- 1Department of Laboratory Medicine, Hanyang University Guri Hospital, Hanyang University College of Medicine, Guri, South Korea
- 2GC Genome, Yongin, South Korea
- 3Department of Laboratory Medicine, Kangbuk Samsung Hospital, Sungkyunkwan University School of Medicine, Seoul, South Korea
Objectives: Hemophagocytic lymphohistiocytosis (HLH) is a clinical syndrome characterized by a life-threatening condition caused by severe hypercytokinemia. The hereditary forms of HLH, also called familial HLH (fHLH), have 4 different genes (PRF1, UNC13D, STX11, and STXBP2) and have been identified as being causative for fHLH. This study aimed to analyze the carrier frequency and expected incidence of fHLH in East Asians and Koreans using exome data from the Genome Aggregation Database (gnomAD).
Methods: We analyzed 9,197 exomes for East Asian populations from gnomAD with 1,909 Korean for four fHLH genes. All identified variants were classified according to 2015 American College of Medical Genetics and Genomics and the Association for Molecular Pathology guideline.
Results: 19 pathogenic variant/likely pathogenic variants (PV/LPVs) were identified in 30 East Asian individuals (0.33%). Among them, 7 PV/LPVs were identified in 17 Korean individuals (0.63%). The estimated incidence of fHLH was 1 in 1,105,652 for East Asians and l in 235,128 for Koreans.
Conclusions: This study is the first to identify carrier frequencies in East Asian and Korean populations for fHLH using gnomAD. It was confirmed that the carrier frequency of fHLH patients was high in Koreans among East Asians and the incidence was also predicted to be higher than that of other East Asians. The variant spectrum of fHLH genes in East Asian and Korean populations differed greatly from those of other ethnic groups.
Introduction
Hemophagocytic lymphohistiocytosis (HLH) is a clinical syndrome characterized by a life-threatening condition caused by severe hypercytokinemia due to an uncontrolled immune response (1, 2). HLH may occur as a hereditary or acquired condition. The hereditary form of HLH, also called familial HLH (fHLH), was first described in 1952 (3). Since the PRF1 gene was first discovered as a causative gene of fHLH in 1999 (4), 4 different genes (PRF1, UNC13D, STX11, and STXBP2) to date have been identified as being causative for fHLH (5). These genes are inherited as autosomal recessive and involved in lymphocyte granule-dependent cytotoxicity (6). In addition, genetic defects for pigmentary disorders, X-linked lymphoproliferative syndromes, and EBV susceptibility disorders are associated with HLH (7).
Although the incidence of HLH has been known to be 0.34–1.8 per 100,000 births, few studies exist with regard to the incidence of HLH (8, 9). Also, carrier frequencies for the four fHLH major genes (PRF1, UNC13D, STX11, and STXBP2) are unknown to our knowledge except for PRF1 (10). According to a recent study, there is a study result that the prognosis can be improved when preemptive allogeneic hematopoietic stem cell transplantation is performed in asymptomatic pediatric patients with pathogenic billeleic variants (11). Therefore, it would be important to identify carrier frequencies and use them to determine the expected incidence.
The Genome Aggregation Database (gnomAD) is a widely used genomic database around the world. The gnomAD V2 is consists of 125,748 exomes and 4,359 genomes (12). The gnomAD V2 contains exome data from 9,197 East Asians, including 1,909 Koreans. Among the open genome databases, it contains the most data on East Asians and Koreans, making it suitable for research on East Asians and Koreans. The four genes (PRF1, UNC13D, STX11, and STXBP2) variants interpreted according to the 2015 American College of Medical Genetics and Genomics and the Association for Molecular Pathology guidelines (2015 ACMG/AMP guidelines), which are currently generally adopted in clinical practice (13). This study aimed to analyze the carrier frequency and expected incidence of fHLH in East Asians and Koreans using exome data from the gnomAD through 2015 ACMG/AMP guidelines.
Materials and methods
GnomAD East Asian population data
The gnomAD data (v2.1.1) for the PRF1, UNC13D, STX11, and STXBP2 genes were obtained from gnomAD (https://gnomad.broadinstitute.org/). We analyzed 9,197 East Asian exomes of which 1,909 were from Koreans, 76 were from Japanese, and 7,212 were from other East Asian populations. The filtered variants that were flagged in gnomAD as failing “InbreedingCoeff,” “AC0,” or “RF” QC filters were excluded from the analysis.
Variant classification and statistical analysis
All variants were interpreted using 2015 ACMG/AMP guidelines and Sequence Variant Interpretation (SVI) general recommendations for ACMG/AMP Criteria by ClinGen (https://clinicalgenome.org/working-groups/sequence-variant-interpretation/, accessed on 1 November, 2021). The 2015 ACMG/AMP guidelines recommend the classification of variants into five categories: pathogenic variants (PV), likely pathogenic variants (LPV), variants of uncertain significance, likely benign variants, and benign variants. REVEL (14), CADD (15), Polyphen-2 (16), SIFT (17) and SpliceAI (18) were used to predict variant pathogenicity. Variants identified from 9,197 East Asian exomes in gnomAD were compared with previously classified disease-causing variants from the Human Gene Mutation Database (HGMD) and ClinVar, which are representative disease databases, to identify overlap. The HGMD professional database (http://www.hgmd.org/, release 2018.03) is a comprehensive collection of germline variants categorized into six categories, of which we analyzed only disease-causing mutations (DM). ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/, accessed on 16 October 2021) is a freely available archive that provides the classification of variants interpreted by clinical laboratories.
fHLH carrier frequency and incidence estimation
East Asian and Korean carrier frequencies were calculated for fHLH genes (PRF1, UNC13D, STX11, and STXBP2) using gnomAD. We used those classified as PV and LPV according to the 2015 ACMG/AMP guidelines, the disease-causing variant (DM) in HGMD and those classified as PV and LPV in ClinVar for carrier frequency analysis. Thereafter, we estimated the incidence of fHLH based on frequency and the Hardy–Weinberg equilibrium principle (1 = p2 + 2pq + q2). The detailed prediction method has been described in a previous study (19). We then summed the estimated carrier frequencies and incidence across all genes. MedCalc ver. 11.5.1.0 (MedCalc Software, Maiakerke, Belgium) was used for statistical analysis and 95% confidence intervals (CIs) were calculated for each value.
Results
9,197 East Asian exomes including 1,909 Korean exomes were analyzed for PRF1, UNC13D, STX11, and STXBP2 gene variants. These variants were classified according to 2015 ACMG/AMP guidelines and two disease classification databases, HGMD and ClinVar (Table 1). According to the 2015 ACMG/AMP guidelines, 19 PV/LPVs were identified in 30 East Asian individuals (30/9,191 = 0.33%). Among them, 7 PV/LPVs were identified in 17 Korean individuals (12/1,909 = 0.63%). The estimated incidence of fHLH was 1 in 1,105,652 in East Asians and l in 235,128 in Koreans. Based on HGMD, the carrier frequency was 6.61% in East Asians and 4.61% in Koreans. Estimated incidence values were 1 in 1,129 for East Asians and l in 2,856 for Koreans. Based on ClinVar, the carrier frequency was 0.27% in East Asians and 0.73% in Koreans. Estimated incidences were 1 in 1,138,896 for East Asians and l in 137,502 for Koreans.
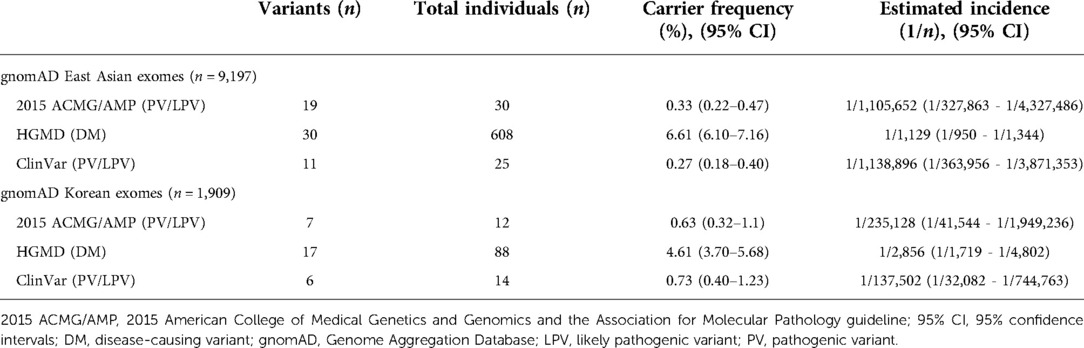
Table 1. Carrier frequency and estimated incidence of familial hemophagocytic lymphohistiocytosis syndrome in east Asian and Korean populations.
PV/LPVs found in East Asians and Koreans in the four fHLH genes are summarized in Table 2 and Supplementary Table S1. A total of 30 alleles were identified in East Asians, among them 14 alleles (14/30, 46.7%) were the PRF1 gene, 9 alleles (9/30, 30.0%) were the UNC13D gene, 2 alleles (2/30, 6.7%) were the STX11 gene, and 5 alleles (5/30, 16.7%) were the STXBP2 gene. In Koreans, 6 alleles (6/12, 50.0%) were the PFR1 gene, 5 alleles (5/12, 41.7%) were the UNC13D gene, and 1 allele (1/12, 8.3%) was the STXBP2 gene. Variants classified as DM in HGMD and variants classified as PV/LPV in ClinVar are summarized in Supplementary Tables S2 and S3.
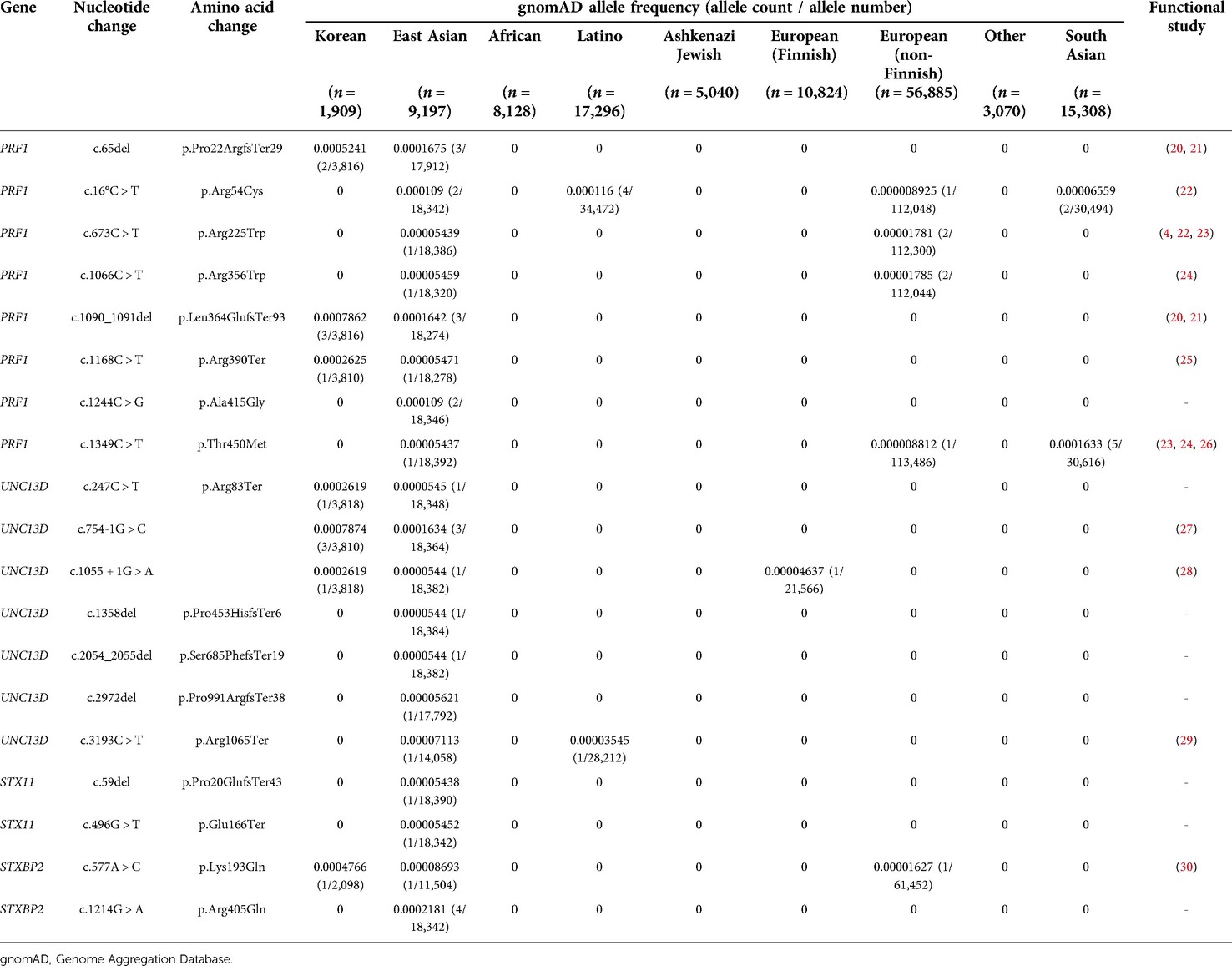
Table 2. Pathogenic variants and likely pathogenic variants of East Asian and Korean populations in gnomAD.
Carrier frequency for each fHLH gene was PRF1 0.15%, UNC13D 0.10%, STXBP2 0.05%, and STX11 0.02% in East Asians, while it was PRF1 0.31%, UNC13D 0.26%, STXBP2 0.05%, and STX11 0% in Koreans (Supplementary Tables S4–7). In East Asians, the estimated incidence was 1/1,726,221 for PRF1, 1/ 4,176,857 for UNC13D, 1/84,555,423 for STX11, and 1/13,531,356 for STXBP2. In Koreans, the estimated incidence was 1/404,920 for PRF1, 1/583,162 for UNC13D, 1/3,641,979 for STX11, and 1/14,579,043 for STXBP2.
STXBP2 c.1214G > A was the most common variant in East Asians but was not found in other ethnicities including Koreans and Japanese. PRF1 c.1090_1091del and UNC13D c.754-1G > A variants were the most common in Koreans. These variants were not identified in other ethnicities and were the only variants found in Koreans. In addition, variants other than PRF1 c.1168C > T, UNC13D c.1055 + 1G > A, and UNC13D c.247C > T were found only in Koreans.
Discussion
In this study, the carrier frequency and estimated incidence of fHLH was analyzed for East Asians and Koreans using gnomAD. The carrier frequency of East Asians was 0.33% and the carrier frequency of Koreans was 0.63%. However, it was difficult to compare because carrier frequency studies of fHLH have not been actively performed; with regard to in practice resources for carrier screening published in ACMG, the maximal carrier frequency of PRF1 was described as 0.67% (10). In this study, the PRF1 carrier frequency was 0.15% in East Asians and 0.31% in Koreans. The carrier frequency of PRF1 was lower than in a prior study. Within East Asians, it was found that the carrier frequency of fHLH was higher in Koreans than in other East Asians.
Through this study, we were able to estimate the approximate incidence of fHLH. When calculated by the Hardy-Weinberg equation, the incidence of fHLH in East Asians was predicted to be 1/1,105,652 (0.09 per 100,000 births) and 1/235,128 (0.43 per 100,000 births) in Koreans. This was lower than the incidence reported in Sweden (1.8 per 100,000 births) and higher than that in Japan (0.34 per 100,000 births), one of the East Asian populations (8, 9). However, since this study was analyzed with exome data registered in gnomAD, the UNC13D c.118–308C > T variant, known as the Korean founder variant, was not included in this study because it is located in the deep intron (31). According to the Korean Reference Genome Database (KRGDB, http://152.99.75.168:9090/KRGDB/menuPages/introKor.jsp), the UNC13D c.118–308C > T variant was identified as a minor allele frequency of 0.001364 in Koreans. Therefore, the actual incidence in East Asians, especially Koreans, was estimated to be higher.
In this study, the fHLH gene carrier frequencies were PRF1 (0.15%), UNC13D (0.10%), STXBP2 (0.05%), and STX11 (0.02%) in the order of East Asians and PRF1 (0.31%), UNC13D (0.26%), STXBP2 (0.05%), and STX11 (0%) in Koreans. The distribution of genes in fHLH patients was also estimated to be similar to that of carriers. According to a large-scale study of 1,892 HLH patients, variants were found in the order of PRF1, STXBP2, UNC13D, and STX11 among 197 genetically confirmed patients (32). Among East Asians, UNC13D, PRF1, and STXBP2 were found in Chinese and PRF1, UNC13D, and STXBP2 were found in Japanese in that order (33, 34). In Koreans, UNC13D was found more frequently than PRF1 (31). In this study, PRF1 was found to be slightly more frequent than UNC13D; if UNC13D c.118–308C > T, which was not included in this study, was included, it could be estimated that UNC13D was similar to PRF1.
The PV/LPVs identified in this study were found to have a significantly different variant spectrum from other ethnicities. In gnomAD, c.1090_1091del in the PRF1 gene and c.754–1G > A in the UNC13D gene was mainly observed in Koreans; however, this variant was not found in other ethnicities. In addition, variants found in Koreans were only found in Koreans, except for PRF1 c.65del, UNC13D c.1055 + 1G > A, and STXBP2 c.1214G > A. All variants identified in East Asians were not found in Ashkenazi Jews or Africans and some PRF1 variants were found in Europeans (non-Finnish). In the largest study of the genetic analysis of HLH patients, c.50del and c.445G > A in the PRF1 gene and c.143 °C > T and c.1247-1G > C in the STXBP2 gene, known to be frequently reported in HLH patients, were not found in East Asians in gnomAD (32). Even among East Asians, Koreans had a different variant spectrum (Supplementary Figure S1). Among the 19 variant fHLH genes found in Koreans, all variants except for the c.65del variant of the PRF1 gene were unique to Koreans. From this, it could be inferred that the variant spectrum of fHLH genes differed between East Asians and other ethnicities, and that Koreans are unique even within East Asians.
When the variants found in Korean fHLH patients were compared with those of Korean carriers identified in gnomAD, it was confirmed that the PRF1 and UNC13D variants found in this study were all major variants in Korean HLH patients (31, 35). PRF1 c.1090_1091del was the variant with the highest carrier frequency in Koreans in gnomAD and was found most frequently in HLH patients where the PRF1 variant was confirmed. The c.118-308C > T variant in UNC13D could not be confirmed in this study; however, the c.754-1G > A, c.1055 + 1G > A, and c.247C > T variants found in UNC13D were all found in Korean fHLH patients. The c.754-1G > A variant, which was the most frequent in UNC13D was also found in patients. Through the results of this study, it was confirmed that predicting the carrier in gnomAD was helpful toward predicting the genetic spectrum in actual patients.
According to data from the Korean Statistical Information Service (http://kosis.kr/; accessed on 26 January 2022) in 2022, the total population of Korea was 51.8 million with 272,337 births. Based on the carrier frequency in this study, the number of carriers is estimated to be 0.3 million with 1,716 in newborns per year. The estimated incidence of fHLH in Korea based on the Hardy–Weinberg equilibrium is 2.7 cases per year.
With the recent development of sequencing technology, studies applying genomic sequencing to healthy or ill newborns have been reported (36, 37). These studies classified categories according to clinical actionability and the age of onset and evaluated whether genomic sequencing would be useful for diseases for which early diagnosis would be beneficial. In one of the representative studies of the BabySeq Project, all four fHLH genes were classified as category A (genes included in the newborn genomic sequencing report with definitive or strong evidence to cause a highly penetrant childhood-onset disorder) (38). In another representative study, the North Carolina Newborn Exome Sequencing for Universal Screening, only UNC13D was classified as category 1 (pediatric onset conditions with high actionability); the rest of the genes were not included in other categories (39). In order to confirm the cost-effectiveness of genomic newborn screening, it would be important to check the carrier frequency and incidence of each disease and gene. In that respect, this study predicting the carrier frequency and incidence of fHLH is considered to be a significant study.
This study has some limitations. We did not analyze structural variations including the large deletion/insertion of the fHLH gene. We also couldn't include the deep intron variant. The c.118-308C > T variant of UNC13D is known to be common in Korean fHLH patients but was not included in this study, it is thus expected that the carrier frequency in Koreans may be underestimated. We identified only four fHLH-related genes. However, in addition to fHLH, there are other HLH predisposition genes such as granule/pigment abnormality genes and X-linked lymphoproliferative disease genes, so further analysis is needed. Nonetheless, this study makes several valuable contributions. This is the largest study among those performed in East Asia with regard to the analysis of four fHLH genes. To the best of our knowledge, there have been no large-scale population studies of carrier frequencies and the estimated fHLH incidence in Koreans. We believe that this study more accurately predicted the carrier frequency of fHLH in East Asians and Koreans. In addition, this study was a meaningful study that could confirm that public data analysis is helpful toward predicting the variant spectrum of actual patients.
Conclusions
This study is the first to identify carrier frequencies in East Asians and Koreans for fHLH using gnomAD. We confirmed that the carrier frequency of fHLH patients was high in Koreans among East Asians and the incidence was also predicted to be higher than in other East Asian populations. The variant spectrum of fHLH genes in East Asian and Korean populations differed greatly from those of other ethnic groups. Our data are expected to serve as a reference for further investigations of fHLH in East Asian and Korean populations.
Data availability statement
The original contributions presented in the study are included in the article/supplementary materials, further inquiries can be directed to the corresponding authors.
Author contributions
JEP participated in analysis and interpretation of the data and the drafting of the manuscript. EHC participated in analysis and interpretation of the data. TL and KH participated in acquisition and analysis of data. CSK and JEP participated in the study concept and design, the drafting of the manuscript and for important intellectual content. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (NRF-2021R1I1A1A01049183).
Acknowledgment
This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (NRF-2021R1I1A1A01049183). The authors are grateful to those responsible for creating and maintaining gnomAD, ClinVar, and HGMD database.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fped.2022.975665/full#supplementary-material.
References
1. Janka GE, Lehmberg K. Hemophagocytic syndromes–an update. Blood Rev. (2014) 28(4):135–42. doi: 10.1016/j.blre.2014.03.002
2. Campo M, Berliner N. Hemophagocytic lymphohistiocytosis in adults. Hematol Oncol Clin North Am. (2015) 29(5):915–25. doi: 10.1016/j.hoc.2015.06.009
3. Farquhar JW, Claireaux AE. Familial haemophagocytic reticulosis. Arch Dis Child. (1952) 27(136):519–25. doi: 10.1136/adc.27.136.519
4. Stepp SE, Dufourcq-Lagelouse R, Le Deist F, Bhawan S, Certain S, Mathew PA, et al. Perforin gene defects in familial hemophagocytic lymphohistiocytosis. Science. (1999) 286(5446):1957–9. doi: 10.1126/science.286.5446.1957
5. Bhatt NS, Oshrine B, An Talano J. Hemophagocytic lymphohistiocytosis in adults. Leuk Lymphoma. (2019) 60(1):19–28. doi: 10.1080/10428194.2018.1482543
6. Canna SW, Marsh RA. Pediatric hemophagocytic lymphohistiocytosis. Blood. (2020) 135(16):1332–43. doi: 10.1182/blood.2019000936
7. Degar B. Familial hemophagocytic lymphohistiocytosis. Hematol Oncol Clin North Am. (2015) 29(5):903–13. doi: 10.1016/j.hoc.2015.06.008
8. Meeths M, Horne A, Sabel M, Bryceson YT, Henter JI. Incidence and clinical presentation of primary hemophagocytic lymphohistiocytosis in Sweden. Pediatr Blood Cancer. (2015) 62(2):346–52. doi: 10.1002/pbc.25308
9. Ishii E, Ohga S, Tanimura M, Imashuku S, Sako M, Mizutani S, et al. Clinical and epidemiologic studies of familial hemophagocytic lymphohistiocytosis in Japan. Japan lch study group. Med Pediatr Oncol. (1998) 30(5):276–83. doi: 10.1002/(sici)1096-911x(199805)30:5%3C276::aid-mpo3%3E3.0.co;2-c
10. Gregg AR, Aarabi M, Klugman S, Leach NT, Bashford MT, Goldwaser T, et al. Screening for autosomal recessive and X-linked conditions during pregnancy and preconception: a practice resource of the American college of medical genetics and genomics (acmg). Genet Med. (2021) 23(10):1793–806. doi: 10.1038/s41436-021-01203-z
11. Lucchini G, Marsh R, Gilmour K, Worth A, Nademi Z, Rao A, et al. Treatment dilemmas in asymptomatic children with primary hemophagocytic lymphohistiocytosis. Blood. (2018) 132(19):2088–96. doi: 10.1182/blood-2018-01-827485
12. Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alföldi J, Wang Q, et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. (2020) 581(7809):434–43. doi: 10.1038/s41586-020-2308-7
13. Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American college of medical genetics and genomics and the association for molecular pathology. Genet Med. (2015) 17(5):405–24. doi: 10.1038/gim.2015.30
14. Ioannidis NM, Rothstein JH, Pejaver V, Middha S, McDonnell SK, Baheti S, et al. Revel: an ensemble method for predicting the pathogenicity of rare missense variants. Am J Hum Genet. (2016) 99(4):877–85.doi: 10.1016/j.ajhg.2016.08.016
15. Kircher M, Witten DM, Jain P, O'Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. (2014) 46(3):310–5. doi: 10.1038/ng.2892
16. Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods. (2010) 7(4):248–9. doi: 10.1038/nmeth0410-248
17. Ng PC, Henikoff S. Predicting deleterious amino acid substitutions. Genome Res. (2001) 11(5):863–74. doi: 10.1101/gr.176601
18. Jaganathan K, Kyriazopoulou Panagiotopoulou S, McRae JF, Darbandi SF, Knowles D, Li YI, et al. Predicting splicing from primary sequence with deep learning. Cell. (2019) 176(3):535–48. e24. doi: 10.1016/j.cell.2018.12.015
19. Park JE, Lee T, Ha K, Ki CS. Carrier frequency and incidence estimation of smith-lemli-opitz syndrome in east Asian populations by genome aggregation database (gnomad) based analysis. Orphanet J Rare Dis. (2021) 16(1):166. doi: 10.1186/s13023-021-01789-2
20. Kwon WK, Choi S, Kim HJ, Huh HJ, Kang JM, Kim YJ, et al. Flow cytometry for the diagnosis of primary immunodeficiency diseases: a single center experience. Allergy Asthma Immunol Res. (2020) 12(2):292–305. doi: 10.4168/aair.2020.12.2.292
21. Kim JY, Shin JH, Sung SI, Kim JK, Jung JM, Ahn SY, et al. A novel Prf1 gene mutation in a fatal neonate case with type 2 familial hemophagocytic lymphohistiocytosis. Korean J Pediatr. (2014) 57(1):50–3. doi: 10.3345/kjp.2014.57.1.50
22. Risma KA, Frayer RW, Filipovich AH, Sumegi J. Aberrant maturation of mutant perforin underlies the clinical diversity of hemophagocytic lymphohistiocytosis. J Clin Invest. (2006) 116(1):182–92. doi: 10.1172/jci26217
23. Trizzino A, zur Stadt U, Ueda I, Risma K, Janka G, Ishii E, et al. Genotype-Phenotype study of familial haemophagocytic lymphohistiocytosis due to perforin mutations. J Med Genet. (2008) 45(1):15–21. doi: 10.1136/jmg.2007.052670
24. Chia J, Yeo KP, Whisstock JC, Dunstone MA, Trapani JA, Voskoboinik I. Temperature sensitivity of human perforin mutants unmasks subtotal loss of cytotoxicity, delayed fhl, and a predisposition to cancer. Proc Natl Acad Sci USA. (2009) 106(24):9809–14. doi: 10.1073/pnas.0903815106
25. Sato H, Kawasaki N, Kawasaki M, Abiko Y, Meguro T, Takahashi N, et al. Three consecutive cases of familial hemophagocytic lymphohistiocytosis, including a case due to maternal uniparental disomy. J Pediatr Hematol Oncol. (2020) 42(8):e819–e21. doi: 10.1097/mph.0000000000001681
26. Ishii E, Ueda I, Shirakawa R, Yamamoto K, Horiuchi H, Ohga S, et al. Genetic subtypes of familial hemophagocytic lymphohistiocytosis: correlations with clinical features and cytotoxic T lymphocyte/natural killer cell functions. Blood. (2005) 105(9):3442–8. doi: 10.1182/blood-2004-08-3296
27. Yamamoto K, Ishii E, Sako M, Ohga S, Furuno K, Suzuki N, et al. Identification of novel Munc13-4 mutations in familial haemophagocytic lymphohistiocytosis and functional analysis of Munc13-4-deficient cytotoxic T lymphocytes. J Med Genet. (2004) 41(10):763–7. doi: 10.1136/jmg.2004.021121
28. Huang JB, Wang J, Jiang L, Wu XJ, Chen C, Xu HG, et al. Familial haemophagocytic lymphohistiocytosis occurs in a Fetus at his third trimester-a case report. Zhongguo Shi Yan Xue Ye Xue Za Zhi. (2017) 25(6):1825–8. doi: 10.7534/j.issn.1009-2137.2017.06.045
29. Sieni E, Cetica V, Santoro A, Beutel K, Mastrodicasa E, Meeths M, et al. Genotype-Phenotype study of familial haemophagocytic lymphohistiocytosis type 3. J Med Genet. (2011) 48(5):343–52. doi: 10.1136/jmg.2010.085456
30. Seo JY, Lee KO, Yoo KH, Sung KW, Koo HH, Kim SH, et al. Prevalence of type 5 familial hemophagocytic lymphohistiocytosis in Korea and novel mutations in Stxbp2. Clin Genet. (2016) 89(2):222–7. doi: 10.1111/cge.12682
31. Seo JY, Song JS, Lee KO, Won HH, Kim JW, Kim SH, et al. Founder effects in two predominant intronic mutations of Unc13d, C.118-308c>T and C.754-1g>C underlie the unusual predominance of type 3 familial hemophagocytic lymphohistiocytosis (Fhl3) in Korea. Ann Hematol. (2013) 92(3):357–64. doi: 10.1007/s00277-012-1628-6
32. Gadoury-Levesque V, Dong L, Su R, Chen J, Zhang K, Risma KA, et al. Frequency and spectrum of disease-causing variants in 1892 patients with suspected genetic hlh disorders. Blood Adv. (2020) 4(12):2578–94. doi: 10.1182/bloodadvances.2020001605
33. Zhang J, Sun Y, Shi X, Zhang R, Wang Y, Xiao J, et al. Genotype characteristics and immunological indicator evaluation of 311 hemophagocytic lymphohistiocytosis cases in China. Orphanet J Rare Dis. (2020) 15(1):112. doi: 10.1186/s13023-020-01390-z
34. Nagai K, Yamamoto K, Fujiwara H, An J, Ochi T, Suemori K, et al. Subtypes of familial hemophagocytic lymphohistiocytosis in Japan based on genetic and functional analyses of cytotoxic T lymphocytes. PLoS One. (2010) 5(11):e14173. doi: 10.1371/journal.pone.0014173
35. Yoon HS, Kim HJ, Yoo KH, Sung KW, Koo HH, Kang HJ, et al. Unc13d is the predominant causative gene with recurrent splicing mutations in Korean patients with familial hemophagocytic lymphohistiocytosis. Haematologica. (2010) 95(4):622–6. doi: 10.3324/haematol.2009.016949
36. Ceyhan-Birsoy O, Murry JB, Machini K, Lebo MS, Yu TW, Fayer S, et al. Interpretation of genomic sequencing results in healthy and ill newborns: results from the babyseq project. Am J Hum Genet. (2019) 104(1):76–93. doi: 10.1016/j.ajhg.2018.11.016
37. Roman TS, Crowley SB, Roche MI, Foreman AKM, O'Daniel JM, Seifert BA, et al. Genomic sequencing for newborn screening: results of the nc nexus project. Am J Hum Genet. (2020) 107(4):596–611. doi: 10.1016/j.ajhg.2020.08.001
38. Ceyhan-Birsoy O, Machini K, Lebo MS, Yu TW, Agrawal PB, Parad RB, et al. A curated gene list for reporting results of newborn genomic sequencing. Genet Med. (2017) 19(7):809–18. doi: 10.1038/gim.2016.193
Keywords: familial hemophagocytic lymphohistiocytosis, gnomAD, carrier frequency, East Asian, Korean
Citation: Park JE, Lee T, Ha K, Cho EH and Ki C (2022) Carrier frequency and incidence estimation of familial hemophagocytic lymphohistiocytosis in East Asian populations by genome aggregation database (gnomAD) based analysis. Front. Pediatr. 10:975665. doi: 10.3389/fped.2022.975665
Received: 22 June 2022; Accepted: 24 October 2022;
Published: 11 November 2022.
Edited by:
Babak Behnam, National Sanitation Foundation International, United StatesReviewed by:
Rui Zhang, Capital Medical University, ChinaHirokazu Kanegane, Tokyo Medical and Dental University, Japan
Franca Fagioli, Regina Margherita Hospital, Italy
© 2022 Park, Lee, Ha, Cho and Ki. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jong Eun Park am9uZ2V1bjgyMEBoYW55YW5nLmFjLmty
Specialty Section: This article was submitted to Genetics of Common and Rare Diseases, a section of the journal Frontiers in Pediatrics
 Jong Eun Park
Jong Eun Park Taeheon Lee
Taeheon Lee Kyeongsu Ha2
Kyeongsu Ha2 Eun Hye Cho
Eun Hye Cho Chang-Seok Ki
Chang-Seok Ki