- 1Fondazione Policlinico Universitario A. Gemelli IRCCS, U.O.C. di Neonatologia, Dipartimento di Scienze della Salute della Donna, del Bambino e di Sanità Pubblica, Rome, Italy
- 2Istituto di Clinica Pediatrica, Università Cattolica del Sacro Cuore, Rome, Italy
- 3Dipartimento di Scienze di Laboratorio e Infettivologiche, Fondazione Policlinico Universitario A. Gemelli IRCCS, Rome, Italy
- 4Dipartimento di Scienze Biotecnologiche di Base, Cliniche Intensivologiche e Perioperatorie, Università Cattolica del Sacro Cuore, Rome, Italy
- 5Fondazione Policlinico Universitario A. Gemelli IRCCS, U.O.C. di Ostetricia e Patologia Ostetrica, Dipartimento di Scienze della Salute della Donna, del Bambino e di Sanità Pubblica, Rome, Italy
- 6Università Cattolica del Sacro Cuore, Istituto di Clinica Ostetrica e Ginecologica, Rome, Italy
- 7Department of Biomedical Sciences, Humanitas University, Milan, Italy
- 8IRCCS Humanitas Research Hospital, Milan, Italy
- 9Dipartimento di Scienze della Vita e Sanitá Pubblica, Universitȧ Cattolica del Sacro Cuore, Rome, Italy
The newborn’s microbiota composition at birth seems to be influenced by maternal microbiota. Maternal vaginal microbiota can be a determining factor of spontaneous Preterm Birth (SPPTB), the leading cause of perinatal mortality. The aim of the study is to investigate the likelihood of a causal relationship between the maternal vaginal microbiota composition and neonatal lung and intestinal microbiota profile at birth, in cases of SPPTB. The association between the lung and/or meconium microbiota with the subsequent development of bronchopulmonary dysplasia (BPD) was also investigated. Maternal vaginal swabs, newborns’ bronchoalveolar lavage fluid (BALF) (1st, 3rd, 7th day of life) and first meconium samples were collected from 20 women and 23 preterm newborns with gestational age ≤ 30 weeks (12 = SPPTB; 11 = Medically Indicated Preterm Birth–MIPTB). All the samples were analyzed for culture examination and for microbiota profiling using metagenomic analysis based on the Next Generation Sequencing (NGS) technique of the bacterial 16S rRNA gene amplicons. No significant differences in alpha e beta diversity were found between the neonatal BALF samples of SPPTB group and the MIPTB group. The vaginal microbiota of mothers with SPPTB showed a significant difference in alpha diversity with a decrease in Lactobacillus and an increase in Proteobacteria abundance. No association was found between BALF and meconium microbiota with the development of BPD. Vaginal colonization by Ureaplasma bacteria was associated with increased risk of both SPPTB and newborns’ BPD occurrence. In conclusion, an increase in α-diversity values and a consequent fall in Lactobacillus in vaginal environment could be associated to a higher risk of SPPTB. We could identify neither a specific neonatal lung or meconium microbiota profiles in preterm infants born by SPPTB nor a microbiota at birth suggestive of subsequent BPD development. Although a strict match has not been revealed between microbiota of SPPTB mother-infant couples, a relationship cannot be excluded. To figure out the reciprocal influence of the maternal-neonatal microbiota and its potential role in the pathogenesis of SPPTB and BPD further research is needed.
Introduction
Spontaneous preterm birth (SPPTB) represents the first cause of perinatal mortality and a huge public health cost in many countries, including Italy.
SPPTB is a complex multifactorial obstetric and neonatological complication, and little is known about its heterogeneous pathogenesis (1). Although a proven infection can only be documented in about 40% of preterm births with premature membrane rupture or spontaneous labor, the colonization of the genital tract by some pathogens and chorioamnionitis have been traditionally considered as the most prevalent causes of SPPTB.
Accumulating evidence suggests that the uterine environment is not sterile but populated by a set of different microbial communities present in the placenta, in the fetal membranes and in the amniotic fluid that could contribute to the formation of the neonatal microbiota before birth (2–4).
The formation of the microbiota begins early at the time of organogenesis and can promote protection or predisposition toward certain insults and pathologies (5–7).
Nevertheless, it is known that fetal and maternal colonization process can be influenced by several factors such as the mode of delivery [vaginal delivery (VD) vs. cesarean section (CS)] (8, 9), maternal bonding, admission to intensive care unit, administration of antibiotic therapy, H2 antagonists, dietary modifications with formulated milk, use of fortifying breast milk and fasting periods (10, 11). These are frequent circumstances in premature infants, who often undergo treatments that can alter the eubiotic colonization process.
The evolution of infants’ gut microbiota was a very investigated topic in recent years, while few researchers have addressed the development of the microbiota of the respiratory tract, particularly lower airways one. Pulmonary colonization starts in utero and can be subsequently influenced by the exposition to the birth canal and the skin of the mother, as well as by microorganisms, introduced through ventilation or micro-aspiration (2, 8, 12–14). The lung microbiota shows changes in the first week of life (15, 16) and an evolution of the colonization pattern is found over the first week of life with evidence of the prevalence of organisms of the species Staphylococcus sp. (Firmicutes) or Ureaplasma spp. (Tenericutes) within 7 days (17).
Furthermore, lung microbiota can be influenced by lung diseases, particularly by the development of bronchopulmonary dysplasia (BPD) (16).
The aim of the study is to investigate the role of maternal and neonatal microbiota in SPPTB cases. We investigated maternal vaginal microbiota, and neonatal lung and intestinal microbiota. We compared preterm infants born by spontaneous birth for suspected chorioamnionitis and/or premature prolonged rupture of the membranes (p-PROM) and/or preterm labor (SPPTB group) with preterm infants born by cesarean section performed for maternal or fetal medical indications, with intact membranes and in the absence of labor (Medically Indicated Preterm Birth–MIPTB).
The identification of a specific maternal and preterm newborns microbiota signature associated to SPPTB and to adverse neonatal outcomes, could open to the investigation of new markers (a) in pregnancy, to select cases at higher risk of SPPTB and (b) in preterm newborns, to early identify cases at higher risk of adverse pulmonary outcomes.
Materials and Methods
Setting
We conducted a prospective observational study at the Department of Woman and Child Health of the Fondazione Policlinico Universitario A. Gemelli IRCCS of Rome (Italy), from August 2019 to August 2020. The study protocol was approved by the Institutional Ethics Committee in July 2019 (study ID 2400, protocol number 0031183/19). Written parental consent was obtained prior to study entry.
Study Population and Inclusion Criteria
The study population included women undergoing PTB and their preterm infants having gestational age (GA) ≤ 30 weeks, who were intubated at birth or in the first 24 h of life.
As this was a pilot study, sample size was not calculated. As in 2018, 58 newborns with a GA ≤ 30 weeks were admitted to the Neonatal Intensive Care Unit (NICU), of them 26 (45%) were born for suspected chorioamnionitis and/or p-PROM and/or preterm labor (SPPTB group) and 32 (55%) by elective C-section performed for maternal or fetal medical indications with intact membranes in the absence of labor (MIPTB group). 70% (18/26) of newborns in SPPTB group and 57% (18/32) in MIPTB group were intubated at birth or in the first 24 h of life; we estimated the possibility of studying up to 36 newborns over a period of about 12 months. The number of infants we intend to study is in line with the available literature’s evidence for a pilot study (18).
The study population was furtherly divided into two groups:
SPPTB group: preterm infants for suspected chorioamnionitis and/or p-PROM and/or preterm labor; and MIPTB group: preterm infants born by CS for maternal or fetal medical indications, with intact membranes and in the absence of labor. BPD was defined as O2-dependence at 36 weeks of post menstrual age (19).
Timing and Technique of Samples Collection
A vaginal swab was collected at the time of delivery for all the enrolled women. Newborns’ bronchoalveolar lavage fluid (BALF) and meconium specimens were collected. BALF were obtained after ensuring that the infant was adequately oxygenated by instilling 1 ml/kg of 0.9% sodium chloride in the endotracheal tube and suctioning the fluid into a sterile mucus trap. The first BALF sample was collected within the first 24 h of life, whereas further collections were performed on the third and seventh day of life in newborns who were still intubated.
Vaginal swab, BALF and meconium were stored at −80°C until further processing.
Data Collection
Data was extracted from electronic medical records. The cause of preterm birth was recorded to distinguish SPPTB infants and MIPTB ones.
Data collection included both prenatal and neonatal data. Among (a) prenatal data: results of previous cultural exams of vaginal swab and/or maternal urine culture; use of antibiotics during pregnancy; prescription of intrapartum antibiotics prophylaxis; prenatal steroids prophylaxis (defined as the administration of two doses of betamethasone within 24 h of each other, with the second dose administered at least 24 h but no more than 7 days before delivery), (b) neonatal data: gestational age at delivery, birth weight, incidence of neonatal BPD (defined as O2-dependence at 36 weeks of post menstrual age), characteristics of the neonatal respiratory pathology (number of the administered doses of surfactant, incidence of persistent pulmonary hypertension requiring treatment, incidence of successful extubation, defined as the lack of need for new tracheal intubation in the 72 h following extubation), duration of ventilatory support (hours of oxygen therapy, hours of mechanical ventilation, hours of non-invasive ventilation), time until initiation of antibiotic therapy, number of antibiotics cycles, time until initiation of oral feeding, incidence of intraventricular hemorrhage (IVH) grade > 2°, length of stay and survival, in addition to the perinatal infectious risk factors.
The incidence of blood stream infections, defined as a positive blood culture, and pneumonia, defined as the worsening respiratory symptoms (increase of FiO2 and/or of the ventilatory setting, increase of secretions from the endotracheal tube, persistent radiographic anomalies ≥ 24 h) with positive BALF culture, were also registered.
Sequencing and Bioinformatic Analysis
For each sample (Vaginal swabs, BALF and meconium), bacterial DNA extraction was performed in a strictly controlled level-2 biological safety workplace using DANAGENE MICROBIOME DNA kits (Danagen-Bioted) according to manufacturer’s instructions (20). DNA was fluorometrically quantified (Qubit dsDNA high-sensitivity assay, Thermo Fisher Scientific), and then subjected to the 16S rRNA V3–V4 region amplification as described above (21, 22). The resulting amplicons were purified using Agencourt AMPure XP beads (Beckman Coulter) and indexed using the Nextera XT Index kit (Illumina). The indexed amplicons were equimolarly diluted and pooled. Sequencing was performed via the 2 × 300-bp paired-end protocol in the MiSeq instrument (Illumina).
Demultiplexed FastQ reads were analyzed using the QIIME2 (v.2020.6) microbiome analysis pipeline (23). Briefly, merged reads were trimmed before filtering and chimera removal to generate amplicon sequence variants (ASVs) using the DADA2 algorithm (24). Both the pre-fitted sklearn-based taxonomy classifier1 and SILVA 132 database2 were used for taxonomic ASVs’ annotation. Final data were pre-processed to remove mitochondrial sequences (25). We used R 4.0.23 and phyloseq (26) statistical packages for downstream analyses of alpha (e.g., Shannon index) and beta (e.g., Jaccard distance) microbial community’s diversity. Before that, we normalized to median sum count each sample in order to restrict uneven sampling effects. Difference between groups according to alpha diversity metric was assessed using the Mann–Whitney U-test, whereas that according to Jaccard distance matrix-computed beta diversity metric was assessed using the permutational multivariate analysis of variance (PERMANOVA).
Statistical Analysis
Statistical analysis was done using GraphPad PRISM Version 8.4.3 considering a value of p < 0.05 as statistically significant.
Categorical variables were compared using a two-tailed Fisher’s exact test, while the differences between the groups for continuous variables were tested by Mann-Whitney U-test for non-parametric data and the Student’s t-test for parametric data.
Results
From August 2019 to August 2020, a total of 30 mothers and 35 newborns were enrolled at birth after receiving informed parental consent. Six mother-newborns couples were excluded because newborns were not intubated in the first 24 h of life. The study group consisted of 29 newborns, including 3 pairs of twins, and 26 mothers.
Twenty-nine BALF, 29 early meconium and 26 vaginal swabs samples were analyzed. Six BALF samples were excluded due to the low bacterial biomass. A total of 20 mothers (11 = SPPTB; 9 = MIPTB) and 23 neonates (12 = SPPTB; 11 = MIPTB), were included in the final statistical analysis.
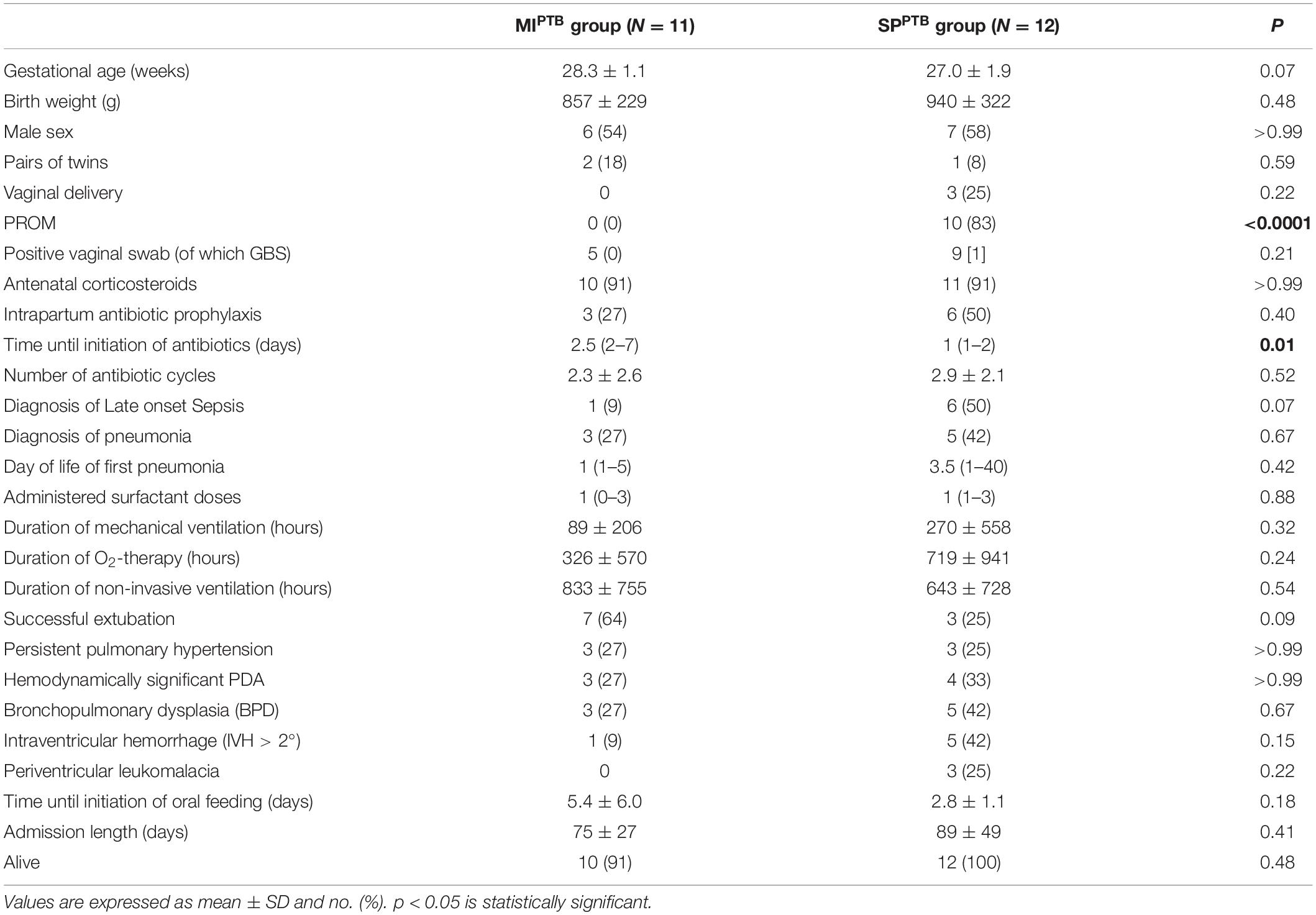
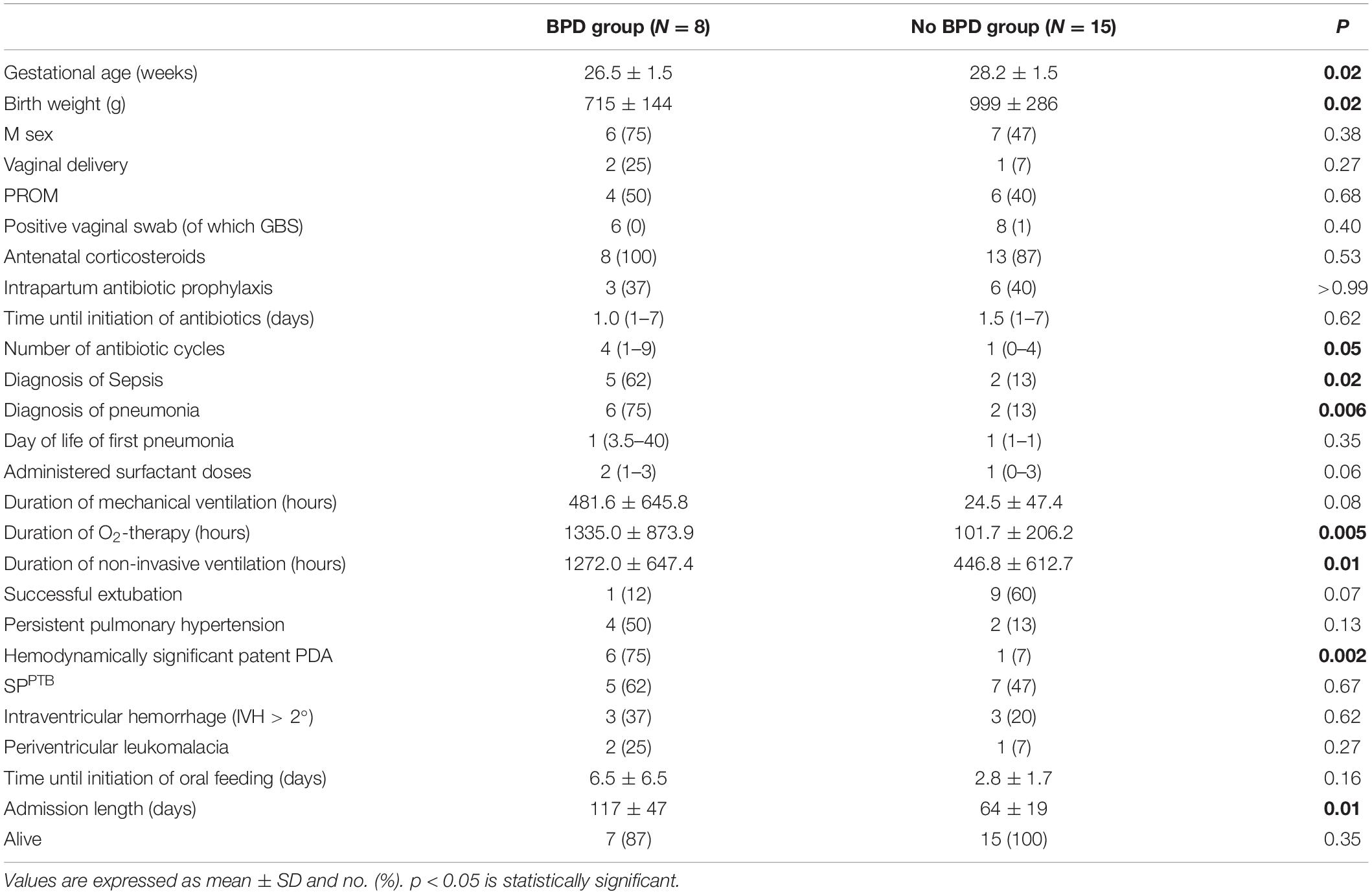
The demographics and clinical characteristics of infants included in the analysis are shown in Table 1. No significant differences were observed between the two groups, except for earlier initiation of antibiotic therapy in infants of the SPPTB group [1 (1–2) vs. 2.5 (2–7) days of life, p = 0.01] and for the occurrence of premature prolonged rupture of the membranes (pPROM) which only registered in the SPPTB group (p < 0.0001). In this group of neonates, only in two cases labor was activated without rupture of the membranes. A trend in terms of lower gestational age (27.0 ± 1.9 vs. 28.3 ± 1.1 weeks, p = 0.07) and of higher incidence of late-onset sepsis [6 (50%) vs. 1 (9%), p = 0.07] was found in the SPPTB group compared to the MIPTB group. All 23 infants survived to 36 weeks’ postmenstrual age (only 1 infant in MIPTB died after 36 weeks postmenstrual age). Based on their pulmonary outcomes, the 23 infants were further classified as those who did not develop BPD (No BPD group, n = 15) or those who later developed BPD (BPD group, n = 8). The demographics and clinical characteristics of patients in these two categories are shown in Table 2, with significant differences in some characteristics and outcomes. In particular, infants in the BPD group had a lower GA and birth weight, had a greater number of episodes of blood stream infection and pneumonia during hospitalization and received more cycles of antibiotic therapy. In addition, infants in the BPD group needed oxygen therapy and non-invasive ventilation for a longer time than infants in the No BPD group, had a hemodynamically significant patent ductus arteriosus (PDA) more frequently, and their hospital stay was significantly longer.
Lung and Meconium Microbiota of Preterm Infants at Birth
A total of 38 BALF samples were collected: 23 were collected during the first 24 h of life, 10 on day 3 of life and 5 on day 7 of life.
We evaluated possible association between birth cause and lung microbiota composition investigating bacterial communities on both SPPTB and MIPTB groups in BALF specimens at day 1 post birth in intubated infants. A total of 1,146,747 high-quality sequences comprising 79 unique ASVs were obtained. All 38 sequenced samples showed yielded ≥ 1,000 sequences with a mean of 30,177 and median of 23,340, range 3,380–141,435 reads/sample. Overall, 6 main phyla were detected: Actinobacteria, Bacteroidetes, Cyanobacteria, Fusobacteria, Firmicutes, and Proteobacteria. In both groups, Firmicutes and Actinobacteria phyla prevailed and showed a comparable relative abundance average of 77 and 15% or 73 and 13% for SPPTB and MIPTB, respectively. Conversely, Proteobacteria and Bacteroidetes were less represented, and Fusobacteria and Cyanobacteria were detected only in few specimens (Figure 1A). Similarly, no significant differences were observed at genus level where Paenibacillus or Staphylococcus genera were predominant (Figure 1B).
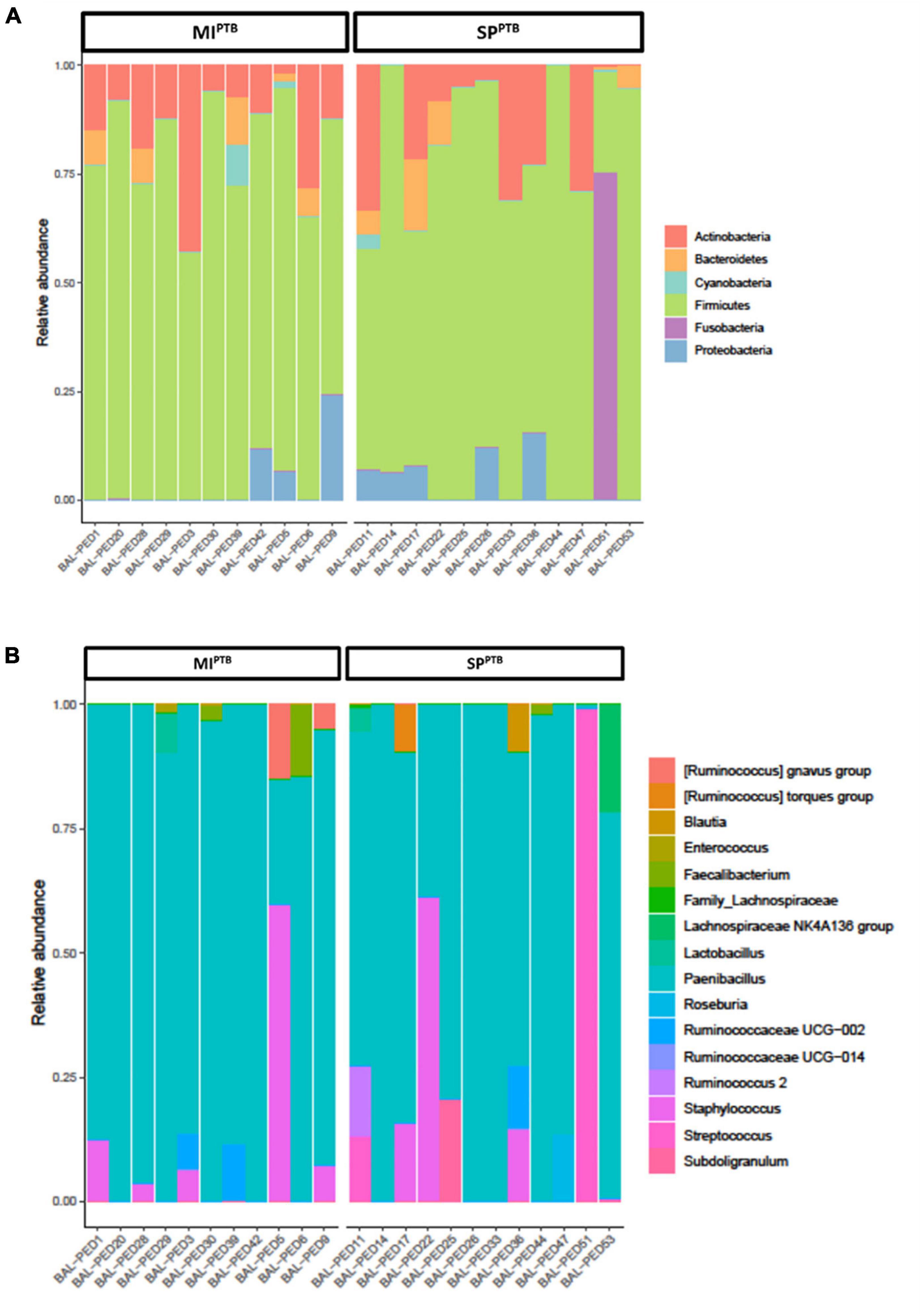
Figure 1. Relative abundances of bacterial taxa composing the lung bacterial communities in hospitalized newborns. For each sample of the MIPTB and SPPTB infants’ groups, proportions for major phyla (A) and major genera (B) were computed, normalized, and presented as stacked bar plots. The statistical significance at a P-value of ≤ 0.05 was assessed using the Mann–Whitney U-test.
Alpha diversity, measured by Shannon Diversity Index, and beta diversity, as Jaccard distance, were evaluated in SPPTB and MIPTB groups, as reported in Figures 2A,B. Shannon index showed comparable values in both groups: 1.85 ± 0.86 and 1.74 ± 0.42 for SPPTB and MIPTB (p > 0.05), respectively. Three couples of twins were included in the study: one in the SPPTB and two in the MIPTB group. Brothers had different values of alpha diversities: 1.22 and 1.69 for twins in the SPPTB group; 1.35 and 0.89 and 1.29 and 0.92 for twins in the MIPTB group. Turning to Beta diversity, these samples showed a different spatial distribution highlighting the importance of other factors in the establishment of a unique microbial composition. No difference in spatial distribution of the SPPTB and MIPTB specimens was described in beta diversity analysis.
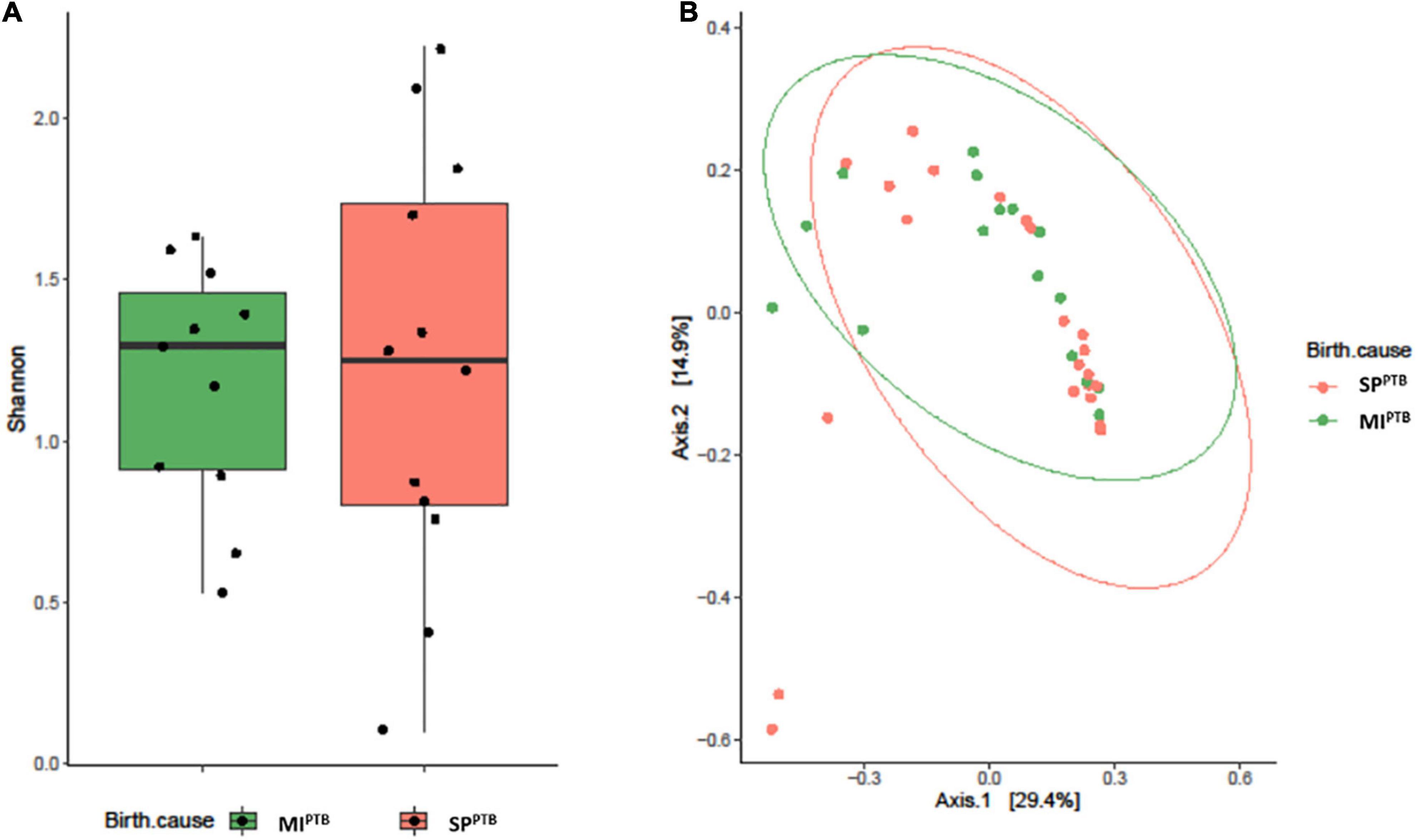
Figure 2. Alveolar microbiota Alpha and Beta diversity analysis of preterm newborns classified on the basis of preterm birth subtype (MIPTB and SPPTB). Alpha diversity was evaluated by Shannon index measure. The values obtained were compared, resulting in no statistically significant difference between MIPTB and SPPTB infants’ groups (Mann–Whitney U-test) (A). Comparison for Beta diversity analysis was made using Jaccard distance. The principal coordinate analysis (PCoA) results are presented as two-dimensional ordination plots, which were generated using two principal coordinates (i.e., axis 1 and axis 2) (B). The statistical significance at a P-value of ≤ 0.05 was assessed using the permutational multivariate analysis of variance (PERMANOVA).
Ten on twenty-three newborns were still intubated on the third day after birth and a second BALF sample was collected (3 in the SPPTB and 7 in the MIPTB group, respectively). Alpha diversity as Shannon index was computed showing no differences between days 1 and 3. At day 3 after birth the average value of Shannon index was equal to 1.80 in the SPPTB group vs. 1.61 MIPTB in the group, p = 0.65) (Figure 3). Likewise, no differences were observed in terms of relative abundances of the detected species (data not shown) suggesting that no main changes occurred after birth at days 1–3.
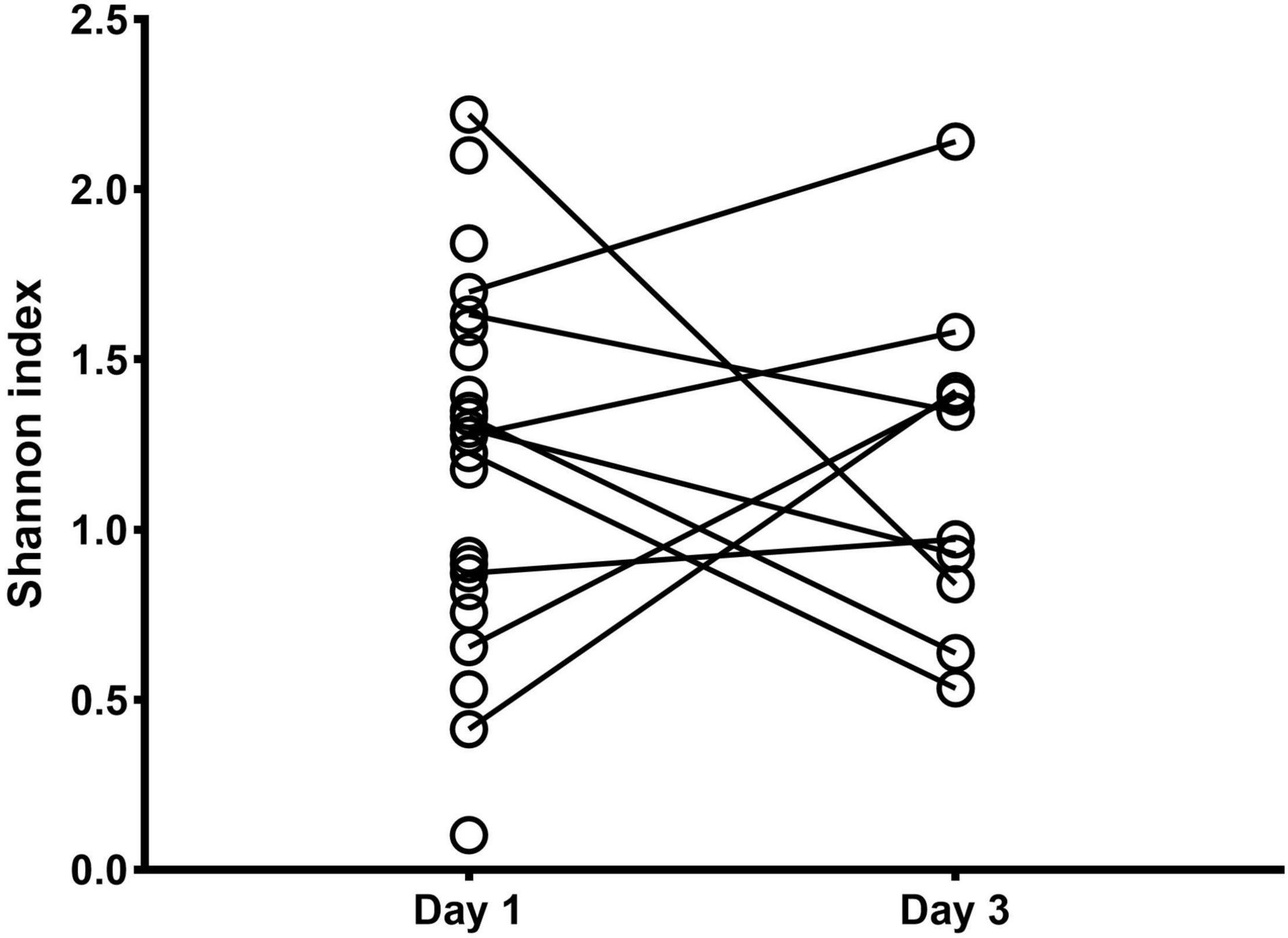
Figure 3. Alpha diversity analysis of the lung bacterial communities in newborns at days 1 and 3 after birth. Shannon index was measured on bronchoalveolar lavage samples at days 1 and 3 after intubation for MIPTB and SPPTB infants’ groups.
The analysis was limited to the BALF samples of the first and third day of life due to the low number of BALF samples at day 7 (a total of five specimens: 3 in the SPPTB and 2 in the MIPTB group, respectively).
In our cohort, eight out of 23 infants developed BPD, so that we investigated lung microbial communities in infants who developed BPD vs. those who did not develop BPD. By analyzing the BALF samples on day 1 of life, both BPD and No BPD groups showed a predominance of Firmicutes and Actinobacteria in addition to the presence of Proteobacteria, Fusobacteria, Cyanobacteria, and Bacteroidetes without a characteristic microbiological signature, linked to disease development (Figure 4A). Similarly, no differences at genus level were detected (Figure 4B). No significant differences were underlined by alpha diversity and beta diversity analysis (Figure 5).
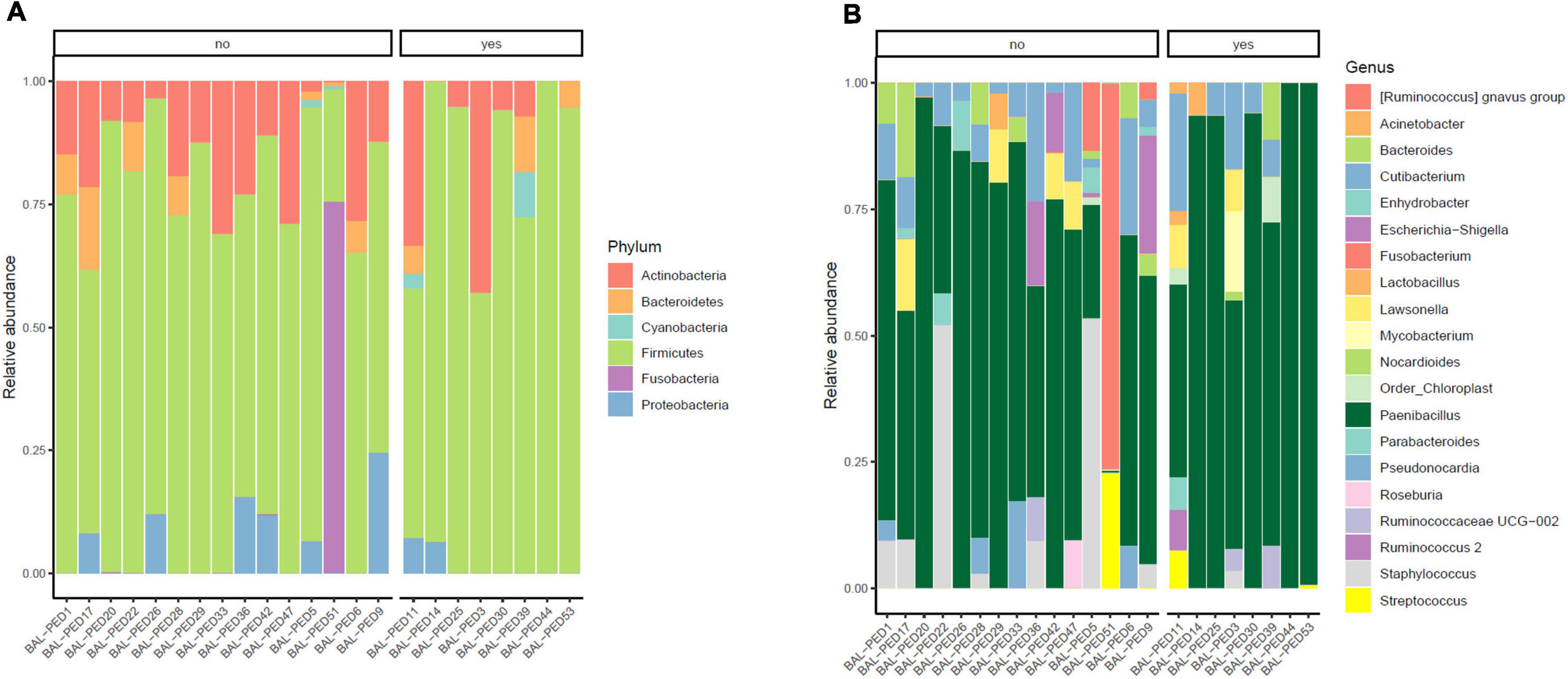
Figure 4. Relative abundances of bacterial taxa composing the lung bacterial communities in hospitalized newborns. For each sample of the BPD and noBPD groups, proportions for major phyla (A) and major genera (B) were computed, normalized, and presented as stacked bar plots. The statistical significance at a P-value of ≤ 0.05 was assessed using the Mann–Whitney U-test.
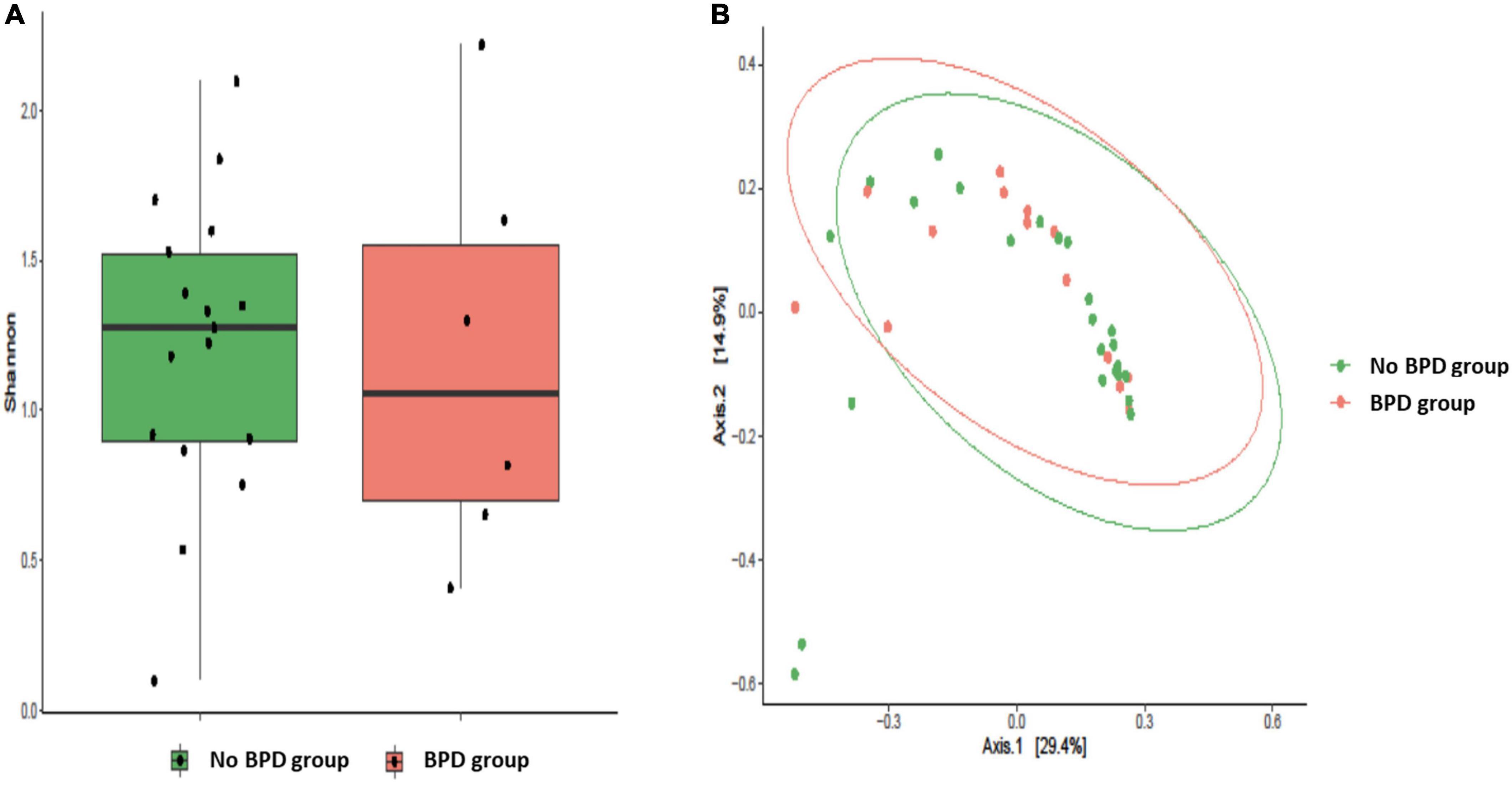
Figure 5. Alveolar microbiota Alpha and Beta diversity analysis of preterm newborns classified on the basis of Bronchopulmonary Dysplasia (BPD) diagnosis. Alpha diversity was evaluated by Shannon index measure. The values obtained were compared, resulting in no statistically significant difference between No BPD and BPD groups (Mann–Whitney U-test) (A). Comparison for Beta diversity analysis was made using Jaccard distance. The principal coordinate analysis (PCoA) results are presented as two-dimensional ordination plots, which were generated using two principal coordinates (i.e., axis 1 and axis 2) (B). The statistical significance at a P-value of ≤ 0.05 was assessed using the permutational multivariate analysis of variance (PERMANOVA).
Finally, we harvested meconium specimens for each infant to search for a microbial signature related to SPPTB and MIPTB. Intriguingly, we were not able to detect any signal after 16S rDNA V3-V4 amplification.
Vaginal Swabs
In order to identify a specific microbial signature in MIPTB and SPPTB, we investigated the mothers’ vaginal microbiota. Twenty vaginal swabs were analyzed for vaginal microbiota composition (9 in the MIPTB due to the presence of two pairs of twins and 11 in the SPPTB due to the presence of a pair of twins). A total of 4,056,491 high-quality sequences comprising 7,145 unique ASVs were obtained. All 20 sequenced samples showed yielded ≥ 17,000 sequences with a mean of 202,824 and median of 224,678, range 17,214–349,108 reads/sample. After removing taxa with prevalence lower than 0.01%, a total of 3,590,745 reads were obtained 511 ASVs.
We found that Shannon Diversity index was significantly higher in SPPTB compared to MIPTB (4.63 ± 0.78 vs. 1.98 ± 0.94, respectively, p < 0.05) suggesting that variation in bacterial population was associated with the two groups (Figure 6A). Particularly, SPPTB group showed an increased Pielou’s evenness (0.79 ± 0.13) in comparison with the MIPTB group (0.34 ± 0.15) that indicated an increasing prevalence of some bacterial species in the latter group (data not shown). To evaluate the similarity of microbiota profiles, Jaccard beta diversity was computed and plotted as PCoA graph. Interestingly, microbial communities appeared diversely distributed with a minimal overlap between SPPTB (red) and MIPTB (green) samples. Nonetheless, individual profiles of the two groups cluster on different spatial level as portrayed by ellipses. PERMANOVA statistic test confirmed our representation (p < 0.05) (Figure 6B).
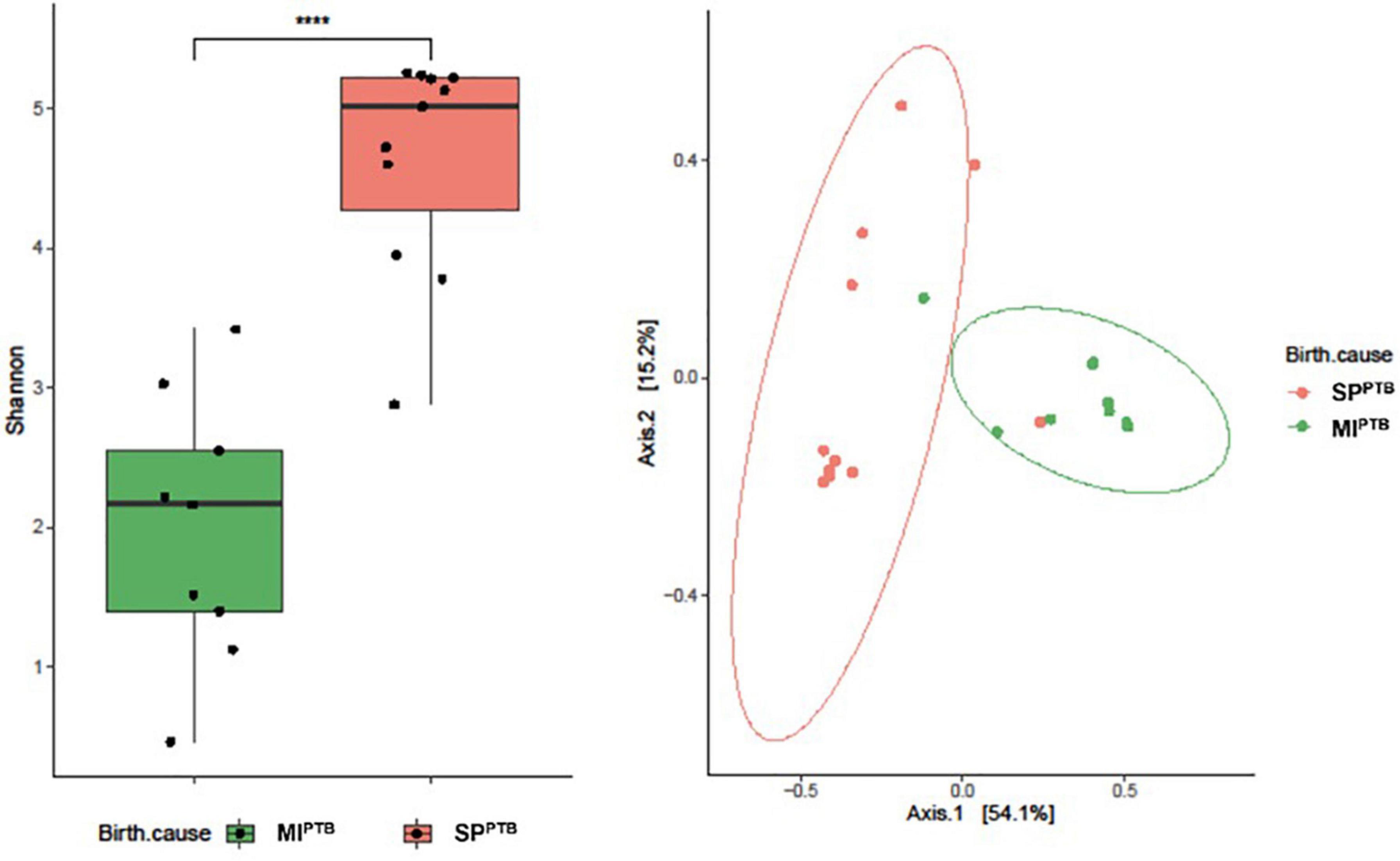
Figure 6. Alpha (A) and Beta (B) diversity analysis of the vaginal bacterial communities of the newborns’ mothers, classified on the basis of preterm birth subtype (MIPTB and SPPTB). Alpha diversity was evaluated by using Shannon index measure, and the values for MIPTB and SPPTB infants were compared (A). The statistical significance at a P-value of ≤ 0.05 (****) was assessed using the Mann–Whitney U-test. Beta diversity analysis was measured using Jaccard distance, computed for MIPTB and SPPTB groups. The principal coordinate analysis (PCoA) results are presented as two-dimensional ordination plots, which were generated using two principal coordinates (i.e., axis 1 and axis 2). The statistical significance at a P-value of ≤ 0.05 was assessed using the permutational multivariate analysis of variance (PERMANOVA).
Both SPPTB and MIPTB groups showed a predominance of Firmicutes, Proteobacteria, Actinobacteria, and Bacteroidetes. However, there was a characteristic feature in the microbiota of women who have had a delivery for SPPTB vs. the microbiota in women who have had a cesarean section for MIPTB. Indeed, SPPTB group showed a significant decreasing in Firmicutes (54% vs. 79%, p = 0.0031) and the consequent increasing Bacteroidetes (31% vs. 9%, p = 0.00002). Intriguingly, a slight increasing in relative abundance of Proteobacteria (5.5% vs. 1.3%), rather than Actinobacteria (showing an average around 10% for both groups), was observed in MIPTB group (Figure 7A).
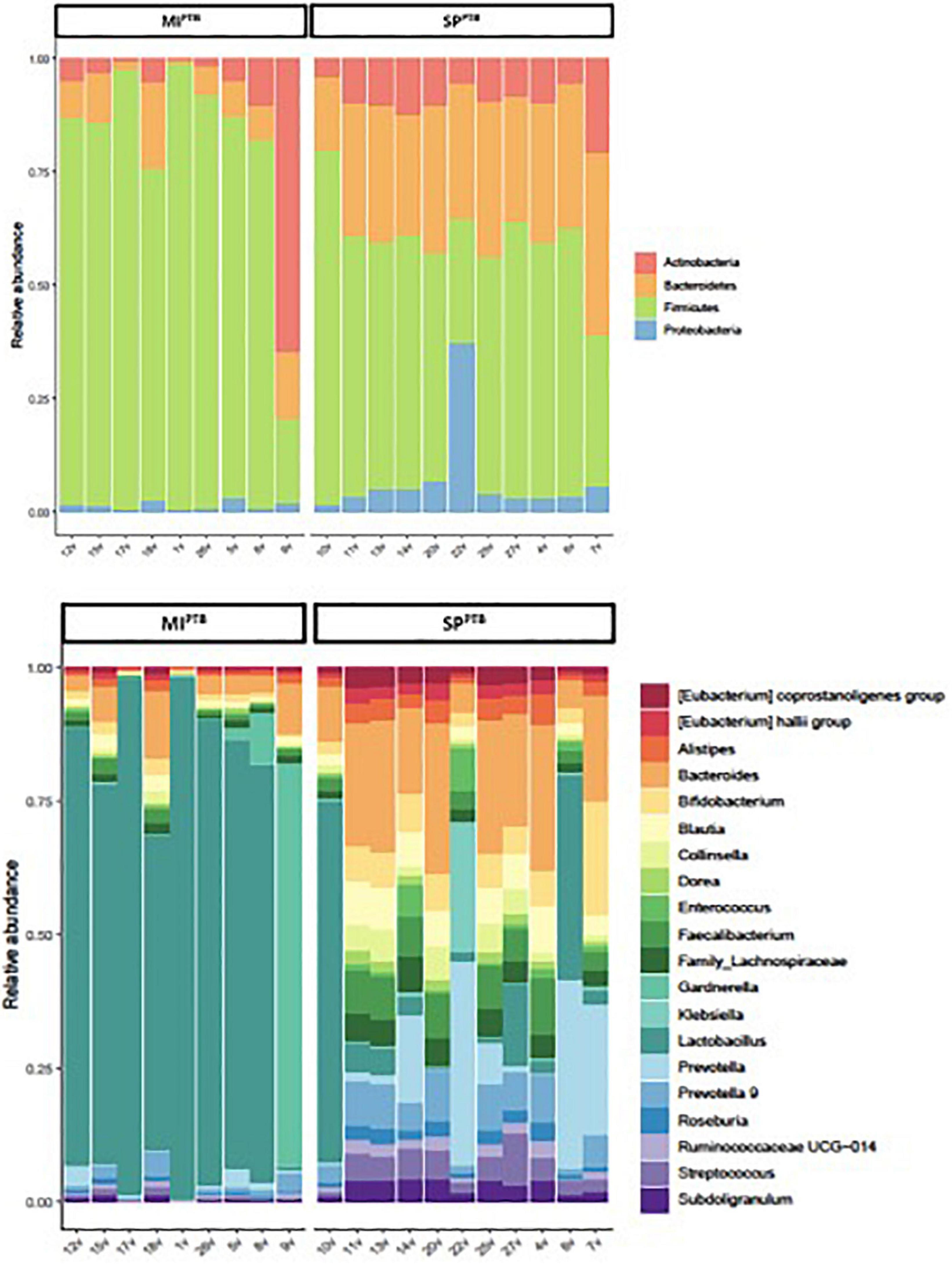
Figure 7. Relative abundances of bacterial taxa composing the newborns’ mothers vaginal flora, classified on the basis of preterm birth type (MIPTB and SPPTB). For MIPTB and SPPTB groups, proportions for major phyla (A) and major genera (B) were computed, normalized, and presented as stacked bar plots, respectively. The statistical significance at a P-value of ≤ 0.05 was assessed using the Mann–Whitney U-test.
As expected, Lactobacillus genus was decreased in SPPTB group vaginal swabs in comparison with MIPTB group (Figure 7B). Relative abundances of the other genera consequently showed an increasing.
Cultural and Molecular Analyses Results
Among BALF samples collected on the first day of life, only 2 of them were positive for Ureaplasma parvum. Ureaplasma parvum was molecularly identified in only one BALF collected on the 7th day of life. All three newborns with positive BALF for Ureaplasma parvum developed BPD, with Ureaplasma parvum BALF positivity statistically correlating to the development of BPD (p = 0.03, data not shown in Table 2). The vaginal swabs of the mothers of Ureaplasma parvum-positive infants also showed positivity for the pathogen.
Culture of all other BALF specimens were negative, except for BALF on days 3 and 7 of a BPD infant who tested positive for Staphylococcus haemolyticus.
Concerning vaginal swab exams, in SPPTB group 9 out of 12 (75%) vaginal swabs were positive (4 = Ureaplasma parvum, 1 = Klebsiella pneumoniae, 2 = Candida albicans, 1 = Streptococcus agalactiae, 1 = Escherichia coli), in MIPTB group 5 out of 11 (45%) vaginal swabs were positive (3 = Ureaplasma parvum, 1 = Escherica coli, 1 = Citrobacter koseri).
Considering BPD/No-BPD groups, 4 vaginal swabs in the BPD group (50%) and 3 in the No-BPD group (20%) were positive for Ureaplasma parvum (p = 0.18, data not shown in Table 2). In the BPD group a vaginal swab was positive for Citrobacter koseri and another one for Klebsiella pneumoniae, while 2 vaginal swabs were negative (25%). In the no-BPD group 7 vaginal swabs were negative (47%), while, in addition to the three Ureaplasma parvum positive vaginal swabs, 2 were positive for Escherichia coli, 1 for Streptococcus agalactiae and 2 for Candida albicans. The cultures of the meconium samples were all negative.
Discussion
The present paper aims to identify specific vaginal and neonatal lung microbiota profiles associated to the occurrence of SPPTB and MIPTB. As preterm births have been historically considered a sign of infection, we compared SPPTB cases to MIPTB cases, to understand if an adverse uterine environment could influence preterm birth and neonatal lung microbiota.
Chorioamnionitis, defined as inflammation of fetal membranes, is one of the mechanisms leading to preterm labor. A clinical diagnosis can be made prenatally by maternal findings (fever, uterine contractions, leucocytosis, high serum levels of C-reactive protein, abnormal vaginal secretions), while a pathological diagnosis is obtained postnatally by histological examination of the placenta and the amniochorionic membranes (27). A proven microorganism infection can be detected only in 50% of cases of chorioamnionitis, probably due to the technical limitations of cultural tests. Microbiota profiling by NGS technique might represent a more sensitive diagnostic tool to identify dysbiosis and increased risk of SPPTB (28, 29).
Recent studies challenged the assumption of a sterile uterine environment, demonstrating the presence of bacteria in the placenta, fetal membranes, and amniotic fluid (2–4). However, these findings are controversial as there is the risk of contamination of low microbial biomass samples.
Vaginal dysbiosis is recognized as a factor that increases the risk of SPPTB. Several species of Lactobacilli promote the maintenance of an acid pH and the production of antimicrobial substances.
During pregnancy there is a progressive increase in abundance of species such as Lactobacillus vaginalis, L. crispatus, L. gasseri, and L. jensenii, belonging to the Phylum of the Firmicutes. Conversely, bacteria such as Prevotella, Sneathia, Gardnerella Vaginalis, Ruminococcaceae, Parvimonas, and Mobiluncus, often associated with bacterial vaginosis and widely present in non-pregnant women of childbearing age, progressively decrease (30). Hence, the microbiota of women in pregnancy becomes progressively more stable, less prone to changes and protective against genitourinary tract infections (31).
Several studies have shown that greater α-diversity of vaginal microbiota is associated with a progressive reduction in the relative abundance of the genus Lactobacillus, which is known for its role in giving stability to the vaginal microbiota and protecting it from pathogenic insults.
Consistently with previous literature, in this study we documented a greater alpha diversity of vaginal microbiota of the SPPTB mothers when compared with mothers of the MIPTB group. Moreover, in the SPPTB group, vaginal swabs revealed a decrease in Firmicutes and an increase in abundance of Bacteroidetes. At Genus analysis, a significant decreased abundance of Lactobacillus was found in SPPTB.
The timeline of lung microbial colonization is highly debated. Mourani and colleagues reported the presence of detectable bacterial DNA in only 2 of 10 tracheal aspirates of intubated preterm infants in the first 72 h of life, whereas all samples from the same newborns were positive at day 7 (17). On the other hand, Lohmann et al. found the presence of bacterial DNA in all tracheal aspirates collected in 25 neonates immediately after orotracheal intubation on the first day of life (16). Lal et al. observed that BALF samples of extremely low birth weight infants exposed to chorioamnionitis showed a decreased genus Lactobacillus at birth, which has been associated with development of BPD.
All cases of chorioamnionitis were determined by positive placental histopathology and prenatal antibiotics administration in mothers (32).
We investigated any possible cause of spontaneous birth and possible adverse uterine environment (2 mothers in SPPTB had chorioamnionitis and 10 had a p-PROM) looking for a microbial signature suggestive of spontaneous birth. However, despite differences observed in maternal vaginal swabs, we did not find in neonatal BALF samples at birth a different pattern of alpha and beta diversity and/or differences either at Phylum and Genus level, comparing SPPTB and MIPTB group.
A problem underlying the analysis of neonatal lung microbiota is the difficulty in retrieving material from the lungs and distal airways in neonatal population, due to the low biomass with bacterial loads close to the detection limit of the assays and to the high risk of contamination from the upper airways (33). In these terms, lung microbiota at first day of life could be significantly impaired by contamination during sampling and by sequencing technique. Moreover, samples derived from sites with a low microbial biomass can give results which are difficult to distinguish from DNA traces present in reagents used for extraction, amplification, and sequence library preparation for molecular microbiology studies (34, 35). Hence, we cannot exclude that our findings are the results of high sensitivity of sequencing process and sampling.
Indeed, the presence of bacterial DNA in BALF samples, while lending support to the hypothesis that the process of microbiota establishment might begin antenatally, does not exclude the possibility of post-birth microbiome development, impaired by environmental factors, or sample contaminations.
On the other hand, the lack of bacterial DNA in the analyzed meconium samples (collected before the start of enteral feeding, with no probiotic intake and in the absence of abdominal pathological processes) supports the idea that microbiota composition is acquired after birth, and it is not related with the cause of preterm birth or transferred from swallowing bacteria previously translocated from uterine niche in the amniotic liquid.
The absence of a specific microbial signature in the BALF samples of the two groups (MIPTB vs. SPPTB) may, at least in part, suggest that the cause of preterm birth was not associated with infant lung microbiota composition.
While the two groups differed for some clinical characteristic as GA and birth weight, the analysis conducted on BALF samples of infants who developed or not BPD, did not show statistically significant differences.
Furthermore, the newborns of the BPD group had a higher incidence of hemodynamically significant PDA, blood stream infections and pneumonia during hospitalization and, consequently, are subjected to a greater number of antibiotic therapy cycles (Table 2). However, the two groups did not differ in the use of intrapartum antibiotic prophylaxis, nor in the time elapsed from birth to the start of antibiotic therapy. This may partly justify the lack of a statistically different expression of the microbiota in BALF samples taken in the first 24 h of life, and certainly more information could have been obtained by having a greater number of BALF samples available on the third, and seventh day and in the more advanced periods of life.
Although not statistically significant, a slight reduction in Shannon diversity index is evident in the BALF samples of BPD infants (Figure 5). This trend confirms the results obtained by Lohmann et al. (16). The authors identified a reduction in the number of species of respiratory microflora and Shannon diversity index in the BALF samples obtained in the first 24 h of life from 10 infants diagnosed with BPD compared to 15 other infants who had not developed the disease. A decrease in microecological diversity of the respiratory tract in BPD was observed also by Lal et al. (32) who analyzed 18 newborns with confirmed BPD, of which 5 infants with an extremely low birth weight.
We did not find a characteristic microbiological signature linked to BPD development both at genus and phylum level. However, the results of the studies by Lohmann et al. and Lal et al. showed opposite trends in Proteobacteria (a downward trend in the results of Lohmann et al. compared to an increase in Lal et al.) and Firmicutes abundances (with an upward trend in Lohmann et al. compared to a decrease in Lal et al.).
Collaterally, from cultural analysis, positive Ureaplasma parvum day-one BALF culture resulted significantly more prevalent in BPD group. Moreover, all neonates with positive day-one BALF were born form mother with positive vaginal swabs for Ureaplasma. This result is in line white previous studies, which associated Ureaplasma colonization in the respiratory tract with the subsequent development of BPD (16, 17, 36, 37).
However, there is still not a total agreement on the relationship between respiratory Ureaplasma colonization and BPD development, as well as there is not agreement on the benefit or risk of antibiotic therapy for respiratory Ureaplasma colonization (38).
We are aware that our study may have several limitations, as the small study population, given the difficulty of obtaining vaginal swabs timely before birth and the difficulty of sampling from the distal airways in the neonatal population. Other limitations of the study are represented by the lack of analysis of the placental microbiota and the lack of a histological diagnosis of chorioamnionitis. The implementation of these analysis, in association with further studies on amniotic fluid microbiota are needed to investigate the presence of a link between maternal and neonatal microbiota.
Conclusion
An increase in α-diversity values and a consequent fall in Lactobacillus in vaginal environment could be associated to a higher risk of SPPTB, probably due to a less stable and protective environment, suggesting that variation in maternal microbiota can predispose to SPPTB.
We could identify neither a specific neonatal lung or meconium microbiota profiles in preterm infants born by SPPTB nor a microbiota at birth suggestive of subsequent BPD development. However, accordingly to previous studies, the presence of Ureaplasma bacteria in newborns’ BALF could be considered a risk factor for the development of BPD.
Although a strict match has not been revealed between microbiota of SPPTB mother-infant couples, a relationship cannot be excluded.
To figure out the reciprocal influence of the maternal-neonatal microbiota and its potential role in the pathogenesis of SPPTB and BPD further research is needed.
Data Availability Statement
The data presented in this study are deposited in the sequence read archive (SRA) and are available as BioProject number PRJNA849991.
Ethics Statement
The studies involving human participants were reviewed and approved by Comitato Etico Fondazione Policlinico Universitario A. Gemelli IRCCS, Rome, Italy. Written informed consent to participate in this study was provided by the participants’ legal guardian/next of kin. Written informed consent was obtained from the individual(s), and minor(s)’ legal guardian/next of kin, for the publication of any potentially identifiable images or data included in this article.
Author Contributions
CTi, AP, and FD conducted the statistical analysis and interpretations of results. CTi designed the study. AP, CTi, and FD wrote the manuscript with the support of CTe, SD’I, ND, ALa, GSc, and GV. FD, FM, GSa, DB, BP, and MS carried out the microbiological analysis. CTi, AP, CTe, SD’I, ND, MT, ALi, and NM collaborated in data collection. All authors read and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank the Italian Ministry of Health for the contribution to the Current Research 2022.
Footnotes
- ^ https://docs.qiime2.org/2021.8/plugins/available/feature-classifier/classify-sklearn/
- ^ https://www.arb-silva.de
- ^ https://www.rstudio.com/
References
1. Goldenberg RL, Culhane JF, Iams JD, Romero R. Epidemiology and causes of preterm birth. Lancet. (2008) 371:75–84. doi: 10.1016/S0140-6736(08)60074-4
2. Aagaard K, Ma J, Antony KM, Ganu R, Petrosino J, Versalovic J. The placenta harbors a unique microbiome. Sci Transl Med. (2014) 6:237ra65. doi: 10.1126/scitranslmed.3008599
3. Jones HE, Harris KA, Azizia M, Bank L, Carpenter B, Hartley JC, et al. Differing prevalence and diversity of bacterial species in fetal membranes from very preterm and term labor. PLoS One. (2009) 4:e8205. doi: 10.1371/journal.pone.0008205
4. Di Giulio DB, Romero R, Amogan HP, Kusanovic JP, Bik EM, Gotsch F, et al. Microbial prevalence, diversity and abundance in amniotic fluid during preterm labor: a molecular and culture-based investigation. PLoS One. (2008) 3:e3056. doi: 10.1371/journal.pone.0003056
5. Morgan XC, Segata N, Huttenhower C. Biodiversity and functional genomics in the human microbiome. Trends Genet. (2013) 29:51–8. doi: 10.1016/j.tig.2012.09.005
6. Macpherson AJ, Harris NL. Interactions between commensal intestinal bacteria and the immune system. Nat Rev Immunol. (2004) 4:475–85. doi: 10.1038/nri1373
7. Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol. (2009) 9:313–23. doi: 10.1038/nri2515
8. Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci U S A. (2010) 107:11971–5. doi: 10.1073/pnas.1002601107
9. Biasucci G, Rubini M, Riboni S, Morelli L, Bessi E, Retetangos C. Mode of delivery affects the bacterial community in the newborn gut. Early Hum Dev. (2010) 86:13–5. doi: 10.1016/j.earlhumdev.2010.01.004
10. Valentine G, Chu DM, Stewart CJ, Aagaard KM. Relationships between perinatal interventions, maternal-infant microbiomes, and neonatal outcomes. Clin Perinatol. (2018) 45:339–55. doi: 10.1016/j.clp.2018.01.008
11. Gupta RW, Tran L, Norori J, Eren AM, Taylor CM, Dowd SE, et al. Histamine-2 receptor blockers alter the fecal microbiota in premature infants. J Pediatr Gastroenterol Nutr. (2013) 56:397–400. doi: 10.1097/MPG.0b013e318282a8c2
12. Cao B, Stout MJ, Lee I, Mysorekar IU. Placental microbiome and its role in preterm birth. Neoreviews. (2014) 15:e537–45. doi: 10.1542/neo.15-12-e537
13. Biasucci G, Benenati B, Morelli L, Bessi E, Boehm G. Cesarean delivery may affect the early biodiversity of intestinal bacteria. J Nutr. (2008) 138:1796S–800S. doi: 10.1093/jn/138.9.1796S
14. Warner BB, Hamvas A. Lungs, microbes and the developing neonate. Neonatology. (2015) 107:337–43. doi: 10.1159/000381124
15. Gollwitzer ES, Saglani S, Trompette A, Yadava K, Sherburn R, McCoy KD, et al. Lung microbiota promotes tolerance to allergens in neonates via PD-L1. Nat Med. (2014) 20:642–7. doi: 10.1038/nm.3568
16. Lohmann P, Luna RA, Hollister EB, Devaraj S, Mistretta TA, Welty SE, et al. The airway microbiome of intubated premature infants: characteristics and changes that predict the development of bronchopulmonary dysplasia. Pediatr Res. (2014) 76:294–301. doi: 10.1038/pr.2014.85
17. Mourani PM, Harris JK, Sontag MK, Robertson CE, Abman SH. Molecular identification of bacteria in tracheal aspirate fluid from mechanically ventilated preterm infants. PLoS One. (2011) 6:e25959. doi: 10.1371/journal.pone.0025959
18. Julious SA. Sample size of 12 per group rule of thumb for a pilot study. Pharmaceut Statist. (2005) 4:287–91. doi: 10.1002/pst.185
19. Jobe AH, Bancalari E. Bronchopulmonary dysplasia. Am J Respir Crit Care Med. (2001) 163:1723–9. doi: 10.1164/ajrccm.163.7.2011060
20. Posteraro P, De Maio F, Menchinelli G, Palucci I, Errico FM, Carbone M, et al. First bloodstream infection caused by Prevotella copri in a heart failure elderly patient with Prevotella-dominated gut microbiota: a case report. Gut Pathog. (2019) 11:44. doi: 10.1186/s13099-019-0325-6
21. Klindworth A, Pruesse E, Schweer T, Peplies J, Quast C, Horn M, et al. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. (2013) 41:e1. doi: 10.1093/nar/gks808
22. De Pascale G, De Maio F, Carelli S, De Angelis G, Cacaci M, Montini L, et al. Staphylococcus aureus ventilator-associated pneumonia in patients with COVID-19: clinical features and potential inference with lung dysbiosis. Crit Care. (2021) 25:197. doi: 10.1186/s13054-021-03623-4
23. Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol. (2019) 37:852–7. doi: 10.1038/s41587-019-0209-9
24. Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, Holmes SP. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods. (2016) 13:581–3. doi: 10.1038/nmeth.3869
25. Karstens L, Asquith M, Davin S, Fair D, Gregory WT, Wolfe AJ, et al. Controlling for contaminants in low-biomass 16S rRNA gene sequencing experiments. mSystems. (2019) 4:e00290–19. doi: 10.1128/mSystems.00290-19
26. McMurdie PJ, Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. (2013) 8:e61217. doi: 10.1371/journal.pone.0061217
27. Jobe A. Effects of chorioamnionitis on the fetal lung. Clin Perinatol. (2012) 39:441–57. doi: 10.1016/j.clp.2012.06.010
28. Theis KR, Romero R, Winters AD, Greenberg JM, Gomez-Lopez N, Alhousseini A, et al. Does the human placenta delivered at term have a microbiota? Results of cultivation, quantitative real-time PCR, 16S rRNA gene sequencing, and metagenomics. Am J Obstet Gynecol. (2019) 220:267.e1–39. doi: 10.1016/j.ajog.2018.10.018
29. de Goffau MC, Lager S, Sovio U, Gaccioli F, Cook E, Peacock SJ, et al. Human placenta has no microbiome but can contain potential pathogens. Nature. (2019) 572:329–34. doi: 10.1038/s41586-019-1451-5
30. Romero R, Hassan SS, Gajer P, Tarca AL, Fadrosh DW, Nikita L, et al. The composition and stability of the vaginal microbiota of normal pregnant women is different from that of non-pregnant women. Microbiome. (2014) 2:4. doi: 10.1186/2049-2618-2-4
31. Verstraelen H, Verhelst R, Claeys G, De Backer E, Temmerman M, Vaneechoutte M. Longitudinal analysis of the vaginal microflora in pregnancy suggests that L. crispatus promotes the stability of the normal vaginal microflora and that L. gasseri and/or L. iners are more conducive to the occurrence of abnormal vaginal microflora. BMC Microbiol. (2009) 9:116. doi: 10.1186/1471-2180-9-116
32. Lal CV, Travers C, Aghai ZH, Eipers P, Jilling T, Halloran B, et al. The airway microbiome at birth. Sci Rep. (2016) 6:31023. doi: 10.1038/srep31023
33. Perez-Muñoz ME, Arrieta M-C, Ramer-Tait AE, Walter J. A critical assessment of the “sterile womb” and “in utero colonization” hypotheses: implications for research on the pioneer infant microbiome. Microbiome. (2017) 5:48. doi: 10.1186/s40168-017-0268-4
34. de Goffau MC, Lager S, Salter SJ, Wagner J, Kronbichler A, Charnock-Jones DS, et al. Recognizing the reagent microbiome. Nat Microbiol. (2018) 3:851–3. doi: 10.1038/s41564-018-0202-y
35. Marsh RL, Nelson MT, Pope CE, Leach AJ, Hoffman LR, Chang AB, et al. How low can we go? The implications of low bacterial load in respiratory microbiota studies. Pneumonia. (2018) 10:7. doi: 10.1186/s41479-018-0051-8
36. Payne MS, Goss KC, Connett GJ, Kollamparambil T, Legg JP, Thwaites R, et al. Molecular microbiological characterization of preterm neonates at risk of bronchopulmonary dysplasia. Pediatr Res. (2010) 67:412–8. doi: 10.1203/PDR.0b013e3181d026c3
37. Wagner BD, Sontag MK, Harris JK, Miller JI, Morrow L, Robertson CE, et al. Airway microbial community turnover differs by BPD severity in ventilated preterm infants. PLoS One. (2017) 12:e0170120. doi: 10.1371/journal.pone.0170120
Keywords: microbiota, vagina, meconium, lung, preterm birth (PTB), bronchopulmonary dysplasia (BPD), bronchoalveolar lavage fluid (BALF)
Citation: Tirone C, Paladini A, De Maio F, Tersigni C, D’Ippolito S, Di Simone N, Monzo FR, Santarelli G, Bianco DM, Tana M, Lio A, Menzella N, Posteraro B, Sanguinetti M, Lanzone A, Scambia G and Vento G (2022) The Relationship Between Maternal and Neonatal Microbiota in Spontaneous Preterm Birth: A Pilot Study. Front. Pediatr. 10:909962. doi: 10.3389/fped.2022.909962
Received: 01 April 2022; Accepted: 17 June 2022;
Published: 22 July 2022.
Edited by:
Arianna Aceti, University of Bologna, ItalyReviewed by:
Flavia Indrio, University of Foggia, ItalyManuela López Azorín, Quirónsalud Madrid University Hospital, Spain
Copyright © 2022 Tirone, Paladini, De Maio, Tersigni, D’Ippolito, Di Simone, Monzo, Santarelli, Bianco, Tana, Lio, Menzella, Posteraro, Sanguinetti, Lanzone, Scambia and Vento. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chiara Tirone, Y2hpYXJhLnRpcm9uZUBwb2xpY2xpbmljb2dlbWVsbGkuaXQ=
†ORCID: Nicoletta Di Simone, orcid.org/0000-0003-1273-3335
This article was submitted to Neonatology, a section of the journal Frontiers in Pediatrics
 Chiara Tirone
Chiara Tirone Angela Paladini
Angela Paladini Flavio De Maio
Flavio De Maio Chiara Tersigni
Chiara Tersigni Silvia D’Ippolito
Silvia D’Ippolito Nicoletta Di Simone
Nicoletta Di Simone Francesca Romana Monzo3
Francesca Romana Monzo3 Giulia Santarelli
Giulia Santarelli Delia Mercedes Bianco
Delia Mercedes Bianco Milena Tana
Milena Tana Alessandra Lio
Alessandra Lio Maurizio Sanguinetti
Maurizio Sanguinetti Giovanni Vento
Giovanni Vento