
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Pediatr. , 17 May 2021
Sec. Genetics of Common and Rare Diseases
Volume 9 - 2021 | https://doi.org/10.3389/fped.2021.668546
Timothy Syndrome (TS) (OMIM #601005) is a rare autosomal dominant syndrome caused by variants in CACNA1C, which encodes the α1C subunit of the voltage-gated calcium channel Cav1.2. TS is classically caused by only a few different genetic changes and characterized by prolonged QT interval, syndactyly, and neurodevelopmental delay; however, the number of identified TS-causing variants is growing, and the resulting symptom profiles are incredibly complex and variable. Here, we aim to review the genetic and clinical findings of all published case reports of TS to date. We discuss multiple possible mechanisms for the variability seen in clinical features across these cases, including mosaicism, genetic background, isoform complexity of CACNA1C and differential expression of transcripts, and biophysical changes in mutant CACNA1C channels. Finally, we propose future research directions such as variant validation, in vivo modeling, and natural history characterization.
Timothy Syndrome (TS) (OMIM #601005) is an ultra-rare, autosomal dominant syndrome that was independently recognized in three case reports in 1992 and 1995 (1–3) and given its name in 2004 when a mutation in the calcium channel gene CACNA1C (12p13.33) was revealed as the molecular cause (4). The molecular defect was first identified in 13 children with syndactyly of the fingers and/or toes, prolonged QT interval as detected by an ECG, and neurological characteristics similar to autism-spectrum disorders. Because this was the eighth LQT syndrome gene to be identified molecularly, it was called LQT8 (4). As new disease-causing alleles were discovered though, the syndrome became much more complex, variably including congenital heart defects (CHDs), hypertrophic cardiomyopathy (HCM), facial dysmorphisms (including small teeth, low set ears, and flat nasal bridge), intermittent hypoglycemia and infections, and epilepsy. This review will explore the full spectrum of symptoms that have been identified in the years since the syndrome's original identification.
Calcium flux is essential for proper function of all cells. Defects in ion channel function, or channelopathies, are responsible for a number of diseases including many that result in cardiac arrhythmias and much more complex syndromes. Mutations in the CACNA1C gene have been linked to multiple arrhythmic syndromes, including Long QT Syndrome Type 8 (LQT8), short QT (SQT), and Brugada Syndrome [OMIM #618447 and 611875 (4–6)]. CACNA1C encodes the α1C subunit of the Cav1.2 voltage-gated calcium channel (VGCC) that is expressed in many tissues of the body (4). The channel also associates with an intracellular β subunit and an extracellular α2δ subunit to form the fully functional channel complex; this review will focus on the α1C subunit since it is the primary pore-forming unit of the channel. The α1C subunit has 4 domains (I–IV), each with six transmembrane (TM)-spanning segments (S1–S6), that make up the large 24-transmembrane-pass channel (Figure 1).
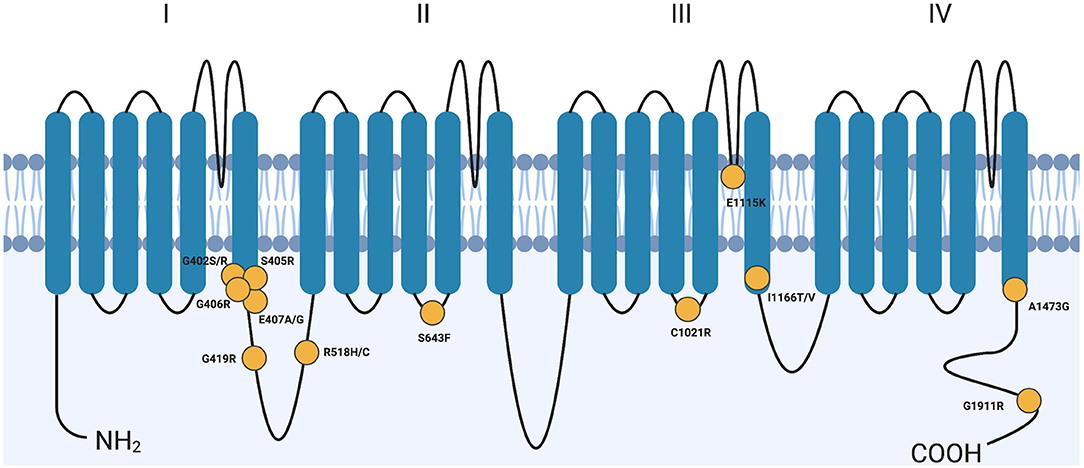
Figure 1. Variants across CACNA1C cause Timothy syndrome. CACNA1C is a 24-transmembrane-pass channel that consists of four repeats (I–IV) of 6 transmembrane segments (SI-S6). Yellow circles denote approximate locations of TS-causing variants. Though G406R is only shown once, it does represent two distinct mutations, one in mutually exclusive exon 8 or exon 8A. G, Glycine; R, Arginine; S, Serine; E, Glutamate; A, Alanine; C, Cysteine; H, Histidine; F, Phenylalanine; K, Lysine; I, Isoleucine; T, Threonine; V, Valine. Created with BioRender.com.
Splawski et al. (4) originally characterized 13 individuals when they reported the molecular cause of Timothy Syndrome. All 13 children had a missense variant in exon 8A of the CACNA1C gene resulting in a Gly406Arg amino acid change. This variant disrupts the 6th transmembrane helix (S6) in domain I of the α1C subunit (Figure 1) and results in a gain-of-function phenotype. This specific syndrome became known as TS Type 1 (TS1) when a second allele was discovered shortly after this initial report. TS was originally described and studied because of its association with a prolonged QT interval and had a relatively simple symptom profile of prolonged QT interval (QTc), syndactyly, autism, tooth decay, and facial dysmorphia.
The CACNA1C gene has numerous splice forms, but important to TS is the alternative splicing of exon 8 and exon 8A. Some confusion in the field exists because different groups published the gene structure of CACNA1C but denoted exon 8 and an alternatively spliced exon 8A in different orientations to each other (GenBank Z26263) (4, 7, 8). The paper by Splawski et al. has been accepted by most, if not all, publications that refer to CACNA1C, putting exon 8 upstream of exon 8A (Figure 2). Exon 8 and 8A are both 104 nucleotides in length, coding for 34 amino acids. They differ in 10 of those amino acids, but both exons code for a Gly at residue 406. mRNAs bearing these alternatively spliced exons are differentially expressed. In organs most notably affected by TS such as brain and heart, exon 8-bearing mRNAs are about 4 times more abundant than exon 8A-bearing mRNAs (9). In 2005, a single child was reported with a Gly406Arg variant in exon 8 (9). The disease caused by Gly406Arg in exon 8 became known as TS Type 2 (TS2). Interestingly, children with an exon 8A variant resulting in Gly406Arg are less-severely affected than children with the same missense variant in exon 8, which likely is due to the difference in expression levels in the heart and brain. TS2 individuals do not present with syndactyly but do variably experience other extra-cardiac symptoms, the only unique symptom of TS2 not seen in TS1 being hip dysplasia. Since the original 2005 publication, more children with Gly406Arg in exon 8 have been identified and these TS2 children also do not present with syndactyly but do present with many symptoms originally associated with TS. These individuals also appear to have worse clinical outcomes than those with the 8A variant, often dying at a younger age. It has come to be accepted that all Gly406Arg variants cause TS regardless of which exon bears this missense variant. Contrary to an opinion expressed in a recent report (10), it still seems clinically useful to note the molecular distinction between exon 8 and 8A, as it may still have some predictive value for the natural history of this disease (i.e., less severe disease and hip dysplasia in individuals with exon 8 Gly406Arg).
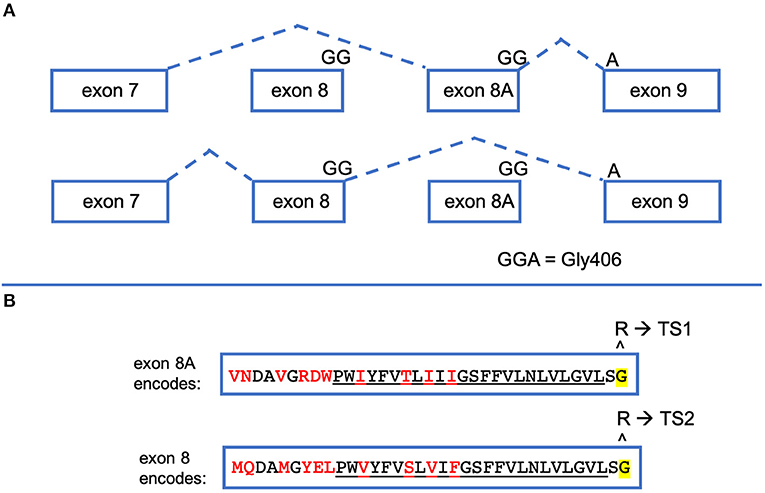
Figure 2. CACNA1C exon 8 and exon 8A are alternatively spliced. (A) When CACNA1C mRNA is processed, exon 7 is spliced to either of two mutually exclusive exons, exon 8 or exon 8A, which is then spliced to exon 9. Exons 8 and 8A are the exact same size (104 bp). The final two nucleotides of exon8/8A and the first nucleotide of exon 9 encode Gly406, the amino acid most commonly implicated in TS. (B) Exons 8 and 8A produce slightly different protein products. Both encode 34 amino acids, differing by the 10 shown in red. The underlined amino acids make up the S6 segment of the first CACNA1C repeat. If the Gly406Arg variant is caused by nucleotide changes in exon 8A, the resulting phenotype is TS1. If the nucleotides in exon 8 are altered to change Gly406 to Arg, TS2 results.
It should be noted that LQT8 also stands alone as a cardiac arrhythmia syndrome. These individuals do have variants in the CACNA1C gene distinct from those described throughout this review. A significant number of individuals with a CACNA1C variant do present with non-syndromic LQT8, but do not have Timothy Syndrome (11–13). To our knowledge, there is no quantification of non-syndromic LQT8 individuals to compare with those affected by TS.
This review serves to highlight the other missense variants that have been identified as TS-causing alleles as well as to stress that sequencing only exons 8A and 8 are no longer the best approach when a TS diagnosis is suspected. These alleles are often referred to as Atypical TS (ATS) alleles (14). This review proposes new nomenclature for how to refer to these distinct new variants as they arise.
In addition to the Gly406Arg variant from exon 8, another missense variant was identified in exon 8 in a single child, resulting in a Gly402Ser change and was initially referred to as TS2 (9). This child did not have syndactyly but did have some neurological deficits similar to TS. It should be noted however that these neurological deficits developed after the child experienced a sudden cardiac arrest. More recently, two siblings with the same Gly402Ser variant were reported to be LQT-only TS2 (15). Their father was mosaic for the variant. Interestingly, they had partial syndactyly. In another report, a Gly402Ser individual was diagnosed as only having the cardiac arrhythmia LQT8 and not a syndromic disorder; alternatively, this individual could be mosaic for the variant (16). So far, this variant has only been observed in exon 8. Since there are only four published cases of Gly402Ser variants in the literature, it remains to be resolved whether this variant is causative of classic TS or a non-syndromic LQT. The Gly402Ser variant has recently been placed into the larger group of ATS alleles. ATS variants essentially include all missense variants that do not result in the single amino acid change Gly406Arg (TS1 and TS2). We agree that Gly402Ser (exon 8) should be classified as an ATS allele since it is quite symptomatically distinct from TS2. Currently, OMIM only lists these two variants for Timothy Syndrome—Gly406Arg and Gly402Ser.
Since 2005, at least 12 other missense variants have been identified as TS-causing alleles (Table 1). These were all published as case reports and as such, the number of individuals with each reported variant is usually one. One theme that seems to arise in ATS cases is variability in disease presentation. This variability could be due to mosaicism, genetic background, or transcriptional regulation leading to variable expression levels, all of which are discussed later in this review. Below is a brief summary of each of these novel variants since the publication of the original TS1 and TS2 papers from 2004 and 2005. Since they each vary in their presentation, it is difficult to assemble them into distinct groups.
In 2012, Gillis et al. reported a single individual with an Ala1473Gly missense variant (14). Like Gly406, this variant resides on the intracellular portion of S6, domain IV (Figure 1). At birth, this child had dysmorphic facial features, sparse hair growth, bilateral hip dislocation, joint contractures in the arms, and syndactyly of fingers and toes on both hands and both feet. The child also failed many neurological and reflex tests at birth. This individual would go on to have seizures, episodes of hypoglycemia and hypocalcemia, apnea, bilateral hip dysplasia, cortical blindness, and severe neurodevelopmental delay (14).
An Ile1166Thr missense variant was identified in two children by three different groups, and like Gly406 and Ala1473, is also on the intracellular portion of S6 but in domain III (11, 31, 32). One child presented with 2:1 AV block shortly after birth with QT-interval prolongation, patent ductus arteriosis (PDA) and left atrial enlargement (32). An ICD was implanted. He was later found to have numerous structural cardiac defects and at 5 months of age began a history of seizures. Consistent with TS, this child had facial dysmorphisms, joint hypermobility, clinodactyly, hypotonia, and severe dental abnormalities leading to the extraction of 20 teeth before the age of 3 (31, 32). This child died at 3 years 8 months after admission for respiratory failure, hypotension, and dehydration. The second child identified suffered sudden cardiac death at age 1 prior to molecular testing, but presented with severely prolonged QT interval, AV block, ductus arteriosis, HCM, and syndactyly. The CACNA1C mutation was identified post-mortem, and a diagnosis of TS was thus presumed (11). Interestingly, the position of the missense variants, Gly406Arg, Gly402Ser, Ala1473Gly, and Ile1166Thr, all occur near the cytoplasmic face of the S6 segment of their respective domains in the CACNA1C structure (Figure 1).
An individual was identified with the missense variant Gly1911Arg in the C-terminal cytoplasmic tail of the CACNA1C channel [Figure 1 (33)]. Unlike the individuals with variants in the S6 segments, this individual developed normally until 30 months of age, when he began experiencing seizures. The seizures were typically accompanied by a loss of consciousness, and at age 5 a seizure was accompanied by monomorphic ventricular tachycardia which required resuscitation. The patient was then diagnosed with prolonged QTc. Follow-up exams at ages 12½ and 17 revealed subtle muscular degeneration, microcephaly, dental abnormalities, and facial dysmorphisms including those common in TS such as flat nasal bridge (33).
The variant Arg518Cys was identified as the disease-causing mutation in a family with multiple affected individuals suffering from QTc prolongation, HCM, SCD, and other congenital heart defects (CHDs) (27). None of these family members had syndactyly, cognitive impairments, facial dysmorphisms, or any extracardiac phenotypes common to a TS diagnosis, so the authors designated this disease presentation “cardiac-only TS” (COTS). Armed with a new, previously-unreported TS-causing variant, they screened samples from a cohort of patients with idiopathic LQTs for this change. Through this screen they identified another individual with the Arg518Cys variant and one with Arg518His in CACNA1C. These individuals had family histories of QTc prolongation and HCM and both variants cosegregated with the cardiac phenotypes (27). The nomenclature assigned to these cases, COTS, is somewhat confusing especially considering there are variants within the CACNA1C gene that cause non-syndromic LQT8, which is also a cardiac-only disease. COTS is distinct from non-syndromic LQT8 in that there are structural heart defects in addition to the prolonged QTc. We propose that cases previously designated COTS be considered ATS to further differentiate them from non-syndromic LQT8. This will also be discussed in terms of mosaicism later in this review.
A 2018 study following the treatment outcomes of many TS individuals identified three previously unreported missense mutations: Gly402Arg, Ser405Arg, and Cys1021Arg (17). The Gly402 and Ser405 variants, like many discussed earlier, reside on the intracellular side of transmembrane segment S6. Gly402Arg was present in two unrelated patients, but symptoms were only described for one. The affected individual presented with severely prolonged QTc and neurodevelopmental delay. Two related individuals were diagnosed with the missense allele Ser405Arg. Both of these cases included prolonged QTc and syndactyly, but only one included neurodevelopmental delay. The individual with the missense variant Cys1021Arg exhibited the most complex symptom profile, presenting with long QTc, syndactyly, neurodevelopmental delay, facial abnormalities, baldness, and intermittent hypoglycemia (17).
The TS symptom profile was further complicated when Kosaki et al. reported an individual with the missense mutation Arg1024Gly who presented with many common TS phenotypes but not QTc prolongation (29). In addition to bilateral cutaneous syndactyly, this individual experienced delayed growth and neurodevelopment, upper limb joint contractures, several seizure-like episodes, thin scalp hair, and facial dysmorphisms (29), all consistent with other cases of TS.
A 14-year-old boy with a prolonged QTc, facial dysmorphisms, neurodevelopmental delay, a history of seizures, but without syndactyly, was diagnosed with TS when his causative mutation, Ser643Phe in CACNA1C, was identified (28). This variant is positioned on the cytoplasmic loop between S4 and S5 of domain II in the CACNA1C channel. Variants associated with LQT8 have also been identified on cytoplasmic loops in domain II, in the loop between domain II and III, and on the C-terminal cytoplasmic tail of this channel (11, 36, 37). This is the first case report of a similarly-located variant that resulted in a TS diagnosis. Although this individual's symptom profile matches that of TS, the relatively late age of diagnosis (14 years old) is notable.
A 14-year-old was identified to have the variant Glu1115Lys after undergoing genetic testing for idiopathic long QT (30). Although the case report does not suggest a TS diagnosis, the patient presented with prolonged QTc, autism spectrum disorder, seizures, and hyperglycemia (rather than hypoglycemia). The Glu1115Lys variant has also been observed in an individual with Brugada Syndrome (38). How some individuals present with such different symptoms and at such a later age than most TS individuals remains unknown, but some possibilities are discussed later.
Two recent case reports suggest that mutations in residues located near Gly406 (that causes TS1) can also cause TS. A Glu407Ala missense variant (25) and a Glu407Gly missense variant (24) were identified in two different children. This last case was most like TS1 with reported syndactyly and LQT, but also included epilepsy. The individual with Glu407Ala presented with mild learning disabilities, epilepsy, and bilateral cutaneous syndactyly, but importantly also has a missense variant in the gene PKP2 (25). PKP2 is a component of desmosomes and has been shown to modulate L-type calcium channels (39). The patient was 22 years old at the time of diagnosis, so the secondary genetic change may somehow modulate disease presentation.
Most recently, a Gly419Arg variant was reported The affected individual presented with bilateral hand and foot syndactyly, PDA, left ventricular non-compaction, ventricular preexcitation, mild neurodevelopmental delay, and prolonged QT interval. Interestingly, after surgical closure of the PDA at age 3 months and correction of hand syndactyly at age 2, this individual experienced no symptoms until he suffered syncope and pre-excited atrial fibrillation at age 15. An ICD was implanted at age 17. The individual is now 23 years old. It is important to note that he has two additional de novo variants: Glu131Gln in TOP1, a DNA topoisomerase, and Gly2493Arg in ITPR1, a channel that releases calcium from the ER upon inositol 1,4,5-triphosphate binding (26). Although the consequences of these variants are unknown, they could provide important context for understanding this case.
Autosomal dominant genetic diseases, like LQT1 for example, are passed on from generation to generation because the presentation is most often sub-lethal and can be managed with medication. Additionally, some may not even recognize that they had the disorder. Thus, the mode of inheritance for such disorders is genetic—one parent passes the mutation onto their progeny with an inheritance likelihood of 50% for each child. Due to the much higher severity of disease, TS does not get passed on by an affected parent in this way because affected individuals rarely survive to reproductive age.
De novo genetic changes are defined as genetic alterations that are present for the first time in one family member as a result of a variant in a germ cell (egg or sperm) of one of the parents, or variants that arise in the fertilized egg itself during early embryogenesis (https://www.cancer.gov/publications/dictionaries/genetics-dictionary/def/de-novo-mutation). de novo variants arise due to errors in DNA replication and account for most of the normal genetic variation that we observe in all organisms. Since TS is such a harmful disease, it is likely that most TS children acquire the variant de novo rather than inherit it from an affected parent. So, de novo mutations may arise during gametogenesis of a parent such that the mutation is present in the mother's egg or father's sperm, and the resulting embryo bears the mutation upon fertilization. de novo mutations acquired in parental germ cells are present in all of the resulting embryo's cells and tissues, leading to a fully heterozygous individual for the variant. The parent can be considered mosaic (having genetically distinct cells within a single individual) for the variant, since only one cell type (gamete) is affected, but the child may not be. It is becoming clear that de novo mutations can also appear post-zygotically, leading to mosaicism of the developing embryo (40). Thus, the affected child is mosaic and expresses the variant in some of their cells and organs. If this error were to happen early, more cell lineages would have the error; if it occurred later in development, fewer cells and organs may contain that mutation. Here we discuss these modes of inheritance and how they could be essential in understanding variability in TS disease presentation.
Parental germline mosaicism appeared to be the case for a Gly406Arg child whose father carried the same variant in 16% of his sperm (41). This low level of mosaicism was possible to detect through cloning and sequencing multiple PCR-amplified products from the father's sperm. The father had QTc prolongation but no other TS-like symptoms. Without being able to biopsy various tissues from a living mosaic individual, we can only presume that this father did not have the TS variant expressed in many other organs other than his heart and gametes.
Since a parent provides a haploid gamete for the creation of a zygote, a mosaic parent either passes on the wildtype copy of the gene or the variant version. In this scenario, the parent is mosaic, but the child is fully heterozygous for the variant from the time of fertilization; that child will maintain the variant in every tissue and organ. We speculate that the TS children born of a mosaic parent likely present with more severe disease than a de novo child who acquires the variant during embryogenesis. In a child who acquires the variant during embryogenesis, fewer organs and tissues may bear this variant. For these reasons, it would be important to determine whether a parent is mosaic and to consult a genetic counselor if the parents are considering additional children in the future. If some of either parents' gametes carry the TS variant, that parent could pass the TS allele onto future progeny.
Post-zygotic point mutations are an under-recognized source of de novo genomic variation (40). Some de novo children may also be mosaic and that may explain, in part, why not all TS children report deficits in all tissues. Recently, a child who was initially negative for any known variants in CACNA1C had their lymphocytes re-analyzed by Next Generation Sequencing (NGS). A Gly406Arg variant was identified in 18% of the sequencing reads. This child had an “incomplete phenotype” in that only LQT and syndactyly were noted, and none of the other common TS phenotypes (23). The authors proposed that there may be additional cases of incomplete TS that go unnoticed by standard gene panel Sanger sequencing. NGS has the benefit of >100-fold coverage of the genome such that mosaicism can be readily detected, even if only present in a few sequencing reads.
There is an incredible amount of phenotypic variability among individuals bearing variants in the CACNA1C gene and TS no longer exclusively applies to children with both cardiac arrhythmias and syndactyly. What remains to be resolved is whether non-syndromic LQT8 is a distinct disease or just another mosaic presentation of TS. The existence of a variant in the CACNA1C gene is not enough to assign a diagnosis of TS. But why is it that all individuals with variants in CACNA1C do not present with identical phenotypes? There are many possibilities for this observation, as is true for many monogenic diseases.
One explanation is through mosaicism as discussed in the previous section. Individuals who inherit a variant at fertilization will likely have a more severe disease presentation than those who acquire it at some point during embryonic development. Additionally, depending on the timing and location of an embryonic de novo variant, certain germ layers or cell lineages could be differentially affected (Figure 3). This could explain how different individuals with TS present with symptoms restricted to different sets of tissues.
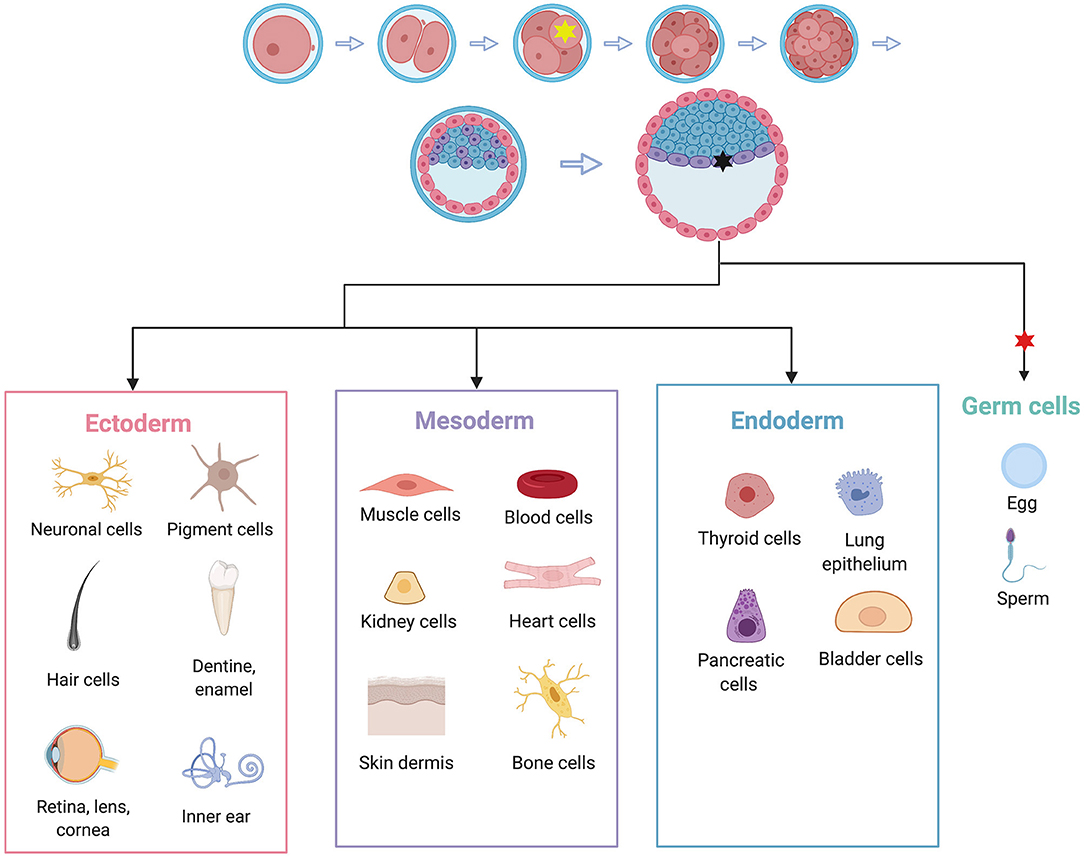
Figure 3. Embryonic de novo mutations can result in highly variable phenotypes due to mosaicism. Individuals who acquire a mutation at the 4-cell stage, represented by the yellow star, may experience symptoms in all cell types (given that the variant in question has equal ability to affect all cell types). A variant acquired at the gastrula stage, represented by the black star, may only affect one germ layer and therefore a more restricted subset of cell types. In this example, the black mutation may only affect heart, muscle, kidney, skin, and bones, but not neuronal tissue. In the case of TS, this is a possible mechanism for cases that have symptoms restricted to LTQ and syndactyly. A variant that arises after germ cells have been set aside, represented by the red star, may only affect germ cells, and therefore may not cause phenotypes in the individual, but may affect the individual's children. Adapted from “Human Embryonic Stem Cell Differentiation,” by BioRender.com (2020). Retrieved from: https://app.Biorender.com/biorender-templates.
A second likely explanation for variability in presentation is that each TS child has a very different genetic background, which can contribute to expressivity of the symptoms. For example, the young woman diagnosed with the CACNA1C variant Glu407Ala at age 22 had a secondary variant in PKP2 (25). Age 22 is much older than most TS individuals are diagnosed, and much older than many survive; it is likely that her presentation of TS is influenced by the presence of a second mutation. Both CACNA1C and PKP2 were included on the rhymologic gene panel run in this individual's case, making it a simple case of two pathogenic mutations. If all individuals with TS underwent whole-exome or whole-genome sequencing in relation to their parents and/or siblings, multiple genetic changes might be revealed that contribute to complex genetic profiles influencing each child's presentation of disease. This possibility exists for most, if not all, inherited diseases.
A third explanation could involve the many different isoforms of the CACNA1C protein. Besides the use of alternative exons 8 and 8A, there is evidence that hundreds of alpha-subunit isoforms exist due to alternative splicing (42). The use of so many different isoforms of CACNA1C make it difficult to establish simple patterns of disease. It may be that ATS variants do not phenocopy the Gly406Arg variant because certain isoforms are expressed more or less abundantly in various tissues, and some threshold of mutant CACNA1C channels is tolerable. Studies of the Gly406Arg allele in adult guinea pig ventricular myocytes (aGPVMs) showed that cells could tolerate a certain proportion of mutant CACNA1C channels. A low proportion of mutant channels caused action potentials (APs) to be only slightly prolonged; once this tolerance threshold was crossed, APs became unstable and pushed cells into an “arrhythmogenic” state. In aGPVMs, this threshold was about 12% for Gly406Arg and about 40% for Gly402Ser (34). This result could be essential in understanding the differences in pathogenesis of TS1 and TS2, and more precise knowledge of transcript expression by tissue could inform the discussion of other, non-exon 8 or 8A alleles.
Alternatively, it may not be the tissue-specific expression levels that matter but that a given tissue, organ, or cell type is more sensitive to an alteration in function of the CACNA1C channel. Each of the missense variants that have been associated with TS compromise the function of the channel, and perhaps to different extents and for different biophysical reasons. It is possible that some cell types are more tolerant to this alteration in function than others, and it is the cell types or organs that are most sensitive that result in disease symptoms. This may explain why numerous variants throughout the CACNA1C gene result in non-syndromic LQT8 and manifest as a disease that has only one symptom. Perhaps the heart is the most sensitive tissue to any alteration in function of the CACNA1C channel and that is why so many seemingly unrelated missense variants throughout the gene are associated with LQT8.
One could hypothesize that the reason so few TS missense variants have been identified to date is that these variants disrupt channel function more dramatically than the non-syndromic LQT8-causing variants. Other organs besides the heart may not be able to tolerate this alteration in function, and this is why we observe syndactyly, neurodevelopmental delays, hip dysplasia, facial abnormalities, hypoglycemia, and other symptoms. We would go so far as to predict that there may be CACNA1C variants that result in any one of these phenotypes without appearing multi-system in nature. For example, there could be hypoglycemic children with CACNA1C variants that have no other TS-like phenotypes. The Arg1024Gly variant (29) supports such a prediction in that this individual has few TS phenotypes, such as syndactyly, but lacks QTc prolongation. This could be a case of mosaicism or a case in which the amino acid change itself is not as detrimental to heart tissue as it is elsewhere. Additionally, cases previously designated as COTS (Arg518Cys, Arg518His) (27) could be interpreted as ATS variants that have effects restricted to the heart, either through embryonic de novo mosaicism or due to higher heart-specific susceptibility to changes to this specific residue.
There may also be CACNA1C variants that are so poorly tolerated that these children die at a very young age and are never diagnosed with TS. That certainly may be true for some exon 8 Gly406Arg children. Genetic variants that result in very early death may 1 day be diagnosed as more clinicians perform Whole Exome Sequencing (WES) on infants that die of SIDS (43).
It can be argued that Katherine Timothy and the cardiologists she worked alongside selected a specific subset of TS children in their landmark 2004 publication in which they first molecularly identified the genetic lesion responsible for TS (4). Because they only sequenced the CACNA1C gene from children presenting with both LQT and syndactyly, it is now clear that they selected for a homogenous group of children. Sixteen years later and after many molecular genetic case reports of ATS children, it is apparent that children presenting with CACNA1C variants and a subset of the “classical” TS phenotypes should also receive the diagnosis of TS. As with many syndromes, not every individual bearing a specific variant presents with all of the same phenotypes, yet most clinicians readily accept that their diagnosis is correct. The acceptance that many distinct variants in CACNA1C can cause TS may prompt clinicians in this field to rethink how they categorize these seemingly different types of TS.
In addition to the previously-discussed sources of variability, biophysical properties of the variant channels themselves could also account for differences in disease presentation. To date, all individuals with TS are heterozygous for their variant. TS-causing mutations are gain-of-function alleles of CACNA1C, prolonging the cardiac action potential (AP) and therefore the QT interval (QTc). Various groups have investigated the electrophysiological effects of different mutations on CACNA1C function and have found a few distinct mechanisms of dysfunction. The classical mechanism of TS pathogenesis is a loss of voltage-dependent inactivation (VDI) of the channel, thereby causing an increase in the maximum flow of Ca2+ through the channel (peak current density) in a given opening event (4, 9, 33). Alternative mechanisms are characterized by a decrease in peak current density and commonly a decrease in degree of depolarization necessary to activate the channel (11, 26–28, 30, 32). Here we review the electrophysiological studies of CACNA1C channel variants in model systems to date (Table 2).
The first descriptions of TS included electrophysiological studies done in Chinese hamster ovary (CHO) cells and Xenopus oocytes (4, 9). Studies in both systems revealed that expression of a transgene containing the Gly406Arg variant in exon 8A results in CACNA1C channels that have impaired VDI, leading to prolonged opening of the channel and subsequently increased Ca2+ flux through the channel (4). The same group later expressed constructs containing the Gly406Arg mutation in exon 8 as well as Gly402Ser in exon 8 in Xenopus oocytes. Both of these mimicked the original exon 8A mutation: loss of VDI which produced sustained inward Ca2+ currents (9).
In addition to these early studies, many groups have since investigated the original TS mutations in great detail and in more systems. Expression of the Gly406Arg variant in HEK293 cells led to a loss of VDI (35). Although they reported that Gly406Arg has no effect on calcium-dependent inactivation (CDI) of CACNA1C, Dick et al. later carefully examined this parameter. They expressed transcripts encoding Gly406Arg-exon 8A, Gly406Arg-exon 8, or Gly402Ser-exon 8 of CACNA1C in HEK293 cells and found that all three of these caused a large decrease in CDI when compared to WT (34). They were also able to elucidate two contrasting methods of CDI loss in the two different missense changes: Gly406Arg in either exon led to a decrease in CDImax, a measure that is a function of channel gating and a result which reflects an increase in channel opening, whereas Gly402Ser led to a decrease in FCDI, a measure that is a function of Ca2+ and a result which reflects a decrease in channel opening (34). While these opposing mechanisms of CDI loss reveal a difference in opening probability (PO) of Gly406Arg- and Gly402Ser-containing channels, both channel types do still suffer from a loss of VDI once they do open, and therefore both experience an increase in inward Ca2+ current, consistent with the model. It is important to mention that although the mechanism is straightforward in its effects on the cardiac AP, the mechanism in other tissues is not yet clear.
Expression of the variant Gly1911Arg in HEK293 cells affected VDI and steady-state inactivation (SSI), thereby significantly increasing the window current compared to WT cells (33). This mutant protein acts in the same way as the classic TS alleles in that it increases Ca2+ flux and thus causes a gain-of-function to the channel. The Gly1911 amino acid resides near the protein's C-terminus, distant from the positioning of other reported TS mutations. Its importance could be a result of its close proximity to a calcineurin binding site or its location on the distal C-terminus. Disruption of either of these domains could be related to impaired VDI (44, 45).
Ile1166Thr, which resides on the intracellular portion of S6 in the third repeat, was investigated in HEK293 cells (32). Functional studies revealed that unlike Gly406Arg and Gly402Ser, Ile1166Thr caused a large reduction in peak current density. Importantly, they also found that Ile1166Thr resulted in a negative shift in V1/2 of activation, or the degree of depolarization necessary for activation of the channel (32). Although less Ca2+ flows through the channel in a given opening event, the shift in activation and the resulting increase in window current confer Ile1166Thr with a net gain-of-function effect on CACNA1C channels. Wemhöner et al. also performed functional studies on this variant in HEK293 cells and Xenopus oocytes. Like Boczek et al., this group found that a negative shift in the V1/2 of activation was responsible for the gain of function conferred by Ile1166Thr. Additionally, Ile1166Thr does not affect VDI but does result in a decrease in steady-state inactivation (SSI) (11).
Boczek et al. performed functional studies on missense alleles Arg518Cys and Arg518His (27). When expressed in HEK293 cells, Arg518Cys resulted in a 55.6% decrease in peak current density. It also resulted in a decreased proportion of channels localized to the membrane vs. the cytoplasm, which could partially account for the reduction in overall Ca2+ flux. Arg518His caused a 63.2% decrease in peak current density and a negative shift in V1/2 of activation, similar to Ile1166Thr. Both variants caused a negative shift in V1/2 of inactivation as well, resulting in an increased window current and consequently, a net gain-of-function. Both Arg518Cys/His also resulted in decreased VDI, more similar to Gly406Arg/Gly402Ser than to Ile1166Thr (27).
Another variant with complex electrophysiological effects, Ser643Phe, was investigated in HEK293 cells (28). This variant resulted in an 86.2% reduction in peak current density. Once again, this variant caused a negative shift in activation and a large decrease in VDI, ultimately causing a net gain-of-function phenotype despite the dramatic reduction in Ca2+ flux (28).
The Gly419Arg variant acts uniquely when expressed in HEK293 cells (26). Despite actually having faster inactivation kinetics (a loss of function) than wild-type cells, peak current density is greatly increased. Like Ile1166Thr, Arg518His, Arg518Cys, and Ser643Phe, Gly419Arg resulted in a leftward, gain-of-function shift in the voltage dependence of activation leading to an increased window current for these channels (26).
Finally, the variant Glu1115Lys decreases Ca2+ flux in a unique way. When expressed in TSA201 cells, this variant converts the channel into a non-selective monovalent cation channel (30). There are four glutamate residues in the S4 helix that collectively form the ion selectivity filter. Altering one of these glutamate residues to a lysine alters the ion selectivity of the channel such that it becomes non-selective for monovalent cations. The result is a Cav1.2 channel with a slowly inactivating inward Na+ current and outward K+ current. The movement of these ions through a channel population not tuned to function in the cardiac action potential (AP) likely creates an environment in which arrhythmias are likely to develop (30).
As one can see from the review of reported ATS variants, some are questionable as to whether they cause TS. Ideally, when new variants arise, it would be useful to carry out validation experiments such as reproducing the variant in human cardiomyocytes, showing that they are arrhythmogenic, and then correcting that variant back to the wild-type sequence. This would provide evidence that a given variant was responsible, at the very least, for the arrhythmic behavior of those cells. Assays don't yet exist to address how these variants specifically perturb the normal physiology of other tissues and organs in the body of a TS individual.
In addition to validating new variants, in vivo models will be useful for further investigation into the mechanism of TS. The various whole animal model studies were recently reviewed by Han et al. (46). Careful characterization of cell and animal models of all known TS variants is essential and will aid in any future development of treatments.
In brief, there are few animal models for TS. Knockout of the CACNA1C gene in mice is embryonic lethal (47) and heterozygotes are viable and fertile with no apparent developmental defects. There are no published reports of a TS1 mouse model other than a mouse line with restricted expression of a mutant CACNA1C gene under a craniofacial and limb bud mesenchymal promoter; such mice display craniofacial defects, specifically in the mandible (48). Defects in the limb buds were not described. This finding is interesting given that many TS1 children have mandible abnormalities and well as syndactyly.
As for TS2, animals expressing an inverted neomycin cassette were generated such that when the cassette was excised by recombination, a TS2 mutation would be expressed. When the cassette is properly recombined out, the homozygotes and heterozygotes are embryonic lethal; only heterozygous animals still bearing an inverted neomycin cassette inserted in the gene survive; these mice are referred to as TS2-neo mice. Presumably these mice survive because of reduced expression of the TS2 allele. The TS2-neo knock-in heterozygous mice have behavioral defects but no obvious cardiac defects (49, 50), however the authors do hypothesize that the TS2 mice lacking the neo cassette die due to cardiac defects. All these models were made before CRISPR gene editing technology was available. No other alleles have been made in the mouse to study any of the many ATS alleles described in this review.
Interestingly, no Timothy Syndrome Drosophila or zebrafish models have been described to date. Truncated alleles in zebrafish revealed a function for the Cav1.2 channel in embryonic heart development (51, 52) and pancreatic islets (53). A C. elegans TS animal was recently created by CRISPR; homozygous animals with the analogous G406R mutation (G369 in C. elegans) have widespread muscle defects and arrest as embryos or young larvae (54). No phenotypes were reported in heterozygous animals. One report of an ATS allele in C. elegans identified some axon targeting phenotypes (55) As there are an increasing number of ATS variants, an animal model that is amenable to inexpensive gene editing is needed to test their phenotypic consequences and investigate their mechanisms. Variants of Unknown Significance (VUS) can be verified in such models as well.
Besides the above models, there are just a few reports using human induced Pluripotent Stem Cells (hiPSCs). These are cells often collected from saliva or blood that can be induced to pluripotency, and then transfected with a cocktail of factors to induce them along a number of different cell fates. Cells collected from TS individuals have been used to study their arrhythmias in cardiomyocytes (CMs) derived from the hiPSCs (27, 56, 57). More recently, with the advent of CRISPR gene editing, “patient-independent” hiPSCs-CMs have been generated, speeding up the process in which cell lines can be generated. In such cases, numerous variants can be made from the same hiPSC control line such that the genetic background is isogenic (58, 59). Though hiPSCs have proved informative for TS and other LQT syndromes (59), it appears as if there is a rising interest in studying TS hiPSCs that have been driven to adapt neuronal fates (60–62). These approaches may highlight some of the neurodevelopmental defects that TS individuals experience.
Currently, gain of function variants in the CACNA1C gene are more likely to result in a diagnosis of non-syndromic LQT8. Variants in this gene have also been found to be associated with neuropsychiatric disorders such as schizophrenia (63, 64), bipolar disorder (65), and major depressive disorder (65). To date, mutations in a limited number of residues have been reported to cause TS.
The nomenclature for TS has been somewhat confusing in that TS1 and TS2 are caused by the same missense mutation but in two alternatively spliced exons. Though it has been argued that there is no clinical utility to distinguishing TS1 from TS2 (10), there is evidence that TS2 children have worse prognoses and differ from TS1 children in that they often develop hip dysplasia. Gly402Ser variants were originally also considered causative of TS2, which adds to the confusion. More recently, others have started to refer to this variant as causative of ATS. A recent proposal was made suggesting all mutations in the CACNA1C gene be considered TS1, and that TS2, TS3, and so on be reserved for other TS-causing genes yet to be discovered (17). After 17 years of TS molecular genetics, it seems unlikely that such other causative genes exist. We thus propose a more straight-forward nomenclature for the many TS variants that now exist. For historical reasons, TS1 and TS2 should remain as is, or perhaps TS1-Gly406Arg (exon 8A) and TS2-Gly406Arg (exon 8). We propose the remaining alleles be called ATS with the missense allele designation, such as ATS-Gly402Ser (exon 8) and ATS-Ser643Phe. Such names immediately indicate the molecular lesion in the CACNA1C gene. Though most ATS alleles are represented by a single case, these designations may prove useful in the future as additional children are identified as having one of these lesions. Alternatively, all the above case reports can be grouped as one syndrome, Timothy Syndrome, with the recognition that there is a broad and variable phenotypic spectrum.
In today's complex regulations involving health insurance, a diagnosis of TS means that a child is able to receive special services and benefits for their symptoms in multiple organs. Some of these services may include physical, occupational, behavioral, and speech therapy. Withholding such a diagnosis because an individual does not have the “classical” variant, or a “complete” TS phenotype, is not warranted at this time. Clinicians and researchers just do not understand this complex syndrome that well, and there are multiple sources of variability (as discussed earlier) that contribute to disease presentations.
The natural history of this syndrome is still being determined. One of us (KWT) has dedicated half her life to acquiring the medical records and family histories of over 60 children with TS, many of whom are no longer alive. To date, there are a number of young adults now in their late twenties thriving with TS. With more research into the progression of this ultra-rare syndrome, we hope that many more children can survive and live a long and productive life. Although much of the research to date has been on the cardiac consequences of these variants, there is an increasing interest in the neurodevelopmental phenotypes (42, 61, 62, 66). It is our hope that a greater number of researchers will also study the cellular mechanisms of these calcium channel variants that contribute to the multisystem nature of Timothy Syndrome (48, 67).
RB and AG wrote sections of the first draft of the manuscript. All authors contributed to overall outline of this review and manuscript revision. All authors read and approved the submitted version.
This work was supported by the Intramural Research Program of the National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors would like to thank Ivy Dick (University of Maryland School of Medicine), Alfred George (Northwestern University), and Michael Sanguinetti (University of Utah) for their critical comments and suggestions of this review.
1. Levin SE, Harrisberg J, Govandrageloo K, Plessis JD. Idiopathic long Q-T syndrome in a black infant. Cardiovasc J Southern Africa. (1992) 3:144–6.
2. Reichenbach H, Meister E-M, Theile H. A new variant exhibiting cardiac conduction disorders and syndactyly including osseous changes in the hands and feet. Kinderärztl Praxis. (1992) 60:54–6.
3. Marks ML, Whisler SL, Clericuzio C, Keating M. A new form of long QT syndrome associated with syndactyly. J Am Coll Cardiol. (1995) 25:59–64. doi: 10.1016/0735-1097(94)00318-K
4. Splawski I, Timothy KW, Sharpe LM, Decher N, Kumar P, Bloise R, et al. CaV1.2 calcium channel dysfunction causes a multisystem disorder including arrhythmia and autism. Cell. (2004) 119:19–31. doi: 10.1016/j.cell.2004.09.011
5. Boczek NJ, Best JM, Tester DJ, Giudicessi JR, Middha S, Evans JM, et al. Exome sequencing and systems biology converge to identify novel mutations in the L-type calcium channel, CACNA1C, linked to autosomal dominant long QT syndrome. Circul Cardiovasc Genet. (2013) 6:279–89. doi: 10.1161/CIRCGENETICS.113.000138
6. Antzelevitch C, Pollevick GD, Cordeiro JM, Casis O, Sanguinetti MC, Aizawa Y, et al. Loss-of-function mutations in the cardiac calcium channel underlie a new clinical entity characterized by ST-segment elevation, short QT intervals, and sudden cardiac death. Circulation. (2007) 115:442–9. doi: 10.1161/CIRCULATIONAHA.106.668392
7. Tang ZZ, Liang MC, Lu S, Yu D, Yu CY, Yue DT, et al. Transcript scanning reveals novel and extensive splice variations in human L-type voltage-gated calcium channel, Cav1.2 α1 subunit. J Biol Chem. (2004) 279:44335–43. doi: 10.1074b39/jbc.M407023200
8. Zühlke RD, Reuter H. Ca2+-sensitive inactivation of L-type Ca2+ channels depends on multiple cytoplasmic amino acid sequences of the α1C subunit. Proc Natl Acad Sci USA. (1998) 95:3287–94. doi: 10.1073/pnas.95.6.3287
9. Splawski I, Timothy KW, Decher N, Kumar P, Sachse FB, Beggs AH, et al. Severe arrhythmia disorder caused by cardiac L-type calcium channel mutations. Proc Natl Acad Sci USA. (2005) 102:8089–96. doi: 10.1073/pnas.0502506102
10. Diep V, Seaver LH. Long QT syndrome with craniofacial, digital, and neurologic features: is it useful to distinguish between timothy syndrome types 1 and 2? Am J Med Genet A. (2015) 167:2780–5. doi: 10.1002/ajmg.a.37258
11. Wemhöner K, Friedrich C, Stallmeyer B, Coffey AJ, Grace A, Zumhagen S, et al. Gain-of-function mutations in the calcium channel CACNA1C (Cav1.2) cause non-syndromic long-QT but not Timothy syndrome. J Mol Cell Cardiol. (2015) 80:186–95. doi: 10.1016/j.yjmcc.2015.01.002
12. Fukuyama M, Ohno S, Ozawa J, Kato K, Makiyama T, Nakagawa Y, et al. High prevalence of late-appearing T-wave in patients with long QT syndrome type 8. Circ J. (2020) 84:559–68. doi: 10.1253/circj.CJ-19-1101
13. Mellor GJ, Panwar P, Lee AK, Steinberg C, Hathaway JA, Bartels K, et al. Type 8 long QT syndrome: pathogenic variants in CACNA1C-encoded Cav1.2 cluster in STAC protein binding site. Ep Europace. (2019) 21:1725–32. doi: 10.1093/europace/euz215
14. Gillis J, Burashnikov E, Antzelevitch C, Blaser S, Gross G, Turner L, et al. Long QT, syndactyly, joint contractures, stroke and novel CACNA1C mutation: expanding the spectrum of Timothy syndrome. Am J Med Genet A. (2012) 158A:182–7. doi: 10.1002/ajmg.a.34355
15. Fröhler S, Kieslich M, Langnick C, Feldkamp M, Opgen-Rhein B, Berger F, et al. Exome sequencing helped the fine diagnosis of two siblings afflicted with atypical Timothy syndrome (TS2). BMC Med Genet. (2014) 15:48. doi: 10.1186/1471-2350-15-48
16. Hiippala A, Tallila J, Myllykangas S, Koskenvuo JW, Alastalo T. Expanding the phenotype of Timothy syndrome type 2: an adolescent with ventricular fibrillation but normal development. Am J Med Genet A. (2015) 167:629–34. doi: 10.1002/ajmg.a.36924
17. Dufendach KA, Timothy K, Ackerman MJ, Blevins B, Pflaumer A, Etheridge S, et al. Clinical outcomes and modes of death in Timothy syndrome a multicenter international study of a rare disorder. JACC Clin Electrophysiol. (2018) 4:459–66. doi: 10.1016/j.jacep.2017.08.007
18. Lo-A-Njoe SM, Wilde AA, Erven L van, Blom NA. Syndactyly and long QT syndrome (CaV1.2 missense mutation G406R) is associated with hypertrophic cardiomyopathy. Heart Rhythm. (2005) 2:1365–8. doi: 10.1016/j.hrthm.2005.08.024
19. Sepp R, Hategan L, Bácsi A, Cseklye J, Környei L, Borbás J, et al. Timothy syndrome 1 genotype without syndactyly and major extracardiac manifestations. Am J Med Genet A. (2017) 173:784–9. doi: 10.1002/ajmg.a.38084
20. Dufendach KA, Giudicessi JR, Boczek NJ, Ackerman MJ. Maternal mosaicism confounds the neonatal diagnosis of Type 1 Timothy syndrome. Pediatrics. (2013) 131:e1991–5. doi: 10.1542/peds.2012-2941
21. Gao Y, Xue X, Hu D, Liu W, Yuan Y, Sun H, et al. Inhibition of late sodium current by Mexiletine. Circul Arrhythmia Electrophysiol. (2018) 6:614–22. doi: 10.1161/CIRCEP.113.000092
22. Krause U, Gravenhorst V, Kriebel T, Ruschewski W, Paul T. A rare association of long QT syndrome and syndactyly: Timothy syndrome (LQT 8). Clin Res Cardiol. (2011) 100:1123–7. doi: 10.1007/s00392-011-0358-4
23. Baurand A, Falcon-Eicher S, Laurent G, Villain E, Bonnet C, Thauvin-Robinet C, et al. Incomplete Timothy syndrome secondary to a mosaic mutation of the CACNA1C gene diagnosed using next-generation sequencing. Am J Med Genet A. (2017) 173:531–6. doi: 10.1002/ajmg.a.38045
24. Po' C, Zordan R, Vecchi M, Cerutti A, Sartori S, Trevisson E, et al. Photosensitive epilepsy and long QT: expanding Timothy syndrome phenotype. Clin Neurophysiol. (2019) 130:2134–6. doi: 10.1016/j.clinph.2019.09.003
25. Colson C, Mittre H, Busson A, Leenhardt A, Denjoy I, Fressard V, et al. Unusual clinical description of adult with Timothy syndrome, carrier of a new heterozygote mutation of CACNA1C. Eur J Med Genet. (2019) 62:103648. doi: 10.1016/j.ejmg.2019.04.005
26. Bisabu KK, Zhao J, Mokrane A-E, Segura É, Marsolais M, Grondin S, et al. Novel gain-of-function variant in CACNA1C associated with Timothy syndrome, multiple accessory pathways, and noncompaction cardiomyopathy. Circ Genom Precis Med. (2020) 13:e003123. doi: 10.1161/CIRCGEN.120.003123
27. Boczek NJ, Ye D, Jin F, Tester DJ, Huseby A, Bos JM, et al. Identification and functional characterization of a novel CACNA1C-mediated cardiac disorder characterized by prolonged QT intervals with hypertrophic cardiomyopathy, congenital heart defects, and sudden cardiac death. Circ Arrhythm Electrophysiol. (2015) 8:1122–32. doi: 10.1161/CIRCEP.115.002745
28. Ozawa J, Ohno S, Saito H, Saitoh A, Matsuura H, Horie M. A novel CACNA1C mutation identified in a patient with Timothy syndrome without syndactyly exerts both marked loss- and gain-of-function effects. Heart Rhythm Case Rep. (2018) 4:273–7. doi: 10.1016/j.hrcr.2018.03.003
29. Kosaki R, Ono H, Terashima H, Kosaki K. Timothy syndrome-like condition with syndactyly but without prolongation of the QT interval. Am J Med Genet A. (2018) 176:1657–61. doi: 10.1002/ajmg.a.38833
30. Ye D, Tester DJ, Zhou W, Papagiannis J, Ackerman MJ. A pore-localizing CACNA1C-E1115K missense mutation, identified in a patient with idiopathic QT prolongation, bradycardia, and autism spectrum disorder, converts the L-type calcium channel into a hybrid nonselective monovalent cation channel. Heart Rhythm. (2019) 16:270–8. doi: 10.1016/j.hrthm.2018.08.030
31. Papineau SD, Wilson S. Dentition abnormalities in a Timothy syndrome patient with a novel genetic mutation: a case report. Pediatr Dent. (2014) 36:245–9.
32. Boczek NJ, Miller EM, Ye D, Nesterenko VV, Tester DJ, Antzelevitch C, et al. Novel Timothy syndrome mutation leading to increase in CACNA1C window current. Heart Rhythm. (2015) 12:211–9. doi: 10.1016/j.hrthm.2014.09.051
33. Hennessey JA, Boczek NJ, Jiang Y-H, Miller JD, Patrick W, Pfeiffer R, et al. A CACNA1C variant associated with reduced voltage-dependent inactivation, increased Cav1.2 channel window current, and arrhythmogenesis. PLoS ONE. (2014) 9:e106982. doi: 10.1371/journal.pone.0106982
34. Dick IE, Joshi-Mukherjee R, Yang W, Yue DT. Arrhythmogenesis in Timothy syndrome is associated with defects in Ca2+-dependent inactivation. Nat Commun. (2016) 7:10370. doi: 10.1038/ncomms10370
35. Barrett CF, Tsien RW. The Timothy syndrome mutation differentially affects voltage- and calcium-dependent inactivation of CaV1.2 L-type calcium channels. Proc Natl Acad Sci USA. (2008) 105:2157–62. doi: 10.1073/pnas.0710501105
36. Fukuyama M, Wang Q, Kato K, Ohno S, Ding W-G, Toyoda F, et al. Long QT syndrome type 8: novel CACNA1C mutations causing QT prolongation and variant phenotypes. Ep Europace. (2014) 16:1828–37. doi: 10.1093/europace/euu063
37. Gardner RJM, Crozier IG, Binfield AL, Love DR, Lehnert K, Gibson K, et al. Penetrance and expressivity of the R858H CACNA1C variant in a five-generation pedigree segregating an arrhythmogenic channelopathy. Mol Genet Genom Med. (2019) 7:e00476. doi: 10.1002/mgg3.476
38. Burashnikov E, Pfeiffer R, Barajas-Martinez H, Delpón E, Hu D, Desai M, et al. Mutations in the cardiac L-type calcium channel associated with inherited J-wave syndromes and sudden cardiac death. Heart Rhythm. (2010) 7:1872–82. doi: 10.1016/j.hrthm.2010.08.026
39. Cerrone M, Montnach J, Lin X, Zhao Y-T, Zhang M, Agullo-Pascual E, et al. Plakophilin-2 is required for transcription of genes that control calcium cycling and cardiac rhythm. Nat Commun. (2017) 8:106. doi: 10.1038/s41467-017-00127-0
40. Acuna-Hidalgo R, Bo T, Kwint MP, van deVorst M, Pinelli M, Veltman JA, et al. Post-zygotic point mutations are an underrecognized source of de novo genomic variation. Am J Hum Genet. (2015) 97:67–74. doi: 10.1016/j.ajhg.2015.05.008
41. Etheridge SP, Bowles NE, Arrington CB, Pilcher T, Rope A, Wilde AAM, et al. Somatic mosaicism contributes to phenotypic variation in Timothy syndrome. Am J Med Genet A. (2011) 155:2578–83. doi: 10.1002/ajmg.a.34223
42. Clark MB, Wrzesinski T, Garcia AB, Hall NAL, Kleinman JE, Hyde T, et al. Long-read sequencing reveals the complex splicing profile of the psychiatric risk gene CACNA1C in human brain. Mol Psychiatr. (2020) 25:37–47. doi: 10.1038/s41380-019-0583-1
43. Liebrechts-Akkerman G, Liu F, Marion R van, Dinjens WNM, Kayser M. Explaining sudden infant death with cardiac arrhythmias: complete exon sequencing of nine cardiac arrhythmia genes in Dutch SIDS cases highlights new and known DNA variants. Forensic Sci Int Genet. (2020) 46:102266. doi: 10.1016/j.fsigen.2020.102266
44. Cohen-Kutner M, Yahalom Y, Trus M, Atlas D. Calcineurin controls voltage-dependent-inactivation (VDI) of the normal and Timothy cardiac channels. Sci Rep UK. (2012) 2:366. doi: 10.1038/srep00366
45. Gao T, Cuadra AE, Ma H, Bünemann M, Gerhardstein BL, Cheng T, et al. C-terminal fragments of the α1C(CaV1.2) subunit associate with and regulate L-type calcium channels containing C-terminal-truncated α1C subunits. J Biol Chem. (2001) 276:21089–97. doi: 10.1074/jbc.M008000200
46. Han D, Xue X, Yan Y, Li G. Dysfunctional Cav1.2 channel in Timothy syndrome, from cell to bedside. Exp Biol Med. (2019) 244:960–71. doi: 10.1177/1535370219863149
47. Seisenberger C, Specht V, Welling A, Platzer J, Pfeifer A, Kühbandner S, et al. Functional embryonic cardiomyocytes after disruption of the L-type α1C (Ca v 1.2) calcium channel gene in the mouse. J Biol Chem. (2000) 275:39193–9. doi: 10.1074/jbc.M006467200
48. Ramachandran KV, Hennessey JA, Barnett AS, Yin X, Stadt HA, Foster E, et al. Calcium influx through L-type CaV1.2 Ca2+ channels regulates mandibular development. J Clin Invest. (2013) 123:1638–46. doi: 10.1172/JCI66903
49. Bader PL, Faizi M, Kim LH, Owen SF, Tadross MR, Alfa RW, et al. Mouse model of Timothy syndrome recapitulates triad of autistic traits. Proc Natl Acad Sci USA. (2011) 108:15432–7. doi: 10.1073/pnas.1112667108
50. Bett GC, Lis A, Wersinger SR, Baizer JS, Duffey ME, Rasmusson RL. A mouse model of Timothy syndrome: a complex autistic disorder resulting from a point mutation in Cav1.2. Am Chin J Med Sci. (2012) 5:135. doi: 10.7156/najms.2012.053135
51. Stainier DY, Fouquet B, Chen JN, Warren KS, Weinstein BM, Meiler SE, et al. Mutations affecting the formation and function of the cardiovascular system in the zebrafish embryo. Development. (1996) 123:285–92.
52. Rottbauer W, Baker K, Wo ZG, Mohideen M-APK, Cantiello HF, Fishman MC. Growth and function of the embryonic heart depend upon the cardiac-specific L-type calcium channel α1 subunit. Dev Cell. (2001) 1:265–75. doi: 10.1016/S1534-5807(01)00023-5
53. Lorincz R, Emfinger CH, Walcher A, Giolai M, Krautgasser C, Remedi MS, et al. In vivo monitoring of intracellular Ca2+ dynamics in the pancreatic β-cells of zebrafish embryos. Islets. (2018) 10:1–18. doi: 10.1080/19382014.2018.1540234
54. Lagoy R, Kim H, Mello C, Albrecht D. A C. elegans model for the rare human channelopathy, Timothy syndrome type 1. microPubl Biol. (2018) 2018. doi: 10.17912/micropub.biology.000081
55. Buddell T, Friedman V, Drozd CJ, Quinn CC. An autism-causing calcium channel variant functions with selective autophagy to alter axon targeting and behavior. PLoS Genet. (2019) 15:e1008488. doi: 10.1371/journal.pgen.1008488
56. Estes SI, Ye D, Zhou W, Dotzler SM, Tester DJ, Bos JM, et al. Characterization of the CACNA1C-R518C missense mutation in the pathobiology of long-QT syndrome using human induced pluripotent stem cell cardiomyocytes shows action potential prolongation and L-type calcium channel perturbation. Circ Genom Precis Med. (2019) 12:e002534. doi: 10.1161/CIRCGEN.119.002534
57. Yazawa M, Dolmetsch RE. Modeling Timothy syndrome with iPS cells. J Cardiovasc Transl. (2013) 6:1–9. doi: 10.1007/s12265-012-9444-x
58. Song L, Park SE, Isseroff Y, Morikawa K, Yazawa M. Inhibition of CDK5 alleviates the cardiac phenotypes in Timothy syndrome. Stem Cell Rep. (2017) 9:50–7. doi: 10.1016/j.stemcr.2017.05.028
59. Hoekstra M, Mummery CL, Wilde AAM, Bezzina CR, Verkerk AO. Induced pluripotent stem cell derived cardiomyocytes as models for cardiac arrhythmias. Front Physiol. (2012) 3:346. doi: 10.3389/fphys.2012.00346
60. Paşca SP, Portmann T, Voineagu I, Yazawa M, Shcheglovitov A, Paşca AM, et al. Using iPSC-derived neurons to uncover cellular phenotypes associated with Timothy syndrome. Nat Med. (2011) 17:1657–62. doi: 10.1038/nm.2576
61. Tian Y, Voineagu I, Pasca SP, Won H, Chandran V, Horvath S, et al. Alteration in basal and depolarization induced transcriptional network in iPSC derived neurons from Timothy syndrome. Genome Med. (2014) 6:75. doi: 10.1186/s13073-014-0075-5
62. Panagiotakos G, Haveles C, Arjun A, Petrova R, Rana A, Portmann T, et al. Aberrant calcium channel splicing drives defects in cortical differentiation in Timothy syndrome. Elife. (2019) 8:e51037. doi: 10.7554/eLife.51037
63. Green EK, Grozeva D, Jones I, Jones L, Kirov G, Caesar S, et al. The bipolar disorder risk allele at CACNA1C also confers risk of recurrent major depression and of schizophrenia. Mol Psychiatr. (2010) 15:1016–22. doi: 10.1038/mp.2009.49
64. Nyegaard M, Demontis D, Foldager L, Hedemand A, Flint TJ, Sørensen KM, et al. CACNA1C (rs1006737) is associated with schizophrenia. Mol Psychiatr. (2010) 15:119–21. doi: 10.1038/mp.2009.69
65. Ferreira MAR, O'Donovan MC, Meng YA, Jones IR, Ruderfer DM, Jones L, et al. Collaborative genome-wide association analysis supports a role for ANK3 and CACNA1C in bipolar disorder. Nat Genet. (2008) 40:1056–8. doi: 10.1038/ng.209
66. Moon AL, Haan N, Wilkinson LS, Thomas KL, Hall J. CACNA1C: association with psychiatric disorders, behavior, and neurogenesis. Schizophr Bull. (2018) 44:958–65. doi: 10.1093/schbul/sby096
Keywords: Timothy syndrome, arrhythmia, congenital heart defect, syndactyly, autism spectrum disorder, CACNA1C, Cav1.2, variant
Citation: Bauer R, Timothy KW and Golden A (2021) Update on the Molecular Genetics of Timothy Syndrome. Front. Pediatr. 9:668546. doi: 10.3389/fped.2021.668546
Received: 16 February 2021; Accepted: 19 April 2021;
Published: 17 May 2021.
Edited by:
Merlin G. Butler, University of Kansas Medical Center, United StatesReviewed by:
Hansen Wang, University of Toronto, CanadaCopyright © 2021 Bauer, Timothy and Golden. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Andy Golden, YW5keWdAbmloLmdvdg==
†Present address: Rosemary Bauer, Driskill Graduate Program, Northwestern University, Chicago, IL, United States
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.