- 1Department of Medicine, Nazarbayev University School of Medicine (NUSOM), Nur-Sultan, Kazakhstan
- 2Department of Pediatrics, National Research Center for Maternal and Child Health, University Medical Center, Nur-Sultan, Kazakhstan
Background and Objective: The gut microbiota plays a role in regulating the host immunity. Therefore, alterations in gut microbiota (or dysbiosis) have been investigated in several gastrointestinal diseases, including Celiac Disease (CD). The aim of this study is to summarize the main characteristics of the gut microbiota in pediatric CD.
Methods: We performed a systematic review to retrieve the available studies investigating the gut microbiota in pediatric CD patients and controls. In detail, after the screening of >2,200 titles from the medical literature, 397 articles were assessed for eligibility based on the abstracts: of those, 114 full-text original articles were considered as eligible according to the aim of this systematic review.
Results: The final search output consisted of 18 articles describing the gut microbiota of CD children and including one or more control groups. Eleven pediatric studies provided information on the duodenal microbiota and as many investigated the fecal microbiota; three articles analyzed the microbiota on both fecal and duodenal samples from the same cohorts of patients.
Conclusion: Due to the heterogeneity of the experimental procedures and study design, it is not possible to evidence any specific celiac signature in the fecal and/or duodenal microbiota of CD children. However, some specific components of the fecal microbiota and, in detail, Bifidobacterium spp. (e.g., Bifidobacterium longum) may deserve additional research efforts, in order to understand their potential value as both probiotic therapy and diagnostic/prognostic biomarker.
Introduction
The microbial communities naturally colonizing the gastrointestinal (GI) tract, namely, gut microbiota, are an essential and physiological component of a healthy human body. In fact, the “whole” deriving from this symbiotic interaction between microbiota and host, has been defined as a “superorganism,” where the former contributes to several metabolic, physiologic, inflammatory, and immunologic functions for the latter (1, 2).
In detail, the microbiota plays a role in regulating the host immunity by influencing the development and homeostasis of the gut epithelial layer and mucosal-associated lymphoid tissue. Therefore, alterations in gut microbiota (or dysbiosis) have been explored and investigated in several GI disorders, including Celiac Disease (CD).
Indeed, CD is a systemic immune-mediated condition, characterized by a variable pattern of GI and extra-GI clinical manifestations, but clearly defined by the presence of gluten-dependent atrophic (small bowel) enteropathy associated with a consistent serological panel (positivity for anti-tissue transglutaminase antibody and/or anti-endomysium antibody) (3, 4). Importantly, gluten can trigger CD in a minority of patients who are carriers of specific HLA-DQ genotypes (HLA-DQ2 and HLA-DQ8): indeed, both these environmental and genetic factors respectively represent necessary, but not sufficient, conditions to develop CD (5).
Each GI tract is characterized by a peculiar microbiota, in terms of qualitative and quantitative composition. In general, the bacterial density progressively increases along the GI tract, ranging from 103 units per gram of luminal content in the duodenum up to values of 1012 order in the colon; concomitantly, the bacterial diversity (in terms of number of different bacterial species) gradually increases from the proximal to the distal GI tracts as well (2, 6).
The knowledge on gut microbiota has greatly improved in the last few years, since culture-independent approaches became available, such as the analysis of bacterial 16S ribosomal RNA (rRNA) gene. Among bacteria (indeed, also yeast and viruses are part of gut microbiota), the dominant phyla are Firmicutes and Bacteroidetes: the former phylum mainly includes Clostridium spp., Bacillus spp., Lactobacillus spp., Ruminococcus spp., and Enterococcus spp.; Bacteroidetes phylum is predominantly composed of Bacteroides spp. and Prevotella spp. The remaining phyla (Actinobacteria, Fusobacteria, Proteobacteria, and Verrucomicrobia) represent around 10% of the bacterial gut population; among them, the most studied genus is Bifidobacterium spp., which belongs to Actinobacteria phylum (7–9).
The inter-individual variability of gut microbiota is high: indeed, its composition is influenced by a multitude of factors, such as genetics, age, diet, hygiene level, and medication exposure (first of all, antibiotics). As mentioned above, deviations in the composition of gut microbiota may also be related to specific pathological conditions, but it is still unclear if these are cause or effect of the comorbid disease, due to the limitations of the available studies so far (10, 11).
In this systematic review, we summarized the main characteristics of the gut microbiota in pediatric CD patients.
Materials and Methods
Protocol
The PRISMA guidelines were used for this systematic review (12). This systematic review includes original articles (such as experimental studies, randomized control trials, case–control studies, cohort studies, and observational studies), providing information on gut microbiota in CD children and age-matched controls. Indeed, the primary endpoint was the description of gut microbiota in pediatric patients affected with CD. This systematic search did not include review and abstract papers.
Search Strategy
The literature search strategy of this review consisted of two stages: (i) an extensive search in five databases, namely, Scopus, Cochrane Central Register of Controlled Trials, PubMed, Ovid, and Web of Science, by using relevant keywords, as described in Table 1; (ii) a search of reference lists from the collected and related articles identified at the previous stage.
Data Extraction
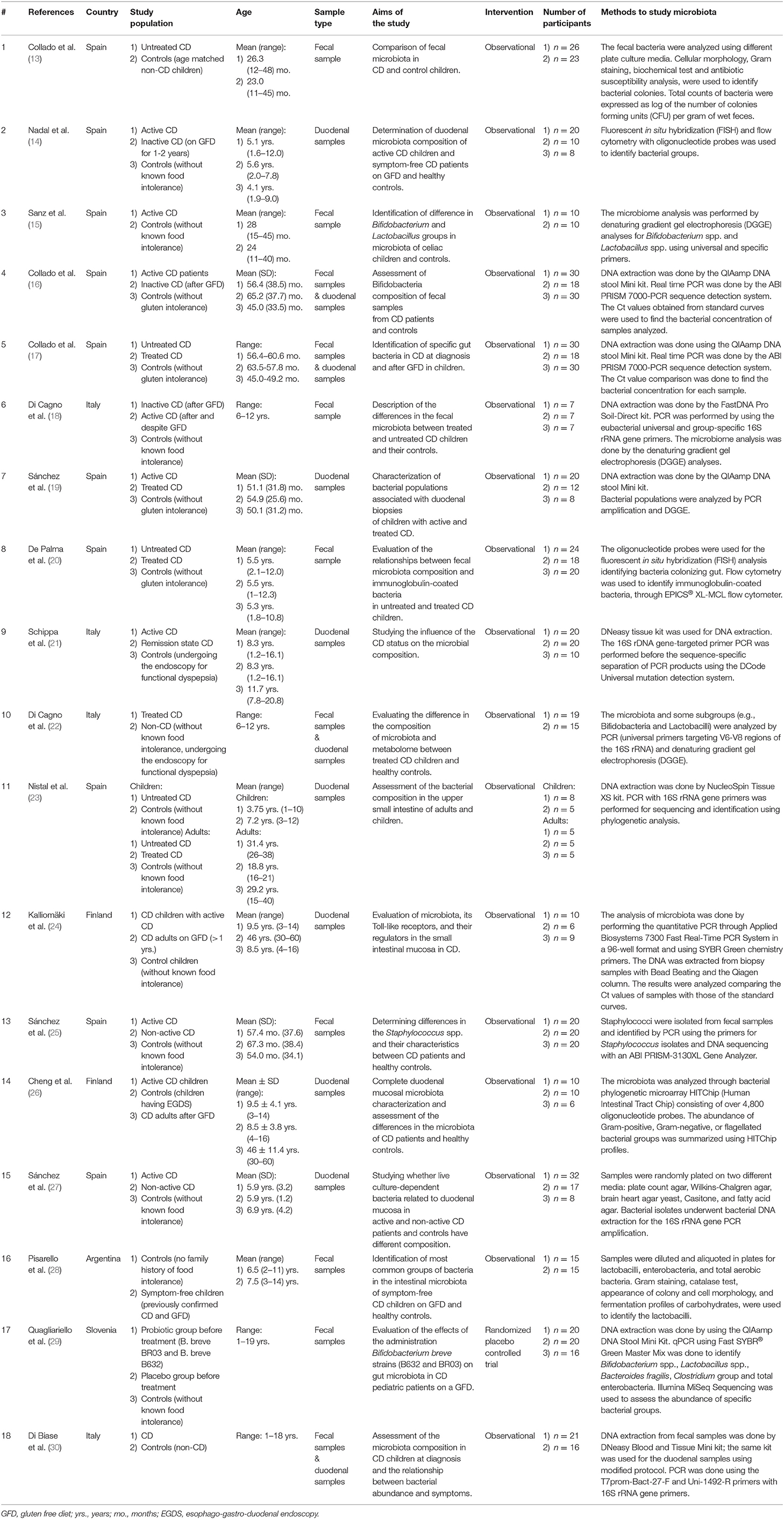
After a critical reading of the articles, data extraction was done by one investigator and then checked by a second investigator following these inclusion criteria: any original articles in which gut microbiota composition was studied in CD patients with clear description of methods and outcomes. In detail, the following items were extracted from each study: first author's last name, publication year, country of origin, study population details, age group, sample type, aims of the study, intervention type, number of participants, and analytical methods to study microbiota.
Results
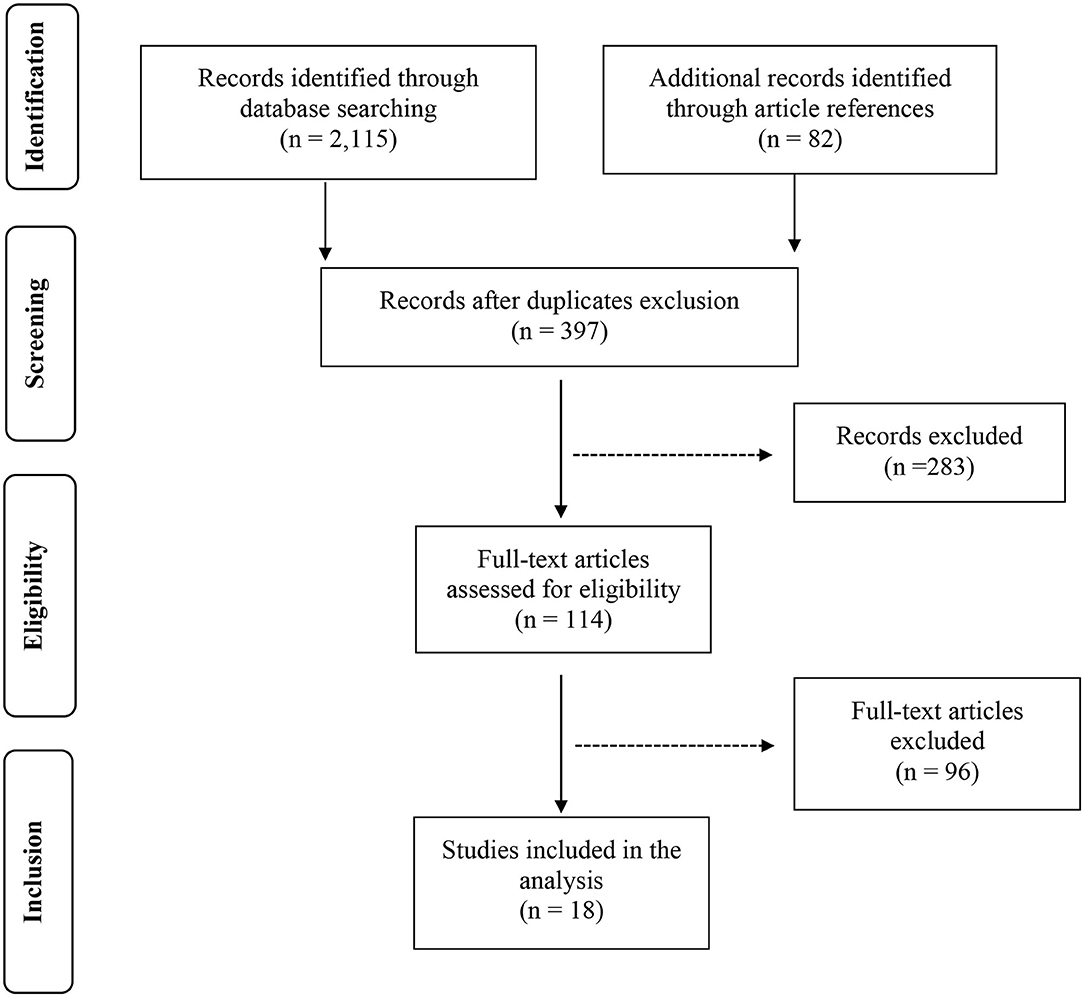
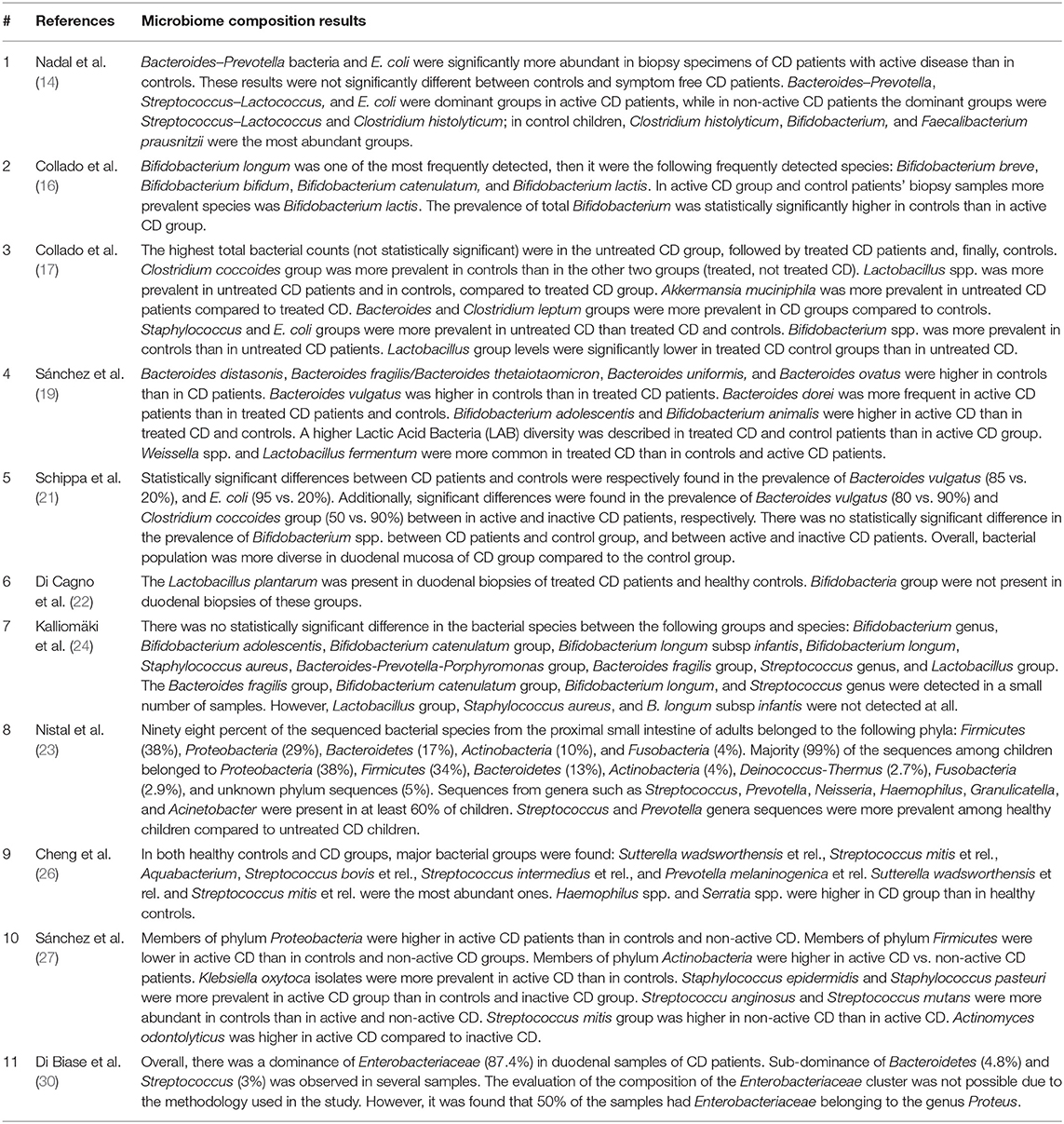
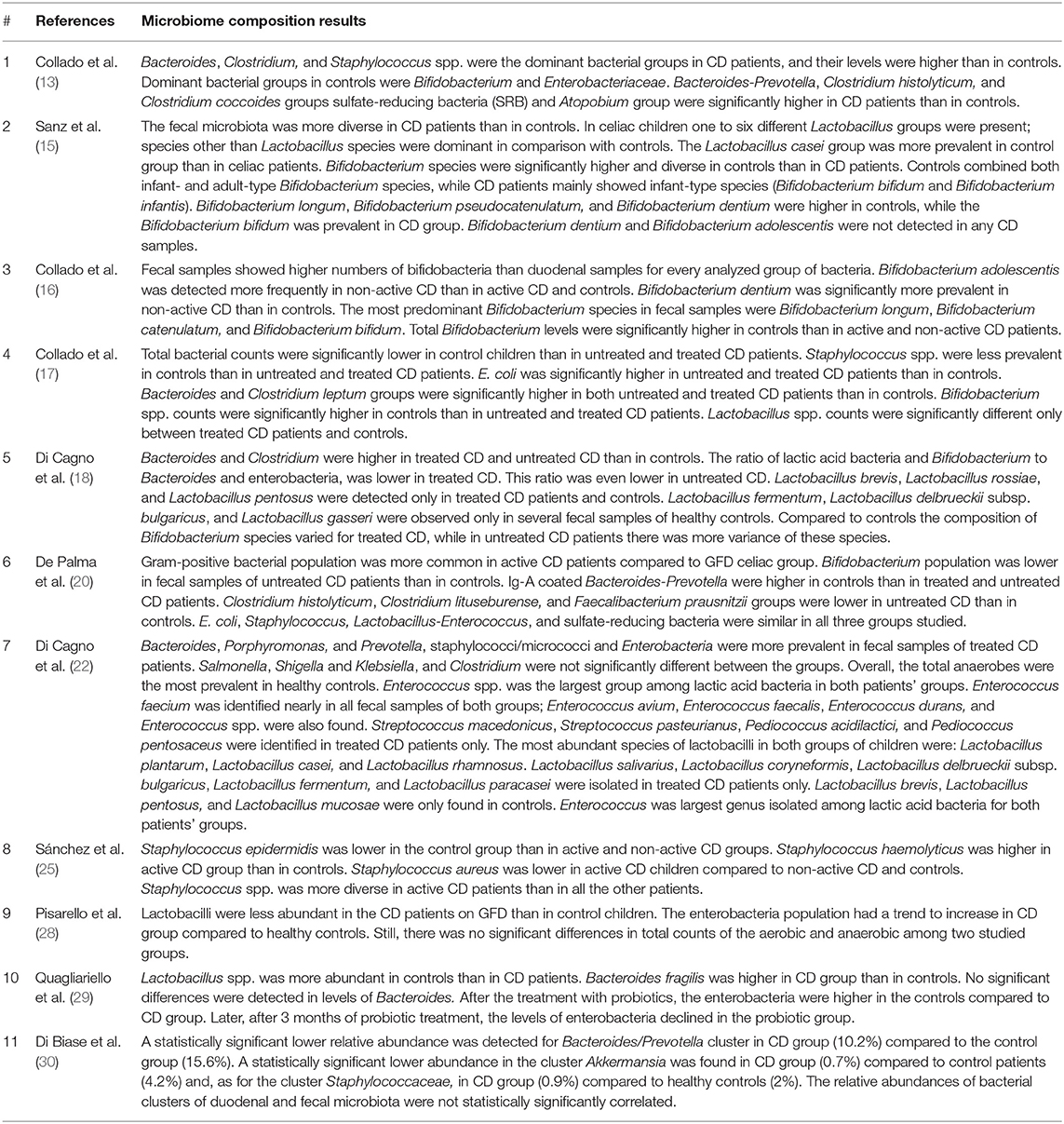
The results of each step of the literature search according to the PRISMA guidelines are schematically summarized in Figure 1. In detail, the literature search resulted in 1,143 papers from Scopus, 44 from the Cochrane Central Register of Controlled Trials, 337 from PubMed, 82 from Ovid, and 509 from Web of Science. Eighty-two additional articles were identified through the references of the previous papers. After the screening by title, the duplicated records were removed, and 397 articles were assessed for eligibility based on the abstracts: of those, 114 full-text original articles were considered as eligible according to the aim of this systematic review. The final search output consisted of 18 articles describing the gut microbiota of CD children and including one or more control groups (13–30). All the articles finally included in this systematic review are listed in Table 2, where the study population, participants' age group, sample type(s), aim of the study, type of intervention, sample size, and detailed laboratory methods are summarized. Importantly, studies focused on HLA-DQ predisposition, which analyzed the gut microbiota in children before becoming celiac (thus, with no microbiome analysis in CD pediatric patients after the diagnosis), were excluded. Moreover, only studies including a control group to be compared to CD children, were included. The specific findings resulting from these selected studies are summarized in Tables 3 and 4, which refer to the gut microbiota studies on duodenal biopsy and stool samples, respectively. In detail, these tables provide the main findings in terms of overall bacterial abundance and/or bacterial diversity and/or specific bacterial composition, according to the variable aims of each study.
Discussion
In the landscape of immune-mediated non-communicable disorders, CD is the only disease for which the necessary HLA genetic background and environmental trigger are known. Briefly, the dietary intake of gluten triggers the development of CD in some of those individuals who are carriers of the specific HLA-DQ allelic variants coding DQ2 and DQ8 heterodimers (DQA1*0501-DQB1*02 and DQA1*0301-DQB1*0302, respectively) (4, 5). Overall, 30–40% of the general population in Europe and North America carry this HLA-DQ genotype, but only 3% of them actually develop CD during the life (despite the dietary exposure to gluten foods), which thus corresponds to around 1% prevalence in the general population (31, 32). Therefore, these HLA-DQ genes and the gluten dietary intake, even if necessary factors, are not sufficient to develop CD; other concomitant (and currently unknown or not well-defined) environmental agents, non-HLA genetic aspects, and maybe epigenetic mechanisms, are supposed to play a critical role in determining which individuals will become celiac within a much larger HLA-predisposed and gluten-exposed population. In this regard, the gut microbiota has been considered among the factors potentially affecting or modulating the risk of developing CD, through its interplay with the intestinal epithelium and/or the host immune system (33, 34).
In this systematic review, we aimed at summarizing and describing the main characteristics of the gut microbiota in CD children, compared to non-celiac controls. In order to achieve this purpose, we analyzed the pediatric studies investigating both fecal samples and duodenal specimens, which present substantial differences, of course. Indeed, as mentioned, the gut microbiota greatly varies along the GI tract, due to the different environmental conditions, in terms of pH, oxygen tension, substrates availability, host secretion, and intestinal motility (35). In the duodenum (which is characterized by a relatively acidic pH, high level of oxygen, and rapid transit time), facultative anaerobic and rapidly growing bacteria able to adhere to the epithelium in the mucus layer, are more likely to survive. On the contrary, the fecal samples are more representative of the colonic environment, which presents more favorable conditions for bacterial survival and, thus, is characterized by a more abundant and diverse microbiota, including mainly anaerobes (36, 37).
Eleven pediatric studies provided information on the duodenal microbiota of untreated CD children and age-matched controls. Among those, eight included only pediatric patients (14, 16, 17, 19, 21, 22, 27, 30), whereas three investigated both children and adults (23, 24, 26).
Nadal et al. (14) mainly found that Bacteroides spp., Prevotella spp., and Escherichia coli populations were significantly more abundant in CD patients with active disease than in controls. Recently, Di Biase et al. (30) showed a “total dominance” of Enterobacteriaceae in the duodenal flora of active CD children; to follow, Bacteroidetes spp. and Streptococcus spp. turned out to be the second most represented category. However, no control group was included for this analysis. Schippa et al. (21) described a significantly higher microbiota biodiversity in CD children, compared to controls; in terms of bacterial composition, these authors highlighted a significant difference in the prevalence of Bacteroides vulgatus (85 vs. 20%) and E. coli (95 vs. 20%) between these two groups. In terms of phyla, as described by Sánchez et al. (27), Firmicutes and Proteobacteria were respectively more and less abundant in children with CD, compared to their controls; importantly, these results were also confirmed with respect to CD patients on gluten-free diet (non-active CD group).
Actually, two of the aforementioned studies also included non-active CD children [Nadal et al. (14) and Schippa et al. (21)]: both found no significant differences between these groups of children and their controls, in terms of bacterial proportions and biodiversity, respectively. Overall, all these three studies on pediatric CD indicated that the bacterial deviations could be normalized after the appropriate gluten-free diet. Conversely, Collado et al. (17) described significantly higher number of Bacteroides and Clostridium leptum in both treated and untreated (active) CD children compared to controls; however, a statistically significant difference between active CD children from one side, and both treated CD and control children, on the other side, was reported for Staphylococcus spp. and E. coli.
Three additional studies included adult CD patients, in addition to children. Actually, Kalliomäki et al. (24) compared CD children to healthy children and non-active CD adults: they found no difference in bacterial counts among these three groups. Again, in the study by Nistal et al., there was no significant difference between active CD and healthy controls, considering the 36 different genera of known bacteria detected by their analysis. Importantly, this study provided a direct comparison between CD children and adults: even though both types of patients were colonized by bacteria mainly belonging to the Firmicutes, Proteobacteria, and Bacteroidetes phyla, the bacterial abundance and diversity were significantly lower in CD children than in CD adults (23). Cheng et al. also reported that the overall composition and diversity of gut microbiota was comparable between CD children and healthy controls; however, the specific profiles of eight bacterial groups were significantly different: in detail, Prevotella spp. (and, in particular, Prevotella melaninogenica), Haemophilus spp., and Serratia spp. (and, in particular, S. marcescens) were more abundant in CD children, whereas Prevotella oralis, Ruminococcus bromii, Papillibacter cinnamivorans, Proteus spp., and Clostridium stercorarium were less abundant (26).
Importantly, several studies focused the attention on some specific bacteria and, in detail, Bifidobacterium spp. and Lactobacillus spp. As for the former group, in both studies by Collado et al. (16), a significantly lower number was reported in CD (and, in particular, untreated) children than in controls; on the contrary, Di Cagno et al. (18) found no significant representation of Bifidobacteria in their CD and control groups, overall. No significant differences were reported in the prevalence of Bifidobacterium spp. between CD patients and controls, and between active and inactive CD, in the study by Schippa et al. (21). As regards Lactobacillus spp., Di Cagno et al. (18) described a relatively homogeneous population in all children, whereas Collado et al. (16) reported that these bacteria were significantly less abundant in controls and treated CD children than in the active CD group.
Therefore, the available data on duodenal microbiota in CD children are not all consistent among themselves. The main limitation of the aforementioned studies is the small sample size, since all these studies included around or no more than 30 children, probably due to technical and ethical issues concerning the invasive procedures required to analyze the duodenal microbiota. This aspect may have also affected the selection and inclusion of an appropriate control group, of course: actually, whereas in the studies by Collado et al. (13) and Nadal et al. (14) these control children are not better defined than as “without gluten intolerance” and “without known food intolerance,” only Schippa et al. (21) disclosed the reason why they underwent upper GI endoscopy, namely, “functional dyspepsia.” Therefore, the unclear description and, anyway, the impossibility to compare these findings with completely healthy children represent additional obstacles to the appropriate interpretation of the microbiota findings obtained from the duodenal mucosa.
These specific research limitations can be lessened by studying the microbiota in fecal samples, whose collection does not imply any invasive procedure, of course. Importantly, this aspect makes this analysis much more attractive as potential non-invasive diagnostic and/or prognostic biomarkers for pediatric CD.
Eleven pediatric studies provided information on the fecal microbiota of untreated CD children and age-matched controls. Among those, all investigated only pediatric patients (13, 16–18, 20, 22, 25, 28–30), except one, which included both children and adults (15).
To start with, Collado et al. (13) showed significantly lower total bacterial counts in stools from the control group compared to treated or untreated CD children, but this specific information was not provided by other studies. In terms of microbiota general diversity, Sanz et al. (15) found it significantly higher in CD children than in controls, but Di Cagno et al. (18) could not confirm this finding.
Some studies mainly analyzed the relative composition of the fecal microbiota. In general, De Palma et al. (20) investigated both active and treated CD children, in addition to healthy controls: they reported a significantly decreased gram+/gram– ratio in both groups of CD children, compared to controls; interestingly, treated CD children showed intermediate values between active CD children and healthy controls, but the difference was statistically significant only with the latter group, suggesting that this specific aspect was not completely restored by the gluten-free diet. In terms of phyla, these authors reported that the Bacteroides/Prevotella cluster was significantly more expressed in untreated CD patients than in healthy controls; this finding was confirmed by Collado et al. (16, 17) and Quagliariello et al. (29).
However, most researchers investigated some specific bacteria of the fecal microbiota, mainly Bifidobacterium spp. and Lactobacillus spp., which differed in terms of relative species abundance between CD patients and healthy controls (15, 22, 28). Sánchez et al. (25) focused on Staphylococcus spp. and observed some peculiarities in species diversity and abundance in CD children; however, the increased presence of S. epidermidis isolates carrying genes conferring resistance to methicillin suggested that these changes may be induced by a greater exposure to antibiotics rather than CD itself. However, Di Biase et al. (30) reported a lower abundance of Staphylococcus spp. in CD children, as well as a significant reduction of the Bacteroides/Prevotella cluster, which is in contrast with some of the aforementioned studies.
Therefore, even though the available fecal microbiota studies in CD children are more than those investigating the duodenal microbiota, their clinical aims and settings were quite heterogeneous: indeed, they provided a qualitative and quantitative information that is not immediately comparable, and the results are often conflicting because of that, probably.
Importantly, 3 of these 11 articles on fecal microbiota also provided data related to duodenal biopsies from the same cohorts of patients (16, 17, 30). In detail, Collado et al. discussed the relation between fecal and duodenal microbiota: they found significant correspondences for Bifidobacterium spp. among all three groups (active CD, treated CD, and controls), whereas other bacterial groups did not correlate in all groups. As regards specifically untreated CD children, Bacteroides spp., Staphylococcus spp., Lactobacillus spp., C. leptum group, Clostridium coccoides group, E. coli, and Akkermansia muciniphila showed a significant correlation between these two types of samples (16, 17). Unfortunately, Di Biase et al. (30) did not analyze such a correlation; however, about this study, it may be worth to emphasize the fact that A. muciniphila was found to be significantly less abundant in CD children's stool, along with Bacteroides spp. and the Staphylococcus group, as previously mentioned. Although Collado et al. (13) described a significant duodenal–fecal relation for that microorganism, they did not actually find statistically significant differences in its content among their study groups. In an additional study, they focused on Bifidobacterium spp. and, once again, they compared duodenal biopsies and fecal samples in the same cohort of patients. The fecal microbiota resulted to include significantly higher numbers of bifidobacteria than the duodenal samples. Interestingly, they found a statistically significant correlation between these sample types in all groups as regards the total number of bifidobacteria and, in terms of species, only Bifidobacterium longum significantly correlated in both samples and all groups. B. longum was the most frequent and abundant species in this study and, importantly, was significantly different among all study groups, both in stools and duodenum, in addition to be the only one to significantly correlate between both types of samples, as previously explained (16).
Indeed, B. longum was the object of some preliminary investigations as potential therapeutic resource and prognostic biomarker. In a double-blind, randomized, and placebo-controlled trial, Olivares et al. (38) showed that the administration of the CECT 7347 strain of B. longum was associated with a significant reduction of the Bacteroides fragilis group in the microbiota of CD patients, in addition to some clinical benefit in terms of anthropometric parameters. Some recent analyses derived from a large perspective cohort study (PROFICEL), comparing the fecal microbiota in children at HLA-DQ genetic risk of CD before the appearance of the disease (exactly, at 4 and 6 months of life), highlighted some microbiota differences between those children who eventually developed CD and those who did not. Interestingly, high-risk genotypes for CD (referring to HLA-DQ2 and HLA-DQ8 heterodimers) were associated to a lower amount of Bifidobacterium spp. and, specifically, B. longum (39–41). More in general, several microbial species and related metabolites (with inflammatory and immunological properties) have been recently suggested as potentially specific to CD, through multi-omics analysis (42).
Even though the interplay between HLA-DQ (and, in detail, HLA-DQB1*02, which is the most frequent allelic variant in CD children) and intestinal microbiota must be precisely elucidated yet, these preliminary observations might provide the background to plan further studies to assess the risk of developing CD in gluten-exposed population and, potentially, to consider additional non-invasive diagnostic tools and prognostic markers for CD (37, 43, 44).
Conclusion
Due to the heterogeneity of the experimental procedures and design of these studies, it is not possible to evidence any specific and absolute celiac signature in the fecal and/or duodenal microbiota of CD children. However, some specific components of the fecal microbiota (e.g., Bifidobacterium spp.) may deserve additional research efforts to understand the potential application as both probiotic therapy and/or diagnostic/prognostic biomarker.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author/s.
Author Contributions
DP and DA conceived the study and wrote the review. DA, DP, and KD performed the literature search. All authors contributed to the article and approved the submitted version.
Funding
This review was funded by the Nazarbayev University Faculty Development Competitive Research Grant 2020-2022 (No. 240919FD3912) and Social Policy Grant (SPG).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Nardone G, Compare D, Rocco A. A microbiota-centric view of diseases of the upper gastrointestinal tract. Lancet Gastroenterol Hepatol. (2017) 2:298–312. doi: 10.1016/S2468-1253(16)30108-X
2. Dieterich W, Schink M, Zopf Y. Microbiota in the gastrointestinal tract. Med Sci. (2018) 6:116. doi: 10.3390/medsci6040116
3. Lindfors K, Ciacci C, Kurppa K, Lundin KEA, Makharia GK, Mearin ML, et al. Coeliac disease. Nat Rev Dis Prim. (2019) 5:3. doi: 10.1038/s41572-018-0054-z
4. Lebwohl B, Sanders DS, Green PHR. Coeliac disease. Lancet. (2018) 391:70–81. doi: 10.1016/S0140-6736(17)31796-8
5. Poddighe D, Rebuffi C, de Silvestri A, Capittini C. Carrier frequency of HLA-DQB1*02 allele in patients affected with celiac disease: a systematic review assessing the potential rationale of a targeted allelic genotyping as a first-line screening. World J Gastroenterol. (2020) 26:1365–81. doi: 10.3748/wjg.v26.i12.1365
6. Rinninella E, Mele MC, Merendino N, Cintoni M, Anselmi G, Caporossi A, et al. The role of diet, micronutrients and the gut microbiota in age-related macular degeneration: new perspectives from the gut–retina axis. Nutrients. (2018) 10:1677. doi: 10.3390/nu10111677
7. Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, et al. Enterotypes of the human gut microbiome. Nature. (2011) 473:174–80. doi: 10.1038/nature09944
8. Thursby E, Juge N. Introduction to the human gut microbiota. Biochem J. (2017) 474:1823–36. doi: 10.1042/BCJ20160510
9. Vuik FER, Dicksved J, Lam SY, Fuhler GM, van der Laan L, van de Winkel A, et al. Composition of the mucosa-associated microbiota along the entire gastrointestinal tract of human individuals. United Eur Gastroenterol J. (2019) 7:897–907. doi: 10.1177/2050640619852255
10. Cho I, Blaser MJ. The human microbiome: at the interface of health and disease. Nat Rev Genet. (2012) 13:260–70. doi: 10.1038/nrg3182
11. Sommer F, Bäckhed F. The gut microbiota-masters of host development and physiology. Nat Rev Microbiol. (2013) 11:227–38. doi: 10.1038/nrmicro2974
12. Moher D, Liberati A, Tetzlaff J, Altman DG. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. J Clin Epidemiol. (2009) 62:1006–12. doi: 10.1016/j.jclinepi.2009.06.005
13. Collado MC, Calabuig M, Sanz Y. Differences between the fecal microbiota of coeliac infants and healthy controls. Curr Issues Intest Microbiol. (2007) 8:9–14.
14. Nadal I, Donant E, Ribes-Koninckx C, Calabuig M, Sanz Y. Imbalance in the composition of the duodenal microbiota of children with coeliac disease. J Med Microbiol. (2007) 56:1669–74. doi: 10.1099/jmm.0.47410-0
15. Sanz Y, Sánchez E, Marzotto M, Calabuig M, Torriani S, Dellaglio F. Differences in faecal bacterial communities in coeliac and healthy children as detected by PCR and denaturing gradient gel electrophoresis. FEMS Immunol Med Microbiol. (2007) 51:562–8. doi: 10.1111/j.1574-695X.2007.00337.x
16. Collado MC, Donat E, Ribes-Koninckx C, Calabuig M, Sanz Y. Imbalances in faecal and duodenal Bifidobacterium species composition in active and non-active coeliac disease. BMC Microbiol. (2008) 8:232. doi: 10.1186/1471-2180-8-232
17. Collado MC, Donat E, Ribes-Koninckx C, Calabuig M, Sanz Y. Specific duodenal and faecal bacterial groups associated with paediatric coeliac disease. J Clin Pathol. (2009) 62:264–9. doi: 10.1136/jcp.2008.061366
18. Di Cagno R, Rizzello CG, Gagliardi F, Ricciuti P, Ndagijimana M, Francavilla R, et al. Different fecal microbiotas and volatile organic compounds in treated and untreated children with celiac disease. Appl Environ Microbiol. (2009) 75:3963–71. doi: 10.1128/AEM.02793-08
19. Sánchez E, Donat E, Ribes-Koninckx C, Calabuig M, Sanz Y. Intestinal bacteroides species associated with coeliac disease. J Clin Pathol. (2010) 63:1105–11. doi: 10.1136/jcp.2010.076950
20. De Palma G, Nadal I, Medina M, Donat E, Ribes-Koninckx C, Calabuig M, et al. Intestinal dysbiosis and reduced immunoglobulin-coated bacteria associated with coeliac disease in children. BMC Microbiol. (2010) 10:63. doi: 10.1186/1471-2180-10-63
21. Schippa S, Iebba V, Barbato M, Di Nardo G, Totino V, Checchi MP, et al. A distinctive “microbial signature” in celiac pediatric patients. BMC Microbiol. (2010) 10:175. doi: 10.1186/1471-2180-10-175
22. Di Cagno R, De Angelis M, De Pasquale I, Ndagijimana M, Vernocchi P, Ricciuti P, et al. Duodenal and faecal microbiota of celiac children: molecular, phenotype and metabolome characterization. BMC Microbiol. (2011) 11:219. doi: 10.1186/1471-2180-11-219
23. Nistal E, Caminero A, Herrán AR, Arias L, Vivas S, de Morales JM, et al. Differences of small intestinal bacteria populations in adults and children with/without celiac disease: effect of age, gluten diet, and disease. Inflamm Bowel Dis. (2012) 18:649–56. doi: 10.1002/ibd.21830
24. Kalliomäki M, Satokari R, Lähteenoja H, Vähämiko S, Grönlund J, Routi T, et al. Expression of microbiota, toll-like receptors, and their regulators in the small intestinal mucosa in celiac disease. J Pediatr Gastroenterol Nutr. (2012) 54:727–32. doi: 10.1097/MPG.0b013e318241cfa8
25. Sánchez E, Ribes-Koninck C, Calabuig M, Sanz Y. Intestinal staphylococcus spp. and virulent features associated with coeliac disease. J Clin Pathol. (2012) 65:830–4. doi: 10.1136/jclinpath-2012-200759
26. Cheng J, Kalliomäki M, Heilig HGHJ, Palva A, Lähteenoja H, de Vos WM, et al. Duodenal microbiota composition and mucosal homeostasis in pediatric celiac disease. BMC Gastroenterol. (2013) 13:113. doi: 10.1186/1471-230X-13-113
27. Sánchez E, Donat E, Ribes-Koninckx C, Fernández-Murga ML, Sanz Y. Duodenal-mucosal bacteria associated with celiac disease in children. Appl Environ Microbiol. (2013) 79:5472–9. doi: 10.1128/AEM.00869-13
28. Pisarello MLJ, Vintiñi EO, González SN, Pagani F, Medina MS. Decrease in lactobacilli in the intestinal microbiota of celiac children with a gluten-free diet, and selection of potentially probiotic strains. Can J Microbiol. (2014) 61:32–7. doi: 10.1139/cjm-2014-0472
29. Quagliariello A, Aloisio I, Bozzi cionci N, Luiselli D, D'Auria G, Martinez-Priego L, et al. Effect of bifidobacterium breve on the intestinal microbiota of coeliac children on a gluten free diet: A pilot study. Nutrients. (2016) 8:660. doi: 10.3390/nu8100660
30. Di Biase AR, Marasco G, Ravaioli F, Dajti E, Colecchia L, Righi B, et al. Gut microbiota signatures and clinical manifestations in celiac disease children at onset: a pilot study. J Gastroenterol Hepatol. (2020) 36:446–54. doi: 10.1111/jgh.15183
31. Ludvigsson JF, Murray JA. Epidemiology of celiac disease. Gastroenterol Clin North Am. (2019) 48:1–8. doi: 10.1016/j.gtc.2018.09.004
32. Poddighe D, Rakhimzhanova M, Marchenko Y, Catassi C. Pediatric celiac disease in central and east Asia: Current knowledge and prevalence. Medicina (Kaunas). (2019) 55:11. doi: 10.3390/medicina55010011
33. Valitutti F, Cucchiara S, Fasano A. Celiac disease and the microbiome. Nutrients. (2019) 11:2403. doi: 10.3390/nu11102403
34. Krishnareddy S. The microbiome in celiac disease. Gastroenterol Clin North Am. (2019) 48:115–26. doi: 10.1016/j.gtc.2018.09.008
35. Flint HJ, Scott KP, Louis P, Duncan SH. The role of the gut microbiota in nutrition and health. Nat Rev Gastroenterol Hepatol. (2012) 9:577–89. doi: 10.1038/nrgastro.2012.156
36. Donaldson GP, Lee SM, Mazmanian SK. Gut biogeography of the bacterial microbiota. Nat Rev Microbiol. (2015) 14:20–32. doi: 10.1038/nrmicro3552
37. Gu S, Chen D, Zhang JN, Lv X, Wang K, Duan LP, et al. Bacterial community mapping of the mouse gastrointestinal tract. PLoS ONE. (2013) 8:e74957. doi: 10.1371/journal.pone.0074957
38. Olivares M, Castillejo G, Varea V, Sanz Y. Double-blind, randomised, placebo-controlled intervention trial to evaluate the effects of Bifidobacterium longum CECT 7347 in children with newly diagnosed coeliac disease. Br J Nutr. (2014) 112:30–40. doi: 10.1017/S0007114514000609
39. Olivares M, Walker AW, Capilla A, Benítez-Páez A, Palau F, Parkhill J, et al. Gut microbiota trajectory in early life may predict development of celiac disease. Microbiome. (2018) 6:36. doi: 10.1186/s40168-018-0415-6
40. Rintala A, Riikonen I, Toivonen A, Pietilä S, Munukka E, Pursiheimo JP, et al. Early fecal microbiota composition in children who later develop celiac disease and associated autoimmunity. Scand J Gastroenterol. (2018) 53:403–9. doi: 10.1080/00365521.2018.1444788
41. De Palma G, Capilla A, Nova E, Castillejo G, Varea V, Pozo T, et al. Influence of milk-feeding type and genetic risk of developing coeliac disease on intestinal microbiota of infants: the PROFICEL study. PLoS ONE. (2012) 7:e30791. doi: 10.1371/journal.pone.0030791
42. Leonard MM, Karathia H, Pujolassos M, Troisi J, Valitutti F, Subramanian P, et al. Multi-omics analysis reveals the influence of genetic and environmental risk factors on developing gut microbiota in infants at risk of celiac disease. Microbiome. (2020) 8:130. doi: 10.1186/s40168-020-00906-w
43. Poddighe D, Turganbekova A, Baymukasheva D, Saduakas Z, Zhanzakova Z, Abdrakhmanova S. Genetic predisposition to celiac disease in Kazakhstan: potential impact on the clinical practice in Central Asia. PLoS ONE. (2020) 15:e0226546. doi: 10.1371/journal.pone.0226546
Keywords: celiac disease, children, gut microbiota, microbiome, HLA-DQ, Bifidobacterium spp
Citation: Abdukhakimova D, Dossybayeva K and Poddighe D (2021) Fecal and Duodenal Microbiota in Pediatric Celiac Disease. Front. Pediatr. 9:652208. doi: 10.3389/fped.2021.652208
Received: 11 January 2021; Accepted: 15 February 2021;
Published: 22 April 2021.
Edited by:
Jorge Amil Dias, Centro Hospitalar de São João, PortugalReviewed by:
Carmen Ribes-Koninckx, La Fe Hospital, SpainFrancesco Valitutti, Ospedali Riuniti San Giovanni di Dio e Ruggi d'Aragona, Italy
Copyright © 2021 Abdukhakimova, Dossybayeva and Poddighe. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dimitri Poddighe, ZGltaXRyaS5wb2RkaWdoZUBudS5lZHUua3o=
 Diyora Abdukhakimova1
Diyora Abdukhakimova1 Dimitri Poddighe
Dimitri Poddighe