- Institute of Biomedical Sciences, Academia Sinica, Taipei, Taiwan
Brown adipose tissue (BAT) is a thermogenic organ owing to its unique expression of uncoupling protein 1 (UCP1), which is a proton channel in the inner mitochondrial membrane used to dissipate the proton gradient and uncouple the electron transport chain to generate heat instead of adenosine triphosphate. The discovery of metabolically active BAT in human adults, especially in lean people after cold exposure, has provoked the “thermogenic anti-obesity” idea to battle weight gain. Because BAT can expend energy through UCP1-mediated thermogenesis, the molecular mechanisms regulating UCP1 expression have been extensively investigated at both transcriptional and posttranscriptional levels. Of note, the 3′-untranslated region (3′-UTR) of Ucp1 mRNA is differentially processed between mice and humans that quantitatively affects UCP1 synthesis and thermogenesis. Here, we summarize the regulatory mechanisms underlying UCP1 expression, report the number of poly(A) signals identified or predicted in Ucp1 genes across species, and discuss the potential and caution in targeting UCP1 for enhancing thermogenesis and metabolic fitness.
Introduction
Brown and white adipocytes possess unique functions in thermogenesis and energy storage, respectively; however, to some extent, one could cover the function of the other. A high fat diet (HFD) can induce brown-to-white adipocyte conversion when brown adipocytes store excess lipids to appear unilocular. Some white adipocytes are known as beige adipocytes, which can be browned in response to adrenergic signaling to become thermogenic (1). Cold-induced metabolic activity of brown adipose tissue (BAT) is positively correlated with resting metabolic rate and negatively with body mass index and body fat percentage in humans (2, 3). Therefore, a “thermogenic anti-obesity” approach has been proposed to combat obesity if the metabolic furnace, BAT, can be pharmacologically activated to burn excess fat (4). Because BAT defends against fat accumulation via uncoupling protein 1 (UCP1)-mediated thermogenesis, extensive efforts have uncovered how UCP1 activity is controlled by allosteric conformation changes and gene expression to affect thermogenesis. Several polymorphisms in the promoter, non-coding, and coding regions of UCP1 gene were found associated with obesity and type 2 diabetes (5–8). Of note, genomic variations in Ucp1 across species indicate that non-shivering thermogenic function and regulation are not necessarily conserved through evolution.
Discovery of Ucp1, Whose 3′-UTR Is Processed Differently Among Species
UCP1 was first isolated from BAT of rats and hamsters in 1980 and named for its heat-producing function by uncoupling the oxidative phosphorylation process in mitochondria (Figure 1A) (9). At the basal status, the binding of purine nucleotide di- and tri-phosphates on the outer facing cavity of UCP1 blocks its proton-translocating activity (10, 11). In response to nutrients or cold environment, thermogenesis is activated via the release of norepinephrine from sympathetic circuits onto β3 adrenergic receptors (β3ARs) in brown adipocytes (4, 12, 13). β3AR signaling elevates cyclic adenosine monophosphate (cAMP) level to activate protein kinase A and lipolysis and acutely enhances UCP1 activity via free fatty acid-induced allosteric changes (14). Chronic activation of β3ARs promotes UCP1 synthesis by multiple transcriptional and posttranscriptional mechanisms to be discussed later.
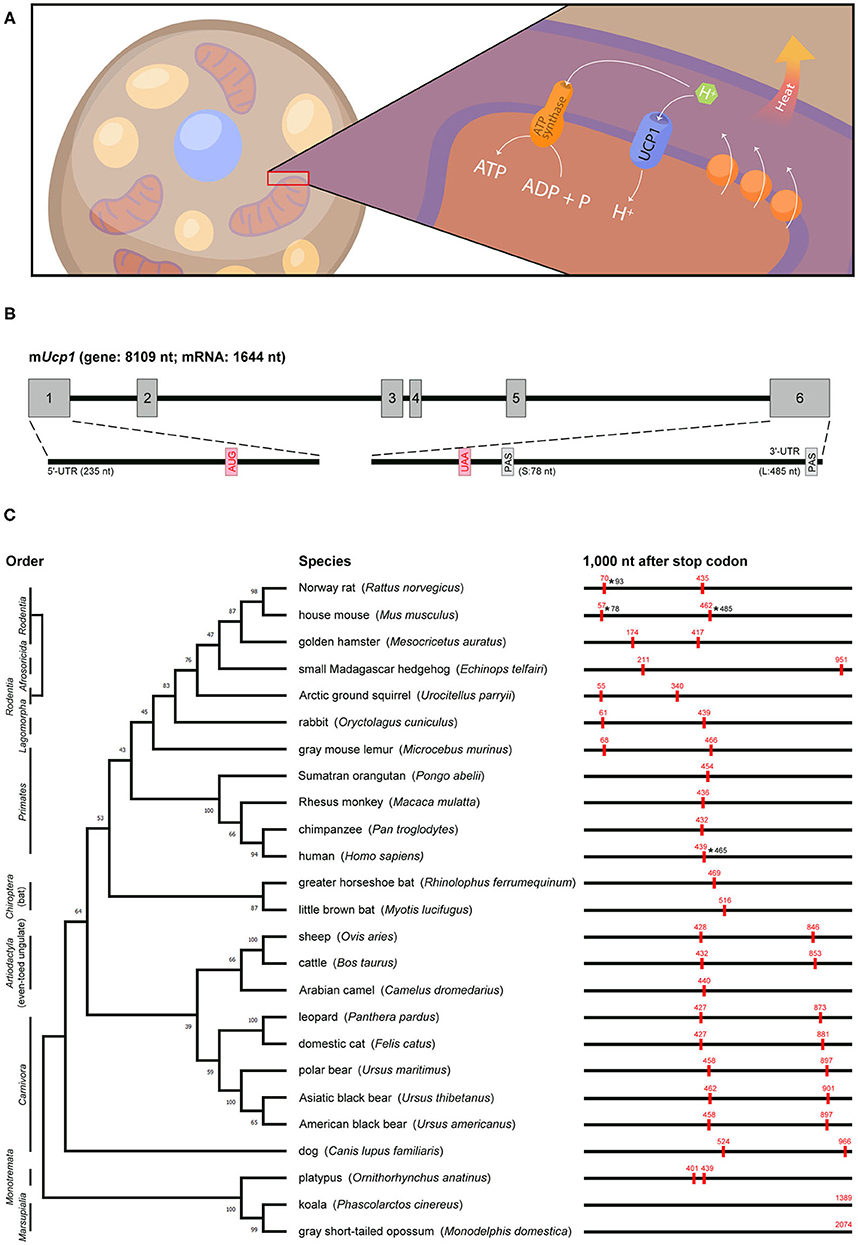
Figure 1. Genomic information, actual or predicted polyadenylation sites in Ucp1 across species. (A) UCP1, a proton channel located in the inner mitochondrial membrane of brown adipocytes, transports protons to the matrix and then disrupts the proton gradient generated by the electron transport chain, thus uncoupling fuel oxidation from ATP synthesis and releasing energy as heat. (B) Mouse Ucp1 spans ~8 kb and contains six exons (gray boxes). The start (AUG) and stop (UAA) codons are labeled in red, and the two polyadenylation sites (PAS) are denoted. The 5′-UTR and 3′-UTR of Ucp1 are located on exons 1 and 6, respectively. (C) Phylogenetic relationship of mammalian Ucp1 by alignment of the nucleotide sequences of coding regions. Animal orders are marked in the left and the percentage of bootstrap support of minimum evolution is shown on the corresponding branch. The BLAST search of 1,000 nt after the Ucp1 stop codon across species involved the Ensembl or NCBI database. The poly(A) signals (AAUAAA) are marked as red boxes with their positions in red. The ends of 3′-UTRs confirmed by cDNA sequencing in rat, mouse, and human are labeled with asterisks and numbers.
Rat and mouse Ucp1 cDNAs were cloned in 1985 (15, 16) and later sequenced to reveal their coding sequences and untranslated regions (UTRs) (17–19). Soon after, the rabbit Ucp1 cDNA sequence was reported (20). The 3′-UTR of Ucp1 in all three species contains two poly(A) signals (i.e., AAUAAA). Northern blot analysis revealed two Ucp1 transcripts of ~1.5 and 1.9 kb (hereafter called Ucp1S and Ucp1L) in mouse and rat BAT (15, 16). The transcripts result from an alternative use of the poly(A) signal to yield 3′-UTRs of 78 and 485 bp in mice, respectively (21) (Figure 1B). By contrast, only the distal poly(A) signal is used to generate Ucp1L in rabbit BAT (20). The human UCP1 coding sequence was identified by screening a genomic library with a rat Ucp1 cDNA probe, and a 1.9-kb UCP1 mRNA was detected in human perirenal BAT (22). More recently, the sequence of human UCP1 cDNA (ENST00000262999) revealed only one poly(A) site in the 3′-UTR, and RT-PCR analysis confirmed that human UCP1 encodes only the long form carrying a 465-bp 3′-UTR (21).
Thermogenic UCP1 Is Not Preserved in All Endothermic Placental Mammals
The functional Ucp1 gene consists of six exons and spans a region of ~8–10 kb among species (an example of mouse Ucp1 is in Figure 1B). The phylogenetic relationship of Ucp1 across species has revealed that the non-thermogenic UCP1 in fish and frogs (23) was derived from a hypothetical proto-UCP hundreds of millions of years ago. Non-thermogenic UCP1 was first lost in Sauropsida (reptiles and birds) around 300 million years ago, then thermogenic UCP1 emerged in endothermic placental mammals about 100 million years ago (24, 25). Although UCP1-mediated non-shivering thermogenesis is believed to provide a survival advantage in the cold and retain the body temperature of eutherian neonates after birth, eight mammalian clades, namely Xenarthra, Pholidota, Cetacea, Sirenia, Proboscidea, Hyracoidea, Equidae, and the most well-known Suidae (pigs), showed inactivation of Ucp1 (26). The loss of exons 3 to 5 in pig Ucp1 occurred ~20 million years ago, so pigs produce no functional UCP1 and depend on shivering for thermoregulation (27).
The previous studies aligned the genomic or amino acid sequences of Ucp1 across species to build up the phylogenetic tree and derive an evolutionary timeline for Ucp1 (24–26). We followed these evolutionary patterns by aligning the nucleotide sequences of the Ucp1 coding regions in selected mammals and analyzed whether alternative polyadenylation could follow any evolutionary path (Figure 1C). The phylogenetic tree of Ucp1 grouped most species following the canonical taxonomy, classifying animals of the same order in the same branch. The three exceptions are Ucp1 in the small Madagascar hedgehog in Afrosoricida more related to rodents, in the gray mouse lemur in Primates more related to rodents and rabbits, and in the dog quite different from other animals in Carnivora (24).
The 3′-UTR information for Ucp1 cDNAs in various species is often lacking or incomplete. We predicted “alternative polyadenylation” by searching the poly(A) signal (i.e., AATAAA) within 1 kb downstream of the stop codon from Ucp1 genomic sequences because the 3′-UTR, including in mouse and human Ucp1 mRNAs, is limited in the last coding exon of most transcripts. Our analyses revealed many representative animals carrying two poly(A) sites, such as mice, rats, and squirrels in Rodentia; rabbits in Lagomorpha; and cats, dogs, cows, leopards, and bears in Laurasiatheria. However, microbats and greater horseshoe bats in Chiroptera are the exceptions, with only one poly(A) motif in the 3′-flanking region. Notably, almost all primates, such as humans, chimpanzees, orangutans, and macaques, have only one poly(A) site in Ucp1, with the exception of mouse lemurs, which carry two poly(A) signals at the position similar to those of rodent Ucp1. The nucleotide sequence of the Ucp1 coding region in mouse lemurs was unexpectedly more similar to those in rodents than primates (Figure 1C). Although the platypus, a unique placental mammal, has two adjacent poly(A) signals in Ucp1, alternative polyadenylation, even if it occurs, would generate two 3′-UTRs of only 32 bp difference. The poly(A) site of Ucp1 in koala and gray short-tailed opossum in Marsupialia is located at 1,389 and 2,074 bp downstream of the stop codon, respectively, implying Ucp1 in both species may be subjected to more complex posttranscriptional regulation. Other than the 3′-UTRs of rat Ucp1S, mouse Ucp1S and Ucp1L, and human UCP1 having been confirmed by cDNA sequencing (17, 18, 21), the actual usage and preference of the two putative polyadenylation sites in other species require further investigation.
Transcriptional Regulation of Ucp1 Gene
Cold exposure markedly increases both Ucp1L and Ucp1S mRNA levels in mouse and rat BAT (15, 16), so elaborative efforts have identified the molecular mechanisms controlling Ucp1 transcription (28–30). DNA footprinting and promoter assays were performed to identify a 212-bp distal enhancer (−2,494 to −2,283 bp from the transcription start site) and a 555-bp proximal promoter (−611 to −57 bp) in the 5′ non-coding region of rat Ucp1 gene (28, 31). Many studies used in vitro assays to identify the trans-acting factors for Ucp1 transcription, including protein kinase A-activated cAMP-response element binding protein (CREB) to compete with the negative regulator, Jun, for binding to the −139- to −122-bp region, and CCAAT/enhancer-binding protein α and β (C/EBPα and β) to activate through regions −457 to −440 and −335 to −318 bp [for details, see the review (32)]. Zfp516 binds to the promoter region (−70 to −45) with PR domain-containing 16 (PRDM16), which is essential for BAT differentiation and promotes browning of beige adipocytes to increase body temperature and energy expenditure (33) (Figure 2A). Notably, the absence of the CCAAT sequence in the human UCP1 promoter suggests evolutionary variations in regulating Ucp1 transcription (22).
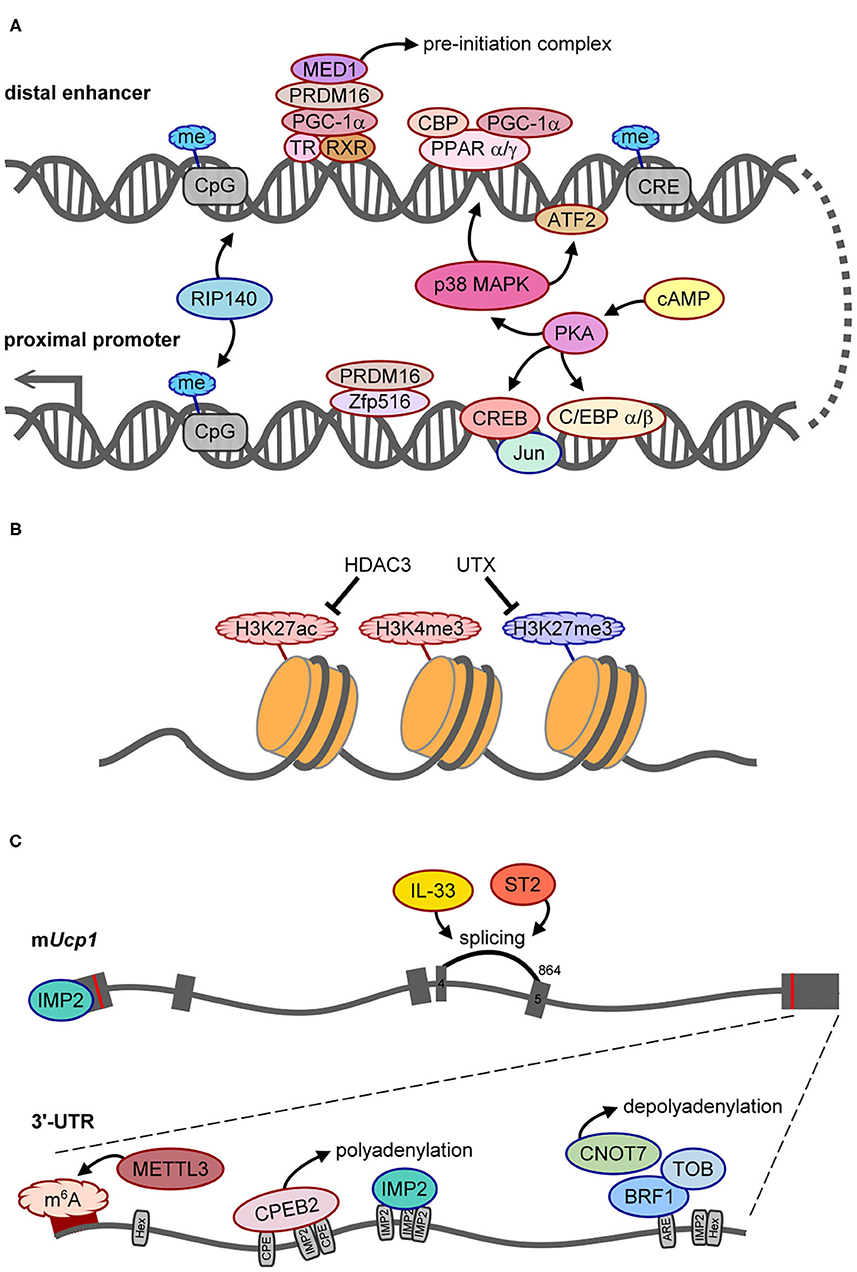
Figure 2. The molecular mechanisms control UCP1 synthesis. Positive and negative regulators of UCP1 expression are outlined in red or blue, respectively. (A) Scheme of regulatory factors acting on the distal enhancer and proximal promoter of Ucp1. (B) The histone modifications, HDAC3 and UTX demethylase, involved in epigenetic regulation of Ucp1 transcription. (C) Illustration of splicing and posttranscriptional regulation of Ucp1 RNA. The gray boxes are six exons of Ucp1 with the start (AUG) and stop (UAA) codons labeled as red lines. The number 864 above exon 5 is the splice acceptor site for generating functional UCP1. The bottom gray line represents the 3′-UTR of mouse Ucp1 with 2 hexanucleotides (Hex, AAUAAA polyadenylation site), denoted regulatory sequences, including CPE (CPEB2-binding site), ARE (AU-rich element for BRF1 binding), and predicted IMP2-binding sites. The dark red band indicates the hot spot area for m6A modification.
When fused to a thymidine kinase promoter-driven reporter in a transgenic mouse line, the 212-bp enhancer sufficiently conferred BAT-specific expression and cold- and β3AR agonist-induced expression (34). Peroxisome proliferator-activated receptor α and γ (PPARα and PPARγ), PPARγ-coactivator 1α (PGC-1α), and CREB-binding protein (CBP) bind to the enhancer (−2,485 to −2,458) together to upregulate Ucp1 transcription (35) (Figure 2A). Phosphorylation of PGC-1α and activating transcription factor 2 (ATF2) by cold exposure-activated p38 mitogen-activated protein kinase can directly or indirectly potentiate Ucp1 transcription because Ppargc1a (i.e., the gene name of PGC-1α) transcription is also enhanced by phosphorylated ATF2 (35, 36). Moreover, PRDM16 recruited by PGC-1α interacts with thyroid receptor/retinoid X receptor heterodimers, and MED1, a component of the Mediator complex, is then recruited by PRDM16 to form a pre-initiation complex on the enhancer to facilitate Ucp1 transcription (37, 38) (Figure 2A). Therefore, the enhancer region of Ucp1 is regulated by signaling of multiple trans-acting factors through β3ARs.
In addition to positive regulators, receptor-interacting protein 140 (RIP140) mediates methylation at CpG sites in both the promoter and enhancer to silence Ucp1 transcription (39) (Figure 2A). In pregnant mice with streptozotocin-induced diabetes, intrauterine exposure to hyperglycemia impaired fetal BAT development by altering methylation in several metabolic genes including Ucp1. Such epigenetic changes can last and affect BAT function and glucose homeostasis later in life (40). Besides CpG methylation, epigenetic modification of histone 3 (H3) affects chromatin structure to regulate transcription. Histone deacetylase 3 (HDAC3) controls the acetylation of H3 on lysine 27 (H3K27ac) in the enhancers of Ucp1 and Pparα (Figure 2B), so its deficiency facilitates Ucp1 and Pparα transcription and browning in beige adipocytes (41). Of note, pregnant mice fed omega-3 polyunsaturated fatty acids resulted in epigenetic changes, including decreased tri-methylation of H3 on lysine 27 (H3K27me3), increased H3K27ac and di-methylation of H3 on lysine 9 (H3K9me2) to affect BAT development, and increased Ucp1 and Ppargc1a transcription in mouse offspring even at 8 weeks after post-weaning (42). H3K9me2 is usually defined as a repressed histone mark (43), so increased H3K9me2 may not directly associate with the Ucp1 promoter. Nevertheless, this study reported that a maternal dietary effect could have long-lasting benefits on the development and function of the offspring's BAT. Moreover, the silencing mark, H3K9me2, was identified at the Ucp1 enhancer in white adipose tissue (WAT), whereas the active mark, H3K4me3, at the Ucp1 promoter was found in BAT in response to cold stimulation (43). The ubiquitously transcribed tetratricopeptide repeat on chromosome X (UTX) is a histone demethylase recruited to Ucp1 upon the activation of β3ARs to reduce H3K27me3 level (Figure 2B), which consequently leads to increased H3K27ac level via unidentified histone acetyltransferases (44). Therefore, epigenetic changes on Ucp1 are dynamically regulated for temporal and spatial control of UCP1 synthesis, and intrauterine nutrient exposure has a long-term effect on metabolic gene expression via epigenetic regulation.
Posttranscriptional Regulation of Ucp1 mRNA
A pre-mRNA undergoes several processing steps, including splicing, 3′-end cleavage, and polyadenylation, to become a mature mRNA. Alternative polyadenylation generates Ucp1 mRNA carrying long (Ucp1L, ~10%) or short (Ucp1S, ~90%) 3′-UTR in mice and rats, but rabbits and humans express only the long form. The regulatory sequences in the 3′-UTR dictate posttranscriptional efficiency for protein production (45), but the effect of different Ucp1 3′-UTRs on UCP1 synthesis and thermogenesis has not been addressed until recently. The study of Ucp1ΔL mice, with 10% Ucp1L converted to Ucp1S, reported that the protein but not the mRNA level of UCP1 was reduced ~50–60% in Ucp1ΔL BAT, so Ucp1L is translated ~15 times more efficiently than Ucp1S in BAT (21). Moreover, the same study also identified that cytoplasmic polyadenylation element binding protein 2 (CPEB2) signaling through β3ARs binds to and activates polyadenylation-induced translation of only Ucp1L but not Ucp1S (Figure 2C), so both CPEB2-knockout (KO) and Ucp1ΔL mice show reduced UCP1 level and impaired BAT thermogenesis (21).
The other posttranscriptional mechanisms identified to date inhibit UCP1 synthesis. Reporter assay revealed that insulin-like growth factor 2 mRNA-binding protein 2 (IGF2BP2/IMP2) is a type 2 diabetes- and obesity-associated gene that suppresses Ucp1 mRNA translation likely via both 5′- and 3′-UTRs. Thus, IMP2-null mice show increased energy expenditure, insulin sensitivity, and glucose tolerance and defend against cold temperature better than wild-type mice (46). HFD reduces the amount of UCP1 in obese adipose tissues by diminishing the stability of mouse Ucp1 mRNA via the CCR4–NOT deadenylase complex. Moreover, HFD induces the expression of several subunits of the CCR4–NOT complex, including Cnot1–3, Cnot6 and Cnot7, and Cnot7-interacting Tob in inguinal WAT (iWAT) of obese mice. The binding of butyrate response factor 1 (BRF1) to the 3′-UTR of Ucp1L mRNA recruits Tob and Cnot7 to cause deadenylation-induced Ucp1 decay (Figure 2C). Thus, Cnot7- or Tob-KO mice are resistant to HFD-induced obesity with increased Ucp1 mRNA level in iWAT and BAT (47). Although IMP2 and BRF1 suppressed the translation and stability, respectively, of a reporter appended to the Ucp1L 3′-UTR in HEK293 cells, both studies did not examine Ucp1L and Ucp1S separately when comparing polysomal distribution (46) or mRNA stability of Ucp1 (47) between wild-type and KO adipose tissues.
MicroRNAs (miRNAs) are a conserved class of small RNAs of about 20–22 nt that regulate the expression of their target mRNAs via the miRNA-induced silencing complex (48, 49). Despite the relatively short 3′-UTR (485 bp) of mouse Ucp1L mRNA, a previous study used miRanda to identify miR-9 and miR-338-3p as putative miRNAs targeting the 3′-UTR of Ucp1L mRNA (50). Activation of β3AR signaling decreased both miRNA levels and increased Ucp1 mRNA level in epididymal WAT (eWAT) but not iWAT and BAT. Despite a negative correlation between miR-9 and miR-338-3p and Ucp1 expression in eWAT, no further experiments were performed to determine whether both miRNAs affect Ucp1 mRNA stability by binding to the predicted target sites in the 3′-UTR (50). Although cold exposure did not change the proportion of Ucp1L and Ucp1S in BAT (21), no studies have measured the proportion of Ucp1L and Ucp1S in WAT and whether it can be changed under HFD or cold temperature. The alternative use of proximal and distal poly(A) signals in mouse Ucp1 mRNA may vary in different adipose tissues.
A recent paper demonstrated that methyltransferase like 3 (METTL3)-catalyzed m6A modifications in Ucp1 mRNA increased its mRNA and protein levels during postnatal BAT development (Figure 2C). In mice with BAT-specific ablation of Mettl3, the loss of m6A-enhanced UCP1 expression reduced energy expenditure and cold tolerance (51). Moreover, these mice showed accelerated HFD-induced obesity, impaired glucose intolerance, and insulin resistance (51). Depending on the reader, m6A modification on mRNA can affect translation efficiency or mRNA stability (52), but how this epitranscriptomic modification mechanistically affects UCP1 expression has not yet been answered.
Besides these posttranscriptional regulations, interleukin 33 (IL-33) and ST2 (also known as IL-1 receptor 4, a receptor of IL-33) are required for accurate splicing of Ucp1 mRNA between exons 4 and 5 by using the splice site at nucleotide 864 on exon 5 (Figure 2C). Without IL-33 or ST2, the splicing machinery prefers the cryptic nucleotide 972 site on exon 5 to produce a non-functional transcript (53). Depletion of either protein results in the loss of UCP1 in brown and beige adipocytes, so IL-33/ST2 signaling is important to register brown and beige adipocytes for uncoupled respiration and thermogenesis during the perinatal period (53).
Discussion
Treating lean healthy men with a specific β3AR agonist, CL316243, increased insulin sensitivity and fat oxidation without affecting body weight (54), but 4-week administration of another β3AR agonist, L796568, in obese men did not confer evident metabolic benefits (55). The lack of chronic effect on shifting energy balance for weight loss may be due to few β3AR-responsive adipose tissues in humans. Indeed, two recent studies found a very low level of β3AR mRNA in human BAT (56, 57). Using cultured human brown adipocytes, one study reported that β2AR activation increased UCP1 level and lipolysis (56), but the other study identified β1AR signaling as responsible for upregulating thermogenic gene expression (57). Because the activation of β1AR and β2AR has a profound impact on cardiopulmonary function, the positive regulatory mechanisms downstream of β-adrenergic signaling must be targeted to promote UCP1 synthesis, energy expenditure, and metabolic fitness in humans. Notably, the mouse model has been used to reveal mechanisms controlling UCP1 synthesis, thermogenesis, and possible implications for human obesity. However, the loss of the CCAAT sequence and Sp1-binding motif in the proximal promoter (22) and alternative polyadenylation in human UCP1 (21) supports that UCP1 expression is regulated by species-dependent variations. For example, despite comparable UCP1 activity in mouse and human BAT (58), we reported that the expression of human Ucp1 mRNA in five different BAT samples is only 1/30–1/300 the level in mice (21). Our result agrees with a Northern blot study, which detected 1.9-kb UCP1 mRNA by using 5 μg poly(A) RNA from human perirenal BAT. Thus, absolute RT-qPCR was used to compare the amount of Ucp1 mRNA between human and mouse BAT (21). Because Ucp1L is translated 15 times more efficiently than Ucp1S in mouse BAT, we predicted that CPEB2-mediated translational activation, if fully functioning in human BAT, can make up the protein level to ~1/2–1/20 of that in mouse BAT (21). Similarly, if the mechanism of BRF1-mediated deadenylation-induced Ucp1L decay (47) is conserved in humans, it is expected that HFD-induced UCP1 mRNA degradation in human beige adipocytes should become more prominent. Thus, humans, and perhaps also other primates, may rely more on posttranscriptional regulation to control UCP1 synthesis because they express a low level of only long 3′-UTR UCP1 mRNA. Moreover, although ectopic expression of mouse Ucp1 in white adipocytes of pigs also enhances thermogenic capacity and lipolysis to decrease fat deposition (59), why UCP1-mediated metabolic plasticity is lost in some mammals through evolution remains unclear. Therefore, the results from studying gene-modified mice in BAT thermogenesis need to be translated with caution into humans and other species.
Author Contributions
W-HL wrote the manuscript and illustrated the figures. Y-MC analyzed the poly(A) sites in Ucp1 across species. Y-SH co-wrote the manuscript and is responsible for its content. All authors contributed to the article and approved the submitted version.
Funding
Research in the lab was funded by the Ministry of Science and Technology, Taiwan (MoST108-2320-B-001-020-MY3), National Health Research Institutes (NHRI-EX109-10719SI), and Aademia Sinica.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank Hao-Wen Chen for the illustration in Figure 1A and Hsi-Nien Chen from the Taipower Research Institute, Taiwan Power Company, for helping with the construction of the phylogenetic tree in Figure 1C.
References
1. Seale P, Bjork B, Yang W, Kajimura S, Chin S, Kuang S, et al. PRDM16 controls a brown fat/skeletal muscle switch. Nature. (2008) 454:961–7. doi: 10.1038/nature07182
2. Cypess AM, Lehman S, Williams G, Tal I, Rodman D, Goldfine AB, et al. Identification and importance of brown adipose tissue in adult humans. N Engl J Med. (2009) 360:1509–17. doi: 10.1056/NEJMoa0810780
3. van Marken Lichtenbelt WD, Vanhommerig JW, Smulders NM, Drossaerts JM, Kemerink GJ, Bouvy ND, et al. Cold-activated brown adipose tissue in healthy men. N Engl J Med. (2009) 360:1500–8. doi: 10.1056/NEJMoa0808718
4. Dulloo AG. Translational issues in targeting brown adipose tissue thermogenesis for human obesity management. Ann N Y Acad Sci. (2013) 1302:1–10. doi: 10.1111/nyas.12304
5. Brondani LA, Assmann TS, Duarte GC, Gross JL, Canani LH, Crispim D. The role of the uncoupling protein 1 (UCP1) on the development of obesity and type 2 diabetes mellitus. Arq Bras Endocrinol Metabol. (2012) 56:215–25. doi: 10.1590/S0004-27302012000400001
6. Chathoth S, Ismail MH, Vatte C, Cyrus C, Al Ali Z, Ahmed KA, et al. Association of Uncoupling Protein 1 (UCP1) gene polymorphism with obesity: a case-control study. BMC Med Genet. (2018) 19:203. doi: 10.1186/s12881-018-0715-5
7. Mori H, Okazawa H, Iwamoto K, Maeda E, Hashiramoto M, Kasuga M. A polymorphism in the 5' untranslated region and a Met229–>Leu variant in exon 5 of the human UCP1 gene are associated with susceptibility to type II diabetes mellitus. Diabetologia. (2001) 44:373–6. doi: 10.1007/s001250051629
8. Nicoletti CF, de Oliveira AP, Brochado MJ, de Oliveira BP, Pinhel MA, Marchini JS, et al. UCP1−3826 A>G polymorphism affects weight, fat mass, and risk of type 2 diabetes mellitus in grade III obese patients. Nutrition. (2016) 32:83–7. doi: 10.1016/j.nut.2015.07.016
9. Lin CS, Klingenberg M. Isolation of the uncoupling protein from brown adipose tissue mitochondria. FEBS Lett. (1980) 113:299–303. doi: 10.1016/0014-5793(80)80613-2
10. Nicholls DG. Hamster brown-adipose-tissue mitochondria. Purine nucleotide control of the ion conductance of the inner membrane, the nature of the nucleotide binding site. Eur J Biochem. (1976) 62:223–8. doi: 10.1111/j.1432-1033.1976.tb10151.x
11. Zhu R, Rupprecht A, Ebner A, Haselgrubler T, Gruber HJ, Hinterdorfer P, et al. Mapping the nucleotide binding site of uncoupling protein 1 using atomic force microscopy. J Am Chem Soc. (2013) 135:3640–6. doi: 10.1021/ja312550k
12. Lowell BB, Spiegelman BM. Towards a molecular understanding of adaptive thermogenesis. Nature. (2000) 404:652–60. doi: 10.1038/35007527
13. Morrison SF, Madden CJ, Tupone D. Central neural regulation of brown adipose tissue thermogenesis and energy expenditure. Cell Metab. (2014) 4:1677–713. doi: 10.1002/cphy.c140013
14. Hankir MK. Loading and firing the brown adipocyte. Adipocyte. (2018) 7:4–11. doi: 10.1080/21623945.2017.1405879
15. Bouillaud F, Ricquier D, Thibault J, Weissenbach J. Molecular approach to thermogenesis in brown adipose tissue: cDNA cloning of the mitochondrial uncoupling protein. Proc Natl Acad Sci USA. (1985) 82:445–8. doi: 10.1073/pnas.82.2.445
16. Jacobsson A, Stadler U, Glotzer MA, Kozak LP. Mitochondrial uncoupling protein from mouse brown fat. Molecular cloning, genetic mapping, and mRNA expression. J Biol Chem. (1985) 260:16250–4. doi: 10.1016/S0021-9258(17)36228-2
17. Bouillaud F, Raimbault S, Ricquier D. The gene for rat uncoupling protein: complete sequence, structure of primary transcript and evolutionary relationship between exons. Biochem Biophys Res Commun. (1988) 157:783–92. doi: 10.1016/S0006-291X(88)80318-8
18. Kozak LP, Britton JH, Kozak UC, Wells JM. The mitochondrial uncoupling protein gene. Correlation of exon structure to transmembrane domains. J Biol Chem. (1988) 263:12274–7. doi: 10.1016/S0021-9258(18)37751-2
19. Ridley RG, Patel HV, Gerber GE, Morton RC, Freeman KB. Complete nucleotide and derived amino acid sequence of cDNA encoding the mitochondrial uncoupling protein of rat brown adipose tissue: lack of a mitochondrial targeting presequence. Nucleic Acids Res. (1986) 14:4025–35. doi: 10.1093/nar/14.10.4025
20. Balogh AG, Ridley RG, Patel HV, Freeman KB. Rabbit brown adipose tissue uncoupling protein MRNA: use of only one of two polyadenylation signals in its processing. Biochem Biophys Res Commun. (1989) 161:156–61. doi: 10.1016/0006-291X(89)91574-X
21. Chen HF, Hsu CM, Huang YS. CPEB2-dependent translation of long 3'-UTR Ucp1 mRNA promotes thermogenesis in brown adipose tissue. EMBO J. (2018) 37:e99071. doi: 10.15252/embj.201899071
22. Cassard AM, Bouillaud F, Mattei MG, Hentz E, Raimbault S, Thomas M, et al. Human uncoupling protein gene: structure, comparison with rat gene, and assignment to the long arm of chromosome 4. J Cell Biochem. (1990) 43:255–64. doi: 10.1002/jcb.240430306
23. Jastroch M, Wuertz S, Kloas W, Klingenspor M. Uncoupling protein 1 in fish uncovers an ancient evolutionary history of mammalian nonshivering thermogenesis. Phys Genom. (2005) 22:150–6. doi: 10.1152/physiolgenomics.00070.2005
24. Saito S, Saito CT, Shingai R. Adaptive evolution of the uncoupling protein 1 gene contributed to the acquisition of novel nonshivering thermogenesis in ancestral eutherian mammals. Gene. (2008) 408:37–44. doi: 10.1016/j.gene.2007.10.018
25. Jastroch M, Oelkrug R, Keipert S. Insights into brown adipose tissue evolution and function from non-model organisms. J Exp Biol. (2018) 221(Pt Suppl 1):jeb169425. doi: 10.1242/jeb.169425
26. Gaudry MJ, Jastroch M, Treberg JR, Hofreiter M, Paijmans JLA, Starrett J, et al. Inactivation of thermogenic UCP1 as a historical contingency in multiple placental mammal clades. Sci Adv. (2017) 3:e1602878. doi: 10.1126/sciadv.1602878
27. Berg F, Gustafson U, Andersson L. The uncoupling protein 1 gene (UCP1) is disrupted in the pig lineage: a genetic explanation for poor thermoregulation in piglets. PLoS Genet. (2006) 2:e129. doi: 10.1371/journal.pgen.0020129
28. Cassard-Doulcier AM, Gelly C, Fox N, Schrementi J, Raimbault S, Klaus S, et al. Tissue-specific and beta-adrenergic regulation of the mitochondrial uncoupling protein gene: control by cis-acting elements in the 5'-flanking region. Mol Endocrinol. (1993) 7:497–506. doi: 10.1210/mend.7.4.8388995
29. Kozak UC, Kopecky J, Teisinger J, Enerback S, Boyer B, Kozak LP. An upstream enhancer regulating brown-fat-specific expression of the mitochondrial uncoupling protein gene. Mol Cell Biol. (1994) 14:59–67. doi: 10.1128/MCB.14.1.59
30. Cummings DE, Brandon EP, Planas JV, Motamed K, Idzerda RL, McKnight GS. Genetically lean mice result from targeted disruption of the RII beta subunit of protein kinase A. Nature. (1996) 382:622–6. doi: 10.1038/382622a0
31. Cassard-Doulcier AM, Larose M, Matamala JC, Champigny O, Bouillaud F, Ricquier D. In vitro interactions between nuclear proteins and uncoupling protein gene promoter reveal several putative transactivating factors including Ets1, retinoid X receptor, thyroid hormone receptor, and a CACCC box-binding protein. J Biol Chem. (1994) 269:24335–42. doi: 10.1016/S0021-9258(19)51087-0
32. Villarroya F, Peyrou M, Giralt M. Transcriptional regulation of the uncoupling protein-1 gene. Biochimie. (2017) 134:86–92. doi: 10.1016/j.biochi.2016.09.017
33. Dempersmier J, Sambeat A, Gulyaeva O, Paul SM, Hudak CS, Raposo HF, et al. Cold-inducible Zfp516 activates UCP1 transcription to promote browning of white fat and development of brown fat. Mol Cell. (2015) 57:235–46. doi: 10.1016/j.molcel.2014.12.005
34. Cassard-Doulcier AM, Gelly C, Bouillaud F, Ricquier D. A 211-bp enhancer of the rat uncoupling protein-1 (UCP-1) gene controls specific and regulated expression in brown adipose tissue. Biochem J. (1998) 333(Pt 2):243–6. doi: 10.1042/bj3330243
35. Barbera MJ, Schluter A, Pedraza N, Iglesias R, Villarroya F, Giralt M. Peroxisome proliferator-activated receptor alpha activates transcription of the brown fat uncoupling protein-1 gene. A link between regulation of the thermogenic and lipid oxidation pathways in the brown fat cell. J Biol Chem. (2001) 276:1486–93. doi: 10.1074/jbc.M006246200
36. Cao W, Daniel KW, Robidoux J, Puigserver P, Medvedev AV, Bai X, et al. p38 mitogen-activated protein kinase is the central regulator of cyclic AMP-dependent transcription of the brown fat uncoupling protein 1 gene. Mol Cell Biol. (2004) 24:3057–67. doi: 10.1128/MCB.24.7.3057-3067.2004
37. Harms MJ, Lim HW, Ho Y, Shapira SN, Ishibashi J, Rajakumari S, et al. PRDM16 binds MED1 and controls chromatin architecture to determine a brown fat transcriptional program. Genes Dev. (2015) 29:298–307. doi: 10.1101/gad.252734.114
38. Iida S, Chen W, Nakadai T, Ohkuma Y, Roeder RG. PRDM16 enhances nuclear receptor-dependent transcription of the brown fat-specific Ucp1 gene through interactions with Mediator subunit MED1. Genes Dev. (2015) 29:308–21. doi: 10.1101/gad.252809.114
39. Kiskinis E, Hallberg M, Christian M, Olofsson M, Dilworth SM, White R, et al. RIP140 directs histone and DNA methylation to silence Ucp1 expression in white adipocytes. EMBO J. (2007) 26:4831–40. doi: 10.1038/sj.emboj.7601908
40. Yu DQ, Lv PP, Yan YS, Xu GX, Sadhukhan A, Dong S, et al. Intrauterine exposure to hyperglycemia retards the development of brown adipose tissue. FASEB J. (2019) 33:5425–39. doi: 10.1096/fj.201801818R
41. Ferrari A, Longo R, Fiorino E, Silva R, Mitro N, Cermenati G, et al. HDAC3 is a molecular brake of the metabolic switch supporting white adipose tissue browning. Nat Commun. (2017) 8:93. doi: 10.1038/s41467-017-00182-7
42. Fan R, Toney AM, Jang Y, Ro SH, Chung S. Maternal n-3 PUFA supplementation promotes fetal brown adipose tissue development through epigenetic modifications in C57BL/6 mice. Biochim Biophys Acta Mol Cell Biol Lipids. (2018) 1863:1488–97. doi: 10.1016/j.bbalip.2018.09.008
43. Shore A, Karamitri A, Kemp P, Speakman JR, Lomax MA. Role of Ucp1 enhancer methylation and chromatin remodelling in the control of Ucp1 expression in murine adipose tissue. Diabetologia. (2010) 53:1164–73. doi: 10.1007/s00125-010-1701-4
44. Zha L, Li F, Wu R, Artinian L, Rehder V, Yu L, et al. The histone demethylase UTX promotes brown adipocyte thermogenic program via coordinated regulation of H3K27 demethylation and acetylation. J Biol Chem. (2015) 290:25151–63. doi: 10.1074/jbc.M115.662650
45. Moore MJ. From birth to death: the complex lives of eukaryotic mRNAs. Science. (2005) 309:1514–8. doi: 10.1126/science.1111443
46. Dai N, Zhao L, Wrighting D, Kramer D, Majithia A, Wang Y, et al. IGF2BP2/IMP2-Deficient mice resist obesity through enhanced translation of Ucp1 mRNA and Other mRNAs encoding mitochondrial proteins. Cell Metab. (2015) 21:609–21. doi: 10.1016/j.cmet.2015.03.006
47. Takahashi A, Adachi S, Morita M, Tokumasu M, Natsume T, Suzuki T, et al. Post-transcriptional Stabilization of Ucp1 mRNA protects mice from diet-induced obesity. Cell Rep. (2015) 13:2756–67. doi: 10.1016/j.celrep.2015.11.056
48. Ameres SL, Zamore PD. Diversifying microRNA sequence and function. Nat Rev Mol Cell Biol. (2013) 14:475–88. doi: 10.1038/nrm3611
49. Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. (2014) 15:509–24. doi: 10.1038/nrm3838
50. Zheng Z, Liu X, Zhao Q, Zhang L, Li C, Xue Y. Regulation of UCP1 in the browning of epididymal adipose tissue by beta3-adrenergic agonist: A Role for MicroRNAs. Int J Endocrinol. (2014) 2014:530636. doi: 10.1155/2014/530636
51. Wang Y, Gao M, Zhu F, Li X, Yang Y, Yan Q, et al. METTL3 is essential for postnatal development of brown adipose tissue and energy expenditure in mice. Nat Commun. (2020) 11:1648. doi: 10.1038/s41467-020-15488-2
52. Huang YS, Lu WH. Decoding hidden messages in neurons: insights from epitranscriptome-controlled and specialized ribosome-controlled translation. Curr Opin Neurobiol. (2018) 48:64–70. doi: 10.1016/j.conb.2017.10.018
53. Odegaard JI, Lee MW, Sogawa Y, Bertholet AM, Locksley RM, Weinberg DE, et al. Perinatal licensing of thermogenesis by IL-33 and ST2. Cell. (2016) 166:841–54. doi: 10.1016/j.cell.2016.06.040
54. Weyer C, Tataranni PA, Snitker S, Danforth E, Ravussin E. Increase in insulin action and fat oxidation after treatment with CL 316,243, a highly selective beta3-adrenoceptor agonist in humans. Diabetes. (1998) 47:1555–61. doi: 10.2337/diabetes.47.10.1555
55. Larsen TM, Toubro S, van Baak MA, Gottesdiener KM, Larson P, Saris WH, et al. Effect of a 28-d treatment with L-796568, a novel beta(3)-adrenergic receptor agonist, on energy expenditure and body composition in obese men. Am J Clin Nutr. (2002) 76:780–8. doi: 10.1093/ajcn/76.4.780
56. Blondin DP, Nielsen S, Kuipers EN, Severinsen MC, Jensen VH, Miard S, et al. Human brown adipocyte thermogenesis is driven by beta2-AR stimulation. Cell Metab. (2020) 32:287–300.e7. doi: 10.1016/j.cmet.2020.07.005
57. Riis-Vestergaard MJ, Richelsen B, Bruun JM, Li W, Hansen JB, Pedersen SB. Beta-1 and not beta-3 adrenergic receptors may be the primary regulator of human brown adipocyte metabolism. J Clin Endocrinol Metab. (2020) 105:e994–1005. doi: 10.1210/clinem/dgz298
58. Porter C, Herndon DN, Chondronikola M, Chao T, Annamalai P, Bhattarai N, et al. Human and mouse brown adipose tissue mitochondria have comparable UCP1 function. Cell Metab. (2016) 24:246–55. doi: 10.1016/j.cmet.2016.07.004
Keywords: alternative polyadenylation, brown adipose tissue, thermogenesis, transcriptional control, translational control, uncoupling protein 1
Citation: Lu W-H, Chang Y-M and Huang Y-S (2021) Alternative Polyadenylation and Differential Regulation of Ucp1: Implications for Brown Adipose Tissue Thermogenesis Across Species. Front. Pediatr. 8:612279. doi: 10.3389/fped.2020.612279
Received: 30 September 2020; Accepted: 30 December 2020;
Published: 09 February 2021.
Edited by:
Ching-Feng Cheng, Taipei Tzu Chi Hospital, TaiwanReviewed by:
Yongguo Li, Technical University of Munich, GermanyHuey-Ling Chen, National Taiwan University Hospital, Taiwan
Copyright © 2021 Lu, Chang and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yi-Shuian Huang, eWlzaHVpYW5AaWJtcy5zaW5pY2EuZWR1LnR3
 Wen-Hsin Lu
Wen-Hsin Lu Yao-Ming Chang
Yao-Ming Chang Yi-Shuian Huang
Yi-Shuian Huang