
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Pediatr. , 30 May 2023
Sec. Pediatric Otolaryngology
Volume 11 - 2023 | https://doi.org/10.3389/fped.2023.1141348
This article is part of the Research Topic Basic and Clinical Research on Children Snoring and Obstructive Sleep Apnea Syndrome View all 11 articles
Background: We investigated the expression and the potential value of plasma transfer RNA-derived fragments (tRFs) of children with obstructive sleep apnea–hypopnea syndrome (OSAHS) as screening biomarkers.
Methods: At first, we randomly selected five plasma samples from the case group and the control group for high-throughput RNA sequencing. Secondly, we screened two tRFs with different expression between the two groups, amplified it by quantitative reverse transcription-PCR (qRT-PCR) on all samples. Then we analyzed the diagnostic value of the tRFs and their correlation with the clinical data.
Results: A total of 50 OSAHS children and 38 healthy controls were included. Our results demonstrated that the plasma levels of tRF-16-79MP9PD and tRF-28-OB1690PQR304 were significantly down-regulated in OSAHS children. Receiver operating characteristic curve (ROC) showed that the area under the curve (AUC) of tRF-16-79MP9PD and tRF-28-OB1690PQR304 was 0.7945 and 0.8276. In addition, the AUC of the combination reached 0.8303 with 73.46% and 76.42% sensitivity and specificity. Correlation analysis showed that the degree of tonsil enlargement, hemoglobin (Hb) and triglyceride (TG). were related to the expression levels of tRF-16-79MP9PD and tRF-28-OB1690PQR304. Multivariable linear regression analysis showed that degree of tonsil enlargement, Hb and TG related to tRF-16-79MP9PD while degree of tonsil enlargement and Hb related to tRF-28-OB1690PQR304.
Conclusions: The expression levels of tRF-16-79MP9PD and tRF-28-OB1690PQR304 in the plasma of OSAHS children decreased significantly which were closely related to the degree of tonsil enlargement, Hb and TG, may become novel biomarkers for the diagnosis of pediatric OSAHS.
Obstructive sleep apnea-hypopnea syndrome (OSAHS) is a common disorder characterized by the snore and repeated episodes of partial or complete collapse of the upper respiratory tract during sleep (1). It has been widely accepted that hypoxia stress caused by upper airway obstruction in OSAHS can lead to systemic multiple system and organ damage (2). The main cause of upper airway obstruction in children is adenoid and/or tonsil hypertrophy (3). OSAHS can occur at all ages, with the peak of incidence occurring between 2 and 8 years old, and the most obvious clinical symptoms and surgical intervention mainly occur between 4 and 7 years old because of tonsil and adenoid hypertrophy accounting for the largest proportion of upper airway volume at this age (4, 5). The incidence of pediatric OSAHS is up to 1%–3% and is on the rise (6). OSAHS may lead to a series of pathological changes such as abnormal behavior, cognitive dysfunction, growth retardation and so on (7–10). In addition, many studies have shown that long-term OSAHS condition can increase the risk of diabetes, hypertension, metabolic diseases and cardiovascular diseases (11–13).
Transfer RNAs (tRNAs) are a class of non-coding RNAs that transport amino acids and assist protein synthesis (14). Under special conditions such as hypoxia, hunger and stress, pre- and mature tRNAs may generate new specific small RNA fragments after enzymatic splicing and chemical modification, which is called tRNA-derived small RNAs (tsRNAs) (15, 16). tsRNAs can also be mainly divided into tRNA halves and tRNA-derived fragments (tRFs) according to the cleavage loci and length (15). It has been reported that tRFs has a variety of biological functions, such as regulating translation level, affecting gene expression and inhibiting cell apoptosis (17). Recently, with the development of high-throughput sequencing and chip technology, novel tRFs have been gradually discovered and attracted researchers' interests. The relationship between tRFs and the occurrence of human diseases such as cancer and metabolic diseases has also been gradually revealed (18, 19).
Intermittent hypoxia is one of the typical characteristics of OSAHS, which can increase the expression of reactive oxygen species, resulting in systemic inflammation state, endothelial dysfunction and metabolic dysregulation (20). Therefore, biomarkers of hypoxia and oxidative stress may also be used to evaluate OSAHS. tsRNAs will be produced when cells are cleaved at specific tRNA sites in response to environmental stresses such as hypoxia, oxidative stress or viral infection (21). In recent years, several OSAHS-related differential microRNAs have been identified to be involved in a variety of pathophysiological processes (22, 23). Taken together, we speculate that tsRNAs may be related to the occurrence and development of OSAHS. In this study, we aim to explore the expression and the potential diagnostic screening value of plasma tRFs in children with OSAHS and further determine the association between tRFs and clinically relevant data in OSAHS children.
This study was an observational clinical study. We collected data from 50 patients aged 4–7 years old who were diagnosed with OSAHS after PSG based on medical history in Ningbo Women and Children's Hospital (Zhejiang, China) from November 2020 to December 2021. The diagnostic criteria of OSAHS refer to Chinese Guideline for the Diagnosis and Treatment of Childhood Obstructive Sleep Apnea (2020) (3), which diagnoses children with obstructive sleep apnea hypopnea index (OAHI) ≥1 events/h as OSAHS. Exclusion criteria for this study were children with congenital abnormalities of the nasolaryngology or airway, craniofacial malformation, anemia, digestive tract malformation, and those who had used antibiotics or probiotics in the last 3 months. In addition, 38 healthy control children were recruited as the control group and underwent free PSG examination.
In this study, basic information such as age, gender, body weight and height of all subjects were collected. Body mass index (BMI) was calculated by dividing weight by height squared.
This study has been approved by the ethical committee of Ningbo Women and Children's Hospital on November 5th, 2020 (Approval No. EC2020-047) and all participants have signed the informed written consent forms.
All blood samples were collected from the peripheral venous blood of the children in Ningbo Women and Children's Hospital. For tsRNA studies, blood was firstly harvested into an ethylenediaminetetraacetic acid anticoagulation tube (EDTA). Within 1 h of blood collection, all specimens were centrifuged at 3,000 rpm/10 mins under 4°C to obtain plasma. The supernatant was transferred to RNase-free Eppendorf tubes and then stored at −80°C until RNA extraction.
Total RNA, including tsRNA, was extracted and isolated from plasma samples by using TRIzol LS reagent (Invitrogen, Carlsbad, CA, USA) according to the instructions of manufacturer. RNA concentration was measured on NanoDrop™ Nd-1000 spectrophotometer (NanoDrop, Thermo Fisher Scientific, Inc., Wilmington, DE, USA) at 260 nm, 280 nm. The acceptance ratio standard for the study is 1.8–2.0. The extracted total RNA was stored at −80°C for subsequent experiments.
In this study, plasma samples of 5 pairs of children with OSAHS and healthy controls were firstly selected. After extracting total RNA, small isolated RNA was processed with the rtStarTM tRF and tiRNA Pretreatment Kit (Arraystar, Rockville, MD, USA) to remove the superfluous modification groups. Furthermore, compared to the original RNA, pretreated RNA had been purified twice to make it meet the concentration requirements for subsequent experiments. Purified RNA was then sent to Shanghai Kangcheng Company (Shanghai, China) for constructing small RNA libraries and performing small RNA sequencing analysis. Briefly, according to the requirements of the commercial kit NEB Next® Multiplex Small RNA Library Prep Set for Illumina (New England BioLabs, Inc., Ipswich, MA, USA), the pretreated RNA was bound with 3′ and 5′ adapters, and small RNA library was established after adding reverse transcription primers to synthesize cDNA. The Illumina NextSeq 500 (Illumina, Inc., San Diego, CA, USA) system was then used for sequencing.
In the original sequencing data, low-quality reads and short reads (<15 nt) should be firstly filtered. After this step, all selected RNA reads were aligned to several non-coding RNA database, such as miRBase database (http://www.mirbase.org/), Genomic tRNA database (http://gtrnadb.ucsc.edu/), tRFdb (http://genome.bioch.virginia.edu/trfdb/) and MintBase (https://cm.jefferson.edu/MINTbase/). In the expression of tsRNAs between OSAHS and healthy controls, the clean data was measured after the expression level was normalized. We believed that tsRNAs with a fold change of ≥2.0 and P < 0.05 are significantly different expressions. We chose tRF-16-79MP9PD and tRF-28-OB1690PQR304 for follow-up research.
Two thousand nanograms of RNA were reverse-transcribed to cDNA according to the instruction of the rtStar™ First-strand cDNA Synthesis Kit (Arraystar, Rockville, MD, USA). First step, input 1 µl of 3′ Adaptor, 0.5 µl of RNA Spike-in and nuclease-free water into 2000 ng RNA to make a total volume of 8.8 µl, then incubate the mixture at 70°C for 2 min. Second, we added 7.2 µl of 3′ Ligation Reaction Buffer, 1 µl 3′ Ligation Enzyme Mix and 1 µl of RNase Inhibitor in sequence, which was then incubated for 1 h at 25°C. Then the procedure involves addition of a 1 µl of nuclease-free water and 1 µl of RT-primer, and incubation for 5 min at 75°C, 15 min at 37°C, 15 min at 25°C. Third, 1 µl of 5′Adaptor (denatured), 2.5 µl of 10 mM ATP, 0.5 µl of 5′ Ligation Reaction Buffer, 1 µl of 5′ Ligation Enzyme Mix and 20 µl samples were mixed and incubated at 25°C at 1 h. Lastly, 25 µl of Adaptor-Ligated RNA, 8 µl of First-Strand synthesis reaction buffer, 3 µl of 0.1 MDTT, 2 µl of 2.5 mM dNTP Mix, 1 µl of RNase Inhibitor and 1 µl of Reverse Transcriptase were mixed in a nuclease-free tube, and the sample was heated for 60 min at 45°C, and then to 75°C for 15 min. qRT-PCR was firstly conducted on plasma samples of the previous 5 pairs of children with OSAHS and healthy controls, and the sequencing results of PCR products showed complete matching with tsRNA original sequence (Figure 1). After that, qRT-PCR was used to detect more differences in the expression levels of OSAHS children and healthy children, so as to verify whether it was consistent with the previous sequencing results. The qRT-PCR reaction was performed on a Mx3005P Real-Time PCR System (Stratagene, Palo Alto, CA, USA). The PCR primer synthesis (Table 1) and sequencing of PCR products were done by Sangon Biotech (Shanghai, China). U6 was used as an internal reference gene, and the relative gene expression was calculated as ΔCT = CTtRF − CTU6, Larger ΔCT values indicate lower expression.
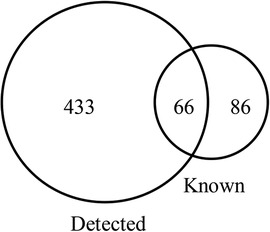
Figure 1. The number of tsRNAs detected in 5 OSAHS children and 5 healthy children plasma samples and the number of tsRNAs known in the tRFs database.
Statistical analysis was performed using SPSS version 24.0 and GraphPad Prism 8.0. The measurement data were expressed as mean ± standard deviation (X ± SD) and the counting data were expressed in percentage (%). independent samples t-test was used to evaluate the significance of measurement data between the two groups. χ2 test was used for comparison of counting data. Receiver operating characteristic (ROC) curve and area under the curve (AUC) were carried out to evaluate the diagnostic value of tRFs in OSAHS. Correlations between tRFs and relevant clinical parameters were analyzed by Pearson and Spearman correlation analysis. Multivariate linear regression analysis was performed to assess the association between plasma tRFs levels and several clinical parameters. All experiments were repeated at least three times independently. P value < 0.05 was considered statistically significant.
A total of 50 OSAHS children and 38 healthy controls were enrolled. Parameters of all subjects are summarized in Table 2. There were significant differences in OAHI and lowest oxygen saturation (LaSO2) between the OSAHS children and the controls. Besides, there were differences in height, serum creatinine (Scr) and total cholesterol (TC) between the two groups.
Among the 10 sequencing libraries, the average number of original reads was about 6.69 million and the average reads ratio of high-quality bases was about 96.97%. Among them, microRNAs accounted for the highest proportion, with an average read number of 935,592.4 (17.64%). The average reads of mature tRNA and precursor tRNA were 156,419.3 (3.04%) and 5,973.1 (0.12%). A total of 499 tsRNAs were detected, of which 66 were recorded in the tRF database (Figure 1). Through the comparison between the two groups, there were 11 tsRNAs that were significantly differentially expressed, of which two tsRNAs were up-regulated and the remaining 9 tsRNAs were down-regulated (Figure 2). Taking into account the fold change, P value and tsRNAs expression difference between two groups, we selected tRF-16-79MP9PD and tRF-28-OB1690PQR304 as our research objects. After that, we designed primers for the two tsRNAs, and the sequencing results of the qRT-PCR products proved the specificity of the primers (Figure 3).
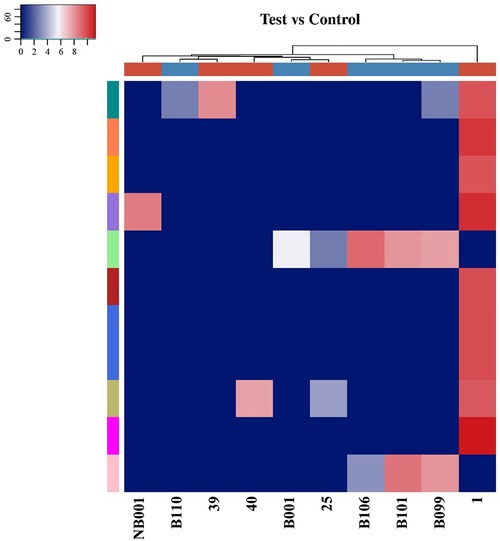
Figure 2. The difference of tsRNAs expression between OSAHS children and healthy children was analyzed by hierarchical clustering heat maps. Each row represented a tsRNA and each column represented a sample. The 25, 1, 39, NB001, and 40 were plasma specimens of children with OSAHS. The B009, B110, B106, B001, and B101 were plasma specimens of normal children. The degree of difference gradually increases from blue to red.
To further investigate the differential expression of tRF-16-79MP9PD and tRF-28-OB1690PQR304 in OSAHS children, we verified the expression of both tRFs in an extended cohort. As expected, our results showed that the expression levels of tRF-16-79MP9PD and tRF-28-OB1690PQR304 were significantly lower in patients with OSAHS than in controls (both, P < 0.0001), which was consistent with the results of high-throughput sequencing (Figure 4).
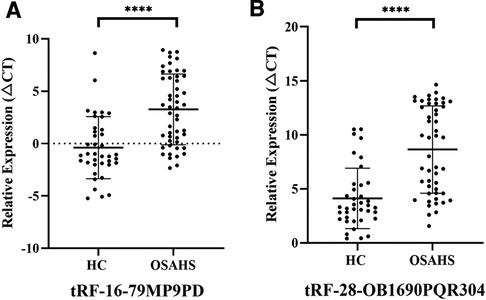
Figure 4. Expression of tRF-16-79MP9PD (A) (t = 5.289, 95% CI: 2.278–5.022) and tRF-28-OB1690PQR304 (B) (t = 5.886, 95% CI: 2.994–6.049) in patients with OSAHS. ****p < 0.0001; CI, Confidence Interval.
The ROC analysis revealed that the AUCs of tRF-16-79MP9PD and tRF-28-OB1690PQR304 were 0.7945 with 52% sensitivity and 94.74% specificity (Figure 5A), 0.8276 with 79.59% sensitivity and 71.05% specificity, respectively (Figure 5B). Furthermore, the AUC of the combination reached 0.8303 with 73.46% sensitivity and 76.32% specificity (Figure 5C).
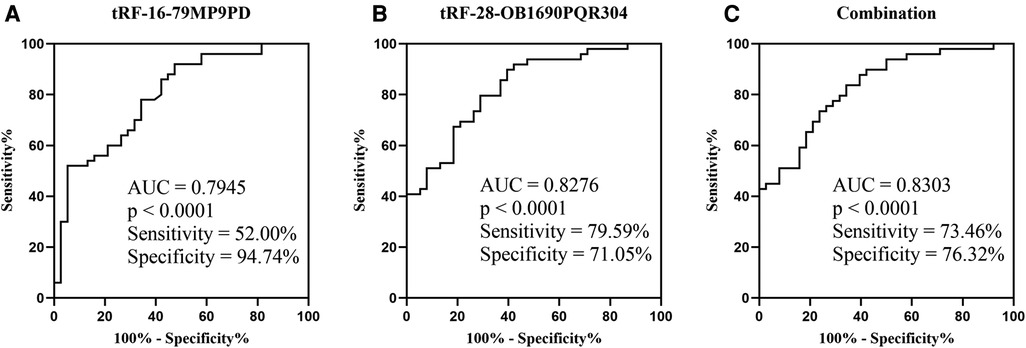
Figure 5. ROC of plasma tRF-16-79MP9PD (A) (SE: 0.0478, 95% CI: 0.7007–0.8882), tRF-28-OB1690PQR304 (B) (SE: 0.0433, 95% CI: 0.7428–0.9124) and the combination (C) (SE: 0.0428, 95% CI: 0.7465–0.9141; OR: tRF-16-79MP9PD: 1.1219, tRF-28-OB1690PQR304: 0.6419). SE, Standard Error; OR, Odds Ratio.
Firstly, we used simple linear correlation to analyze the correlation between tRF-16-79MP9PD and tRF-28-OB1690PQR304 and the clinical parameters we collected. The visual correlation heat map (Figure 6) showed the correlation between all parameters. We found that the level of tRF-16-79MP9PD was significantly correlated with the degree of tonsil enlargement, Hb, TG and CK-MB level (Figures 7A–D) while tRF-16-79MP9PD was correlated with the degree of tonsil enlargement, Hb and TG (Figures 7E–G).
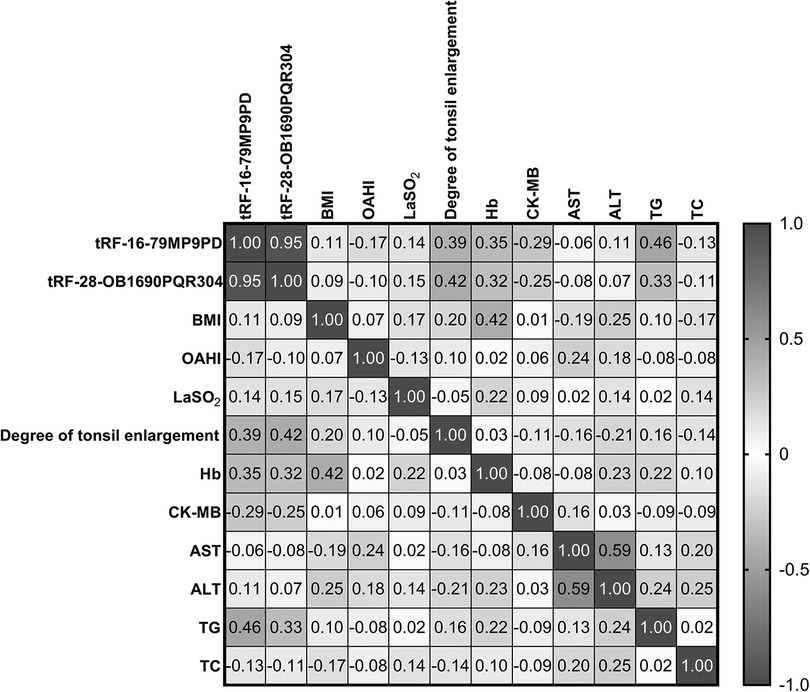
Figure 6. The visual correlation heat map of correlation between tRF-16-79MP9PD and tRF-28-OB1690PQR304 and the clinical parameters.
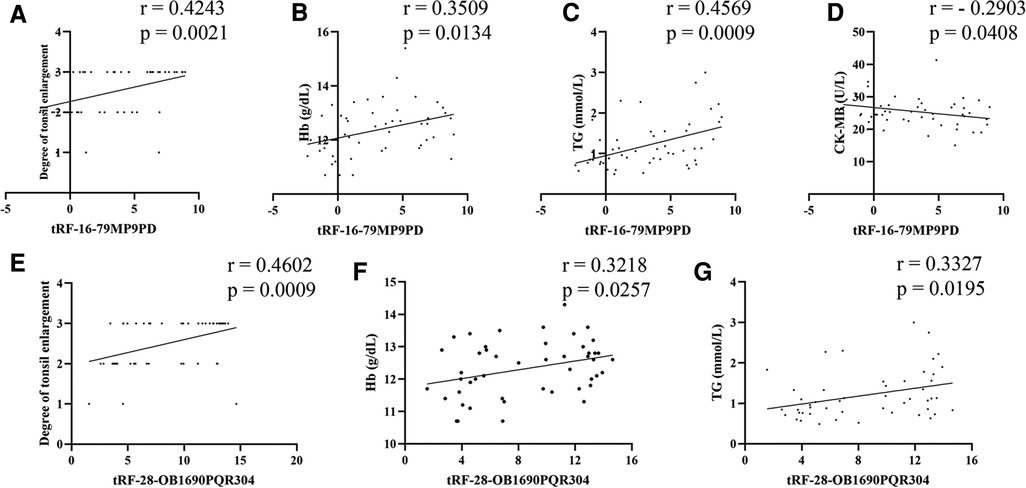
Figure 7. Correlation between tRF-16-79MP9PD and the degree of tonsil enlargement (A), Hb (B), TG (C) and CK-MB (D) level, tRF-28-OB1690PQR304 and the degree of tonsil enlargement (E), Hb (F), TG (G). Hb, hemoglobin, TG, triglyceride, CK-MB, creatine kinase-MB.
Subsequently, we performed multiple linear regression using the regression method. The results showed that degree of tonsil enlargement, Hb and TG levels were related to the tRF-16-79MP9PD expression (Table 3) whose mathematic model was y = −10.309 + 0.902x1 + 1.887x2 − 0.160x3 + 1.656x4. Differently, Hb and the degree of tonsil enlargement were related to tRF-28-OB1690PQR304 (Table 4) whose mathematic model was y = −14.383 + 1.353x1 + 2.503x2.
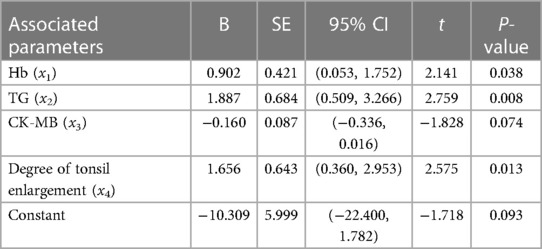
Table 3. Multivariate linear regression analysis of plasma tRF-16-79MP9PD level and clinical parameters.
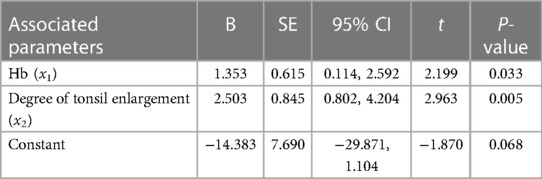
Table 4. Multivariate linear regression analysis of plasma tRF-28-OB1690PQR304 level and clinical parameters.
As one of the most serious diseases among childhood sleep disordered breathing, OSAHS is characterized by hypoventilation, decreased blood oxygen saturation and disrupted sleep structure (24). Currently, guidelines recommend PSG as a standard diagnostic method for pediatric OSAHS. However, standard PSG monitoring is limited by the equipment, operation, personnel and cost of standard PSG monitoring. Although many efforts have been made to explore biomarkers of OSAHS, there is currently no definite biomarker available in the clinic.
tsRNAs exist widely in tissues and cells of various organisms with tissue specificity and disease correlation and can play roles in cell proliferation, regulation of gene expression, RNA processing, modulation of DNA damage response and tumor suppression (25). tRFs can be expressed and measured in human body fluids such as serum, plasma and urine. In addition, they are not easily to be degraded by RNase due to their short length. Thus, tRFs have great potential to serve as a non-invasive biomarker for various diseases (18). Accumulating evidence has demonstrated tRFs serve as ideal biomarkers and therapeutic targets in a variety of diseases, such as prostate cancer (26), pancreatic ductal adenocarcinoma (27) and systemic lupus erythematosus (28).
To our knowledge, this study was the first to investigate the plasma tRFs levels in children with OSAHS. Specifically, we first analyzed general baseline data of the OSAHS and HC groups. The results showed that the height of children with OSAHS was lower than that of HC group, which was consistent with the research of Johnson C (29) and may due to the decrease of growth hormone secretion caused by poor sleep quality of children with OSAHS. Similarly, the two groups had statistical differences in Scr and TC, which was consistent with the studies of other scholars (30, 31). Secondly, we performed small RNA sequencing to detect the expression profile of tRFs in 5 pairs of OSAHS children and control specimens. The results showed that the expression of 11 tsRNAs was significantly different between the two groups. Two dysregulated tRFs: tRF-16-79MP9PD and tRF-28-OB1690PQR304 were screened out based on fold changes, P value and tsRNAs expression differences. Subsequently, qRT-PCR was performed in the extended cohort to verify the authenticity of the sequencing results. It was proved that the expression levels of tRF-16-79MP9PD and tRF-28-OB1690PQR304 were down-regulated in the OSAHS group compared with the HC group, which was consistent with the results of small RNA sequencing, indicating that the decreased expression of tRF-16-79MP9PD and tRF-28-OB1690PQR304 were closely related to OSAHS.
In addition, we further evaluated the diagnostic efficacy of tRF-16-79MP9PD and tRF-28-OB1690PQR304 by ROC curve analysis, and the results demonstrated that tRF-16-79MP9PD and tRF-28-OB1690PQR304 had high sensitivity and specificity to distinguish OSAHS from healthy children, processing AUCs of 0.7945 with 52% sensitivity and 94.74% specificity, 0.8276 with 79.59% sensitivity and 71.05% specificity, respectively. More importantly, the AUC of their combination increased to 0.8303 with 73.46% sensitivity and 76.32% specificity, suggesting the great potential of tRF-16-79MP9PD and tRF-28-OB1690PQR304 as diagnosis biomarkers for childhood OSAHS. At present, studies on OSAHS in children have not obtained clear biomarkers, and existing studies also have some limitations, such as lack of specificity and sensitivity analysis (32). In our study, the AUC of the joint diagnosis of two tRFs exceeds 0.83, and the sensitivity and specificity exceed 70%, which has certain clinical screening diagnostic value. As a clinical OSAHS screening indicator, tRF can serve as a new research direction, but its diagnostic value needs to be evaluated in multi-center studies with larger sample sizes.
Furthermore, we analyzed the correlation between tRFs levels and laboratory parameters in OSAHS children. We found that the expression levels of tRF-16-79MP9PD and tRF-28-OB1690PQR304 were closely related to the degree of tonsil enlargement. At the same time, we found that the expression level of tRF-16-79MP9PD was also significantly correlated with Hb, TG and CK-MB level. Multivariable linear regression analysis showed that Hb, TG and degree of tonsil enlargement were related to tRF-16-79MP9PD. In addition, tRF-28-OB1690PQR304 was significantly correlated with Hb and TG. The degree of tonsil enlargement and Hb were related to the tRF-28-OB1690PQR304 level through multivariable linear regression analysis. Thus, the plasma levels of tRF-16-79MP9PD and tRF-28-OB1690PQR304 are related to a variety of laboratory indicators in OSHAS. More experiments are needed to investigate the role of tRF-16-79MP9PD and tRF-28-OB1690PQR304 in OSAHS.
Nevertheless, there are still some limitations to our study. Firstly, the sample size of this study was 50 OSAHS children and 38 healthy controls. Further expansion of sample size is needed to evaluate the diagnostic value of tRFs. Secondly, this study did not explore the function of tRFs by combining the pathological and physiological mechanisms of OSAHS. In the future, we will explore the role of tRFs in the pathogenesis of OSAHS based on this research. Thirdly, this study was limited to children in Ningbo City. Considering regional and racial differences, future multicenter and larger sample size studies are needed to evaluate the value of tRF as a biomarker.
In conclusion, our study identified that the plasma levels of tRF-16-79MP9PD and tRF-28-OB1690PQR304 were down-regulated in 4- to 7-year-old children with OSAHS and were closely related to the clinical parameters of OSAHS children. tRF-16-79MP9PD and tRF-28-OB1690PQR304 may be used as a new potential biomarker for the diagnosis of pediatric OSAHS.
The data presented in the study are deposited in the NCBI BioProject repository, accession number PRJNA862251.
The studies involving human participants were reviewed and approved by medical Ethics Committee of Ningbo Women and Children's Hospital. Written informed consent to participate in this study was provided by the participants’ legal guardian/next of kin.
JW and ZS conceived and designed the study, and reviewed and revised the manuscript. XC and YL analyzed and drafted the initial manuscript. YS completed the PCR experiment in this study and wrote the method of manuscript. QL coordinated and supervised data analyses, and critically reviewed the manuscript for important intellectual content. All authors approved the final manuscript as submitted and agree to be accountable for all aspects of the work. JW and XC contributed equally to this work. ZS and QL contributed equally to this work. All authors contributed to the article and approved the submitted version.
This study was supported by the Ningbo Clinical Research Center for Children's Health and Diseases (No. 2019A21002), the Science and Technology Projects of Medicine and Health of Zhejiang (No. 2021KY1056), the Ningbo public welfare science and technology plan project (No. 2021S100), Ningbo key discipline Pediatrics (No. 2022-B17), Ningbo Top Medical and Health Research Program (No. 2022020405) and Ningbo Clinical Research Center for Otolaryngology Head and Neck Disease (No. 2022L005).
The authors thank all the children and their parents who participated in this study.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
tRNAs, transfer RNAs; tRFs, transfer RNA related fragments; tsRNAs, tRNA-derived small RNAs; OSAHS, obstructive sleep apnea–hypopnea syndrome; qRT-PCR, quantitative reverse transcription- Polymerase Chain Reaction; BMI, Body mass index; OAHI, Obstructive apnea hypopnea index; ROC, Receiver operating characteristic curve; AUC, area under the curve; HB, hemoglobin; TG, triglyceride; LaSO2, lowest oxygen saturation; SBP, systolic blood pressure; DBP, diastolic blood pressure; Hb, hemoglobin; ALT, alanine aminotransferase; AST, aspartate aminotransferase; CK-MB, creatine kinase-MB; UA, uric acid; BUN, blood urea nitrogen; TG, triglyceride; TC, total cholesterol.
1. Bitners AC,, Arens R. Evaluation and management of children with obstructive sleep apnea syndrome. Lung. (2020) 198(2):257–70. doi: 10.1007/s00408-020-00342-5
2. Ji L, Liu Y, Liu P, Ji G, He J, Gan Y, et al. Serum periostin and TNF-α levels in patients with obstructive sleep apnea-hypopnea syndrome. Sleep Breath. (2021) 25(1):331–7. doi: 10.1007/s11325-020-02124-y
3. Ni X, Gao Z, Han D, Wang C, Wang Q, Wei J, et al. Chinese Guideline for the diagnosis and treatment of childhood obstructive sleep apnea (2020). Chin J Otorhinolaryngol Head Neck Surg. (2020) 55(8):729–46.
4. Yang DZ, Liang J, Zhang F, Yao HB, Shu Y. Clinical effect of montelukast sodium combined with inhaled corticosteroids in the treatment of OSAS children. Medicine (Baltimore). (2017) 96(19):e6628. doi: 10.1097/MD.0000000000006628
5. Gipson K, Lu M, Kinane TJ. Sleep-disordered breathing in children. Pediatr Rev. (2019) 40(1):3–13. doi: 10.1542/pir.2018-0142
6. Akkari M, Yildiz S, Marianowski R, Monteyrol PJ, Chalumeau F, Fayoux P, et al. Role of the ENT specialist in the diagnosis of pediatric obstructive sleep apnea-hypopnea syndrome (POSAHS). part 3: sleep recordings. Eur Ann Otorhinolaryngol Head Neck Dis. (2020) 137(5):405–10. doi: 10.1016/j.anorl.2020.02.001
7. Zhao J, Han S, Zhang J, Wang G, Wang H, Xu Z, et al. Association between mild or moderate obstructive sleep apnea-hypopnea syndrome and cognitive dysfunction in children. Sleep Med. (2018) 50:132–6. doi: 10.1016/j.sleep.2018.04.009
8. Shi Y, Feng Y, Zhang Y, Liu H, Luo H, Shang L, et al. Assessment of event-related evoked potentials and China-Wechsler intelligence scale for cognitive dysfunction in children with obstructive sleep apnea. Int J Pediatr Otorhinolaryngol. (2021) 150:110901. doi: 10.1016/j.ijporl.2021.110901
9. Esteller E, Villatoro JC, Agüero A, Lopez R, Matiñó E, Argemi J, et al. Obstructive sleep apnea syndrome and growth failure. Int J Pediatr Otorhinolaryngol. (2018) 108:214–8. doi: 10.1016/j.ijporl.2018.03.011
10. Lagravère MO, Zecca PA, Caprioglio A, Fastuca R. Metabolic effects of treatment in patients with obstructive sleep apnea: a systematic review. Minerva Pediatr. (2019) 71(4):380–9. doi: 10.23736/S0026-4946.18.05223-4
11. Song SO, He K, Narla RR, Kang HG, Ryu HU, Boyko EJ. Metabolic consequences of obstructive sleep apnea especially pertaining to diabetes mellitus and insulin sensitivity. Diabetes Metab J. (2019) 43(2):144–55. doi: 10.4093/dmj.2018.0256
12. Kario K, Hettrick DA, Prejbisz A, Januszewicz A. Obstructive sleep apnea-induced neurogenic nocturnal hypertension: a potential role of renal denervation?. Hypertension. (2021) 77(4):1047–60. doi: 10.1161/HYPERTENSIONAHA.120.16378
13. Strausz S, Havulinna AS, Tuomi T, Bachour A, Groop L, Mäkitie A, et al. Obstructive sleep apnoea and the risk for coronary heart disease and type 2 diabetes: a longitudinal population-based study in Finland. BMJ Open. (2018) 8(10):e022752. doi: 10.1136/bmjopen-2018-022752
14. Sun C, Fu Z, Wang S, Li J, Li Y, Zhang Y, et al. Roles of tRNA-derived fragments in human cancers. Cancer Lett. (2018) 414:16–25. doi: 10.1016/j.canlet.2017.10.031
15. Yu X, Xie Y, Zhang S, Song X, Xiao B, Yan Z tRNA-derived fragments: mechanisms underlying their regulation of gene expression and potential applications as therapeutic targets in cancers and virus infections. Theranostics. (2021) 11(1):461–9. doi: 10.7150/thno.51963
16. Zhu L, Ge J, Li T, Shen Y, Guo J. tRNA-derived fragments and tRNA halves: the new players in cancers. Cancer Lett. (2019) 452:31–7. doi: 10.1016/j.canlet.2019.03.012
17. Zeng T, Hua Y, Sun C, Zhang Y, Yang F, Yang M, et al. Relationship between tRNA-derived fragments and human cancers. Int J Cancer. (2020) 147(11):3007–18. doi: 10.1002/ijc.33107
18. Zhang Y, Bi Z, Dong X, Yu M, Wang K, Song X, et al. tRNA-derived fragments: tRF-gly-CCC-046, tRF-tyr-GTA-010 and tRF-pro-TGG-001 as novel diagnostic biomarkers for breast cancer. Thorac Cancer. (2021) 12(17):2314–23. doi: 10.1111/1759-7714.14072
19. Han X, Cai L, Lu Y, Li D, Yang J. Identification of tRNA-derived fragments and their potential roles in diabetic cataract rats. Epigenomics. (2020) 12(16):1405–18. doi: 10.2217/epi-2020-0193
20. Labarca G, Gower J, Lamperti L, Dreyse J, Jorquera J. Chronic intermittent hypoxia in obstructive sleep apnea: a narrative review from pathophysiological pathways to a precision clinical approach. Sleep Breath. (2020) 24(2):751–60. doi: 10.1007/s11325-019-01967-4
21. Li S, Xu Z, Sheng J. tRNA-derived small RNA: a novel regulatory small non-coding RNA. Genes (Basel). (2018) 9(5):246. doi: 10.3390/genes9050246
22. Shao H, Shen P, Chen J. Expression profile analysis and image observation of miRNA in serum of patients with obstructive sleep apnea-hypopnea syndrome. Contrast Media Mol Imaging. (2021) 2021:9731502. doi: 10.1155/2021/9731502
23. Pinilla L, Barbé F, De Gonzalo-Calvo D. MicroRNAs to guide medical decision-making in obstructive sleep apnea: a review. Sleep Med Rev. (2021) 59:101458. doi: 10.1016/j.smrv.2021.101458
24. Al-Shamrani A, Alharbi AS. Diagnosis and management of childhood sleep-disordered breathing. Clinical approach. Saudi Med J. (2020) 41(9):916–29. doi: 10.15537/smj.2020.9.25262
25. Kumar P, Kuscu C, Dutta A. Biogenesis and function of transfer RNA-related fragments (tRFs). Trends Biochem Sci. (2016) 41(8):679–89. doi: 10.1016/j.tibs.2016.05.004
26. Olvedy M, Scaravilli M, Hoogstrate Y, Visakorpi T, Jenster G, Martens-Uzunova ES. A comprehensive repertoire of tRNA-derived fragments in prostate cancer. Oncotarget. (2016) 7(17):24766–77. doi: 10.18632/oncotarget.8293
27. Pan L, Huang X, Liu ZX, Ye Y, Li R, Zhang J, et al. Inflammatory cytokine-regulated tRNA-derived fragment tRF-21 suppresses pancreatic ductal adenocarcinoma progression. J Clin Invest. (2021) 131(22):e148130. doi: 10.1172/JCI148130
28. Geng G, Wang H, Xin W, Liu Z, Chen J, Danting Z, et al. tRNA derived fragment (tRF)-3009 participates in modulation of IFN-α-induced CD4(+) T cell oxidative phosphorylation in lupus patients. J Transl Med. (2021) 19(1):305. doi: 10.1186/s12967-021-02967-3
29. Johnson C, Leavitt T, Daram SP, Johnson RF, Mitchell RB Obstructive sleep apnea in underweight children. Otolaryngol Head Neck Surg. (2021) 167(3):566–72. doi: 10.1177/01945998211058722
30. Xu H, Guan J, Yi H, Zou J, Meng L, Tang X, et al. Elevated low-density lipoprotein cholesterol is independently associated with obstructive sleep apnea: evidence from a large-scale cross-sectional study. Sleep Breath. (2016) 20(2):627–34. doi: 10.1007/s11325-015-1262-3
31. Dong M, Guo F, Zhou T, Wei Q. Association of diabetic nephropathy with the severity of obstructive sleep apnea-hypopnea syndrome in patients with type 2 diabetes mellitus[J]. Endocr J. (2020) 67(5):515–22. doi: 10.1507/endocrj.EJ19-0324
Keywords: obstructive sleep apnea-hypopnea syndrome, tRNA-derived fragments, biomarker, pediatrics, diagnostic screening
Citation: Wu J, Cai X, Lu Y, Shen Y, Shen Z and Lyv Q (2023) Plasma tRF-16-79MP9PD and tRF-28-OB1690PQR304 as potential biomarkers for 4- to 7-year-old children with obstructive sleep apnea-hypopnea syndrome. Front. Pediatr. 11:1141348. doi: 10.3389/fped.2023.1141348
Received: 10 January 2023; Accepted: 12 May 2023;
Published: 30 May 2023.
Edited by:
Zhifei Xu, Capital Medical University, ChinaReviewed by:
Elton Lambert, Texas Children’s Hospital, United States© 2023 Wu, Cai, Lu, Shen, Shen and Lyv. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhisen Shen c3pzNzIxNkAxNjMuY29t Qin Lyv YXJ0b3NlQHNpbmEuY29t
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.