- 1Department of Clinical Laboratory, Children's Hospital of Fudan University, National Children's Medical Center, Shanghai, China
- 2School of Laboratory Medicine and Life Sciences, Wenzhou Medical University, Wenzhou, China
- 3Shanghai Institute of Infectious Disease and Biosecurity, Fudan University, Shanghai, China
Background: Cytomegalovirus (CMV) is the leading cause of congenital infections worldwide and contributes to long-term sequelae in neonates and children. CMV envelope glycoproteins play a vital role in virus entry and cell fusion. The association between CMV polymorphisms and clinical outcomes remains controversial. The present study aims to demonstrate the distribution of glycoprotein B (gB), H (gH) and N (gN) genotypes in congenitally CMV (cCMV) infected symptomatic infants and attempts to figure out the association between viral glycoprotein genotypes and clinical outcomes.
Methods: Genotyping of gB, gH and gN was performed in 42 cCMV symptomatic infants and 149 infants with postnatal CMV (pCMV) infection in Children's hospital of Fudan university. Nested PCR, gene sequencing and phylogenetic analyses were used to identify the genotypes.
Results: Our study demonstrated that: 1. The CMV gB1, gH1 and gN1 were the predominant genotypes among symptomatic cCMV infected infants, while gB1, gH1 and gN3a were more prevalent in pCMV group. gH1 genotype has a significant association with symptomatic cCMV infection (p = 0.006). 2. No significant correlation was found between CMV genotypes and hearing impairment. However, gH1 was more prevalent among cCMV infected infants with moderate/severe hearing loss although without statistical difference (p = 0.130). 3. gB3 was more prevalent among infants with skin petechiae (p = 0.049) and found to be associated with an increased risk of skin petechiae (OR = 6.563). The gN4a subtype was significantly associated with chorioretinitis due to cCMV infection (p = 0.007). 4. Urine viral loads were not significantly associated with different genotypes or hearing impairment among symptomatic cCMV infected infants.
Conclusions: Our findings demonstrated the overall distribution of gB, gH and gN genotypes in infants with symptomatic cCMV infection in Shanghai for the first time. The findings in our study may suggest a possible association between gH1 genotype and early infancy hearing loss. gB3 genotype was associated with a 6.5-fold increased risk of petechiae while gN4a strongly correlated with chorioretinitis due to cCMV infection. No significant correlation was found between urine viral loads and CMV genotypes or hearing impairment in cCMV infected infants.
1. Introduction
Human cytomegalovirus (CMV) belongs to the ß-herpesvirus family infecting most individuals and immunocompromised patients, including the fetus, organ transplant recipients and AIDS population. After primary infection, CMV will establish a lifelong latent infection and could periodically reactivate from latency or reinfection with a new strain (1–4). Cytomegalovirus is the leading cause of congenital infections worldwide while vast majority (90%) of infants with congenital CMV (cCMV) infection is asymptomatic at birth. Approximately 40%–60% of infants with symptomatic and 10%–15% of children with asymptomatic CMV infection will develop long-term neurological sequelae, particularly sensorineural hearing loss (SNHL) (5–12). Congenital CMV infection is the major cause of nongenetic SNHL that could be present at birth or appears later (13). cCMV infection contributes more to the permanent disabilities of infants and young children than other congenital diseases (14–16). Neonates with symptomatic CMV infections are at even higher risk for the adverse neurodevelopmental sequelae and proved to be associated with many severe clinical manifestations (17).
CMV is able to replicate in varieties of cell types, including epithelial cells, endothelial cells, fibroblasts and smooth muscle cells, which facilitate the virus spread within the host and inter-host transmission (18, 19). The broad cell tropism of CMV requires the coordinated interaction of envelope glycoproteins with cell surface receptors (20). The CMV envelope glycoproteins, important targets of virus neutralizing antibodies, are involved in viral entry and cell fusion (21). CMV requires glycoprotein complex gH/gL to fuse with plasma membrane of fibroblasts cells, while entry into epithelial and endothelial cells requires the gH/gL/UL128–131 complex that involves the macropinocytosis and endosomes fusion. Different pathways and glycoprotein complexes play a vital role on the CMV entry into different cell types (22).
Glycoprotein B (gB), encoded by the UL55 gene and classified into 4 genotypes (gB1, gB2, gB3 and gB4), is an abundant and the most highly conserved glycoprotein of CMV. Previous studies have determined that gB is essential for the virus entry and cell-to-cell spread (20, 22–27). The gB variant mediates CMV initial adsorption onto heparin sulfate glycosaminoglycans and interacts with multiple cellular receptors to trigger entry fusion (22, 28). The glycoprotein H (gH), divided into two major genotypes (gH1 and gH2), is an 86-kDa protein and encoded by the UL75 gene. The AD169 and related strains formed gH1 with 743 codons, while Towne and related strains formed gH2 with 742 codons with a deletion of codon 36 (29, 30). gH and gL formed the complex gH/gL, a dimer that was essential to virus entry (31). Besides, the dimer gH/gL together with gO comprise the trimeric glycoprotein complex III (gC-III), or form a pentameric complex which was named as gH/gL/pUL128–131A with three other viral protein UL128, UL130 and UL131. Both of the complexes play a vital role on virus entry as well as inducing virus-neutralizing antibodies (32–38). The highly polymorphic gene UL73 encodes the viral glycoprotein N (gN), a component of the gC-II complex that is involved in virus attachment to the host cell and spread (39–41). The UL73 locus has 7 identified genotypes which were named as gN1, gN2, gN3a, gN3b, gN4a, gN4b and gN4c (42, 43). The UL73 gene possesses highly hypervariable regions (approximately 50% variability), while the nucleotide variations is lower (5%–10%) in gB and gH genes (19, 44). Thus, the polymorphism of gN may enable the virus to evade from neutralizing-antibody response as well as facilitate CMV reinfection in seropositive individuals (45).
Many previous studies have focused on the variability within gB, gH and gN genes owing to their significant role of being major targets of virus neutralizing antibodies. To date, studies on the association between viral glycoprotein polymorphisms and the outcome of symptomatic cCMV infection is controversial (39, 46–51). Hence, the present study aims to determine the distribution of gB, gH and gN genotypes and attempts to ascertain the association between viral glycoprotein polymorphisms and clinical outcomes among symptomatic cCMV infected patients.
2. Methods
2.1. Study population
A total of 191 infants with symptomatic CMV infection were enrolled from September 2012 to March 2022 at Children's Hospital of Fudan University in Shanghai, China. The geographical origins of the patients were scattered in 16 different districts across the city of Shanghai. Patients were divided into two groups: cCMV infection group and the postnatal CMV (pCMV) infection group. CMV infection was diagnosed by means of real-time PCR performed on urine or plasma. Forty-two infants suffered a cCMV infection (positive test within 21 days from birth), while pCMV infection was diagnosed in 149 infants after three weeks of life (negative test within 21 days of life) (52, 53). The two subpopulations did not differ in their date of infection and geographical area.
The children were classified as having symptomatic infection with any of the following clinical manifestations: jaundice, petechiae, intrauterine growth retardation, hepatosplenomegaly, hepatitis, cholestasis, hearing loss, microcephaly, neurological dysfunction (tremor, hypotonia/hypertonia, or poor sucking reflex), CNS damage in neuroimaging (cerebral calcifications, germinal matrix cysts, ventriculomegaly, and cerebellar hypoplasia), chorioretinitis, pneumonia or laboratory findings, including thrombocytopenia, granulocytopenia, anemia (19, 54).
Our study was reviewed and approved by the Ethics Committee of the Children's Hospital of Fudan University.
2.2. Clinical examinations
Demographic data and clinical findings were retrospectively collected from the medical histories of the infants. The clinical manifestations were diagnosed during both hospital stay and outpatient follow-up.
2.2.1. Audiological assessment
All the cCMV infected infants underwent the audiological assessment within the first month of life, by means of Otoacoustic Emission (OAE), Auditory Brainstem Response Audiometry (ABR) and Acoustic impedance test. Hearing thresholds were assessed by means of ABR. Infants with a hearing threshold > 25 decibels (dB) on ABR were considered to have SNHL and scheduled for follow-up of OAE, ABR and acoustic impedance test once a month after discharge. While the cCMV infected infants with a normal hearing on ABR were scheduled for follow-up of OAE, ABR and acoustic impedance test at 3, 6, 12, and 18 months of life after discharge. All the cCMV infected infants were generally followed up until the age of 3 years old. Degree of hearing loss was characterized as normal (≤25 dB), mild (26–40 dB), moderate (41–60 dB), severe (61–80 dB) and profound (over 81 dB) according to the classification of the WHO. Infants diagnosed with monolateral or bilateral hearing loss were classified according to the worst ear threshold.
2.2.2. Ophthalmologic evaluation
Chorioretinitis caused by congenital CMV infection is mainly characterized by yellow-white punctate or patchy exudative changes in the fundus, retinal calcifications, and optic nerve hypoplasia. Ophthalmologic examination was undertaken in the neonatal period and scheduled at 3, 6, 12, and 18 months and up to 3 years of life.
2.2.3. Developmental assessment
Developmental delay in this study refers to infants who did not achieve the appropriate scores in ≥2 performance areas (gross or fine motor, language, cognitive, social and social adjustment, etc.) according to the pediatrician's diagnosis during follow-up. Children's developmental status was investigated by means of the Developmental Screening Test (DST) and the Gesell scales. Griffiths scale is performed in part of clinical patients according to their parents’ opinion. Psychomotor development was assessed at least every 6 months of life and scheduled for a long-term follow-up.
2.2.4. Other definitions
Hyperbilirubinemia was diagnosed when bilirubin value, according to the reference curve of the American Academy of Pediatrics, was over the 95th percentile for the gestational age and for the days of life.
Infantile cholestasis was defined as serum conjugated bilirubin >17.1 μmol/l or direct/total bilirubin ratio >20%.
In the present study, in addition to infants with cyanogenic or non-cyanogenic heart diseases, infants with a Patent Ductus Arteriosus (PDA) and/or a Patent Foramen Ovale (PFO) were considered to have a congenital heart disease (CDH).
2.3. CMV detection by real-time PCR
The CMV DNA of urine and plasma were extracted using a commercial diagnostic kit (DaAn Gene Co., Ltd, China) according to the manufacturer's instruction. Saliva samples were extracted using the QIAamp DNA Blood Mini Kit (Qiagen, Germany) according to the manufacturer's protocol.
We used a commercial kit for CMV viral loads measurement (DaAn Gene Co., Ltd, China). The highly conserved non-coding region of IE1 gene in CMV AD169 genome was selected as the amplification target. A 7,500 Real-Time PCR System (Applied Biosystems, Foster City, CA, United States) was used to determine the CMV DNA copy numbers.
2.4. CMV glycoprotein genotyping
A nested PCR with two pairs of primers were used to amplify the UL55, UL75 and UL73 gene fragment as described previously (55). All the positive products of nested PCR were sent out for sequencing (Sangon or Tsingke Biotechnology Co., Ltd.). The phylogenetic analysis was conducted by MEGA 11.0 using the neighbor-joining (NJ) method (1,000 bootstrap replications for branch support).
2.5. Statistical analysis
All statistical analyses were performed using the SPSS 25.0 software. The data were analyzed using descriptive statistics, including the median, range, and 95% confidence intervals (CIs). Continuous variables are described as medians and categorical data as percentages. The χ2 test or Fisher's exact test were used to compare the proportions of categorical variables. Independent group t-test was performed on the continuous variables that were normally distributed, while the Mann-Whitney U test or Kruskal-Wallis test was used for continuous variables not normally distributed. Two-sided p-values of less than 0.05 were considered to be statistically significant.
3. Results
3.1. Study population
Forty-two newborns (26 males and 16 females) with symptomatic cCMV infection were enrolled in the study. The 149 symptomatic pCMV infection group was consisted of 93 males and 56 females. The median age of cCMV group was 9 days (range 0.13–21 days), while the median age of pCMV infected infants was 3 months (range 0.87–48 months).
In the present study, all the enrolled infants had a symptomatic CMV infection. The distribution and frequency of clinical manifestations in symptomatic neonates with cCMV infection are as follows: Hepatosplenomegaly (4/42, 9.5%), hyperbilirubinemia (25/42, 59.5%), infantile cholestasis (5/42, 11.9%), pneumonia (8/42, 19.0%), thrombocytopenia (16/42, 38.1%), petechiae (10/42, 23.8%), anemia (15/42, 35.7%), neonatal respiratory distress syndrome (NRDS) (7/42, 16.7%), chorioretinitis (12/38, 31.6%), hearing loss (21/39, 53.8%), developmental delay (4/42, 9.5%), congenital heart disease (CHD) (30/42, 71.4%), ventriculomegaly (9/39, 23.1%) and microcephaly (1/42, 2.4%).
3.2. Genotype distribution and prevalence of CMV variants
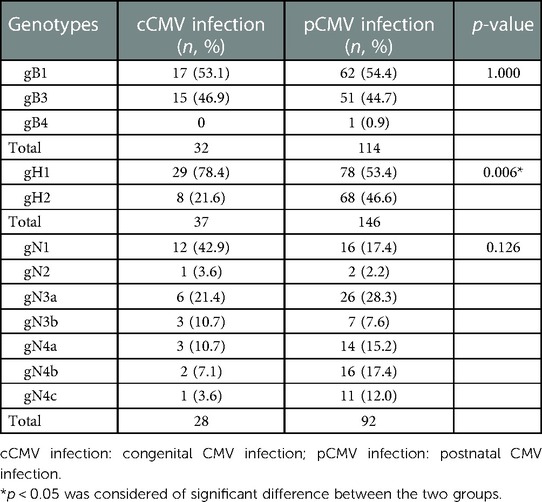
52 specimens from 42 symptomatic cCMV infected neonates were analysed to determine the gB, gH and gN genotypes. The UL55 (gB), UL75 (gH) and UL73 (gN) gene were amplified and sequenced successfully for 32/42 (76.2%), 37/42 (88.1%) and 28/42 (66.7%) of the neonates, respectively. Two out of four genotypes of gB were represented and distributed as follows: gB1 in 17/32 (53.1%) and gB3 in 15/32 (46.9%) of infants. gB2 and gB4 were absent in this group (Figure 1A). With regard to UL75 gene, gH1 was detected in 29/37 (78.4%) of the infants followed by gH2 in 8/37 (21.6%, Figure 1B). UL73 genotyping was accomplished in 28 newborns with gN1 (12/28, 42.9%) being the most dominant genotype, followed by gN3a (6/28, 21.4%), gN3b (3/28, 10.7%), gN4a (3/28, 10.7%), gN4b (2/28, 7.1%), gN4c (1/28, 3.6%) and gN2 (1/28, 3.6%). All seven genotypes of gN were represented in this cohort although some types were available in very small numbers (Figure 1C). All the expected genotypes were present except gB2 and gB4 among symptomatic cCMV infection group (Table 1).
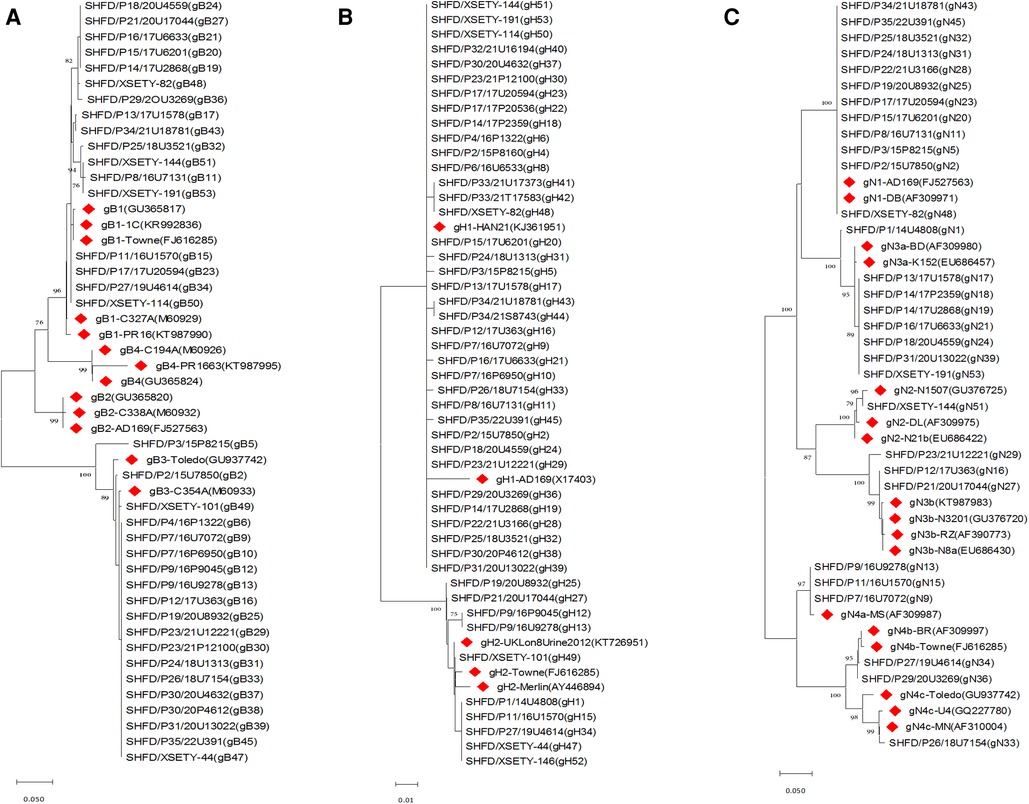
Figure 1. Phylogenetic trees of CMV gB, gH and gN genotypes among symptomatic cCMV infected infants. Phylogenetic trees were generated using MEGA software with method of neighbor-joining and branch supported with 1,000 bootstrap iterations. Reference sequences from GenBank were identified by accession number with red diamonds. (A) Phylogenetic trees of gB genotypes. (B) Phylogenetic trees of gH genotypes. (C) Phylogenetic trees of gN genotypes.
Out of 149 infants with symptomatic pCMV infection, 76.5% (114/149) of gB, 98.0% (146/149) of gH, and 61.7% (92/149) of gN accomplished the genotyping. The CMV gB1 (62/114, 54.4%), gB3 (51/114, 44.7%) and gB4 (1/114, 0.9%) were detected except the genotype of gB2. gH1 was found in 78/146 (53.4%) of the infected infants followed by gH2 (68/146, 46.6%). All seven genotypes of gN were presented in this group and distributed as follows: gN1 in 16/92 (17.4%), gN2 in 2/92 (2.2%), gN3a in 26/92 (28.3%), gN3b in 7/92 (7.6%), gN4a in 14/92 (15.2%), gN4b in 16/92 (17.4%) and gN4c in 11/92 (12.0%) of infants, respectively (Table 1).
Genotype distribution of gH was significantly different between the cCMV and pCMV groups. The predominance of gH1 in cCMV infected infants was much more pronounced compared to the pCMV infected children (p = 0.006). There was no statistical difference in the distribution of gB and gN genotypes between the two groups (Table 1).
3.3. CMV glycoprotein polymorphisms and neonatal hearing loss
The hearing test was available in 39/42 (93%) of symptomatic cCMV infected neonates in our study. 18 patients (46%) passed the hearing test while 21 cases (54%) failed. Of the 21 infants who failed the hearing test, the gB genotyping was completed for 15 infants with 7 (46.7%) for gB1 and 8 (53.3%) for gB3. The gH genotyping could be accomplished in 19 newborns with 15 (78.9%) for gH1 and 4 (21.1%) for gH2. Genotyping of gN was completed in 13 infants and distributed as follows: gN1 in 5/13 (38.5%), gN2 in 1/13 (7.7%), gN3a in 3/13 (23.1%), gN3b in 1/13 (7.7%), gN4a in 2/13 (15.4%) and gN4c in 1/13 (7.7%). There was no statistical difference in the distribution of gB, gH and gN genotypes among symptomatic cCMV infected neonates with or without hearing loss (Table 2).
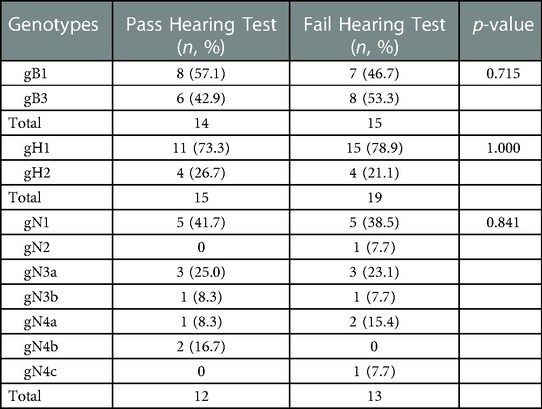
Table 2. Distributions of gB, gH and gN genotypes among symptomatic cCMV infected neonates with or without hearing loss.
To further explore the hearing impairment among neonates, we analyzed the gB, gH and gN genotypes depending on the hearing loss severity among infants who failed the test (Table 3). We found that among these infants with hearing impairment, 11 out of 13 ears from 8 newborns with moderate/severe hearing loss all exhibited the gH1 genotype rather than gH2. Two ears from one child with moderate hearing loss failed the sequencing. gH1 was the predominant genotype among symptomatic cCMV infected infants with moderate/severe hearing impairment although without statistical difference (p = 0.130, Table 3).
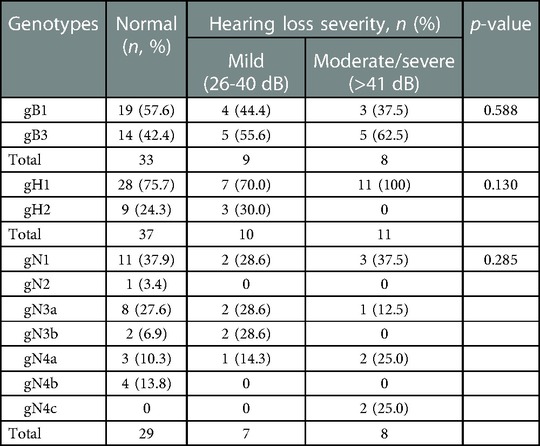
Table 3. Distribution of gB, gH and gN genotypes among symptomatic cCMV infected children with different hearing loss severities.
3.4. Viral loads
The CMV DNA was detected in all the 42 newborns with confirmed cCMV infection. The log values of CMV DNA concentration in 38 urine specimens ranged from 3.03 to 7.64 copies/ml (median 5.34 copies/ml; mean 5.34 ± 1.31 copies/ml). In 13 blood samples, the viral loads ranged from 3.13 to 5.94 copies/ml (median 4.41 copies/mL; mean 4.47 ± 0.87 copies/ml). The median viral loads in urine samples were significantly higher than those in blood samples (p = 0.03, data not shown). We did not find the association between CMV gB, gH and gN genotypes with urine viral loads among symptomatic infants (Table 4).
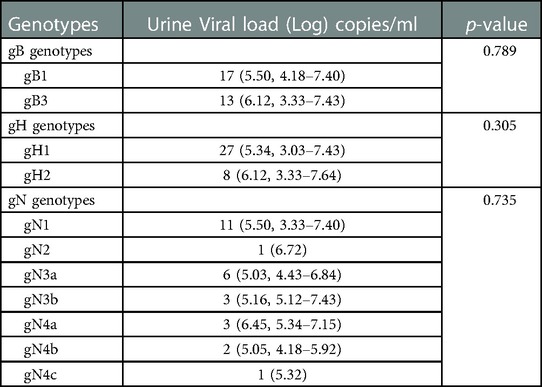
Table 4. Comparison of viral loads of gB, gH and gN genotypes in urine samples in infants with symptomatic cCMV infection.
Besides, we compared the urine viral loads in cCMV infected infants with or without hearing impairment. The log values of CMV DNA concentration in 16 infants with normal hearing ranged from 3.33 to 7.43 copies/ml (median 5.24 copies/ml; mean 5.12 ± 1.16 copies/ml). In 19 infants with hearing impairment, the viral loads ranged from 3.03 to 7.64 copies/ml (median 5.58 copies/ml; mean 5.42 ± 1.46 copies/ml). The hearing impairment group has a higher urine viral loads than the normal hearing group although without statistical difference (p = 0.512, data not shown).
3.5. gB, gH, gN genotypes vs. clinical indicators and cCMV-related symptoms/outcomes
In the present study, no significant association was found between a specific gB, gH or gN genotype and different clinical indicators (Table 5). gB3 (7/15, 46.7%) was more prevalent among newborns with skin petechiae compared with gB1 (2/17, 11.8%; p = 0.049, Table 6). Infection with gB3 genotype was associated with a 6.5-fold increased risk of skin petechiae (OR= 6.563 [95% CI, 1.095–39.324]; p = 0.049, data not shown). Besides, the gN4a subtype was significantly correlated with chorioretinitis diagnosis (3/3, 100%; p = 0.007, Table 6).
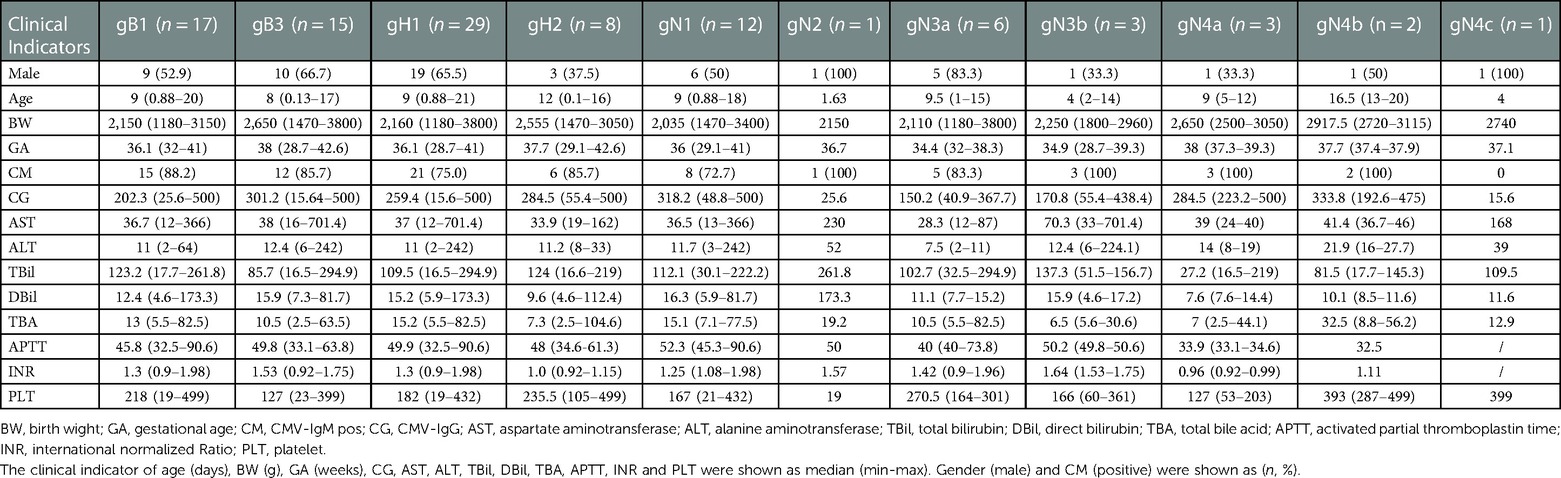
Table 5. Distribution of gB, gH and gN genotypes in different clinical indicators among infants with symptomatic cCMV infection.
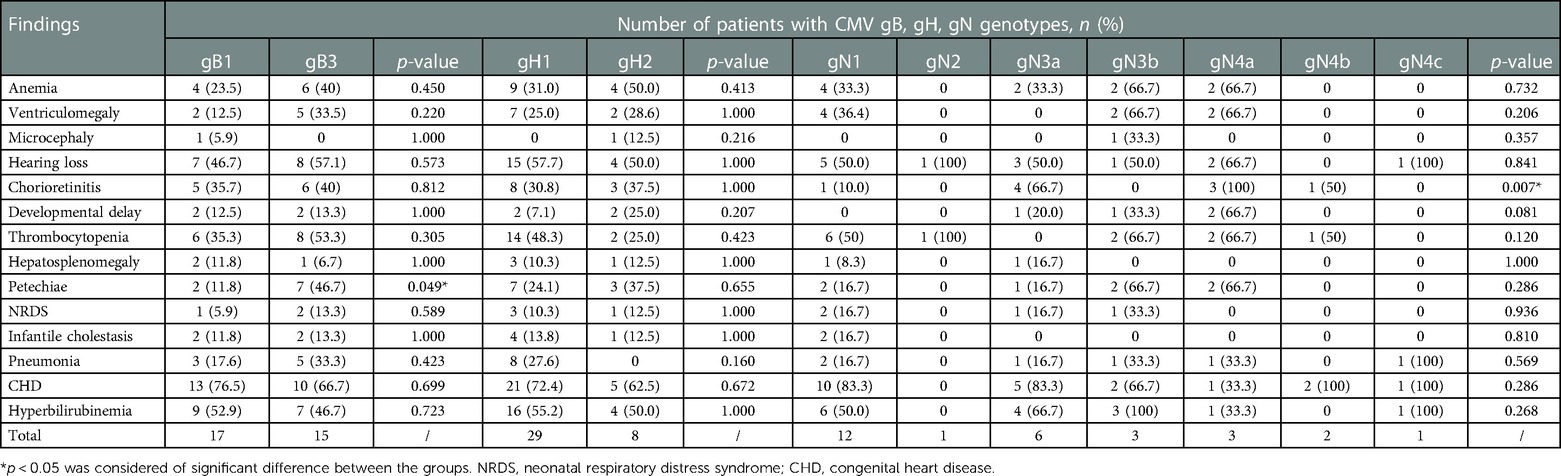
Table 6. Association between the gB, gH and gN genotypes and clinical symptoms/outcomes among infants with symptomatic cCMV infection.
4. Discussion
Congenital cytomegalovirus infection in newborns may cause severe sequelae such as central nervous system (CNS) damage, SNHL, intellectual disability and various congenital malformations. Infants born with symptomatic cCMV infection are at a higher risk for developing adverse long-term outcomes. The present study focused on the polymorphisms of UL55, UL75 and UL73 gene in infants with symptomatic CMV infection and evaluated the possible association between CMV genotypes and clinical outcomes in neonates with symptomatic cCMV infection.
Overall, our study indicated that gB1 (17/32, 53.1%), gH1 (29/37, 78.4%) and gN1 (12/28, 42.9%) were the most common genotypes among cCMV group, while gB1 (62/114, 54.4%), gH1 (78/146, 53.4%) and gN3a (26/92, 28.3%) were more prevalent in pCMV group. The cCMV genotypic distribution in our study was similar to the reports from previous studies. A research from Puhakka et al. (56) in cCMV infected infants confirmed our findings: they demonstrated that gB1 was the most common genotype (19/37, 51%) followed by gB3 (24%), gB2 (19%) and gB4 (5%). The most common genotype of gN was gN1 (7/24, 29%) followed by gN4c (25%), gN3b (21%), gN4a (13%), gN3a and gN4b (8%). Another cohort from Pakistan came to a similar result demonstrating gB1 (4/10, 40%), gH1 (7/11, 63.7%) and gN1 (3/15, 20%) to be the most common genotypes among cCMV infected infants (57). Previous studies from different geographical regions further indicated that the gB1 (47, 58–64), gH1 (19, 65) and gN1 (64) were the most prevalent genotypes in newborns with cCMV infection.
To date, the dominant genotypes of UL55, UL75 and UL73 gene among different cCMV cohorts worldwide are still controversial. Paradowska et al. demonstrated that the gB2 genotype was prevalent in Polish newborns with symptomatic cCMV infection (48, 66). However, Sarkar et al. (17) demonstrated that gB1 was the most widespread genotype among Indian neonates with symptomatic cCMV infection. Another two cohorts also confirmed our findings that gB1 was the most common genotype in neonates and infants with symptomatic cCMV infection (57, 67). Moreover, a cohort from India (68) revealed that gB3 was the most prevalent genotype in symptomatic infants. The controversial results were also observed in gH and gN genotypes. Pati et al. showed that the most predominant genotype among cCMV infected infants in American individuals were gH2 (59%) and gN3a (27%), respectively (62). Contrary to our results, Paradowska et al. (69) demonstrated that the gH2 variant occurred more frequently compared with gH1 in newborns with symptomatic cCMV infection. It was observed in a study of Pignatelli et al. that gN4a (66.7%), gN4b (60%) and gN4c (52.9%) were more prevalent among symptomatic newborns in Italy (70). Furthermore, Paradowska et al. (71) demonstrated that gN3b (14/42, 33.3%), gN4b (12/42, 28.6%), and gN4c (11/42, 26.2%) were more prevalent and supported a potential role of gN as the virological marker in newborns with symptomatic CMV infection. All these discrepancies may be attributed to geographical distribution, population/sample selection bias, genotyping method and/or CMV tissue tropism.
The majority of previous studies indicated that the genotypic distribution was similar between cCMV and pCMV infected infants (19, 66, 69). A study in Italy (44) found a significant association of gN4c and gO3 genotype with congenital infection (p = 0.037 and 0.045, respectively). In our study, no significant difference was observed on the distribution of gB and gN genotype between the two groups. Notably, we revealed that the gH1 genotype has a significant association with symptomatic cCMV infection for the first time (p = 0.006).
In this cohort, we were not able to demonstrate a significant association between CMV glycoprotein polymorphisms and neonatal hearing loss. However, infants who suffered moderate or more severe hearing loss all exhibited the gH1 genotype. Paradowska et al. demonstrated for the first time that SNHL has a significant correlation with gH1 genotype (p = 0.032) and suggest gH2 could diminish the risk of hearing impairment in infants (69). These results were confirmed by the same group in a larger cohort (19). A chinese study (65) also found a predominance of gH1 genotype in cCMV children suffering SNHL. We observed in a previous study that the gH1 genotype was predominant in infants with active CMV infection, while gH2 was more prevalent in children with latent infection (55). All these observations suggest that gH1 may be involved in early infancy hearing loss due to cCMV infection. However, a larger cohort of neonates is needed to clarify the trends observed in our study.
We observed that the median viral load in urine was significantly higher than that in blood samples, as other author also reported previously (19). We did not find a significant association between CMV genotypes and urine viral loads. The hearing impairment group had a higher urine viral loads when compared to unaffected group but without statistical significance (p = 0.512). Previous studies from different cohorts have demonstrated that a higher CMV DNAemia during early infancy was associated with long-term sequelae (72–75). Ross et al. also indicated that a virus burden of <3500 ge/ml in blood is found to be at lower risk of hearing loss in asymptomatic cCMV infected infants (76). We did not analyze the correlation between blood viral loads and CMV genotypes or hearing impairment due to the limited sample size. A larger cohort is needed to illustrate the correlation between blood viral loads and CMV genotypes or hearing impairment.
Previous reports have demonstrated with controversial results that the gN4 genotype represented the most virulent variant and was associated with severe manifestations compared with gN1 and gN3a (70, 77,78). These studies indicated that gN1 or gN3a could reduce the risk of CMV related sequelae 5 folds, whereas the gN4 genotypes increase the risk of sequelae 8 folds. Paradowska et al. (71) confirmed these findings and demonstrated that gN4 genotype was significantly associated with neurological disorders (p = 0.045). They suggest that gN2 or gN4 genotypes might be an indicator of serious manifestation in children, while gN1 and gN3b might represent less virulent strains. Similar to their findings, we found that the gN4a subtype was associated significantly with chorioretinitis (p = 0.007) due to cCMV infection.
In the present study, CHD incidence (approximately 70%) is much higher than currently reported. Two reasons may explain the huge incidence of CHD in this study. First, the majority of our population consisted of preterm infant and thus the rate of PDA at the first echocardiographic screening was high. Second, PFO was also included between CHD and it is well known that PFO has an extremely high incidence in early infancy. Due to the lack of the results of the echocardiographic follow up proving, in all likelyhood, the resolution of a great part of these cases, all PDA and PFO cases were then included as CHD.
To date, this is the first study focusing on three CMV genotypes and trying to figure out the association between the genotypes and symptomatic cCMV symptoms/outcomes in Shanghai. However, there are several limitations in our study as well. Firstly, the cCMV sample size is small and the results observed in our study should be interpreted with caution. Additional investigation with a larger population of symptomatic cCMV infection is needed to confirm our findings. Secondly, the successful sequencing rate was not satisfactory. Some of the specimens have been stored for a long time and the viral loads may degrade to some extent. The primer specificity of gB, gH and gN may vary and cause the different sequencing results (gH > gB > gN). Further, a successful sequencing greatly depends on the type of sample used. In our study, urine and saliva samples achieved a higher rate of sequencing than blood samples. Thirdly, we did not have the data on mixed infections due to the limitation of the method. We used the direct PCR-sequencing method and the primers were designed to amplify the variable regions of CMV genome. This genotyping method allows detection of the predominant genotype in single specimen and the mixed infections may have been missed. As genotypic tests can only detect viral genotypes when these comprise at least 20%–25% of the total viral population (66), it is likely that only the dominant genotypes were detected. Finally, it should be underlined that we are focusing on the symptomatic cCMV infection population. The absence of a population of asymptomatic infants limits the stratification of disease risk (based on different genotypes) only to the type of disease rather than to the possible occurrence of the disease itself.
In summary, our study showed that CMV gB1, gH1 and gN1 were the predominant genotypes among symptomatic cCMV infected infants in Shanghai. The findings in our study suggest a possible correlation between gH1 genotype and SNHL due to cCMV infection. The gB3 genotype was associated with a 6.5-fold increased risk of skin petechiae and gN4a significantly correlated with chorioretinitis. No significant association was established between CMV genotypes or hearing impairment and the urine viral loads.
Data availability statement
The data presented in the study are deposited in the GenBank repository, accession number OQ453042-OQ453154.
Ethics statement
The studies involving human participants were reviewed and approved by the Ethical Committee of Children's Hospital of Fudan University. Written informed consent from the participants’ legal guardian/next of kin was not required to participate in this study in accordance with the national legislation and the institutional requirements.
Author contributions
NND and JX conceived and designed the study; NND, LFC, LYS, LJL, ZQD and MHX collected the clinical samples of CMV from patients; NND and DNZ collected the clinical data of infants; NND performed the experiments; NND, DNZ and JX analyzed the data; NND and JX wrote the paper. All authors contributed to the article and approved the submitted version.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Griffiths P, Baraniak I, Reeves M. The pathogenesis of human cytomegalovirus. J Pathol. (2015) 235:288–97. doi: 10.1002/path.4437
2. Cannon MJ, Schmid DS, Hyde TB. Review of cytomegalovirus seroprevalence and demographic characteristics associated with infection. Rev Med Virol. (2010) 20:202–13. doi: 10.1002/rmv.655
3. Gandhi MK, Khanna R. Human cytomegalovirus: clinical aspects, immune regulation, and emerging treatments. Lancet Infect Dis. (2004) 4:725–38. doi: 10.1016/S1473-3099(04)01202-2
4. Kenneson A, Cannon MJ. Review and meta-analysis of the epidemiology of congenital cytomegalovirus (CMV) infection. Rev Med Virol. (2007) 17:253–76. doi: 10.1002/rmv.535
5. Manicklal S, Emery VC, Lazzarotto T, Boppana SB, Gupta RK. The “silent” global burden of congenital cytomegalovirus. Clin Microbiol Rev. (2013) 26:86–102. doi: 10.1128/CMR.00062-12
6. Dollard SC, Grosse SD, Ross DS. New estimates of the prevalence of neurological and sensory sequelae and mortality associated with congenital cytomegalovirus infection. Rev Med Virol. (2007) 17:355–63. doi: 10.1002/rmv.544
7. Fowler KB, Boppana SB. Congenital cytomegalovirus infection. Semin Perinatol. (2018) 42:149–54. doi: 10.1053/j.semperi.2018.02.002
8. Fowler KB, Stagno S, Pass RF, Britt WJ, Boll TJ, Alford CA. The outcome of congenital cytomegalovirus infection in relation to maternal antibody status. N Engl J Med. (1992) 326:663–7. doi: 10.1056/NEJM199203053261003
9. Fowler KB, Dahle AJ, Boppana SB, Pass RF. Newborn hearing screening: will children with hearing loss caused by congenital cytomegalovirus infection be missed? J Pediatr. (1999) 135:60–4. doi: 10.1016/s0022-3476(99)70328-8
10. Dahle AJ, Fowler KB, Wright JD, Boppana SB, Britt WJ, Pass RF. Longitudinal investigation of hearing disorders in children with congenital cytomegalovirus. J Am Acad Audiol. (2000) 11:283–90. doi: 10.1016/s0022-3476(97)70248-8
11. Fowler KB, McCollister FP, Dahle AJ, Boppana S, Britt WJ, Pass RF. Progressive and fluctuating sensorineural hearing loss in children with asymptomatic congenital cytomegalovirus infection. J Pediatr. (1997) 130:624–30. doi: 10.1016/s0022-3476(97)70248-8
12. Fowler KB, Boppana SB. Congenital cytomegalovirus (CMV) infection and hearing deficit. J Clin Virol. (2006) 35:226–31. doi: 10.1016/j.jcv.2005.09.016
13. Morton CC, Nance WE. Newborn hearing screening–a silent revolution. N Engl J Med. (2006) 354:2151–64. doi: 10.1056/NEJMra050700
14. Cannon MJ. Congenital cytomegalovirus (CMV) epidemiology and awareness. J Clin Virol. (2009) 46(Suppl 4):S6–10. doi: 10.1016/j.jcv.2009.09.002
15. Fowler KB, Pass RF. Sexually transmitted diseases in mothers of neonates with congenital cytomegalovirus infection. J Infect Dis. (1991) 164:259–64. doi: 10.1093/infdis/164.2.259
16. Ross DS, Dollard SC, Victor M, Sumartojo E, Cannon MJ. The epidemiology and prevention of congenital cytomegalovirus infection and disease: activities of the centers for disease control and prevention workgroup. J Womens Health (Larchmt). (2006) 15:224–9. doi: 10.1089/jwh.2006.15.224
17. Sarkar A, Das D, Ansari S, Chatterjee RP, Mishra L, Basu B, et al. Genotypes of glycoprotein B gene among the Indian symptomatic neonates with congenital CMV infection. BMC Pediatr. (2019) 19:291. doi: 10.1186/s12887-019-1666-5
18. Sinzger C, Digel M, Jahn G. Cytomegalovirus cell tropism. Curr Top Microbiol Immunol. (2008) 325:63–83. doi: 10.1007/978-3-540-77349-8_4
19. Paradowska E, Jabłońska A, Studzińska M, Kasztelewicz B, Wiśniewska-Ligier M, Dzierżanowska-Fangrat K, et al. Distribution of the CMV glycoprotein gH/gL/gO and gH/gL/pUL128/pUL130/pUL131A complex variants and associated clinical manifestations in infants infected congenitally or postnatally. Sci Rep. (2019) 9:16352. doi: 10.1038/s41598-019-52906-y
20. Compton T. Receptors and immune sensors: the complex entry path of human cytomegalovirus. Trends Cell Biol. (2004) 14:5–8. doi: 10.1016/j.tcb.2003.10.009
21. Rasmussen L, Geissler A, Winters M. Inter- and intragenic variations complicate the molecular epidemiology of human cytomegalovirus. J Infect Dis. (2003) 187:809–19. doi: 10.1086/367900
22. Vanarsdall AL, Johnson DC. Human cytomegalovirus entry into cells. Curr Opin Virol. (2012) 2:37–42. doi: 10.1016/j.coviro.2012.01.001
23. Chou SW, Dennison KM. Analysis of interstrain variation in cytomegalovirus glycoprotein B sequences encoding neutralization-related epitopes. J Infect Dis. (1991) 163:1229–34. doi: 10.1093/infdis/163.6.1229
24. Chee MS, Bankier AT, Beck S, Bohni R, Brown CM, Cerny R, et al. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr Top Microbiol Immunol. (1990) 154:125–69. doi: 10.1007/978-3-642-74980-3_6
25. Cranage MP, Kouzarides T, Bankier AT, Satchwell S, Weston K, Tomlinson P, et al. Identification of the human cytomegalovirus glycoprotein B gene and induction of neutralizing antibodies via its expression in recombinant vaccinia virus. EMBO J. (1986) 5:3057–63. doi: 10.1002/j.1460-2075.1986.tb04606.x
26. Varnum SM, Streblow DN, Monroe ME, Smith P, Auberry KJ, Pasa-Tolic L, et al. Identification of proteins in human cytomegalovirus (HCMV) particles: the HCMV proteome. J Virol. (2004) 78:10960–6. doi: 10.1128/JVI.78.20.10960-10966.2004
27. Isaacson MK, Compton T. Human cytomegalovirus glycoprotein B is required for virus entry and cell-to-cell spread but not for virion attachment, assembly, or egress. J Virol. (2009) 83:3891–903. doi: 10.1128/JVI.01251-08
28. Boyle KA, Compton T. Receptor-binding properties of a soluble form of human cytomegalovirus glycoprotein B. J Virol. (1998) 72:1826–33. doi: 10.1128/JVI.72.3.1826-1833.1998
29. Chou S. Molecular epidemiology of envelope glycoprotein H of human cytomegalovirus. J Infect Dis. (1992) 166:604–7. doi: 10.1093/infdis/166.3.604
30. Keay S, Merigan TC, Rasmussen L. Identification of cell surface receptors for the 86-kilodalton glycoprotein of human cytomegalovirus. Proc Natl Acad Sci U S A. (1989) 86:10100–3. doi: 10.1073/pnas.86.24.10100
31. Wille PT, Wisner TW, Ryckman B, Johnson DC. Human cytomegalovirus (HCMV) glycoprotein gB promotes virus entry in trans acting as the viral fusion protein rather than as a receptor-binding protein. mBio. (2013) 4:e00332–13. doi: 10.1128/mBio.00332-13
32. Ryckman BJ, Rainish BL, Chase MC, Borton JA, Nelson JA, Jarvis MA, et al. Characterization of the human cytomegalovirus gH/gL/UL128-131 complex that mediates entry into epithelial and endothelial cells. J Virol. (2008) 82:60–70. doi: 10.1128/JVI.01910-07
33. Hahn G, Revello MG, Patrone M, Percivalle E, Campanini G, Sarasini A, et al. Human cytomegalovirus UL131-128 genes are indispensable for virus growth in endothelial cells and virus transfer to leukocytes. J Virol. (2004) 78:10023–33. doi: 10.1128/JVI.78.18.10023-10033.2004
34. Wang D, Shenk T. Human cytomegalovirus virion protein complex required for epithelial and endothelial cell tropism. Proc Natl Acad Sci U S A. (2005) 102:18153–8. doi: 10.1073/pnas.0509201102
35. Straschewski S, Patrone M, Walther P, Gallina A, Mertens T, Frascaroli G. Protein pUL128 of human cytomegalovirus is necessary for monocyte infection and blocking of migration. J Virol. (2011) 85:5150–8. doi: 10.1128/JVI.02100-10
36. Fouts AE, Chan P, Stephan JP, Vandlen R, Feierbach B. Antibodies against the gH/gL/UL128/UL130/UL131 complex comprise the majority of the anti-cytomegalovirus (anti-CMV) neutralizing antibody response in CMV hyperimmune globulin. J Virol. (2012) 86:7444–7. doi: 10.1128/JVI.00467-12
37. Macagno A, Bernasconi NL, Vanzetta F, Dander E, Sarasini A, Revello MG, et al. Isolation of human monoclonal antibodies that potently neutralize human cytomegalovirus infection by targeting different epitopes on the gH/gL/UL128-131A complex. J Virol. (2010) 84:1005–13. doi: 10.1128/JVI.01809-09
38. Rasmussen L, Geissler A, Cowan C, Chase A, Winters M. The genes encoding the gCIII complex of human cytomegalovirus exist in highly diverse combinations in clinical isolates. J Virol. (2002) 76:10841–8. doi: 10.1128/jvi.76.21.10841-10848.2002
39. Pignatelli S, Dal Monte P, Rossini G, Landini MP. Genetic polymorphisms among human cytomegalovirus (HCMV) wild-type strains. Rev Med Virol. (2004) 14:383–410. doi: 10.1002/rmv.438
40. Kari B, Gehrz R. Structure, composition and heparin binding properties of a human cytomegalovirus glycoprotein complex designated gC-II. J Gen Virol. (1993) 74:255–64. doi: 10.1099/0022-1317-74-2-255
41. Mach M, Kropff B, Dal Monte P, Britt W. Complex formation by human cytomegalovirus glycoproteins M (gpUL100) and N (gpUL73). J Virol. (2000) 74:11881–92. doi: 10.1128/jvi.74.24.11881-11892.2000
42. Pignatelli S, Dal Monte P, Landini MP. gpUL73 (gN) genomic variants of human cytomegalovirus isolates are clustered into four distinct genotypes. J Gen Virol. (2001) 82:2777–84. doi: 10.1099/0022-1317-82-11-2777
43. Pignatelli S, Dal Monte P, Rossini G, Chou S, Gojobori T, Hanada K, et al. Human cytomegalovirus glycoprotein N (gpUL73-gN) genomic variants: identification of a novel subgroup, geographical distribution and evidence of positive selective pressure. J Gen Virol. (2003) 84:647–55. doi: 10.1099/vir.0.18704-0
44. Arcangeletti MC, Vasile Simone R, Rodighiero I, De Conto F, Medici MC, Martorana D, et al. Combined genetic variants of human cytomegalovirus envelope glycoproteins as congenital infection markers. Virol J. (2015) 12:202. doi: 10.1186/s12985-015-0428-8
45. Burkhardt C, Himmelein S, Britt W, Winkler T, Mach M. Glycoprotein N subtypes of human cytomegalovirus induce a strain-specific antibody response during natural infection. J Gen Virol. (2009) 90:1951–61. doi: 10.1099/vir.0.010967-0
46. Bale JF Jr, Murph JR, Demmler GJ, Dawson J, Miller JE, Petheram SJ. Intrauterine cytomegalovirus infection and glycoprotein B genotypes. J Infect Dis. (2000) 182:933–6. doi: 10.1086/315770
47. Barbi M, Binda S, Caroppo S, Primache V, Didò P, Guidotti P, et al. CMV gB genotypes and outcome of vertical transmission: study on dried blood spots of congenitally infected babies. J Clin Virol. (2001) 21:75–9. doi: 10.1016/s1386-6532(00)00188-8
48. Paradowska E, Studzińska M, Nowakowska D, Wilczyński J, Rycel M, Suski P, et al. Distribution of UL144, US28 and UL55 genotypes in Polish newborns with congenital cytomegalovirus infections. Eur J Clin Microbiol Infect Dis. (2012) 31:1335–45. doi: 10.1007/s10096-011-1447-z
49. Arav-Boger R, Battaglia CA, Lazzarotto T, Gabrielli L, Zong JC, Hayward GS, et al. Cytomegalovirus (CMV)-encoded UL144 (truncated tumor necrosis factor receptor) and outcome of congenital CMV infection. J Infect Dis. (2006) 194:464–73. doi: 10.1086/505427
50. Bale JF Jr, Petheram SJ, Robertson M, Murph JR, Demmler G. Human cytomegalovirus a sequence and UL144 variability in strains from infected children. J Med Virol. (2001) 65:90–6. doi: 10.1002/jmv.2006
51. Picone O, Costa JM, Chaix ML, Ville Y, Rouzioux C, Leruez-Ville M. Human cytomegalovirus UL144 gene polymorphisms in congenital infections. J Clin Microbiol. (2005) 43:25–9. doi: 10.1128/JCM.43.1.25-29.2005
52. Rawlinson WD, Boppana SB, Fowler KB, Kimberlin DW, Lazzarotto T, Alain S, et al. Congenital cytomegalovirus infection in pregnancy and the neonate: consensus recommendations for prevention, diagnosis, and therapy. Lancet Infect Dis. (2017) 17:e177–88. doi: 10.1016/S1473-3099(17)30143-3
53. Kelly MS, Benjamin DK, Puopolo KM, Laughon MM, Clark RH, Mukhopadhyay S, et al. Postnatal cytomegalovirus infection and the risk for bronchopulmonary dysplasia. JAMA Pediatr. (2015) 169:e153785. doi: 10.1001/jamapediatrics.2015.3785
54. Boppana SB, Pass RF, Britt WJ, Stagno S, Alford CA. Symptomatic congenital cytomegalovirus infection: neonatal morbidity and mortality. Pediatr Infect Dis J. (1992) 11:93–9. doi: 10.1097/00006454-199202000-00007
55. Dong N, Cao L, Su L, Lu L, Dong Z, Xu M, et al. Human cytomegalovirus envelope glycoprotein B, H, and N polymorphisms among infants of Shanghai area in China. J Med Virol. (2020) 92:3674–81. doi: 10.1002/jmv.26210
56. Puhakka L, Pati S, Lappalainen M, Lönnqvist T, Niemensivu R, Lindahl P, et al. Viral shedding, and distribution of cytomegalovirus glycoprotein H (UL75), glycoprotein B (UL55), and glycoprotein N (UL73) genotypes in congenital cytomegalovirus infection. J Clin Virol. (2020) 125:104287. doi: 10.1016/j.jcv.2020.104287
57. Mujtaba G, Khurshid A, Sharif S, Alam MM, Aamir UB, Shaukat S, et al. Distribution of cytomegalovirus genotypes among neonates born to infected mothers in Islamabad, Pakistan. PLoS One. (2016) 11:e0156049. doi: 10.1371/journal.pone.0156049
58. Zawilińska B, Szostek S, Kopeć J, Koprynia M, Kosz-Vnenchak M. UL55 Genotype diversity of cytomegalovirus strains isolated from newborns and infants hospitalized in southern Poland. Przegl Epidemiol. (2011) 65:409–13.
59. Lukácsi A, Taródi B, Endreffy E, Bábinszki A, Pál A, Pusztai R. Human cytomegalovirus gB genotype 1 is dominant in congenital infections in south Hungary. J Med Virol. (2001) 65:537–42. doi: 10.1002/jmv.2070
60. Yu ZS, Zou CC, Zheng JY, Zhao ZY. Cytomegalovirus gB genotype and clinical features in Chinese infants with congenital infections. Intervirology. (2006) 49:281–5. doi: 10.1159/000093458
61. Nijman J, Mandemaker FS, Verboon-Maciolek MA, Aitken SC, van Loon AM, de Vries LS, et al. Genotype distribution, viral load and clinical characteristics of infants with postnatal or congenital cytomegalovirus infection. PLoS One. (2014) 9:e108018. doi: 10.1371/journal.pone.0108018
62. Pati SK, Pinninti S, Novak Z, Chowdhury N, Patro RK, Fowler K, et al. NIDCD CHIMES study investigators. Genotypic diversity and mixed infection in newborn disease and hearing loss in congenital cytomegalovirus infection. Pediatr Infect Dis J. (2013) 32:1050–4. doi: 10.1097/INF.0b013e31829bb0b9
63. de Vries JJ, Wessels E, Korver AM, van der Eijk AA, Rusman LG, Kroes AC, et al. Rapid genotyping of cytomegalovirus in dried blood spots by multiplex real-time PCR assays targeting the envelope glycoprotein gB and gH genes. J Clin Microbiol. (2012) 50:232–7. doi: 10.1128/JCM.05253-11
64. Brañas P, Blázquez-Gamero D, Galindo A, Prieto C, Olabarrieta I, Cuadrado I, et al. Cytomegalovirus genotype distribution among congenitally and postnatally infected patients: association of particular glycoprotein (g)B and gN types with symptomatic disease. Open Forum Infect Dis. (2015) 2:ofv151. doi: 10.1093/ofid/ofv151
65. Chen LY, Li W, Xu JL, Tao R, Li HM, Liu LF, et al. Relationship between gH genotyping and clinical characteristics of children with congenital cytomegalovirus infection. Zhonghua Er Ke Za Zhi. (2019) 57:597–602. doi: 10.3760/cma.j.issn.0578-1310.2019.08.005
66. Paradowska E, Studzińska M, Suski P, Kasztelewicz B, Wiśniewska-Ligier M, Zawilińska B, et al. Human cytomegalovirus UL55, UL144, and US28 genotype distribution in infants infected congenitally or postnatally. J Med Virol. (2015) 87:1737–48. doi: 10.1002/jmv.24222
67. Yan H, Koyano S, Inami Y, Yamamoto Y, Suzutani T, Mizuguchi M, et al. Genetic variations in the gB, UL144 and UL149 genes of human cytomegalovirus strains collected from congenitally and postnatally infected Japanese children. Arch Virol. (2008) 153:667–74. doi: 10.1007/s00705-008-0044-7
68. Gandhoke I, Hussain SA, Pasha ST, Chauhan LS, Khare S. Glycoprotein B genotyping in congenital/perinatal cytomegalovirus infection in symptomatic infants. Indian Pediatr. (2013) 50:663–7. doi: 10.1007/s13312-013-0199-5
69. Paradowska E, Jabłońska A, Studzińska M, Kasztelewicz B, Zawilińska B, Wiśniewska-Ligier M, et al. Cytomegalovirus glycoprotein H genotype distribution and the relationship with hearing loss in children. J Med Virol. (2014) 86:1421–7. doi: 10.1002/jmv.23906
70. Pignatelli S, Lazzarotto T, Gatto MR, Dal Monte P, Landini MP, Faldella G, et al. Cytomegalovirus gN genotypes distribution among congenitally infected newborns and their relationship with symptoms at birth and sequelae. Clin Infect Dis. (2010) 51:33–41. doi: 10.1086/653423
71. Paradowska E, Jabłońska A, Studzińska M, Suski P, Kasztelewicz B, Zawilińska B, et al. Distribution of cytomegalovirus gN variants and associated clinical sequelae in infants. J Clin Virol. (2013) 58:271–5. doi: 10.1016/j.jcv.2013.05.024
72. Waters A, Hassan J, De Gascun C, Kissoon G, Knowles S, Molloy E, et al. Human cytomegalovirus UL144 is associated with viremia and infant development sequelae in congenital infection. J Clin Microbiol. (2010) 48:3956–62. doi: 10.1128/JCM.01133-10
73. Lanari M, Lazzarotto T, Venturi V, Papa I, Gabrielli L, Guerra B, et al. Neonatal cytomegalovirus blood load and risk of sequelae in symptomatic and asymptomatic congenitally infected newborns. Pediatrics. (2006) 117:e76–83. doi: 10.1542/peds.2005-0629
74. Walter S, Atkinson C, Sharland M, Rice P, Raglan E, Emery VC, et al. Congenital cytomegalovirus: association between dried blood spot viral load and hearing loss. Arch Dis Child Fetal Neonatal Ed. (2008) 93:F280–5. doi: 10.1136/adc.2007.119230
75. Bradford RD, Cloud G, Lakeman AD, Boppana S, Kimberlin DW, Jacobs R, et al. National institute of allergy and infectious diseases collaborative antiviral study group. Detection of cytomegalovirus (CMV) DNA by polymerase chain reaction is associated with hearing loss in newborns with symptomatic congenital CMV infection involving the central nervous system. J Infect Dis. (2005) 191:227–33. doi: 10.1086/426456
76. Ross SA, Novak Z, Fowler KB, Arora N, Britt WJ, Boppana SB. Cytomegalovirus blood viral load and hearing loss in young children with congenital infection. Pediatr Infect Dis J. (2009) 28:588–92. doi: 10.1097/INF.0b013e3181979a27
77. Pignatelli S, Dal Monte P, Rossini G, Lazzarotto T, Gatto MR, Landini MP. Intrauterine cytomegalovirus infection and glycoprotein N (gN) genotypes. J Clin Virol. (2003) 28:38–43. doi: 10.1016/s1386-6532(02)00236-6
78. Rossini G, Pignatelli S, Dal Monte P, Camozzi D, Lazzarotto T, Gabrielli L, et al. Monitoring for human cytomegalovirus infection in solid organ transplant recipients through antigenemia and glycoprotein N (gN) variants: evidence of correlation and potential prognostic value of gN genotypes. Microbes Infect. (2005) 7:890–6. doi: 10.1016/j.micinf.2005.01.016
Keywords: congenital, cytomegalovirus, glycoprotein B, glycoprotein H, glycoprotein N, hearing loss
Citation: Dong N, Cao L, Zheng D, Su L, Lu L, Dong Z, Xu M and Xu J (2023) Distribution of CMV envelope glycoprotein B, H and N genotypes in infants with congenital cytomegalovirus symptomatic infection. Front. Pediatr. 11:1112645. doi: 10.3389/fped.2023.1112645
Received: 30 November 2022; Accepted: 20 February 2023;
Published: 15 March 2023.
Edited by:
Qing Ye, Zhejiang University, China© 2023 Dong, Cao, Zheng, Su, Lu, Dong, Xu and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jin Xu amlueHVfMTI1QDE2My5jb20=
Specialty Section: This article was submitted to General Pediatrics and Pediatric Emergency Care, a section of the journal Frontiers in Pediatrics
 Niuniu Dong
Niuniu Dong Lingfeng Cao
Lingfeng Cao Danni Zheng2
Danni Zheng2 Lijuan Lu
Lijuan Lu Menghua Xu
Menghua Xu Jin Xu
Jin Xu