- 1Department of Pediatric Hematology/Oncology and Cell and Gene Therapy, Istituto di Ricovero e Cura a Carattere Scientifico Bambino Gesù Children's Hospital, Rome, Italy
- 2Laboratory of Medical Genetics, Istituto di Ricovero e Cura a Carattere Scientifico Bambino Gesù Children's Hospital, Rome, Italy
- 3Greenwood Genetic Center, Greenwood, SC, United States
- 4Clemson University School of Health Research, Clemson, SC, United States
- 5Sapienza, University of Rome, Rome, Italy
- 6Neurosurgery Unit, Department of Neurological and Psychiatric Sciences, Istituto di Ricovero e Cura a Carattere Scientifico Bambino Gesù Children's Hospital, Rome, Italy
Pediatric High-Grade Gliomas (pHGG) are among the deadliest childhood brain tumors and can be associated with an underlying cancer predisposing syndrome. The thorough understanding of these syndromes can aid the clinician in their prompt recognition, leading to an informed genetic counseling for families and to a wider understanding of a specific genetic landscape of the tumor for target therapies. In this review, we summarize the main pHGG-associated cancer predisposing conditions, providing a guide for suspecting these syndromes and referring for genetic counseling.
Introduction
Central Nervous System (CNS) tumors are the most common pediatric solid tumors and represent the second most frequent neoplasm in pediatric age, second only to leukemias. They count for 1.12–5.14 cases per 100,000 people in individuals aged 0–19 years, with variable incidence rates across different countries, the highest being in the USA (1). Management of pediatric CNS tumors is challenging and requires specific oncological training.
Among brain tumors in the pediatric age, gliomas are the most represented. Approximately 21% of all primary pediatric gliomas are high-grade tumors (2, 3). Even though from a histopathological point of view pediatric high-grade gliomas (pHGGs) are similar to their adult counterpart, their genetic and epigenetic features reflect intrinsic differences compared to adult HGGs. Despite an increased understanding of their biological basis, therapeutic options for these tumors are still very limited, and the long-term prognosis remains poor, with high levels of both morbidity and mortality (3, 4) and a 5-year survival rate of <20% (4).
Risk factors for pHGG seem to be mostly genetic in nature, even though some predisposing environmental factors such as irradiation have been described (5). In contrast to adult population, where cancer associated mutations are mostly somatic and resulting from external causes, germline mutations are frequently encountered in children.
Several cancer predisposing syndromes (CPS) associated with an increased risk of developing to pHGG have been identified so far, including Neurofibromatosis type 1 (NF1), Turcot syndrome and Li-Fraumeni syndrome. In this review we will address the impact of these syndromes for the management of pHGG.
Methods
The authors conducted a literature search describing CNS tumors and cancer predisposing syndromes. Selection of studies were based on research topics (such as cancer predisposition syndrome AND/OR brain tumor genetics, brain tumor cancer predisposition syndrome, HGG predisposition syndromes, HGG in childhood) found in the PubMed. Only papers written in the English language and those published from the year 2000 up to May 2020 were selected. We included reviews, case series and research studies that were classified according to their relevance. No abstracts were included. The information found in the selected studies was carefully evaluated, which is described and discussed in the following sections.
LI-Fraumeni Syndrome
Li-Fraumeni Syndrome (LFS) (OMIM #151623) was reported for the first time in 1969 by Frederick Li and Joseph Fraumeni (6). LFS is an autosomal dominant, highly penetrant cancer predisposition syndrome associated with germline mutations in the TP53 gene. It lacks additional clinical features and is only characterized by the high frequency of malignancies in multiple organs, making it a difficult syndrome to diagnose in the absence of a significant family history of multiple cancers (7). The involved gene encodes the TP53 transcription factor, tumor protein p53, also known as the “guardian of the genome” (8). TP53 is involved in cellular growth control by regulating the expression of several genes causing cell-cycle arrest and apoptosis in response to DNA damage.
Epidemiology and Cancer Spectrum
LFS prevalence is estimated between 1/5.000 and 1/20.000 of the population (9, 10), even if the estimated prevalence of pathogenic and likely pathogenic germline TP53 variants seems to be higher, as described by Andrade et al. (9).
LFS is characterized by a high lifetime cancer risk and, due to its extremely high penetrance, by a familiar clustering of tumors. Cancer types are variable and often present during childhood. Osteosarcoma, soft-tissue sarcomas, brain tumors, early-onset breast cancer, leukemia, and adrenocortical tumors are the most frequently observed tumors (10). It can also be associated with myelodysplastic syndromes, lymphoma and other benign and malignant tumors (11, 12). In children with LFS, brain tumors are the second most common malignancies following adrenocortical carcinoma. A quarter of childhood tumors involved CNS compared to only 13% of adult LFS related tumors (13). In LFS, the median age of onset of brain tumors is 16 years, compared to 57 years in the general population.
CNS tumors related to LFS have a prevalence ranging from 9 to 14% (14) and the most frequent types are glioblastoma and astrocytoma. Nonetheless, medulloblastoma, ependymoma, choroid plexus carcinomas, and other embryonal tumors are also described.
Etiopathology
The main gene disrupted in LSF is TP53, a tumor suppressor gene encoding the p53 protein, fundamental for the transcription of target genes involved in cell cycle arrest, DNA repair and response to DNA damage (15). TP53 gene is located on chromosome 17p13.1 and more than 250 different germline alterations have been reported in medical literature to date. In brain tumors, most mutations reside within the DNA binding domain, even though all the genotypic-phenotypic correlations are not fully understood (16). Despite genetic lesions in LFS have been widely studied, not all the underlying genetic defects responsible for LFS have been found. In fact, several families fulfill the definition of classical LFS without the recognition of any known TP53 defect being found (16). Although few LFS cases have been reported with germline mutations in the CHK2 gene, no pediatric CNS tumors have been detected in these patients, suggesting a genotype-phenotype correlation between such malignancies and TP53 mutations (17, 18). See Figure 1 for details.
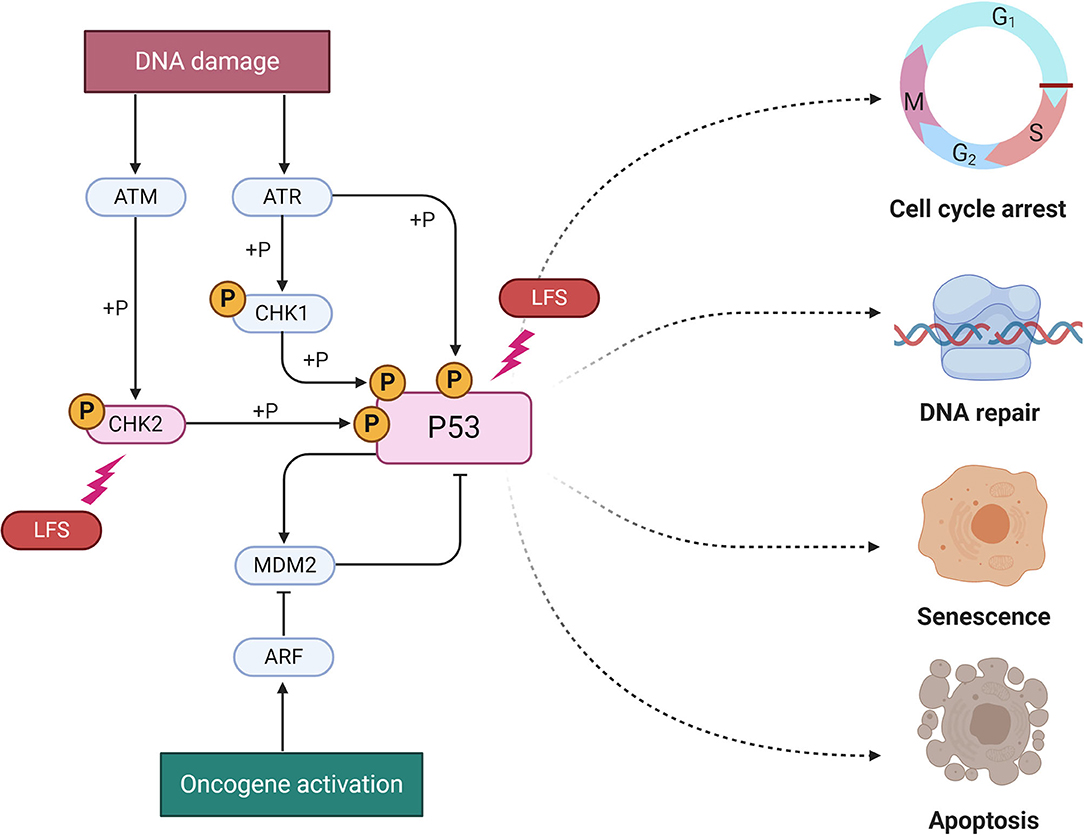
Figure 1. Molecular pathways of Li-Fraumeni (LFS) Syndrome. The two known mutation for LFS are represented here (P53 and CHK2) as lighting bolt. A group of protein kinases such as ATM, ATR, CHK1, CHK2 is implicated in the genome integrity checkpoint, a molecular cascade that detects and responds to several forms of DNA damage caused by genotoxic stress. Oncogenes also stimulate p53 activation, mediated by the protein ARF. In a normal cell, p53 is inactivated by its negative regulator, MDM2. Upon oncogene activation, various pathways will lead to the dissociation of the P53 and MDM2 complex. Once activated, p53 will induce a cell cycle arrest to allow either repair and survival of the cell or apoptosis to discard the damaged cell. Adapted from “P53 Regulation and Signaling,” by BioRender.com (2020). Available online at: https://app.biorender.com/biorender-templates.
Clinical and Therapeutic Considerations
As already mentioned, there are no clinical characteristics associated with LFS other than an increased cancer risk. Considering this and the highly penetrance of LFS, clinical and familial diagnostic criteria are essential for the diagnosis. Classic diagnostic criteria and revised Chompret criteria for LFS are reported in Supplementary Table 1 (19). It is essential to individuate families where LFS might be present as it has been demonstrated that intense tumor surveillance leads to increased survival (20).
It has been proven that TP53 mutations are a negative prognostic factor in several tumor types, including pHGG (21). Despite the high risk of secondary malignancies after exposure to ionizing radiation, no specific treatment is available for LFS pHGG patients. Treatment strategies in these patients can be challenging, since mutations in the TP53 gene have been associated with resistance to both chemotherapy and radiation (22). Also, LFS patients with CNS tumors show an overall worse outcome if compared to non-affected patients (22).
Neurofibromatosis
Neurofibromatosis type 1 (NF1) (OMIM #162200), also known as von Recklinghausen disease, is a common autosomal dominant disorder with a prevalence of 1:4.000 individuals due to mutations of the NF1 gene on chromosome 17q11.2 (23). The protein product of the NF1 gene, neurofibromin, regulates several intracellular processes, including the RAS/ERK/MAP kinase cascade and cytoskeletal assembly. Loss-of-function mutations of NF1 gene lead to a high risk of tumor development due to decreased RAS signaling inhibition (24). Clinically, NF1 is characterized by café au lait macules, skin fold freckling, optic pathway gliomas, neurofibromas and plexiform neurofibromas, osseous lesions, and iris hamartomas (Lisch nodules) (23). The clinical diagnosis requires the fulfillment of at least two of the criteria as listed in Supplementary Table 2, however there are other possible manifestations that are not included in the diagnostic criteria but that can be present in patients harboring the mutation, such as macrocephaly, learning disabilities, vasculopathies and scoliosis. NF1 is associated with some CNS neoplasms in infancy, namely optic pathway gliomas and brainstem gliomas.
Epidemiology and Cancer Spectrum
NF1 (von Recklinghausen disease) is one of the most common CPS (13). It is an autosomal dominant inherited condition and about 50% of cases are found de novo with no associated family history (25).
CNS neoplasms predominantly associated with NF1 are optic pathway gliomas (15–20%) and brainstem -gliomas (1–2%). Other malignant tumors can be observed in these patients such as malignant peripheral nerve sheath tumors (MPNST) and juvenile myelomonocytic leukemia (JMML) (26).
Etiopathology
The gene involved in the pathogenesis of this syndrome is NF1, an onco-suppressor located on chromosome 17q11.2. The protein encoded by this gene is called Neurofibromin and is a GTPase activating protein that inhibits the product of the RAS oncogene, mediating the passage from GTP-RAS to GDP-RAS. RAS, in turn, is an activator of cell-cycle signaling pathways such as MAPK (RAF-MEK-ERK) and PI3K/AKT/mTOR pathways (27). NF1 loss-of-function mutations remove this inhibition on RAS and the downstream pathways, leading to abnormal cell proliferation and tumorigenesis.
Clinical and Therapeutic Considerations
NF1 brain tumors are considered more indolent than same histology counterparts observed in patients without NF1, and can even regress over time without treatment (28). Histologically, most of them are low-grade gliomas (LGG), with a smaller representation of pHGG (81). It is notable that NF1 associated pHGG exhibit the same genetic alterations found in sporadic pHGG (such as P53 and CDKN2A alterations) (29). On the other hand, NF1 alterations are frequently found as somatic genetic lesions in sporadic HGGs of childhood (30).
Apart from LGG, differential diagnosis of pHGG in NF1 children has to include the frequent finding of Focal Areas of Signal Intensity (FASI) in these patients. These are benign lesions, usually multiple and radiologically characterized as non-enhancing, small areas without mass effect or edema. They can be found in around 70% of NF1 pediatric cases and must be differentiated from gliomas (31).
Being pHGGs very uncommon in NF1, surveillance neuroimaging is controversial and not universally recommended (24). Regardless, families should be instructed to recognize the warning signs of brain tumors.
Treatment of pHGG in NF1 is similar to sporadic cases, some reports suggest that prognosis might be better than sporadic pHGG (32, 33). As for target specific therapies, MEK inhibitors have shown promising results in NF1 patients with low grade gliomas, this result may pose the basis for future treatment strategies also in NF1-pHGG (34).
Radiotherapy is generally part of the treatment protocol, despite increased complications, namely secondary malignancies and stroke (35).
Constitutional Mismatch-Repair Deficiency Syndrome
Constitutional mismatch repair deficiency (CMMRD) syndrome (OMIM #276300) is a childhood autosomal recessive cancer predisposition syndrome caused by a biallelic germline mutation in the DNA mismatch repair (MMR) genes, namely mutL homolog1 (MLH1), mutS homolog1 (MSH2), pms2 c-terminal like pseudogene (PMS2), or mutS homolog6 (MSH6) (36). Patients with monoallelic mutations in the MMR genes develop hereditary non-polyposis colorectal carcinoma (HNPCC), also known as Lynch syndrome, an autosomal dominant genetic disorder associated with increased risk of colorectal cancer, endometrial carcinoma, and other gastrointestinal and genitourinary malignancies in the fourth and fifth decades of life (37).
Epidemiology and Cancer Spectrum
CMMRD is a rare disease with roughly 200 cases reported to date (38, 39). However, its prevalence might be underestimated and a consistent number of cases might go undiagnosed in South Asian and Middle Eastern countries where consanguinity is more prevalent (40). In CMMRD, the tumor spectrum is very broad including CNS (glioblastoma, oligodendroglioma, low-grade glioma, medulloblastoma, and other embryonal tumors), hematological, genitourinary and intestinal tract tumors (41). Among brain tumors, malignant gliomas are the most frequent CMMRD-associated tumors, typically presenting within the first 2 decades of life and accounting for 25–40% of CMMRD cancers (41). Overall, there is a high degree of consanguinity within the family, indicating that inbreeding is a major risk factor for this otherwise rare disorder.
Etiopathology
MSH2, MSH6, MLH1, and PMS2 genes are involved in the mismatch repair mechanisms, one of the most important DNA repair machinery of the cell (36). Its main role is to correct errors arising during DNA replication, thus tumors arising in the context of CMMRD exhibit an extraordinary number of DNA mutations. The most common type of defects found in these “hypermutated cancers” are point mutations (single nucleotide variations) and microsatellite instability (MSI) where repetitive sequences (microsatellites) are not adequately repaired. Recently, new genetic alterations affecting this machinery have been described, such as MSH3 variants (42), deletions of the EPCAM gene (43), and mutations in DNA polymerases epsilon and delta 1 (POLE, POLD1) (44). See Figure 2 for details.
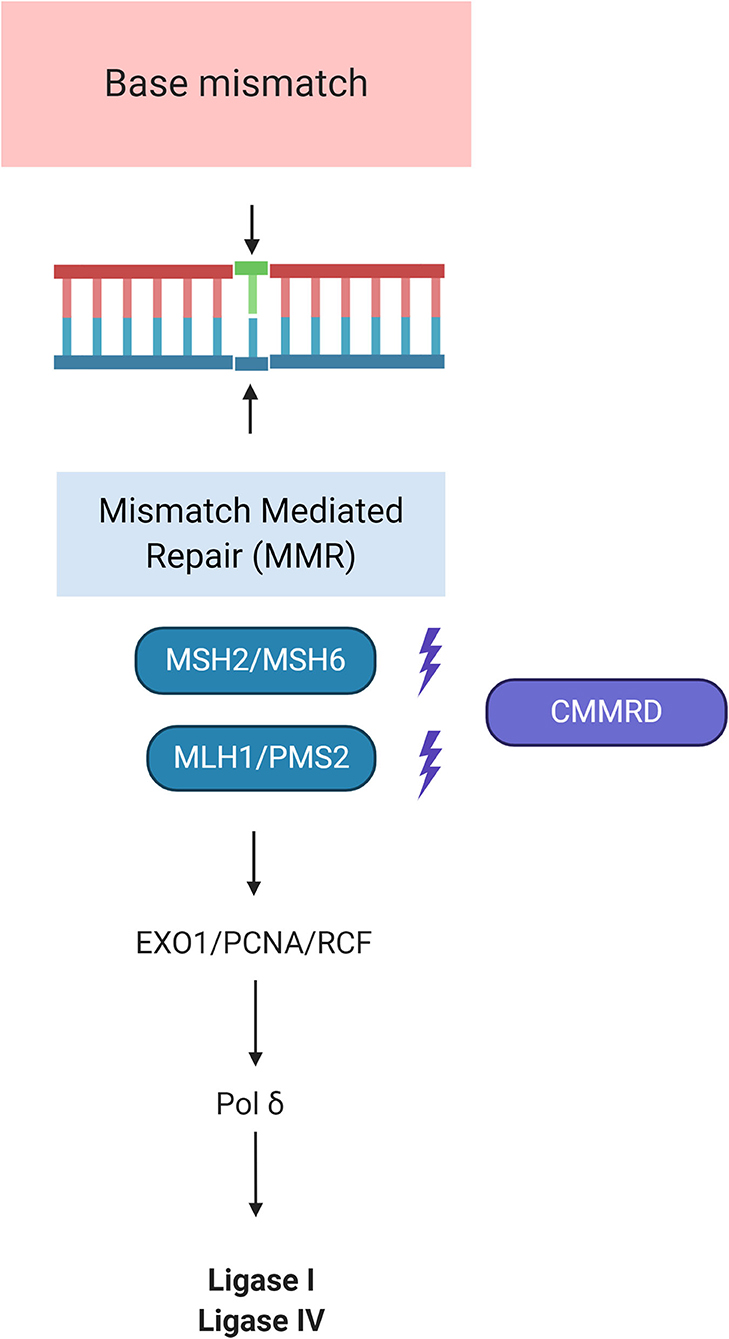
Figure 2. Molecular pathways of Constitutional Mismatch Repair Deficiency Syndrome (CMMRD). MSH2 dimerizes with MSH6 to form the MutSα complex, which is involved in base mismatch repair and short insertion/deletion loops. The formation of the MSH2-MSH6 heterodimer accommodates a second heterodimer of MLH1 and PMS2. This protein complex formed between the 2 sets of heterodimers enables initiation of repair of the mismatch defect by recruiting PCN/EXO1/RCF. RFC is essential for PCNA loading and function in DNA replication. PCNA loads onto double-strand breaks and promotes Exo1 damage association through direct interaction with Exo1. By tethering Exo1 to the DNA substrate, PCNA confers processivity to Exo1 in resection. This role of PCNA in DNA resection is analogous to its function in DNA replication where PCNA serves as a processivity co-factor for DNA polymerases such as polymerases δ. DNA Pol δ is an enzyme used for both leading and lagging strand synthesis by engaging Ligase I and IV. Adapted from “DNA Repair Mechanisms by BioRender.com (2020). Available online at: https://app.biorender.com/biorender-templates.
Clinical and Therapeutic Considerations
In addition to cancer, CMMRD patients frequently have other physical features such as cutaneous café-au-lait spots and hypo- or hyperpigmented spots that may mimic some of the skin features usually observed in NF1. Also neurofibromas, Lisch nodules and freckling have been reported, although less frequently than in NF1 (39, 45). Other findings have occasionally been described in these patients such as vascular anomalies, pilomatrixomas, agenesis of the corpus callosum (46), and decreased levels of immunoglobulins IgG2/4 and IgA (39). However, none of these features are mandatory to diagnose the syndrome. The penetrance of the disease is very high, reaching more than 90% by the first two decades of life. Most patients will have childhood cancer and more than one tumor, often presenting synchronously (13).
Initial screenings can be performed by immunohistochemistry showing loss of MMR protein both in normal and malignant cells. Diagnosis can be confirmed by genetic testing for the presence of biallelic mutations in one of the four MMR genes. Evidence of low grade glial lesions and premalignant, dysplastic polyps advocates for surveillance protocols to intercept asymptomatic tumors at early stages, when they are more amenable to complete resection (47). Current protocols suggest annual whole-body MRI (WBMRI) from the age of 6 years. In addition, it is recommended to start colon surveillance by colonoscopy from 6 years of age. Treatment of CMMRD tumors is complicated by resistance to standard therapies for pHGG such as temozolomide, since it requires adequate mismatch repair to perform its action.
Interestingly, immunotherapy has proved to be a promising strategy in these tumors. One of the main mechanisms through which tumors escape immune recognition and induce immunosuppression is PD-L1 overexpression of cancers that acts as a binding site for PD1. The binding of PD1 to PDL1 activates PD1 signaling that inhibits T cells allowing the tumor to evade immune attack (48). These principles have been used to develop drugs named checkpoint inhibitors that counteract the interactions of the PD1 protein. It has been demonstrated that CMMRD tumors are more responsive to PD1 blockers than MMR proficient tumors. In particular, in children with CMMRD with recurrent glioblastoma, shrinking of tumors was observed on MRI, suggesting these tumors as ideal candidates for such therapies (49).
Ollier Disease and Maffucci Syndrome
Ollier disease (OD, OMIM 166000) and Maffucci syndrome (MS, OMIM 6145692) are related conditions characterized by multiple endochondromas and caused by somatic mutations in the IDH1 and IDH2 genes, respectively (50, 51). The main difference between the two conditions is the presence of hemangiomata in MS, moreover, while OD presents with multiple enchondromas, typically unilateral in distribution with a predilection for the appendicular skeleton, MS is often characterized by multiple enchondromas bilaterally distributed (51).
Epidemiology and Cancer Spectrum
Most cases of OD and MS have been reported as sporadic, with an estimated prevalence of 1 out of 100,000 individuals, although the description of few familial cases of OD suggests a possible autosomal dominant pattern of inheritance (51). About half of the individuals with OD or MS develop a malignancy, such as chondrosarcoma (with a prevalence of 30% in both conditions), glioma, and ovarian juvenile granulosa cell tumor, accompanied by other clinical features, such as multiple swellings on the extremity, deformity around the joints, limitations in joint mobility, scoliosis, bone shortening, leg-length discrepancy, gait disturbances, pain, loss of function, and pathological fractures (51).
Etiopathology
Mutations in the IDH1 or IDH2 genes have been detected in a large number of adult diffuse grade II and grade III gliomas; such high frequency has suggested a possible role for those variants as the earliest oncogenic event in these malignancies (52). It has been proven that pathogenic variants in these two genes cause an abnormal production of 2-hydroxyglutarate (2-HG), a structural analog of alpha-ketoglutarate, a key intermediate of the Krebs cycle. 2-HG competitively inhibits the active sites of multiple alpha-ketoglutarate enzymes, resulting in hypermethylation of histones and DNA, altered cell differentiation, and activation of a series of downstream enzymes (53, 54). Some of these enzymes are involved in the degradation of HIF-1 (hypoxia-induced factor 1), a key player in the cellular adaptation to low oxygen and nutrient-deprived environment and in the progression to malignancy in human solid cancers, and in the overexpression of platelet-derived growth factor receptor A (PDGFRA), implicated in the pathogenesis of leukemias, lymphomas, gastrointestinal stromal tumors, and various types of brain tumors (53–55).
Clinical and Therapeutic Considerations
The clinical management of individuals with OD and MS is mostly focused on treating via surgery the complications arising from the enchondromas, such as fractures, growth defects, and tumors. The prevailing strategy aims to treat and remove any extraneous bone tissue preserving the limb function (51). Although gliomas are not the most frequent types of malignancies reported in OD and MS, imaging surveillance is recommended. The gliomas described in these conditions are similar to the ones caused by sporadic variants in IDH1 or IDH2 for their frequent location in the frontal lobe and their prevalent histological type: more commonly diffuse low-grade or anaplastic gliomas than glioblastomas (53). However, they present some substantial differences as compared to the sporadic forms: they are diagnosed at an earlier age and involve more frequently the brainstem, hinting toward an earlier origin of gliomas associated with enchondromatosis.
Other Syndromes and pHGG
Some less-known syndromes have been associated with pHGG with lower frequency than the afore-mentioned syndromes.
One of those is the Familial Melanoma Astrocytoma Syndrome (56, 57). It is caused by germline inactivating deletion of the CDKN2A tumor suppressor gene. Affected individuals have a predisposition to develop melanoma and CNS tumors, most commonly astrocytoma.
Since familial predisposition to glioma has been consistently observed within non-syndromic families, an international consortium named GLIOGENE was formed in order to collect such non-syndromic glioma families, and possibly identify new genes involved in the pathogenesis of these tumors. One of the genomic regions identified by the consortium lies in chromosome 17q. According to these linkage studies the MYO19 and KIF18B genes and rare variants in SPAG9 and RUNDC1 have been identified as candidates worthy of further investigation (58). Also, whole exome sequencing allowed the identification of mutations in POT1 (p.G95C, p.E450X), a member of the telomere shelterin complex (59). These new findings may not only have a leading role in identifying new pathogenic pathways in gliomas but may also contribute to improve targeted treatment of this disease.
Mutations in BRCA1 and BRCA2, tumor suppressor genes involved in DNA repair, have been traditionally associated with an increased risk of breast and ovarian cancer. More recently, they have been recognized to also play a role in CNS tumors (60). In particular, germline variants of BRCA2 which is also essential for normal neurogenesis (61) have been described in individuals with brain tumors including glial tumors, meningioma and medulloblastoma (62–64).
There have been some anecdotal reports of pHGG in other syndromes (65), such as tuberous sclerosis (66), Beckwith-Wiedemann and Fanconi Anemia (67). However, these case reports do not prove a real increased risk for pHGG.
Molecular Diagnostics of CPSs
Genetic testing in pediatric oncology is of great interest for the investigation into potentially underlying CPSs. Molecular diagnosis of a CPS can influence cancer surveillance program initiation or frequency, and directly impact treatment decisions. Genetic diagnostic laboratories have introduced next-generation sequencing (NGS) technologies into their practices. NGS has specific advantages over traditional Sanger sequencing, considered the gold standard for mutation analysis for many years, as multiple genes in several patients can be tested simultaneously. Different approaches are being used, and currently, most laboratories that use these technologies are performing targeted gene panel testing or clinical whole exome sequencing (WES), more rarely whole genome sequencing (WGS). These revolutionary technological advances have drastically reduced sequencing costs and shortened the turnaround time, increasing the detection rate (68). Multi-gene panels usually include high and moderate penetrance genes, and sometimes, some low or unknown risk genes, that offer the advantage of identifying germline pathogenic variants in genes that would not normally be tested based on the patient's diagnosis (69). Unfortunately, depending on the disease, between 70 and 92% of the patients remain mutation-negative or undiagnosed after gene-panel testing (70). It is possible that variants in genes not included in these panels contribute to the cancer risk and WES or WGS can explore the genetic basis of familial syndromes in a more extensive way, permitting to identify new high- and moderate-risk cancer predisposition genes. WES of parent-child trios has become a widely used strategy to identify presumably pathogenetic genetic variants in children with rare diseases. However, it has not yet been routinely implemented in pediatric oncology, with few exceptions (71). Genome-wide approaches generate huge amounts of genetic data and it remains challenging to interpret the identified variants. Such data interpretation needs close collaboration among bioinformaticians, molecular geneticists and clinicians. However, as sequencing costs are decreasing and computer and technological resources are expanding, genome-wide analysis will become more common in the clinical practice and hopefully help to advance on the path of personalized medicine, by providing more precise genetic diagnoses and better molecular information for more effective treatments.
DNA Methylation Profiling
Recently, a machine learning approach for classification of CNS tumors based on the analysis of global DNA methylation profiling has been developed and introduced to reach a histopathological-molecular integrated diagnosis, discriminating tumor classes and ameliorating diagnostic precision (72, 73). In detail, the developed “Classifer” provides a methylation-based classification assigning a subgroup score for an index tumor compared to 91 different brain tumor entities. Furthermore, it also provides a chromosomal copy-number variation (CNV) analysis.
Interestingly, Capper and colleagues found that a high proportion of unclassifiable CNS tumors were associated with various hereditary tumor syndromes, and/or diagnosed in childhood (73). Additional chromothripsis and unusual complex chromosomal changes should also be considered as a cue for Li–Fraumeni syndrome-associated tumors.
Conclusions
Pediatric HGG cancer predisposition syndromes are rare and diverse pathological conditions that may be present in children with CNS tumors and deserve consideration.
Knowing when to suspect one of these predisposing syndromes is essential for the pediatric oncologist, not only to make the correct diagnosis, but also to formulate a more accurate prognostic judgment and provide an adequate treatment. Moreover, it is mandatory to refer the family for genetic counseling when such conditions are suspected. This latter aspect is of particular relevance since it has been demonstrated that close surveillance can decrease the morbidity and mortality in these patients.
The ever-growing knowledge of the genetic mechanisms underlying cancer is a key tool in the understanding of this disease, opening new scenarios for the introduction of molecular target therapy.
Since these conditions are extremely rare, several patients' associations have been created to help families find the nearest structure for follow-up and to raise funds and consciousness for these diseases.
Author Contributions
GC, GD, AM, and AC designed the study. GC, GD, MV, LB, RC, and EM cured the literature research and its organization. GC, EA, FD, and MR cured the literature research focusing on the genetics aspect. GC, GD, EM, and LB wrote the final version of the manuscript. AM, AC, FL, and LB critically revised the manuscript for intellectual content. Finally, all authors approved the version to be published and agreed to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Funding
We thank the association Heal for the support.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The author thanks Megan Eckley for the english language editing.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fped.2020.561487/full#supplementary-material
References
1. Johnson KJ, Cullen J, Barnholtz-Sloan JS, Ostrom QT, Langer CE, Turner MC, et al. Childhood brain tumor epidemiology: a brain tumor epidemiology consortium review. Cancer Epidemiol Biomarkers Prev. (2014) 23:2716–36. doi: 10.1158/1055-9965.EPI-14-0207
2. Sturm D, Pfister SM, Jones DTW. Pediatric gliomas: current concepts on diagnosis, biology, and clinical management. J Clin Oncol. (2017) 35:2370–7. doi: 10.1200/JCO.2017.73.0242
3. Juratli TA, Qin N, Cahill DP, Filbin MG. Molecular pathogenesis and therapeutic implications in pediatric high-grade gliomas. Pharmacol Ther. (2018) 182:70–9. doi: 10.1016/j.pharmthera.2017.08.006
4. Braunstein S, Raleigh D, Bindra R, Mueller S, Haas-Kogan D. Pediatric high-grade glioma: current molecular landscape and therapeutic approaches. J Neurooncol. (2017) 134:541–9. doi: 10.1007/s11060-017-2393-0
5. Pettorini BL, Park Y-S, Caldarelli M, Massimi L, Tamburrini G, Di Rocco C. Radiation-induced brain tumours after central nervous system irradiation in childhood: a review. Childs Nerv Syst. (2008) 24:793–805. doi: 10.1007/s00381-008-0631-7
6. Li FP, Fraumeni JF. Rhabdomyosarcoma in children: epidemiologic study and identification of a familial cancer syndrome. J Natl Cancer Inst. (1969) 43:1365–73.
7. Malkin D, Li FP, Strong LC, Fraumeni JF, Nelson CE, Kim DH, et al. Germ line p53 mutations in a familial syndrome of breast cancer, sarcomas, and other neoplasms. Science. (1990) 250:1233–8. doi: 10.1126/science.1978757
8. Royds JA, Iacopetta B. p53 and disease: when the guardian angel fails. Cell Death Differ. (2006) 13:1017–26. doi: 10.1038/sj.cdd.4401913
9. de Andrade KC, Frone MN, Wegman-Ostrosky T, Khincha PP, Kim J, Amadou A, et al. Variable population prevalence estimates of germline TP53 variants: a gnomAD-based analysis. Hum Mut. (2018) 40:97–105. doi: 10.1002/humu.23673
10. Mai PL, Best AF, Peters JA, DeCastro RM, Khincha PP, Loud JT, et al. Risks of first and subsequent cancers among TP53 mutation carriers in the National Cancer Institute Li-Fraumeni syndrome cohort. Cancer. (2016) 122:3673–81. doi: 10.1002/cncr.30248
11. Nichols KE, Malkin D, Garber JE, Fraumeni J, Li FP. Germ-line p53 mutations predispose to a wide spectrum of early-onset cancers. Cancer Epidemiol Biomarkers Prev. (2001) 10:83–7.
12. Nichols KE, Malkin D, Garber JE, Fraumeni JF, Li FP. Germ-line p53 mutations predispose to a wide spectrum of early-onset cancers. Cancer Epidemiol Biomarkers Prev. (2001) 10:83–7.
13. Michaeli O, Tabori U. Pediatric high grade gliomas in the context of cancer predisposition syndromes. J Korean Neurosurg Soc. (2018) 61:319–32. doi: 10.3340/jkns.2018.0031
14. Bougeard G, Renaux-Petel M, Flaman J-M, Charbonnier C, Fermey P, Belotti M, et al. Revisiting Li-fraumeni syndrome from TP53 mutation carriers. J Clin Oncol. (2015) 33:2345–52. doi: 10.1200/JCO.2014.59.5728
15. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. (2011) 144:646–74. doi: 10.1016/j.cell.2011.02.013
16. Olivier M, Goldgar DE, Sodha N, Ohgaki H, Kleihues P, Hainaut P, et al. Li-Fraumeni and related syndromes: correlation between tumor type, family structure, and TP53 genotype. Cancer Res. (2003) 63:6643–50.
17. Bell DW, Varley JM, Szydlo TE, Kang DH, Wahrer DC, Shannon KE, et al. Heterozygous germ line hCHK2 mutations in Li-Fraumeni syndrome. Science. (1999) 286:2528–31. doi: 10.1126/science.286.5449.2528
18. Vahteristo P, Tamminen A, Karvinen P, Eerola H, Eklund C, Aaltonen LA, et al. p53, CHK2, and CHK1 genes in Finnish families with Li-Fraumeni syndrome: further evidence of CHK2 in inherited cancer predisposition. Cancer Res. (2001) 61:5718–22.
19. Ruijs MWG, Verhoef S, Rookus MA, Pruntel R, van der Hout AH, Hogervorst FBL, et al. TP53 germline mutation testing in 180 families suspected of Li-Fraumeni syndrome: mutation detection rate and relative frequency of cancers in different familial phenotypes. J Med Genet. (2010) 47:421–8. doi: 10.1136/jmg.2009.073429
20. Villani A, Tabori U, Schiffman J, Shlien A, Beyene J, Druker H, et al. Biochemical and imaging surveillance in germline TP53 mutation carriers with Li-Fraumeni syndrome: a prospective observational study. Lancet Oncol. (2011) 12:559–67. doi: 10.1016/S1470-2045(11)70119-X
21. Eisenstat DD, Pollack IF, Demers A, Sapp MV, Lambert P, Weisfeld-Adams JD, et al. Impact of tumor location and pathological discordance on survival of children with midline high-grade gliomas treated on children's cancer group high-grade glioma study CCG-945. J Neurooncol. (2015) 121:573–81. doi: 10.1007/s11060-014-1669-x
22. Tabori U, Shlien A, Baskin B, Levitt S, Ray P, Alon N, et al. TP53 alterations determine clinical subgroups and survival of patients with choroid plexus tumors. J Clin Oncol. (2010) 28:1995–2001. doi: 10.1200/JCO.2009.26.8169
23. Gutmann DH, Ferner RE, Listernick RH, Korf BR, Wolters PL, Johnson KJ. Neurofibromatosis type 1. Nat Rev Dis Primers. (2017) 3:17004. doi: 10.1038/nrdp.2017.4
24. Evans DGR, Salvador H, Chang VY, Erez A, Voss SD, Schneider KW, et al. Cancer and central nervous system tumor surveillance in pediatric neurofibromatosis 1. Clin Cancer Res. (2017) 23:e46–53. doi: 10.1158/1078-0432.CCR-17-0589
25. Ferner RE, Huson SM, Thomas N, Moss C, Willshaw H, Evans DG, et al. Guidelines for the diagnosis and management of individuals with neurofibromatosis 1. J Med Genet. (2007) 44:81–8. doi: 10.1136/jmg.2006.045906
26. Jett K, Friedman JM. Clinical and genetic aspects of neurofibromatosis 1. Genet Med. (2010) 12:1–11. doi: 10.1097/GIM.0b013e3181bf15e3
27. Scheffzek K, Ahmadian MR, Wiesmüller L, Kabsch W, Stege P, Schmitz F, et al. Structural analysis of the GAP-related domain from neurofibromin and its implications. EMBO J. (1998) 17:4313–27. doi: 10.1093/emboj/17.15.4313
28. Listernick R, Charrow J, Greenwald MJ, Esterly NB. Optic gliomas in children with neurofibromatosis type 1. J Pediatr. (1989) 114:788–92. doi: 10.1016/S0022-3476(89)80137-4
29. Gutmann DH, James CD, Poyhonen M, Louis DN, Ferner R, Guha A, et al. Molecular analysis of astrocytomas presenting after age 10 in individuals with NF1. Neurology. (2003) 61:1397–400. doi: 10.1212/WNL.61.10.1397
30. Cancer Genome Atlas Research Networ. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. (2008) 455:1061–8. doi: 10.1038/nature07385
31. Khatua S, Gutmann DH, Packer RJ. Neurofibromatosis type 1 and optic pathway glioma: molecular interplay and therapeutic insights. Pediatr Blood Cancer. (2018) 65. doi: 10.1002/pbc.26838
32. Huttner AJ, Kieran MW, Yao X, Cruz L, Ladner J, Quayle K, et al. Clinicopathologic study of glioblastoma in children with neurofibromatosis type 1. Pediatr Blood Cancer. (2010) 54:890–6. doi: 10.1002/pbc.22462
33. Shibahara I, Sonoda Y, Suzuki H, Mayama A, Kanamori M, Saito R, et al. Glioblastoma in neurofibromatosis 1 patients without IDH1, BRAF V600E, and TERT promoter mutations. Brain Tumor Pathol. (2018) 35:10–8. doi: 10.1007/s10014-017-0302-z
34. Banerjee A, Jakacki RI, Onar-Thomas A, Wu S, Nicolaides T, Young Poussaint T, et al. A phase I trial of the MEK inhibitor selumetinib (AZD6244) in pediatric patients with recurrent or refractory low-grade glioma: a Pediatric Brain Tumor Consortium (PBTC) study. Neuro-oncology. (2017) 19:1135–44. doi: 10.1093/neuonc/now282
35. Sharif S, Ferner R, Birch JM, Gillespie JE, Gattamaneni HR, Baser ME, et al. Second primary tumors in neurofibromatosis 1 patients treated for optic glioma: substantial risks after radiotherapy. J Clin Oncol. (2006) 24:2570–5. doi: 10.1200/JCO.2005.03.8349
36. Wimmer K, Etzler J. Constitutional mismatch repair-deficiency syndrome: have we so far seen only the tip of an iceberg? Hum Genet. (2008) 124:105–22. doi: 10.1007/s00439-008-0542-4
37. Lynch HT, Lynch PM, Lanspa SJ, Snyder CL, Lynch JF, Boland CR. Review of the Lynch syndrome: history, molecular genetics, screening, differential diagnosis, and medicolegal ramifications. Clin Genet. (2009) 76:1–18. doi: 10.1111/j.1399-0004.2009.01230.x
38. Bakry D, Aronson M, Durno C, Rimawi H, Farah R, Alharbi QK, et al. Genetic and clinical determinants of constitutional mismatch repair deficiency syndrome: report from the constitutional mismatch repair deficiency consortium. Eur J Cancer. (2014) 50:987–96. doi: 10.1016/j.ejca.2013.12.005
39. Wimmer K, Kratz CP, Vasen HFA, Caron O, Colas C, Entz-Werle N, et al. Diagnostic criteria for constitutional mismatch repair deficiency syndrome: suggestions of the European consortium “care for CMMRD” (C4CMMRD). J Med Genet. (2014) 51:355–65. doi: 10.1136/jmedgenet-2014-102284
40. Amayiri N, Tabori U, Campbell B, Bakry D, Aronson M, Durno C, et al. High frequency of mismatch repair deficiency among pediatric high grade gliomas in Jordan. Int J Cancer. (2016) 138:380–5. doi: 10.1002/ijc.29724
41. Kim B, Tabori U, Hawkins C. An update on the CNS manifestations of brain tumor polyposis syndromes. Acta Neuropathol. (2020) 139:703–15. doi: 10.1007/s00401-020-02124-y
42. Adam R, Spier I, Zhao B, Kloth M, Marquez J, Hinrichsen I, et al. Exome sequencing identifies biallelic MSH3 germline mutations as a recessive subtype of colorectal adenomatous polyposis. Am J Hum Genet. (2016) 99:337–51. doi: 10.1016/j.ajhg.2016.06.015
43. Ligtenberg MJL, Kuiper RP, Chan TL, Goossens M, Hebeda KM, Voorendt M, et al. Heritable somatic methylation and inactivation of MSH2 in families with Lynch syndrome due to deletion of the 3' exons of TACSTD1. Nat Genet. (2009) 41:112–7. doi: 10.1038/ng.283
44. POLE and POLD1 Screening in 155 Patients With Multiple Polyps and Early-Onset Colorectal Cancer. Available online at: https://www-ncbi-nlm-nih-gov.opbg.clas.cineca.it/pubmed/?term=POLE+and+POLD1+screening+in+155+patients+with+multiple+polyps+and+early-onset+colorectal+cancer (accessed February 11, 2020).
45. Connections between constitutional mismatch repair deficiency syndrome neurofibromatosis type 1. Available online at: https://www-ncbi-nlm-nih-gov.opbg.clas.cineca.it/pubmed/?term=Connections+between+constitutional+mismatch+repair+deficiency+syndrome+and+neurofibromatosis+type+1 (accessed February 11, 2020).
46. Baas AF, Gabbett M, Rimac M, Kansikas M, Raphael M, Nievelstein RA, et al. Agenesis of the corpus callosum and gray matter heterotopia in three patients with constitutional mismatch repair deficiency syndrome. Eur J Hum Genet. (2013) 21:55–61. doi: 10.1038/ejhg.2012.117
47. Durno CA, Aronson M, Tabori U, Malkin D, Gallinger S, Chan HSL. Oncologic surveillance for subjects with biallelic mismatch repair gene mutations: 10 year follow-up of a kindred. Pediatr Blood Cancer. (2012) 59:652–6. doi: 10.1002/pbc.24019
48. Abedalthagafi M. Constitutional mismatch repair-deficiency: current problems and emerging therapeutic strategies. Oncotarget. (2018) 9:35458–69. doi: 10.18632/oncotarget.26249
49. Bouffet E, Larouche V, Campbell BB, Merico D, de Borja R, Aronson M, et al. Immune checkpoint inhibition for hypermutant glioblastoma multiforme resulting from germline biallelic mismatch repair deficiency. J Clin Oncol. (2016) 34:2206–11. doi: 10.1200/JCO.2016.66.6552
50. Amary MF, Damato S, Halai D, Eskandarpour M, Berisha F, Bonar F, et al. Ollier disease and Maffucci syndrome are caused by somatic mosaic mutations of IDH1 and IDH2. Nat Genet. (2011) 43:1262–5. doi: 10.1038/ng.994
51. El Abiad JM, Robbins SM, Cohen B, Levin AS, Valle DL, Morris CD, et al. Natural history of Ollier disease and Maffucci syndrome: patient survey and review of clinical literature. Am J Med Genet A. (2020) 182:1093–103. doi: 10.1002/ajmg.a.61530
52. Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. (2009) 360:765–73. doi: 10.1056/NEJMoa0808710
53. Bonnet C, Thomas L, Psimaras D, Bielle F, Vauléon E, Loiseau H, et al. Characteristics of gliomas in patients with somatic IDH mosaicism. Acta Neuropathol Commun. (2016) 4:31. doi: 10.1186/s40478-016-0302-y
54. Lu C, Ward PS, Kapoor GS, Rohle D, Turcan S, Abdel-Wahab O, et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature. (2012) 483:474–8. doi: 10.1038/nature10860
55. Turcan S, Rohle D, Goenka A, Walsh LA, Fang F, Yilmaz E, et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. (2012) 483:479–83. doi: 10.1038/nature10866
56. Kaufman DK, Kimmel DW, Parisi JE, Michels VV. A familial syndrome with cutaneous malignant melanoma and cerebral astrocytoma. Neurology. (1993) 43:1728–31. doi: 10.1212/WNL.43.9.1728
57. Randerson-Moor JA, Harland M, Williams S, Cuthbert-Heavens D, Sheridan E, Aveyard J, et al. A germline deletion of p14(ARF) but not CDKN2A in a melanoma-neural system tumour syndrome family. Hum Mol Genet. (2001) 10:55–62. doi: 10.1093/hmg/10.1.55
58. Jalali A, Amirian ES, Bainbridge MN, Armstrong GN, Liu Y, Tsavachidis S, et al. Targeted sequencing in chromosome 17q linkage region identifies familial glioma candidates in the Gliogene Consortium. Sci Rep. (2015) 5:8278. doi: 10.1038/srep08278
59. Bainbridge MN, Armstrong GN, Gramatges MM, Bertuch AA, Jhangiani SN, Doddapaneni H, et al. Germline mutations in shelterin complex genes are associated with familial glioma. J Natl Cancer Inst. (2015) 107:384. doi: 10.1093/jnci/djv173
60. Mersch J, Jackson MA, Park M, Nebgen D, Peterson SK, Singletary C, et al. Cancers associated with BRCA1 and BRCA2 mutations other than breast and ovarian. Cancer. (2015) 121:269–75. doi: 10.1002/cncr.29041
61. Frappart P-O, Lee Y, Lamont J, McKinnon PJ. BRCA2 is required for neurogenesis and suppression of medulloblastoma. EMBO J. (2007) 26:2732–42. doi: 10.1038/sj.emboj.7601703
62. Wilson BT, Douglas SF, Polvikoski T. Astrocytoma in a breast cancer lineage: part of the BRCA2 phenotype? J Clin Oncol. (2010) 28:e596–8. doi: 10.1200/JCO.2010.28.9173
63. Bayrakli F, Akgun B, Soylemez B, Kaplan M, Gurelik M. Variation in the BRCA2 gene in a child with medulloblastoma and a family history of breast cancer. J Neurosurg Pediatr. (2011) 8:476–8. doi: 10.3171/2011.8.PEDS11210
64. Miele E, Mastronuzzi A, Po A, Carai A, Alfano V, Serra A, et al. Characterization of medulloblastoma in fanconi anemia: a novel mutation in the BRCA2 gene and SHH molecular subgroup. Biomark Res. (2015) 3:13. doi: 10.1186/s40364-015-0038-z
65. Ranger A, Szymczak A, Hammond RR, Zelcer S. Pediatric thalamic glioblastoma associated with Ollier disease (multiple enchondromatosis): a rare case of concurrence. J Neurosurg Pediatr. (2009) 4:363–7. doi: 10.3171/2009.5.PEDS08422
66. Reyes D, Prayson R. Glioblastoma in the setting of tuberous sclerosis. J Clin Neurosci. (2015) 22:907–8. doi: 10.1016/j.jocn.2014.12.001
67. Bleeker FE, Hopman SMJ, Merks JHM, Aalfs CM, Hennekam RCM. Brain tumors and syndromes in children. Neuropediatrics. (2014) 45:137–61. doi: 10.1055/s-0034-1368116
68. Zhang J, Chiodini R, Badr A, Zhang G. The impact of next-generation sequencing on genomics. J Genet Genomics. (2011) 38:95–109. doi: 10.1016/j.jgg.2011.02.003
69. Tsaousis GN, Papadopoulou E, Apessos A, Agiannitopoulos K, Pepe G, Kampouri S, et al. Analysis of hereditary cancer syndromes by using a panel of genes: novel and multiple pathogenic mutations. BMC Cancer. (2019) 19:535. doi: 10.1186/s12885-019-5756-4
70. Kamps R, Brandão RD, Bosch BJ van den, Paulussen ADC, Xanthoulea S, Blok MJ, et al. Next-Generation sequencing in oncology: genetic diagnosis, risk prediction and cancer classification. Int J Mol Sci. (2017) 18:308. doi: 10.3390/ijms18020308
71. Brozou T, Taeubner J, Velleuer E, Dugas M, Wieczorek D, Borkhardt A, et al. Genetic predisposition in children with cancer - affected families' acceptance of Trio-WES. Eur J Pediatr. (2018) 177:53–60. doi: 10.1007/s00431-017-2997-6
72. Capper D, Jones DTW, Sill M, Hovestadt V, Schrimpf D, Sturm D, et al. DNA methylation-based classification of central nervous system tumours. Nature. (2018) 555:469–74. doi: 10.1038/nature26000
Keywords: brain tumors, cancer predisposition, genetics of cancer, pediatric neuro- oncology, high grade gliomas
Citation: Ceglie G, Del Baldo G, Agolini E, Rinelli M, Cacchione A, Del Bufalo F, Vinci M, Carta R, Boccuto L, Miele E, Mastronuzzi A, Locatelli F and Carai A (2020) Cancer Predisposition Syndromes Associated With Pediatric High-Grade Gliomas. Front. Pediatr. 8:561487. doi: 10.3389/fped.2020.561487
Received: 12 May 2020; Accepted: 26 October 2020;
Published: 12 November 2020.
Edited by:
Anat Erdreich-Epstein, Children's Hospital of Los Angeles, United StatesReviewed by:
Joshua John Breunig, Cedars Sinai Medical Center, United StatesGeorge Michaiel, University of Southern California, United States
Copyright © 2020 Ceglie, Del Baldo, Agolini, Rinelli, Cacchione, Del Bufalo, Vinci, Carta, Boccuto, Miele, Mastronuzzi, Locatelli and Carai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Angela Mastronuzzi, YW5nZWxhLm1hc3Ryb251enppQG9wYmcubmV0
 Giulia Ceglie
Giulia Ceglie Giada Del Baldo
Giada Del Baldo Emanuele Agolini
Emanuele Agolini Martina Rinelli
Martina Rinelli Antonella Cacchione
Antonella Cacchione Francesca Del Bufalo
Francesca Del Bufalo Maria Vinci
Maria Vinci Roberto Carta
Roberto Carta Luigi Boccuto
Luigi Boccuto Evelina Miele1
Evelina Miele1 Angela Mastronuzzi
Angela Mastronuzzi Franco Locatelli
Franco Locatelli Andrea Carai
Andrea Carai