
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Pediatr., 28 September 2022
Sec. Pediatric Hematology and Hematological Malignancies
Volume 10 - 2022 | https://doi.org/10.3389/fped.2022.911093
This article is part of the Research TopicHot Topics in PediatricsView all 50 articles
Acute monoblastic/monocytic leukemia (AMoL), previously defined as M5 according to FAB classification, is one of the most common subtypes of Acute Myeloid Leukemia (AML) in children, representing ~15–24% of all pediatric AMLs. Currently, the characterization of monocytic-lineage neoplasia at diagnosis includes cytomorphology, cytochemistry, immunophenotyping by multiparametric flow cytometry, cytogenetics, and molecular biology. Moreover, measurable residual disease (MRD) detection is critical in recognizing residual blasts refractory to chemotherapy. Nonetheless, diagnosis and MRD detection may still be challenging in pediatric AMoL since the morphological and immunophenotypic features of leukemic cells potentially overlap with those of normal mature monocytic compartment, as well as differential diagnosis can be troublesome, particularly with Juvenile Myelomonocytic Leukemia and reactive monocytosis in infants and young children. A failure or delay in diagnosis and inaccuracy in MRD assessment may worsen the AMoL prognosis. Therefore, improving diagnosis and monitoring techniques is mandatory to stratify and tailor therapies to the risk profile. This Mini Review aims to provide an updated revision of the scientific evidence on pediatric AMoL diagnostic tools.
Acute monoblastic/monocytic leukemia (AMoL) belongs to the heterogeneous group of Acute Myeloid Leukemias (AMLs), accounting for about 15–24% of AMLs in children (1–3).
The French-American-British (FAB) classification defines AMoL as AML-M5 in the presence of at least 30% of blast cells in bone marrow (BM) or peripheral blood (PB) specimens, 80% or more of those belonging to the monocytic lineage, generally evaluated on Wright-Giemsa or May-Grünwald-Giemsa-stained smears (4–6). The 2016 Revision of the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemias lowers the level of blast cells defining an AML to 20%, at least 80% of those being of monocytic derivation, and includes AMoL in the category AML, not otherwise specified (NOS) (1). Regardless, AMoL morphology has been described as associated with specific genetic abnormalities not included in the WHO classification yet, as well as genetic alterations belonging to the category AML with recurrent genetic abnormalities, which prevail on morphological and immunophenotypic features for the final AML group assignment, as per WHO classification (1).
According to morphological features and maturation stage of monocytic-lineage blast cells, AMoL may be further divided into acute monoblastic leukemia (AML-M5a as per FAB classification) when at least 80% of the monocytic cells are monoblasts and acute monocytic leukemia (AML-M5b as per FAB classification) when blasts at a more mature stage are prevalent (predominantly promonocytes) (4–6).
From a clinical point of view, AMoL is characterized by high leukocyte counts (7–10), major propensity for extramedullary infiltrates as compared with other AMLs, involving skin, gum, and central nervous system (11–18), and possible association with disseminated intravascular coagulation along with other bleeding disorders (19–21).
Since leukemic cells morphology and immunophenotypic features may substantially overlap with those of normal monocytic compartment, AMoL diagnosis and Measurable Residual Disease (MRD) detection are challenging.
Nowadays, the main diagnostic tools for monocytic cell compartment characterization include standard cytomorphology and cytochemistry, immunophenotyping by multiparametric flow cytometry (MFC), cytogenetics, and molecular biology (22, 23).
This review highlights standard diagnostic tools in AMoL.
The morphological evaluation is the first step of the process that leads to AMoL diagnosis.
Both PB and BM smears are traditionally stained with May-Grünwald-Giemsa or Wright-Giemsa stains, allowing proper cytosolic granules and nuclear chromatin discrimination. Cytomorphology allows detecting monocytic lineage cells at different maturation stages (monoblasts, promonocytes, immature or abnormal monocytes, mature monocytes, and reactive monocytes) as well as blast cells. Nevertheless, in clinical practice, the discrimination of blast cells from normal precursors or reactive monocytes is troublesome due to their relevant morphological overlap.
In the presence of a morphological picture suggesting AMoL, monoblasts and promonocytes should be considered as blasts/blast equivalents in acute monoblastic (M5a) and monocytic (M5b) leukemia.
In acute monoblastic leukemia (M5a), BM is usually hypercellular, showing a predominant population of large (up to 30 μm in diameter), poorly differentiated blasts with a rounded to oval nucleus containing reticular and immature chromatin pattern and one to four light-blue nucleoli. The cytoplasm is usually abundant and basophilic, with rare scattered azurophilic granules, fine vacuolizations, and the absence of Auer rods. The presence of translucent pseudopod formation may erroneously be misinterpreted as a double membrane (24–26).
In acute monocytic leukemia (M5b), promonocytes predominate in BM specimens, whereas mature monocytes are prevalent in PB samples. Extramedullary lesions can be composed of both two cell types. Promonocytes have less basophilic cytoplasm with a grayish ground-glass appearance and occasional large azurophilic granules and vacuoles (27). They have large, irregular-shaped, and folded nuclei, often containing nucleoli, with nuclear segmentations. This aspect allows differentiating them from monoblasts (Table 1) (28). Auer rods are rare. Associated hemophagocytes (erythrophagocytosis) may be observed, sometimes in case of positivity for t (8; 16) (p11.2; p13.3)/KAT6A-CREBBP translocation (1, 28, 29). Mature monocytes are usually characterized by a large overall size, deeply folded or convoluted nuclei with condensed, mature-appearing chromatin, absence of prominent nucleoli, and abundant gray-blue cytoplasm, often with a few vacuoles and azurophilic granules (Table 1) (28).
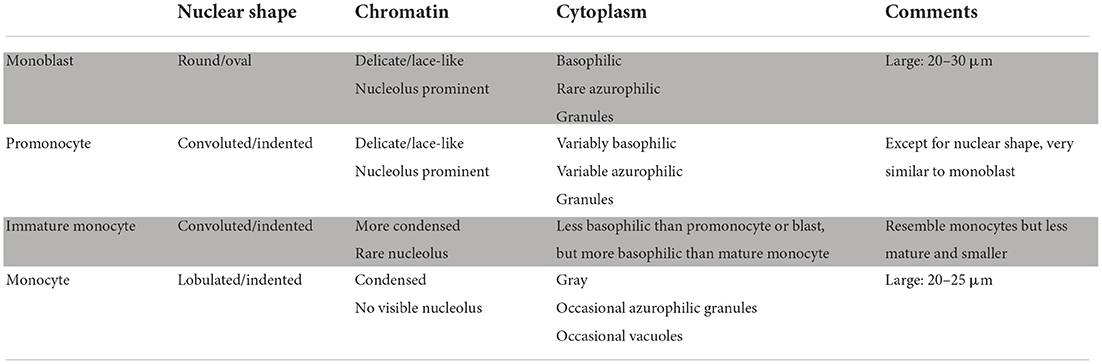
Table 1. Recommendations for monocyte evaluation in the blood or bone marrow smears by Goasguen et al. (28).
Immature monocytes, also defined as “abnormal” or “atypical” monocytes, can be found in normal BM smears as well as in Chronic Myelomonocytic Leukemia (CMML) and Juvenile Myelomonocytic Leukemia (JMML) (30). They are similar but smaller and more basophilic than mature monocytes, showing immature-appearing chromatin, prominent nuclear folds or convolutions, and, rarely, small nucleoli (Table 1) (28).
Reactive monocytes, usually encountered in response to inflammatory or infectious disorders [systemic lupus erythematosus, rheumatoid arthritis, sarcoidosis, Epstein-Barr virus (EBV), cytomegalovirus (CMV), human herpes virus-6 (HHV-6), histoplasma, mycobacteria, and toxoplasma] can show a range of morphologic patterns, including variable cell size (12–20 μm), increased nuclear to cytoplasmic ratio, less condensed or immature chromatin, small nucleoli, and prominent cytoplasmic vacuolization, basophilia, and granularity (Table 1) (28).
Therefore, morphological AMoL diagnosis requires expert operators, constant inter- and intra-laboratory training, updating, and confrontation, to minimize interobserver variability and standardize the final interpretation of blood smears.
Cytochemistry represents a helpful tool in discriminating immature monocytic- from myeloid-lineage cells and identifying monocytic cells at different maturation stages.
The myeloperoxidase (MPO) staining, a fundamental tool of the diagnostic process, is usually negative in immature monocytic cells (monoblasts) and mature monocytes, whereas promonocytes may show slight and scattered positivity. Conversely, granulocytic cells typically show strong MPO positivity (1, 29).
Intense non-specific esterase activity (NSE) [naphthyl acetate esterase, naphthol AS-D acetate esterase (CAE), and alpha-naphthyl butyrate esterase] represents one of the most specific hallmarks of cytochemical staining in monocytic lineage leukemia, resulting strongly positive in both pathological monoblasts and promonocytes, even if 10–20% of AMoL cases show NSE negativity or weak positivity. Moreover, NSE is negative or only weakly positive in myeloid-lineage cells and allowing their discrimination from monocytic ones (1, 29).
Nevertheless, NSE requires a complex and time-consuming multistep preparation and is associated with variability in staining results and subsequent interpretation. Therefore, it should be routinely performed only in laboratories with expert morphologists to obtain accurate results.
The addition of sodium fluoride to NSE staining allows further discriminating monocytic- from myeloid-lineage cells since it inhibits NSE reaction only in the first group (1, 31).
Regarding other cytochemical stainings, used only exceptionally in the clinical routine process, Sudan black and naphthol AS-D chloroacetate esterase are typically negative in monoblasts, whereas tartrate-sensitive acid phosphatase is usually positive. Variable results are described with the periodic acid-Schiff stain, 3-glucuronidase, and oil red O stain (24–26, 32–35).
Immunophenotyping by MFC accurately detects and characterizes any potential pathological cells in biological samples. In the presence of an abnormal proliferation of monocytic-lineage cells, at diagnosis MFC is critical in discriminating reactive from dysplastic or leukemic origin. Indeed, it allows overcoming the well-known overlap existing between monocytic-lineage blasts cells and normal counterparts, identifying asynchronous monocytic marker combinations as well as any aberrant expression of myeloid and lymphoid markers (Figure 1). Immunophenotyping at diagnosis relies on a careful selection of monoclonal antibodies (MoAb) conjugated with specific fluorochromes to be used in multiple combinations (36–38).
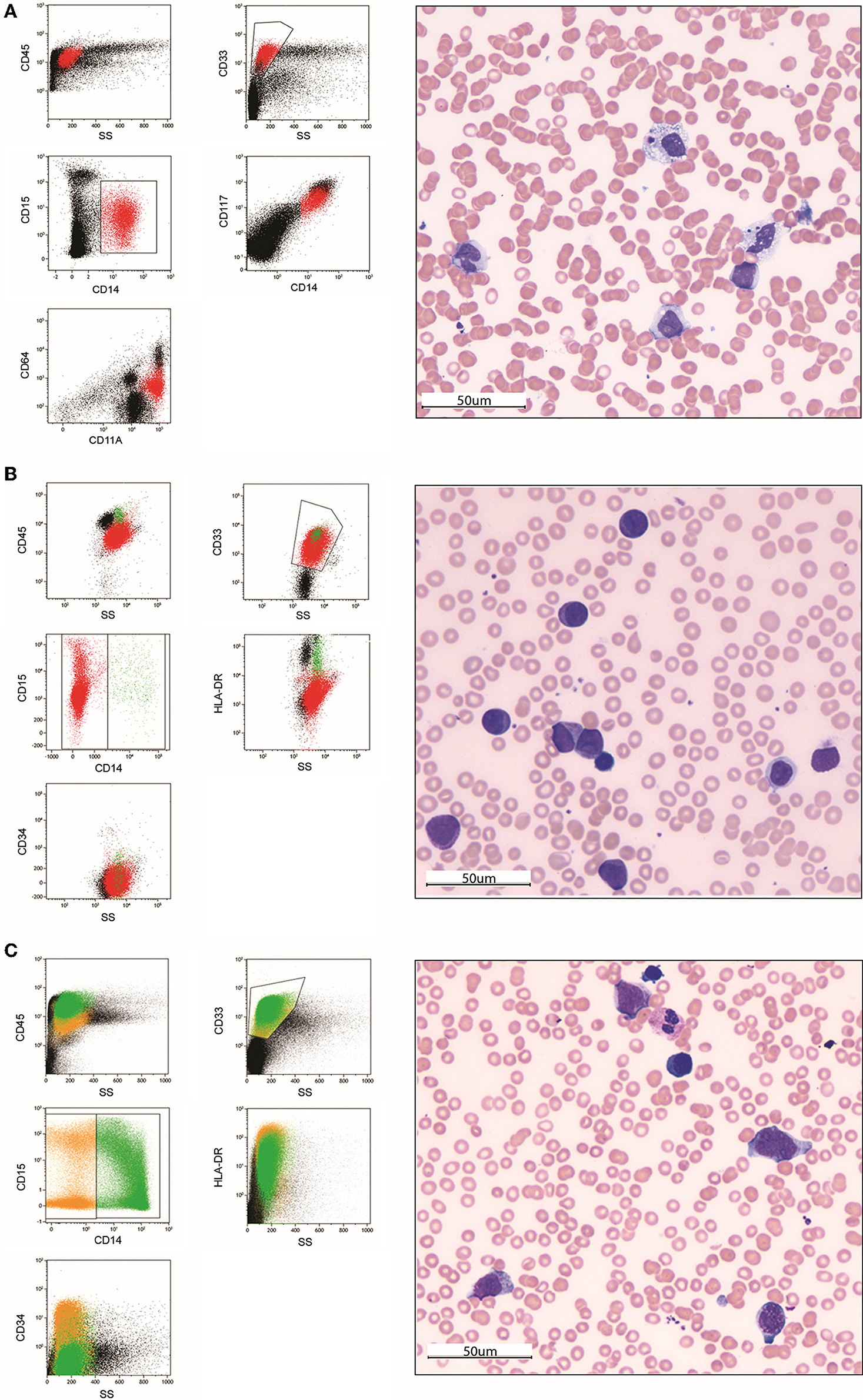
Figure 1. Morphological and immunophenotypic analysis of two pediatric acute monoblastic/monocytic leukemia cases at diagnosis (A,B) and after the first induction course (C). In the first case (A), immunophenotyping at diagnosis on a bone marrow sample allows identifying a population of blast cells characterized by an asynchronous expression of early myelomonocytic-lineage markers CD33, CD117, and CD64 together with the maturing monocytic-lineage marker CD15 and the mature monocytes marker CD14. The May-Grünwald-Giemsa (MGG)-stained bone marrow smear (40× magnification) shows a population of large cells with abundant grayish ground-glass cytoplasm containing rare vacuoles and irregular-shaped and folded nuclei with nucleoli. Two images of hematophagocytosis are also present. In the second case at diagnosis (B), blast cells show an immunophenotype overlapping with immature monocytic-lineage cells (positivity of CD33 and CD15, negativity of CD14) in the absence of CD34 expression. The MGG-stained smear shows a homogeneous population of large, poorly differentiated blasts with a rounded nucleus, reticular chromatin pattern and one to multiple nucleoli, and basophilic cytoplasm, including several azurophilic granules (monoblasts). After the first induction course (C), standard morphology identifies a population of immature monocytes not clearly discriminable from blast cells. Conversely, MFC-MRD analysis is able to define them as regenerating cells and exclude the presence of residual blasts. (40 × magnification). Color legend: red (monoblasts), green (monocytes), orange (monocyte precursors).
Several markers are useful to discriminate monocytic precursors at different maturation stages and monocytic-lineage blasts from myeloid counterpart, among those the most significant are: CD45, CD11a, CD38, CD99 (pan-leukocytic markers) (38–41); CD34 (hematopoietic stem cells marker) (42, 43); CD64, CD13, CD33, CD123, cyMPO, CD117 (early myelomonocytic-lineage markers) (43, 44); CD11b, CD15, CD36 (maturing monocytic-lineage markers); CD14, CD4, CD35, CD300e (mature monocytes markers); HLA-DR (pan-monocytic lineage marker) (43, 45); cyLYZO (pan-myelomonocytic lineage marker) (46); CD66b (47), CD16 (maturing and mature granulocytic-lineage marker) (48); CD371 (granulocyte-macrophage-lineage marker) (49). Additionally, a complete immunophenotypic study requires association with B- (CD19, CD10, cyCD79A, cyCD22) and T-(CD7, CD3, CD2, CD56, CD4, CD8, CD45RA) lymphoid- lineage markers (38, 43, 50, 51).
Regarding normal precursors, monoblasts are characterized by strong positivity of CD34, CD117, and HLA-DR and progressive upregulation of CD64 (Fc receptor/Fc gamma receptor 1). Even if CD117 is a tyrosine kinase receptor widely expressed on hematopoietic, endothelial, and immature cardiomyocytes, its association with CD34 and CD64 accurately identifies monoblasts precursors. Maturing promonocytes keep strong positivity of CD64 and HLA-DR but show a progressive downregulation of CD34 and CD117 and acquisition of CD14 (belonging to the family of leucine-rich repeat proteins), CD36 (fatty acid translocase/platelet glycoprotein 4/glycoprotein IIIb/glycoprotein PAS-4/scavenger receptor class B member 3), and CD35 (C3b-C4b receptor/complement receptor type 1). Finally, mature monocytes present strong positivity of CD64, CD14, and CD35, downregulation of HLA-DR, negativity of CD34 and CD117, and acquisition of CD300c (CMRF35-like molecule 6). Unlike CD64 and CD14, CD36, CD35, and CD300c are not mainly committed to the monocytic lineage. Regardless, they play a crucial role in identifying monocytic precursors in the described associations (43).
At diagnosis, the knowledge of monocytic maturation sequence of antigen expression critically helps detect AMoL blast cells, potentially escaping from this schema. In acute monoblastic leukemia, blasts may show a more immature immunophenotype, presenting a heterogeneous positivity of CD34, strong positivity of CD64 and HLA-DR, downregulation of maturating monocytic-associated markers CD36 and CD11b, and absence of mature monocytic-associated antigens CD14, CD35, and CD300c, together with downregulation of myelomonocytic markers cyMPO, CD13, CD123. Conversely, acute monocytic leukemia may be associated with a more mature immunophenotype, characterized by the expression of mature monocytic-associated markers CD14, CD35, and CD300c and the absence of CD34 (43).
AMoL blast cells may show aberrant expression of markers like CD56, CD7, CD19, cyCD79a (lymphoid antigens), and NG2 (neural/glial antigen 2), the positivity of which is potentially associated with the presence of KMT2A rearrangements. Whether the expression of NG2, CD7, CD19, and cyCD79a on monocytic lineage cells undoubtfully indicates the presence of blast cells (43, 52–54), CD56 may be aberrantly positive on reactive and normal monocytes (55–57).
An accurate definition of blast immunophenotype at diagnosis is also critical for measurable residual disease monitoring by MFC (MFC-MRD) during therapy. MFC-MRD relies on the Leukemia-Associated Immunophenotype (LAIP) approach, in which leukemic blasts immunophenotype are characterized at diagnosis and tracked at re-evolution points, and the Different-from-Normal (DfN) approach, based on discrimination between cells with aberrant immunophenotypes and normal counterpart during follow-up (58).
In AMoL, MFC-MRD is usually more challenging than in other AML subtypes. First, the blast population may have an immunophenotypic heterogeneity at diagnosis potentially hidden by a predominant LAIP. Second, blast immunophenotype may change during therapy, hampering the accuracy of the LAIP method. Third, in the absence of aberrant lymphoid marker expression, AMoL blast LAIPs frequently overlap the immunophenotype of monocytic-lineage precursors in regenerating BM, making MFC-MRD detection very challenging. Therefore, the DfN approach may be more useful in these cases, even if more complex and requires a deep knowledge of normal hematopoietic precursors immunophenotypic maturation curves (58–66).
Consequently, it is critical to identify new markers specific for monocytic-lineage blasts and possibly not expressed on normal hematopoietic cells. In the last few years, several antigens have been studied to detect AMoL blasts. In 2015, Pereira and colleagues demonstrated the aberrant expression of B lymphoid-lineage antigen CD37, a transmembrane protein of the tetraspanin superfamily, in 15 different AML cell lines, 5 of those AMoL, and confirmed it in a cohort of 26 patients'-derived AML samples (67). In 2019, Lo et al. showed the positivity of antigen CD302, a type I transmembrane C-type lectin receptor usually expressed on myeloid-lineage cells, in all the 6 AMoL out of 33 analyzed AML samples (68). CD157, a glycosylphosphatidylinositol-anchored glycoprotein, is another marker associated with AMoL, but also with normal myelomonocytic lineage cells (69). Additionally, in 2020 Churchill et al. described the coexpression of the two inhibitory leukocyte Ig-like receptors, LILRB1 and LILRB4, belonging to a family of immunoregulatory receptors, specific for AMoL (70).
To summarize, MFC is an essential step of AMoL diagnosis in childhood, allowing blasts discrimination from normal monocytic counterpart even when their cytomorphological definition is challenging. In diagnostics practice, an accurate MFC diagnosis of AMoL relies on the following steps: (i) negativity of B- (at least 2 among CD19, CD10, cyCD22, and cyCD79) or T-lineage (s/cyCD3 and CD7) acute lymphoblastic leukemia criteria; (ii) blasts assignment to the myeloid/monocytic lineage (positivity of at least 2 among MPO, CD13, CD33, CD64, CD65, CD117, cy-LYZO, CD14, CD11c); (iii) investigation of specific monocytic-lineage markers (CD11b, CD15, HLA-DR, CD371), (iv) hematopoietic stem cell marker (CD34), and (v) potential aberrant expression of CD7, CD56, CD19, cyCD79a, NG2 (1, 38, 71).
In the last few decades, genetic characterization has shown to be critical in AML clinicopathological evaluation, refining diagnosis, assigning patients to a risk class, predicting prognosis, and promoting target therapy (72, 73). Genetic characterization mainly relies on cytogenetic (conventional karyotyping and FISH analysis) and molecular studies [next-generation sequencing (NGS) and real-time reverse transcriptase-polymerase chain reaction (RT-PCR)] (72).
Pediatric AMoL is a heterogeneous disease as concerns genetics and is associated with different outcomes regarding the displayed aberrations. At cytogenetic analyses, it presents a normal karyotype in about 20% (74). Among somatic cytogenetic aberrations, chromosomal translocations involving the KMT2A gene on 11q23 are the most frequent, accounting for about 20% of the genetic anomalies in AMoLs (74). Previously known as MLL (Mixed Lineage Leukemia), the KMT2A gene encodes an important histone-H3 lysine-4 (H3K4) methyltransferase involved in the epigenetic regulation of hematopoietic stem/progenitor cells development.
A total of 135 KMT2A rearrangements have been described in leukemias, mostly resulting in KMT2A-fusion proteins capable of transforming hematopoietic stem cells into leukemic blasts with stem cell-like properties (72, 75–78). The most frequent fusion partners in AMLs are AF9/MLLT3, AF10/MLLT10, ELL, ENL/MLLT1, AF6/MLLT4, and MLL-PTDs (77, 79). Nowadays, only AML with t (9; 11) (p21.3; q23.3)/KMT2A-MLLT3 is included in the category AML with recurrent genetic abnormalities of the WHO classification (1).
Of note, Balgobind et al. reported a significantly better prognosis of t (9; 11) (p22; q23) positive AMLs in the presence of M5 FAB rather than non-M5 FAB. Moreover, Balgobind et al. described new risk subgroups related to KMT2A translocations: t (1; 11) (q21; q23) showed favorable outcomes regardless of other risk factors, whereas t (6; 11) (q27; q23), t (10; 11) (p12; q23), and t (10; 11) (p11.2; q23) turned out to be an independent risk factor of poor clinical outcome (80).
Another clinically relevant genetic aberration in AMoLs, not detectable with cytogenetic studies, is the FMS-like tyrosine kinase 3 (FLT3) gene mutation, described in about 5.5–26.5% of AMoLs in different pediatric cohorts (71, 74, 81–83). FLT3 belongs to the class III tyrosine kinase receptor family expressed on hematopoietic progenitors and promotes cell survival, proliferation, and differentiation being activated by an extracellular ligand (FLT3 ligand) (71).
FLT3 mutations may be found in normal karyotype AMLs and associated with additional cytogenetic lesions. They are classified into two major groups: internal tandem duplications (ITDs-about two-thirds of FLT3 mutations) and point mutations in the tyrosine kinase domain (TKD-about one-third of FLT3 mutations), both leading to the constitutive activation of the FLT3 kinase. The prognostic role of FLT3-TKD remains unclear, whereas FLT3-ITDs have been associated with an unfavorable prognosis, particularly in the presence of a high allelic ratio (> 0.51) (1, 74, 84–91). Regardless, it was recently demonstrated that the association with additional genetic aberrations, like WT1 and NPM1 mutations and NUP98 translocations, modulates the outcome (91).
AMoL may be also rarely associated with Core Binding Factor (CBF) mutations involving chromosome 16 [inv (16) (p13.1q22) and t (16; 16) (p13.1; q22)/CBFB-MYH11], and t (8; 21) (q22; q22.1); RUNX1-RUNX1T1, both included in the category AML with recurrent genetic abnormalities according to WHO classification (1, 3, 91, 92). To note, the presence of any CBF abnormalities allows diagnosing an AML independently from blast count (1).
AMoL could also be associated with somatic mutations of the nucleophosmin gene 1 (NPM1), encoding a nuclear pleiotropic protein that regulates cell growth and proliferation, protein chaperoning, maintenance of genomic stability, and activation of tumor suppressor p53. NPM1 mutations involve about 6.5% of pediatric AMLs, may be associated with normal karyotype and FLT3/ITD mutations, and show a wide range of morphological subtypes, of those FAB-M4 and M5 are the most frequent (93–95). NPM1 mutations seem to confer a favorable outcome in childhood AML, in the absence of FLT3-ITD mutations (91, 95).
Finally, AMoL may display activating mutations of NRAS and KRAS genes, encoding two G-proteins involved in proliferation and survival signals transmission among cells (96). NRAS and KRAS mutations are detectable in about 18–20% and 28% of pediatric AMoLs, respectively, being more common in younger patients (74, 96). RAS mutations may be associated with normal karyotype, as well as with other genetic aberrations, like NPM1 mutation and MLL-PTD (91, 96). The prognostic role of RAS mutation is still unclear, being variable in different pediatric and adult cohorts (96).
Beyond their critical role in pediatric AMoL diagnostics, MFC and molecular biology allow identifying potential targets for a precision medicine approach. Among MFC-detectable markers, CD33 is the target of the antibody-drug conjugated gemtuzumab ozogamicin, approved by the FDA in 2018 for the treatment of relapsed/refractory pediatric AMLs (97) and adopted in several first- and additional-line pediatric therapeutic trials (98–104). CD123-positive AMoLs may potentially benefit from flotetuzumab, a CD123/CD3 bispecific antibody, and antibody-drug conjugated IMGN632 and tagraxofusp-erzs, all under investigation for pediatric relapsed/refractory AMLs (105, 106). Additionally, CD33, together with CD123 and CD371, is under investigation for generating chimeric antigen-receptor T cells (CAR-T) directed against AML (106–108).
Among molecular alterations, AMoL harboring FLT3-ITD mutation may benefit from targeted therapy with FLT3 inhibitors (sorafenib, midostaurin, sunitinib, lestaurtinib, quizartinib, gilterinib) (109, 110). In the presence of KMT2A rearrangements, AMoL may potentially benefit from targeted therapy with DOT1L-inhibitor (pinometostat) and menin-inhibitors (KO-539, SNDX-5613), actually under investigation (109, 111, 112).
An accurate characterization of monocytic-lineage cells in PB and BM samples is crucial to avoid misdiagnosis. Indeed, AMoL needs to be differentiated from other morphological AMLs subtypes (AML without maturation, AML with minimal differentiation, acute megakaryoblastic leukemia, acute myelomonocytic leukemia, microgranular acute promyelocytic leukemia, CMML, JMML), and reactive causes of monocytosis.
Moreover, several infections can result in myelomonocytosis along with persistent fever, failure to thrive, hepatosplenomegaly, skin lesions, anemia, and thrombocytopenia, mimicking leukemia.
Developing new diagnostic markers is required to obtain unequivocal discrimination between leukemic and reactive monocytes. Particularly, the identification of new specific MFC antigens may help recognize monocytic blasts even in the absence of specific leukemia-related genetic markers.
BB and EV designed the study. EM, FS, FD, EV, and BB wrote the manuscript. All authors contributed to the article and approved the submitted version.
This project is part of a training activity supported by the residency program in Pediatrics of the Department of Woman and Child's Health, University of Padua, Padua, Italy.
The authors would like to thank Dr. Samuela Francescato, Dr. Andrea Zangrando, and Dr. Giovanni Faggin who prepared the figure.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Swerdlow SH, Campo E, Harris NL, Jaffe E, Pileri S, Stein H, et al. WHO Classification of Tumours of Haematopoietic and Lymphoid tissues, 4th edn. Lyon: IARC Press. (2017).
2. Webb DK, Harrison G, Stevens RF, Gibson BG, Hann IM, Wheatley K, et al. Relationships between age at diagnosis, clinical features, and outcome of therapy in children treated in the Medical Research Council AML 10 and 12 trials for acute myeloid leukemia. Blood. (2001) 98:1714–20. doi: 10.1182/blood.V98.6.1714
3. Pession A, Masetti R, Rizzari C, Putti MC, Casale F, Fagioli F, et al. Results of the AIEOP AML 2002/01 multicenter prospective trial for the treatment of children with acute myeloid leukemia. Blood. (2013) 122:170–8. doi: 10.1182/blood-2013-03-491621
4. Bennett JM, Catovsky D, Daniel MT, Flandrin G, Galton DA, Gralnick HR, et al. Proposed revised criteria for the classification of acute myeloid leukemia. A report of the French-American–British cooperative group. Ann Intern Med. (1985) 103:620–5. doi: 10.7326/0003-4819-103-4-620
5. Brunning RD, Matutes E, Flandrin G, Vardiman J. Pathology and Genetics of Tumours of Haematopoietic and Lymphoid Tissues. Lyon: IARC Press. (2001).
6. Hossfeld DK. Book reviews. World health organization classification of tumours: pathology and genetics of tumours of haematopoietic and lymphoid tissues. Ann Onc. (2002) 13:490–1. doi: 10.1093/annonc/mdf146
7. Creutzig U, Ritter J, Budde M, Sutor A, Schellong G. Early deaths due to hemorrhage and leukostasis in childhood acute myelogenous leukemia. Associations with hyperleukocytosis and acute monocytic leukemia. Cancer. (1987) 60:3071–9.3.
8. Porcu P, Cripe LD, Ng EW, Bhatia S, Danielson CM, Orazi A, et al. Hyperleukocytic leukemias and leukostasis: a review of pathophysiology, clinical presentation and management. Leuk Lymphoma. (2000) 39:1–18. doi: 10.3109/10428190009053534
9. Schwonzen M, Kuehn N, Vetten B, Diehl V, Pfreundschuh M. Phenotyping of acute myelomonocytic (AMMOL) and monocytic leukemia (AMOL): association of T-cell-related antigens and skin-infiltration in AMOL. Leuk Res. (1989) 13:893–8. doi: 10.1016/0145-2126(89)90042-8
10. Cuttner J, Conjalka MS, Reilly M, Goldberg J, Reisman A, Meyer RJ, et al. Association of monocytic leukemia in patients with extreme leukocytosis. Am J Med. (1980) 69:555–8. doi: 10.1016/0002-9343(80)90467-2
11. Peterson L, Dehner LP, Brunning RD. Extramedullary masses as presenting features of acute monoblastic leukemia. Am J Clin Pathol. (1981) 75:140–8. doi: 10.1093/ajcp/75.2.140
12. Font RL, Mackay B, Tang R. Acute monocytic leukemia recurring as bilateral perilimbal infiltrates. Immunohistochemical and ultrastructural confirmation. Ophthalmology. (1985) 92:1681–5. doi: 10.1016/S0161-6420(85)34091-5
13. Hertler AA, Rosenwasser GO, Gluck WL. Isolated anterior chamber relapse in acute monoblastic leukemia. Am J Hematol. (1986) 23:401–3. doi: 10.1002/ajh.2830230412
14. Krendel DA, Albright RE, Graham DG. Infiltrative polyneuropathy due to acute monoblastic leukemia in hematologic remission. Neurology. (1987) 37:474–7. doi: 10.1212/WNL.37.3.474
15. Peterson BA, Brunning RD, Bloomfield CD, Hurd DD, Gau JA, Peng GT, et al. Central nervous system involvement in acute nonlymphocytic leukemia. A prospective study of adults in remission. Am J Med. (1987) 83:464–70. doi: 10.1016/0002-9343(87)90756-X
16. Gottesfeld E, Silverman RA, Coccia PF, Jacobs G, Zaim MT. Transient blueberry muffin appearance of a newborn with congenital monoblastic leukemia. J Am Acad Dermatol. (1989) 21:347–51. doi: 10.1016/S0190-9622(89)80032-5
17. Roberts JP, Atwell JD. Testicular enlargement as a presenting feature of monocytic leukemia in an infant. J Pediatr Surg. (1989) 24:1306–7. doi: 10.1016/S0022-3468(89)80573-1
18. Jones D, Dorfman DM, Barnhill RL, Granter SR. Leukemic vasculitis: a feature of leukemia cutis in some patients. Am J Clin Pathol. (1997) 107:637–42. doi: 10.1093/ajcp/107.6.637
19. Bussel JB, Steinherz PG, Miller DR, Hilgartner MW A. heparin-like anticoagulant in an 8-month-old boy with acute monoblastic leukemia. Am J Hematol. (1984) 16:83–90. doi: 10.1002/ajh.2830160111
20. Auger MJ Mackie MJ A A quantitative study of monocyte procoagulant activity in acute monoblastic and chronic myelomonocytic leukaemias. Acta Haematol. (1987) 78:37–40. doi: 10.1159/000205833
21. Alvarenga B, Manzo E, Arca G, Higashi M, Mattos E, Melo I, et al. Disseminated intravascular coagulation and concomitant central nervous system bleeding in acute monoblastic leukemia: case report. Hematol Transfusion Cell Therapy. (2020) 42:308–9. doi: 10.1016/j.htct.2020.10.513
22. Vardiman JW, Thiele J, Arber DA, Brunning RD, Borowitz MJ, Porwit A, et al. The 2008 revision of the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia: rationale and important changes. Blood. (2009) 114:937–51. doi: 10.1182/blood-2009-03-209262
23. Dunphy CH. Comparative analysis of detecting monocytic cells and their aberrancy. Appl Immunohistochem Mol Morphol. (2011) 19:336–40. doi: 10.1097/PAI.0b013e3182052805
24. McKenna RW, Bloomfield CD, Dick F, Nesbit ME, Brunning RD. Acute monoblastic leukemia: diagnosis and treatment of ten cases. Blood. (1975) 46:481–94. doi: 10.1182/blood.V46.4.481.481
25. Brynes RK, Golomb HM, Desser RK, Recant W, Reese C, Rowley J. Acute monocytic leukemia. Cytologic, histologic, cytochemical, ultrastructural, and cytogenetic observations. Am J Clin Pathol. (1976) 65:471–82. doi: 10.1093/ajcp/65.4.471
26. Shaw MT, Nordquist RE. “Pure” monocytic or histiomonocytic leukemia: a revised concept. Cancer. (1975) 35:208–14.
27. Bennett JM, Catovsky D, Daniel MT, Flandrin G, Galton DA, Gralnick HR, et al. Proposals for the classification of the acute leukaemias. French–American–British (FAB) co-operative group. Br J Haematol. (1976) 33:451–8. doi: 10.1111/j.1365-2141.1976.tb03563.x
28. Goasguen JE, Bennett JM, Bain BJ, Vallespi T, Brunning R, Mufti GJ, et al. Morphological evaluation of monocytes and their precursors. Haematologica. (2009) 94:994–7. doi: 10.3324/haematol.2008.005421
29. Medeiros L, Miranda R. Diagnostic Pathology: Lymph Nodes and Extranodal Lymphomas- 2nd Edition. Elsevier Inc.: Amsterdam (2018).
30. Gupta AK, Meena JP, Chopra A, Tanwar P, Seth R. Juvenile myelomonocytic leukemia-A comprehensive review and recent advances in management. Am J Blood Res. (2021) 11:1–21.
31. Wrotnowski U, Innes DJ, Hobson AS. Nonspecific esterase staining patterns in acute monocytic leukemia. Am J Clin Pathol. (1987) 87:515–8. doi: 10.1093/ajcp/87.4.515
32. Sultan C, Imbert M, Ricard MF, Sebaoun G, Marquet M, Brun B, et al. Pure acute monocytic leukemia. A study of 12 cases. Am J Clin Pathol. (1977) 68:752–7. doi: 10.1093/ajcp/68.6.752
33. Flandrin G, Daniel MT. Practical value of cytochemical studies for the classification of acute leukemias. In: Mathé, G, Pouillart, P, Schwarzenberg L, editors. Nomenclature, Methodology and Results of Clinical Trials in Acute Leukemias Recent Results in Cancer Research/Fortschritte der Krebsforschung/Progrès dans les recherches sur le cancer. Springer: Berlin, Heidelberg (1973). doi: 10.1007/978-3-642-80776-3_8
34. Tobelem G, Jacquillat C, Chastang C, Auclerc MF, Lechevallier T, Weil M, et al. Acute monoblastic leukemia: a clinical and biologic study of 74 cases. Blood. (1980) 55:71–6. doi: 10.1182/blood.V55.1.71.bloodjournal55171
35. Daniel MT, Flandrin G, Lejeune F, Liso P, Lortholary PI. Specific esterases of monocytes: their use in the classification of acute leukemias. Nouv Rev Fr Hematol. (1971) 11:233–9.
36. Greig B, Oldaker T, Warzynski M, Wood B. 2006 bethesda international consensus recommendations on the immunophenotypic analysis of hematolymphoid neoplasia by flow cytometry: recommendations for training and education to perform clinical flow cytometry. Cytometry B Clin Cytom. (2007) 72:S23–33. doi: 10.1002/cyto.b.20364
37. Porwit A, Béné M-C. Multiparameter Flow Cytometry in the Diagnosis of Hematologic Malignancies. Cambridge, United Kingdom; New York, NY: Cambridge University Press (2018). p. 234. doi: 10.1017/9781316218549
38. Dworzak MN, Buldini B, Gaipa G, Ratei R, Hrusak O, Luria D, et al. AIEOP-BFM consensus guidelines 2016 for flow cytometric immunophenotyping of Pediatric acute lymphoblastic leukemia. Cytometry B Clin Cytom. (2018) 94:82–93. doi: 10.1002/cyto.b.21518
39. Naeim F, Rao P, Grody W. Hematopathology, Morphology, Immunophenotype, Cytogenetics, and Molecular Approaches, 1st Edn. Elsevier (2008).
40. Campana D, Suzuki T, Todisco E, Kitanaka A. CD38 in Hematopoiesis. In: Mehta KMF, editor. Human CD38 and Related Molecules. 75. Basel: Karger (2000). p. 169–88. doi: 10.1159/000058768
41. Dworzak MN, Fritsch G, Buchinger P, Fleischer C, Printz D, Zellner A, et al. Flow cytometric assessment of human MIC2 expression in bone marrow, thymus, and peripheral blood. Blood. (1994) 83:415–25. doi: 10.1182/blood.V83.2.415.415
42. Krause DS, Fackler MJ, Civin CI, May WS. CD34: structure, biology, and clinical utility. Blood. (1996) 87:1–13. doi: 10.1182/blood.V87.1.1.1
43. Matarraz S, Almeida J, Flores-Montero J, Lécrevisse Q, Guerri V, López A, et al. Introduction to the diagnosis and classification of monocytic-lineage leukemias by flow cytometry. Cytometry B Clin Cytom. (2017) 92:218–27. doi: 10.1002/cyto.b.21219
44. Escribano L, Ocqueteau M, Almeida J, Orfao A, San Miguel JF. Expression of the c-kit (CD117) molecule in normal and malignant hematopoiesis. Leuk Lymphoma. (1998) 30:459–66. doi: 10.3109/10428199809057558
45. van Lochem EG, van der Velden VH, Wind HK. te Marvelde JG, Westerdaal NA, van Dongen JJ. Immunophenotypic differentiation patterns of normal hematopoiesis in human bone marrow: reference patterns for age-related changes and disease-induced shifts. Cytometry B Clin Cytom. (2004) 60:1–13. doi: 10.1002/cyto.b.20008
46. Merlini G, Bellotti V. Lysozyme: a paradigmatic molecule for the investigation of protein structure, function and misfolding. Clin Chim Acta. (2005) 357:168–72. doi: 10.1016/j.cccn.2005.03.022
47. Gustafson MP, Lin Y, Maas ML, Van Keulen VP, Johnston PB, Peikert T, et al. A method for identification and analysis of non-overlapping myeloid immunophenotypes in humans. PLoS ONE. (2015) 10:e0121546. doi: 10.1371/journal.pone.0121546
48. Arnoulet C, Béné MC, Durrieu F, Feuillard J, Fossat C, Husson B, et al. Four- and five-color flow cytometry analysis of leukocyte differentiation pathways in normal bone marrow: a reference document based on a systematic approach by the GTLLF and GEIL. Cytometry B Clin Cytom. (2010) 78:4–10. doi: 10.1002/cyto.b.20484
49. Bill MB, van Kooten Niekerk PS, Woll P, Laine Herborg L, Stidsholt Roug A, Hokland P, et al. Mapping the CLEC12A expression on myeloid progenitors in normal bone marrow; implications for understanding CLEC12A-related cancer stem cell biology. J Cell Mol Med. (2018) 22:2311–8. doi: 10.1111/jcmm.13519
50. Courville EL, Lawrence MG. Characteristic CD45RA/CD45RO maturation pattern by flow cytometry associated with the CD45 C77G polymorphism. Cytometry B Clin Cytom. (2021) 100:602–5. doi: 10.1002/cyto.b.21993
51. Dunphy CH, Orton SO, Mantell J. Relative contributions of enzyme cytochemistry and flow cytometric immunophenotyping to the evaluation of acute myeloid leukemias with a monocytic component and of flow cytometric immunophenotyping to the evaluation of absolute monocytoses. Am J Clin Pathol. (2004) 122:865–74. doi: 10.1309/BH588HVG6UHN2RF2
52. Zerkalenkova E, Mikhaylova E, Lebedeva S, Illarionova O, Baidun L, Kashpor S, et al. Quantification of NG2-positivity for the precise prediction of KMT2A gene rearrangements in childhood acute leukemia. Genes Chromosomes Cancer. (2021) 60:88–99. doi: 10.1002/gcc.22915
53. Liu YR, Zhu HH, Ruan GR, Qin YZ, Shi HX, Lai YY, et al. NPM1-mutated acute myeloid leukemia of monocytic or myeloid origin exhibit distinct immunophenotypes. Leuk Res. (2013) 37:737–41. doi: 10.1016/j.leukres.2013.03.009
54. Behm FG, Smith FO, Raimondi SC, Pui CH, Bernstein ID. Human homologue of the rat chondroitin sulfate proteoglycan, NG2, detected by monoclonal antibody 7. 1, identifies childhood acute lymphoblastic leukemias with t (4; 11) (q21; q23) or t (11; 19) (q23; p13) and MLL gene rearrangements. Blood. (1996) 87:1134–9. doi: 10.1182/blood.V87.3.1134.bloodjournal8731134
55. Dutt TS, LaVergne SM, Webb TL, Baxter BA, Stromberg S, McFann K, et al. Comprehensive Immune Profiling Reveals CD56. J Immunol. (2022) 208:685–96. doi: 10.4049/jimmunol.2100830
56. Langebrake C, Klusmann JH, Wortmann K, Kolar M, Puhlmann U, Reinhardt D. Concomitant aberrant overexpression of RUNX1 and NCAM in regenerating bone marrow of myeloid leukemia of Down's syndrome. Haematologica. (2006) 91:1473–80.
57. Sconocchia G, Keyvanfar K, El Ouriaghli F, Grube M, Rezvani K, Fujiwara H, et al. Phenotype and function of a CD56+ peripheral blood monocyte. Leukemia. (2005) 19:69–76. doi: 10.1038/sj.leu.2403550
58. Schuurhuis GJ, Heuser M, Freeman S, Béné MC, Buccisano F, Cloos J, et al. Minimal/measurable residual disease in AML: a consensus document from the European LeukemiaNet MRD Working Party. Blood. (2018) 131:1275–91. doi: 10.1182/blood-2017-09-801498
59. Al-Mawali A, Gillis D, Lewis I. The role of multiparameter flow cytometry for detection of minimal residual disease in acute myeloid leukemia. Am J Clin Pathol. (2009) 131:16–26. doi: 10.1309/AJCP5TSD3DZXFLCX
60. van der Velden VH, van der Sluijs-Geling A, Gibson BE. te Marvelde JG, Hoogeveen PG, Hop WC, et al. Clinical significance of flowcytometric minimal residual disease detection in pediatric acute myeloid leukemia patients treated according to the DCOG ANLL97/MRC AML12 protocol. Leukemia. (2010) 24:1599–606. doi: 10.1038/leu.2010.153
61. van Dongen JJ, Orfao A, Consortium E. EuroFlow: Resetting leukemia and lymphoma immunophenotyping. Basis for companion diagnostics and personalized medicine. Leukemia. (2012) 26:1899–907. doi: 10.1038/leu.2012.121
62. Gaipa G, Buracchi C, Biondi A. Flow cytometry for minimal residual disease testing in acute leukemia: opportunities and challenges. Expert Rev Mol Diagn. (2018) 18:775–87. doi: 10.1080/14737159.2018.1504680
63. Inaba H, Coustan-Smith E, Cao X, Pounds SB, Shurtleff SA, Wang KY, et al. Comparative analysis of different approaches to measure treatment response in acute myeloid leukemia. J Clin Oncol. (2012) 30:3625–32. doi: 10.1200/JCO.2011.41.5323
64. Buldini B, Maurer-Granofszky M, Varotto E, Dworzak MN. Flow-cytometric monitoring of minimal residual disease in pediatric patients with acute myeloid leukemia: recent advances and future strategies. Front Pediatr. (2019) 7:412. doi: 10.3389/fped.2019.00412
65. Zeijlemaker W, Gratama JW, Schuurhuis GJ. Tumor heterogeneity makes AML a “moving target” for detection of residual disease. Cytometry B Clin Cytom. (2013). doi: 10.1002/cytob.21134
66. Buldini B, Rizzati F, Masetti R, Fagioli F, Menna G, Micalizzi C, et al. Prognostic significance of flow-cytometry evaluation of minimal residual disease in children with acute myeloid leukaemia treated according to the AIEOP-AML 2002/01 study protocol. Br J Haematol. (2017) 177:116–26. doi: 10.1111/bjh.14523
67. Pereira DS, Guevara CI, Jin L, Mbong N, Verlinsky A, Hsu SJ, et al. AGS67E, an anti-CD37 monomethyl Auristatin E antibody-drug conjugate as a potential therapeutic for B/T-cell malignancies and AML: a new role for CD37 in AML. Mol Cancer Ther. (2015) 14:1650–60. doi: 10.1158/1535-7163.MCT-15-0067
68. Lo TH, Abadir E, Gasiorowski RE, Kabani K, Ramesh M, Orellana D, et al. Examination of CD302 as a potential therapeutic target for acute myeloid leukemia. PLoS ONE. (2019) 14:e0216368. doi: 10.1371/journal.pone.0216368
69. Yakymiv Y, Augeri S, Fissolo G, Peola S, Bracci C, Binaschi M, et al. CD157: from myeloid cell differentiation marker to therapeutic target in acute myeloid leukemia. Cells. (2019) 8:1580. doi: 10.3390/cells8121580
70. Churchill HRO, Fuda FS, Xu J, Deng M, Zhang CC, An Z, et al. Leukocyte immunoglobulin-like receptor B1 and B4 (LILRB1 and LILRB4): Highly sensitive and specific markers of acute myeloid leukemia with monocytic differentiation. Cytometry B Clin Cytom. (2021) 100:476–87. doi: 10.1002/cyto.b.21952
71. Creutzig U, van den Heuvel-Eibrink MM, Gibson B, Dworzak MN, Adachi S, de Bont E, et al. Diagnosis and management of acute myeloid leukemia in children and adolescents: recommendations from an international expert panel. Blood. (2012) 120:3187–205. doi: 10.1182/blood-2012-03-362608
72. Narayanan D, Weinberg OK. How I investigate acute myeloid leukemia. Int J Lab Hematol. (2020) 42:3–15. doi: 10.1111/ijlh.13135
73. Szczepański T, Harrison CJ, van Dongen JJ. Genetic aberrations in paediatric acute leukaemias and implications for management of patients. Lancet Oncol. (2010) 11:880–9. doi: 10.1016/S1470-2045(09)70369-9
74. Liu LP, Zhang AL, Ruan M, Chang LX, Liu F, Chen X, et al. Prognostic stratification of molecularly and clinically distinct subgroup in children with acute monocytic leukemia. Cancer Med. (2020) 9:3647–55. doi: 10.1002/cam4.3023
75. Li X, Song Y. Structure, function and inhibition of critical protein-protein interactions involving mixed lineage leukemia 1 and its fusion oncoproteins. J Hematol Oncol. (2021) 14:56. doi: 10.1186/s13045-021-01057-7
76. Arber DA. Acute Myeloid Leukemia. In: Hsi ED, editor. Foundations in Diagnostic Pathology, Hematopathology (Third Edition). Elsevier: Amsterdam (2018). p. 429–66.e5. doi: 10.1016/B978-0-323-47913-4.00014-8
77. Meyer C, Burmeister T, Gröger D, Tsaur G, Fechina L, Renneville A, et al. The MLL recombinome of acute leukemias in 2017. Leukemia. (2018) 32:273–84. doi: 10.1038/leu.2017.213
78. Meyer C, Hofmann J, Burmeister T, Gröger D, Park TS, Emerenciano M, et al. The MLL recombinome of acute leukemias in 2013. Leukemia. (2013) 27:2165–76. doi: 10.1038/leu.2013.135
79. Pigazzi M, Masetti R, Bresolin S, Beghin A, Di Meglio A, Gelain S, et al. MLL partner genes drive distinct gene expression profiles and genomic alterations in pediatric acute myeloid leukemia: an AIEOP study. Leukemia. (2011) 25:560–3. doi: 10.1038/leu.2010.316
80. Balgobind BV, Raimondi SC, Harbott J, Zimmermann M, Alonzo TA, Auvrignon A, et al. Novel prognostic subgroups in childhood 11q23/MLL-rearranged acute myeloid leukemia: results of an international retrospective study. Blood. (2009) 114:2489–96.
81. Choi S, Kim BK, Ahn HY, Hong KT, Choi JY, Shin HY, et al. Outcomes of pediatric acute myeloid leukemia patients with FLT3-ITD mutations in the pre-FLT3 inhibitor era. Blood Res. (2020) 55:217–24. doi: 10.5045/br.2020.2020127
82. Meshinchi S, Alonzo TA, Stirewalt DL, Zwaan M, Zimmerman M, Reinhardt D, et al. Clinical implications of FLT3 mutations in pediatric AML. Blood. (2006) 108:3654–61. doi: 10.1182/blood-2006-03-009233
83. Iwai T, Yokota S, Nakao M, Okamoto T, Taniwaki M, Onodera N, et al. Internal tandem duplication of the FLT3 gene and clinical evaluation in childhood acute myeloid leukemia. The Children's Cancer and Leukemia Study Group, Japan. Leukemia. (1999) 13:38–43. doi: 10.1038/sj.leu.2401241
84. Daver N, Schlenk RF, Russell NH, Levis MJ. Targeting FLT3 mutations in AML: review of current knowledge and evidence. Leukemia. (2019) 33:299–312. doi: 10.1038/s41375-018-0357-9
85. Koh Y, Park J, Ahn KS, Kim I, Bang SM, Lee JH, et al. Different clinical importance of FLT3 internal tandem duplications in AML according to FAB classification: possible existence of distinct leukemogenesis involving monocyte differentiation pathway. Ann Hematol. (2009) 88:1089–97. doi: 10.1007/s00277-009-0733-7
86. Thiede C, Steudel C, Mohr B, Schaich M, Schäkel U, Platzbecker U, et al. Analysis of FLT3-activating mutations in 979 patients with acute myelogenous leukemia: association with FAB subtypes and identification of subgroups with poor prognosis. Blood. (2002) 99:4326–35. doi: 10.1182/blood.V99.12.4326
87. Schlenk RF, Kayser S, Bullinger L, Kobbe G, Casper J, Ringhoffer M, et al. Differential impact of allelic ratio and insertion site in FLT3-ITD-positive AML with respect to allogeneic transplantation. Blood. (2014) 124:3441–9. doi: 10.1182/blood-2014-05-578070
88. Manara E, Basso G, Zampini M, Buldini B, Tregnago C, Rondelli R, et al. Characterization of children with FLT3-ITD acute myeloid leukemia: a report from the AIEOP AML-2002 study group. Leukemia. (2017) 31:18–25. doi: 10.1038/leu.2016.177
89. Annesley CE, Brown P. The Biology and Targeting of FLT3 in Pediatric Leukemia. Front Oncol. (2014) 4:263. doi: 10.3389/fonc.2014.00263
90. Marceau-Renaut A, Duployez N, Ducourneau B, Labopin M, Petit A, Rousseau A, et al. Molecular profiling defines distinct prognostic subgroups in childhood AML: a report from the French ELAM02 study group. Hemasphere. (2018) 2:e31. doi: 10.1097/HS9.0000000000000031
91. Bolouri H, Farrar JE, Triche T, Ries RE, Lim EL, Alonzo TA, et al. The molecular landscape of pediatric acute myeloid leukemia reveals recurrent structural alterations and age-specific mutational interactions. Nat Med. (2018) 24:103–12. doi: 10.1038/nm.4439
92. Pigazzi M, Manara E, Buldini B, Beqiri V, Bisio V, Tregnago C, et al. Minimal residual disease monitored after induction therapy by RQ-PCR can contribute to tailor treatment of patients with t (8; 21) RUNX1-RUNX1T1 rearrangement. Haematologica. (2015) 100:e99–101. doi: 10.3324/haematol.2014.114579
93. Rau R, Brown P. Nucleophosmin (NPM1) mutations in adult and childhood acute myeloid leukaemia: towards definition of a new leukaemia entity. Hematol Oncol. (2009) 27:171–81. doi: 10.1002/hon.904
94. Cazzaniga G. Dell'Oro MG, Mecucci C, Giarin E, Masetti R, Rossi V, et al. Nucleophosmin mutations in childhood acute myelogenous leukemia with normal karyotype. Blood. (2005) 106:1419–22. doi: 10.1182/blood-2005-03-0899
95. Brown P, McIntyre E, Rau R, Meshinchi S, Lacayo N, Dahl G, et al. The incidence and clinical significance of nucleophosmin mutations in childhood AML. Blood. (2007) 110:979–85. doi: 10.1182/blood-2007-02-076604
96. Sano H, Shimada A, Taki T, Murata C, Park MJ, Sotomatsu M, et al. RAS mutations are frequent in FAB type M4 and M5 of acute myeloid leukemia, and related to late relapse: a study of the Japanese Childhood AML cooperative study group. Int J Hematol. (2012) 95:509–15. doi: 10.1007/s12185-012-1033-x
97. Norsworthy KJ, Ko CW, Lee JE, Liu J, John CS, Przepiorka D, et al. FDA approval summary: mylotarg for treatment of patients with relapsed or refractory CD33-positive acute myeloid leukemia. Oncologist. (2018) 23:1103–8. doi: 10.1634/theoncologist.2017-0604
98. Zahler S, Bhatia M, Ricci A, Roy S, Morris E, Harrison L, et al. A phase i study of reduced-intensity conditioning and allogeneic stem cell transplantation followed by dose escalation of targeted consolidation immunotherapy with gemtuzumab ozogamicin in children and adolescents with CD33+ acute myeloid leukemia. Biol Blood Marrow Transplant. (2016) 22:698–704. doi: 10.1016/j.bbmt.2016.01.019
99. Tarlock K, Alonzo TA, Gerbing RB, Raimondi SC, Hirsch BA, Sung L, et al. Gemtuzumab ozogamicin reduces relapse risk in FLT3/ITD acute myeloid leukemia: a report from the children's oncology group. Clin Cancer Res. (2016) 22:1951–7. doi: 10.1158/1078-0432.CCR-15-1349
100. Niktoreh N, Lerius B, Zimmermann M, Gruhn B, Escherich G, Bourquin JP, et al. Gemtuzumab ozogamicin in children with relapsed or refractory acute myeloid leukemia: a report by Berlin-Frankfurt-Münster study group. Haematologica. (2019) 104:120–7. doi: 10.3324/haematol.2018.191841
101. Penel-Page M, Plesa A, Girard S, Marceau-Renaut A, Renard C, Bertrand Y. Association of fludarabin, cytarabine, and fractioned gemtuzumab followed by hematopoietic stem cell transplantation for first-line refractory acute myeloid leukemia in children: A single-center experience. Pediatr Blood Cancer. (2020) 67:e28305. doi: 10.1002/pbc.28305
102. Tomizawa D, Tsujimoto SI, Tanaka S, Matsubayashi J, Aoki T, Iwamoto S, et al. A phase III clinical trial evaluating efficacy and safety of minimal residual disease-based risk stratification for children with acute myeloid leukemia, incorporating a randomized study of gemtuzumab ozogamicin in combination with post-induction chemotherapy for non-low-risk patients (JPLSG-AML-20). Jpn J Clin Oncol. (2022). doi: 10.1093/jjco/hyac105
103. Pollard JA, Guest E, Alonzo TA, Gerbing RB, Loken MR, Brodersen LE, et al. Gemtuzumab ozogamicin improves event-free survival and reduces relapse in pediatric. J Clin Oncol. (2021) 39:3149–60. doi: 10.1200/JCO.20.03048
104. Dhunputh C, Strullu M, Petit A, Merched M, Pasquet M, Azarnoush S, et al. Single-dose (4. 5 mg/m. Br J Haematol. (2022) 198:373–81. doi: 10.1111/bjh.18203
105. Brivio E, Baruchel A, Beishuizen A, Bourquin JP, Brown PA, Cooper T, et al. Targeted inhibitors and antibody immunotherapies: novel therapies for paediatric leukaemia and lymphoma. Eur J Cancer. (2022) 164:1–17. doi: 10.1016/j.ejca.2021.12.029
106. Pearson ADJ, Zwaan CM, Kolb EA, Karres D, Guillot J, Kim SY, et al. Paediatric Strategy Forum for medicinal product development for acute myeloid leukaemia in children and adolescents: ACCELERATE in collaboration with the European medicines agency with participation of the food and drug administration. Eur J Cancer. (2020) 136:116–29. doi: 10.1016/j.ejca.2020.04.038
107. Willier S, Rothämel P, Hastreiter M, Wilhelm J, Stenger D, Blaeschke F, et al. CLEC12A and CD33 coexpression as a preferential target for pediatric AML combinatorial immunotherapy. Blood. (2021) 137:1037–49. doi: 10.1182/blood.2020006921
108. Zhang H, Wang P, Li Z, He Y, Gan W, Jiang H. Anti-CLL1 chimeric antigen receptor T-cell therapy in children with relapsed/refractory acute myeloid leukemia. Clin Cancer Res. (2021) 27:3549–55. doi: 10.1158/1078-0432.CCR-20-4543
109. Lonetti A, Pession A, Masetti R. Targeted therapies for pediatric AML: gaps and perspective. Front Pediatr. (2019) 7:463. doi: 10.3389/fped.2019.00463
110. Yu J, Jiang PYZ, Sun H, Zhang X, Jiang Z, Li Y, et al. Advances in targeted therapy for acute myeloid leukemia. Biomark Res. (2020) 8:17. doi: 10.1186/s40364-020-00196-2
111. Rubnitz JE, Kaspers GJL. How I treat pediatric acute myeloid leukemia. Blood. (2021) 138:1009–18. doi: 10.1182/blood.2021011694
Keywords: pediatric acute monoblastic/monocytic leukemia, immunophenotype, FAB-M5 AML, children, multiparametric flow cytometry, cytogenetics, molecular biology, cytomorphology
Citation: Varotto E, Munaretto E, Stefanachi F, Della Torre F and Buldini B (2022) Diagnostic challenges in acute monoblastic/monocytic leukemia in children. Front. Pediatr. 10:911093. doi: 10.3389/fped.2022.911093
Received: 01 April 2022; Accepted: 01 September 2022;
Published: 28 September 2022.
Edited by:
Alfonso Galderisi, Hôpital Necker-Enfants Malades, FranceReviewed by:
Marie C. Bene, Centre Hospitalier Universitaire de Nantes, FranceCopyright © 2022 Varotto, Munaretto, Stefanachi, Della Torre and Buldini. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Barbara Buldini, YmFyYmFyYS5idWxkaW5pQHVuaXBkLml0
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.