
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Pediatr. , 25 October 2021
Sec. Pediatric Infectious Diseases
Volume 9 - 2021 | https://doi.org/10.3389/fped.2021.756043
 Nisreen Khambati1*
Nisreen Khambati1* Laura Olbrich1,2,3
Laura Olbrich1,2,3 Jerrold Ellner4
Jerrold Ellner4 Padmini Salgame4
Padmini Salgame4 Rinn Song1,5†
Rinn Song1,5† Else Margreet Bijker1†
Else Margreet Bijker1†The diagnosis of pulmonary tuberculosis (TB) in children remains a significant challenge due to its paucibacillary nature, non-specificity of symptoms and suboptimal sensitivity of available diagnostic methods. In young children particularly, it is difficult to obtain high-quality sputum specimens for testing, with this group the least likely to be diagnosed, while most at risk of severe disease. The World Health Organization (WHO) has prioritized research into rapid biomarker-based tests for TB using easily obtainable non-sputum samples, such as saliva. However, the role of biomarkers in saliva for diagnosing TB in children has not been fully explored. In this mini-review, we discuss the value of saliva as a diagnostic specimen in children given its ready availability and non-invasive nature of collection, and review the literature on the use of host-based biomarkers in saliva for diagnosing active pulmonary TB in adults and children. Based on available data from adult studies, we highlight that combinations of cytokines and other proteins show promise in reaching WHO-endorsed target product profiles for new TB triage tests. Given the lack of pediatric research on host biomarkers in saliva and the differing immune response to TB infection between children and adults, we recommend that pediatric studies are now performed to discover and validate salivary host biosignatures for diagnosing pulmonary TB in children. Future directions for pediatric saliva studies are discussed, with suggestions for technologies that can be applied for salivary biomarker discovery and point-of-care test development.
In 2019, there were 192,000 deaths in children due to tuberculosis (TB), although most experts agree this figure is underestimated (1). Over 90% of child TB deaths occurred in those not receiving treatment, predominantly due to difficulties in diagnosis (2). The majority of missed TB cases occurs in children under five, the group with the highest risk of severe disease (3). Diagnosis of pulmonary TB in children is challenging due to its paucibacillary nature, non-specific symptoms, and limitations of the available diagnostic tests (4–6). Sputum microscopy is positive in <15% of children with probable TB and culture has a long turn-around time (7). Molecular DNA-based methods such as GeneXpert® are faster but require a stable electricity supply, limiting useability in low resource settings (8). Moreover, these tests require high-quality respiratory samples, which is problematic in young children who cannot expectorate sputum on demand, necessitating collection of gastric aspirates, nasopharyngeal aspirates, or induced sputum (9). These procedures are invasive and require trained staff (10), potentially missing children not presenting to secondary healthcare. In the absence of microbiological confirmation, diagnostic scoring systems based on clinical and radiological information are often used with variability in performance (7), and the risk of misdiagnosis (11).
The development of non-sputum-based point-of-care (POC) tests on easily obtainable samples like blood, urine, stool, and saliva has been prioritized by the World Health Organization (WHO) (12). These diagnostic approaches ideally should be feasible in primary care settings with the purpose of starting therapy during the same clinical encounter (13). Specific target product profiles (TPP) relating to the performance of new tests in children have been outlined by the WHO, with minimal targets of 66% sensitivity and 98% specificity for a diagnostic test and 90% sensitivity and 70% specificity for a triage test (12).
Diagnostic tests based on biomarkers have gained attention due to their potential for translation into non-sputum POC technologies (14). Biomarkers are characteristics that objectively indicate a normal biological or pathogenic process (15). Biomarker tests can be host-based, measuring the immune response following infection (16), or pathogen-based, identifying components of Mycobacterium tuberculosis (Mtb) (17). They may be able to identify culture-negative TB in early stages (18), which could help target the high number of children with microbiologically unconfirmed TB.
The role of saliva as a diagnostic specimen for pulmonary TB in children has not been fully explored. Detection of pathogen DNA using GeneXpert MTB/RIF in saliva from adults demonstrated low sensitivity (39%) compared to sputum (19), and sensitivity in children is likely to be lower due to the paucibacillary nature of pediatric disease. Moreover, the fact that TB disease in young children is often more severe and more frequently disseminated than in adults (20) conceivably makes children more suited to host-based diagnostics. This review therefore focuses on host-based salivary biomarkers. We outline the evidence on host biomarkers in saliva for diagnosing active pulmonary TB in relation to the WHO-endorsed TPP. We demonstrate the research gaps in children and based on available data from adult studies, discuss the potential for salivary biomarkers for children with recommendations for future pediatric studies.
Saliva contains various components enabling its potential as a sample for diagnosing infectious diseases (21). For example, detection of specific IgM in saliva has been used to confirm infection with measles, mumps and rubella (22). Congenital cytomegalovirus infection in neonates can be diagnosed through detection of viral DNA in saliva (21), and there has been interest in diagnosing SARS-CoV-2 infection in children using saliva instead of nasopharyngeal swabs (23). There is also a growing body of pediatric research demonstrating the clinical utility of salivary biomarkers for predicting metabolic syndrome (24) and diagnosing and monitoring chronic conditions like chronic kidney disease (25, 26) and hypertension (27). Whilst salivary assay development for adult conditions has increased, diagnostics specifically for pediatrics remain comparatively limited (21).
Saliva offers specific advantages compared to other sample types. It can be collected non-invasively and painlessly which is attractive for children and allows for repeated sampling (28). Importantly for TB, collection does not require highly skilled personnel, enabling useability in primary healthcare settings (29). As a mucosal and airway associated specimen, saliva may be better for studying host biomarkers in pulmonary TB compared to blood (30). Subbian et al. found that gene expression signatures in lung biopsies from TB patients only partly corresponded to those identified in their blood (31), suggesting that the lung-specific host response is not fully captured in blood-based biomarkers (32). With the first pathogen-host contact in pulmonary TB occurring in the oral and nasal passages, saliva may more accurately represent the immune response in the respiratory system (33).
Various collection systems are available for children. Whole saliva collected through passive drool into a collection tube is recognized as the optimal method (34). This avoids the potential issue of absorbent material interfering with analytes or the volume of saliva collected (35). It also may allow for a more consistent sample by avoiding saliva collection from specific salivary glands (36). However, this technique is only possible for older children who can cooperate (37). For infants and young children, absorbent oral swabs or eye spears which have a cellulose sponge on a shaft that can be inserted into the mouth, are feasible and safe (38, 39). Saliva is extracted from the absorbent device through centrifugation or compression (34).
We conducted a search to identify studies which reported the diagnostic performance of salivary host-based biomarkers for active pulmonary TB. PubMed and Web of Science databases were searched for diagnostic studies published up to 8th August 2021 using the search text words: “saliva” and “(tuberculos* OR TB OR Mtb)” and associated medical subject headings: “saliva,” “mycobacterium tuberculosis,” and “tuberculosis.” Animal studies were excluded. We did not specifically include “children” as a search term in line with the Cochrane Handbook for Systematic Reviews of Diagnostic Test Accuracy (40) to maximize sensitivity of the search. We found nine studies on host salivary diagnostic biomarkers, published between 1973 and 2021. Since only one study involved children (41), we also reviewed the evidence from eight adult studies (30, 42–48). Supplementary Figure 1 outlines the search procedure and Supplementary Table 1 summarizes the included studies.
A study in the USA in 1973 evaluated levels of hemagglutinating IgA to tuberculoprotein and tuberculopolysaccharide in saliva (47). Salivary antibodies were detected in only one of 20 TB cases, and also in one of 20 COPD patients, indicating very poor sensitivity and specificity. Later studies found more promising results. Araujo et al. evaluated secretory IgA in saliva against the 38 kDa antigen of Mtb in Venezuelan children using ELISA (41). Based on 34 children with TB and 46 healthy controls, sensitivity was only 36%, but specificity was 91%. Raras et al. also evaluated secretory IgA levels, although they measured antibodies against a recombinant semi-purified 38 kDa antigen in saliva from 30 TB adult cases and 30 healthy controls in Indonesia using dot blot (42). In contrast to Araujo et al., sensitivity was higher at 80%, but specificity was lower at 37% (42). Differences in population, geography and type of antibody assay may account for some of the variation between the two studies. The reference standard also differed: while Raras et al. defined a positive TB case based on sputum smear positivity (i.e., high bacillary burden) (42), Araujo et al. relied on clinical, radiological or microbiological information with only 3 of 34 positive cases confirmed bacteriologically (41).
Anti-38 kDa antibodies in blood generally offer good specificity but their low sensitivity has limited their potential as serological tests in diagnosing TB (49). Measurement in saliva does not appear to perform any better. In both studies of anti-38 kDa antibodies in saliva, diagnostic performance fell considerably short of TPP criteria. Moreover, both studies used a case control design instead of including a spectrum of patients in whom the test would be applied in clinical settings. This approach can overestimate diagnostic performance (50). In summary, despite their potential for translation into simple assays (16), the data on TB-specific antibodies in saliva as stand-alone tests for diagnostic purposes have not been encouraging to date.
Five studies measured salivary levels of selected cytokines, growth factors, enzymes and acute phase proteins in adults with confirmed TB or symptoms of TB using Luminex multiplex assays (30, 43–46). Of the 54 biomarkers analyzed in more than one study, eight showed statistically significant differences (p < 0.05) between TB patients and negative controls, albeit only one study reported correction for multiple comparisons (46). These eight biomarkers included alpha 2 macroglobulin (A2M), serum amyloid P (SAP), interferon gamma-induced protein (IP-10), vascular endothelial growth factor (VEGF), interleukin 6 (IL-6), monocyte chemoattractant protein-1 (MCP-1), fraktaline, and c-reactive protein (CRP). Sensitivities and specificities of these individual biomarkers ranged between 36 and 83% and between 52 and 96%, respectively (Table 1). Crucially, six biomarkers showed opposing trends between studies. For example, IP-10 was higher in TB patients in one study (46) whereas it was higher in healthy controls in another (45). Only salivary CRP and fraktaline showed a similar pattern across studies and were raised in TB cases in more than one study. No individual biomarkers met TPP criteria for a diagnostic or triage test.
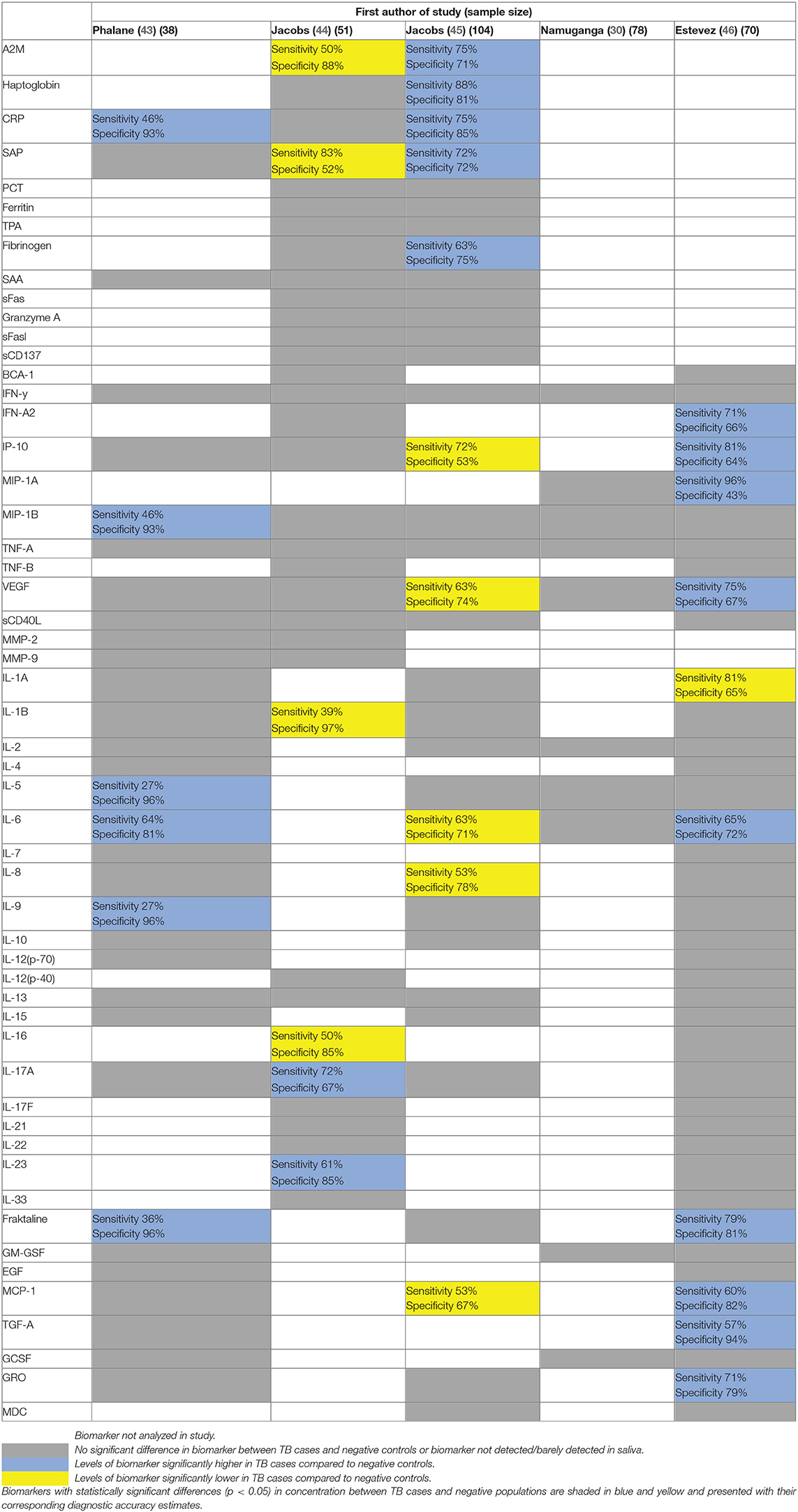
Table 1. Summary of individual biomarkers analyzed in more than one study using Luminex bead-based multiplex assays.
Compared to individual biomarkers, combinations of markers in biosignatures showed more potential. Table 2 summarizes the best performing biosignatures from each study. The most promising biosignatures originated from a prospective cohort study in South Africa with a low risk of bias by Jacobs and colleagues (44). They recruited 51 adults based on symptoms of pulmonary TB and compared biomarker levels between those with culture positive TB and with other respiratory diseases in a blinded manner. An eight-marker biosignature involving salivary granzyme A, growth differentiation factor 15 (GDF-15), serum amyloid A (SAA), epithelial-neutrophil activating peptide 78 (ENA-78), IL-12(p40), IL-13, IL-21, and plasminogen activator inhibitor-1 (PAI-1) had a sensitivity of 93% and specificity of 100% (40). This biosignature met TPP for both a triage and diagnostic test. Another eight-marker biosignature involving extracellular matrix protein 1 (ECM1), myoglobulin, hemofiltrate CC chemokine 1 (HCC1), tissue plasminogen activator (TPA), ENA-78, IL-12 (p40), IL-13, and IL-21 had a sensitivity of 100% and a specificity of 95%, reaching TPP for a triage test (44). Both these biosignatures were defined in adults who were HIV negative. The optimal biosignature evaluated in both HIV positive and negative individuals, included IL-1β, IL-23, ECM-1, HCC1, and fibrinogen, and had a sensitivity of 89% and specificity of 90% (44). Biosignatures from the other Luminex studies performed less well, but were overall closer in approaching the WHO-endorsed TPP than individual biomarkers (Table 2).
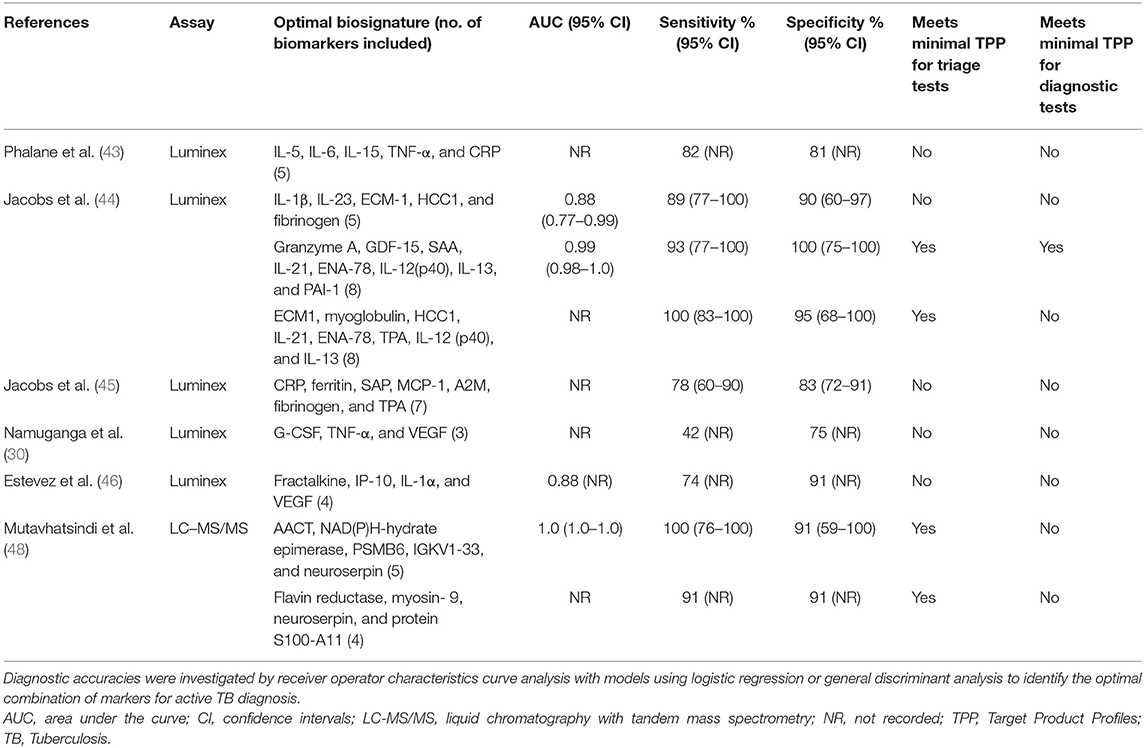
Table 2. Optimal biosignatures in relation to WHO-endorsed target product profiles for triage tests (90% sensitivity and 70% specificity) and diagnostic tests (66% sensitivity and 98% specificity).
There was little overlap in the optimal biosignatures identified across the Luminex studies. Only tumor necrosis factor α (TNF-α), CRP, fibrinogen, and VEGF appeared in the optimal biosignatures from more than one study. There are various possible explanations for this heterogeneity. All studies except one (46) reported cross validation methods to reduce overfitting of the data in their models. However, they did not validate results in an independent cohort to demonstrate generalizability. The diagnostic thresholds to define sensitivity and specificity for each biomarker and the statistical methods to determine these thresholds also varied amongst the studies. In terms of population, studies recruited from different geographic regions. Host genetics, microbiome, and coinfections may therefore vary, also attributing to heterogeneity in host response (32). Many of the promising biomarkers were acute phase reactants which could be affected by infections other than TB and otherwise altered immune states (43). Apart from HIV status, no study included information on other diseases that could have influenced biomarker levels. Lastly, two studies included healthy individuals as their negative controls (43, 46). This could overestimate diagnostic performance compared to those using clinically relevant negative populations (30, 44, 45).
In a discovery study, Mutavhatsindi et al. performed label-free liquid chromatography with tandem mass spectrometry on saliva from 22 adults with symptoms of TB (48). Following correction for multiple testing, they identified 26 differentially expressed proteins between adults with culture-positive TB and those with other respiratory diseases. Five proteins diagnosed TB with an area under the ROC curve above 0.8 (macrophage-capping protein, plasminogen, profilin-1, f-actin-capping protein subunit beta, and alpha-1-antichymotrysin). Most of these 26 proteins have not previously been investigated in this context, however, many are involved in inflammation, activation of the immune system and enzyme regulation (48). Similarly to the Luminex studies described above, TPP criteria were only achieved when markers were used in combination. After leave-one out cross validation, a five protein biosignature diagnosed TB with a sensitivity of 100% and specificity of 90.9% and a four protein biosignature showed a sensitivity and specificity of 90.9% (Table 2). Whilst validation in larger studies is necessary, these results further demonstrate the potential of multiple salivary proteins as diagnostic biomarkers.
We reviewed the current evidence on salivary host-based biomarkers for diagnosing pulmonary TB. Based on adult studies, combinations of cytokines and other proteins appear more promising than single proteins or antibodies. Despite the heterogeneity, many of these proteins are involved in the inflammatory response and may be elevated in diseases other than TB (51), therefore, specificity may be an issue in saliva host-based tests. This is important in children in whom respiratory infections are common. Saliva host diagnostics therefore primarily have potential as triage tests, distinguishing between those unlikely to have TB and those requiring confirmatory testing. Triage tests have a lower specificity requirement (>70% minimally) compared to diagnostic tests (98%) (12), but they need a high sensitivity and negative predictive value given the high fatality rate in young children. Ahmad and colleagues found that addition of serum antibodies against the TB antigen Ag85B to a four-cytokine blood-based triage test increased sensitivity from 80 to 86% and specificity from 65 to 69% (52). Although this fell just short of TPP criteria, it illustrates a useful strategy for enhancing diagnostic performance by expansion of protein-based tests with another diagnostic assay.
With the exception of one study on immunoglobulins in children, all studies on salivary biomarkers were conducted in adults. Despite the lack of pediatric research, salivary proteins in children are worth exploring. Host cytokine biosignatures in blood have showed promise in reaching TPP for triage tests in both adults (53) and children (54), although the blood cytokine expression profile differed between the two populations. Disappointing salivary cytokine biomarkers from adult studies do not necessary preclude value for children. Given the complex differences in the TB immune response between children and adults (32, 55), candidate salivary biomarkers for diagnostic assays may vary. Therefore, early phase pediatric studies should first be targeted to identify biosignatures which can identify symptomatic children with microbiologically confirmed TB (56).
Various technologies exist for biomarker discovery in saliva. High-throughput multiplex assays with bead-based (Luminex) or planar (Mesoscale) technology can measure multiple proteins simultaneously, with minimal sample requirements compared to traditional methods like ELISA (57). However, such platforms rely on targeted antibody capture-based strategies and issues relating to antibody specificity and cross-reactivity can limit the number of multiplexable targets (58). Shotgun proteomics, although more expensive and resource-intensive, may be a more comprehensive and unbiased method to biomarker discovery (48). A proteomics approach also has potential to discover novel proteins within extracellular vesicles, such as exosomes, which are found in saliva and have shown promise as non-invasive diagnostic markers for other pulmonary diseases (59, 60).
Subsequently, biosignatures identified from discovery studies should be independently validated in well-characterized cohorts where the test is clinically indicated (51). These cohorts should include children with the full spectrum of suspected TB and with other respiratory diseases presenting similarly to TB (56). Further validation in multiple populations under real-life field conditions will further assess robustness and accuracy and inform selection of biomarkers for potential implementation into POC tests (61). Conducting studies in diverse geographical locations will also help capture how genetic heterogeneity and location-specific exposure to microorganisms impact upon host responses to TB (32).
Measurement of several biomarkers using multiplex immunoassays or shotgun proteomics is not feasible at lower levels of care. However, recent developments have highlighted the possibility of translating validated biosignatures into affordable POC technology. Application of a lateral flow assay detecting two blood chemokines was feasible in a multi-center study in Africa, without requiring a cold chain for storage or distribution (62). Three- and six-biomarker POC tests using fingerstick blood are currently being validated as triage tests at multiple African sites (63). Whilst the viscosity of saliva and potentially low concentration of analytes present specific challenges for saliva-based POC platforms, electrochemical immunosensors have been suggested as possible solutions (64). Such biosensors contain an electrochemical transducer and have been used successfully to detect cytokines in saliva at low detection limits (65). Unique advantages, including high sensitivity, low cost, simple operation, rapid response and functionality in turbid media (64, 66) mark them as potential tools for future saliva POC diagnostic devices.
Future pediatric studies should prioritize recruitment of those under five (67). Given that pediatric TB is heterogeneous, approval of diagnostics for children under five should ideally be based on data specifically from this age group. Collecting clinical information on comorbidities, particularly HIV, malnutrition and respiratory coinfections, would help appreciate how disease phenotype can influence the host response. Unlike in adults, sputum-based reference tests are often not appropriate in young children. Microbiological reference standards based on combinations of alternative samples such as gastric or nasopharyngeal aspirates are therefore recommended (68). To ensure comparability of results across studies, classification of children should conform to the National Institutes of Health consensus definitions for childhood pulmonary tuberculosis (i.e., confirmed, unconfirmed, and unlikely TB) (69). Finally, the significant upfront investment in setting up clinical pediatric cohorts has led to recommendations to establish pediatric specimen biorepositories (67). As improved diagnostics become available, storage of saliva may allow future biomarker testing from cohorts with different geographical locations, age groups, and comorbidities.
Saliva could be a valuable specimen for testing host biomarkers to diagnose pulmonary TB in children, however very little research in this population exists. Discovery studies in adults show that host salivary proteins, when used in combination as biosignatures, demonstrate considerable promise as triage tests. Given the differing TB immune response in children, studies in pediatric populations are now needed. The ready availability of saliva and non-invasive nature of collection is appealing for children, particularly those under five. Late inclusion of children into clinical studies of new technologies is a major barrier to progress in childhood TB (3). The COVID-19 pandemic is expected to have worsened TB control, with 6.3 additional million cases estimated to occur between 2020 and 2025, and children are especially vulnerable (1). Novel and optimized diagnostics in children are now more than ever urgently required.
NK, RS, and EB devised the review and the main conceptual ideas. NK drafted the manuscript. LO, JE, PS, RS, and EB critically revised the work. All authors approved the final version of the manuscript.
Research reported in this publication was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under Award Number R01AI152159. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We would like to thank Dr. Jayne Sutherland at the Medical Research Council Unit in the Gambia, for her helpful comments and insights in this article.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fped.2021.756043/full#supplementary-material
A2M, alpha 2 macroglobulin; AACT, alpha-1- antichymotrypsin; AUC, area under the curve; BCA-1, B cell-attracting chemokine 1; CI, confidence intervals; CRP, c-reactive protein; EGF, epidermal growth factor; ENA-78, epithelial-neutrophil activating peptide 78; ECM1, extracellular matrix protein 1; GCSF, granulocyte-colony stimulating factor; GM-CSF, granulocyte-macrophage colony-stimulating factor; GDF-15, growth differentiation factor 15; HCC1, hemofiltrate CC chemokine 1; IFN-y, interferon gamma; IFN-A2, interferon alpha-2; IP-10, interferon gamma-induced protein 10; IL-1A, interleukin 1 alpha; IL-1B, interleukin 1 beta; IL-2, interleukin 2; IL-4, interleukin 4; IL-5, interleukin 5; IL-6, interleukin 6; IL-7, interleukin 7; IL-8, interleukin 8; IL-9, interleukin 9; IL-10, interleukin 10; IL-12(p-40), interleukin 12(p-40); IL-12(p-70), interleukin 12(p-70); IL-13, interleukin 13; IL-15, interleukin 15; IL-16, interleukin 16; IL-17A, interleukin 17A; IL-17F, interleukin17F; IL-21, interleukin 21; IL-22, interleukin 22; IL-23, interleukin 23; IL-33, interleukin 33; IGKV1-33, immunoglobulin kappa variable 1–33; LC-MS/MS, liquid chromatography with tandem mass spectrometry; MDC, macrophage-derived chemokine; MIP-1A, macrophage inflammatory protein 1-alpha; MIP-1B, macrophage inflammatory protein 1-beta; MMP-2, matrix metalloproteinase-2; MMP-9, matrix metalloproteinase-9; MCP-1, monocyte chemoattractant protein-1; MGIT, Mycobacteria growth indicator tube; Mtb, Mycobacterium tuberculosis; NR, not recorded; PCT, procalcitonin; PAI-1, plasminogen activator inhibitor-1; PSMB6, proteasome subunit beta type-6; SAA, serum amyloid A; SAP, serum amyloid P component; sCF40L, soluble CD40 ligand; sCD137, soluble CD137; sFAS, soluble Fas; sFASl, soluble Fas ligand; SD, standard deviation; TPA, tissue plasminogen activator; TB, Tuberculosis; TPP, Target Product Profile; TGF-A, transforming growth factor-alpha; TST, tuberculin skin test; TNF-A, tumor necrosis factor alpha; TNF-B, tumor necrosis factor beta, VEGF, vascular endothelial growth factor; WHO, World Health Organization; ZN, Ziehl Neelson.
2. Dodd PJ, Yuen CM, Sismanidis C, Seddon JA, Jenkins HE. The global burden of tuberculosis mortality in children: a mathematical modelling study. Lancet Glob Health. (2017) 5:e898–906. doi: 10.1016/S2214-109X(17)30289-9
3. The World Health Organization. Roadmap Towards Ending TB in Children and Adolescents. 2nd ed. Geneva: The WHO (2018).
4. Nkereuwem E, Kampmann B, Togun T. The need to prioritise childhood tuberculosis case detection. Lancet. (2021) 397:1248–9. doi: 10.1016/S0140-6736(21)00672-3
5. Tebruegge M, Ritz N, Curtis N, Shingadia D. Diagnostic tests for childhood tuberculosis: past imperfect, present tense and future perfect? Pediatr Infect Dis J. (2015) 34:1014–9. doi: 10.1097/INF.0000000000000796
6. Dunn JJ, Starke JR, Revell PA. Laboratory diagnosis of mycobacterium tuberculosis infection and disease in children. J Clin Microbiol. (2016) 54:1434–41. doi: 10.1128/JCM.03043-15
7. Nicol MP, Zar HJ. New specimens and laboratory diagnostics for childhood pulmonary TB: progress and prospects. Paediatr Respir Rev. (2011) 12:16–21. doi: 10.1016/j.prrv.2010.09.008
8. MacLean E, Kohli M, Weber SF, Suresh A, Schumacher SG, Denkinger CM, et al. Advances in molecular diagnosis of tuberculosis. J Clin Microbiol. (2020) 58:e01582–19. doi: 10.1128/JCM.01582-19
9. Perez-Velez CM, Roya-Pabon CL, Marais BJ. A systematic approach to diagnosing intra-thoracic tuberculosis in children. J Infect. (2017) 74 (Suppl. 1):S74–83. doi: 10.1016/S0163-4453(17)30195-0
10. Zar HJ, Hanslo D, Apolles P, Swingler G, Hussey G. Induced sputum versus gastric lavage for microbiological confirmation of pulmonary tuberculosis in infants and young children: a prospective study. Lancet. (2005) 365:130–4. doi: 10.1016/S0140-6736(05)17702-2
11. Schumacher SG, van Smeden M, Dendukuri N, Joseph L, Nicol MP, Pai M, et al. Diagnostic test accuracy in childhood pulmonary tuberculosis: a bayesian latent class analysis. Am J Epidemiol. (2016) 184:690–700. doi: 10.1093/aje/kww094
12. Denkinger CM, Kik SV, Cirillo DM, Casenghi M, Shinnick T, Weyer K, et al. Defining the needs for next generation assays for tuberculosis. J Infect Dis. (2015) 211 (Suppl. 2):S29–38. doi: 10.1093/infdis/jiu821
13. The World Health Organization. High-Priority Target Product Profiles or New Tuberculosis Diagnostics: Report of a Consensus Meeting. Geneva: The WHO (2014).
14. Togun T, Hoggart CJ, Agbla SC, Gomez MP, Egere U, Sillah AK, et al. A three-marker protein biosignature distinguishes tuberculosis from other respiratory diseases in Gambian children. EBioMedicine. (2020) 58:102909. doi: 10.1016/j.ebiom.2020.102909
15. Biomarkers Definitions Working Group. Biomarkers and surrogate endpoints: preferred definitions and conceptual framework. Clin Pharmacol Ther. (2001) 69:89–95. doi: 10.1067/mcp.2001.113989
16. Yong YK, Tan HY, Saeidi A, Wong WF, Vignesh R, Velu V, et al. Immune biomarkers for diagnosis and treatment monitoring of tuberculosis: current developments and future prospects. Front Microbiol. (2019) 10:2789. doi: 10.3389/fmicb.2019.02789
17. Tucci P, Gonzalez-Sapienza G, Marin M. Pathogen-derived biomarkers for active tuberculosis diagnosis. Front Microbiol. (2014) 5:549. doi: 10.3389/fmicb.2014.00549
18. Drain PK, Gardiner J, Hannah H, Broger T, Dheda K, Fielding K, et al. Guidance for studies evaluating the accuracy of biomarker-based nonsputum tests to diagnose tuberculosis. J Infect Dis. (2019) 220 (220 Suppl. 3):S108–15. doi: 10.1093/infdis/jiz356
19. Shenai S, Amisano D, Ronacher K, Kriel M, Banada PP, Song T, et al. Exploring alternative biomaterials for diagnosis of pulmonary tuberculosis in HIV-negative patients by use of the GeneXpert MTB/RIF assay. J Clin Microbiol. (2013) 51:4161–6. doi: 10.1128/JCM.01743-13
20. Roya-Pabon CL, Perez-Velez CM. Tuberculosis exposure, infection and disease in children: a systematic diagnostic approach. Pneumonia. (2016) 8:23. doi: 10.1186/s41479-016-0023-9
21. Hassaneen M, Maron JL. Salivary diagnostics in pediatrics: applicability, translatability, and limitations. Front Public Health. (2017) 5:83. doi: 10.3389/fpubh.2017.00083
22. Public Health England. Measles: The Green Book, chapter 21. London: Public Health England (2019).
23. Al Suwaidi H, Senok A, Varghese R, Deesi Z, Khansaheb H, Pokasirakath S, et al. Saliva for molecular detection of SARS-CoV-2 in school-age children. Clin Microbiol Infect. (2021) 27:1330–5. doi: 10.1016/j.cmi.2021.02.009
24. Goodson JM, Kantarci A, Hartman ML, Denis GV, Stephens D, Hasturk H, et al. Metabolic disease risk in children by salivary biomarker analysis. PLoS ONE. (2014) 9:e98799. doi: 10.1371/journal.pone.0098799
25. Maciejczyk M, Szulimowska J, Skutnik A, Taranta-Janusz K, Wasilewska A, Wisniewska N, et al. Salivary biomarkers of oxidative stress in children with chronic kidney disease. J Clin Med. (2018) 7:209. doi: 10.3390/jcm7080209
26. Kovalcikova AG, Pavlov K, Liptak R, Hladova M, Renczes E, Boor P, et al. Dynamics of salivary markers of kidney functions in acute and chronic kidney diseases. Sci Rep. (2020) 10:21260. doi: 10.1038/s41598-020-78209-1
27. Maciejczyk M, Taranta-Janusz K, Wasilewska A, Kossakowska A, Zalewska A. A case-control study of salivary redox homeostasis in hypertensive children. Can salivary uric acid be a marker of hypertension? J Clin Med. (2020) 9:837. doi: 10.3390/jcm9030837
28. Pappa E, Kousvelari E, Vastardis H. Saliva in the “Omics” era: a promising tool in paediatrics. Oral Dis. (2019) 25:16–25. doi: 10.1111/odi.12886
29. Sapkota D, Soland TM, Galtung HK, Sand LP, Giannecchini S, To KKW, et al. COVID-19 salivary signature: diagnostic and research opportunities. J Clin Pathol. (2020) 74:344–9. doi: 10.1136/jclinpath-2020-206834
30. Namuganga AR, Chegou NN, Mubiri P, Walzl G, Mayanja-Kizza H. Suitability of saliva for Tuberculosis diagnosis: comparing with serum. BMC Infect Dis. (2017) 17:600. doi: 10.1186/s12879-017-2687-z
31. Subbian S, Tsenova L, Kim MJ, Wainwright HC, Visser A, Bandyopadhyay N, et al. Lesion-specific immune response in granulomas of patients with pulmonary tuberculosis: a pilot study. PLoS ONE. (2015) 10:e0132249. doi: 10.1371/journal.pone.0132249
32. Burel JG, Babor M, Pomaznoy M, Lindestam Arlehamn CS, Khan N, Sette A, et al. Host transcriptomics as a tool to identify diagnostic and mechanistic immune signatures of tuberculosis. Front Immunol. (2019) 10:221. doi: 10.3389/fimmu.2019.00221
33. Mateos J, Estevez O, Gonzalez-Fernandez A, Anibarro L, Pallares A, Reljic R, et al. High-resolution quantitative proteomics applied to the study of the specific protein signature in the sputum and saliva of active tuberculosis patients and their infected and uninfected contacts. J Proteomics. (2019) 195:41–52. doi: 10.1016/j.jprot.2019.01.010
34. Bellagambi FG, Lomonaco T, Salvo P, Vivaldi F, Hangouët M, Ghimenti S, et al. Saliva sampling: methods and devices. An overview. TrAC Trends Anal Chem. (2020) 124:115781. doi: 10.1016/j.trac.2019.115781
35. Groschl M. Saliva: a reliable sample matrix in bioanalytics. Bioanalysis. (2017) 9:655–68. doi: 10.4155/bio-2017-0010
36. Salimetrics. Saliva Collection Handbook. (2020). Available online at: https://salimetrics.com/saliva-collection-handbook/#saliva-collection-devices-for-adults (accessed May 7, 2021).
37. Keil MF. Salivary cortisol: a tool for biobehavioral research in children. J Pediatr Nurs. (2012) 27:287–9. doi: 10.1016/j.pedn.2012.02.003
38. Yap TL, Fan JD, Ho MF, Choo CSC, Ong LY, Chen Y. Salivary biomarker for acute appendicitis in children: a pilot study. Pediatr Surg Int. (2020) 36:621–7. doi: 10.1007/s00383-020-04645-9
39. Orami T, Ford R, Kirkham LA, Thornton R, Corscadden K, Richmond PC, et al. Pneumococcal conjugate vaccine primes mucosal immune responses to pneumococcal polysaccharide vaccine booster in Papua New Guinean children. Vaccine. (2020) 38:7977–88. doi: 10.1016/j.vaccine.2020.10.042
40. de Vet HCW, Eisinga A, Riphagen II, Aertgeerts B, Pewsner D. Chapter 7: searching for studies. In: Eising A, editor. Cochrane Handbook for Systematic Reviews of Diagnostic Test Accuracy Version 0.4. The Cochrane Collaboration (2008).
41. Araujo Z, de Waard JH, Fernández de Larrea C, López D, Fandiño C, Maldonado A, et al. Study of the antibody response against Mycobacterium tuberculosis antigens in Warao Amerindian children in Venezuela. Mem Inst Oswaldo Cruz. (2004) 99:517–24. doi: 10.1590/S0074-02762004000500011
42. Raras TY, Sholeh G, Lyrawati D. Salivary sIg-A response against the recombinant Ag38 antigen of Mycobacterium tuberculosis Indonesian strain. Int J Clin Exp Med. (2014) 7:129–35.
43. Phalane KG, Kriel M, Loxton AG, Menezes A, Stanley K, van der Spuy GD, et al. Differential expression of host biomarkers in saliva and serum samples from individuals with suspected pulmonary tuberculosis. Mediators Inflamm. (2013) 2013:981984. doi: 10.1155/2013/981984
44. Jacobs R, Maasdorp E, Malherbe S, Loxton AG, Stanley K, van der Spuy G, et al. Diagnostic potential of novel salivary host biomarkers as candidates for the immunological diagnosis of tuberculosis disease and monitoring of tuberculosis treatment response. PLoS ONE. (2016) 11:e0160546. doi: 10.1371/journal.pone.0160546
45. Jacobs R, Tshehla E, Malherbe S, Kriel M, Loxton AG, Stanley K, et al. Host biomarkers detected in saliva show promise as markers for the diagnosis of pulmonary tuberculosis disease and monitoring of the response to tuberculosis treatment. Cytokine. (2016) 81:50–6. doi: 10.1016/j.cyto.2016.02.004
46. Estevez O, Anibarro L, Garet E, Pallares A, Pena A, Villaverde C, et al. Identification of candidate host serum and saliva biomarkers for a better diagnosis of active and latent tuberculosis infection. PLoS ONE. (2020) 15:e0235859. doi: 10.1371/journal.pone.0235859
47. Barlow PB, Daniel TM. The immunoglobulin response to tuberculosis. 3. Studies of hemagglutinating antibody to tuberculoprotein and tuberculopolysaccharide in serum, sputum, and saliva of patients with tuberculosis. Am Rev Respir Dis. (1973) 108:675–6. doi: 10.1164/arrd.1973.108.3.675
48. Mutavhatsindi H, Calder B, McAnda S, Malherbe ST, Stanley K, Kidd M, et al. Identification of novel salivary candidate protein biomarkers for tuberculosis diagnosis: a preliminary biomarker discovery study. Tuberculosis. (2021) 130:102118. doi: 10.1016/j.tube.2021.102118
49. Welch RJ, Lawless KM, Litwin CM. Antituberculosis IgG antibodies as a marker of active Mycobacterium tuberculosis disease. Clin Vaccine Immunol. (2012) 19:522–6. doi: 10.1128/CVI.05573-11
50. Togun TO, MacLean E, Kampmann B, Pai M. Biomarkers for diagnosis of childhood tuberculosis: a systematic review. PLoS ONE. (2018) 13:e0204029. doi: 10.1371/journal.pone.0204029
51. MacLean E, Broger T, Yerlikaya S, Fernandez-Carballo BL, Pai M, Denkinger CM. A systematic review of biomarkers to detect active tuberculosis. Nat Microbiol. (2019) 4:748–58. doi: 10.1038/s41564-019-0380-2
52. Ahmad R, Xie L, Pyle M, Suarez MF, Broger T, Steinberg D, et al. A rapid triage test for active pulmonary tuberculosis in adult patients with persistent cough. Sci Transl Med. (2019) 11: eaaw8287. doi: 10.1126/scitranslmed.aaw8287
53. Chegou NN, Sutherland JS, Malherbe S, Crampin AC, Corstjens PL, Geluk A, et al. Diagnostic performance of a seven-marker serum protein biosignature for the diagnosis of active TB disease in African primary healthcare clinic attendees with signs and symptoms suggestive of TB. Thorax. (2016) 71:785–94. doi: 10.1136/thoraxjnl-2015-207999
54. Kumar NP, Hissar S, Thiruvengadam K, Banurekha VV, Suresh N, Shankar J, et al. Discovery and validation of a three-cytokine plasma signature as a biomarker for diagnosis of pediatric tuberculosis. Front Immunol. (2021) 12:653898. doi: 10.3389/fimmu.2021.653898
55. Basu Roy R, Whittaker E, Seddon JA, Kampmann B. Tuberculosis susceptibility and protection in children. Lancet Infect Dis. (2019) 19:e96–108. doi: 10.1016/S1473-3099(18)30157-9
56. Cuevas LE, Browning R, Bossuyt P, Casenghi M, Cotton MF, Cruz AT, et al. Evaluation of tuberculosis diagnostics in children: 2. Methodological issues for conducting and reporting research evaluations of tuberculosis diagnostics for intrathoracic tuberculosis in children. Consensus from an expert panel. J Infect Dis. (2012) 205 (Suppl. 2):S209–15. doi: 10.1093/infdis/jir879
57. Gunther A, Becker M, Gopfert J, Joos T, Schneiderhan-Marra N. Comparison of bead-based fluorescence versus planar electrochemiluminescence multiplex immunoassays for measuring cytokines in human plasma. Front Immunol. (2020) 11:572634. doi: 10.3389/fimmu.2020.572634
58. Carlyle BC, Trombetta BA, Arnold SE. Proteomic approaches for the discovery of biofluid biomarkers of neurodegenerative dementias. Proteomes. (2018) 6:32. doi: 10.3390/proteomes6030032
59. Biadglegne F, Konig B, Rodloff AC, Dorhoi A, Sack U. Composition and clinical significance of exosomes in tuberculosis: a systematic literature review. J Clin Med. (2021) 10:145. doi: 10.3390/jcm10010145
60. Sun Y, Huo C, Qiao Z, Shang Z, Uzzaman A, Liu S, et al. Comparative proteomic analysis of exosomes and microvesicles in human saliva for lung cancer. J Proteome Res. (2018) 17:1101–7. doi: 10.1021/acs.jproteome.7b00770
61. Mulenga H, Zauchenberger CZ, Bunyasi EW, Mbandi SK, Mendelsohn SC, Kagina B, et al. Performance of diagnostic and predictive host blood transcriptomic signatures for Tuberculosis disease: a systematic review and meta-analysis. PLoS ONE. (2020) 15:e0237574. doi: 10.1371/journal.pone.0237574
62. Corstjens PL, Tjon Kon Fat EM, de Dood CJ, van der Ploeg-van Schip JJ, Franken KL, Chegou NN, et al. Multi-center evaluation of a user-friendly lateral flow assay to determine IP-10 and CCL4 levels in blood of TB and non-TB cases in Africa. Clin Biochem. (2016) 49:22–31. doi: 10.1016/j.clinbiochem.2015.08.013
63. TriageTB. TriageTB- Activties. (2021). Available online at: https://www.triagetb.com/activities (accessed May 5, 2021).
64. Chen X, Dong T, Wei X, Yang Z, Matos Pires NM, Ren J, et al. Electrochemical methods for detection of biomarkers of chronic obstructive pulmonary disease in serum and saliva. Biosens Bioelectron. (2019) 142:111453. doi: 10.1016/j.bios.2019.111453
65. Aydin EB, Sezginturk MK. An impedimetric immunosensor for highly sensitive detection of IL-8 in human serum and saliva samples: a new surface modification method by 6-phosphonohexanoic acid for biosensing applications. Anal Biochem. (2018) 554:44–52. doi: 10.1016/j.ab.2018.05.030
66. Golichenari B, Nosrati R, Farokhi-Fard A, Faal Maleki M, Gheibi Hayat SM, Ghazvini K, et al. Electrochemical-based biosensors for detection of Mycobacterium tuberculosis and tuberculosis biomarkers. Crit Rev Biotechnol. (2019) 39:1056–77. doi: 10.1080/07388551.2019.1668348
67. Nicol MP, Gnanashanmugam D, Browning R, Click ES, Cuevas LE, Detjen A, et al. A blueprint to address research gaps in the development of biomarkers for pediatric tuberculosis. Clin Infect Dis. (2015) 61 (Suppl. 3):S164–72. doi: 10.1093/cid/civ613
68. Song R, Click ES, McCarthy KD, Heilig CM, McHembere W, Smith JP, et al. Sensitive and feasible specimen collection and testing strategies for diagnosing tuberculosis in young children. JAMA Pediatr. (2021) 175:e206069. doi: 10.1001/jamapediatrics.2020.6069
Keywords: saliva, tuberculosis, biomarker, diagnosis, host-specificity
Citation: Khambati N, Olbrich L, Ellner J, Salgame P, Song R and Bijker EM (2021) Host-Based Biomarkers in Saliva for the Diagnosis of Pulmonary Tuberculosis in Children: A Mini-Review. Front. Pediatr. 9:756043. doi: 10.3389/fped.2021.756043
Received: 09 August 2021; Accepted: 27 September 2021;
Published: 25 October 2021.
Edited by:
Mateusz Maciejczyk, Medical University of Bialystok, PolandReviewed by:
Sri Tjahajawati, Universitas Padjadjaran, IndonesiaCopyright © 2021 Khambati, Olbrich, Ellner, Salgame, Song and Bijker. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nisreen Khambati, bmlzcmVlbi5raGFtYmF0aUBjb250ZWQub3guYWMudWs=
†These authors have contributed equally to this work and share last authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.