
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
SYSTEMATIC REVIEW article
Front. Pediatr., 02 February 2021
Sec. Pediatric Gastroenterology, Hepatology and Nutrition
Volume 9 - 2021 | https://doi.org/10.3389/fped.2021.626232
This article is part of the Research TopicPediatric Inflammatory Bowel Disease: the Next FrontierView all 26 articles
Background and Aim: Accumulating evidence have implicated gut microbiota alterations in pediatric and adult patients with inflammatory bowel disease (IBD); however, the results of different studies are often inconsistent and even contradictory. It is believed that early changes in new-onset and treatment-naïve pediatric patients are more informative. We performed a systematic review to investigate the gut microbiota profiles in pediatric IBD and identify specific microbiota biomarkers associated with this disorder.
Methods: Electronic databases were searched from inception to 31 July 2020 for studies that observed gut microbiota alterations in pediatric patients with IBD. Study quality was assessed using the Newcastle–Ottawa scale.
Results: A total of 41 original studies investigating gut microbiota profiles in pediatric patients with IBD were included in this review. Several studies have reported a decrease in α-diversity and an overall difference in β-diversity. Although no specific gut microbiota alterations were consistently reported, a gain in Enterococcus and a significant decrease in Anaerostipes, Blautia, Coprococcus, Faecalibacterium, Roseburia, Ruminococcus, and Lachnospira were found in the majority of the included articles. Moreover, there is insufficient data to show specific microbiota bacteria associated with disease activity, location, and behavior in pediatric IBD.
Conclusions: This systematic review identified evidence for differences in the abundance of some bacteria in pediatric patients with IBD when compared to patients without IBD; however, no clear overall conclusion could be drawn from the included studies due to inconsistent results and heterogeneous methodologies. Further studies with large samples that follow more rigorous and standardized methodologies are needed.
Inflammatory bowel disease (IBD) includes a group of complicated, long-lasting, and relapsing-remitting inflammatory disorders, of which ulcerative colitis (UC) and Crohn's disease (CD) are the two most prevalent subtypes (1, 2). The incidence of IBD has increased steadily in the Western world, but the rising incidence identified in newly industrialized countries is expected to continue to climb (3). IBD often occurs in young adulthood, but pediatric patients are also increasingly affected with 4% of patients with IBD younger than 5 years of age and 18% younger than 10 years of age (4). The epidemiologic characteristics, clinical presentation, and natural history differ depending on the age-of-diagnosis of IBD, while the potential mechanisms for disease heterogeneity remain poorly understood (5). Approximately 25% of patients with IBD demonstrate clinical manifestations during adolescence and young adulthood, and may experience lifelong disease courses with potential negative effects on growth, development, psychosocial function, and overall well-being (6). In addition, children with IBD exhibit a more severe course, especially those with a very early onset. Although the detailed pathogenesis of IBD remains unexplained, genetic predisposition, dietary patterns, inappropriate immune responses, and environmental factors have been reported to be closely associated with IBD (7–10). Though genetic factors seem to play a great role in patients with pediatric-onset IBD, environmental and microbial factors show a more prominent role in the occurrence and development of this disease in pediatric patients (11–13).
The human gut microbiota colonization is dynamic during early life and development owing to numerous factors, which may influence multiple physiological and pathophysiological processes throughout a patient's life (14, 15). The disruption of the ecological balance of gut microbiota composition and bacterial function triggers an aberrant immune response, leading to chronic intestinal inflammation. With advances in multi-omics methods, the study of gut microbial composition and function alterations in various diseases has emerged as a new potential area of clinical significance (16, 17). Gut microbiota imbalances identified in pediatric and adult patients from clinical and experimental studies over the past decade have shed light on the pathogenesis of IBD; however, the results differ and are sometimes contradictory owing to small sample sizes or heterogeneous methods (18–20). A growing number of studies have reported that the early human gut microbiota composition/development may affect adult health conditions, indicating that the gut microbiota profile in pediatric-onset IBD patients is associated with long-term consequences (14). However, comprehensive reviews of gut microbiota alterations in pediatric patients with IBD are rare, which may limit the development of new therapeutic options and the improvement of existing therapies for pediatric IBD (21, 22).
In order to facilitate the use of gut microbiota-targeted diagnoses and treatment methods in pediatric-onset IBD, a better understanding of what microbiota are associated with this disorder and how they affect the occurrence and development of IBD is urgently needed. However, most studies paid attention to the alterations of intestinal bacteria rather than viruses, protozoa and helminths. Thus, the aim of this systematic review was to investigate and describe the current findings relating to altered gut bacteria composition in pediatric patients with IBD, and to summarize specific bacteria taxa profiles associated with this disorder that distinguish pediatric IBD from adult IBD.
The protocol for this systematic review is registered on the International Prospective Register of Systematic Reviews (PROSPERO) and the Preferred Reporting Items for Systematic Review and Meta-Analyses (PRISMA) checklist was used as a guideline. A comprehensive search was performed on public databases, including PubMed, Web of Science, Embase, Scopus, and the Cochrane Library (last search: 31 July 2020), with no date or language restrictions. The MeSH terms and free-text word combinations used in the search included: “Crohn's disease,” “CD,” “Ulcerative colitis,” “UC,” “Inflammatory bowel disease,” “IBD,” “microbiota,” “microbiome,” “microflora,” “bacterial flora,” “bacteria,” “children,” and “pediatric.”
Articles were selected based on the following criteria: original studies performed in pediatric patients with IBD, CD, or UC diagnosis; assessment of the microbial communities of pediatric patients from different samples; data for bacterial profiles at different taxonomic levels provided; and the inclusion of a control group to which the gut microbiota was compared. Articles obtained from the initial literature searches were combined, and the duplicates were automatically removed. Titles and abstracts were screened by two independent investigators in the first round of the screening process. Next, potential studies were arranged for whole-paper reading, and their accompanying references were screened to identify additional eligible articles. Any discrepancies between the two investigators were resolved through consensus-based discussion, and a third reviewer was involved if necessary.
Data extracted from the included studies included demographic and clinical characteristics (such as year of publication, country, gender, IBD subtype, number of patients, disease activity, and treatment status), bacterial richness and diversity, taxonomic bacterial composition, and methodology of microbiota collection and evaluation (such as type of specimen, DNA extraction method, microbiota assessment techniques, reference database, and multiple comparison correction).
The quality of the included case-control studies was evaluated using the Newcastle-Ottawa Scale (NOS) (23). NOS contains three domains: selection (adequacy of case definition, representativeness of the cases, selection of controls, and definition of controls); comparability (comparability of baseline characteristics); and exposure (ascertainment of exposure, consistent method of ascertainment for cases and controls, and attrition rate). A final score of 0–3 indicated low quality, 4–6 indicated medium quality, and 7–9 indicated high quality.
The initial database search yielded 809 citations. The removal of duplicate articles resulted in 516 unique records. Subsequently, 63 studies were assigned to a whole-paper review after a secondary screening based on titles and abstracts. Finally, 41 original articles investigating the gut microbiota profile in pediatric patients with IBD were included for further review according to the inclusion criteria (Figure 1). As shown in Supplementary Table 1, all included studies received a moderate NOS score between 6 and 8. Studies that received a score of 8 strictly controlled multiple variables (in contrast to only controlling age). Ten studies received a score of 6 (moderate) due to control groups consisting of pediatric patients with functional gastrointestinal disorders, which is not representative of the community, or control groups being older or younger than IBD groups.
Four studies reported data in patients with UC, 19 studies evaluated patients with CD, 11 studies included both CD and UC, and 7 studies assessed patients with IBD and did not differentiate subtypes (Table 1). Nineteen studies recruited patients with active disease, 11 with the combination of active and remissive disease, 10 with unreported disease activity, and only one study investigated patients in remission. Geographically, the included studies were performed in Asia (China, Japan, and Saudi Arabia), Europe (UK, Sweden, Poland, Australia, Italy, Czech Republic, Croatia, Norway, Denmark, Germany, and Finland), and North America (USA and Canada). This review included 2,188 pediatric patients with IBD (59.0% males) and 1,233 controls (51.8% males) without IBD. Non-related participants were chosen as controls in most of the included studies, while three studies compared patients to their siblings, and two compared cases to a control group consisting of relatives and non-relatives of patients. The majority of studies investigated patients that received medication, whereas 14 studies explored the treatment-naïve microbiome in new-onset patients.
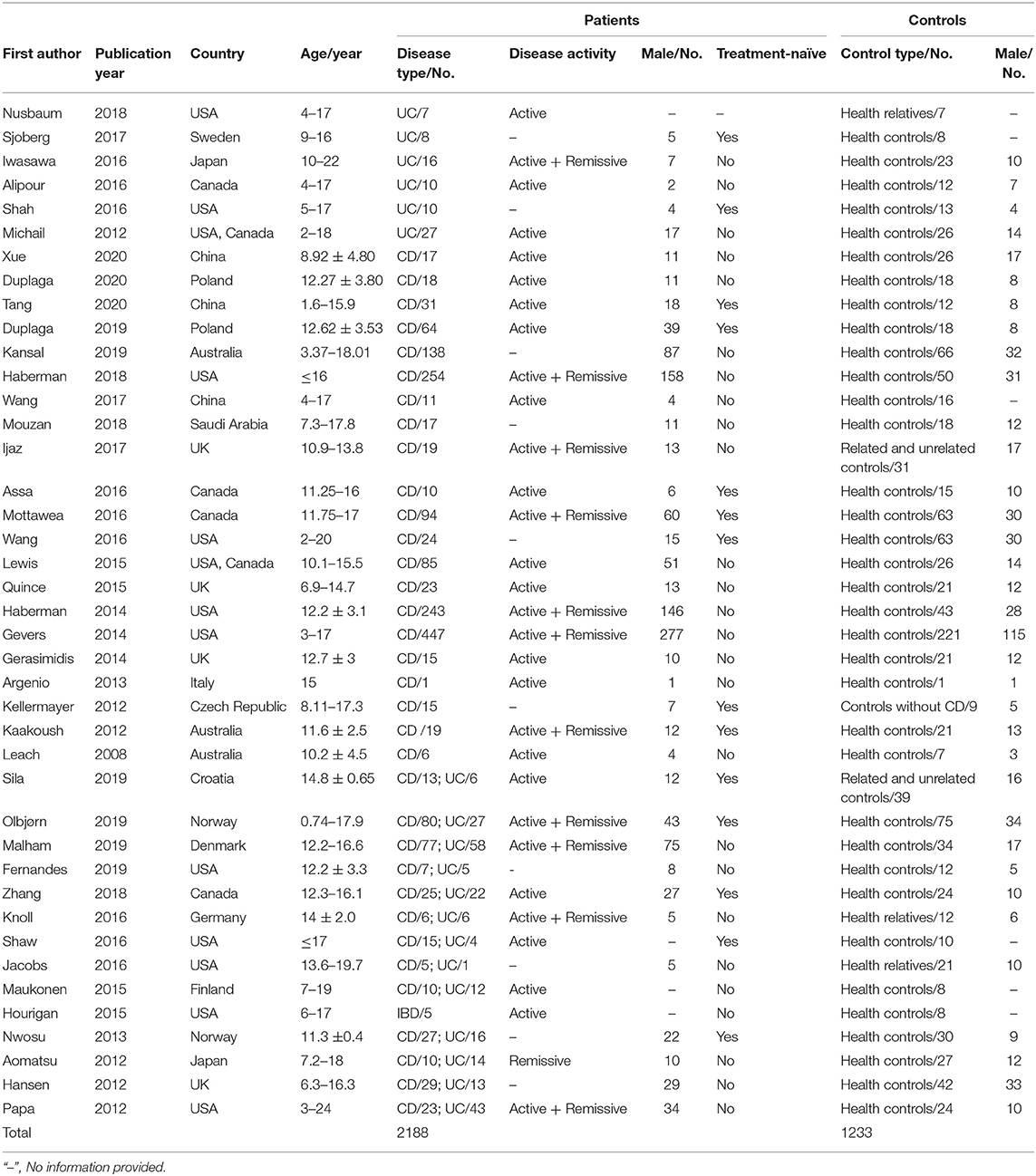
Table 1. Characteristics of included studies investigating gut microbiota profiles in pediatric patients with IBD and controls.
The sample handling and data analysis from individual studies are described in Table 2. Overall, 28 studies observed the gut microbiome from stool, 9 studies evaluated intestinal tissue from different segments, 2 studies used both stool and mucosal tissue, 1 study analyzed duodenal fluid, and 1 study investigated the mucosal–luminal interface aspirate. Most studies stored samples at either −70 or −80°C, after collection while seven studies did not provide information on sample storage. More than half of the studies implemented commercial spin column-based extraction kits according to the manufacturer's protocol for DNA extraction. Thirty-one studies assessed the gut microbiota by 16S rRNA gene sequencing targeting different variable regions and two used metagenome sequencing, while the remaining studies employed terminal restriction fragment length polymorphism, denaturing gradient gel electrophoresis, temporal temperature gradient gel electrophoresis, and GA-Map™ technology. Taxonomy was assessed using a variety of reference databases, most commonly Greengenes, RDP, or SILVA. Furthermore, approximately half of the studies (58.5%) performed multiple comparison tests when comparing gut microbiota alterations between IBD patients and controls.
Thirty studies investigated gut microbiota profiles from pediatric patients with CD, and a majority reported microbial data from fecal samples. The remaining studies received microbial data from biopsy specimens, mucosal–luminal interface aspirate, and duodenal fluid. As shown in Table 3, the gut microbial communities of pediatric patients with CD were compared to controls, assessing α- and β-diversity as well as individual bacterial alterations at different taxonomic levels. The majority of studies reported that the richness and diversity (α-diversity) of gut microbiota in pediatric patients with CD were decreased and their gut microbiota β-diversity differed from controls. Intriguingly, these altered trends were observed to be more consistent in fecal microbiota than in mucosal microbiota. However, several studies found no difference in the α- or β-diversity of the gut microbiota between subjects with and without IBD. Consistent microbiota alterations at various taxonomic levels from fecal or mucosal samples in pediatric patients with CD had been highlighted (with a more comprehensive summary in Supplementary Table 2), though highly heterogeneous results existed in the included studies.
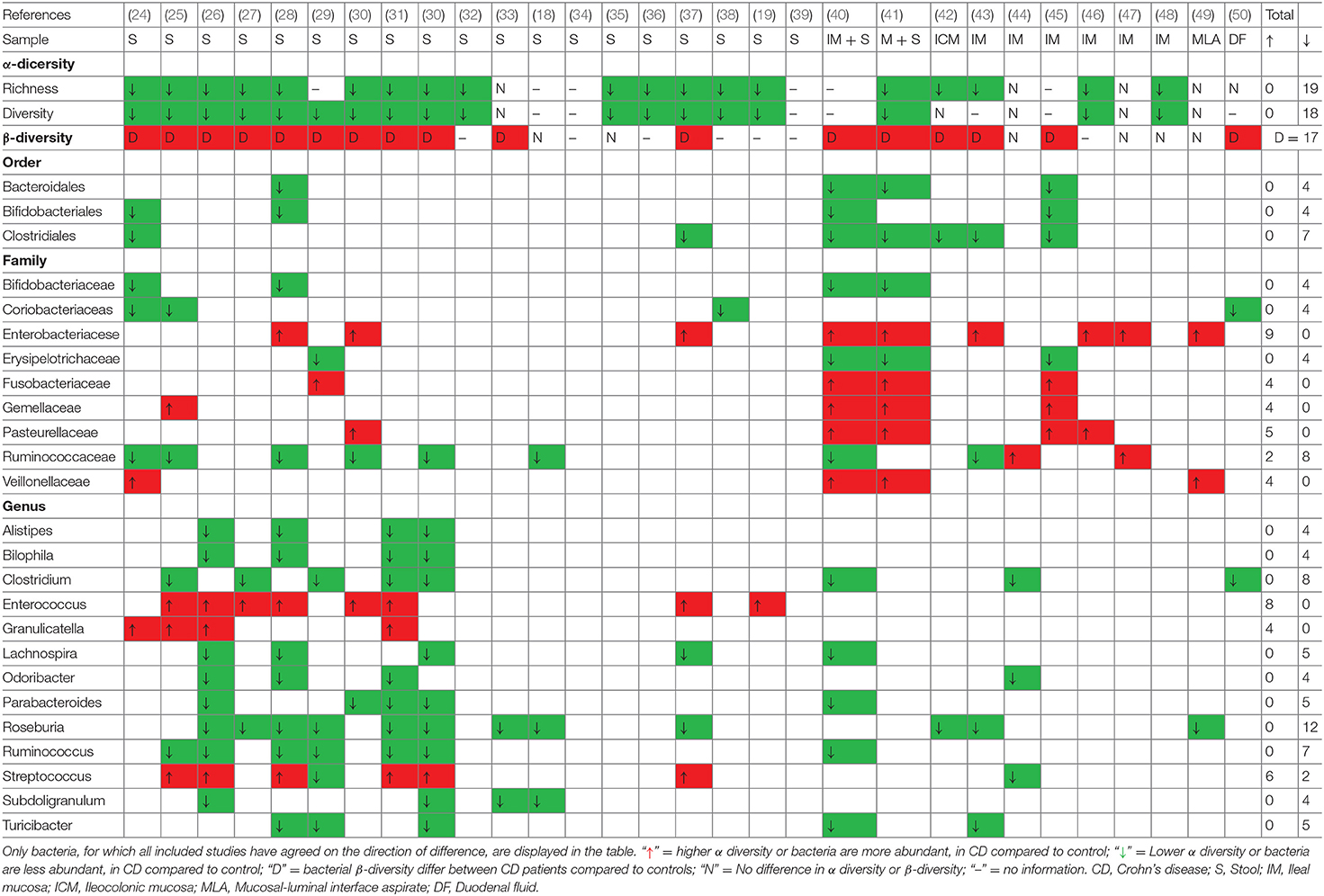
Table 3. Consistent gut microbiota alterations in pediatric patients with CD compared to controls at different taxonomies.
At the phylum level, a decreased abundance of Actinobacteria and Bacteroidetes, and an increased abundance of Proteobacteria were reported in pediatric patients with CD, which is similar to the results of studies that assessed adult patients. However, these consistent results were reported in a very small number of studies. At the class level, no clear overall conclusion about gut microbiota alterations could be drawn from the included studies, but decreased amounts of Clostridia and increased amounts of Gammaproteobacteria were noted. Additionally, several studies have demonstrated a significant decrease in the orders Bacteroidales, Clostridiales, Bifidobacteriales, and Erysipelotrichales as well as an increase in Enterobacteriales and Fusobacteriales. In terms of lower taxonomic levels, previous studies have demonstrated inconclusive results of gut microbiota alterations in pediatric patients with CD. Even so, a decreased relative abundance of several genera belonging to the families Lachnospiraceae (Anaerostipes, Blautia, Coprococcus, Lachnospira, and Roseburia), Ruminococcaceae (Anaerotruncus, Faecalibacterium, and Ruminococcus), Clostridium, Holdemania, Odoribacter, Parabacteroides, and Turicibacter were reported in most included studies. In addition, the abundance of the genera Actinomyces, Corynebacterium, Enterococcus, Escherichia, Fusobacterium, Granulicatella, Sutterella, and Veillonella were assessed in several studies. However, some bacteria from duodenal fluid showed an opposite change trend when compared to feces-associated and mucosa-associated microbiota, with a decrease in Proteobacteria and Enterobacteriaceae, and an increase in the family Erysipelotrichaceae.
Fifteen studies evaluated the gut microbiota profile in pediatric patients with UC, with nine studies assessing feces-associated microbiota, five studies using mucosa-associated microbiota, and only one study analyzing duodenal fluid-associated microbiota. Nine of 15 UC studies showed a significant decrease in the α-diversity indexes and 5 studies revealed β-diversity differences between the microbiota of patients with UC, while the remaining study found no differences. A comprehensive list of gut microbiota alterations in pediatric patients with UC is shown in Table 4 (with a more comprehensive summary in Supplementary Table 3). Similarly, patients with UC tended to have a decreased abundance of bacteria belonging to the phylum Firmicutes, though this was only reported in two studies. In addition, the differences in Proteobacteria and Verrucomicrobiae were not consistent among the studies. Additionally, five studies reported decreased bacteria in the class Clostridia and three studies reported decreased bacteria in the order Clostridiales in pediatric patients with UC compared to controls. Bacteria in the family Lachnospiraceae were decreased in six studies, and bacteria in the family Enterobacteriacese was decreased in three studies. No clear overall conclusions could be drawn from the included studies regarding alterations at the genus level. Several studies have reported an increased abundance of Haemophilus and Veillonella as well as a reduced abundance of Alistipes, Anaerostipes, Blautia, Oscillospira, Roseburia, and Ruminococcus. The genera Bacteroides, Bifidobacterium, Clostridium, Faecalibacterium, and Parabacteroides showed no consistent changes in the included studies, which is distinct from the findings in pediatric patients with CD.
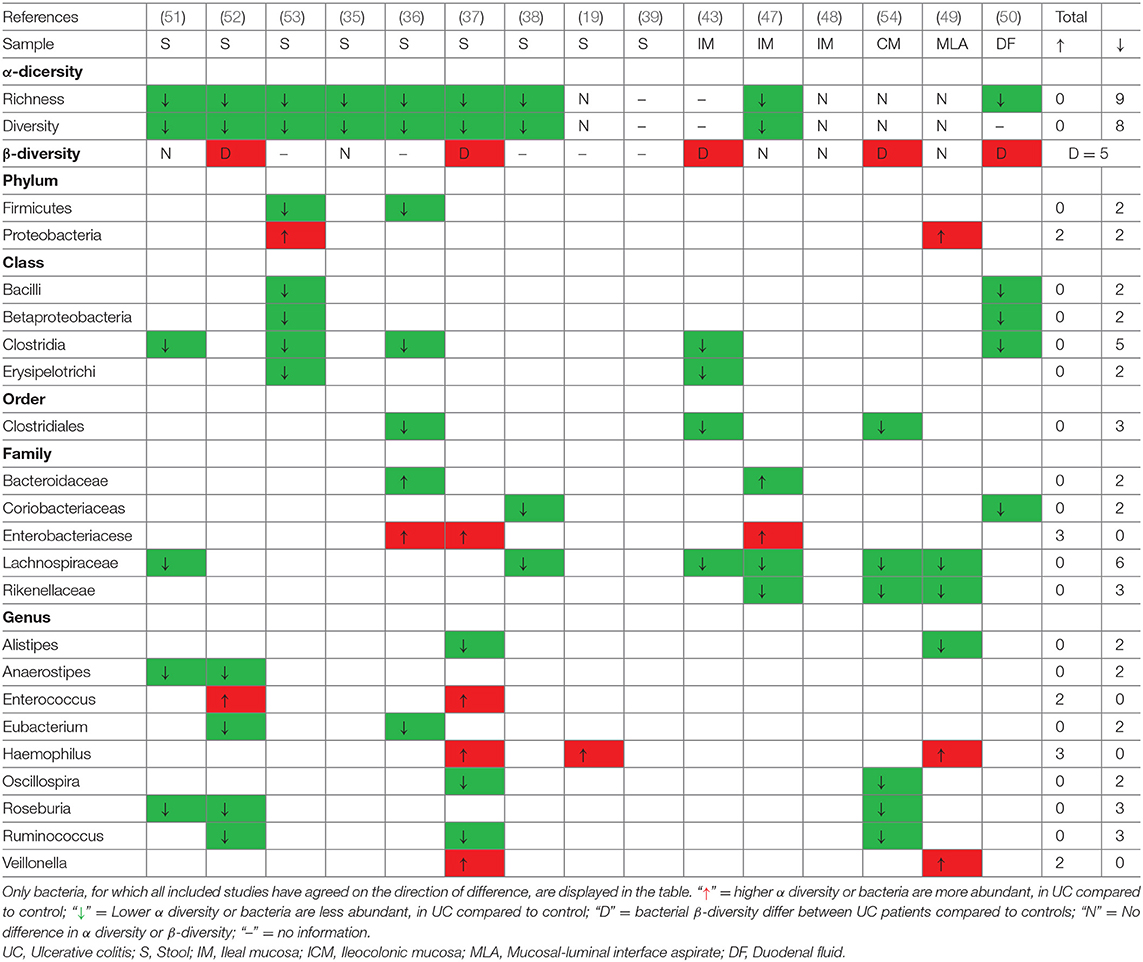
Table 4. Consistent gut microbiota alterations in pediatric patients with UC compared to controls at different taxonomies.
In an attempt to define the specific gut microbiota profiles in pediatric patients with IBD, several studies have compared IBD cases with controls without IBD (Table 5). However, the evaluation of these studies is limited due to the different compositions of mucosal and luminal microbiota and small sample sizes. Eight studies aimed to identify specific gut microbiota alterations in pediatric patients with IBD using case-control studies, while no consistent and definitive conclusions could be drawn from the included studies (Table 5). Seven studies assessed intestinal microbiota from feces, one study evaluated mucosal–luminal interface aspirates, and one study analyzed duodenal fluid. All studies investigating feces-associated microbiota showed a significant decrease in the α-diversity in patients with IBD, however, only three studies reported that the microbiota differed between patients with IBD and controls. The most consistent finding among the studies is that the potentially harmful bacteria Enterobacter and Escherichia within the family Enterobacteriacese are increased in patients with IBD. Furthermore, increased amounts of Actinobacillus, Prevotella, Streptococcus, and Veillonella were found in pediatric patients with IBD, while the amounts of Eubacterium, Lactobacillus, and Parabacteroides were reported to be decreased. Conflicting results for changes in the genera Bacillus, Bacteroides, Citrobacter, and Odoribacter were reported. A more comprehensive list of gut microbiota alterations in pediatric patients with IBD at the phylum, class, order, family, and genus levels is presented in Supplementary Table 4.
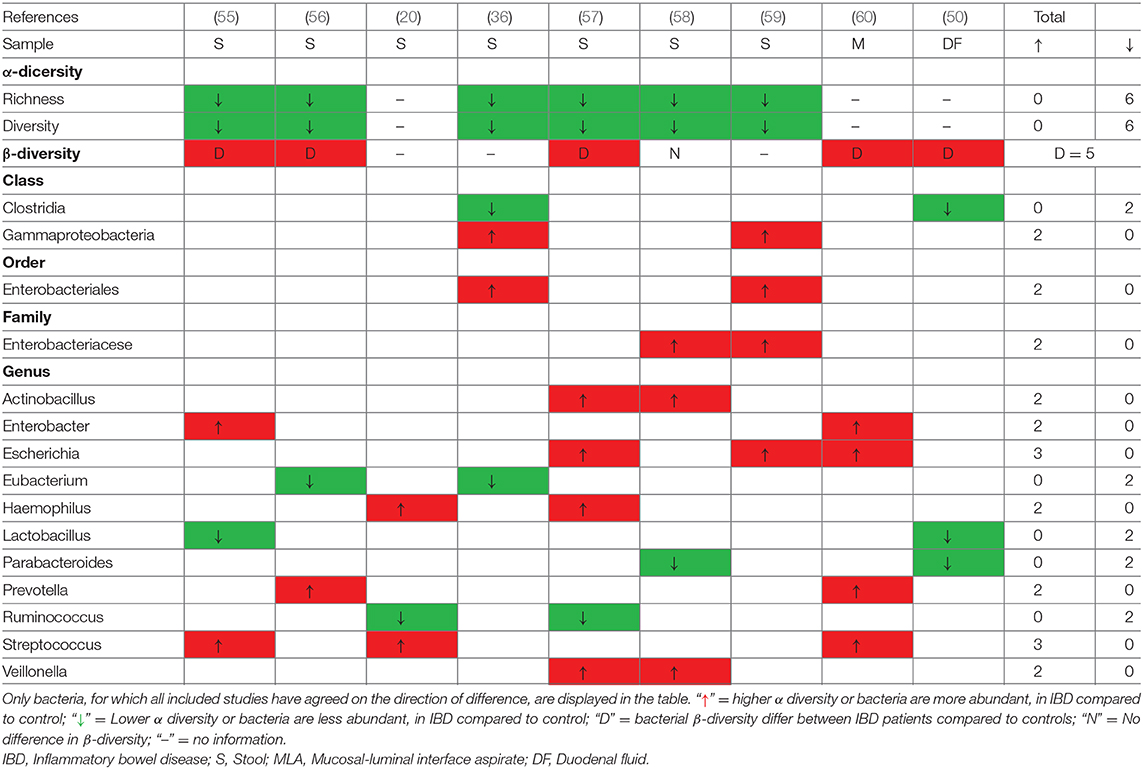
Table 5. Consistent gut microbiota alterations in pediatric patients with IBD compared to controls at different taxonomies.
The importance of the interaction between intestinal microbiota and specific medication used in IBD has been reported. The unique gut microbiota characteristics collected prior to treatment in new-onset cases may result in an early diagnosis and administration of specific medications in pediatric patients with IBD. Fourteen studies assessed the treatment-naïve microbiome in new-onset IBD, however, the reported changes were inconsistent. In patients with treatment-naïve CD, decreased amounts of Clostridium, Coprococcus, Faecalibacterium, Roseburia, Ruminococcus, and Subdoligranulum, and an increased amount of Enterococcus were reported. However, such alterations were also found in patients with CD who had been treated. Bacteria in the genus Dialister increased in pediatric patients with treatment-naïve CD and decreased in patients who received treatment prior to sample collection. There is insufficient data to determine the differences between the microbiota of patients with treatment-naïve UC and patients undergoing treatment for UC, due to the small number of studies and various locations of samples. A decrease in the abundance of bacteria in the family Lachnospiraceae and the genera Clostridium, Roseburia, and Ruminococcus were found consistently in pediatric patients with UC regardless of therapeutic interventions. Haemophilus bacteria increased in both non-treated and treated pediatric patients with UC. However, no signature of gut microbiota could be determined for pediatric patients with treatment-naïve UC.
Although the gut microbiome changes throughout a patient's life, no systematic changes were found in patients with CD with different ages of diagnosis, suggesting that CD-associated dysbiosis is already established in younger CD patients. Five studies compared the gut microbiota of pediatric patients with IBD with those of healthy relatives and found significantly decreased microbial diversity with a distinct bacteria profile in pediatric patients with IBD. However, no consistent conclusions could be drawn. The group of healthy relatives had a significantly higher microbiota diversity richness and evenness than pediatric patients with IBD in some studies. Nevertheless, three studies reported that the relatives of pediatric patients with IBD had a distinct bacterial composition that differed from non-related controls, rather than a characteristically altered gut microbiota. Jacobs et al. reported that healthy first-degree relatives of pediatric patients with IBD may have a fecal microbial and metabolomic profile with a pre-disease susceptibility state or subclinical inflammation, which may be found in patients with quiescent IBD, suggesting the potential risk of this disease. In addition, several studies reported that healthy relatives might be suitable donors for fecal microbiota transplantation as they share genetic and environmental factors with the patients.
The majority of studies included in this report used fecal samples to assess the gut microbial diversity and composition in pediatric patients with IBD, which is a taxonomically distinct mucosal microbiome. Due to the high availability of feces-associated microbiota, consistent findings were found between the studies, though the mucosa-associated microbiota may be more likely to play a role in IBD. For pediatric patients with CD, the alterations in feces-associated microbiota differed from those in mucosa-associated microbiota (such as decreased amounts of Alistipes, Bacteroides, Bilophila, Dialister, Faecalibacterium, and Subdoligranulum, and an increased amount of Enterococcus). In addition, several taxa showed opposite trends between fecal samples and mucosal samples. For example, a low amount of bacteria in the family Porphyromonadaceae in mucosa-associated microbiota was reported and a high amount was reported in feces-associated microbiota. Moreover, an increased abundance of Dialister was found in the mucosal microbiome, while a decreased amount was reported in fecal samples. For pediatric patients with UC, similar observations of gut microbiota alterations were found in fecal and mucosal samples. An increase in the amount of bacteria in the family Enterobacteriaceae and a decrease in Roseburia and Ruminococcus belonging to the family Lachnospiraceae were reported in both the mucosal and fecal samples. However, there is insufficient data to show distinctive gut microbiota profiles in pediatric patients with UC due to the small number of studies.
Knowledge regarding the geographic and environmental factors (dietary lifestyle) that are relevant for shaping the gut microbiota has increased considerably in recent years. The previously reported dysbiosis in adult-onset IBD patients may be already established during the pediatric period. Most of the studies included in this report were conducted in Western countries, which may limit the possibility of determining differences between studies from the East and West. Twenty-five studies assessed the microbiota of Western patients with CD, and 5 studies included Eastern patients (3 studies from China, 1 from Japan, and 1 from Saudi Arabia). The gut microbiota alterations in pediatric patients with CD that are consistent between Eastern and Western patients include decreased amounts of Alistipes, Anaerotruncus, Bilophila, Clostridium, Lachnospira, Parabacteroides, Roseburia, Ruminococcus, and Turicibacter, and increased amounts of Enterobacter and Enterococcus. Thirteen studies assessed the microbiota of Western patients with UC, and only two studies included Eastern patients (both from Japan). The data from these studies were insufficient to evaluate consistent findings between Eastern and Western patients. In this review, gut microbiota alterations from Eastern patients were similar to that from Western pediatric patients, suggesting that conclusive results could not obtained when different geographical locations or dietary lifestyles are considered.
Not all included studies reported the disease activity of patients. Nineteen studies evaluated the gut microbiota in patients with active disease and 11 studies assessed the gut microbiota for patients with both active and remissive disease. A strong association between microbiota diversity and disease activity has been reported in several studies independent of the type of disease (CD or UC). Although no conclusive determinations can be made, a diminished richness of the taxa and alterations of several specific bacteria have been reported to be associated with disease activity. Kaakoush et al. reported that the detection frequencies of Bacteroidetes and Firmicutes correlated (positively and negatively, respectively) with the calculated pediatric CD activity index (PCDAI) scores of patients. However, findings from Haberman et al. demonstrated that PCDAI scores were positively associated with Gammaproteobacteria and Enterobacteriaceae and negatively associated with Clostridiales and Bacteroides. Furthermore, Mottawea et al. found that major short-chain fatty acids (SCFA) producers such as Lachnospiraceae, Blautia, Roseburia, Ruminococcus, Clostridium, and Faecalibacterium were negatively correlated with disease severity, while some H2S producers such as Atopobium, Fusobacterium, Veillonella, Prevotella, Streptoccocus, and Leptotrichia were positively correlated with disease severity. Additionally, Gevers et al. assessed that the microbial dysbiosis index showed a strong positive correlation with PCDAI, and the levels of Fusobacterium and Haemophilus were positively correlated with PCDAI, which was further confirmed in a study by Shaw et al. Maukonen et al. found that Bacteroides was the only bacterial group in which lower disease activity was associated with higher bacterial numbers in both UC and CD. In addition, Xue et al. found a significant correlation between PCDAI and Simplified Endoscopic Score for Crohn's Disease (SES-CD) scores and the dysbiosis indices (Lactobacillales, Micrococcales, Veillonellaceae, Clostridiales, and Selenomonadales). Overall, conclusive results of specific gut microbiota alterations associated with disease activity could not be determined due to inconsistent data and methods among the included studies.
Generalized dysbiosis independent of gut location has been previously demonstrated in a number of studies. Despite the limitation of a small sample size, several studies have attempted to investigate the relationship between disease location and gut microbiota alterations. However, the results of these studies are inconsistent and sometimes contradictory. Olbjørn et al. found that patients with CD with upper gastrointestinal involvement had an increased abundance of Veillonella compared to patients without upper gastrointestinal lesions. Gevers et al. reported that ileal and rectal biopsies have similar discriminatory power for classifying diseases, regardless of the disease location. Haberman et al. reported a similar microbial shift in the ilea between patients with ileal CD (iCD) and colonic CD (cCD), irrespective of histologic involvement in cCD, showing a decrease in the amount of Lachnospiraceae, Bifidobacteriaceae, Clostridiales, and Erysipelotrichaceae and an increase in the amount of Veillonellaceae, Pasteurellaceae, Neisseriaceae, Gemellaceae, Fusobacteriaceae, and Enterobacteriaceae. In addition, no significant differences in the bacterial community between inflamed and uninflamed mucosa in pediatric patients with CD has been reported.
Data regarding disease behavior and gut microbiota alterations was limited, as few studies have focused on this association. One pediatric study evaluated the relationship between disease behavior and gut microbiota alterations. Olbjørn et al. found that higher abundance of Proteobacteria in pediatric patients with CD means more likely to have complicated disease behavior (structuring or penetrating disease) as compared to patients with lower levels of these bacteria.
Identifying characteristic gut microbiota changes may provide insight into the role of the gut microbiome in the etiology and treatment methods of pediatric IBD. Several reviews attempting to evaluate the gut microbiota profiles in adult patients with IBD found that patients with IBD tend to have a reduced abundance of bacteria belonging to the Firmicutes phylum and an increased abundance of bacteria from the Proteobacteria phylum (61–63). However, there is no review that identifies specific bacteria that can distinguish adult and pediatric patients with IBD. Several studies have reported that the microbiota composition changes gradually with time, and that even though the gut microbiota begins to resemble the adult flora by 3 years of age, it is crucial to study the gut microbiota of pediatric patients with IBD (64, 65). Additionally, IBD is dynamic and little is known about the individual nature of microbiome dynamics in IBD from childhood through adulthood (66). In this systematic review, we firstly sought to provide a unique framework for understanding microbial dysbiosis and to summarize specific gut microbiota alterations in pediatric patients with IBD compared to controls. Collectively, no specific bacteria consistently differed between pediatric patients with IBD and controls in each of the included studies due to the heterogeneity of the type of samples, sample handling, and microbiome assessment methods of the included studies. Even though pediatric CD and UC have unique gut microbiota alterations, no bacteria consistently differed in these patients among the studies included in this review. Furthermore, there is insufficient data to evaluate the gut microbiota associated with disease activity, lesion location, or disease behavior in pediatric patients with IBD.
In the majority of the studies included in this review, a decreased microbial biodiversity with alterations in the composition of the gut microbiota community was observed in fecal and mucosal samples from pediatric patients with IBD. In addition, a number of differences were found between patients with IBD and controls when comparing the relative abundance of individual bacterial taxonomy, although no distinct bacteria were consistently identified in all of the included studies. Consistent alterations in gut microbiota were identified among studies that used fecal samples. Similar to previous studies investigating adult patients with IBD, pediatric patients were reported to have an increased amount of Enterococcus and a significant decrease in Anaerostipes, Blautia, Coprococcus, Faecalibacterium, Roseburia, Ruminococcus, and Lachnospira. These findings suggest that the previously reported dysbiosis in patients with adult-onset IBD may be established during the pediatric period. In recent years, there is accumulating evidence suggesting that alterations in the metabolites of the gut microbiota and specific bacterial metabolic pathways may help elucidate the precise cause-effect mechanistic relationships between gut microbiota and IBD, which may result in new discoveries regarding the pathogenesis and therapeutic methods for this disease (67–69). In the majority of studies, a decreased abundance of Anaerostipes, Lachnospira, Roseburia, Faecalibacterium, Coprococcus, Ruminococcus, and Roseburia bacteria were observed in pediatric patients with CD. These bacteria produce SCFA, which may be the primary source of energy for colonocytes, create an environment for proper intestinal colonization, and show immunomodulatory and anti-inflammatory properties (70, 71). SCFA play essential roles in maintaining the intestinal barrier function via promoting Treg cell development and enhancing mucus production from goblet cells as well as in inflammation-associated immunosuppression via regulating chemokine and cytokine production (72–75). Additionally, vanished SCFA-producing bacteria may favor a shift toward an inflammation-promoting microbiome, thereby enhancing host inflammation and worsening the condition of CD (28). A depletion of several Clostridium species that adhere to epithelial cells can mediate diverse effects on mucosal immunity via altering the differentiation of T-helper 17 cells and regulatory T cells (76, 77). Other bacteria that were found to be decreased in pediatric patients with CD include Akkermansia, which may play a role in the intestinal barrier function by providing crucial anti-inflammatory responses (78), Alistipes may act as a colitis-attenuating bacteria with unclear mechanisms (79), while Parabacteroides may have anti-inflammatory and epithelium-reinforcing capacities (80), Turicibacter may cause alterations in the bacterial diversity of the host and subsequent changes in steroid and lipid metabolism by interacting with serotonin (81). Still, more research is required to determine the mechanisms of bacteria involved in IBD.
Faecalibacterium prausnitzii, a potentially protective bacterium associated with disease activity, was found to be decreased in pediatric patients with IBD only in studies analyzing fecal sampling. Gram-negative pathobionts including Enterobacteriaceae species (Enterobacter and Escherichaia), Enterococcus and Fusobacterium were increased in pediatric patients with IBD, and have been reported to have the ability to adhere to and invade intestinal epithelial cells, leading to an inflammatory immune response (82–85). In addition, alterations in the abundance of H2S-producing bacteria (Atopobium, Fusobacterium, Veillonella, Prevotella, and Streptoccocus) suggest a possible role for these bacteria in the pathogenesis of CD (49). H2S, an important mediator of many physiological and pathological processes, can damage the gastrointestinal epithelium and exerts influence on the gut microbiota and on mucus and biofilm interactions in the context of intestinal inflammation (86, 87). Furthermore, depletion of butyrate-producing microbes from the colonic microbiota may dampen host H2S defense systems, which exacerbates inflammation and markedly alters the intestinal microbiota biofilm (88). In addition, alterations in gut microbiota with other microbial metabolites (such as bile acid and tryptophan metabolites) may have important and diverse effects on immune maturation, immune homoeostasis, host energy metabolism, and maintenance of mucosal integrity (67, 69, 89–93). Although the gut microbiota profile in pediatric patients with IBD showed similar patterns to adult patients with IBD, it is believed that gut microbiota characterisations of pediatric patients without extraneous influences of adult behaviors are likely to be more informative and useful for the treatment of pediatric IBD.
Changes in gut microbiota composition and function are thought to be associated with disease activity, disease behavior, and lesion location in patients with IBD. A systematic review showed that patients with active IBD had a lower abundance of Clostridium coccoides, Clostridium leptum, Faecalibacterium prausnitzii, and Bifidobacterium (94). Although no specific bacteria were consistently associated with disease activity of pediatric IBD in this review, a negative association between the activity of IBD and the abundance of Clostridiales was found in several studies. There are insufficient data to evaluate differences in gut microbiota associated with the location of lesions as most samples were collected from the ileum. Haberman et al. reported a persistent decrease in Lachnospiraceae, Bifidobacteriaceae, Clostridiales, and Erysipelotrichaceae in all forms of CD, with an increase in Veillonellaceae, Pasteurellaceae, Neisseriaceae, Gemellaceae, Fusobacteriaceae, and Enterobacteriaceae bacteria, independent of cecal involvement (43), which differs from previously reported results that Enterococcus faecalis is more abundant when CD is localized in ileum compared with the ileocolon (95). A prospective inception cohort study found that Ruminococcus is associated with stricturing complications and Veillonella is associated with penetrating complications (96). However, a similar relationship in pediatric patients was not reported in the studies included in this review.
Several studies have explored the association between oral microbiota and CD as oral manifestations are very common. Kelsen et al. reported that Capnocytophaga, Rothia, and TM7 were more abundant in subgingival plaques samples of pediatric patients with CD than in healthy controls (97). Iwasawa found that the abundance of Streptococcus from salivary samples was significantly lower in patients with UC than in healthy controls (98). Nevertheless, data on specific oral-associated microbiota changes involved in pediatric IBD is limited.
There are some limitations to this study that warrant discussion. It is widely known that the composition and function of the human gut microbiome is shaped by multiple factors, including host genetics, geographic location, demographics, dietary patterns, and life behaviors, which may explain discrepancies between different studies (99–102). The methodologies of these studies differ according to the type of IBD (both CD and UC, only CD or only UC), age of control group (healthy minor or related siblings or adult), disease activity (active or inactive), sample type (feces, mucosal biopsy, mucosal–luminal interface aspirate, or duodenal fluid), and methods used for microbiota analysis (DNA extraction, sequencing methodology, or data analysis), which may also result in highly heterogeneous gut microbiota compositions between studies. Although it has long been proposed that an altered gut microbiome plays an important role in IBD, a direct causal relationship between dysbiosis and the disease has not been definitively established in humans. A comprehensive understanding of gut microbiota profiles in pediatric patients with new-onset IBD prior to therapeutic intervention is required to understand gut microbiota alterations associated with the occurrence and development of IBD and whether potential relationships exist between specific bacteria and disease subtype, activity, and location.
Altered gut microbiota composition in pediatric patients with IBD are associated with this disorder. Moreover, the gut microbiota profiles of pediatric patients with IBD are similar to those of adult patients. In addition, specific gut microbiota is associated with disease characteristics (subtype, activity, and location), however, the studies included in this review varied widely in geography and methodology, leading to considerable heterogeneity in our findings. Future studies should consider bacteria species or strain levels with gut microbiota functions to elucidate the causative or correlative relationships between microbial changes and pediatric IBD. Furthermore, prospective, well-designed trials of adults and newly-diagnosed pediatric patients are required to reveal the typical gut microbiota changes associated with the onset of IBD. Determining specific gut microbiota associated with pediatric IBD may allow for the development of microbiome-targeted interventions, which may influence the natural course of disease or prevent the disease from developing. Future research using rigorous and standardized methodologies will lead to more consistent results.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
MC and ZZ are the guarantor of the article. ZZ and XZ designed the study. XZ wrote the manuscript. CL and SZ collected the data. ZT and NL analyzed the data. RM, ZZ, and MC revised the manuscript. All authors approved the final version.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fped.2021.626232/full#supplementary-material
1. Torres J, Mehandru S, Colombel JF, Peyrin-Biroulet L. Crohn's disease. Lancet. (2017) 389:1741–55. doi: 10.1016/S0140-6736(16)31711-1
2. Ungaro R, Mehandru S, Allen PB, Peyrin-Biroulet L, Colombel JF. Ulcerative colitis. Lancet. (2017) 389:1756–70. doi: 10.1016/S0140-6736(16)32126-2
3. Ng SC, Shi HY, Hamidi N, Underwood FE, Tang W, Benchimol EI, et al. Worldwide incidence and prevalence of inflammatory bowel disease in the 21st century: a systematic review of population-based studies. Lancet. (2018) 390:2769–78. doi: 10.1016/S0140-6736(17)32448-0
4. Rosen MJ, Dhawan A, Saeed SA. Inflammatory bowel disease in children and adolescents. JAMA Pediatr. (2015) 169:1053–60. doi: 10.1001/jamapediatrics.2015.1982
5. Ruel J, Ruane D, Mehandru S, Gower-Rousseau C, Colombel JF. IBD across the age spectrum: is it the same disease? Nat Rev Gastroenterol Hepatol. (2014) 11:88–98. doi: 10.1038/nrgastro.2013.240
6. Kaplan GG. The global burden of IBD: from 2015 to 2025. Nat Rev Gastroenterol Hepatol. (2015) 12:720–27. doi: 10.1038/nrgastro.2015.150
7. Liu TC, Stappenbeck TS. Genetics and pathogenesis of inflammatory bowel disease. Annu Rev Pathol. (2016) 11:127–48. doi: 10.1146/annurev-pathol-012615-044152
8. de Souza HS, Fiocchi C. Immunopathogenesis of IBD: current state of the art. Nat Rev Gastroenterol Hepatol. (2016) 13:13–27. doi: 10.1038/nrgastro.2015.186
9. Ananthakrishnan AN. Epidemiology and risk factors for IBD. Nat Rev Gastroenterol Hepatol. (2015) 12:205–17. doi: 10.1038/nrgastro.2015.34
10. Peloquin JM, Goel G, Villablanca EJ, Xavier RJ. Mechanisms of pediatric inflammatory bowel disease. Annu Rev Immunol. (2016) 34:31–64. doi: 10.1146/annurev-immunol-032414-112151
11. Ananthakrishnan AN, Bernstein CN, Iliopoulos D, Macpherson A, Neurath MF, Ali RAR, et al. Environmental triggers in IBD: a review of progress and evidence. Nat Rev Gastroenterol Hepatol. (2018) 15:39–49. doi: 10.1038/nrgastro.2017.136
12. Shouval DS, Rufo PA. The role of environmental factors in the pathogenesis of inflammatory bowel diseases: a review. JAMA Pediatr. (2017) 171:999–1005. doi: 10.1001/jamapediatrics.2017.2571
13. Rothschild D, Weissbrod O, Barkan E, Kurilshikov A, Korem T, Zeevi D, et al. Environment dominates over host genetics in shaping human gut microbiota. Nature. (2018) 555:210–15. doi: 10.1038/nature25973
14. Milani C, Duranti S, Bottacini F, Casey E, Turroni F, Mahony J, et al. The first microbial colonizers of the human gut: composition, activities, and health implications of the infant gut microbiota. Microbiol Mol Biol Rev. (2017) 81:e00036-17. doi: 10.1128/MMBR.00036-17
15. Gilbert JA, Blaser MJ, Caporaso JG, Jansson JK, Lynch SV, Knight R. Current understanding of the human microbiome. Nat Med. (2018) 24:392–400. doi: 10.1038/nm.4517
16. Lloyd-Price J, Arze C, Ananthakrishnan AN, Schirmer M, Avila-Pacheco J, Poon TW, et al. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature. (2019) 569:655–62. doi: 10.1038/s41586-019-1237-9
17. Segal JP, Mullish BH, Quraishi MN, Acharjee A, Williams HRT, Iqbal T, et al. The application of omics techniques to understand the role of the gut microbiota in inflammatory bowel disease. Therap Adv Gastroenterol. (2019) 12:1756284818822250. doi: 10.1177/1756284818822250
18. Kaakoush NO, Day AS, Huinao KD, Leach ST, Lemberg DA, Dowd SE, et al. Microbial dysbiosis in pediatric patients with Crohn's disease. J Clin Microbiol. (2012) 50:3258–66. doi: 10.1128/JCM.01396-12
19. Nwosu FC, Thorkildsen LT, Avershina E, Ricanek P, Perminow G, Brackmann S, et al. Age-dependent fecal bacterial correlation to inflammatory bowel disease for newly diagnosed untreated children. Gastroenterol Res Pract. (2013) 2013:302–98. doi: 10.1155/2013/302398
20. Fernandes MA, Verstraete SG, Phan TG, Deng X, Stekol E, LaMere B, et al. Enteric virome and bacterial microbiota in children with ulcerative colitis and Crohn disease. J Pediatr Gastroenterol Nutr. (2019) 68:30–36. doi: 10.1097/MPG.0000000000002140
21. Serban DE. Microbiota in inflammatory bowel disease pathogenesis and therapy: is it all about diet? Nutr Clin Pract. (2015) 30:760–79. doi: 10.1177/0884533615606898
22. Brusaferro A, Cavalli E, Farinelli E, Cozzali R, Principi N, Esposito S. Gut dysbiosis and paediatric Crohn's disease. J Infect. (2019) 78:1–7. doi: 10.1016/j.jinf.2018.10.005
23. Wells GA, Shea B, O'Connell D, Peterson J, Welch V, Losos M, et al. The Newcastle-Ottawa Scale (NOS) for Assessing the Quality of Nonrandomised Studies in Meta-Analyses. Available online at: http://www.ohri.ca/programs/clinical_epidemiology/oxford.asp (accessed September 1, 2020).
24. Xue AJ, Miao SJ, Sun H, Qiu XX, Wang SN, Wang L, et al. Intestinal dysbiosis in pediatric Crohn's disease patients with IL10RA mutations. World J Gastroenterol. (2020) 26:3098–109. doi: 10.3748/wjg.v26.i22.3098
25. Kowalska-Duplaga K, Gosiewski T, Kapusta P, Sroka-Oleksiak A, Wedrychowicz A, Pieczarkowski S, et al. Differences in the intestinal microbiome of healthy children and patients with newly diagnosed Crohn's disease. Sci Rep. (2019) 9:18880. doi: 10.1038/s41598-019-55290-9
26. Tang W, Huang Y, Shi P, Wang Y, Zhang Y, Xue A, et al. Effect of exclusive enteral nutrition on the disease process, nutrition status, and gastrointestinal microbiota for Chinese children with Crohn's disease. JPEN J Parenter Enteral Nutr. (2020). doi: 10.1002/jpen.1938. [Epub ahead of print].
27. Kowalska-Duplaga K, Kapusta P, Gosiewski T, Sroka-Oleksiak A, Ludwig-Słomczyńska AH, Wołkow PP, et al. Changes in the intestinal microbiota are seen following treatment with infliximab in children with Crohn's disease. J Clin Med. (2020) 9:687. doi: 10.3390/jcm9030687
28. Wang Y, Gao X, Ghozlane A, Hu H, Li X, Xiao Y, et al. Characteristics of faecal microbiota in paediatric Crohn's disease and their dynamic changes during infliximab therapy. J Crohns Colitis. (2018) 12:337–46. doi: 10.1093/ecco-jcc/jjx153
29. El Mouzan MI, Winter HS, Assiri AA, Korolev KS, Al Sarkhy AA, Dowd SE, et al. Microbiota profile in new-onset pediatric Crohn's disease: data from a non-Western population. Gut Pathog. (2018) 10:49. doi: 10.1186/s13099-018-0276-3
30. Ijaz UZ, Quince C, Hanske L, Loman N, Calus ST, Bertz M, et al. The distinct features of microbial 'dysbiosis' of Crohn's disease do not occur to the same extent in their unaffected, genetically-linked kindred. PLoS ONE. (2017) 12:e0172605. doi: 10.1371/journal.pone.0172605
31. Lewis JD, Chen EZ, Baldassano RN, Otley AR, Griffiths AM, Lee D, et al. Inflammation, antibiotics, and diet as environmental stressors of the gut microbiome in pediatric Crohn's disease. Cell Host Microbe. (2015) 18:489–500. doi: 10.1016/j.chom.2015.09.008
32. Gerasimidis K, Bertz M, Hanske L, Junick J, Biskou O, Aguilera M, et al. Decline in presumptively protective gut bacterial species and metabolites are paradoxically associated with disease improvement in pediatric Crohn's disease during enteral nutrition. Inflamm Bowel Dis. (2014) 20:861–71. doi: 10.1097/MIB.0000000000000023
33. Kellermayer R, Mir SA, Nagy-Szakal D, Cox SB, Dowd SE, Kaplan JL, et al. Microbiota separation and C-reactive protein elevation in treatment-naïve pediatric granulomatous Crohn disease. J Pediatr Gastroenterol Nutr. (2012) 55:243–50. doi: 10.1097/MPG.0b013e3182617c16
34. Leach ST, Mitchell HM, Eng WR, Zhang L, Day AS. Sustained modulation of intestinal bacteria by exclusive enteral nutrition used to treat children with Crohn's disease. Aliment Pharmacol Ther. (2008) 28:724–33. doi: 10.1111/j.1365-2036.2008.03796.x
35. Malham M, Lilje B, Houen G, Winther K, Andersen PS, Jakobsen C. The microbiome reflects diagnosis and predicts disease severity in paediatric onset inflammatory bowel disease. Scand J Gastroenterol. (2019) 54:969–75. doi: 10.1080/00365521.2019.1644368
36. Knoll RL, Forslund K, Kultima JR, Meyer CU, Kullmer U, Sunagawa S, et al. Gut microbiota differs between children with inflammatory bowel disease and healthy siblings in taxonomic and functional composition: a metagenomic analysis. Am J Physiol Gastrointest Liver Physiol. (2017) 312:327–39. doi: 10.1152/ajpgi.00293.2016
37. Jacobs JP, Goudarzi M, Singh N, Tong M, McHardy IH, Ruegger P, et al. A disease-associated microbial and metabolomics state in relatives of pediatric inflammatory bowel disease patients. Cell Mol Gastroenterol Hepatol. (2016) 2:750–66. doi: 10.1016/j.jcmgh.2016.06.004
38. Maukonen J, Kolho KL, Paasela M, Honkanen J, Klemetti P, Vaarala O, et al. Altered fecal microbiota in paediatric inflammatory bowel disease. J Crohns Colitis. (2015) 9:1088–95. doi: 10.1093/ecco-jcc/jjv147
39. Aomatsu T, Imaeda H, Fujimoto T, Takahashi K, Yoden A, Tamai H, et al. Terminal restriction fragment length polymorphism analysis of the gut microbiota profiles of pediatric patients with inflammatory bowel disease. Digestion. (2012) 86:129–35. doi: 10.1159/000339777
40. Wang F, Kaplan JL, Gold BD, Bhasin MK, Ward NL, Kellermayer R, et al. Detecting microbial dysbiosis associated with pediatric crohn disease despite the high variability of the gut microbiota. Cell Rep. (2016) 14:945–55. doi: 10.1016/j.celrep.2015.12.088
41. Gevers D, Kugathasan S, Denson LA, Vázquez-Baeza Y, Van Treuren W, Ren B, et al. The treatment-naive microbiome in new-onset Crohn's disease. Cell Host Microbe. (2014) 15:382–92. doi: 10.1016/j.chom.2014.02.005
42. Kansal S, Catto-Smith AG, Boniface K, Thomas S, Cameron DJ, Oliver M, et al. The microbiome in paediatric Crohn's disease-a longitudinal, prospective, single-centre study. J Crohns Colitis. (2019) 13:1044–54. doi: 10.1093/ecco-jcc/jjz016
43. Haberman Y, Tickle TL, Dexheimer PJ, Kim MO, Tang D, Karns R, et al. Pediatric Crohn disease patients exhibit specific ileal transcriptome and microbiome signature. J Clin Invest. (2014) 124:3617–33. doi: 10.1172/JCI75436
44. Assa A, Butcher J, Li J, Elkadri A, Sherman PM, Muise AM, et al. Mucosa-associated ileal microbiota in new-onset pediatric Crohn's disease. Inflamm Bowel Dis. (2016) 22:1533–9. doi: 10.1097/MIB.0000000000000776
45. Haberman Y, Schirmer M, Dexheimer PJ, Karns R, Braun T, Kim MO, et al. Age-of-diagnosis dependent ileal immune intensification and reduced alpha-defensin in older versus younger pediatric Crohn disease patients despite already established dysbiosis. Mucosal Immunol. (2019) 12:491–502. doi: 10.1038/s41385-018-0114-4
46. D'Argenio V, Precone V, Casaburi G, Miele E, Martinelli M, Staiano A, et al. An altered gut microbiome profile in a child affected by Crohn's disease normalized after nutritional therapy. Am J Gastroenterol. (2013) 108:851–2. doi: 10.1038/ajg.2013.46
47. Alipour M, Zaidi D, Valcheva R, Jovel J, Martínez I, Sergi C, et al. Mucosal barrier depletion and loss of bacterial diversity are primary abnormalities in paediatric ulcerative colitis. J Crohns Colitis. (2016) 10:462–71. doi: 10.1093/ecco-jcc/jjv223
48. Hansen R, Russell RK, Reiff C, Louis P, McIntosh F, Berry SH, et al. Microbiota of de-novo pediatric IBD: increased Faecalibacterium prausnitzii and reduced bacterial diversity in Crohn's but not in ulcerative colitis. Am J Gastroenterol. (2012) 107:1913–22. doi: 10.1038/ajg.2012.335
49. Mottawea W, Chiang CK, Mühlbauer M, Starr AE, Butcher J, Abujamel T, et al. Altered intestinal microbiota-host mitochondria crosstalk in new onset Crohn's disease. Nat Commun. (2016) 7:13419. doi: 10.1038/ncomms13419
50. Sjöberg F, Barkman C, Nookaew I, Östman S, Adlerberth I, Saalman R, et al. Low-complexity microbiota in the duodenum of children with newly diagnosed ulcerative colitis. PLoS ONE. (2017) 12:e0186178. doi: 10.1371/journal.pone.0186178
51. Nusbaum DJ, Sun F, Ren J, Zhu Z, Ramsy N, Pervolarakis N, et al. Gut microbial and metabolomic profiles after fecal microbiota transplantation in pediatric ulcerative colitis patients. FEMS Microbiol Ecol. (2018) 94:fiy133. doi: 10.1093/femsec/fiy133
52. Iwasawa K, Suda W, Tsunoda T, Oikawa-Kawamoto M, Umetsu S, Inui A, et al. Characterisation of the faecal microbiota in Japanese patients with paediatric-onset primary sclerosing cholangitis. Gut. (2017) 66:1344–46. doi: 10.1136/gutjnl-2016-312533
53. Michail S, Durbin M, Turner D, Griffiths AM, Mack DR, Hyams J, et al. Alterations in the gut microbiome of children with severe ulcerative colitis. Inflamm Bowel Dis. (2012) 18:1799–808. doi: 10.1002/ibd.22860
54. Shah R, Cope JL, Nagy-Szakal D, Dowd S, Versalovic J, Hollister EB, et al. Composition and function of the pediatric colonic mucosal microbiome in untreated patients with ulcerative colitis. Gut Microbes. (2016) 7:384–96. doi: 10.1080/19490976.2016.1190073
55. Sila S, Jelić M, Trivić I, Tambić Andrašević A, Hojsak I, Kolaček S. Altered gut microbiota is present in newly diagnosed pediatric patients with inflammatory bowel disease. J Pediatr Gastroenterol Nutr. (2020) 70:497–502. doi: 10.1097/MPG.0000000000002611
56. Olbjørn C, Cvancarova Småstuen M, Thiis-Evensen E, Nakstad B, Vatn MH, Jahnsen J, et al. Fecal microbiota profiles in treatment-naïve pediatric inflammatory bowel disease - associations with disease phenotype, treatment, and outcome. Clin Exp Gastroenterol. (2019) 12:37–49. doi: 10.2147/CEG.S186235
57. Shaw KA, Bertha M, Hofmekler T, Chopra P, Vatanen T, Srivatsa A, et al. Dysbiosis, inflammation, and response to treatment: a longitudinal study of pediatric subjects with newly diagnosed inflammatory bowel disease. Genome Med. (2016) 8:75. doi: 10.1186/s13073-016-0331-y
58. Hourigan SK, Chen LA, Grigoryan Z, Laroche G, Weidner M, Sears CL, et al. Microbiome changes associated with sustained eradication of Clostridium difficile after single faecal microbiota transplantation in children with and without inflammatory bowel disease. Aliment Pharmacol Ther. (2015) 42:741–52. doi: 10.1111/apt.13326
59. Papa E, Docktor M, Smillie C, Weber S, Preheim SP, Gevers D, et al. Non-invasive mapping of the gastrointestinal microbiota identifies children with inflammatory bowel disease. PLoS ONE. (2012) 7:e39242. doi: 10.1371/journal.pone.0039242
60. Zhang X, Deeke SA, Ning Z, Starr AE, Butcher J, Li J, et al. Metaproteomics reveals associations between microbiome and intestinal extracellular vesicle proteins in pediatric inflammatory bowel disease. Nat Commun. (2018) 9:2873. doi: 10.1038/s41467-018-05357-4
61. Ni J, Wu GD, Albenberg L. Gut microbiota and IBD: causation or correlation? Nat Rev Gastroenterol Hepatol. (2017) 14:573–84. doi: 10.1038/nrgastro.2017.88
62. Glassner KL, Abraham BP, Quigley EMM. The microbiome and inflammatory bowel disease. J Allergy Clin Immunol. (2020) 145:16–27. doi: 10.1016/j.jaci.2019.11.003
63. Nishida A, Inoue R, Inatomi O, Bamba S, Naito Y, Andoh A. Gut microbiota in the pathogenesis of inflammatory bowel disease. Clin J Gastroenterol. (2018) 11:1–10. doi: 10.1007/s12328-017-0813-5
64. An R, Wilms E, Masclee AAM, Smidt H, Zoetendal EG, Jonkers D. Age-dependent changes in GI physiology and microbiota: time to reconsider? Gut. (2018) 67:2213–22. doi: 10.1136/gutjnl-2017-315542
65. Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, et al. Human gut microbiome viewed across age and geography. Nature. (2012) 486:222–7. doi: 10.1038/nature11053
66. Schirmer M, Franzosa EA, Lloyd-Price J, McIver LJ, Schwager R, Poon TW, et al. Dynamics of the human gut microbiome in inflammatory bowel disease. Nat Microbiol. (2017) 2:17004. doi: 10.1038/nmicrobiol.2017.4
67. Lavelle A, Sokol H. Gut microbiota-derived metabolites as key actors in inflammatory bowel disease. Nat Rev Gastroenterol Hepatol. (2020) 17:223–37. doi: 10.1038/s41575-019-0258-z
68. Schirmer M, Garner A, Vlamakis H, Xavier RJ. Microbial genes and pathways in inflammatory bowel disease. Nat Rev Microbiol. (2019) 17:497–511. doi: 10.1038/s41579-019-0213-6
69. Franzosa EA, Sirota-Madi A, Avila-Pacheco J, Fornelos N, Haiser HJ, Reinker S, et al. Gut microbiome structure and metabolic activity in inflammatory bowel disease. Nat Microbiol. (2019) 4:293–305. doi: 10.1038/s41564-018-0306-4
70. Parada Venegas D, De la Fuente MK, Landskron G, González MJ, Quera R, Dijkstra G, et al. Short Chain Fatty Acids (SCFAs)-mediated gut epithelial and immune regulation and its relevance for inflammatory bowel diseases. Front Immunol. (2019) 10:277. doi: 10.3389/fimmu.2019.01486
71. Sun M, Wu W, Liu Z, Cong Y. Microbiota metabolite short chain fatty acids, GPCR, and inflammatory bowel diseases. J Gastroenterol. (2017) 52:1–8. doi: 10.1007/s00535-016-1242-9
72. Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly-Y M, et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. (2013) 341:569–73. doi: 10.1126/science.1241165
73. Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, deRoos P, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. (2013) 504:451–5. doi: 10.1038/nature12726
74. Yang W, Yu T, Huang X, Bilotta AJ, Xu L, Lu Y, et al. Intestinal microbiota-derived short-chain fatty acids regulation of immune cell IL-22 production and gut immunity. Nat Commun. (2020) 11:4457. doi: 10.1038/s41467-020-18262-6
75. Silva JPB, Navegantes-Lima KC, Oliveira ALB, Rodrigues DVS, Gaspar SLF, Monteiro VVS, et al. Protective mechanisms of butyrate on inflammatory bowel disease. Curr Pharm Des. (2018) 24:4154–66. doi: 10.2174/1381612824666181001153605
76. Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, Nishikawa H, et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. (2013) 500:232–6. 78. doi: 10.1038/nature12331
77. Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. (2011) 331:337–41. doi: 10.1126/science.1198469
78. Ring C, Klopfleisch R, Dahlke K, Basic M, Bleich A, Blaut M. Akkermansia muciniphila strain ATCC BAA-835 does not promote short-term intestinal inflammation in gnotobiotic interleukin-10-deficient mice. Gut Microbes. (2019) 10:188–203. doi: 10.1080/19490976.2018.1511663
79. Parker BJ, Wearsch PA, Veloo ACM, Rodriguez-Palacios A. The genus alistipes: gut bacteria with emerging implications to inflammation, cancer, and mental health. Front Immunol. (2020) 11:906. doi: 10.3389/fimmu.2020.00906
80. Hiippala K, Kainulainen V, Suutarinen M, Heini T, Bowers JR, Jasso-Selles D, et al. Isolation of anti-inflammatory and epithelium reinforcing Bacteroides and Parabacteroides spp. from a healthy fecal donor. Nutrients. (2020) 12:935. doi: 10.3390/nu12040935
81. Hoffman JM, Margolis KG. Building community in the gut: a role for mucosal serotonin. Nat Rev Gastroenterol Hepatol. (2020) 17:6–8. doi: 10.1038/s41575-019-0227-6
82. Palmela C, Chevarin C, Xu ZL, Torres J, Sevrin G, Hirten R, et al. Adherent-invasive Escherichia coli in inflammatory bowel disease. Gut. (2018) 67:574–87. doi: 10.1136/gutjnl-2017-314903
83. Mirsepasi-Lauridsen HC, Vallance BA, Krogfelt KA, Petersen AM. Escherichia coli pathobionts associated with inflammatory bowel disease. Clin Microbiol Rev. (2019) 32:e00060-18. doi: 10.1128/CMR.00060-18
84. Seishima J, Iida N, Kitamura K, Yutani M, Wang ZY, Seki A, et al. Gut-derived Enterococcus faecium from ulcerative colitis patients promotes colitis in a genetically susceptible mouse host. Genome Biol. (2019) 20:252. doi: 10.1186/s13059-019-1879-9
85. Chen Y, Chen Y, Cao P, Su W, Zhan N, Dong W. Fusobacterium nucleatum facilitates ulcerative colitis through activating IL-17F signaling to NF-κB via the upregulation of CARD3 expression. J Pathol. (2020) 250:170–82. doi: 10.1002/path.5358
86. Wallace JL, Motta JP, Buret AG. Hydrogen sulfide: an agent of stability at the microbiome-mucosa interface. Am J Physiol Gastrointest Liver Physiol. (2018) 314:143–9. doi: 10.1152/ajpgi.00249.2017
87. Ijssennagger N, Belzer C, Hooiveld GJ, Dekker J, van Mil SW, Müller M, et al. Gut microbiota facilitates dietary heme-induced epithelial hyperproliferation by opening the mucus barrier in colon. Proc Natl Acad Sci USA. (2015) 112:10038–43. doi: 10.1073/pnas.1507645112
88. De Preter V, Arijs I, Windey K, Vanhove W, Vermeire S, Schuit F, et al. Decreased mucosal sulfide detoxification is related to an impaired butyrate oxidation in ulcerative colitis. Inflamm Bowel Dis. (2012) 18:2371–80. doi: 10.1002/ibd.22949
89. Duboc H, Rajca S, Rainteau D, Benarous D, Maubert MA, Quervain E, et al. Connecting dysbiosis, bile-acid dysmetabolism and gut inflammation in inflammatory bowel diseases. Gut. (2013) 62:531–9. doi: 10.1136/gutjnl-2012-302578
90. Heinken A, Ravcheev DA, Baldini F, Heirendt L, Fleming RMT, Thiele I. Systematic assessment of secondary bile acid metabolism in gut microbes reveals distinct metabolic capabilities in inflammatory bowel disease. Microbiome. (2019) 7:75. doi: 10.1186/s40168-019-0689-3
91. Connors J, Dunn KA, Allott J, Bandsma R, Rashid M, Otley AR, et al. The relationship between fecal bile acids and microbiome community structure in pediatric Crohn's disease. ISME J. (2020) 14:702–13. doi: 10.1038/s41396-019-0560-3
92. Nikolaus S, Schulte B, Al-Massad N, Thieme F, Schulte DM, Bethge J, et al. Increased tryptophan metabolism is associated with activity of inflammatory bowel diseases. Gastroenterology. (2017) 153:1504–16. doi: 10.1053/j.gastro.2017.08.028
93. Bosch S, de Meij TGJ, de Boer NK. Altered tryptophan levels in patients with inflammatory bowel disease owing to colonic leakage, metabolism, or malabsorption? Gastroenterology. (2018) 154:1855–6. doi: 10.1053/j.gastro.2018.01.071
94. Prosberg M, Bendtsen F, Vind I, Petersen AM, Gluud LL. The association between the gut microbiota and the inflammatory bowel disease activity. Scand J Gastroenterol. (2016) 51:1407–15. doi: 10.1080/00365521.2016.1216587
95. Pascal V, Pozuelo M, Borruel N, Casellas F, Campos D, Santiago A, et al. A microbial signature for Crohn's disease. Gut. (2017) 66:813–22. doi: 10.1136/gutjnl-2016-313235
96. Kugathasan S, Denson LA, Walters TD, Kim MO, Marigorta UM, Schirmer M, et al. Prediction of complicated disease course for children newly diagnosed with Crohn's disease: a multicentre inception cohort study. Lancet. (2017) 389:1710–8. doi: 10.1016/S0140-6736(17)30317-3
97. Kelsen J, Bittinger K, Pauly-Hubbard H, Posivak L, Grunberg S, Baldassano R, et al. Alterations of the subgingival microbiota in pediatric Crohn's disease studied longitudinally in discovery and validation cohorts. Inflamm Bowel Dis. (2015) 21:2797–805. doi: 10.1097/MIB.0000000000000557
98. Iwasawa K, Suda W, Tsunoda T, Oikawa-Kawamoto M, Umetsu S, Takayasu L, et al. Dysbiosis of the salivary microbiota in pediatric-onset primary sclerosing cholangitis and its potential as a biomarker. Sci Rep. (2018) 8:5480. doi: 10.1038/s41598-018-23870-w
99. Bonder MJ, Kurilshikov A, Tigchelaar EF, Mujagic Z, Imhann F, Vila AV, et al. The effect of host genetics on the gut microbiome. Nat Genet. (2016) 48:1407–12. doi: 10.1038/ng.3663
100. Zuo T, Kamm MA, Colombel JF, Ng SC. Urbanization and the gut microbiota in health and inflammatory bowel disease. Nat Rev Gastroenterol Hepatol. (2018) 15:440–52. doi: 10.1038/s41575-018-0003-z
101. Ng SC, Tang W, Leong RW, Chen M, Ko Y, Studd C, et al. Environmental risk factors in inflammatory bowel disease: a population-based case-control study in Asia-Pacific. Gut. (2015) 64:1063–71. doi: 10.1136/gutjnl-2014-307410
Keywords: inflammatory bowel disease, Crohn's disease, ulcerative colitis, pediatric, gut microbiota
Citation: Zhuang X, Liu C, Zhan S, Tian Z, Li N, Mao R, Zeng Z and Chen M (2021) Gut Microbiota Profile in Pediatric Patients With Inflammatory Bowel Disease: A Systematic Review. Front. Pediatr. 9:626232. doi: 10.3389/fped.2021.626232
Received: 05 November 2020; Accepted: 05 January 2021;
Published: 02 February 2021.
Edited by:
Jeff Critch, Memorial University of Newfoundland, CanadaReviewed by:
Jan De Laffolie, University of Giessen, GermanyCopyright © 2021 Zhuang, Liu, Zhan, Tian, Li, Mao, Zeng and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Minhu Chen, Y2hlbm1pbmh1QG1haWwuc3lzdS5lZHUuY24=; Zhirong Zeng, emVuZ3poaXJvbmdAbWFpbC5zeXN1LmVkdS5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.