study of human IS-related gene mutations. By combining zebrafish IS models with high-throughput small-molecule screening, advanced genome-editing tools enable robust precision medical services for IS investigation.
4.2. Research hotspots and frontiers
Keywords can express existing research questions or concepts intensively, whereas burst keywords can reflect emerging trends and research frontiers in this field. The two frontiers of relevant research are as follows: extracellular matrix (2020–2022) and rare variant (2018–2022).
4.2.1. Extracellular matrix
Long-term deformation and changes in mechanical load on scoliosis intervertebral disc tissue affect cells in several ways. As determined by gene expression analyses using cDNA arrays and RT-PCR, extracellular matrix components, such as polymerins, contain large quantities of mRNA (78). In contrast to physiological aging, morphological disc degeneration in scoliosis is accelerated by approximately 22 years (79). In response to changes in the mechanical environment of the tissue, patients with scoliosis synthesize a more robust metabolic matrix at a young age (80).
Spinal developmental disorders increase the risk of neurological and other physiological dysfunctions in adolescents with scoliosis. The structural integrity of the extracellular matrix and the variation in homeostasis factors, for example, have a non-negligible impact on human cartilage biogenesis and intervertebral disc development, which may be one of the causes of AIS (81). Furthermore, studies (82) revealed that MAPK7 has a variety of functions in cell migration and proliferation, and osteoblast abnormalities caused by MAPK7 variants have been implicated in zebrafish and human IS.
4.2.2. Rare variant
Numerous AIS candidate genes have been identified using high-throughput sequencing techniques in candidate gene association studies. The validity of these candidate genes has been questioned because there is insufficient evidence that they can frequently be replicated in various populations (46). As rare genotypes are unexpectedly identified in the general population, it is generally assumed that rare variants have a more significant impact on the genetics of complex diseases than common variants. Patten SA et al. (83) reported three rare variants of POC5 that are related to AIS function using WES. Subsequently, rare variants of HSPG2 were reported as potential factors for susceptibility to AIS in Caucasians (49). Haller G et al. (84) reported that rare variants in extracellular matrix genes substantially affect the risk of AIS in patients of European ancestry. In a recent study, Xia C et al. (85) validated the effect of the rare variant HSPG2 on IS progression. Overall, IS is a complex disease, and it is worthwhile to identify and validate rare variants associated with IS and investigate their impact on patient phenotypes.
4.3. Limitations
Although the highly regarded WoSCC database was utilized in this study to identify the knowledge map and hotspots of IS gene research, there are crucial limitations that must be considered. Firstly, WoSCC only includes certain journals and publications, resulting in limited coverage and an inability to represent all subject areas. Secondly, WoSCC largely concentrates on English-language literature, potentially overlooking non-English language literature that could greatly influence idiopathic scoliosis-related gene research. Additionally, the search time frame may not be extensive enough to fully capture short-term research trends. Finally, while bibliometric analysis constitutes a relatively objective method for analyzing literature, authors, and topics through network science, machine learning, among others, these methodologies also possess their own limitations. For instance, keyword-based analysis may disregard semantically complex literature, leading to inaccuracies in analysis results. Moreover, this study failed to comprehensively evaluate some large cohort studies or specific cases, potentially missing cutting-edge discoveries such as new IS gene identification. The functional annotation and enrichment analysis of well-known IS genes might not have been administered to tools like DAVID, thereby possibly constraining the discovery of new mechanisms of IS disease. Hence, future research needs to continue refining databases, technologies, and methodologies to thoroughly and expansively explore the forefront of IS gene research.
5. Conclusions
This study provides a visual analysis and review of the IS gene-related literature using a bibliometric analysis method. In addition to identifying current research hotspots, the report provides an objective and detailed overview of the research found over the last two decades. Furthermore, it envisions a future development model for the IS gene field, which can be a reference for scholars and clinicians engaged in clinical research and treatment in this field.
The level of interest in IS-related gene analysis is higher and higher in recent years. Extracellular matrices and rare variants are becoming the hotspots and frontiers of research. Meanwhile, early prevention and treatment of sensitive groups were positively affected by identifying molecular markers in IS development. The discovery of recovery patterns of ciliary motility can lead to the development of new therapeutic agents for IS. Further functional analysis may clarify the downstream pathways of the identified susceptibility genes in the pathogenesis and mechanism of action of IS. Understanding the mechanisms and mutation patterns of the downstream pathways of susceptibility genes in IS pathogenesis could be significantly enhanced by optimizing the functional analysis. In the future, more in-depth studies on IS should be carried out from the aspects of extracellular matrices, rare variants and molecular markers, in order to provide insights for understanding the pathogenesis of IS and the development of anti-IS drugs. Ultimately, and most significantly, it can be argued that the prevention and intervention of curve progression rely predominantly on empirical evidence. Therefore, it is imperative to explore the integration of empirical approaches with epigenetics mechanisms to establish more efficacious preventive measures.
Author contributions
LR, HZ, and WL contributed to the study conception and design. LR organized the database. WJ performed the statistical analyses. LR wrote the first draft of the manuscript. LR, HZ, SZ, and QF wrote the manuscript sections. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the Special Project of Scientific Research on Traditional Chinese Medicine in the Henan Province Assignment (2021JDZY049) and Chinese Children's Growth and Development Academic Exchange Special Fund (Z-2019-41-2101-02).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fped.2023.1177983/full#supplementary-material
References
1. Negrini S, Donzelli S, Aulisa AG, Czaprowski D, Schreiber S, de Mauroy JC, et al. 2016 SOSORT guidelines: orthopaedic and rehabilitation treatment of idiopathic scoliosis during growth. Scoliosis Spinal Disord. (2018) 13:3. doi: 10.1186/s13013-017-0145-8
2. Comite Nacional de Adolescencia SAP. Comite de diagnostico por imagenes SAP, sociedad Argentina de ortopedia y traumatologia I, sociedad Argentina de patologia de la Columna V, comite de diagnostico por I, colaboradores. Adolescent idiopathic scoliosis. Arch Argent Pediatr. (2016) 114(6):585–94.27869435
3. Kou I, Takahashi Y, Johnson TA, Takahashi A, Guo L, Dai J, et al. Genetic variants in GPR126 are associated with adolescent idiopathic scoliosis. Nature Genet. (2013) 45(6):676. doi: 10.1038/ng.2639
4. Riseborough EJ, Wynne-Davies R. A genetic survey of idiopathic scoliosis in Boston, Massachusetts. J Bone Joint Surg Am. (1973) 55(5):974–82. doi: 10.2106/00004623-197355050-00006
5. Zhao LL, Roffey DM, Chen S. Genetics of adolescent idiopathic scoliosis in the post-genome-wide association study era. Ann Transl Med. (2015) 3:2. doi: 10.3978/j.issn.2305-5839.2015.03.54
6. Wang YJ, Liu ZH, Yang GT, Gao QL, Xiao LG, Li J, et al. Coding variants coupled with rapid modeling in zebrafish implicate dynein genes, dnaaf1 and zmynd10, as adolescent idiopathic scoliosis candidate genes. Front Cell Dev Biol. (2020) 8:14. doi: 10.3389/fcell.2020.00014
7. Simony A, Carreon LY, Hojmark K, Kyvik KO, Andersen MO. Concordance rates of adolescent idiopathic scoliosis in a danish twin population. Spine. (2016) 41(19):1503–7. doi: 10.1097/BRS.0000000000001681
8. Borysiak K, Janusz P, Andrusiewicz M, Chmielewska M, Kozinoga M, Kotwicki T, et al. CHD7 Gene polymorphisms in female patients with idiopathic scoliosis. BMC Musculoskelet Disord. (2020) 21(1):11. doi: 10.1186/s12891-019-3031-0
9. Montanaro L, Parisini P, Greggi T, Di Silvestre M, Campoccia D, Rizzi S, et al. Evidence of a linkage between matrilin-1 gene (MATN1) and idiopathic scoliosis. Scoliosis. (2006) 1:21. doi: 10.1186/1748-7161-1-21
10. Perez-Machado G, Berenguer-Pascual E, Bovea-Marco M, Rubio-Belmar PA, Garcia-Lopez E, Garzon MJ, et al. From genetics to epigenetics to unravel the etiology of adolescent idiopathic scoliosis. Bone. (2020) 140:12. doi: 10.1016/j.bone.2020.115563
11. Nikolova S, Dikova M, Dikov D, Djerov A, Savov A, Kremensky I, et al. Positive association between TGFB1 gene and susceptibility to idiopathic scoliosis in Bulgarian population. Anal Cell Pathol (Amsterdam). (2018) 2018:6836092. doi: 10.1155/2018/6836092
12. Bae JW, Cho CH, Min WK, Kim UK. Associations between matrilin-1 gene polymorphisms and adolescent idiopathic scoliosis curve patterns in a Korean population (vol 39, pg 5561, 2012). Mol Biol Rep. (2012) 39(9):9275. doi: 10.1007/s11033-012-1859-6
13. Broadus RN. Toward a definition of “bibliometrics”. Scientometrics. (1987) 12:373–9. doi: 10.1007/BF02016680
14. Chen C. A glimpse of the first eight months of the COVID-19 literature on microsoft academic graph: themes, citation contexts, and uncertainties. Front Res Met Anal. (2020) 5:607286. doi: 10.3389/frma.2020.607286
15. Chen C, Dubin R, Kim MC. Emerging trends and new developments in regenerative medicine: a scientometric update (2000–2014). Expert Opin Biol Ther. (2014) 14(9):1295–317. doi: 10.1517/14712598.2014.920813
16. Wise CA, Barnes R, Gillum J, Herring JA, Bowcock AM, Lovett M. Localization of susceptibility to familial idiopathic scoliosis. Spine. (2000) 25(18):2372–80. doi: 10.1097/00007632-200009150-00017
17. Kesling KL, Reinker KA. Scoliosis in twins—a meta-analysis of the literature and report of six cases. Spine. (1997) 22(17):2009–14. doi: 10.1097/00007632-199709010-00014
18. Marosy B, Justice CM, Nzegwu N, Kumar G, Wilson AF, Miller NH. Lack of association between the aggrecan gene and familial idiopathic scoliosis. Spine. (2006) 31(13):1420–5. doi: 10.1097/01.brs.0000219944.18223.52
19. Londono D, Kou I, Johnson TA, Sharma S, Ogura Y, Tsunoda T, et al. A meta-analysis identifies adolescent idiopathic scoliosis association with LBX1 locus in multiple ethnic groups. J Med Genet. (2014) 51(6):401–6. doi: 10.1136/jmedgenet-2013-102067
20. Baschal EE, Terhune EA, Wethey CI, Baschal RM, Robinson KD, Cuevas MT, et al. Idiopathic scoliosis families highlight actin-based and microtubule-based cellular projections and extracellular matrix in disease etiology. G3-Genes Genomes Genet. (2018) 8(8):2663–72. doi: 10.1534/g3.118.200290
21. Fan YH, Song YQ, Chan D, Takahashi Y, Ikegawa S, Matsumoto M, et al. SNP Rs11190870 near LBX1 is associated with adolescent idiopathic scoliosis in southern Chinese. J Hum Genet. (2012) 57(4):244–6. doi: 10.1038/jhg.2012.11
22. Takahashi Y, Kou I, Takahashi A, Johnson TA, Kono K, Kawakami N, et al. A genome-wide association study identifies common variants near LBX1 associated with adolescent idiopathic scoliosis. Nature Genet. (2011) 43(12):1237–U96. doi: 10.1038/ng.974
23. Terhune EA, Cuevas MT, Monley AM, Wethey CI, Chen XM, Cattell MV, et al. Mutations in KIF7 implicated in idiopathic scoliosis in humans and axial curvatures in zebrafish. Hum Mutat. (2021) 42(4):392–407. doi: 10.1002/humu.24162
24. Justice CM, Miller NH, Marosy B, Zhang J, Wilson AF. Familial idiopathic scoliosis: evidence of an X-linked susceptibility locus. Spine. (2003) 28(6):589–94. doi: 10.1097/01.BRS.0000049940.39801.E6
25. Gorman KF, Tredwell SJ, Breden F. The mutant guppy syndrome curveback as a model for human heritable spinal curvature. Spine. (2007) 32(7):735–41. doi: 10.1097/01.brs.0000259081.40354.e2
26. Chan V, Fong GCY, Luk KDK, Yip B, Lee MK, Wong MS, et al. A genetic locus for adolescent idiopathic scoliosis linked to chromosome 19p13.3. Am J Hum Genet. (2002) 71(2):401–6. doi: 10.1086/341607
27. Wu J, Yong Q, Zhang L, Sun Q, Qiu XS, He YX. Association of estrogen receptor gene polymorphisms with susceptibility to adolescent idiopathic scoliosis. Spine. (2006) 31(10):1131–6. doi: 10.1097/01.brs.0000216603.91330.6f
28. Qiu XS, Tang NLS, Yeung HY, Qiu Y, Cheng JCY. Association study between adolescent idiopathic scoliosis and the DPP9 gene which is located in the candidate region identified by linkage analysis. Postgrad Med J. (2008) 84(995):498–501. doi: 10.1136/pgmj.2007.066639
29. Inoue M, Minami S, Nakata Y, Kitahara H, Otsuka Y, Isobe K, et al. Association between estrogen receptor gene polymorphisms and curve severity of idiopathic scoliosis. Spine. (2002) 27(21):2357–62. doi: 10.1097/00007632-200211010-00009
30. Qiu XS, Tang NLS, Yeung HY, Lee KM, Hung VWY, Ng BKW, et al. Melatonin receptor 1B (MTNR1B) gene polymorphism is associated with the occurrence of adolescent idiopathic scoliosis. Spine. (2007) 32(16):1748–53. doi: 10.1097/BRS.0b013e3180b9f0ff
31. Gorman KF, Julien C, Moreau A. The genetic epidemiology of idiopathic scoliosis. Eur Spine J. (2012) 21(10):1905–19. doi: 10.1007/s00586-012-2389-6
32. Moreau A, Wang DS, Forget S, Azeddine B, Angeloni D, Fraschini F, et al. Melatonin signaling dysfunction in adolescent idiopathic scoliosis. Spine. (2004) 29(16):1772–81. doi: 10.1097/01.BRS.0000134567.52303.1A
33. Zhu ZZ, Tang NLS, Xu LL, Qin XD, Mao SH, Song YM, et al. Genome-wide association study identifies new susceptibility loci for adolescent idiopathic scoliosis in Chinese girls. Nat Commun. (2015) 6:6. doi: 10.1038/ncomms9355
34. Buchan JG, Alvarado DM, Haller GE, Cruchaga C, Harms MB, Zhang TX, et al. Rare variants in FBN1 and FBN2 are associated with severe adolescent idiopathic scoliosis. Hum Mol Genet. (2014) 23(19):5271–82. doi: 10.1093/hmg/ddu224
35. Morocz M, Czibula A, Grozer ZB, Szecsenyi A, Almos PZ, Rasko I, et al. Association study of BMP4, IL6, leptin, MMP3, and MTNR1B gene promoter polymorphisms and adolescent idiopathic scoliosis. Spine. (2011) 36(2):E123–E30. doi: 10.1097/BRS.0b013e318a511b0e
36. Qiu XS, Tang NLS, Yeung HY, Cheng JCY, Qiu Y. Lack of association between the promoter polymorphism of the MTNR1A gene and adolescent idiopathic scoliosis. Spine. (2008) 33(20):2204–7. doi: 10.1097/BRS.0b013e31817e0424
37. Yang MY, Chen K, Hou CL, Yang YL, Zhai X, Wei XZ, et al. RHOA Inhibits chondrogenic differentiation of mesenchymal stem cells in adolescent idiopathic scoliosis. Connect Tissue Res. (2022) 63(5):475–84. doi: 10.1080/03008207.2021.2019247
38. Nelson LM, Ward K, Ogilvie JW. Genetic variants in melatonin synthesis and signaling pathway are not associated with adolescent idiopathic scoliosis. Spine. (2011) 36(1):37–40. doi: 10.1097/BRS.0b013e3181e8755b
39. Nikolova ST, Yablanski VT, Vlaev EN, Stokov LD, Savov AS, Kremensky IM, et al. Association between IL-6 and MMP3 common genetic polymorphisms and idiopathic scoliosis in Bulgarian patients. Spine. (2016) 41(9):785–91. doi: 10.1097/BRS.0000000000001360
40. Wang WG, Chen TL, Liu YB, Wang SS, Yang NN, Luo M. Predictive value of single-nucleotide polymorphisms in curve progression of adolescent idiopathic scoliosis. Eur Spine J. (2022) 31(9):2311–25. doi: 10.1007/s00586-022-07213-y
41. Gluckman PD, Hanson MA, Low FM. The role of developmental plasticity and epigenetics in human health. Birth Defects Res Part C Embryo Today: Rev. (2011) 93(1):12–8. doi: 10.1002/bdrc.20198
42. Grivas TB, Vasiliadis E, Savvidou O, Mouzakis V, Koufopoulos G. Geographic latitude and prevalence of adolescent idiopathic scoliosis. Stud Health Technol Inform. (2006) 123:84–9.17108408
43. Day JJ, Sweatt JD. Epigenetic modifications in neurons are essential for formation and storage of behavioral memory. Neuropsychopharmacology. (2011) 36(1):357–8. doi: 10.1038/npp.2010.125
44. Wang WJ, Yeung HY, Chu WCW, Tang NLS, Lee KM, Qiu Y, et al. Top theories for the etiopathogenesis of adolescent idiopathic scoliosis. J Pediat Orthop. (2011) 31:S14–27. doi: 10.1097/BPO.0b013e3181f73c12
45. Liu G, Wang LL, Wang XY, Yan ZH, Yang XZ, Lin M, et al. Whole-Genome methylation analysis of phenotype discordant monozygotic twins reveals novel epigenetic perturbation contributing to the pathogenesis of adolescent idiopathic scoliosis. Front Bioeng Biotechnol. (2019) 7:8. doi: 10.3389/fbioe.2019.00008
46. Liu Z, Tang NLS, Cao XB, Liu WJ, Qiu XS, Cheng JCY, et al. Lack of association between the promoter polymorphisms of MMP-3 and IL-6 genes and adolescent idiopathic scoliosis A case-control study in a Chinese han population. Spine. (2010) 35(18):1701–5. doi: 10.1097/BRS.0b013e3181c6ba13
47. Xu LL, Dai ZC, Xia C, Wu ZC, Feng ZH, Sun X, et al. Asymmetric expression ofWnt/B-cateninPathway in AIS primary or secondary to the curve? Spine. (2020) 45(12):E677–E83. doi: 10.1097/BRS.0000000000003409
48. Grauers A, Wang JW, Einarsdottir E, Simony A, Danielsson A, Akesson K, et al. Candidate gene analysis and exome sequencing confirm LBX1 as a susceptibility gene for idiopathic scoliosis. Spine J. (2015) 15(10):2239–46. doi: 10.1016/j.spinee.2015.05.013
49. Baschal EE, Wethey CI, Swindle K, Baschal RM, Gowan K, Tang NLS, et al. Exome sequencing identifies a rare HSPG2 variant associated with familial idiopathic scoliosis. G3-Genes Genomes Genet. (2015) 5(2):167–74. doi: 10.1534/g3.114.015669
50. Rose CD, Pompili D, Henke K, Van Gennip JLM, Meyer-Miner A, Rana R, et al. SCO-Spondin defects and neuroinflammation are conserved mechanisms driving spinal deformity across genetic models of idiopathic scoliosis. Curr Biol. (2020) 30(12):2363. doi: 10.1016/j.cub.2020.04.020
51. Karner CM, Long F, Solnica-Krezel L, Monk KR, Gray RS. Gpr126/Adgrg6 deletion in cartilage models idiopathic scoliosis and pectus excavatum in mice. Hum Mol Genet. (2015) 24(15):4365–73. doi: 10.1093/hmg/ddv170
52. Cheng JC, Castelein RM, Chu WC, Danielsson AJ, Dobbs MB, Grivas TB, et al. Adolescent idiopathic scoliosis. Nat Rev Dis Primers. (2015) 1:15030. doi: 10.1038/nrdp.2015.30
53. Wang QF, Yang JL, Lin X, Huang ZF, Xie CF, Fan HW. Spot14/Spot14R expression may be involved in MSC adipogenic differentiation in patients with adolescent idiopathic scoliosis. Mol Med Rep. (2016) 13(6):4636–42. doi: 10.3892/mmr.2016.5109
54. Hui SY, Yang Y, Li J, Li N, Xu PC, Li HL, et al. Differential miRNAs profile and bioinformatics analyses in bone marrow mesenchymal stem cells from adolescent idiopathic scoliosis patients. Spine J. (2019) 19(9):1584–96. doi: 10.1016/j.spinee.2019.05.003
55. Kresovich JK, Park YM, Keller JA, Sandler DP, Taylor JA. Healthy eating patterns and epigenetic measures of biological age. Am J Clin Nutr. (2022) 115(1):171–9. doi: 10.1093/ajcn/nqab307
56. Allison J, Kaliszewska A, Uceda S, Reiriz M, Arias N. Targeting DNA methylation in the adult brain through diet. Nutrients. (2021) 13(11):3979. doi: 10.3390/nu13113979
57. Clare CE, Brassington AH, Kwong WY, Sinclair KD. One-Carbon metabolism: linking nutritional biochemistry to epigenetic programming of long-term development. Annu Rev Anim Biosci. (2019) 7:263–87. doi: 10.1146/annurev-animal-020518-115206
58. Enwonwu CO, Sanders C. Nutrition: impact on oral and systemic health. Compendium of continuing education in dentistry (Jamesburg, NJ: 1995). Compend contin educ dent. (2001) 22(3 Spec No):12–8.11913248
59. Worthington V, Shambaugh P. Nutrition as an environmental factor in the etiology of idiopathic scoliosis. J Manipulative Physiol Ther. (1993) 16(3):169–73.8492060
60. Stevens AJ, Rucklidge JJ, Kennedy MA. Epigenetics, nutrition and mental health. Is there a relationship? Nutr Neurosci. (2018) 21(9):602–13. doi: 10.1080/1028415X.2017.1331524
61. Weinstein SL, Dolan LA, Wright JG, Dobbs MB. Effects of bracing in adolescents with idiopathic scoliosis. N Engl J Med. (2013) 369(16):1512–21. doi: 10.1056/NEJMoa1307337
62. Burwell RG, Dangerfield PH, Moulton A, Grivas TB. Adolescent idiopathic scoliosis (AIS), environment, exposome and epigenetics: a molecular perspective of postnatal normal spinal growth and the etiopathogenesis of AIS with consideration of a network approach and possible implications for medical therapy. Scoliosis Spinal Disord. (2011) 6:20. doi: 10.1186/1748-7161-6-26
63. Asakura K, Michikawa T, Takaso M, Minami S, Soshi S, Tsuji T, et al. Dietary habits had No relationship with adolescent idiopathic scoliosis: analysis utilizing quantitative data about dietary intakes. Nutrients. (2019) 11(10):13. doi: 10.3390/nu11102327
64. Margalit A, McKean G, Constantine A, Thompson CB, Lee RJ, Sponseller PD. Body mass hides the curve: thoracic scoliometer readings vary by body mass Index value. J Pediatr Orthop. (2017) 37(4):e255–e60. doi: 10.1097/BPO.0000000000000899
65. Lee WT, Cheung CS, Tse YK, Guo X, Qin L, Ho SC, et al. Generalized low bone mass of girls with adolescent idiopathic scoliosis is related to inadequate calcium intake and weight bearing physical activity in peripubertal period. Osteoporos Int. (2005) 16(9):1024–35. doi: 10.1007/s00198-004-1792-1
66. Greene DA, Naughton GA. Calcium and vitamin-D supplementation on bone structural properties in peripubertal female identical twins: a randomised controlled trial. Osteoporos Int. (2011) 22(2):489–98. doi: 10.1007/s00198-010-1317-z
67. Kaati G, Bygren LO, Edvinsson S. Cardiovascular and diabetes mortality determined by nutrition during parents’ and grandparents’ slow growth period. Eur J Hum Genet. (2002) 10(11):682–8. doi: 10.1038/sj.ejhg.5200859
68. Burwell RG. Aetiology of idiopathic scoliosis: current concepts. Pediatr Rehabil. (2003) 6(3–4):137–70. doi: 10.1080/13638490310001642757
69. Weiss HR. Adolescent idiopathic scoliosis (AIS)—an indication for surgery? A systematic review of the literature. Disabil Rehabil. (2008) 30(10):799–807. doi: 10.1080/09638280801889717
70. Lai CQ, Parnell LD, Smith CE, Guo T, Sayols-Baixeras S, Aslibekyan S, et al. Carbohydrate and fat intake associated with risk of metabolic diseases through epigenetics of CPT1A. Am J Clin Nutr. (2020) 112(5):1200–11. doi: 10.1093/ajcn/nqaa233
71. Pérez-Machado G, Berenguer-Pascual E, Bovea-Marco M, Rubio-Belmar PA, García-López E, Garzón MJ, et al. From genetics to epigenetics to unravel the etiology of adolescent idiopathic scoliosis. Bone. (2020) 140:115563. doi: 10.1016/j.bone.2020.115563
72. Peterson LE, Nachemson AL. Prediction of progression of the curve in girls who have adolescent idiopathic scoliosis of moderate severity. Logistic regression analysis based on data from the brace study of the scoliosis research society. J Bone Joint Surg Am. (1995) 77(6):823–7. doi: 10.2106/00004623-199506000-00002
73. Roye BD, Wright ML, Matsumoto H, Yorgova P, McCalla D, Hyman JE, et al. An independent evaluation of the validity of a DNA-based prognostic test for adolescent idiopathic scoliosis. J Bone Joint Surg-Am Vol. (2015) 97A(24):1994–8. doi: 10.2106/JBJS.O.00217
74. Meng YC, Lin T, Liang SL, Gao R, Jiang H, Shao W, et al. Value of DNA methylation in predicting curve progression in patients with adolescent idiopathic scoliosis. EBioMedicine. (2018) 36:489–96. doi: 10.1016/j.ebiom.2018.09.014
75. Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. (2003) 33:245–54. doi: 10.1038/ng1089
76. McMaster ME. Heated indoor swimming pools, infants, and the pathogenesis of adolescent idiopathic scoliosis: a neurogenic hypothesis. Environ Health. (2011) 10:86. doi: 10.1186/1476-069X-10-86
77. McMaster MJ. Spinal growth and congenital deformity of the spine. Spine. (2006) 31(20):2284–7. doi: 10.1097/01.brs.0000238975.90422.c4
78. Bertram H, Steck E, Zimmermann G, Chen BH, Carstens C, Nerlich A, et al. Accelerated intervertebral disc degeneration in scoliosis versus physiological ageing develops against a background of enhanced anabolic gene expression. Biochem Biophys Res Commun. (2006) 342(3):963–72. doi: 10.1016/j.bbrc.2006.02.048
79. Bernstein P, Hentschel S, Platzek I, Hühne S, Ettrich U, Hartmann A, et al. Thoracal flat back is a risk factor for lumbar disc degeneration after scoliosis surgery. Spine J. (2014) 14(6):925–32. doi: 10.1016/j.spinee.2013.07.426
80. Zloof Y, Ankory R, Braun AE, Braun M, Abuhasira S, Schwartz N, et al. The hereditary nature of adolescent spinal deformities a study of over 600,000 adolescents. Spine. (2022) 47(12):841–6. doi: 10.1097/BRS.0000000000004355
81. Wise CA, Sepich D, Ushiki A, Khanshour AM, Kidane YH, Makki N, et al. The cartilage matrisome in adolescent idiopathic scoliosis. Bone Res. (2020) 8(1):13. doi: 10.1038/s41413-020-0089-0
82. Xie HB, Li MZ, Kang YS, Zhang JJ, Zhao CT. Zebrafish: an important model for understanding scoliosis. Cell Mol Life Sci. (2022) 79(9):16. doi: 10.1007/s00018-022-04534-5
83. Patten SA, Margaritte-Jeannin P, Bernard JC, Alix E, Labalme A, Besson A, et al. Functional variants of POC5 identified in patients with idiopathic scoliosis. J Clin Invest. (2015) 125(3):1124–8. doi: 10.1172/JCI77262
84. Haller G, Alvarado D, McCall K, Yang P, Cruchaga C, Harms M, et al. A polygenic burden of rare variants across extracellular matrix genes among individuals with adolescent idiopathic scoliosis. Hum Mol Genet. (2016) 25(1):202–9. doi: 10.1093/hmg/ddv463
Keywords: idiopathic scoliosis, genes, data visualization, bibliometrics, Citespace, VOSviewer
Citation: Ru L, Zheng H, Lian W, Zhao S and Fan Q (2023) Knowledge mapping of idiopathic scoliosis genes and research hotspots (2002–2022): a bibliometric analysis. Front. Pediatr. 11:1177983. doi: 10.3389/fped.2023.1177983
Received: 2 March 2023; Accepted: 6 November 2023;
Published: 4 December 2023.
Edited by:
Daniel Robert Schlatterer, WellStar Health System, United States© 2023 Ru, Zheng, Lian, Zhao and Fan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hong Zheng ZHJ6aGVuZ2hzQDEyNi5jb20=
 Like Ru
Like Ru Hong Zheng
Hong Zheng Wenjun Lian1
Wenjun Lian1