- 1Department of Pediatric Surgery, Guangzhou Institute of Pediatrics, Guangzhou Women and Children’s Medical Center, Guangzhou Medical University, Guangzhou, Guangdong, China
- 2Guangzhou Women and Children's Medical Center, Guangdong Provincial Clinical Research Center for Child Health, Guangzhou Medical University, Guangzhou, China
- 3Changan Hospital of Dongguan, Dongguan, China
Background: Hirschsprung's disease (HSCR) is currently considered to be a congenital gastrointestinal malformation caused mainly by genetic factors. Endothelin Converting Enzyme-1 (ECE1) has been reported to be associated with HSCR. However, the relationship between ECE1 single nucleotide polymorphism (SNP) rs169884 and HSCR in the southern Chinese population remains unknown.
Methods: 1,470 HSCR patients and 1,473 controls from a southern Chinese population were recruited. The intronic SNP rs169884 in ECE1 was genotyped in all samples. We tested the association between rs169884 and HSCR under various genetic models. We also evaluated the effect of rs169884 on HSCR subtypes, including short-segment HSCR (S-HSCR), long-segment HSCR (L-HSCR) and total colonic aganglionosis (TCA). External epigenetic data were integrated to investigate the potential biological function of rs169884.
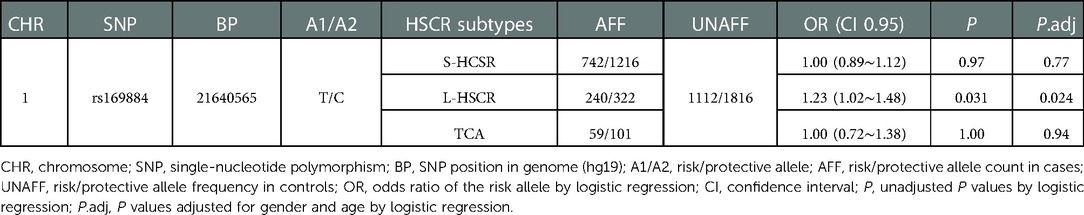
Results: Chromatin states data from derived neuron cells or fetal colon tissue revealed that rs169884 might control ECE1 expression through regulating its enhancer function. We did not find a significant association between rs169884 and HSCR. For HSCR subtypes, although no significant associations were detected between rs169884 and S-HSCR (OR = 1.00, 95% CI: 0.89∼1.12, Padj = 0.77) or TCA (OR = 1.00, 95% CI: 0.72∼1.38, Padj = 0.94), we found that rs169884 could increase the risk of L-HSCR (OR = 1.23, 95% CI 1.02∼1.45, Padj = 0.024).
Conclusion: These results suggested that rs169884 might play a regulatory role for ECE1 expression and increase susceptibility of L-HSCR in southern Chinese children.
Introduction
Hirschsprung's disease (HSCR) is a common gastrointestinal malformation worldwide (1). The abnormal development of neural crest cells (NCC) in embryo is the main cause of the disease (2). If NCC fail to colonize at the end of the digestive tract during development, functionally mature ganglia is lacking, resulting in intestinal lesions (3). The diseased bowel segment that lacks ganglia shows spastic stricture, causing abdominal distension and constipation (4). According to the current statistics, the incidence of HSCR has a difference in races and genders, with a global incidence of about 1 in 5,000 and a higher incidence of about 1.4 in 5,000 in Asia (5). The incidence of HSCR is 4 to 1 between males and females (6). Clinicians have classified HSCR into short-segment HSCR (S-HSCR), long-segment HSCR (L-HSCR) and total colonic aganglionosis (TCA), accounting for about 60%–80%, 10%–15% and 5% of HSCR cases, respectively (7). From a genetic perspective, only 10%–15% of HSCR patients have familial inheritance, while more than 70% are sporadic cases (8). Increasing studies have shown that HSCR is a highly polygenic disease. At present, nearly 20 genes (such as RET, EDNRB, SOX10, etc 9, 10). have been reported to be associated with HSCR. Among these genes, GDNF/RET comprise a signaling pathway which is a validated pathogenic mechanism underlying HSCR (11, 12). Abnormal signal transduction affect the differentiation, proliferation and migration of enteric neural crest cells (ENCCs), which leads to colon aganglionosis and onset of HSCR (3).
Endothelin Converting Enzyme-1 (ECE1), a peptide-chain endonuclease that transforms large endothelin (ET) into active ET in vivo, is a highly glycosylated glycoprotein that plays an extremely important role in the regulation of endothelin bioactivity (13). ECE1 has been found to be widely expressed in intestinal tissues, controlling intestinal motor and secretory function by promoting the production of endothelin (14). More importantly, there is abundant ECE1 in the endosome of intestinal myenteric neurons, which plays an important role in degrading some neuropeptides in the endosome, thereby regulating the transport of important receptors and signal transmission (15, 16). Further investigation revealed that ECE1 knockout mouse models exhibited a phenotype of intestinal neuron depletion, suggesting an important role in neuronal development (17).
According to previous studies, ECE1 is involved in the pathogenesis of HSCR (9, 18). Considering that rs169884 locates at a regulatory site in the genomic region of ECE1, we conducted a case–control study to explore the association between ECE1 rs169884 polymorphism and HSCR susceptibility in Southern Chinese population.
Materials and methods
Study subjects
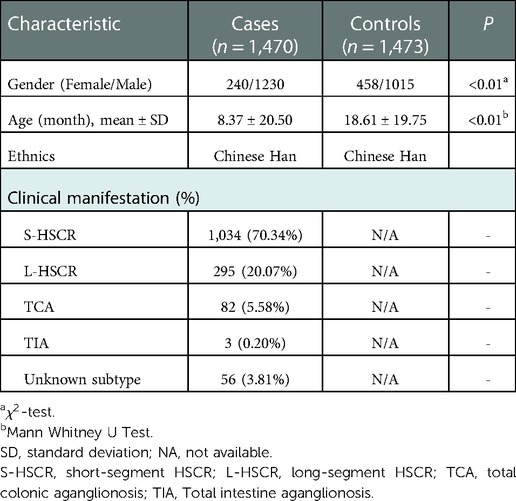
All participants in this study were recruited from Guangzhou Women and Children's Medical Center. Signed informed consent had been obtained from guardians of all participants prior to enrollment in the study. In the case group, patients were diagnosed with HSCR by histological examination of colon biopsies after surgical resection, and were classified into S-HSCR, L-HSCR, and TCA according to the length of the aganglionosis colon by pathologists. Control samples were obtained from patients without HSCR, enteritis, neurological disease or HSCR-related familial history based on the medical files at the time of enrollment. No environmental factors (e.g., toxic or drug exposure) that strongly associated with predisposition to HSCR have been identified in control samples from the medical records. Data of age, gender and ethnic were collected from the medical files for both HSCR and control samples. The significance test of differences in characteristics between cases and controls were analyzed with -test or Mann Whitney U Test based on data properties. All participants were unrelated Chinese Han. Ethical approval was obtained from the Institutional Review Board of Guangzhou Women and Children's Medical Center (Ethical Approval Number: 2018052406).
Chromatin states data
The chromatin states were obtained from ROADMAP project (http://www.roadmapepigenomics.org). We selected data from cells/tissues related with HSCR: (1) H1 derived neuronal progenitor cultured cells; (2) H9 derived neuronal progenitor cultured cells; (3) H9 derived neuron cultured cells; (4) Fetal large intestine. The result was visualized using WashU Epigenome Browser (http://epigenomegateway.wustl.edu/browser/).
SNP genotyping
Genomic DNA were extracted from venous blood of all samples. The SNP rs169884 was genotyped by Sequenom MassARRAY iPLEX Gold system (San Diego, CA, USA).
Statistical analysis
The association test was performed using PLINK1.9 software (https://www.cog-genomics.org/plink/1.9/). The Hardy–Weinberg equilibrium (HWE) was calculated for this SNP in the control group and P > 0.05 was considered a good genotyping quality. Various genetic models were investigated, including the allelic model (ALLELIC), the 2df genotypic model (GENO), the additive model (ADD), the dominant model (DOM) and the recessive model (REC). Given the controls and HSCR cases, we used “–logistic” option to calculate the odds ratio (OR) and its significance for the rs169884 risk on HSCR. The age and gender were used as covariates to calculate the adjusted OR and P value. For the allelic model, we used “–assoc” option to run the 1df chi-square allelic test. Then we performed similar analysis for HSCR subtypes, i.e., S-HSCR, L-HSCR, and TCA, respectively.
Results
Regulatory potential of rs169884
To investigate the potential regulatory function of rs169884, we collected data of chromatin states along the ECE1 genomic region from three derived neuron-related cells and the fetal colon tissue (Figure 1). Interestingly, we noticed that the genomic region nearby rs169884 showed obvious enhancer signals in all the selected chromatin states data. This suggests that rs169884 might play a role for ECE1 expression through regulating the enhancer function.
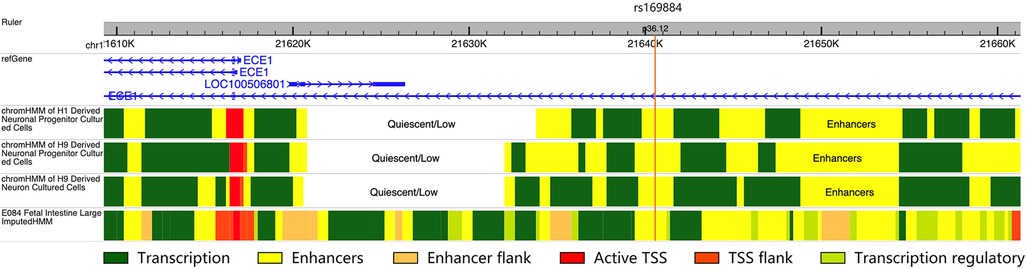
Figure 1. The chromatin states of the genomic region surrounding rs169884. The chromatin states were obtained from H1 or H9 derived neuronal progenitors, H9 derived neurons as well fetal large intestine. The genomic region nearby rs169884, which belongs to the ECE1 intron, was enriched with enhancer signals across the selected cell types or tissue. The genomic version was hg19.
Association of ECE1 SNP with HSCR susceptibility
A total of 1,470 HSCR patients and 1,473 controls were included in the study. The clinical information of the participants was summarized in Table 1. The genotype frequency of rs169884 and the association analysis between rs169884 and HSCR were calculated, as shown in Table 2. As shown, a total of 1,398 cases and 1,464 controls were successfully genotyped. HWE was confirmed in the control group (P > 0.05). Association between rs169884 and HSCR was evaluated under the allelic model (ALLELIC), the 2df genotypic model (GENO), the additive model (ADD), the dominant model (DOM) and the recessive model (REC), respectively. Since we noticed significant difference for age and gender between the case and control group (Table 1), we further run the logistic regression adjusted for the two covariates. In all five genetic models, we could not verify a significant association between rs169884 and HSCR (Table 2).
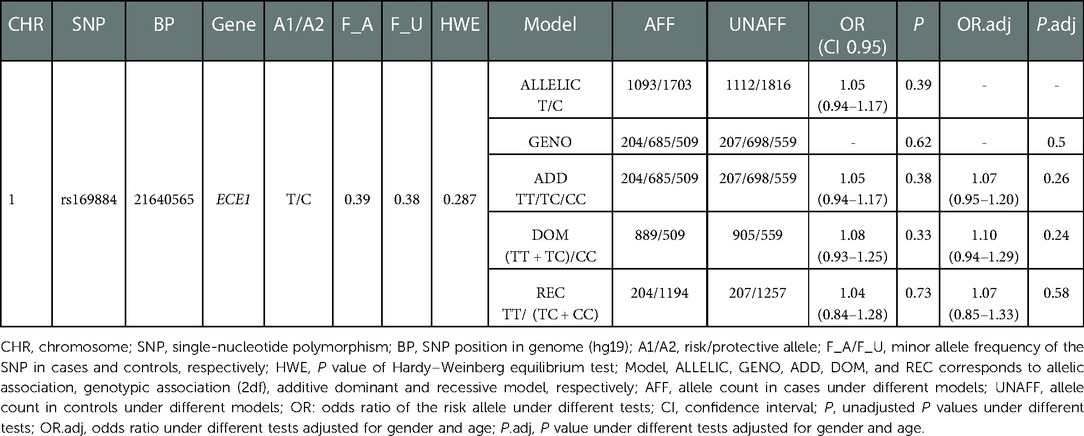
Table 2. Results of SNP rs160844 in a southern Chinese population of 1,470 cases and 1,473 controls.
Stratification analysis of ECE1 rs169884 with HSCR subtypes
Further stratification analyses were carried out to assess whether rs169884 is associated with HSCR subtypes (S-HSCR, L-HSCR, and TCA). The results revealed that rs169884 was significantly associated with L-HSCR (OR = 1.23, 95% CI: 1.02∼1.48, Padj = 0.024). Evidence of association between rs169884 and S-HSCR or TCA is insufficient (OR = 1.00, 95% CI: 0.89∼1.12, Padj = 0.77; OR = 1.00, 95% CI: 0.72∼1.38, Padj = 0.94) (Table 3).
Discussion
As a congenital digestive defect, HSCR is a life-threatening disease. Although surgery can largely relieve a patient's condition, complications after surgery can be lifelong (19), and the suffering caused by the disease is enormous (20). However, the knowledge of the pathogenesis of HSCR is still very limited.
In our study, we focused on the HSCR susceptibility of ECE1 gene, a key component of endothelin signaling pathway. This pathway is critical in regulating the development of the cardiovascular system, kidney, and pulmonary processes, as well as vertebrate-specific neural crest cell populations and their derivatives, especially at an early stage (21). The endothelin system has been found to be essential in the development of neural crest-derived tissues, including intestinal neurons (22). At the embryonic stage, endothelin signaling is required for the migration of enteric neural progenitors from the foregut to the hindgut. On the other hand, lack of appropriate endothelin signaling in mice and human leads to HSCR phenotype (23). At present, several members in the endothelin signaling pathway have been proved to be closely related to the development of Hirschsprung's disease (22, 24, 25). ECE1, an important member in endothelin signaling pathway, can regulate neurokinin 1 receptor (NK1R) trafficking and signal transduction in endosomes of myoenteric neurons, attenuating NK1R-mediated ERK1/2 activation in myoenteric neurons by promoting NK1R re-sensitivity, leading to intestinal neuron depletion (26). In addition, loss-of-function mutations in ECE1 are associated with Hirschsprung's disease (17, 27). Together, these may explain the pathogenic role of ECE1 for HSCR (Figure 2).
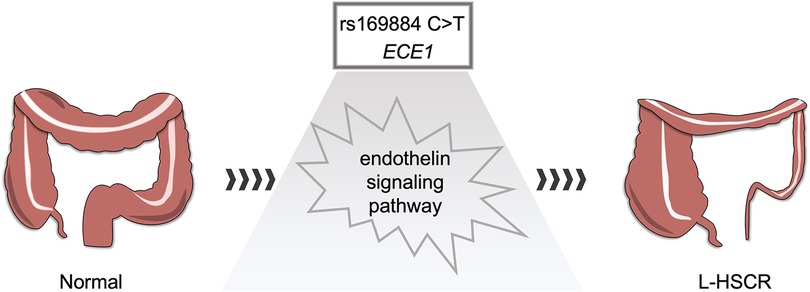
Figure 2. Hypothesis of rs169884 risk on L-HSCR. rs169884 may regulate the expression of ECE1, which further causes abnormal development of enteric nervous system through the endothelin signaling pathway.
In this study, we found that SNP rs169884 C > T in ECE1 intron increases the risk of L-HSCR. Gene introns could enhance gene expression; however, the mechanism remains unclear (28). SNPs in introns could cause multiple regulatory consequences, including effects on splicing and promoter-enhancer interaction (29, 30). One recent study has reported that an intronic SNP in BCL2 showed allele-specific enhancer activity for BCL2 expression by affecting the transcription factor binding (31). By integrating chromatin states data, we found an enhancer signal surrounding the intronic rs169884, indicating rs169884 may have the ability to regulate ECE1 expression. Similar as previous studies (32) which EDNRB and EDN3 lead to L-HSCR as well as syndromic HSCR, we fail to explain the susceptibility for S-HSCR, which is the most common subtype in HSCR. So far, the genes responsible for the L-HSCR include SLC6A20 (33) and miR-618 (34). Genetic models of different genes leading to different HSCR subtypes still need to be elucidated. We recruited nearly 300 L-HSCR patients in this study; however, more patients are needed to explore more risk SNPs. Besides, environmental factors (such as toxic or drug exposure) affect birth defects should be considered to study the effect on prevention of HSCR.
Although our study is the first discovery of association between ECE1 rs169884 and HSCR, some limitations should be noted. First, despite of the large sample size for all HSCR cases, the sample size of each subtype, especially for TCA, was still limited. Hence, we need to validate the conclusion that rs169884 is associated with an increased susceptibility of L-HSCR in larger sample size. Second, only one intronic SNP was selected in this study. The effects of other ECE1 SNPs or SNPs in other endothelin signaling-related genes have not been explored, which might also play roles in the predisposition to HSCR. Third, although we found regulatory evidence of rs169884 in ECE1 enhancer region in various derived neuron cells or fetal colon tissue, functional experiments are still needed to verify these results. Lastly, our study only represents the characteristic inheritance of the southern Chinese population. Further studies should focus on genotype-phenotype relationships in multi-ethnic populations.
Conclusion
To summarize, in this HSCR case-control study from a southern Chinese population, we found an association between L-HSCR (one subtype of HSCR) and rs169884. This SNP may have the potential to regulate the expression of ECE1, which plays a critical role in endothelin signaling pathway during the development of enteric nervous system.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Ethics statement
The studies involving human participants were reviewed and approved by Institutional Review Board of Guangzhou Women and Children's Medical Center (Ethical Approval Number: 2018052406). Written informed consent to participate in this study was provided by the participants’ legal guardian/next of kin.
Author contributions
ZL and WZ designed the experiment. CL, YL, XW, BW, SX and QH collected samples and conducted the study. CL, ZL and WZ analyzed the data. CL and Yanqing Liu wrote the paper. All authors contributed to the article and approved the submitted version.
Funding
This study was funded by grants from the Natural Science Foundation of Guangdong Province (NO. 2022A1515012254, 2018A030313570), and the Science and Technology Project of Guangzhou (NO. 201903010074, 202206080002), Dongguan Social Development Technology Project (NO. 20221800904962).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Chen WC, Chang SS, Sy ED, Tsai MC. A De Novo novel mutation of the EDNRB gene in a Taiwanese boy with hirschsprung disease. J Formos Med Assoc. (2006) 105(4):349–54. doi: 10.1016/S0929-6646(09)60128-5
2. Beltman L, Windster JD, Roelofs J, van der Voorn JP, Derikx JPM, Bakx R. Diagnostic accuracy of calretinin and acetylcholinesterase staining of rectal suction biopsies in hirschsprung disease examined by unexperienced pathologists. Virchows Arch. (2022) 481(2):245–52. doi: 10.1007/s00428-022-03334-3
3. Mueller JL, Goldstein AM. The science of hirschsprung disease: what we know and where we are headed. Semin Pediatr Surg. (2022) 31(2):151157. doi: 10.1016/j.sempedsurg.2022.151157
4. Klein M, Varga I. Hirschsprung's disease-recent understanding of embryonic aspects, etiopathogenesis and future treatment avenues. Medicina (Kaunas). (2020) 56(11):611. doi: 10.3390/medicina56110611
5. Amiel J, Sproat-Emison E, Garcia-Barcelo M, Lantieri F, Burzynski G, Borrego S, et al. Hirschsprung disease, associated syndromes and genetics: a review. J Med Genet. (2008) 45(1):1–14. doi: 10.1136/jmg.2007.053959
6. Bahrami A, Joodi M, Moetamani-Ahmadi M, Maftouh M, Hassanian SM, Ferns GA, et al. Genetic background of hirschsprung disease: a bridge between basic science and clinical application. J Cell Biochem. (2018) 119(1):28–33. doi: 10.1002/jcb.26149
7. Ji Y, Tam PK, Tang CS. Roles of enteric neural stem cell niche and enteric nervous system development in hirschsprung disease. Int J Mol Sci. (2021) 22(18):9659. doi: 10.3390/ijms22189659
8. Torroglosa A, Villalba-Benito L, Luzon-Toro B, Fernandez RM, Antinolo G, Borrego S. Epigenetic mechanisms in hirschsprung disease. Int J Mol Sci. (2019) 20(13):3123. doi: 10.3390/ijms20133123
9. Jiang Q, Ho YY, Hao L, Nichols Berrios C, Chakravarti A. Copy number variants in candidate genes are genetic modifiers of hirschsprung disease. PLoS One. (2011) 6(6):e21219. doi: 10.1371/journal.pone.0021219
10. Diposarosa R, Bustam NA, Sahiratmadja E, Susanto PS, Sribudiani Y. Literature review: enteric nervous system development, genetic and epigenetic regulation in the etiology of Hirschsprung's Disease. Heliyon. (2021) 7(6):e07308. doi: 10.1016/j.heliyon.2021.e07308
11. Chatterjee S, Chakravarti A. A gene regulatory network explains RET-EDNRB epistasis in Hirschsprung disease. Hum Mol Genet. (2019) 28(18):3137–47. doi: 10.1093/hmg/ddz149
12. Tomuschat C, Puri P. RET Gene is a major risk factor for Hirschsprung's Disease: a meta-analysis. Pediatr Surg Int. (2015) 31(8):701–10. doi: 10.1007/s00383-015-3731-y
13. Kohan DE, Rossi NF, Inscho EW, Pollock DM. Regulation of blood pressure and salt homeostasis by endothelin. Physiol Rev. (2011) 91(1):1–77. doi: 10.1152/physrev.00060.2009
14. Escrig C, Bishop AE, Inagaki H, Moscoso G, Takahashi K, Varndell IM, et al. Localisation of endothelin like immunoreactivity in adult and developing human gut. Gut. (1992) 33(2):212–7. doi: 10.1136/gut.33.2.212
15. Cattaruzza F, Cottrell GS, Vaksman N, Bunnett NW. Endothelin-converting enzyme 1 promotes re-sensitization of neurokinin 1 receptor-dependent neurogenic inflammation. Br J Pharmacol. (2009) 156(5):730–9. doi: 10.1111/j.1476-5381.2008.00039.x
16. Cottrell GS, Padilla BE, Amadesi S, Poole DP, Murphy JE, Hardt M, et al. Endosomal endothelin-converting enzyme-1: a regulator of beta-arrestin-dependent ERK signaling. J Biol Chem. (2009) 284(33):22411–25. doi: 10.1074/jbc.M109.026674
17. Yanagisawa H, Yanagisawa M, Kapur RP, Richardson JA, Williams SC, Clouthier DE, et al. Dual genetic pathways of endothelin-mediated intercellular signaling revealed by targeted disruption of endothelin converting enzyme-1 gene. Development. (1998) 125(5):825–36. doi: 10.1242/dev.125.5.825
18. Vohra BP, Planer W, Armon J, Fu M, Jain S, Heuckeroth RO. Reduced endothelin converting enzyme-1 and endothelin-3 mRNA in the developing bowel of Male mice may increase expressivity and penetrance of hirschsprung disease-like distal intestinal aganglionosis. Dev Dyn. (2007) 236(1):106–17. doi: 10.1002/dvdy.21028
19. Ahmad H, Yacob D, Halleran DR, Gasior AC, Lorenzo CD, Wood RJ, et al. Evaluation and treatment of the post pull-through Hirschsprung patient who is not doing well; update for 2022. Semin Pediatr Surg. (2022) 31(2):151164. doi: 10.1016/j.sempedsurg.2022.151164
20. Tham SW, Rollins MD, Reeder RW, Lewis KE, Calkins CM, Avansino JR, et al. Health-related quality of life in children with hirschsprung disease and children with functional constipation: parent-child variability. J Pediatr Surg. (2022) 57(8):1694–700. doi: 10.1016/j.jpedsurg.2022.04.009
21. Korth P, Bohle RM, Corvol P, Pinet F. Cellular distribution of endothelin-converting enzyme-1 in human tissues. J Histochem Cytochem. (1999) 47(4):447–62. doi: 10.1177/002215549904700403
22. Baynash AG, Hosoda K, Giaid A, Richardson JA, Emoto N, Hammer RE, et al. Interaction of endothelin-3 with endothelin-B receptor is essential for development of epidermal melanocytes and enteric neurons. Cell. (1994) 79(7):1277–85. doi: 10.1016/0092-8674(94)90018-3
23. Druckenbrod NR, Powers PA, Bartley CR, Walker JW, Epstein ML. Targeting of endothelin receptor-B to the neural crest. Genesis. (2008) 46(8):396–400. doi: 10.1002/dvg.20415
24. Cantrell VA, Owens SE, Chandler RL, Airey DC, Bradley KM, Smith JR, et al. Interactions between Sox10 and EdnrB modulate penetrance and severity of aganglionosis in the Sox10Dom mouse model of Hirschsprung disease. Hum Mol Genet. (2004) 13(19):2289–301. doi: 10.1093/hmg/ddh243
25. Carrasquillo MM, McCallion AS, Puffenberger EG, Kashuk CS, Nouri N, Chakravarti A. Genome-wide association study and mouse model identify interaction between RET and EDNRB pathways in Hirschsprung disease. Nat Genet. (2002) 32(2):237–44. doi: 10.1038/ng998
26. Pelayo JC, Poole DP, Steinhoff M, Cottrell GS, Bunnett NW. Endothelin-converting enzyme-1 regulates trafficking and signalling of the neurokinin 1 receptor in endosomes of myenteric neurones. J Physiol. (2011) 589(Pt 21):5213–30. doi: 10.1113/jphysiol.2011.214452
27. Hofstra RM, Valdenaire O, Arch E, Osinga J, Kroes H, Loffler BM, et al. A loss-of-function mutation in the endothelin-converting enzyme 1 (ECE-1) associated with hirschsprung disease, cardiac defects, and autonomic dysfunction. Am J Hum Genet. (1999) 64(1):304–8. doi: 10.1086/302184
28. Shaul O. How introns enhance gene expression. Int J Biochem Cell Biol. (2017) 91(Pt B):145–55. doi: 10.1016/j.biocel.2017.06.016
29. Wang GS, Cooper TA. Splicing in disease: disruption of the splicing code and the decoding machinery. Nat Rev Genet. (2007) 8(10):749–61. doi: 10.1038/nrg2164
30. Hua JT, Ahmed M, Guo H, Zhang Y, Chen S, Soares F, et al. Risk SNP-mediated promoter-enhancer switching drives prostate cancer through lncRNA PCAT19. Cell. (2018) 174(3):564–75. doi: 10.1016/j.cell.2018.06.014
31. Dong SS, Zhu DL, Zhou XR, Rong Y, Zeng M, Chen JB, et al. An intronic risk SNP rs12454712 for central obesity acts as an allele-specific enhancer to regulate BCL2 expression. Diabetes. (2021) 70(8):1679–88. doi: 10.2337/db20-1151
32. Pusch CM, Sasiadek MM, Hirschsprung BN. RET-SOX and beyond: the challenge of examining non-Mendelian traits (review). Int J Mol Med. (2002) 10(4):367–70. PMID: 12239580.
33. Lee JS, Oh JT, Kim JH, Seo JM, Kim DY, Park KW, et al. Association analysis of SLC6A20 polymorphisms with Hirschsprung disease. J Pediatr Gastroenterol Nutr. (2016) 62(1):64–70. doi: 10.1097/MPG.0000000000000880
Keywords: Hirschsprung's disease, Endothelin Converting Enzyme-1, single nucleotide polymorphism, epigenetic regulation, HSCR subtypes
Citation: Lan C, Liu Y, Wu X, Wang B, Xin S, He Q, Zhong W and Liu Z (2022) Susceptibility of ECE1 polymorphisms to Hirschsprung's disease in southern Chinese children. Front. Pediatr. 10:1056938. doi: 10.3389/fped.2022.1056938
Received: 29 September 2022; Accepted: 5 December 2022;
Published: 22 December 2022.
Edited by:
Pedro Gutierrez-Castrellon, Hospital General Dr. Manuel Gea Gonzalez, MexicoReviewed by:
Ariadna Gonzalez-Del Angel, National Institute of Pediatrics, MexicoGabriel López-Velázquez, National Institute of Pediatrics, Mexico
© 2022 Lan, Liu, Wu, Wang, Xin, He, Zhong and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zipeng Liu enB6cGxpdUAxNjMuY29t Wei Zhong emhvbmd3ZWlAZ3djbWMub3Jn
†These authors have contributed equally to this work
Specialty Section: This article was submitted to Pediatric Gastroenterology, Hepatology and Nutrition, a section of the journal Frontiers in Pediatrics
 Chaoting Lan
Chaoting Lan Yanqing Liu1,†
Yanqing Liu1,† Bingtong Wang
Bingtong Wang Qiuming He
Qiuming He Wei Zhong
Wei Zhong Zipeng Liu
Zipeng Liu