- 1Department of Hematology and Oncology, Children's Hospital of Soochow University, Suzhou, China
- 2Department of Hematology and Oncology, Institute of Biomedical Sciences, Children's Hospital of Fudan University, Shanghai, China
- 3Department of Pediatrics, Subei People's Hospital of Jiangsu Province, Yangzhou, China
- 4Department of Pediatrics, Yiyuan People’s Hospital, Zibo, China
- 5Department of Emergency, Children’s Hospital of Soochow University, Suzhou, China
- 6Institute of Pediatric Research, Children’s Hospital of Soochow University, Suzhou, China.
- 7Department of Hematology and Oncology, Fujian Branch of Shanghai Children’s Medical Center, Fujian Children’s Hospital, Fuzhou, China
- 8Department of Hematology and Oncology, Shanghai Children’s Medical Center, Shanghai Jiao Tong University School of Medicine, Shanghai, China
Purpose: To evaluate the influence of GATA3 rs3824662 on pre-B-cell acute lymphoblastic leukemia (pre-B-cell ALL) susceptibility and long-term prognosis in Han Chinese children with pre-B-cell ALL treated with the CCLG-2008 protocol at the Children’s Hospital of Soochow University.
Methods: A total of 256 patients with childhood pre-B-cell ALL under the CCLG-2008 protocol were enrolled in this study, and 174 healthy children were used as case controls. GATA3 rs3824662 genotyping was performed using a polymerase chain reaction, followed by Sanger sequencing. The association of genotype with clinical characteristics, treatment response, adverse events, and outcomes were analyzed.
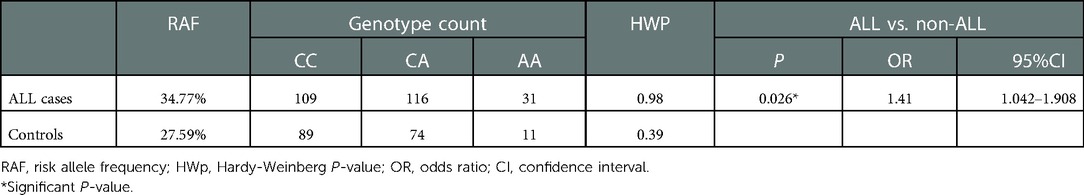
Results: The A allele frequency of GATA3 rs3824662 in patients with pre-B cell ALL was significantly higher than that in healthy children (OR = 1.41, 95% CI = 1.042–1.908; P = 0.026). Among patients with pre-B-cell ALL, the GATA3 rs3824662 AA genotype was associated with poor prednisolone response and high blast cell burden on day 15 of the induction therapy (P = 0.011 and 0.007, respectively). Patients with the rs3824662 AA variant suffered more episodes of sepsis than those with the CC or CA variants (P = 0.021). The GATA3 rs3824662 AA genotype was significantly associated with sepsis [hazard ratio (HR) = 3.375; P = 0.01]. No significant differences were found in the cumulative incidence of relapse, overall survival, and event-free survival among all genotypes.
Conclusion: GATA3 rs3824662 was associated with susceptibility in Han Chinese children with pre-B-cell ALL and could be a possible risk factor for poor early treatment response and treatment-related sepsis.
Introduction
The occurrence of leukemia is believed to be influenced by both environmental and genetic factors. Inherited genetic variation strongly influences both susceptibility and treatment outcomes of childhood acute lymphoblastic leukemia (ALL). Over the past few years, large-scale genome-wide association studies have identified a few susceptibility genes for pediatric ALL, including IKZF1, CEBPE, PIP4K2A, CDKN2A, TP63, and GATA3 (1, 2). The association between GATA3 rs3824662 and B-cell ALL susceptibility was first identified among Hispanic people in 2013 (3). Acute lymphoblastic leukemia susceptibility has been confirmed by multiple studies among different ethnicities. Recent reports have shown that the rs3824662 A allele is associated with a high initial white blood cell (WBC) count, old age, and Ph-like ALL profiling (4–6). In addition, the risk allele A confers high minimal residual disease (MRD) levels by the end of induction therapy and an increased risk of recurrence via JAK-STAT-autophagy activation, demonstrating its critical role in ALL pathogenesis and prognosis (5–7).
Immunosuppression has been demonstrated to be an unavoidable accompanied side effect during leukemia therapy. However, immune reconstitution in leukemia therapy depends on the type of lymphocytes, residual thymic function, treatment intensity, and individual genetic factors (8). GATA3 has been regarded as a master regulator in both innate and adaptive immunity because of its crucial role in hematopoietic cells. The function of GATA3 as a key regulator in T cell development, differentiation, and maintenance, especially for T helper 2 (Th2) cells, is well established (9–11). The rs3824662 locus is located in the enhancer region of GATA3, leading to increased GATA3 transcript levels (7, 12). However, the association between rs3824662 and chemotherapy-related adverse events has not yet been investigated.
With this in mind, we systemically analyzed the association between GATA3 rs3824662 and clinical features and found that GATA3 rs3824662 was associated with susceptibility in Chinese children of Han nationality with pre-B-cell ALL and poor early treatment response. In addition, patients with the AA variant had more episodes of sepsis than those with the CC or CA genotypes.
Patients and methods
Patients
A total of 256 newly diagnosed children with pre-B-cell ALL and 174 healthy controls were enrolled in this study between November 2012 and August 2015 at the Children's Hospital of Soochow University. All patients were treated according to the Chinese Children Leukemia Group-ALL 2008 (CCLG-ALL 2008) protocol (13). Informed consent forms were signed, and the study was conducted following the principles of the Declaration of Helsinki. The characteristics of patients were presented in Supplementary Table S1.
Response evaluation
The National Comprehensive Cancer Network guidelines version 1.2021 on ALL was used to assess treatment response. Briefly, poor prednisolone response was defined as an absolute count of leukemia cells ≥1,000/µl in the peripheral blood after induction therapy with prednisone for seven consecutive days. The bone marrow morphology was categorized as M1 (bone marrow blasts <5%), M2 (≥5% and <25%), and M3 (≥25%). Complete remission (CR) was defined as no symptoms of leukemia cell infiltration in the body, no circulating blasts after induction therapy, and M1 in the bone marrow, with normal levels of platelets and absolute neutrophil counts. Induction failure was defined as failure to achieve CR at the end of induction. Relapse was defined as primitive and naive lymphocytes >20% in the bone marrow or any extramedullary site (central nervous system, testis, or skin) after CR or primary or naive lymphocytes >5%–20% in the bone marrow associated with evidence of a positive conversion in molecular biology. The stages of relapse were divided into three stages: very early relapse (recurrence <18 months from initial diagnosis), early relapse (recurrence ≥18 months but <36 months from initial diagnosis), and late relapse (recurrence from initial diagnosis ≥36 months). Overall survival (OS) calculation started from the date of diagnosis and ended at the date of death. Missed follow-up data were censored at the last follow-up. Event-free survival (EFS) was defined as the time interval between the date of achieving CR and recurrence or death. Relapse-free survival (RFS) was calculated from the date of CR achievement to the date of relapse.
Adverse side effects
The adverse effects of CCLG-2008 therapy were evaluated and graded using the Common Terminology Criteria for Adverse Events 5.0. Sepsis, myelosuppression, pneumonia, and liver function impairment were included in the analysis of adverse effects.
Genotyping
Genomic DNA was extracted from the peripheral blood of patients in the CR stage or healthy controls using the QIAamp DNA Mini Kit (QIAGEN, Germany) and stored at −20 °C until use. The touchdown polymerase chain reaction was used to amplify the intron fragment of rs3824662. The corresponding DNA fragments were amplified by a polymerase chain reaction using the following primers: forward, 5′-TATCACCCTCCCCACCA-3′; reverse, 5′-GGAAAGCCCCAGATCAA-3′, and then sent for sequencing (Suzhou Jinweizhi Biotechnology Co., Ltd.). Mutation Surveyor V4.0.8 software was used to analyze the SNP genotypes.
Statistical analysis
SPSS statistics 22 and GraphPad Prism 5 were used to analyze the data. The chi-square test was used for the comparison of qualitative data (frequency and percentage) and the Hardy–Weinberg equilibrium test. Logistic regression analysis was used to perform the susceptibility analysis. The association between genotype and the number of adverse events was based on the Kruskal–Wallis test. Survival analysis was performed using the Kaplan–Meier method. The log-rank test was used to compare survival among groups. All P-values were obtained from two-sided tests, and statistical significance was set at P < 0.05.
Results
Baseline characteristics of patients with pre-B-cell ALL
The baseline characteristics of the patients with pre-B-cell ALL were summarized in Supplementary Table S1. The last follow-up period was August 2022, and the median follow-up period was 100.32 months. Among the 256 patients with pre-B-cell ALL, 108 (42.2%) were female and 148 (57.8%) were male; the male/female ratio was 1.4:1. There were 38 (14.8%) patients who were older than 10 years old, and 54 (21.1%) had high WBC (>50 × 109/L). The most frequent molecular abnormality subtypes were TEL/AML1 (n = 46, 18%) and hypodiploidy (n = 41, 16%). There were 32 (12.5%) patients who responded poorly to prednisone and 48 (18.8%) patients remained at M3 status on day 15 of induction therapy. The frequency of MRD positivity was 18.8% (n = 48) on day 33 and 27.7% (n = 71) on week 12. Thus, the standard-, intermediate-, and high-risk classifications were 25.8% (n = 66), 41% (n = 105), and 33.2% (n = 85), respectively.
GATA3 rs3824662 of pre-B-cell leukemia susceptibility
The ALL susceptibility of rs3824662 has been reported previously; however, its role in Han Chinese children remains unclear. To address this issue, we applied logistic regression analysis to investigate the association between GATA3 rs3824662 and pre-B-cell ALL susceptibility in our study cohort. The P-values of the Hardy–Weinberg equilibrium in both patients and controls were >0.05, which indicated that the surveyed population had reached a genetic balance (Table 1). The A allele frequency accounted for 34.7% and 27.59% in patients with B-cell leukemia and healthy controls, respectively. The AA genotype was significantly higher in patients with pre-B-cell ALL than that in the non-ALL children (OR = 1.41, 95% CI = 1.042–1.908, P = 0.026; Table 1), indicating the susceptibility of GATA3 rs3824662 in pre-B-cell leukemia. No correlation was observed between the risk allele A and other clinical features, such as sex, age, WBC count at diagnosis, and genetic abnormalities (Table 2).
Effect of GATA3 rs3824662 genotype on the treatment response
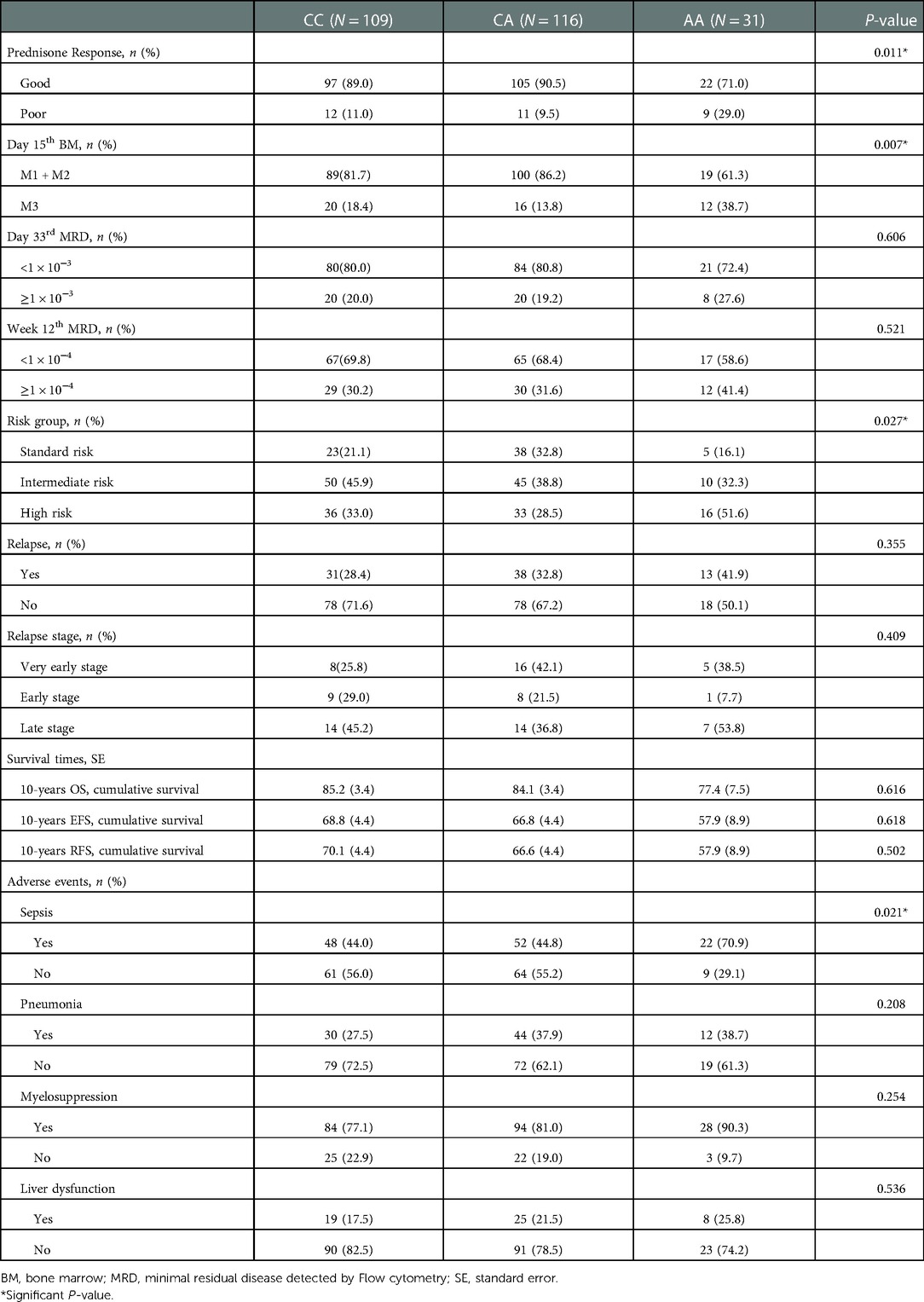
Overall, the estimated 10-year OS, RFS, and EFS were 83.8 ± 2.3%, 66.5 ± 3.0%, and 67.4 ± 3.0%, respectively (Figure 1), and the cumulative relapse rate was 32.0% (82/256). According to risk classification, the estimated 10-year OS and EFS in SR, IR, and HR were 92.4 ± 3.3% and 87.4 ± 3.3% vs. 72.7 ± 4.9% and 81.7 ± 4.8% and 72.1 ± 4.4% vs. 48.1 ± 5.4%, respectively (Figure 1). To evaluate the influence of rs3824662 on long-term prognosis, we associated the genotype with the survival and relapse data. Patients with the AA genotype had a shorter EFS term and a higher rate of relapse than that observed in patients with other genotypes. However, no significant differences were observed between the groups. The estimated 10-year OS and EFS for the CC, CA, and AA genotypes were 85.2 ± 3.4% and 84.1 ± 3.4% vs. 77.4 ± 7.5%, and 66.8 ± 4.4% and 66.8 ± 4.4% vs. 57.9 ± 8.9%, respectively. The P-values were 0.616 and 0.618, respectively (Figure 2). The estimated 10-year RFS in the AA genotype was lower than that in the CC and CA genotypes (57.9 ± 8.9% vs. 66.6 ± 4.4% and 70.1 ± 4.4%, respectively, P = 0.370), and the distribution of relapse in the AA and CC + CA genotypes was 41.9% (13/31) and 30.6% (69/225), respectively (Table 3 and Figure 2).
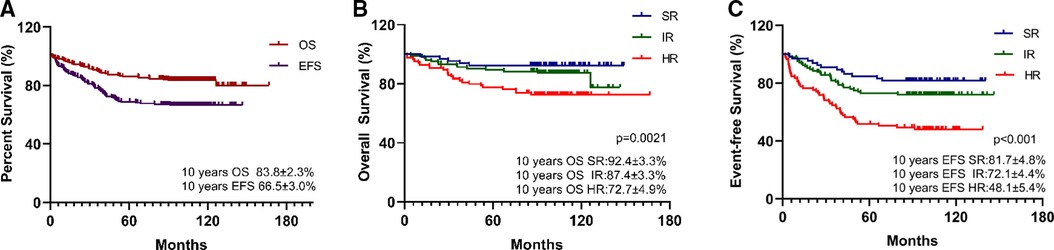
Figure 1. Survival analysis of pre-B-ALL treated by CCLG-2008 protocol (A) and the impact of risk group on long-term survival: 10-year OS (B), 10-year EFS (C), with P-values estimated by log-rank test (P = 0.0021 and P < 0.001), suggesting that risk group associated with poor survival.
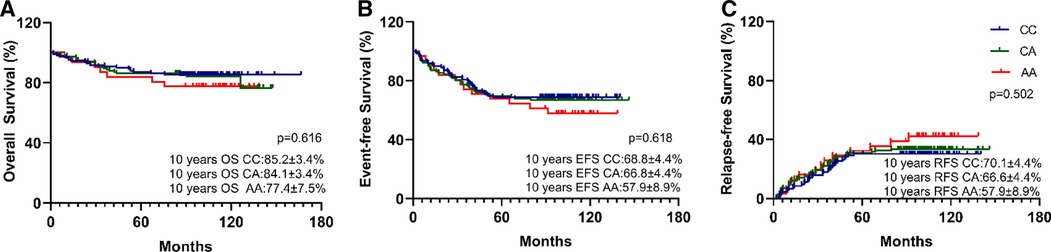
Figure 2. Kaplan-Meier curves for 10-year OS (A), 10-year EFS (B), and 10-year RFS (C) by GATA3 SNP rs3824662 genotype (AA, AC, or CC). Differences were assessed using a log-rank test. P-value was 0.616, 0.618, and 0.502 respectively. The A allele has no impact on long-term survival in B-ALL.
To further investigate the effect on treatment response, we analyzed the association between rs3824662 and morphologic and MRD responses. As shown in Table 3, patients with the AA genotype had a poorer prednisone treatment response than those with the CC and CA genotypes (9/23 vs. 22/202, P = 0.011), as reflected by peripheral blood blast counts. On day 15 of induction therapy, the M3 status was more frequent in the AA genotype than the M1 or M2 status (12/36 vs. 19/189, P = 0.007; Table 3). Although a slightly high MRD level was observed in patients with the AA genotype, the difference was not statistically significant (P = 0.606 and P = 0.521, respectively; Table 3). Interestingly, the high-risk ALL patients were more frequent in the AA genotype group than that in the SR or IR patients (P = 0.027; Table 3). To further clarify whether the prognosis of GATA3 SNP was affected by risk-based treatment, we analyzed the prognosis of the GATA3 rs3824662 genotype in the same risk group. The results showed that GATA3 rs3824662 had less influence on prognosis than the risk group did (Supplementary Figure S1).
GATA3 rs3824662 associated with susceptibility to sepsis
GATA3 is a master transcription factor involved in immune regulation. Thus, we investigated whether rs3824662 could increase the occurrence of chemotherapy-related infections. To test this hypothesis, we performed an association analysis between the rs3824662 genotype and treatment-related adverse effects. The risk allele A was associated with susceptibility to sepsis. The AA genotype had a higher occurrence of sepsis than the other genotypes (22/31 vs. 100/125, P = 0.021, Table 3). In addition, some patients experienced more than one adverse event during chemotherapy. Therefore, we performed a statistical analysis of the number of adverse events. As shown in Figure 3, patients with the AA genotype had more episodes of sepsis than those with the CC or CA genotype (P = 0.019). However, we did not identify a significant association between the rs3824662 genotype and other side effects, such as myelosuppression, pneumonia, and liver dysfunction (Table 3 and Figure 3).
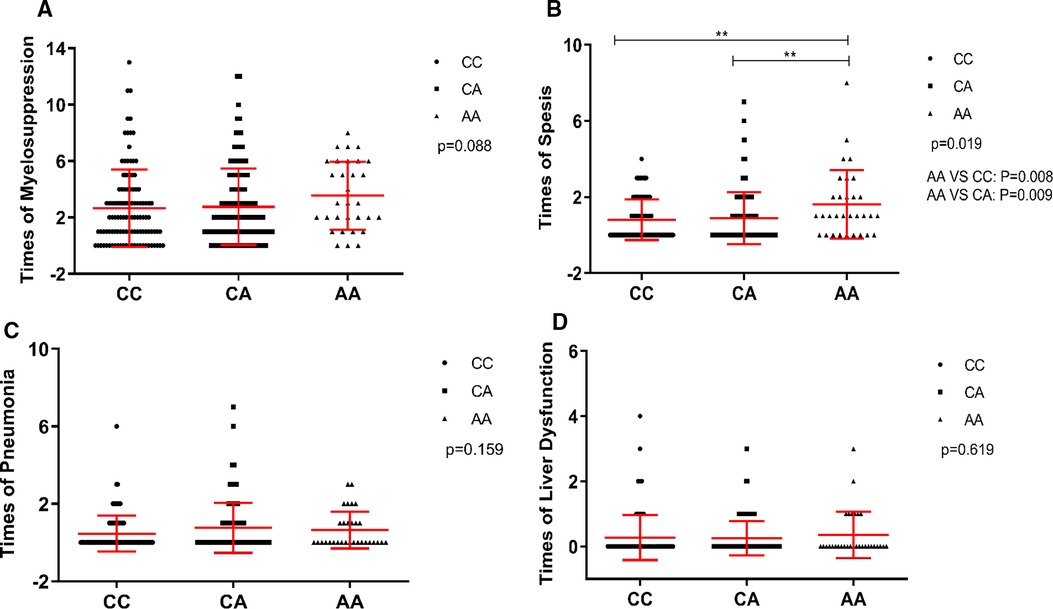
Figure 3. Association of times of myelosuppression (A), sepsis (B), pneumonia (C), and liver dysfunction (D) with genotype at rs3824662 in CCLG-2008 protocol, with P-values estimated by Kruskal-Wallis test. Indicating that the GATA3 SNP rs3824662 genotype has sepsis susceptibility.
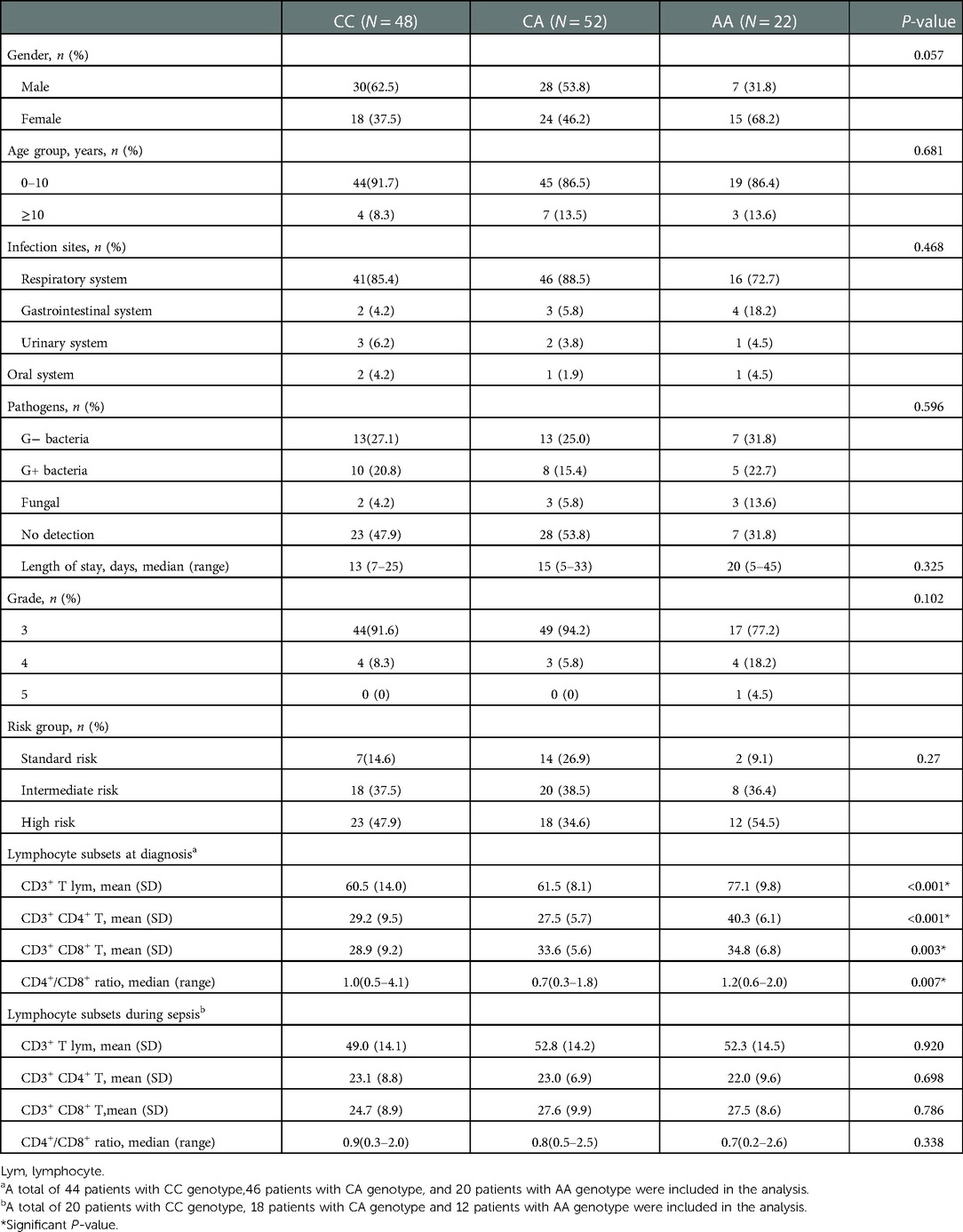
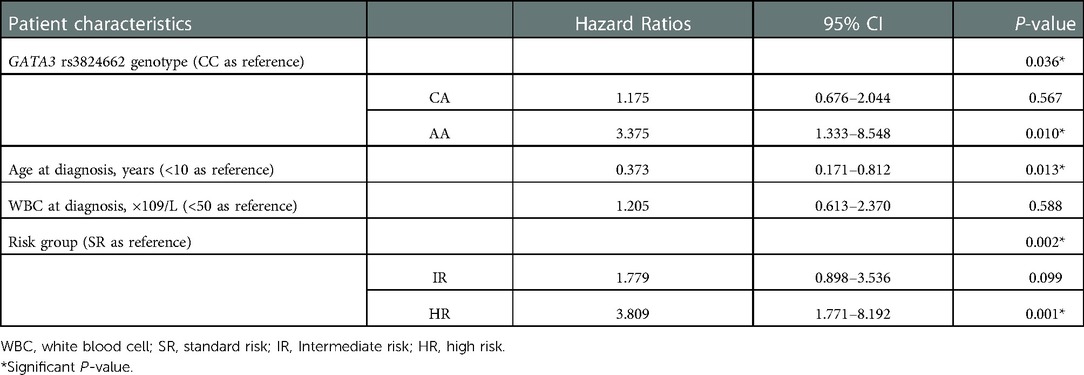
Next, we analyzed the features of sepsis among the rs3824662 genotypes. In general, based on our data, sepsis was most often caused by respiratory system infection, with rates of 85.4%, 88.5%, and 72.7% in the CC, CA, and AA genotypes, respectively. Bacteria were common pathogens in sepsis, whereas fungal infection was often accompanied by a longer course of treatment. High-risk group treatment may also have contributed to the incidence of sepsis, accounting for 43.4% of patients with sepsis. As most sepsis occurred during the period of bone marrow suppression after chemotherapy, advanced antibiotics, such as imipenem and vancomycin, were mainly used to cover most bacteria in early empirical treatment. There were no significant differences in sex, age, pathogens, length of stay, risk group, and grade of sepsis among the three groups (Table 4). Importantly, AA genotype remained statistically significantly associated with sepsis after adjusting clinical variables (i.e., age, white blood cell count, and risk group) (HR = 3.375; P = 0.01), as shown in Table 5.
Notably, patients with AA genotype had a higher percentage of CD3+ T cells than patients with CC and CA genotypes, mainly represented by the higher proportion of CD4+ T cells (P < 0.001) in peripheral blood at diagnosis (Table 4 and Supplementary Figure S2). Although there was no significant difference in the percentages of lymphocyte subsets among the rs3824662 genotypes in the early stage of sepsis. Compared with CC and CA genes, patients with AA genotype showed a more significant downward trend of CD3+ T cells and CD4+ T cells from diagnosis to sepsis (Supplementary Figure S3).
Discussion
GATA3 rs3824662 has been described as an ALL-susceptibility locus in several pediatric pre-B-cell ALL cohorts. Recent studies have also shown its impact on diabetes and systemic lupus erythematosus because of its crucial role in immunity (14, 15). In our ALL patients, the frequency of the AA genotype was close to the frequency tested in European populations and higher than that reported in African populations (16). Consistent with previous findings, we showed that the AA genotype was associated with poor early treatment outcomes in induction therapy. In addition, for the first time, a relationship between the AA genotype and the risk of sepsis during chemotherapy was documented. However, GATA3 rs3824662 did not show a significant difference in long-term prognosis.
Gabriele et al. identified GATA3 rs3824662 as a novel susceptibility locus for European childhood ALL through genome-wide association studies and linked it to poor early treatment outcomes (16). Similarly, Joanna et al. verified that an intronic mutation of GATA3 led to the risk of ALL in Polish pediatric patients, and the risk allele contributed to a high burden of MRD at the end of induction therapy (12). Consistent with the above studies, our study also revealed an increased frequency of the risk allele in pediatric pre-B-cell ALL compared to that in healthy controls (OR = 1.41, 95%, CI = 1.042–1.908, P = 0.026), which suggests that the A allele may be a risk factor for the development of pediatric pre-B-cell ALL in Chinese populations. The AA genotype had a poor prednisone response and a high level of bone marrow blasts on day 15 (P = 0.011 and 0.007, respectively), indicating its possible role in drug resistance in early chemotherapy. Therefore, GATA3 rs3824662 may be related to leukemogenesis and drug resistance in ALL. Aberrant GATA3 expression in different types of leukemia reveals its special role in oncogenesis. GATA3 is conventionally used as a tumor suppressor in early T-cell precursor ALL (ETP-ALL). Inactive mutations in GATA3, resulting in low GATA3 expression in ETP-ALL, are related to high DNA methylation and FLT3 mutations (17). However, a test of GATA3 mRNA expression in different types of leukemia demonstrated that compared to ETP-ALL patients, some B-ALL and non-ETP ALL patients showed extremely high GATA3 expression, suggesting that GATA3 may also act as an oncogene (18). In NOTCH1-induced T-ALL, GATA3 modulated the NOTCH1-enhancer by driving nucleosome eviction for the initiation and maintenance of tumors (19). However, the role of GATA3 in B-ALL remains unclear. Qianqian et al. reported that GATA3 may influence the development of B-ALL via epigenetic regulation of several pathways associated with leukemia (20). The GATA3 rs3824662 locus was identified with significantly high expression of GATA3 mRNA and DNase I hypersensitivity in lymphoblast cell lines (5). Moreover, PU.1 and P300 were found to bind to the rs3824662 loci in ChIp-seq data of lymphoid cells, which further confirmed the possible enhancer activity, indicating that the A allele affects local chromium access and transcription activity to improve GATA3 expression (5). Therefore, further research is required to explore the role of GATA3 rs3824662 in pre-B-cell ALL.
Our study demonstrated that the AA genotype of rs3824662 was associated with the occurrence of sepsis during chemotherapy. Several genetic variations in genes with immunological functions, such as CTL4, IL1-B, IL6, CD40, and HMGB1, have been reported to be associated with sepsis (21–25). GATA3 also plays an important role in immunology, particularly in the differentiation of Th2 cells (11). In our study, a significantly high percentage of CD3+ T cells, caused mainly by the high proportion of CD4+ T cells, was observed at diagnosis in patients with AA genotype. The transcription factor GATA3 has emerged as a critical regulator of both innate and adaptive immunities. Several clinical studies have shown that most pediatric ALL patients have a lower proportion of CD4+ and CD8+ T cells in the peripheral blood at diagnosis than healthy children of the same age stage. After cessation of chemotherapy, defects in immunity can recover to a certain level (26–28). Although GATA3 was originally identified as a master regulator of Th2 differentiation in CD4+ T cells, increasing evidence suggests that it is critical for the maintenance of mature T cells in the periphery (11). Therefore, we considered that the promotion of GATA3 expression by the rs3824662 site could, to a certain extent, compensate for the inhibition of tumor immunity to T cells in peripheral blood, contributing to the maintenance of T cells in peripheral blood at a stable level in the early stage of leukemia. In addition, the onset and development of sepsis involve many types of immune cells and cytokines, which can change the adaptive immune response, ultimately inducing Th1 cell inhibition and Th2 cell polarization (29). Huang et al. confirmed that GATA3 plays an important role in the early stages of sepsis. Triggered by TCR, upregulation of GATA3 causes the Th1/Th2 ratio to skew toward Th2, which promotes immunosuppression. GATA3 could also decrease the expression of the membrane adhesive protein annexin-A1, an anti-inflammatory protein, leading to an early imbalance of proinflammatory function (30). Although our data were limited in terms of Th cell subsets, risk genotype AA showed a more significant downward trend in the proportion of CD3+ T cells and CD4+ T cells from diagnosis to sepsis. Therefore, we hypothesized that the risk allele may accelerate the differentiation of Th2 cells and inhibit Th1 cells by increasing the expression of GATA3, ultimately leading to the balance disorder of Th1/Th2, which is more likely to cause sepsis. The GATA3 SNP rs3824662 may have also contributed to the apoptosis of CD4+ T cells in the onset of sepsis, accelerating the progress of sepsis. However, relevant mechanisms still need further research.
In summary, we identified risk loci at rs3824662 for pre-B-cell ALL in Han Chinese individuals, which could be a possible risk factor for poor early treatment response and sepsis. Further studies are needed to elucidate its role in the pathogenesis of pre-B-cell ALL and in the high incidence of sepsis. These analyses will help to provide a theoretical basis for evaluating clinical prognosis and risk stratification, which will also provide molecular targets for the prevention and treatment of chemotherapy-related complications.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Ethics statement
The studies involving human participants were reviewed and approved by Ethics Committee of Children's Hospital of Soochow University (No. 2020CS010). Written informed consent to participate in this study was provided by the participants’ legal guardian/next of kin.
Author contributions
All the authors contributed to the conception and design of the study. Material preparation, data collection, and analysis were performed by XC, DW, JG, and LC. FF guided the statistical methods. MQ and HZ designed the primers and the reaction conditions for the experiments. JP and SH modified the text. The first draft of the manuscript was written by XC, and all the authors commented on the previous versions of the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by a Translational Research Grant of NCRCH (No. 2020ZKPB02) and the Suzhou (GSWS2020039 and SZS201615) and Jiangsu (CXTDA2017014 and BE2021654) projects.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fped.2022.1044866/full#supplementary-material.
References
1. Walsh KM, de Smith AJ, Welch TC, Smirnov I, Cunningham MJ, Ma X, et al. Genomic ancestry and somatic alterations correlate with age at diagnosis in Hispanic children with B-cell acute lymphoblastic leukemia. Am J Hematol. (2014) 89(7):721–5. doi: 10.1002/ajh.23727
2. Xu H, Yang W, Perez-Andreu V, Devidas M, Fan Y, Cheng C, et al. Novel susceptibility variants at 10p12.31-12.2 for childhood acute lymphoblastic leukemia in ethnically diverse populations. J Natl Cancer Inst. (2013) 105(10):733–42. doi: 10.1093/jnci/djt042
3. Walsh KM, de Smith AJ, Chokkalingam AP, Metayer C, Roberts W, Barcellos LF, et al. GATA3 Risk alleles are associated with ancestral components in Hispanic children with ALL. Blood. (2013) 122(19):3385–7. doi: 10.1182/blood-2013-08-524124
4. Perez-Andreu V, Roberts KG, Xu H, Smith C, Zhang H, Yang W, et al. A genome-wide association study of susceptibility to acute lymphoblastic leukemia in adolescents and young adults. Blood. (2015) 125(4):680–6. doi: 10.1182/blood-2014-09-595744
5. Perez-Andreu V, Roberts KG, Harvey RC, Yang W, Cheng C, Pei D, et al. Inherited GATA3 variants are associated with Ph-like childhood acute lymphoblastic leukemia and risk of relapse. Nat Genet. (2013) 45(12):1494–8. doi: 10.1038/ng.2803
6. Clay-Gilmour AI, Hahn T, Preus LM, Onel K, Skol A, Hungate E, et al. Genetic association with B-cell acute lymphoblastic leukemia in allogeneic transplant patients differs by age and sex. Blood Adv. (2017) 1(20):1717–28. doi: 10.1182/bloodadvances.2017006023
7. Moriyama T, Relling MV, Yang JJ. Inherited genetic variation in childhood acute lymphoblastic leukemia. Blood. (2015) 125(26):3988–95. doi: 10.1182/blood-2014-12-580001
8. Wiegering V, Frank J, Freudenberg S, Morbach H, Schlegel PG, Eyrich M, et al. Impaired B-cell reconstitution in children after chemotherapy for standard or medium risk acute precursor B-lymphoblastic leukemia. Leuk Lymphoma. (2014) 55(4):870–5. doi: 10.3109/10428194.2013.816423
9. Gao J, Chen YH, Peterson LC. GATA Family transcriptional factors: emerging suspects in hematologic disorders. Exp Hematol Oncol. (2015) 4:28. doi: 10.1186/s40164-015-0024-z
10. Nakata Y, Brignier AC, Jin S, Shen Y, Rudnick SI, Sugita M, et al. c-Myb, Menin, GATA-3, and MLL form a dynamic transcription complex that plays a pivotal role in human T helper type 2 cell development. Blood. (2010) 116(8):1280–90. doi: 10.1182/blood-2009-05-223255
11. Wan YY. GATA3: a master of many trades in immune regulation. Trends Immunol. (2014) 35(6):233–42. doi: 10.1016/j.it.2014.04.002
12. Madzio J, Pastorczak A, Sedek L, Braun M, Taha J, Wypyszczak K, et al. GATA3 Germline variant is associated with CRLF2 expression and predicts outcome in pediatric B-cell precursor acute lymphoblastic leukemia. Genes Chromosomes Cancer. (2019) 58(9):619–26. doi: 10.1002/gcc.22748
13. Hu YX, Lu J, He HL, Wang Y, Li JQ, Xiao PF, et al. A prospective evaluation of minimal residual disease as risk stratification for CCLG-ALL-2008 treatment protocol in pediatric B precursor acute lymphoblastic leukemia. Eur Rev Med Pharmacol Sci. (2016) 20(9):1680–90.27212157
14. Mosaad YM, Hammad A, Elghzaly AA, Tawhid ZME, Hammad EM, Showma A, et al. GATA3 Rs3824662 gene polymorphism as possible risk factor for systemic lupus erythematosus. Lupus. (2018) 27(13):2112–9. doi: 10.1177/0961203318804894
15. Huda N, Hosen MI, Yasmin T, Sarkar PK, Hasan A, Nabi A. Genetic variation of the transcription factor GATA3, not STAT4, is associated with the risk of type 2 diabetes in the Bangladeshi population. PLoS One. (2018) 13(7):e0198507. doi: 10.1371/journal.pone.0198507
16. Pui CH, Yang JJ, Bhakta N, Rodriguez-Galindo C. Global efforts toward the cure of childhood acute lymphoblastic leukaemia. Lancet Child Adolesc Health. (2018) 2(6):440–54. doi: 10.1016/S2352-4642(18)30066-X
17. Fattizzo B, Rosa J, Giannotta JA, Baldini L, Fracchiolla NS. The physiopathology of T- cell acute lymphoblastic leukemia: focus on molecular aspects. Front Oncol. (2020) 10:273. doi: 10.3389/fonc.2020.00273
18. Fransecky L, Neumann M, Heesch S, Schlee C, Ortiz-Tanchez J, Heller S, et al. Silencing of GATA3 defines a novel stem cell-like subgroup of ETP-ALL. J Hematol Oncol. (2016) 9(1):95. doi: 10.1186/s13045-016-0324-8
19. Belver L, Yang AY, Albero R, Herranz D, Brundu FG, Quinn SA, et al. GATA3-controlled nucleosome eviction drives MYC enhancer activity in T-cell development and leukemia. Cancer Discov. (2019) 9(12):1774–91. doi: 10.1158/2159-8290.CD-19-0471
20. Hou Q, Liao F, Zhang S, Zhang D, Zhang Y, Zhou X, et al. Regulatory network of GATA3 in pediatric acute lymphoblastic leukemia. Oncotarget. (2017) 8(22):36040–53. doi: 10.18632/oncotarget.16424
21. Mewes C, Büttner B, Hinz J, Alpert A, Popov AF, Ghadimi M, et al. The CTLA-4 rs231775 GG genotype is associated with favorable 90-day survival in Caucasian patients with sepsis. Sci Rep. (2018) 8(1):15140. doi: 10.1038/s41598-018-33246-9
22. Varljen T, Sekulovic G, Rakic O, Maksimovic N, Jekic B, Novakovic I, et al. Genetic variant rs16944 in IL1B gene is a risk factor for early-onset sepsis susceptibility and outcome in preterm infants. Inflamm Res. (2020) 69(2):155–7. doi: 10.1007/s00011-019-01301-4
23. Zhang Y, Li M, Bao L, Hu P. A case-control study on the relationship between miRNAs single nucleotide polymorphisms and sepsis risk. Medicine (Baltimore). (2019) 98(33):e16744. doi: 10.1097/MD.0000000000016744
24. Liu ZL, Hu J, Xiao XF, Peng Y, Zhao SP, Xiao XZ, et al. The CD40 rs1883832 polymorphism affects sepsis susceptibility and sCD40l levels. Biomed Res Int. (2018) 2018:7497314. doi: 10.1155/2018/7497314.29780830
25. Qiu P, Wang L, Ni J, Zhang Y. Associations between HMGB1 gene polymorphisms and susceptibility and clinical outcomes in Chinese Han sepsis patients. Gene. (2019) 687:23–9. doi: 10.1016/j.gene.2018.11.027
26. Alanko S, Salmi TT, Pelliniemi TT. Recovery of blood T-cell subsets after chemotherapy for childhood acute lymphoblastic leukemia. Pediatr Hematol Oncol. (1994) 11(3):281–92. doi: 10.3109/08880019409141671
27. Ek T, Mellander L, Andersson B, Abrahamsson J. Immune reconstitution after childhood acute lymphoblastic leukemia is most severely affected in the high risk group. Pediatr Blood Cancer. (2005) 44(5):461–8. doi: 10.1002/pbc.20255
28. Haining WN, Neuberg DS, Keczkemethy HL, Evans JW, Rivoli S, Gelman R, et al. Antigen-specific T-cell memory is preserved in children treated for acute lymphoblastic leukemia. Blood. (2005) 106(5):1749–54. doi: 10.1182/blood-2005-03-1082
29. Delano MJ, Scumpia PO, Weinstein JS, Coco D, Nagaraj S, Kelly-Scumpia KM, et al. MyD88-dependent expansion of an immature GR-1(+)CD11b(+) population induces T cell suppression and Th2 polarization in sepsis. J Exp Med. (2007) 204(6):1463–74. doi: 10.1084/jem.20062602
Keywords: GATA3 rs3824662, pre-B-cell acute lymphoblastic leukemia, pediatric, early treatment, sepsis
Citation: Chu X, Qian M, Yang J, Wu D, Gao J, Cao L, Fang F, Pan J, Zhang H and Hu S (2023) Effect of GATA3 rs3824662 gene polymorphism in Han Chinese children with pre-B-cell acute lymphoblastic leukemia with 10 years follow-up. Front. Pediatr. 10:1044866. doi: 10.3389/fped.2022.1044866
Received: 15 September 2022; Accepted: 21 December 2022;
Published: 11 January 2023.
Edited by:
Tomasz Szczepanski, Medical University of Silesia, PolandReviewed by:
Giovanni Cazzaniga, University of Milano Bicocca, ItalyShouyue Zhang, Tsinghua University, China
© 2023 Chu, Qian, Yang, Wu, Gao, Cao, Fang, Pan, Zhang and Hu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shaoyan Hu aHVzaGFveWFuQHN1ZGEuZWR1LmNu Hui Zhang emhhbmdodWlyamhAZ3djbWMub3Jn
†These authors have contributed equally to this work and share first authorship
Specialty Section: This article was submitted to Pediatric Hematology and Hematological Malignancies, a section of the journal Frontiers in Pediatrics
 Xinran Chu
Xinran Chu Maoxiang Qian
Maoxiang Qian Jin Yang3
Jin Yang3 Jian Pan
Jian Pan Hui Zhang
Hui Zhang Shaoyan Hu
Shaoyan Hu