
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Pediatr., 20 March 2023
Sec. Children and Health
Volume 11 - 2023 | https://doi.org/10.3389/fped.2023.1090048
 Trisha Rahman1,†,§
Trisha Rahman1,†,§ Prioty F. Sarwar1,†,§
Prioty F. Sarwar1,†,§ Cassie Potter1,§
Cassie Potter1,§ Sarah S. Comstock2*‡,§
Sarah S. Comstock2*‡,§ Vanja Klepac-Ceraj1*‡,§
Vanja Klepac-Ceraj1*‡,§
Despite affecting up to 20% of infants in the United States, there is no cure for atopic dermatitis (AD), also known as eczema. Atopy usually manifests during the first six months of an infant's life and is one predictor of later allergic health problems. A diet of human milk may offer protection against developing atopic dermatitis. One milk component, human milk oligosaccharides (HMOs), plays an important role as a prebiotic in establishing the infant gut microbiome and has immunomodulatory effects on the infant immune system. The purpose of this review is to summarize the available information about bacterial members of the intestinal microbiota capable of metabolizing HMOs, the bacterial genes or metabolic products present in the intestinal tract during early life, and the relationship of these genes and metabolic products to the development of AD/eczema in infants. We find that specific HMO metabolism gene sets and the metabolites produced by HMO metabolizing bacteria may enable the protective role of human milk against the development of atopy because of interactions with the immune system. We also identify areas for additional research to further elucidate the relationship between the human milk metabolizing bacteria and atopy. Detailed metagenomic studies of the infant gut microbiota and its associated metabolomes are essential for characterizing the potential impact of human milk-feeding on the development of atopic dermatitis.
Atopic dermatitis (AD), commonly referred to as eczema, is an inflammatory skin condition that affects up to 20% of infants. This condition is a growing problem in the United States, as higher percentages have been reported in more recent years (1–4). Development of atopy in early life is also one of the predictors of later allergic or other hyperinflammatory health problems, such as asthma, rhinitis or food allergies (5). Symptoms usually begin during the first six months of an infant's life and include itchy and painful skin and sleep disruption (6–8). Not only does this disease decrease quality of life for infants with AD/eczema and their families (9, 10), children with AD/eczema also have more hospital visits and hospitalizations than those without AD/eczema (11). Families also experience substantial financial burden; the annual cost for both adults and children in the United States with AD/eczema was estimated to be $5.3 billion in 2015 (12), with out-of-pocket expenses disproportionately affecting Black Americans (13). Thus, this is a highly prevalent immune condition that negatively impacts infants and their families, where early interventions may prevent later negative health outcomes.
There is a genetic predisposition for AD/eczema, but genetics can only explain a percentage of AD/eczema cases. Mutations in the FLG gene are known to be associated with the development of AD/eczema because the lack of the structural protein filaggrin leads to impairment of the barrier function of the skin. FLG gene mutations, however, explain up to only 50% of AD/eczema cases (14, 15). In addition to genetics, gut microorganisms have been implicated in the development of AD/eczema (16–18). The gut-associated microorganisms affect the development of the immune system by providing substrates and metabolites that induce and recruit a repertoire of innate immune cells and train long-term adaptive immune responses (17). Gut microbiota-mediated stimulation of regulatory immune pathways leads to many changes in the host, such as changes in gene expression that can lead to the recruitment and proliferation of immune cells, changes in intestinal barrier integrity and the release of antimicrobial peptides, to name a few (19–23). Even though the infant gut microbiome is largely transient, the impact of the early-life gut microbiome on the immune system has been shown to have both positive and negative effects on the development of inflammatory diseases during infancy (24).
Human milk shapes the early infant gut microbiota, and in this way, plays key roles in modulating the development of the innate and adaptive mucosal immune systems and regulating gut barrier integrity (25). One important component of human milk is human milk oligosaccharides (HMOs), a group of prebiotic oligosaccharides and the third most abundant component in human milk after lactose and lipids. An HMO is a soluble glycan consisting of five basic units: one acid monosaccharide, one amino sugar and three monosaccharides. The three major categories of HMOs include fucosylated, sialylated and neutral oligosaccharides (26), and the immunomodulatory effects of each type on innate and systemic immunity has been extensively reviewed by Kulinich and Liu (27), and Donovan and Comstock (17), respectively. HMOs cannot be digested by the human infant and, instead, serve as a carbon and energy source for bacteria (28, 29). There are over 200 known structures of HMOs (30). Several genetic and non-genetic factors result in vastly different HMO repertoires in the milk of lactating individuals (31). Thus, the most abundant HMO varies by lactating person and has been reviewed by others (32, 33).
There is conflicting evidence regarding the associations between human milk and AD/eczema (Table 1). Several studies and meta-analyses have reported a protective role against AD/eczema when infants are fed a diet of human milk, especially in the first 4 months of life (37–40). Yet several others have found compounding factors that can reduce the effectiveness of a human milk diet in the prevention of AD/eczema, like atopic heredity and the infant's age (40–44). The observed human milk diet-specific protections against AD/eczema potentially stem from differences in the composition of the infant gut microbiota and its ability to completely metabolize HMOs (16). Only a handful of taxa are known to possess HMO metabolizing enzymes, and the prevalence of metabolic machinery capable of metabolizing HMOs is low among microbial taxa residing within the human gastrointestinal tract. Notably, several species from the genus Bifidobacterium have the ability to metabolize HMOs, and these organisms have been known to influence neonatal mucosal and systemic immunity (17, 45) via the production of short-chain fatty acids (SCFAs) or direct modulation of the immune system (Figure 1). Hence, differences in types and abundances of HMO metabolizing bacteria may impact host immune development (47, 48). The differential ability of infant intestinal bacteria to metabolize specific HMOs may explain why a diet of human milk protects against AD/eczema in some but not all infants fed human milk.
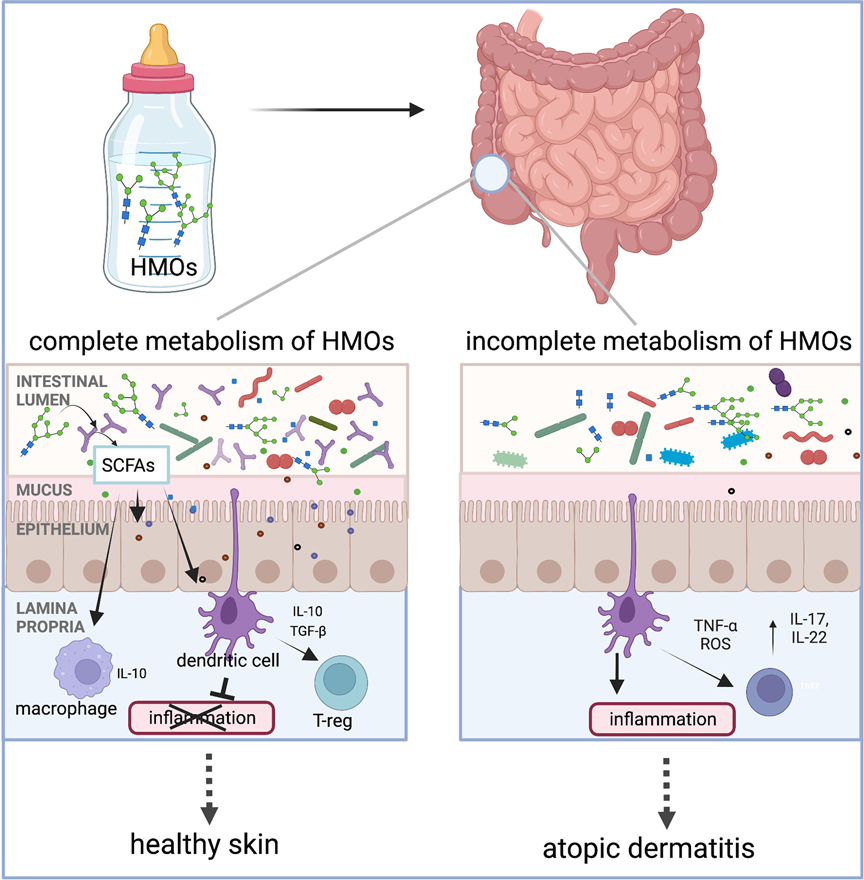
Figure 1. Human milk oligosaccharides (HMOs) are the third most abundant component in human milk after lactose and lipids. Humans cannot digest HMOs. Instead, these carbohydrates are a food source for infant gut microorganisms such as B. infantis, Bacteroides spp., and others. In the gut, bacteria with HMO metabolizing genes break down complex oligosaccharides into simpler metabolites, such as short-chain fatty acids (SCFAs). These metabolites either signal the immune system through interactions with dendritic cells whose dendrites are sampling the gut lumen or by crossing the epithelial barrier to interact with immune cells in the lamina propria. Interaction between the dendritic cells and specific HMO metabolites results in dendritic cells releasing interleukins, such as IL-10, which are involved in regulating inflammation. We hypothesize that infants with microorganisms that have the complete set of HMO metabolizing genes are protected from the development of AD/eczema and/or experience reduced severity of AD/eczema. Intestinal cells and immune cells have receptors for SCFAs which are not pictured in this simplified schematic but have been reviewed by elsewhere (46). The figure was created in BioRender.com.
To understand the development of atopy in early life, it is imperative to investigate the links between human milk exposure, variations in HMO metabolic capacity of commensal bacteria and health outcomes such as atopic dermatitis. Atopy, including the development of AD/eczema, is the first step toward a lifetime of disease in many children. Halting the progression of the atopic march (5, 49) is of utmost importance to have positive health outcomes for US infants and children. Herein, we: 1) summarize what is currently known about the microorganisms and genes involved in HMO metabolism, 2) describe patterns of HMO metabolites associated with AD/eczema, and 3) evaluate whether the abundance and diversity of HMO metabolizing genes in breastfed infants can be a predictor of an infant's risk for AD/eczema. We also identify the gaps in our understanding of how microorganisms or their metabolites modulate the infant immune system during atopic disease. To address the topic, we followed the classical methodology for a narrative review (50). A list of keywords such as human milk oligosaccharide metabolizing genes, atopy, atopic march, Bifidobacterium infantis, metabolites, short chain fatty acids, immunity, and infant gut microbiome were initially identified. Then, keyword combinations adding the term “eczema” or “atopy” or “atopic dermatitis” were used. We used Google Scholar, PubMed, and Web of Knowledge to search for available literature published in English and indexed either as original articles or reviews. We focused on publications published in the last 5 years, but did not limit our search to a specific time range.
Humans and their microbiomes have co-evolved for tens of thousands of years, resulting in a tight partnership between the microorganisms and their host. HMOs have likely played a role in this co-evolutionary relationship as they cannot be digested by infants but instead support the colonization and growth of beneficial microbes in the infant gut (51). In fact, longitudinal sampling of fecal HMOs matched with the infant gut microbiota shows an inverse relationship between the levels of fecal HMOs and the abundance of HMO metabolizing bacteria (52). Therefore, it is not surprising that the infant gut microbial community composition differs in infants fed exclusively human milk compared to those fed no human milk or mixed diets, especially early in life (53–57). Early colonization of the infant gut is variable between individual infants, but human milk fed infants have higher abundances of bifidobacterial species compared to formula fed infants (47, 58). Bifidobacterium longum subsp. infantis, (B. infantis), one of a few microorganisms capable of metabolizing HMOs and one of the pioneer colonizers of the infant gut in breastfed infants, generates metabolites that protect against inflammation, such as HMO-derived SCFAs and aromatic lactic acids (59, 60). Decreased levels of Bifidobacterium spp. in the infant gut have been associated with increased incidence of obesity, allergies, and diabetes (61–63). In addition to B. infantis, only a few microbes from the Bifidobacterium, Lactobacillus, Enterococcus and Bacteroides genera have been shown to encode a substantial number of genes to either partially or fully metabolize HMOs (47, 64–69). These HMO-utilizing bacteria are also routinely detected in the fecal microbiota of neonates (66, 67, 70, 71). Several of these early colonizers of the gut of human milk fed infants affect human immune system development and function (72, 73).
Some surveys state that the microbiomes of healthy, human milk fed infants in countries where breastfeeding is widespread are dominated by Bifidobacterium at abundances that often exceed 60% (74–77). On the other hand, a generational loss of Bifidobacterium can be observed in regions like the US and Europe where there is a high prevalence of formula feeding and disruption of the fecal-oral transfer of maternal gut bacteria to infants due to higher incidence of cesarean births and perinatal antibiotic use (78). It is unclear whether the purported disappearance of specific Bifidobacterium taxa from the infant gut coincides with an overall decline of HMO metabolizing microorganisms, but it is clear that functional differences in the gut microbiota can be attributed to lifestyle, with the presence of more complex HMO degradation machinery associated with non-industrialized infants (79).
However, some studies suggest a low relative abundance of Bifidobacterium in infants, at levels of ∼30% or lower, regardless of geography, industrialization and breastfeeding status (80, 81). Reduced abundance and prevalence of Bifidobacterium may be due to the presence of other HMO metabolizing microbes from the genera Lactobacillus or Bacteroides that compete for the same resources as members of the genus Bifidobacterium (68, 71). Alternatively, discrepancies in reports may also be tied to a lack of sensitivity in the detection of Bifidobacterium taxa (82–84). For example, B. infantis is challenging to detect and identify by PCR-based methods using the 16S rRNA gene as its 16S rRNA gene is almost identical to that of the closely related B. longum and B. suis strains (85). Both the sequencing technique (52) and the use of different 16S rRNA gene primer pairs (83) lead to a varying degree of sensitivity in detecting and identifying B. infantis. More recently, studies using metagenomic approaches have indeed observed a decline of B. infantis in industrialized populations as compared to non-industrialized populations (79). Given a true decline in HMO metabolizing capacity in the infant gut, restoring these missing microorganisms could prevent pediatric diseases such as islet autoimmunity, type I diabetes, and intestinal disorders (86, 87). Despite these reports that suggest the importance of HMO metabolizing bacteria in pediatric diseases, our understanding of the prevalence of HMO metabolizing bacteria in present infant populations is limited. Furthermore, the association of these bacteria with specific intensities of human milk exposure has only been evaluated to a limited extent (29).
Only a subset of bacteria possess the enzymatic machinery that is required to break down, metabolize, and grow on HMOs (Table 2) (86). These microbes are highly specialized such that they typically disappear when HMOs are removed from the diet (53, 108). B. infantis has the majority of HMO metabolizing genes that have been discovered and characterized to date (60, 109), and these genes can be classified into three categories: extracellular glycosidases, transporters, and intracellular enzymes. Bacteria use multiple combinations of these HMO metabolizing gene types in two different HMO metabolizing strategies (110). One strategy involves the hydrolysis of the HMO extracellularly into mono- and disaccharides using cell wall-anchored secretory glycosidases, followed by saccharide transport into the cells (111). The second strategy is to directly import HMOs with ≥3 sugar subunits into the cells via ATP-binding cassette (ABC) transporters followed by intracellular HMO hydrolysis by exoglycosidases (112). HMO metabolizing gene cassettes also have been found in some species of Bacteroides (66). Some members of the genus Bifidobacterium or Lactobacillus possess incomplete metabolic machinery and may rely on other strains for completion of HMO digestion or may cross-feed on digested or partially digested metabolites from other bacteria (65).
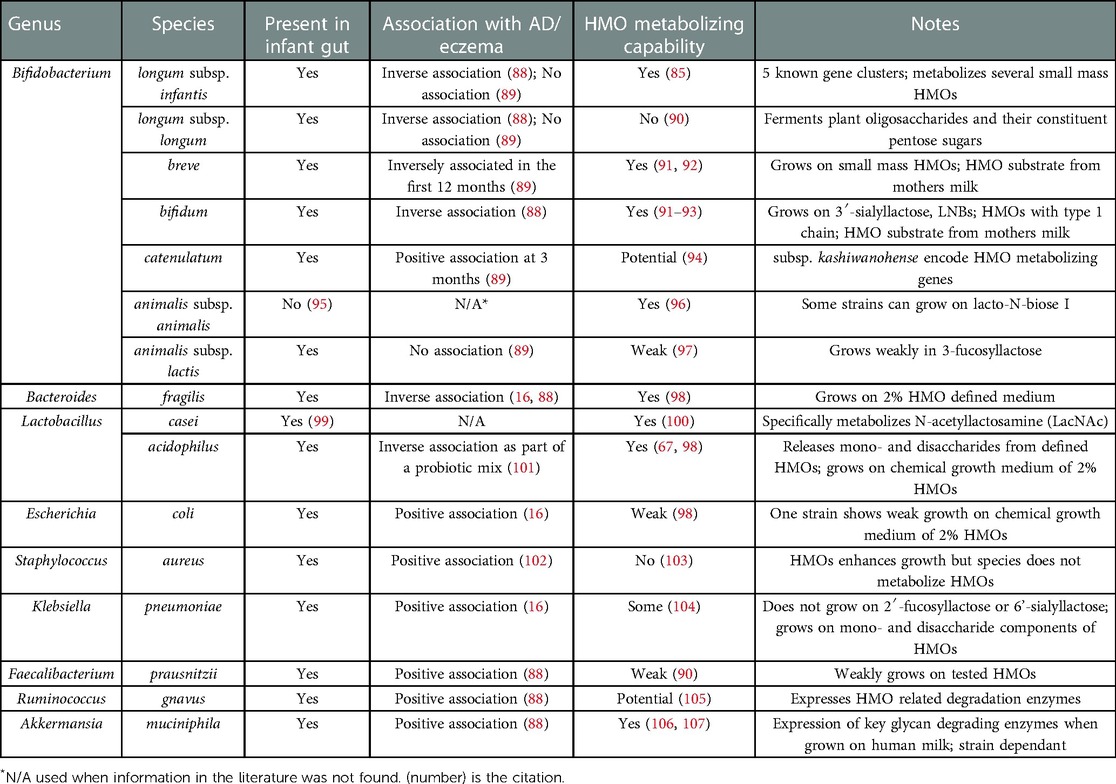
Table 2. Select bacterial species and their association with AD/eczema and HMO metabolizing capacity.
B. infantis is unique for its presence of five gene clusters that encode enzymes that bind, cleave and transport HMOs (109, 113), but little is known about the specific targets of each gene cluster. We have recently explored publicly available genomes within the B. longum group: B. longum infantis, B. longum longum and B longum suis. Using average nucleotide identity (ANI) which is a measure of nucleotide-level genomic similarity between the coding regions of two genomes, we were able to show that B. infantis clustered separately from other relatives of the B. longum group, and individual B. infantis genomes contained all or almost all HMO metabolizing genes associated with the five clusters. B. longum and B. suis were missing these genes, although several B. longum genomes contained a complete set of genes from cluster four but lacked genes from the other clusters (85). Notably, the metabolites resulting from HMO degradation processes can feed other members of the gut bacterial community or signal to the host via immune cells, enterocytes or other cells in the gastrointestinal tract (17, 65, 114).
Microbial species are formed by acquisition and loss of functional traits by horizontal gene transfer (HGT), gene loss, duplication, and selection. These events are important in the evolution of microbial genomes (115). Thus, even in closely related microorganisms belonging to the same species or species group, gene content can vary significantly (116, 117). The B. longum group comprising of three subspecies is a good example of such variation. Additionally, the gene content can also vary significantly across metagenomes (47, 69). In fact, two different microorganisms can use different metabolic pathways to perform the same function. For example, Bacteroides and Bifidobacterium are both present in the infant gut and can digest HMOs. However, Bacteroides use the set of genes they typically use to metabolize host mucus glycans to metabolize HMOs (66) and these genes do not overlap with the HMO metabolism genes encoded by B. infantis (66). Moreover, even within Bacteroides spp., the genes for HMO utilization are not conserved, and B. fragilis and B. thetaiotaomicron use different sets of genes to metabolize identical HMO (66, 98). Low conservation of HMO metabolizing gene sets across genera and even within genera illustrates an important challenge in identifying other HMO utilizing bacteria. It is important to note that the lack of known HMO utilization genes in a species is not indicative of a lack of HMO metabolizing capacity as the species may use an entirely different set of genes for HMO metabolism. Thus, analyses of the presence and abundance of specific genes rather than cataloging specific bacterial taxa will be required to fully understand any associations between HMOs and the development of AD/eczema in infants.
HMOs are known to influence neonatal mucosal and systemic immunity directly and indirectly (17, 45, 51). For example, HMOs increase intestinal cell maturation and barrier function (118, 119), and directly interact with immune cells to influence proliferation and cytokine production (17). Some HMOs, particularly those carrying fucosyl groups, are associated with a Th2 promoting anti-inflammatory response (27). The loss of anti-inflammatory signals can indicate a shift in the homeostatic environment of the GI tract to favor chronic inflammation (120). Furthermore, differences in types and abundances of bacteria capable of metabolizing HMOs may also play a role in host immune development (47, 48), but understanding how these microorganisms or their metabolites modulate the immune system is still in its infancy. Both human milk feeding (17, 51) and the presence of Bifidobacterium spp. (121, 122), have been previously reported to protect children from AD/eczema. However, few studies to date have conducted a detailed analysis of (1) HMO metabolizing genes or (2) HMO metabolites and their patterns of association with AD/eczema.
Once in the gut, the HMOs are metabolized by gut microbiota, most commonly by a member from the genus Bifidobacterium, into simpler carbohydrates and SCFAs (123). SCFAs interact with the immune system in various beneficial ways. For instance, they assist with maintaining intestinal epithelial barrier integrity, increase key gene expression and modulate dendritic cells and T cells (114). Butyrate, propionate, and acetate are the most notable SCFAs that influence immune health, with isobutyrate, isovalerate, valeric acid, and n-butyric acid also being present in infant guts (124). The presence of Bifidobacterium species in the infant gut, associated predominantly with breastfeeding, leads to the production of aromatic lactic acids from HMOs, specifically indolelactic acid, phenyllactic acid and 4-hydroxyphenyllactic acid. These Bifidobacterium species also produce amino acids, specifically tryptophan, phenylalanine and tyrosine (123, 125, 126). In particular, it has been shown with both in vitro and ex vivo methods that fecal concentrations of Bifidobacterium-derived indolelactic acid can activate the aryl hydrocarbon receptor important for controlling intestinal homeostasis and immune responses (125). Furthermore, two metabolites of tryptophan, an amino acid product of HMO degradation nicotinamide and 3-hydrokynurenine, are associated with increased atopy. But the association of increased levels of nicotinamide in maternal plasma and the development of infant AD/eczema conflict in the literature (127, 128).
Several studies have reported associations of bacterial taxa with AD/eczema as summarized in Table 2. At the phylum level, reduced diversity of Bacteroidetes has been observed in one-month old infants with AD/eczema, while reduced diversity of Proteobacteria has been associated with AD/eczema in 12 month old infants (129). Although not known to metabolize HMOs, several bacterial genera have been found in greater abundance in infants with AD/eczema and these include Streptococcus, Escherichia, Shigella, Veillonella, Faecalibacterium, Lachnospiraceae incertae sedis and Clostridium XlVa (88, 130). In terms of specific species, Faecalibacterium prausnitzii, Ruminococcus gnavus, Bacteroides clarus, Bacteroides plebeius, Parabacteroides merdae, Prevotella buccae, Gemmiger formicillis, Akkermansia muciniphila, Hungatella hathewayi and several members of the families Bacteroidaceae, Clostridiaceae, and Enterobacteriaceae, have been found in greater abundance in infants with AD/eczema (88, 131, 132). These associations can provide the first step in identifying candidates for further study of HMO metabolizing activity since our understanding of HMO metabolism and the identity of HMO metabolizing genes is still relatively nascent.
In contrast, Bifidobacterium and Bacteroides genera are present in higher abundances in infants without AD/eczema. Specifically, Bifidobacterium breve is associated with a decreased risk of AD/eczema development especially in the first 12 months of life (88, 89, 129). A few Bifidobacterium species, such as B. adolescentis, B. lactis and B. longum subsp. longum were found in comparable abundance in infants with and without AD/eczema while B. catenulatum was associated with a higher risk of developing AD/eczema (89). None of these bifidobacteria are known to metabolize HMOs completely, and at best, partially metabolize HMOs. There are also non-bifidobacterial species found in lower abundance in infants with AD/eczema, like Bacteroides fragilis and Streptococcus salivarius, with B. fragilis able to partially metabolize HMOs (16, 88). A few members of the genera Lactobacillus have also been found to have the ability to metabolize HMOs, namely L. casei and L. acidophilus. Even though studies of microbiomes associated with AD/eczema do not identify any associations between Lactobacillus in the infant gut and the development of AD/eczema, some prenatal or neonatal probiotic interventions with Lactobacillus species, like L. acidophilus or L. rhamnosus, find reduced eczema cases in treatment groups, although with varying levels of efficacy (101, 133–136).
The association between AD/eczema and HMO metabolites is still poorly described and is complicated by the variation in the amounts of HMO derived SCFAs at different stages of infancy. Wopereis et al. (137) noted that stool samples of infants with AD/eczema have decreased amounts of lactate and increased propionate and butyrate at 12 weeks. However, the opposite was observed with increased lactate and decreased propionate and butyrate at 26 weeks, with another decrease in butyrate levels by 12 months of age. Ta et al. (16) noticed a similar pattern of low butyrate and propionate levels in the stools of infants with AD/eczema, with AD/eczema infants in their cohort exhibiting consistently low butyrate and propionate levels even at 12 weeks compared to infants with healthy skin. Increasing severity of AD/eczema correlated with a decrease in butyrate-producing bacteria, suggesting there is less butyrate in the gastrointestinal tracts of infants with severe AD/eczema compared to those with milder or no AD/eczema (138). However, other studies have observed similar levels of butyrate in stool samples from infants with or without AD/eczema (130, 139). Another metabolite that is decreased in infants with AD/eczema is valerate/valeric acid, and infants with higher valerate are also protected from later development of AD/eczema (130, 139, 140). Butyrate and valerate were negatively correlated with the genus Streptococcus and were decreased in infants with AD/eczema (130). Several studies investigated the fecal metabolites from human milk fed infants (Table 3). In these studies, HMO metabolites in stool easily differentiated formula fed from human milk fed infants. To some extent, the presence of specific SCFAs in feces could differentiate human milk fed from formula fed infants. Dotz et al. (143) found that their cohort of exclusively human milk fed infants could be clustered into three groups characterized by their fecal HMO metabolites as compared to the HMO profiles in mother's milk. These three profiles include (1) infant fecal HMOs with similar or higher complexity than the HMOs in mother's milk, (2) reduced HMO diversity and intensity but increased number and intensity of non-HMOs than mother's milk, or (3) decreased relative intensities of fucosylated HMOs. The acetylated neutral HMOs identified by Dotz et al. (143) in infant and maternal urine have been determined by others to be important in human immunology (150), making this metabolite a candidate metabolite important for the development of atopy and of specific interest to the present review. Similarly, a relationship between Bifidobacterium spp., which encodes many HMO metabolizing genes, is found in higher abundance in infants without AD/eczema (60, 89), and fewer fecal HMO metabolites have been observed in infants lacking B. infantis (151). Other studies have reported that infants consuming lacto-N-fucopentaose-rich human milk (152) and infants whose mothers have the gene responsible for making 2'fucosyllactose (FUT2) (153) are protected from atopic diseases.
There is some evidence that bacteria which use an extracellular glycosidase-dependent strategy share HMOs and metabolites with other microbes, whereas those bacteria which use an oligosaccharide transporter-dependent method for metabolizing HMOs may inhibit other bacteria from using the HMOs or the resulting HMO metabolites (64). Further, when B. bifidum, B. breve, and B. infantis were present in combination and grown on HMOs, growth was improved compared to that achieved when grown as single strains. Other recent work has demonstrated that HMO metabolizing Bifidobacterium spp. can also provide substrates for the growth of non-HMO metabolizing microbes (91). We hypothesize that the diversity of HMO metabolizing genes present in the infant gut microbiome plays an important role in the ability of human milk feeding to reduce the risk for AD/eczema. Thus, approaches which address the full complement of such genes are needed to advance the field.
In addition to their indirect role in immune development by shaping the infant gut microbiome, HMOs also have direct effects on intestinal maturation and immune development. HMOs can suppress cell cycle progression in intestinal cells to either induce differentiation or cause apoptosis (154). HMOs can also protect the gut against colonization by pathogenic bacteria due to their structural similarities to cell surface glycoconjugates that are recognized by microbes (45). HMOs further interfere with the adhesion of pathogenic bacteria to the intestinal epithelium by modifying the extracellular glycosylation of epithelial cells (154).
HMOs interact with the host's immune system through binding to lectin and TLR receptors. The presence of lectin receptors on a variety of immune cells like macrophages, dendritic cells, neutrophils, eosinophils, monocytes and natural killer (NK) cells (155) and also the detection of HMOs in the plasma of infants consuming human milk diets suggest a direct interaction between HMOs and immune cells both in the GI tract and in the blood (27). Even though direct evidence of the immunomodulatory effect of HMOs during AD/eczema development is sparse, the anti-inflammatory properties of HMOs and their ability to affect tolerogenic factors important for the prevention of allergic diseases have been described in several in vitro and in vivo studies. In particular, fucosylated HMOs have been associated with a Th2-promoting anti-inflammatory response (27, 154), while sialylated HMOs assist in lymphocyte maturation and maintain the balance between cytokine production related to Th1/Th2 type T-helper responses (155) and also inhibit TLR4/NLRP3 inflammosome pathways (156).
At the intersection of the immunomodulatory effects of HMOs and HMO utilizing bacteria is the effect of HMO derived SCFAs on the infant immune system. While the direct relation between SCFAs and AD/eczema development is still unclear, we can hypothesize that the immunomodulatory effects of metabolites representing unique AD/eczema metabolic signatures may play a role in the development of eczema. SCFAs, specifically propionate and butyrate which are often found in lower levels in individuals with eczema, have been shown to protect individuals from food allergies. The protective effect of SCFAs is thought to be via the development of a regulatory T cell network in the GI tract (157, 158). Butyrate, in particular, can regulate several immunomodulatory pathways by inhibiting histone deacetylases and altering epigenomic signatures (159). Epigenetic modulation is one of the ways that has been identified for microbes to have a long lasting effect on immunity from early childhood exposures (19, 20).
This review addresses fundamental gaps in our understanding of the ecology and evolution of HMO metabolizing microorganisms as well as their association with the prevalent disease of infancy and early childhood, AD/eczema. Though current evidence suggests a protective effect of human milk, enough conflicting studies exist to question the association. Herein, we presented evidence for diversity of HMOs, microbes, and metabolites among even exclusively breastfed infants and suggested that specific repertoires of microbial genes and gene products may be required in order for human milk to protect infants from AD/eczema. In the discussion, we propose possible future research which could elucidate the role of these specific genes and metabolites in human health and disease.
Up to one in five infants in the US experiences AD/eczema. While genetics, such as modifications in the filaggrin gene, can explain a fraction of eczema cases, other factors like the immunomodulatory effect of gut microbes also can play a part in AD/eczema development (16–18). Furthermore, oligosaccharides in human milk serve as a prebiotic to shape the infant gut microbiota, promote growth of commensals and confer immunomodulatory effects to the infant immune system both directly and indirectly. Future research should take an innovative approach to defining factors associated with the developing gut microbiota that could protect from AD/eczema. Although diet is likely an important contributor to this disease, the composition of the microbiota and its ability to digest a specific component of the diet is potentially just as important in protecting against AD/eczema. In fact, the ability of the intestinal microbiota to metabolize distinct sets of HMOs (Table 2) may explain why human milk feeding protects against AD/eczema in some but not all infants fed human milk. Within the context of the host, both the microbial genetic potential for HMO metabolism as well as the resulting metabolites should be examined. While human milk exposure has been suggested to be protective from AD/eczema in observational studies, the evidence presented in this review suggests that specific HMO metabolizing phenotypes may underlie the inconsistent effect of human milk on the prevention of atopy. The HMO metabolites are likely important as these metabolites are known to protect against pathogens and to interact with the infant's developing immune system (51, 160).
In addition to HMOs, there are other immunomodulatory components in human milk, and these can add complexity to how we interpret any observed immunoprotective effects of HMOs. Human milk contains immunoglobulins like secretory IgA and IgG, enzymes such as lysozymes and other factors like lactoferrin and these molecules provide protection from pathogens. Maternal antibodies in the milk can also interact with infant T and B cell programming to influence the adaptive immune response in infants (161). Heterogeneity in the composition of these other human milk components, varying between individuals and even pregnancies in the same individual, may also have a role in the development of atopy.
Measuring HMOs and the by-products of HMO metabolism in infant fecal samples is critical in order to understand their link to AD/eczema. However, assessing the metabolite content of the fecal samples has challenges and limitations. SCFAs are volatile. Their concentrations may be affected by fecal water content, and SCFAs are also readily adsorbed by the infant upon production (162, 163). This means that fecal SCFA concentrations might not reflect the true SCFA production resulting from HMO metabolism in the gut. Despite these challenges, understanding the metabolic signatures in the infant gut provides important insight into the factors affecting the development of AD/eczema.
Specifically, future work should (1) identify the diversity and abundance of HMO metabolizing genes in human atopic infants, (2) link HMO metabolizing genes to specific microbes, (3) evaluate whether there are geographically specific HMO metabolizing repertoires, (4) determine if specific HMO metabolizing genes are associated with an increased or decreased risk of AD/eczema, (5) determine if specific HMO metabolites are associated with the risk of developing AD/eczema. Such studies must control for a variety of covariates including the infant's non-milk diet, mode of delivery, antibiotic use, maternal pre-pregnancy body mass index, as well as additional medical and behavioral outcomes. Those undertaking such research should have combined expertise in microbial ecology, bacterial genomics, bioinformatics, immunology, atopy and allergy, HMOs, and human milk. Detailed genomic, metagenomic and metabolomic studies of the infant gut microbiota, especially during the human milk-feeding stage, are essential for elucidating the impact of early exposures on health outcomes and development of atopy.
Host-associated microbes are important determinants of health. Through their interactions with dietary intake or other environmental exposures, these microbes can exacerbate or protect their host from negative health outcomes. As demonstrated in this literature review, human milk may protect against AD/eczema. However, it is the specific genes of, and thus metabolites produced by, microbes consuming HMOs that are likely the most important determinants of the protective role for human milk. Future work must elucidate the diversity of bacterial metabolism and bacterial metabolites associated with AD/eczema in human infants. Through such work, we can gain an understanding of the mechanisms by which microbes and microbial metabolites modulate the developing immune system. Then we can begin identifying metabolic components and metabolites involved in protection against AD/eczema. Ultimately, this will enable the development of biotherapeutics to upregulate or block identified microbial pathways.
Conception or design of the work: SSC, VK. Data collection: all authors. Data analysis and interpretation: TR, PS, CP. Drafting the article: all authors. Critical revision of the article: SSC, VK. Final approval of the version to be published: SSC, VK. All authors contributed to the article and approved the submitted version.
This work was supported by the National Institutes of Health (R15AI160139, UH3OD023313, and UH3OD023285) and Wellcome LEAP 1KD.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Eichenfield LF, Tom WL, Chamlin SL, Feldman SR, Hanifin JM, Simpson EL, et al. Guidelines of care for the management of atopic dermatitis: section 1. Diagnosis and assessment of atopic dermatitis. J Am Acad Dermatol. (2014) 70(2):338–51. doi: 10.1016/j.jaad.2013.10.010
2. Fishbein AB, Silverberg JI, Wilson EJ, Ong PY. Update on atopic dermatitis: diagnosis, severity assessment, and treatment selection. J Allergy Clin Immunol Pract. (2020) 8(1):91–101. doi: 10.1016/j.jaip.2019.06.044
3. Nutten S. Atopic dermatitis: global epidemiology and risk factors. Ann Nutr Metab. (2015) 66(Suppl 1):8–16. doi: 10.1159/000370220
4. Shaw TE, Currie GP, Koudelka CW, Simpson EL. Eczema prevalence in the United States: data from the 2003 national survey of children’s health. J Invest Dermatol. (2011) 131(1):67–73. doi: 10.1038/jid.2010.251
5. Dharmage SC, Lowe AJ, Matheson MC, Burgess JA, Allen KJ, Abramson MJ. Atopic dermatitis and the atopic march revisited. Allergy. (2014) 69(1):17–27. doi: 10.1111/all.12268
6. Dawn A, Papoiu ADP, Chan YH, Rapp SR, Rassette N, Yosipovitch G. Itch characteristics in atopic dermatitis: results of a web-based questionnaire. Br J Dermatol. (2009) 160(3):642–4. doi: 10.1111/j.1365-2133.2008.08941.x
7. Silverberg JI. Associations between atopic dermatitis and other disorders. F1000Res. (2018) 7:303. doi: 10.12688/f1000research.12975.1
8. Silverberg JI, Gelfand JM, Margolis DJ, Boguniewicz M, Fonacier L, Grayson MH, et al. Pain is a common and burdensome symptom of atopic dermatitis in United States adults. J Allergy Clin Immunol Pract. (2019) 7(8):2699–2706.e7. doi: 10.1016/j.jaip.2019.05.055
9. Djurović MR, Janković J, Ćirković A, Spirić VT, Maksimović N, Timotijević ZS, et al. Quality of life in infants with atopic dermatitis and their families. Postepy Dermatol Alergol. (2020) 37(1):66–72. doi: 10.5114/ada.2020.93385
10. Reed B, Blaiss MS. The burden of atopic dermatitis. Allergy Asthma Proc. (2018) 39(6):406–10. doi: 10.2500/aap.2018.39.4175
11. Hua T, Silverberg JI. Atopic dermatitis is associated with increased hospitalization in US children. J Am Acad Dermatol. (2019) 81(3):862–5. doi: 10.1016/j.jaad.2019.05.019
12. Drucker AM, Wang AR, Li WQ, Sevetson E, Block JK, Qureshi AA. The burden of atopic dermatitis: summary of a report for the national eczema association. J Invest Dermatol. (2017) 137(1):26–30. doi: 10.1016/j.jid.2016.07.012
13. Smith Begolka W, Chovatiya R, Thibau IJ, Silverberg JI. Financial burden of atopic dermatitis out-of-pocket health care expenses in the United States. Dermatitis. (2021) 32(1S):S62–70. doi: 10.1097/DER.0000000000000715
14. Rasheed Z, Zedan K, Saif GB, Salama RH, Salem T, Ahmed AA, et al. Markers of atopic dermatitis, allergic rhinitis and bronchial asthma in pediatric patients: correlation with filaggrin, eosinophil major basic protein and immunoglobulin E. Clin Mol Allergy. (2018) 16(1):23. doi: 10.1186/s12948-018-0102-y
15. van den Oord RAHM, Sheikh A. Filaggrin gene defects and risk of developing allergic sensitisation and allergic disorders: systematic review and meta-analysis. Br Med J. (2009) 339(jul08 4):b2433. doi: 10.1136/bmj.b2433
16. Ta LDH, Chan JCY, Yap GC, Purbojati RW, Drautz-Moses DI, Koh YM, et al. A compromised developmental trajectory of the infant gut microbiome and metabolome in atopic eczema. Gut Microbes. (2020) 12(1):1801964. doi: 10.1080/19490976.2020.1801964
17. Donovan SM, Comstock SS. Human milk oligosaccharides influence neonatal mucosal and systemic immunity. ANM. (2016) 69(Suppl. 2):41–51. doi: 10.1159/000452818
18. Komatsu Y, Kumakura D, Seto N, Izumi H, Takeda Y, Ohnishi Y, et al. Dynamic associations of milk components with the infant gut microbiome and fecal metabolites in a mother–infant model by microbiome, NMR metabolomic, and time-series clustering analyses. Front Nutr. (2022) 8:813690. doi: 10.3389/fnut.2021.813690
19. Belkaid Y, Hand TW. Role of the microbiota immunity and inflammation. Cell. (2014) 157(1):121–41. doi: 10.1016/j.cell.2014.03.011
20. Lambrecht BN, Hammad H. The immunology of the allergy epidemic and the hygiene hypothesis. Nat Immunol. (2017) 18(10):1076–83. doi: 10.1038/ni.3829
21. Martinez-García MA, Oscullo G, Posadas T, Zaldivar E, Villa C, Dobarganes Y, et al. Pseudomonas aeruginosa and lung function decline in patients with bronchiectasis. Clin Microbiol Infect. (2021) 27(3):428–34. doi: 10.1016/j.cmi.2020.04.007
22. Widhiati S, Purnomosari D, Wibawa T, Soebono H. The role of gut microbiome in inflammatory skin disorders: a systematic review. Dermatol Rep. (2021) 14(1):9188. doi: 10.4081/dr.2022.9188
23. Macpherson AJ, Geuking MB, McCoy KD. Immune responses that adapt the intestinal mucosa to commensal intestinal bacteria. Immunology. (2005) 115(2):153–62. doi: 10.1111/j.1365-2567.2005.02159.x
24. Healy DB, Ryan CA, Ross RP, Stanton C, Dempsey EM. Clinical implications of preterm infant gut microbiome development. Nat Microbiol. (2021) 7(1):22–33. doi: 10.1038/s41564-021-01025-4
25. Dawod B, Marshall JS, Azad MB. Breastfeeding and the developmental origins of mucosal immunity: how human milk shapes the innate and adaptive mucosal immune systems. Curr Opin Gastroenterol. (2021) 37(6):547–56. doi: 10.1097/MOG.0000000000000778
26. Zhang S, Li T, Xie J, Zhang D, Pi C, Zhou L, et al. Gold standard for nutrition: a review of human milk oligosaccharide and its effects on infant gut microbiota. Microb Cell Fact. (2021) 20(1):108. doi: 10.1186/s12934-021-01599-y
27. Kulinich A, Liu L. Human milk oligosaccharides: the role in the fine-tuning of innate immune responses. Carbohydr Res. (2016) 432:62–70. doi: 10.1016/j.carres.2016.07.009
28. Sugino KY, Ma T, Paneth N, Comstock SS. Effect of environmental exposures on the gut microbiota from early infancy to two years of age. Microorganisms. (2021) 9(10):2140. doi: 10.3390/microorganisms9102140
29. Ferro LE, Sugino KY, Klepac-Ceraj V, Comstock SS. The abundance of human milk oligosaccharide (HMO)-metabolizing genes in fecal samples from six-month-old human infants. Microorganisms. (2021) 9(7):1352. doi: 10.3390/microorganisms9071352
30. Masi AC, Stewart CJ. Untangling human milk oligosaccharides and infant gut microbiome. iScience. (2022) 25(1):103542. doi: 10.1016/j.isci.2021.103542
31. Han SM, Derraik JGB, Binia A, Sprenger N, Vickers MH, Cutfield WS. Maternal and infant factors influencing human milk oligosaccharide composition: beyond maternal genetics. J Nutr. (2021) 151(6):1383–93. doi: 10.1093/jn/nxab028
32. Thurl S, Munzert M, Boehm G, Matthews C, Stahl B. Systematic review of the concentrations of oligosaccharides in human milk. Nutr Rev. (2017) 75(11):920–33. doi: 10.1093/nutrit/nux044
33. Walsh C, Lane JA, van Sinderen D, Hickey RM. Human milk oligosaccharides: shaping the infant gut microbiota and supporting health. J Funct Foods. (2020) 72:104074. doi: 10.1016/j.jff.2020.104074
34. Parazzini F, Cipriani S, Zinetti C, Chatenoud L, Frigerio L, Amuso G, et al. Perinatal factors and the risk of atopic dermatitis: a cohort study. Pediatr Allergy Immunol. (2014) 25(1):43–50. doi: 10.1111/pai.12165
35. Lee KS, Rha YH, Oh IH, Choi YS, Kim YE, Choi SH. Does breast-feeding relate to development of atopic dermatitis in young Korean children?: based on the fourth and fifth Korea national health and nutrition examination survey 2007–2012. Allergy Asthma Immunol Res. (2017) 9(4):307. doi: 10.4168/aair.2017.9.4.307
36. Schoetzau A, Filipiak-Pittroff B, Franke K, Koletzko S, Von Berg A, Gruebl A, et al. Effect of exclusive breast-feeding and early solid food avoidance on the incidence of atopic dermatitis in high-risk infants at 1 year of age: breast-feeding and atopic dermatitis. Pediatr Allergy Immunol. (2002) 13(4):234–42. doi: 10.1034/j.1399-3038.2002.01050.x
37. Gdalevich M, Mimouni D, David M, Mimouni M. Breast-feeding and the onset of atopic dermatitis in childhood: a systematic review and meta-analysis of prospective studies. J Am Acad Dermatol. (2001) 45(4):520–7. doi: 10.1067/mjd.2001.114741
38. Laubereau B, Brockow I, Zirngibl A, Koletzko S, Gruebl A, Von Berg A, et al. Effect of breast-feeding on the development of atopic dermatitis during the first 3 years of life - results from the GINI-birth cohort study. J Pediatr. (2004) 144(5):602–7. doi: 10.1016/j.jpeds.2003.12.029
39. Kull I, Böhme M, Wahlgren CF, Nordvall L, Pershagen G, Wickman M. Breast-feeding reduces the risk for childhood eczema. J Allergy Clin Immunol. (2005) 116(3):657–61. doi: 10.1016/j.jaci.2005.04.028
40. Lodge C, Tan D, Lau M, Dai X, Tham R, Lowe A, et al. Breastfeeding and asthma and allergies: a systematic review and meta-analysis. Acta Paediatr. (2015) 104:38–53. doi: 10.1111/apa.13132
41. Kramer MS, Kakuma R. Optimal duration of exclusive breastfeeding. Cochrane Database Syst. Rev. (2012) 8:CD003517. doi: 10.1002/14651858.CD003517.pub2
42. Yang YW, Tsai CL, Lu CY. Exclusive breastfeeding and incident atopic dermatitis in childhood: a systematic review and meta-analysis of prospective cohort studies. Br J Dermatol. (2009) 161(2):373–83. doi: 10.1111/j.1365-2133.2009.09049.x
43. Björkstén B, Aït-Khaled N, Innes Asher M, Clayton TO, Robertson C. Global analysis of breast feeding and risk of symptoms of asthma, rhinoconjunctivitis and eczema in 6–7 year old children: iSAAC phase three. Allergol Immunopathol (Madr). (2011) 39(6):318–25. doi: 10.1016/j.aller.2011.02.005
44. Lin B, Dai R, Lu L, Fan X, Yu Y. Breastfeeding and atopic dermatitis risk: a systematic review and meta-analysis of prospective cohort studies. Dermatology. (2020) 236(4):345–60. doi: 10.1159/000503781
45. Plaza-Díaz J, Fontana L, Gil A. Human milk oligosaccharides and immune system development. Nutrients. (2018) 10(8):1038. doi: 10.3390/nu10081038
46. Parada Venegas D, De la Fuente MK, Landskron G, González MJ, Quera R, Dijkstra G, et al. Short chain fatty acids (SCFAs)-mediated gut epithelial and immune regulation and its relevance for inflammatory bowel diseases. Front Immunol. (2019) 10:277. doi: 10.3389/fimmu.2019.00277
47. Vatanen T, Franzosa EA, Schwager R, Tripathi S, Arthur TD, Vehik K, et al. The human gut microbiome in early-onset type 1 diabetes from the TEDDY study. Nature. (2018) 562(7728):589–94. doi: 10.1038/s41586-018-0620-2
48. Wang W, Luo X, Zhang Q, He X, Zhang Z, Wang X. Bifidobacterium infantis relieves allergic asthma in mice by regulating Th1/Th2. Med Sci Monit. (2020) 26:e920583. doi: 10.12659/MSM.920583
49. Selene KB, Zu Z, Zheng T. The atopic march: progression from atopic dermatitis to allergic rhinitis and asthma. J Clin Cell Immunol. (2014) 05(02):202. doi: 10.4172/2155-9899.1000202
50. Grant MJ, Booth A. A typology of reviews: an analysis of 14 review types and associated methodologies. Health Info Libr J. (2009) 26(2):91–108. doi: 10.1111/j.1471-1842.2009.00848.x
51. Le Doare K, Holder B, Bassett A, Pannaraj PS. Mother’s milk: a purposeful contribution to the development of the infant microbiota and immunity. Front Immunol. (2018) 9:361. doi: 10.3389/fimmu.2018.00361
52. De Leoz MLA, Kalanetra KM, Bokulich NA, Strum JS, Underwood MA, German JB, et al. Human milk glycomics and gut microbial genomics in infant feces show a correlation between human milk oligosaccharides and gut microbiota: a proof-of-concept study. J Proteome Res. (2015) 14(1):491–502. doi: 10.1021/pr500759e
53. Bäckhed F, Roswall J, Peng Y, Feng Q, Jia H, Kovatcheva-Datchary P, et al. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe. (2015) 17(5):690–703. doi: 10.1016/j.chom.2015.04.004
54. Forbes JD, Azad MB, Vehling L, Tun HM, Konya TB, Guttman DS, et al. Association of exposure to formula in the hospital and subsequent infant feeding practices with gut microbiota and risk of overweight in the first year of life. JAMA Pediatr. (2018) 172(7):e181161. doi: 10.1001/jamapediatrics.2018.1161
55. Raspini B, Porri D, De Giuseppe R, Chieppa M, Liso M, Cerbo RM, et al. Prenatal and postnatal determinants in shaping offspring’s microbiome in the first 1000 days: study protocol and preliminary results at one month of life. Ital J Pediatr. (2020) 46(1):45. doi: 10.1186/s13052-020-0794-8
56. Sugino KY, Paneth N, Comstock SS. Michigan Cohorts to determine associations of maternal pre-pregnancy body mass index with pregnancy and infant gastrointestinal microbial communities: late pregnancy and early infancy. PLoS One. (2019) 14(3):e0213733. doi: 10.1371/journal.pone.0213733
57. Yasmin F, Tun HM, Konya TB, Guttman DS, Chari RS, Field CJ, et al. Cesarean section, formula feeding, and infant antibiotic exposure: separate and combined impacts on gut microbial changes in later infancy. Front Pediatr. (2017) 5:200. doi: 10.3389/fped.2017.00200
58. Palmer C, Bik EM, DiGiulio DB, Relman DA, Brown PO. Development of the human infant intestinal Microbiota. Ruan Y, editor. PLoS Biol. (2007) 5(7):e177. doi: 10.1371/journal.pbio.0050177
59. Guo S, Guo Y, Ergun A, Lu L, Walker WA, Ganguli K. Secreted metabolites of Bifidobacterium infantis and Lactobacillus acidophilus protect immature human enterocytes from IL-1β-induced inflammation: a transcription profiling analysis. PLoS One. (2015) 10(4):e0124549. doi: 10.1371/journal.pone.0124549
60. Underwood MA, German JB, Lebrilla CB, Mills DA. Bifidobacterium longum subspecies infantis: champion colonizer of the infant gut. Pediatr Res. (2015) 77(1):229–35. doi: 10.1038/pr.2014.156
61. Arboleya S, Stanton C, Ryan CA, Dempsey E, Ross PR. Bosom buddies: the symbiotic relationship between infants and Bifidobacterium longum ssp. longum and ssp. infantis. Genetic and probiotic features. Annu Rev Food Sci Technol. (2016) 7(1):1–21. doi: 10.1146/annurev-food-041715-033151
62. Sedighi M, Razavi S, Navab-Moghadam F, Khamseh ME, Alaei-Shahmiri F, Mehrtash A, et al. Comparison of gut microbiota in adult patients with type 2 diabetes and healthy individuals. Microb Pathog. (2017) 111:362–9. doi: 10.1016/j.micpath.2017.08.038
63. Gong H, Gao H, Ren Q, He J. The abundance of bifidobacterium in relation to visceral obesity and serum uric acid. Sci Rep. (2022) 12(1):13073. doi: 10.1038/s41598-022-17417-3
64. Gotoh A, Katoh T, Sakanaka M, Ling Y, Yamada C, Asakuma S, et al. Sharing of human milk oligosaccharides degradants within bifidobacterial communities in faecal cultures supplemented with Bifidobacterium bifidum. Sci Rep. (2018) 8(1):13958. doi: 10.1038/s41598-018-32080-3
65. Lawson MAE, O’Neill IJ, Kujawska M, Gowrinadh Javvadi S, Wijeyesekera A, Flegg Z, et al. Breast milk-derived human milk oligosaccharides promote Bifidobacterium interactions within a single ecosystem. ISME J. (2020) 14(2):635–48. doi: 10.1038/s41396-019-0553-2
66. Marcobal A, Barboza M, Sonnenburg ED, Pudlo N, Martens EC, Desai P, et al. Bacteroides in the infant gut consume milk oligosaccharides via mucus-utilization pathways. Cell Host Microbe. (2011) 10(5):507–14. doi: 10.1016/j.chom.2011.10.007
67. Schwab C, Gänzle M. Lactic acid bacteria fermentation of human milk oligosaccharide components, human milk oligosaccharides and galactooligosaccharides: lAB fermentation of HMOs and GOSs. FEMS Microbiol Lett. (2011) 315(2):141–8. doi: 10.1111/j.1574-6968.2010.02185.x
68. Yu ZT, Chen C, Newburg DS. Utilization of major fucosylated and sialylated human milk oligosaccharides by isolated human gut microbes. Glycobiology. (2013) 23(11):1281–92. doi: 10.1093/glycob/cwt065
69. Zeng S, Patangia D, Almeida A, Zhou Z, Mu D, Paul Ross R, et al. A compendium of 32,277 metagenome-assembled genomes and over 80 million genes from the early-life human gut microbiome. Nat Commun. (2022) 13(1):5139. doi: 10.1038/s41467-022-32805-z
70. Martín R, Heilig GHJ, Zoetendal EG, Smidt H, Rodríguez JM. Diversity of the Lactobacillus group in breast milk and vagina of healthy women and potential role in the colonization of the infant gut. J Appl Microbiol. (2007) 103(6):2638–44. doi: 10.1111/j.1365-2672.2007.03497.x
71. Borewicz K, Gu F, Saccenti E, Arts ICW, Penders J, Thijs C, et al. Correlating infant fecal microbiota composition and human milk oligosaccharide consumption by microbiota of 1-month-old breastfed infants. Mol Nutr Food Res. (2019) 63(13):1801214. doi: 10.1002/mnfr.201801214
72. Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science. (2012) 336(6086):1268–73. doi: 10.1126/science.1223490
73. Rivière A, Selak M, Lantin D, Leroy F, De Vuyst L. Bifidobacteria and butyrate-producing colon bacteria: importance and strategies for their stimulation in the human gut. Front Microbiol. (2016) 7:979. doi: 10.3389/fmicb.2016.00979
74. Davis JCC, Lewis ZT, Krishnan S, Bernstein RM, Moore SE, Prentice AM, et al. Growth and morbidity of gambian infants are influenced by maternal milk oligosaccharides and infant gut microbiota. Sci Rep. (2017) 7(1):40466. doi: 10.1038/srep40466
75. Lei WT, Huang KY, Jhong JH, Chen CH, Weng SL. Metagenomic analysis of the gut microbiome composition associated with vitamin D supplementation in Taiwanese infants. Sci Rep. (2021) 11(1):2856. doi: 10.1038/s41598-021-82584-8
76. Oyedemi OT, Shaw S, Martin JC, Ayeni FA, Scott KP. Changes in the gut microbiota of Nigerian infants within the first year of life. Zoetendal EG, editor. PLoS One. (2022) 17(3):e0265123. doi: 10.1371/journal.pone.0265123
77. Stewart CJ, Ajami NJ, O’Brien JL, Hutchinson DS, Smith DP, Wong MC, et al. Temporal development of the gut microbiome in early childhood from the TEDDY study. Nature. (2018) 562(7728):583–8. doi: 10.1038/s41586-018-0617-x
78. Henrick BM, Hutton AA, Palumbo MC, Casaburi G, Mitchell RD, Underwood MA, et al. Elevated fecal pH indicates a profound change in the breastfed infant gut microbiome due to reduction of Bifidobacterium over the past century. mSphere. (2018) 3(2):e00041–18. doi: 10.1128/mSphere.00041-18
79. Olm MR, Dahan D, Carter MM, Merrill BD, Yu FB, Jain S, et al. Robust variation in infant gut microbiome assembly across a spectrum of lifestyles. Science. (2022) 376(6598):1220–3. doi: 10.1126/science.abj2972
80. Lackey KA, Williams JE, Meehan CL, Zachek JA, Benda ED, Price WJ, et al. What’s normal? Microbiomes in human milk and infant feces are related to each other but vary geographically: the INSPIRE study. Front Nutr. (2019) 6:45. doi: 10.3389/fnut.2019.00045
81. Lewis ZT, Mills DA. Differential establishment of bifidobacteria in the breastfed infant gut. Nestle Nutr Inst Workshop Ser. (2017) 88:149–59. doi: 10.1159/000455399
82. Jansen GJ, Wildeboer-Veloo AC, Tonk RH, Franks AH, Welling GW. Development and validation of an automated, microscopy-based method for enumeration of groups of intestinal bacteria. J Microbiol Methods. (1999) 37(3):215–21. doi: 10.1016/S0167-7012(99)00049-4
83. Sim K, Cox MJ, Wopereis H, Martin R, Knol J, Li MS, et al. Improved detection of bifidobacteria with optimised 16S rRNA-gene based pyrosequencing. PLoS One. (2012) 7(3):e32543. doi: 10.1371/journal.pone.0032543
84. Zoetendal EG, Ben-Amor K, Akkermans ADL, Abee T, de Vos WM. DNA Isolation protocols affect the detection limit of PCR approaches of bacteria in samples from the human gastrointestinal tract. Syst Appl Microbiol. (2001) 24(3):405–10. doi: 10.1078/0723-2020-00060
85. Tso L, Bonham KS, Fishbein A, Rowland S, Klepac-Ceraj V. Targeted high-resolution taxonomic identification of Bifidobacterium longum subsp. infantis using human milk oligosaccharide metabolizing genes. Nutrients. (2021) 13(8):2833. doi: 10.3390/nu13082833
86. Garrido D, Barile D, Mills DA. A molecular basis for bifidobacterial enrichment in the infant gastrointestinal tract. Adv Nutr. (2012) 3(3):415S–21S. doi: 10.3945/an.111.001586
87. Insel R, Knip M. Prospects for primary prevention of type 1 diabetes by restoring a disappearing microbe. Pediatr Diabetes. (2018) 19(8):1400–6. doi: 10.1111/pedi.12756
88. Zheng H, Liang H, Wang Y, Miao M, Shi T, Yang F, et al. Altered gut microbiota composition associated with eczema in infants. PLoS One. (2016) 11(11):e0166026. doi: 10.1371/journal.pone.0166026
89. Ismail IH, Boyle RJ, Licciardi PV, Oppedisano F, Lahtinen S, Robins-Browne RM, et al. Early gut colonization by Bifidobacterium breve and B. catenulatum differentially modulates eczema risk in children at high risk of developing allergic disease. Pediatr Allergy Immunol. (2016) 27(8):838–46. doi: 10.1111/pai.12646
90. Cheng L, Kiewiet MBG, Logtenberg MJ, Groeneveld A, Nauta A, Schols HA, et al. Effects of different human milk oligosaccharides on growth of bifidobacteria in monoculture and co-culture with Faecalibacterium prausnitzii. Front Microbiol. (2020) 11:569700. doi: 10.3389/fmicb.2020.569700
91. Walsh C, Lane JA, van Sinderen D, Hickey RM. Human milk oligosaccharide-sharing by a consortium of infant derived Bifidobacterium species. Sci Rep. (2022) 12(1):4143. doi: 10.1038/s41598-022-07904-y
92. Ward RE, Niñonuevo M, Mills DA, Lebrilla CB, German JB. In vitro fermentability of human milk oligosaccharides by several strains of bifidobacteria. Mol Nutr Food Res. (2007) 51(11):1398–405. doi: 10.1002/mnfr.200700150
93. Wada J, Ando T, Kiyohara M, Ashida H, Kitaoka M, Yamaguchi M, et al. Bifidobacterium bifidum lacto- N -biosidase, a critical enzyme for the degradation of human milk oligosaccharides with a type 1 structure. Appl Environ Microbiol. (2008) 74(13):3996–4004. doi: 10.1128/AEM.00149-08
94. Liu J, Li W, Yao C, Yu J, Zhang H. Comparative genomic analysis revealed genetic divergence between Bifidobacterium catenulatum subspecies present in infant versus adult guts. BMC Microbiol. (2022) 22(1):158. doi: 10.1186/s12866-022-02573-3
95. Gavini F, Delcenserie V, Kopeinig K, Pollinger S, Beerens H, Bonaparte C, et al. Bifidobacterium species isolated from animal feces and from beef and pork meat. J Food Prot. (2006) 69(4):871–7. doi: 10.4315/0362-028X-69.4.871
96. Xiao JZ, Takahashi S, Nishimoto M, Odamaki T, Yaeshima T, Iwatsuki K, et al. Distribution of in vitro fermentation ability of lacto- N -biose I, a major building block of human milk oligosaccharides, in bifidobacterial strains. Appl Environ Microbiol. (2010) 76(1):54–9. doi: 10.1128/AEM.01683-09
97. Salli K, Hirvonen J, Siitonen J, Ahonen I, Anglenius H, Maukonen J. Selective utilization of the human milk oligosaccharides 2′-fucosyllactose, 3-fucosyllactose, and difucosyllactose by various probiotic and pathogenic bacteria. J Agric Food Chem. (2021) 69(1):170–82. doi: 10.1021/acs.jafc.0c06041
98. Marcobal A, Barboza M, Froehlich JW, Block DE, German JB, Lebrilla CB, et al. Consumption of human milk oligosaccharides by gut-related microbes. J Agric Food Chem. (2010) 58(9):5334–40. doi: 10.1021/jf9044205
99. Cox MJ, Huang YJ, Fujimura KE, Liu JT, McKean M, Boushey HA, et al. Lactobacillus casei abundance is associated with profound shifts in the infant gut microbiome. Bell T, editor. PLoS One. (2010) 5(1):e8745. doi: 10.1371/journal.pone.0008745
100. Bidart GN, Rodríguez-Díaz J, Pérez-Martínez G, Yebra MJ. The lactose operon from Lactobacillus casei is involved in the transport and metabolism of the human milk oligosaccharide core-2 N-acetyllactosamine. Sci Rep. (2018) 8(1):7152. doi: 10.1038/s41598-018-25660-w
101. Kim JY, Kwon JH, Ahn SH, Lee SI, Han YS, Choi YO, et al. Effect of probiotic mix (Bifidobacterium bifidum, Bifidobacterium lactis, Lactobacillus acidophilus) in the primary prevention of eczema: a double-blind, randomized, placebo-controlled trial. Pediatr Allergy Immunol. (2010) 21(2p2):e386–93. doi: 10.1111/j.1399-3038.2009.00958.x
102. Zimmermann P, Messina N, Mohn WW, Finlay BB, Curtis N. Association between the intestinal microbiota and allergic sensitization, eczema, and asthma: a systematic review. J Allergy Clin Immunol. (2019) 143(2):467–85. doi: 10.1016/j.jaci.2018.09.025
103. Hunt KM, Preuss J, Nissan C, Davlin CA, Williams JE, Shafii B, et al. Human milk oligosaccharides promote the growth of Staphylococci. Appl Environ Microbiol. (2012) 78(14):4763–70. doi: 10.1128/AEM.00477-12
104. Hoeflinger JL, Davis SR, Chow J, Miller MJ. In vitro impact of human milk oligosaccharides on Enterobacteriaceae growth. J Agric Food Chem. (2015) 63(12):3295–302. doi: 10.1021/jf505721p
105. Nilsen M, Madelen Saunders C, Leena Angell I, Arntzen MØ, Lødrup Carlsen KC, Carlsen KH, et al. Butyrate levels in the transition from an infant to an adult-like gut microbiota correlate with bacterial networks associated with Eubacterium rectale and Ruminococcus gnavus. Genes (Basel). (2020) 11(11):1245. doi: 10.3390/genes11111245
106. Kostopoulos I, Elzinga J, Ottman N, Klievink JT, Blijenberg B, Aalvink S, et al. Akkermansia muciniphila uses human milk oligosaccharides to thrive in the early life conditions in vitro. Sci Rep. (2020) 10(1):14330. doi: 10.1038/s41598-020-71113-8
107. Luna E, Parkar SG, Kirmiz N, Hartel S, Hearn E, Hossine M, et al. Utilization efficiency of human milk oligosaccharides by human-associated Akkermansia is strain dependent. McBain AJ, editor. Appl Environ Microbiol. (2022) 88(1):e01487–21. doi: 10.1128/AEM.01487-21
108. Galazzo G, van Best N, Bervoets L, Dapaah IO, Savelkoul PH, Hornef MW, et al. Development of the microbiota and associations with birth mode, diet, and atopic disorders in a longitudinal analysis of stool samples, collected from infancy through early childhood. Gastroenterology. (2020) 158(6):1584–96. doi: 10.1053/j.gastro.2020.01.024
109. LoCascio RG, Desai P, Sela DA, Weimer B, Mills DA. Broad conservation of milk utilization genes in Bifidobacterium longum subsp. infantis as revealed by comparative genomic hybridization. Appl Environ Microbiol. (2010) 76(22):7373–81. doi: 10.1128/AEM.00675-10
110. Sakanaka M, Gotoh A, Yoshida K, Odamaki T, Koguchi H, Xiao JZ, et al. Varied pathways of infant gut-associated Bifidobacterium to assimilate human milk oligosaccharides: prevalence of the gene set and its correlation with bifidobacteria-rich microbiota formation. Nutrients. (2019) 12(1):71. doi: 10.3390/nu12010071
111. Katayama T. Host-derived glycans serve as selected nutrients for the gut microbe: human milk oligosaccharides and bifidobacteria. Biosci, Biotechnol, Biochem. (2016) 80(4):621–32. doi: 10.1080/09168451.2015.1132153
112. Thomson P, Medina DA, Garrido D. Human milk oligosaccharides and infant gut bifidobacteria: molecular strategies for their utilization. Food Microbiol. (2018) 75:37–46. doi: 10.1016/j.fm.2017.09.001
113. Sela DA, Chapman J, Adeuya A, Kim JH, Chen F, Whitehead TR, et al. The genome sequence of Bifidobacterium longum subsp. infantis reveals adaptations for milk utilization within the infant microbiome. Proc Natl Acad Sci USA. (2008) 105(48):18964–9. doi: 10.1073/pnas.0809584105
114. Zuurveld M, van Witzenburg NP, Garssen J, Folkerts G, Stahl B, van’t Land B, et al. Immunomodulation by human milk oligosaccharides: the potential role in prevention of allergic diseases. Front Immunol. (2020) 11:801. doi: 10.3389/fimmu.2020.00801
115. Cohan FM. Bacterial species and speciation. Syst Biol. (2001) 50(4):513–24. doi: 10.1080/10635150118398
116. Acinas SG, Klepac-Ceraj V, Hunt DE, Pharino C, Ceraj I, Distel DL, et al. Fine-scale phylogenetic architecture of a complex bacterial community. Nature. (2004) 430(6999):551–4. doi: 10.1038/nature02649
117. Thompson JR, Pacocha S, Pharino C, Klepac-Ceraj V, Hunt DE, Benoit J, et al. Genotypic diversity within a natural coastal bacterioplankton population. Science. (2005) 307(5713):1311–3. doi: 10.1126/science.1106028
118. Holscher HD, Davis SR, Tappenden KA. Human milk oligosaccharides influence maturation of human intestinal caco-2Bbe and HT-29 cell lines. J Nutr. (2014) 144(5):586–91. doi: 10.3945/jn.113.189704
119. Holscher HD, Bode L, Tappenden KA. Human milk oligosaccharides influence intestinal epithelial cell maturation in vitro. J Pediatr Gastroenterol Nutr. (2017) 64(2):296–301. doi: 10.1097/MPG.0000000000001274
120. Ruhaak LR, Stroble C, Underwood MA, Lebrilla CB. Detection of milk oligosaccharides in plasma of infants. Anal Bioanal Chem. (2014) 406(24):5775–84. doi: 10.1007/s00216-014-8025-z
121. Akay HK, Bahar Tokman H, Hatipoglu N, Hatipoglu H, Siraneci R, Demirci M, et al. The relationship between bifidobacteria and allergic asthma and/or allergic dermatitis: a prospective study of 0-3 years-old children in Turkey. Anaerobe. (2014) 28:98–103. doi: 10.1016/j.anaerobe.2014.05.006
122. Kalliomäki M, Kirjavainen P, Eerola E, Kero P, Salminen S, Isolauri E. Distinct patterns of neonatal gut microflora in infants in whom atopy was and was not developing. J Allergy Clin Immunol. (2001) 107(1):129–34. doi: 10.1067/mai.2001.111237
123. Stewart CJ. Breastfeeding promotes bifidobacterial immunomodulatory metabolites. Nat Microbiol. (2021) 6(11):1335–6. doi: 10.1038/s41564-021-00975-z
124. Cait A, Cardenas E, Dimitriu PA, Amenyogbe N, Dai D, Cait J, et al. Reduced genetic potential for butyrate fermentation in the gut microbiome of infants who develop allergic sensitization. J Allergy Clin Immunol. (2019) 144(6):1638–1647.e3. doi: 10.1016/j.jaci.2019.06.029
125. Laursen MF, Sakanaka M, von Burg N, Mörbe U, Andersen D, Moll JM, et al. Bifidobacterium species associated with breastfeeding produce aromatic lactic acids in the infant gut. Nat Microbiol. (2021) 6(11):1367–82. doi: 10.1038/s41564-021-00970-4
126. Laursen MF, Pekmez CT, Larsson MW, Lind MV, Yonemitsu C, Larnkjær A, et al. Maternal milk microbiota and oligosaccharides contribute to the infant gut microbiota assembly. ISME COMMUN. (2021) 1(1):21. doi: 10.1038/s43705-021-00021-3
127. El-Heis S, Crozier SR, Robinson SM, Harvey NC, Cooper C, Inskip HM, et al. Higher maternal serum concentrations of nicotinamide and related metabolites in late pregnancy are associated with a lower risk of offspring atopic eczema at age 12 months. Clin Exp Allergy. (2016) 46(10):1337–43. doi: 10.1111/cea.12782
128. Lau HX, El-Heis S, Yap QV, Chan YH, Tan CPT, Karnani N, et al. Role of maternal tryptophan metabolism in allergic diseases in the offspring. Clin Exp Allergy. (2021) 51(10):1346–60. doi: 10.1111/cea.13953
129. Abrahamsson TR, Jakobsson HE, Andersson AF, Björkstén B, Engstrand L, Jenmalm MC. Low diversity of the gut microbiota in infants with atopic eczema. J Allergy Clin Immunol. (2012) 129(2):434–440.e2. doi: 10.1016/j.jaci.2011.10.025
130. Kang MJ, Lee SY, Park YM, Kim BS, Lee MJ, Kim JH, et al. Interactions between IL-17 variants and Streptococcus in the gut contribute to the development of atopic dermatitis in infancy. Allergy Asthma Immunol Res. (2020) 13(3):404–19. doi: 10.4168/aair.2021.13.3.404
131. Zimmermann J, Macpherson AJ. Breast milk modulates transgenerational immune inheritance. Cell. (2020) 181(6):1202–4. doi: 10.1016/j.cell.2020.05.030
132. Chan CWH, Leung TF, Choi KC, Tsui SKW, Wong CL, Chow KM, et al. Association of early-life gut microbiome and lifestyle factors in the development of eczema in Hong Kong infants. Exp Dermatol. (2021) 30(6):859–64. doi: 10.1111/exd.14280
133. Gore C, Custovic A, Tannock GW, Munro K, Kerry G, Johnson K, et al. Treatment and secondary prevention effects of the probiotics Lactobacillus paracasei or Bifidobacterium lactis on early infant eczema: randomized controlled trial with follow-up until age 3 years. Clin Exp Allergy. (2012) 42(1):112–22. doi: 10.1111/j.1365-2222.2011.03885.x
134. Kalliomäki M, Salminen S, Poussa T, Isolauri E. Probiotics during the first 7 years of life: a cumulative risk reduction of eczema in a randomized, placebo-controlled trial. J Allergy Clin Immunol. (2007) 119(4):1019–21. doi: 10.1016/j.jaci.2006.12.608
135. Wickens K, Stanley TV, Mitchell EA, Barthow C, Fitzharris P, Purdie G, et al. Early supplementation with Lactobacillus rhamnosus HN001 reduces eczema prevalence to 6 years: does it also reduce atopic sensitization? Clin Exp Allergy. (2013) 43(9):1048–57. doi: 10.1111/cea.12154
136. Boyle RJ, Ismail IH, Kivivuori S, Licciardi PV, Robins-Browne RM, Mah LJ, et al. Lactobacillus GG treatment during pregnancy for the prevention of eczema: a randomized controlled trial. Allergy. (2011) 66(4):509–16. doi: 10.1111/j.1398-9995.2010.02507.x
137. Wopereis H, Sim K, Shaw A, Warner JO, Knol J, Kroll JS. Intestinal microbiota in infants at high risk for allergy: effects of prebiotics and role in eczema development. J Allergy Clin Immunol. (2018) 141(4):1334–1342.e5. doi: 10.1016/j.jaci.2017.05.054
138. Nylund L, Nermes M, Isolauri E, Salminen S, de Vos WM, Satokari R. Severity of atopic disease inversely correlates with intestinal microbiota diversity and butyrate-producing bacteria. Allergy. (2015) 70(2):241–4. doi: 10.1111/all.12549
139. Gio-Batta M, Sjöberg F, Jonsson K, Barman M, Lundell AC, Adlerberth I, et al. Fecal short chain fatty acids in children living on farms and a link between valeric acid and protection from eczema. Sci Rep. (2020) 10(1):22449. doi: 10.1038/s41598-020-79737-6
140. Gio-Batta M, Spetz K, Barman M, Bråbäck L, Norin E, Björkstén B, et al. Low concentration of fecal valeric acid at 1 year of age is linked with eczema and food allergy at 13 years of age: findings from a Swedish birth cohort. IAA. (2022) 183(4):398–408. doi: 10.1159/000520149
141. Chow J, Panasevich MR, Alexander D, Vester Boler BM, Rossoni Serao MC, Faber TA, et al. Fecal metabolomics of healthy breast-fed versus formula-fed infants before and during in vitro batch culture fermentation. J Proteome Res. (2014) 13(5):2534–42. doi: 10.1021/pr500011w
142. Martin FPJ, Moco S, Montoliu I, Collino S, Da Silva L, Rezzi S, et al. Impact of breast-feeding and high- and low-protein formula on the metabolism and growth of infants from overweight and obese mothers. Pediatr Res. (2014) 75(4):535–43. doi: 10.1038/pr.2013.250
143. Dotz V, Rudloff S, Meyer C, Lochnit G, Kunz C. Metabolic fate of neutral human milk oligosaccharides in exclusively breast-fed infants. Mol Nutr Food Res. (2015) 59(2):355–64. doi: 10.1002/mnfr.201400160
144. Kim HK, Rutten NBMM, Besseling-van der Vaart I, Niers LEM, Choi YH, Rijkers GT, et al. Probiotic supplementation influences faecal short chain fatty acids in infants at high risk for eczema. Benefic Microbes. (2015) 6(6):783–90. doi: 10.3920/BM2015.0056
145. Martin R, Makino H, Cetinyurek Yavuz A, Ben-Amor K, Roelofs M, Ishikawa E, et al. Early-life events, including mode of delivery and type of feeding, siblings and gender, shape the developing gut microbiota. PLoS One. (2016) 11(6):e0158498. doi: 10.1371/journal.pone.0158498
146. Bazanella M, Maier TV, Clavel T, Lagkouvardos I, Lucio M, Maldonado-Gòmez MX, et al. Randomized controlled trial on the impact of early-life intervention with bifidobacteria on the healthy infant fecal microbiota and metabolome. Am J Clin Nutr. (2017) 106(5):1274–86. doi: 10.3945/ajcn.117.157529
147. Bridgman SL, Azad MB, Field CJ, Haqq AM, Becker AB, Mandhane PJ, et al. Fecal short-chain fatty acid variations by breastfeeding status in infants at 4 months: differences in relative versus absolute concentrations. Front Nutr. (2017) 4:11. doi: 10.3389/fnut.2017.00011
148. Roduit C, Frei R, Ferstl R, Loeliger S, Westermann P, Rhyner C, et al. High levels of butyrate and propionate in early life are associated with protection against atopy. Allergy. (2019) 74(4):799–809. doi: 10.1111/all.13660
149. Brink LR, Mercer KE, Piccolo BD, Chintapalli SV, Elolimy A, Bowlin AK, et al. Neonatal diet alters fecal microbiota and metabolome profiles at different ages in infants fed breast milk or formula. Am J Clin Nutr. (2020) 111(6):1190–202. doi: 10.1093/ajcn/nqaa076
150. Muthana SM, Campbell CT, Gildersleeve JC. Modifications of glycans: biological significance and therapeutic opportunities. ACS Chem Biol. (2012) 7(1):31–43. doi: 10.1021/cb2004466
151. Davis JCC, Totten SM, Huang JO, Nagshbandi S, Kirmiz N, Garrido DA, et al. Identification of oligosaccharides in feces of breast-fed infants and their correlation with the gut microbial community. Mol Cell Proteomics. (2016) 15(9):2987–3002. doi: 10.1074/mcp.M116.060665
152. Seppo AE, Autran CA, Bode L, Järvinen KM. Human milk oligosaccharides and development of cow’s milk allergy in infants. J Allergy Clin Immunol. (2017) 139(2):708–711.e5. doi: 10.1016/j.jaci.2016.08.031
153. Sprenger N, Odenwald H, Kukkonen AK, Kuitunen M, Savilahti E, Kunz C. FUT2-dependent Breast milk oligosaccharides and allergy at 2 and 5 years of age in infants with high hereditary allergy risk. Eur J Nutr. (2017) 56(3):1293–301. doi: 10.1007/s00394-016-1180-6
154. Rousseaux A, Brosseau C, Le Gall S, Piloquet H, Barbarot S, Bodinier M. Human milk oligosaccharides: their effects on the host and their potential as therapeutic agents. Front Immunol. (2021) 12:680911. doi: 10.3389/fimmu.2021.680911
155. Singh RP, Niharika J, Kondepudi KK, Bishnoi M, Tingirikari JMR. Recent understanding of human milk oligosaccharides in establishing infant gut microbiome and roles in immune system. Food Res Int. (2022) 151:110884. doi: 10.1016/j.foodres.2021.110884
156. Zhang W, He-Yang J, Tu W, Zhou X. Sialylated human milk oligosaccharides prevent intestinal inflammation by inhibiting toll like receptor 4/NLRP3 inflammasome pathway in necrotizing enterocolitis rats. Nutr Metab (Lond). (2021) 18(1):5. doi: 10.1186/s12986-020-00534-z
157. Luu M, Monning H, Visekruna A. Exploring the molecular mechanisms underlying the protective effects of microbial SCFAs on intestinal tolerance and food allergy. Front Immunol. (2020) 11:1225. doi: 10.3389/fimmu.2020.01225
158. Tan J, McKenzie C, Vuillermin PJ, Goverse G, Vinuesa CG, Mebius RE, et al. Dietary fiber and bacterial SCFA enhance oral tolerance and protect against food allergy through diverse cellular pathways. Cell Rep. (2016) 15(12):2809–24. doi: 10.1016/j.celrep.2016.05.047
159. Paparo L, Nocerino R, Ciaglia E, Di Scala C, De Caro C, Russo R, et al. Butyrate as a bioactive human milk protective component against food allergy. Allergy. (2021) 76(5):1398–415. doi: 10.1111/all.14625
160. Triantis V, Bode L, van Neerven RJJ. Immunological effects of human milk oligosaccharides. Front Pediatr. (2018) 6:190. doi: 10.3389/fped.2018.00190
161. Dimitroglou M, Iliodromiti Z, Christou E, Volaki P, Petropoulou C, Sokou R, et al. Human breast milk: the key role in the maturation of immune, gastrointestinal and central nervous systems: a narrative review. Diagnostics. (2022) 12(9):2208. doi: 10.3390/diagnostics12092208
162. Jones J, Reinke SN, Ali A, Palmer DJ, Christophersen CT. Fecal sample collection methods and time of day impact microbiome composition and short chain fatty acid concentrations. Sci Rep. (2021) 11(1):13964. doi: 10.1038/s41598-021-93031-z
Keywords: atopic dermatitis, eczema, human milk oligosaccharides (HMO), infant gut microbiome, bifidobacterium infantis, metagenome, metabolome, HMO metabolizing genes
Citation: Rahman T, Sarwar PF, Potter C, Comstock SS and Klepac-Ceraj V (2023) Role of human milk oligosaccharide metabolizing bacteria in the development of atopic dermatitis/eczema. Front. Pediatr. 11:1090048. doi: 10.3389/fped.2023.1090048
Received: 4 November 2022; Accepted: 23 February 2023;
Published: 20 March 2023.
Edited by:
Tim S. Nawrot, University of Hasselt, BelgiumReviewed by:
Thessa Van Pee, University of Hasselt, Belgium© 2023 Rahman, Sarwar, Potter, Comstock and Klepac-Ceraj. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Vanja Klepac-Ceraj dmtsZXBhY2NAd2VsbGVzbGV5LmVkdQ== Sarah S. Comstock Y29tc3RvMzdAbXN1LmVkdQ==
†These authors have contributed equally to this work and share first authorship
‡These authors have contributed equally to this work and share senior authorship
§ORCID Trisha Rahman orcid.org/0000-0002-9456-4063 Prioty F. Sarwar orcid.org/0000-0002-1852-6401 Cassie Potter orcid.org/0000-0002-5966-9855 Sarah S. Comstock orcid.org/0000-0003-3107-7652 Vanja Klepac-Ceraj orcid.org/0000-0001-5387-5706
Specialty Section: This article was submitted to Children and Health, a section of the journal Frontiers in Pediatrics
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.