- 1Department of Clinical and Experimental Medicine, University of Catania, Catania, Italy
- 2Unit of Pediatric Emergency, Department of Human Pathology of the Adult and Developmental Age “Gaetano Barresi”, University of Messina, Messina, Italy
- 3Department of Pediatrics, Foundation IRCCS Policlinico San Matteo, University of Pavia, Pavia, Italy
During the last several years, the interest in the role of microbiota in human health has grown significantly. For many years, the lung was considered a sterile environment, and only recently, with the use of more sophisticated techniques, has it been demonstrated that colonization by a complex population of microorganisms in lower airways also occurs in healthy subjects; a predominance of some species of Proteobacteria, Firmicutes, and Bacteroidetes phyla and with a peculiar composition in some disease conditions, such as asthma, have been noted. Lung microbiota derives mainly from the higher airways microbiota. Although we have some information about the role of gut microbiota in modulation of immune system, less it is known about the connection between lung microbiota and local and systemic immunity. There is a correlation between altered microbiota composition and some diseases or chronic states; however, despite this correlation, it has not been clearly demonstrated whether the lung microbiota dysbiosis could be a consequence or a cause of these diseases. We are far from a scientific approach to the therapeutic use of probiotics in airway diseases, but we are only at the starting point of a knowledge process in this fascinating field that could reveal important surprises, and randomized prospective studies in future could reveal more about the clinical possibilities for controlling lung microbiota. This review was aimed at updating the current knowledge in the field of airway microbiota.
Introduction
The “microbiota” consists of different species of microorganisms that live in a defined environment. In our body, microbiota are present in organs that are in contact with the outside environment, mainly the gut, but now we know that microbiota is also present in lung. Microbiota differs among various individuals and also varies according to pathological events or the person's health state, and it can modulate immune responses. The term “microbiome” is considered to include the complete set of microorganisms (bacteria, viruses, and fungi) with their genomes (1). There are numerous mutually beneficial interactions between the human body and microbiota with metabolic reactions, which are important for our health and can contribute to the pathogenesis of some diseases (2). The first contact with bacteria seems to happen prenatally (in utero) via a microbial transfer at the fetomaternal interface at which point microbial DNA has been discovered (3). In fact the maternal–fetal unit is not sterile as was previously believed. Fetal colonization occurs through vertical transfer in the placenta of non-pathogenic commensal microbiota from the phyla Firmicutes, Tenericutes, Proteobacteria, Bacteroidetes, and Fusobacteria (4). During birth, babies have their first important contact with the external world and subsequently, begin to undergo colonization by external bacteria (5). There is a potential relationship between microbial composition that is formed during the initial time after birth for the early life environment and health outcomes, such as allergic diseases (6, 7).
The aim of this review was to understand the lung microbiota function and the interactions between pathogens and commensal microbes and to improve the possibility of prevention and treatment of airway infections in children. A futuristic and innovative project about prevention of pediatric airway infections was initiated in 2007 by the United States National Institutes of Health in order to better understand the microbial communities in our body and their role in health and disease (8).
Methods
This review was conducted using two databases: (1) PubMed and (2) Science Direct. Using these websites, we searched for articles in English using the following key words: (1) airway microbiota; (2) lung microbiota; (3) asthma and microbiota; (4) infections and microbiota; (5) microbiota and immunity; and (6) probiotics in airway diseases. As a rule of thumb, we decided to use the abstracts of articles to assess whether the articles fit the topic. We also reviewed the references of the selected articles and read those with titles that might be of interest for the topic.
Airway Microbiota
The lower airways were previously considered to be a sterile environment, but this has been demonstrated to be false, and recently, using culture-independent techniques, the presence of Actinobacteria, Proteobacteria, Bacteroidetes, and Firmicutes ribosomal DNA has been shown to exist in healthy people's lungs (9). Bacteria have been identified using sensitive identification techniques, including the 16S rRNA gene, which is specific for bacterial cells (10–12). Lung microbiota is composed of about of 2,000 bacterial genomes per cm2. Next generation sequencing (NGS) of 16S rRNA has allowed accurate identification of bacterial genetic material in the lung of healthy subjects, and a significant increase of Proteobacteria presence was found in asthmatic children (13).
The main contributor to the lower airway microbiota composition seems to be the upper airway microbiota (14). Direct contact between the upper and lower airway microbiota can happen more frequently than previously thought, also for aspiration which commonly occurs in healthy young subjects during sleep (15). Aspiration of oropharyngeal secretions, micro aspiration, or direct dispersal by contiguous mucosa creates the microbiome environment in the lung (16). Lung colonization starts immediately after birth (17), and the gut microbiota is established in the first years. Thereafter, it maintains this initial composition stability throughout life (18).
Role of Environment
The microbiota is enriched by the environment, especially during childhood during which time specific immunity and tolerance to antigens are developed. A protective role against asthma and allergies emerges in children who grow up on a farm, showing the importance of exposing children to different microbes from animals and plants in the environment (19). Limited exposure to environmental bacteria and fungi from different conditions in life and overuse of antibiotics are responsible for the increase of autoimmune diseases in the last decades (20). Children who live in cities have less exposure to environmental microbes, and they present a higher incidence of allergy and asthma than children who were born in a rural environment, who have a lower probability of having asthma. There are different mechanisms of this protective role in inflammatory disease; one of them could be the activation of mucosal invariant natural killer T cell (iNKT) tolerance, which is caused by early contact with these antigens (21).
The Gut/lung Axis
The metabolites produced by the gut microbiome in the intestinal microenvironment consist of short-chain fatty acid (SCFAs) that reach other organs and influence lung respiratory disease (22–24). There is clear evidence concerning the contribution to lung immunity by the gut microbiota in the gut–lung immunity axis, such as during pneumococcal pneumonia in which macrophage functions increase at alveolar sites (25). Dietary changes influence the composition of the gut microbiota and asthma and allergic disease statuses (26). The influence of nutrition, in particular dietary composition, on lung immunity was examined, and it was shown that a diet rich in fiber causes an increase in circulating SCFAs levels, which are produced by fiber fermentation in the gut. This fermentation process prevents allergy and asthma-related inflammation of the lower airway (27). Lung SCFAs are produced by gut bacteria, and these are the main metabolic products of anaerobic bacteria fermentation. SCFAs promote recruitment and activation of leukocytes and immune regulation within the inflammatory process (28), and B cell differentiation occurs through regulated gene expression that supports the antibody production (29, 30).
Microbiota and Asthma
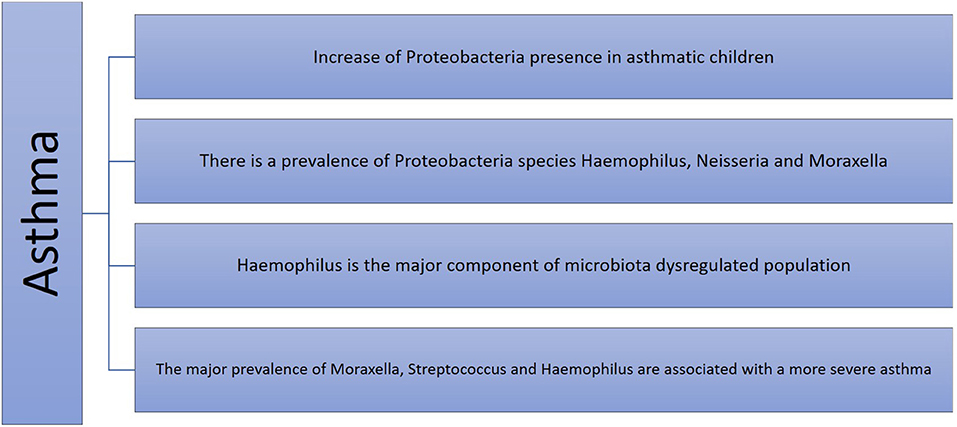
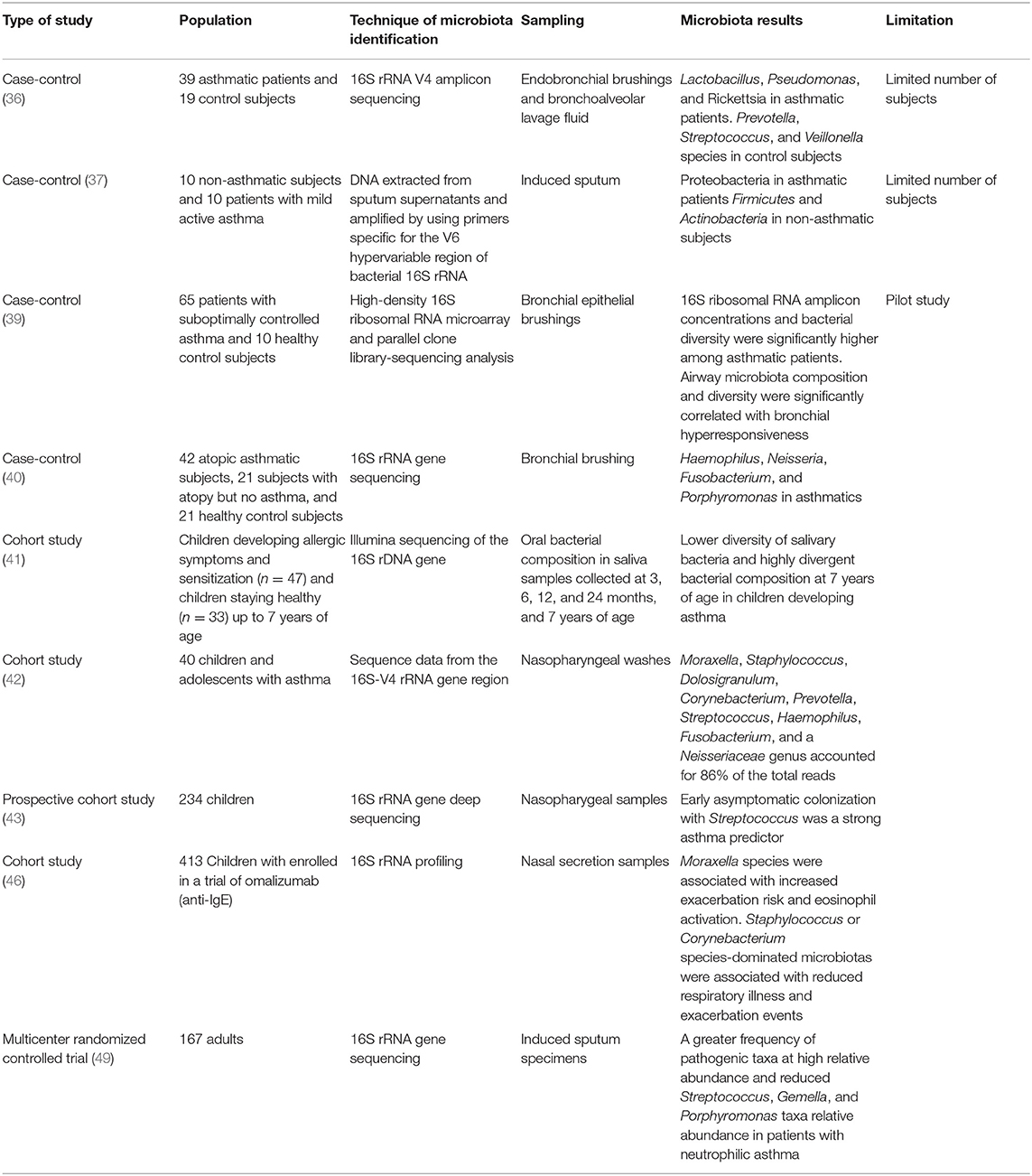
Asthma is the most frequent chronic respiratory disease in children, and its beginning and development is influenced in the first years of life by genetic predisposition and environmental factors, such as antibiotic exposure, breastfeeding, and contact with animals and the natural environment, which play an important role in asthma and other inflammatory disease development (31). In mice, the positive or negative influence of specific bacteria have been investigated with respect to detecting the susceptibility to allergic asthma (32). In humans, microbiota dysbiosis of some bacterial communities has been strongly associated with asthma (33–35). In fact, a difference in microbiota composition between asthmatic patients and healthy control subjects has been demonstrated. The former have a predominance of Lactobacillus, Pseudomonas, Rickettsiae, and Proteobacteria species, and the latter are characterized by population of Prevotella, Streptococcus, Veillonella, Firmicutes, and Actinobacteria (Figure 1). However, studies show different results according to sampling, technique isolation, and examined population (age, asthma severity, and control) (36–40). Patients with more severe airway obstruction and those who require higher doses of inhaled corticosteroids or oral corticosteroids have higher pathogenetic species than asthmatic patients with better-controlled disease (36). Species belonging to Proteobacteria phylum (Comamonadaceae, Sphingomonadaceae, and Oxalobacteraceae) are considered markers of worse disease control and are predictors of bronchial hyperresponsiveness (39, 40).
Lower airway microbiota in asthmatic patients is also related to a different upper airway microbiota composition. In fact, early alterations in oral microbial composition emerged in children who were developing asthma and who had a lower diversity of salivary bacteria together with highly divergent bacterial composition. These early changes seem to influence immune maturation and allergy development (41). Beyond the mouth, nasopharyngeal microbiome diversity can change over time in children with asthma; in particular, the relative proportions of Haemophilus, Moraxella, Staphylococcus, and Corynebacterium genus change over time (42). The results from a prospective cohort of 234 children show that children with early asymptomatic Streptococcus nasopharynx colonization in the first months of life have sensitization to allergens and a major risk of asthma development (43, 44). Also the nasal microbiota seems to be important; in fact, in children with asthma, the microbiome study from nasal secretion samples shows a distinct nasal airway microbiota-dominated Moraxella species associated with increased exacerbation risk (45, 46).
The asthma phenotype derives from an endotype that is based on the prevalence of a specific inflammatory cells and cytokines, which can be influenced by the microbiota (47). The neutrophilic phenotype, which is associated with a more severe clinical course and is refractory to corticosteroids (48), has been associated with a specific microbiota in the airway environment (49). An altered bacterial profile with more Neisseria, Bacteroides, and Rothia species was found in patients with low eosinophilic levels (50). The main prevalence of Moraxella, Streptococcus, and Haemophilus genera were associated with more severe asthma, and the phylum Proteobacteria was the most prevalent in Th17 cell-mediated asthma (51). Table 1 summarizes the main associations between microbiota and asthma.
In patients with severe asthma, antibiotics can change the microbiota composition and also influence the clinical state; an example of such an antibiotic is azithromycin, which can reduce exacerbations and improve the quality of life in patients with uncontrolled asthma undergoing long-term treatment for their disease (52). Inflammation of the airway surface could be an important primary factor that changes the microbiota composition; however, the interdependent dynamics between the asthmatic host and the microbiota are not fully understood (53).
Early Life Events and Asthma
The environment could be a risk or a protective factor, and the farming environment has been suggested to be protective against asthma and other allergic diseases that are likely affected by microorganisms, which influence the innate immunity and determine the early gene expression that will remain throughout life (54). Children who live in a rural environment, such as a farm, have a significantly reduced risk of asthma and atopic dermatitis (55). The decreased exposure to microorganisms during early life predispose children to develop asthma (56). The first year is a critical time for microbiota maturation, and an immature gut microbiota composition is linked to an increased risk of asthma at 5 years of age (57). For example, early colonization by Clostridium difficile at 1 month of age has been associated with later asthma or wheezing development (58). The use of some antibiotics, such as macrolides, in early life creates a compositional shift in the intestinal microbiota with depletion of Actinobacteria and an increase in Proteobacteria and Bacteroides. This shift has been correlated with an increased risk of immunological diseases, such as asthma, in children (59). The use of antibiotics in the prenatal period seems to be associated with an increased risk of asthma. Altered metabolic short-chain fatty acid (SFCA) levels were found in children who are at risk of developing asthma (60) and the mother's exposure to a rural environment during pregnancy with a connected increase to circulating SCFAs has also been associated with a decrease in the rate of asthma development in children with the possible induction of regulatory T lymphocytes in the fetal lung as was demonstrated in mice (61).
Infections and Pathology
The healthy lung is colonized only by a limited population of bacteria that are maintained by an equilibrium among immigration, elimination, and growth. Some changes in the local environment during pathology can permit an increase in some bacterial populations that could became pathological, especially in chronic conditions (62). Lung microbiota could be involved in wheezing development because of the association between lower airway infections during the first years of life and an increase in the risk of wheezing. The number of episodes of airway infections in the first year of life has been associated with the later risk of developing asthma (63). Susceptibility to pulmonary infections depends on the stability of microbiota composition built in the first years as seen in the early life profiles that contain more Moraxella and Corynebacterium/Dolosigranulum in the upper respiratory tract of children (17). There is a direct effect of viral infections in the overgrowth of some pathogenic bacteria, such as a significant increase in the nasopharyngeal load of Streptococcus pneumoniae in children with influenza (64). After influenza A infection, there is an alteration in the microbiota equilibrium that results from direct physical mucosal damage. However, there are also changes in some host-produced immune modulating molecules or cytokines, which can alter the fine interaction between different colonizing bacteria and biofilm formation with their microenvironment. This permits pathogenic invasion as seen between S. pneumoniae and Staphylococcus aureus in the transition to secondary pneumonia (65). Moreover, in patients with influenza, there is an overgrowth of Proteobacteria, such as Enterobacter and Moraxella (66). Rhinovirus infection can influence the microbiota composition as demonstrated in chronic obstructive pulmonary disease patients in whom there is a significant growth in the Haemophilus influenzae population (67). Viral infections, such as respiratory syncytial virus (RSV) infection with a documented increase in Moraxella and Haemophilus members of the phylum Proteobacteria, alter the microbiome composition and influence the susceptibility to asthma (68).
The presence of a particular microbiota signature during upper respiratory tract infections in children compared to healthy patients shows that children with viral infections have a higher density and frequency of colonization with S. pneumoniae, M. catarrhalis, and H. influenzae in the nasopharynx (69).
Some changes in microbiota composition, such as a role for fungal microbiota in inflammation, have been observed in patients with chronic rhinosinusitis, allergy, cystic fibrosis (CF), and asthma (70). There is a crucial relationship between the airway microbiome and the stage and clinical progression of CF that is connected to the reduction in bacterial population diversity (71). In sputum samples from CF children, chronic colonization of specific pathogens, such as Pseudomonas aeruginosa was identified (72, 73), and in these patients it may be possible to use the microbiome as a primary target of prevention. In fact, a reduction in the exacerbation after the administration of the probiotics Lactobacillus casei and L. rhamnosus has been shown (74).
Microbiota and Immunity
There are more bacterial (prokaryotic) cells than eukaryotic cells in our body. The cells are interconnected with the organs, and they form an important part in the regulation of immune system function (75). The microbiota is directly connected to the immune system, and it undergoes metabolic and antigenic interactions. The gut and lung are interconnected, and dysbiosis in the gut microbiota is also associated with lung diseases because the microbiota participates in the development and maintenance of the immune system. Because dysbiosis can permit disease development, immunity can also influence the microbiota composition, which provides resistance to colonization by respiratory pathogens that have a reciprocal influence on maturation and health maintenance (76). Regulation of the balance between Th-mediated inflammation and the T-regulatory response to environmental allergens can be controlled by skin commensals, such as Acinetobacter (77), and the same important role may be played by other microorganisms in the lung. The overgrowth of some species of lung microbiota with a reduction in species diversity could cause an inflammatory cell-mediated host response that is connected with alveolar tissue remodeling with consequent chronic changes (78).
The metabolites produced by bacteria can activate alveolar macrophages through nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) and also activate the cell response through the Toll-like receptor (TLR) link, thereby influencing airway immunity function (79).
The creation of an airway microbiota within the first weeks of life is connected with early development of immune system tolerance and includes the induction of regulatory cells with specific genetic activation of innate immune cytokines (interleukins-4, -5, and -13) and cell ligands that are observed in mice (80). The microbe stimuli can then influence maturation of the lung mucosal barrier with homeostasis of the immune regulatory surface that is in contact with the environment, thus expanding the regulatory lung interstitial macrophages and influencing the susceptibility to allergic asthma (81). The segmented filamentous bacteria that are present in the gut microbiota induce autoantibodies in the lung through a Th17 T-cell-receptor-mediated inflammatory response (82).
For the possible role of immunity-related genes in microbiome composition, the role of genetic variations in mucosal immunity pathways on the upper airway microbiome has been investigated. Previous findings have shown an interesting association between the relative abundance of Dermacoccus and the variant 8 kb upstream of TINCR, a long non-coding RNA that binds to peptidoglycan recognition protein 3 (PGLYRP3) mRNA, which is a gene encoding a known antimicrobial protein. Moreover, the association between a missense variant in PGLYRP4 (rs3006458) and the relative abundance of an unclassified genus of family Micrococcaceae (phylum Actinobacteria) has also emerged (83).
Probiotics
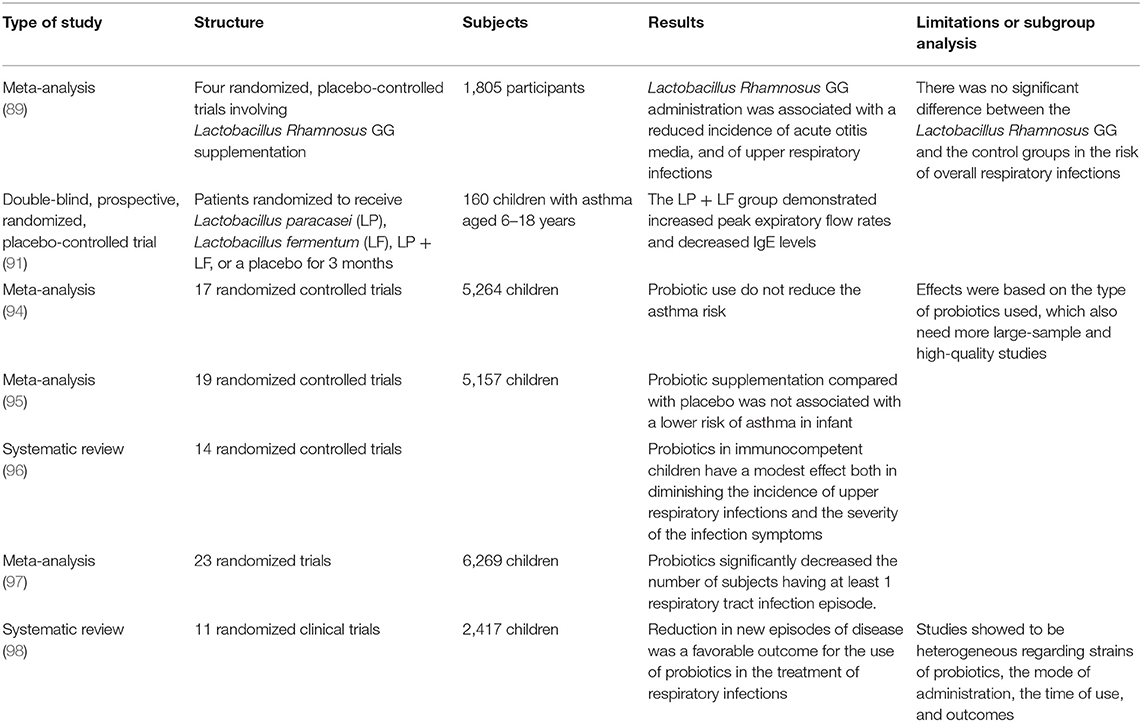
There is a growing interest in the potential future use of probiotics for promoting stronger immune lung function by manipulating microbiota interactions in airways health. The immune function derived by immune system homeostasis represents the mucosal barrier and its microbiome environment interactions in the intact barrier surface; when this unit is disrupted, immunity is compromised for a long time after an acute infection (84). Probiotics could play a role in promoting immune modulation in innate immune function, such as via recruitment of natural killer lymphocytes (85). The efficacy of the immune response in the lung seems to correlate with the microbiota composition and connect with metabolism products, which are derived from these living organisms. There are microbiota profiles that are distinct in composition and distinct from the upper airways with respect to composition and structure in the lower airways, and these are associated with different local immune responses and peripheral metabolic reprogramming (86). In infant mice, intranasal administration of the commensal C. pseudodiphteriticum was shown to provide a protective influence through innate immunity activation that was mediated by TLR-3 against RSV infection and secondary pneumococcal pneumonia infection (87). In a prospective pilot study of children with CF, the administration of probiotics, particularly Lactobacillus GG, was shown to cause a reduction in the pulmonary exacerbation rate (88). Four randomized trials, in which the administration of L. Rhamnosus GG in children was studied, reported a reduction in the incidence of acute otitis and upper respiratory infections (89). Probiotic administration, in particular L. rhamnosus, within the first 2 years of life, has been demonstrated to reduce the incidence of allergy in children (90). Probiotic supplementary therapy has been investigated in many randomized trials for investigating clinical efficacy in common respiratory childhood diseases, such as asthma, rhinitis, or wheezing. Singular randomized placebo-controlled studies sometimes have shown an interesting effect on asthma after the administration of probiotics, such as L. paracasei (LP) and L. fermentum (LF) (91). In two randomized double-blind placebo-controlled trials of 472 hospitalized children and 281 children attending day care centers, Lactobacillus GG administration caused a decrease in the risk of respiratory tract infections (92, 93). A recent meta-analysis showed an insignificant association of probiotic use with the reduction in asthma risk; emerging from these studies, was a heterogeneity in the type of probiotics used and quality of bacterial identification technique (94–96). However, in other two meta-analyses from 2016 and 2015, a decrease in the respiratory tract infection rate valuated on the number of new episodes and number of days of fever was demonstrated despite the heterogeneity of the probiotics administration (97, 98). Table 2 summarizes the clinical trials evaluating probiotic administration.
There is a lack of favorable evidence for probiotic use in preventing subsequent asthma or allergy because the complexity of the microbiome–human interaction is greater than a simple cause-and-effect relationship of probiotic administration. The influence of diet in late adolescence on the microbiome composition with its consequent influences in lung also seems to be an important primary preventive factor for allergic disease (99). The use of probiotics with live microorganisms for preventing or curing respiratory infections has not been clearly defined, and there are no scientific recommendations about the use of probiotics because the quality of evidence is low, but there are some trials that seem to provide important positive evidence for a future clinical scientific approach of probiotic use.
Conclusion
A strong relationship exists between the lung and intestinal microbiota, the environment, and the effects of early life exposure to non-pathogenic microbes of the natural environment, which are important for immune system development. The axis between the gut and lung is important for immune tolerance, which can determine the susceptibility to developing asthma or allergy, in particular, during the early phases of immune system structuring. Despite important correlations between microbiota and inflammation or immune response homeostasis in preclinical studies and in specific group of patients, there is not enough robust evidence, except for a few efficacious results from randomized clinical trials, which are heterogeneous for probiotics dose and sampling analysis, to recommend the general use of probiotics to prevent asthma or allergy. Further studies are needed to better understand the role of the microbiota in respiratory diseases and to define the possibility of a therapeutic intervention with probiotics or prebiotics or simply, an early life with more rural experiences.
Author Contributions
AL developed the original idea and the final revision. GP and GFP wrote the manuscript. AG, SM, and MP revised firstly the manuscript and contributed to English revision and references update. SS, GM, and SL made the final analysis and critical revision of the manuscript. All authors read and approved the final manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Marchesi JR, Ravel J. The vocabulary of microbiome research: a proposal. Microbiome. (2015) 3:31. doi: 10.1186/s40168-015-0094-5
2. Riiser A. The human microbiome, asthma, and allergy. Allergy Asthma Clin Immunol. (2015) 11:35. doi: 10.1186/s13223-015-0102-0
3. Collado MC, Rautava S, Aakko J, Isolauri E, Salminen S. Human gut colonisation may be initiated in utero by distinct microbial communities in the placenta and amniotic fluid. Sci Rep. (2016) 6:23129. doi: 10.1038/srep23129
4. Aagaard K, Ma J, Antony KM, Ganu R, Petrosino J, Versalovic J. The placenta harbors a unique microbiome. Sci Transl Med. (2014) 6:237ra65. doi: 10.1126/scitranslmed.3008599
5. Mueller NT, Bakacs E, Combellick J, Grigoryan Z, Dominguez-Bello MG. The infant microbiome development: mom matters. Trends Mol Med. (2015) 21:109–17. doi: 10.1016/j.molmed.2014.12.002
6. Prescott SL. Early-life environmental determinants of allergic diseases and the wider pandemic of inflammatory noncommunicable diseases. J Allergy Clin Immunol. (2013) 131:23–30. doi: 10.1016/j.jaci.2012.11.019
7. Palm NW, de Zoete MR, Flavell RA. Immune-microbiota interactions in health and disease. Clin Immunol. (2015) 159:122–7. doi: 10.1016/j.clim.2015.05.014
8. Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI. The human microbiome project. Nature. (2007) 449:804–10. doi: 10.1038/nature06244
9. Segal LN, Blaser MJ. A brave new world: the lung microbiota in an era of change. Ann Am Thorac Soc. (2014) 11 (Suppl. 1):S21–7. doi: 10.1513/AnnalsATS.201306-189MG
10. Charlson ES, Bittinger K, Haas AR, Fitzgerald AS, Frank I, Yadav A, et al. Topographical continuity of bacterial populations in the healthy human respiratory tract. Am J Respir Crit Care Med. (2011) 184:957–63. doi: 10.1164/rccm.201104-0655OC
11. Dickson RP, Erb-Downward JR, Freeman CM, McCloskey L, Beck JM, Huffnagle GB, et al. Spatial variation in the healthy human lung microbiome and the adapted island model of lung biogeography. Ann Am Thorac Soc. (2015) 12:821–30. doi: 10.1513/AnnalsATS.201501-029OC
12. Morris A, Beck JM, Schloss PD, Campbell TB, Crothers K, Curtis JL, et al. Comparison of the respiratory microbiome in healthy nonsmokers and smokers. Am J Respir Crit Care Med. (2013) 187:1067–75. doi: 10.1164/rccm.201210-1913OC
13. Hilty M, Burke C, Pedro H, Cardenas P, Bush A, Bossley C, et al. Disordered microbial communities in asthmatic airways. PLoS ONE. (2010) 5:e8578. doi: 10.1371/journal.pone.0008578
14. Carmody LA, Caverly LJ, Foster BK, Rogers MAM, Kalikin LM, Simon RH, et al. Fluctuations in airway bacterial communities associated with clinical states and disease stages in cystic fibrosis. PLoS ONE. (2018) 13:e0194060. doi: 10.1371/journal.pone.0194060
15. Gleeson K, Eggli DF, Maxwell SL. Quantitative aspiration during sleep in normal subjects. Chest. (1997) 111:1266–72. doi: 10.1378/chest.111.5.1266
16. Dickson RP, Erb-Downward JR, Freeman CM, McCloskey L, Falkowski NR, Huffnagle GB, et al. Bacterial topography of the healthy human lower respiratory tract. MBio. (2017) 8:e02287–16. doi: 10.1128/mBio.02287-16
17. Biesbroek G, Tsivtsivadze E, Sanders EA, Montijn R, Veenhoven RH, Keijser BJ, et al. Early respiratory microbiota composition determines bacterial succession patterns and respiratory health in children. Am J Respir Crit Care Med. (2014) 190:1283–92. doi: 10.1164/rccm.201407-1240OC
18. Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, et al. Human gut microbiome viewed across age and geography. Nature. (2012) 486:222–7. doi: 10.1038/nature11053
19. Ege MJ, Mayer M, Normand AC, Genuneit J, Cookson WO, Braun-Fahrländer C, et al. Exposure to environmental microorganisms and childhood asthma. N Engl J Med. (2011) 364:701–9. doi: 10.1056/NEJMoa1007302
20. Abrahamsson TR, Jakobsson HE, Andersson AF, Björkstén B, Engstrand L, Jenmalm MC. Low gut microbiota diversity in early infancy precedes asthma at school age. Clin Exp Allergy. (2014) 44:842–50. doi: 10.1111/cea.12253
21. Olszak T, An D, Zeissig S, Vera MP, Richter J, Franke A, et al. Microbial exposure during early life has persistent effects on natural killer T cell function. Science. (2012) 336:489–93. doi: 10.1126/science.1219328
22. Marsland BJ, Trompette A, Gollwitzer ES. The gut-lung axis in respiratory disease. Ann Am Thorac Soc. (2015) 12 (Suppl. 2):S150–6. doi: 10.1513/AnnalsATS.201503-133AW
23. Bingula R, Filaire M, Radosevic-Robin N, Bey M, Berthon JY, Bernalier-Donadille A, et al. Desired turbulence? Gut-lung axis, immunity, and lung cancer. J Oncol. (2017) 2017:5035371. doi: 10.1155/2017/5035371
24. Tomkovich S, Jobin C. Microbiota and host immune responses: a love-hate relationship. Immunology. (2016) 147:1–10. doi: 10.1111/imm.12538
25. Schuijt TJ, Lankelma JM, Scicluna BP, de Sousa e Melo F, Roelofs JJ, de Boer JD, et al. The gut microbiota plays a protective role in the host defence against pneumococcal pneumonia. Gut. (2016) 65:575–83. doi: 10.1136/gutjnl-2015-309728
26. Fujimura KE, Lynch SV. Microbiota in allergy and asthma and the emerging relationship with the gut microbiome. Cell Host Microbe. (2015) 17:592–602. doi: 10.1016/j.chom.2015.04.007
27. Halnes I, Baines KJ, Berthon BS, MacDonald-Wicks LK, Gibson PG, Wood LG. Soluble fibre meal challenge reduces airway inflammation and expression of GPR43 and GPR41 in asthma. Nutrients. (2017) 9:E57. doi: 10.3390/nu9010057
28. Vinolo MA, Rodrigues HG, Nachbar RT, Curi R. Regulation of inflammation by short chain fatty acids. Nutrients. (2011) 3:858–76. doi: 10.3390/nu3100858
29. Kim M, Qie Y, Park J, Kim CH. Gut microbial metabolites fuel host antibody responses. Cell Host Microbe. (2016) 20:202–14. doi: 10.1016/j.chom.2016.07.001
30. Li M, van Esch BCAM, Henricks PAJ, Folkerts G, Garssen J. The anti-inflammatory effects of short chain fatty acids on lipopolysaccharide- or tumor necrosis factor α-stimulated endothelial cells via activation of GPR41/43 and inhibition of HDACs. Front Pharmacol. (2018) 9:533. doi: 10.3389/fphar.2018.00533
31. McKenzie C, Tan J, Macia L, Mackay CR. The nutrition-gut microbiome-physiology axis and allergic diseases. Immunol Rev. (2017) 278:277–95. doi: 10.1111/imr.12556
32. Remot A, Descamps D, Noordine ML, Boukadiri A, Mathieu E, Robert V, et al. Bacteria isolated from lung modulate asthma susceptibility in mice. ISME J. (2017) 11:1061–74. doi: 10.1038/ismej.2016.181
33. Chung KF. Airway microbial dysbiosis in asthmatic patients: a target for prevention and treatment? J Allergy Clin Immunol. (2017) 139:1071–81. doi: 10.1016/j.jaci.2017.02.004
34. Bosch AATM, de Steenhuijsen Piters WAA, van Houten MA, Chu MLJN, Biesbroek G, Kool J, et al. Maturation of the infant respiratory microbiota, environmental drivers, and health consequences. a prospective cohort study. Am J Respir Crit Care Med. (2017) 196:1582–90. doi: 10.1164/rccm.201703-0554OC
35. De Steenhuijsen Piters WA, Huijskens EG, Wyllie AL, Biesbroek G, van den Bergh MR, Veenhoven RH, et al. Dysbiosis of upper respiratory tract microbiota in elderly pneumonia patients. ISME J. (2016) 10:97–108. doi: 10.1038/ismej.2015.99
36. Denner DR, Sangwan N, Becker JB, Hogarth DK, Oldham J, Castillo J, et al. Corticosteroid therapy and airflow obstruction influence the bronchial microbiome, which is distinct from that of bronchoalveolar lavage in asthmatic airways. J Allergy Clin Immunol. (2016) 137:1398–1405.e3. doi: 10.1016/j.jaci.2015.10.017
37. Marri PR, Stern DA, Wright AL, Billheimer D, Martinez FD. Asthma-associated differences in microbial composition of induced sputum. J Allergy Clin Immunol. (2013) 131:346–52. doi: 10.1016/j.jaci.2012.11.013
38. Singanayagam A, Ritchie A, Johnston SL. Role of microbiome in the pathophysiology and disease course of asthma. Curr Opin Pulm Med. (2017) 23:41–47. doi: 10.1097/MCP.0000000000000333
39. Huang YJ, Nelson CE, Brodie EL, Desantis TZ, Baek MS, Liu J, et al. Airway microbiota and bronchial hyperresponsiveness in patients with suboptimally controlled asthma. J Allergy Clin Immunol. (2011) 127:372–81.e1–3. doi: 10.1016/j.jaci.2010.10.048
40. Durack J, Lynch SV, Nariya S, Bhakta NR, Beigelman A, Castro M, et al. Features of the bronchial bacterial microbiome associated with atopy, asthma, and responsiveness to inhaled corticosteroid treatment. J Allergy Clin Immunol. (2017) 140:63–75. doi: 10.1016/j.jaci.2016.08.055
41. Dzidic M, Abrahamsson TR, Artacho A, Collado MC, Mira A, Jenmalm MC. Oral microbiota maturation during the first 7 years of life in relation to allergy development. Allergy. (2018) 73:2000–11. doi: 10.1111/all.13449
42. Pérez-Losada M, Alamri L, Crandall KA, Freishtat RJ. Nasopharyngeal microbiome diversity changes over time in children with asthma. PLoS ONE. (2017) 12:e0170543. doi: 10.1371/journal.pone.0170543
43. Teo SM, Mok D, Pham K, Kusel M, Serralha M, Troy N, et al. The infant nasopharyngeal microbiome impacts severity of lower respiratory infection and risk of asthma development. Cell Host Microbe. (2015) 17:704–15. doi: 10.1016/j.chom.2015.03.008
44. Sullivan A, Hunt E, MacSharry J, Murphy DM. The microbiome and the pathophysiology of asthma. Respir Res. (2016) 17:163. doi: 10.1186/s12931-016-0479-4
45. Pérez-Losada M, Authelet KJ, Hoptay CE, Kwak C, Crandall KA, Freishtat RJ. Pediatric asthma comprises different phenotypic clusters with unique nasal microbiotas. Microbiome. (2018) 6:179. doi: 10.1186/s40168-018-0564-7
46. McCauley K, Durack J, Valladares R, Fadrosh DW, Lin DL, Calatroni A, et al. Distinct nasal airway bacterial microbiotas differentially relate to exacerbation in pediatric patients with asthma. J Allergy Clin Immunol. (2019) 2019:35. doi: 10.1016/j.jaci.2019.05.035
47. Adami AJ, Bracken SJ. Breathing better through bugs: asthma and the microbiome. Yale J Biol Med. (2016) 89:309–24.
48. Ray A, Kolls JK. Neutrophilic inflammation in asthma and association with disease severity. Trends Immunol. (2017) 38:942–54. doi: 10.1016/j.it.2017.07.003
49. Taylor SL, Leong LEX, Choo JM, Wesselingh S, Yang IA, Upham JW, et al. Inflammatory phenotypes in patients with severe asthma are associated with distinct airway microbiology. J Allergy Clin Immunol. (2018) 141:94–103.e15. doi: 10.1016/j.jaci.2017.03.044
50. Sverrild A, Kiilerich P, Brejnrod A, Pedersen R, Porsbjerg C, Bergqvist A, et al. Eosinophilic airway inflammation in asthmatic patients is associated with an altered airway microbiome. J Allergy Clin Immunol. (2017) 140:407–17.e11. doi: 10.1016/j.jaci.2016.10.046
51. Noval Rivas M, Crother TR, Arditi M. The microbiome in asthma. Curr Opin Pediatr. (2016) 28:764–71. doi: 10.1097/MOP.0000000000000419
52. Gibson PG, Yang IA, Upham JW, Reynolds PN, Hodge S, James AL, et al. Effect of azithromycin on asthma exacerbations and quality of life in adults with persistent uncontrolled asthma (AMAZES): a randomised, double-blind, placebo-controlled trial. Lancet. (2017) 390:659–68. doi: 10.1016/S0140-6736(17)31281-3
53. Huang YJ, Marsland BJ, Bunyavanich S, O'Mahony L, Leung DY, Muraro A, et al. The microbiome in allergic disease: current understanding and future opportunities-2017 PRACTALL document of the American Academy of Allergy, Asthma & Immunology and the European Academy of Allergy and Clinical Immunology. J Allergy Clin Immunol. (2017) 139:1099–110. doi: 10.1016/j.jaci.2017.02.007
54. Loss G, Bitter S, Wohlgensinger J, Frei R, Roduit C, Genuneit J, et al. Prenatal and early-life exposures alter expression of innate immunity genes: the PASTURE cohort study. J Allergy Clin Immunol. (2012) 130:523–30.e9. doi: 10.1016/j.jaci.2012.05.049
55. Illi S, Depner M, Genuneit J, Horak E, Loss G, Strunz-Lehner C, et al. Protection from childhood asthma and allergy in Alpine farm environments-the GABRIEL Advanced Studies. J Allergy Clin Immunol. (2012) 129:1470–7.e6. doi: 10.1016/j.jaci.2012.03.013
56. Smits HH, Hiemstra PS, Prazeres da Costa C, Ege M, Edwards M, Garn H, et al. Microbes and asthma: opportunities for intervention. J Allergy Clin Immunol. (2016) 137:690–7. doi: 10.1016/j.jaci.2016.01.004
57. Stokholm J, Blaser MJ, Thorsen J, Rasmussen MA, Waage J, Vinding RK, et al. Maturation of the gut microbiome and risk of asthma in childhood. Nat Commun. (2018) 9:141. doi: 10.1038/s41467-018-03150-x
58. van Nimwegen FA, Penders J, Stobberingh EE, Postma DS, Koppelman GH, Kerkhof M, et al. Mode and place of delivery, gastrointestinal microbiota, and their influence on asthma and atopy. J Allergy Clin Immunol. (2011) 128:948–55. doi: 10.1016/j.jaci.2011.07.027
59. Korpela K, Salonen A, Virta LJ, Kekkonen RA, Forslund K, Bork P, et al. Intestinal microbiome is related to lifetime antibiotic use in Finnish pre-school children. Nat Commun. (2016) 7:10410. doi: 10.1038/ncomms10410
60. Arrieta MC, Stiemsma LT, Dimitriu PA, Thorson L, Russell S, Yurist-Doutsch S, et al. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci Transl Med. (2015) 7:307ra152. doi: 10.1126/scitranslmed.aab2271
61. Gray LE, O'Hely M, Ranganathan S, Sly PD, Vuillermin P. The maternal diet, gut bacteria, and bacterial metabolites during pregnancy influence offspring asthma. Front Immunol. (2017) 8:365. doi: 10.3389/fimmu.2017.00365
62. Dickson RP, Martinez FJ, Huffnagle GB. The role of the microbiome in exacerbations of chronic lung diseases. Lancet. (2014) 384:691–702. doi: 10.1016/S0140-6736(14)61136-3
63. Bønnelykke K, Vissing NH, Sevelsted A, Johnston SL, Bisgaard H. Association between respiratory infections in early life and later asthma is independent of virus type. J Allergy Clin Immunol. (2015) 136:81–6.e4. doi: 10.1016/j.jaci.2015.02.024
64. Vu HT, Yoshida LM, Suzuki M, Nguyen HA, Nguyen CD, Nguyen AT, et al. Association between nasopharyngeal load of Streptococcus pneumoniae, viral coinfection, and radiologically confirmed pneumonia in Vietnamese children. Pediatr Infect Dis J. (2011) 30:11–8. doi: 10.1097/INF.0b013e3181f111a2
65. Reddinger RM, Luke-Marshall NR, Sauberan SL, Hakansson AP, Campagnari AA. Streptococcus pneumoniae modulates Staphylococcus aureus biofilm dispersion and the transition from colonization to invasive disease. MBio. (2018) 9:e02089-17. doi: 10.1128/mBio.02089-17
66. Yildiz S, Mazel-Sanchez B, Kandasamy M, Manicassamy B, Schmolke M. Influenza A virus infection impacts systemic microbiota dynamics and causes quantitative enteric dysbiosis. Microbiome. (2018) 6:9. doi: 10.1186/s40168-017-0386-z
67. Molyneaux PL, Mallia P, Cox MJ, Footitt J, Willis-Owen SA, Homola D, et al. Outgrowth of the bacterial airway microbiome after rhinovirus exacerbation of chronic obstructive pulmonary disease. Am J Respir Crit Care Med. (2013) 188:1224–31. doi: 10.1164/rccm.201302-0341OC
68. Rosas-Salazar C, Shilts MH, Tovchigrechko A, Chappell JD, Larkin EK, Nelson KE, et al. Nasopharyngeal microbiome in respiratory syncytial virus resembles profile associated with increased childhood asthma risk. Am J Respir Crit Care Med. (2016) 193:1180–3. doi: 10.1164/rccm.201512-2350LE
69. DeMuri GP, Gern JE, Eickhoff JC, Lynch SV, Wald ER. Dynamics of bacterial colonization with Streptococcus pneumoniae, Haemophilus influenzae, and Moraxella catarrhalis during symptomatic and asymptomatic viral upper respiratory tract infection. Clin Infect Dis. (2018) 66:1045–53. doi: 10.1093/cid/cix941
70. Zhang I, Pletcher SD, Goldberg AN, Barker BM, Cope EK. Fungal microbiota in chronic airway inflammatory disease and emerging relationships with the host immune response. Front Microbiol. (2017) 8:2477. doi: 10.3389/fmicb.2017.02477
71. Zhao J, Schloss PD, Kalikin LM, Carmody LA, Foster BK, Petrosino JF, et al. Decade-long bacterial community dynamics in cystic fibrosis airways. Proc Natl Acad Sci USA. (2012) 109:5809–14. doi: 10.1073/pnas.1120577109
72. Lipuma JJ. The changing microbial epidemiology in cystic fibrosis. Clin Microbiol Rev. (2010) 23:299–323. doi: 10.1128/CMR.00068-09
73. McKone EF, Emerson SS, Edwards KL, Aitken ML. Effect of genotype on phenotype and mortality in cystic fibrosis: a retrospective cohort study. Lancet. (2003) 361:1671–6. doi: 10.1016/S0140-6736(03)13368-5
74. Plaza-Díaz J, Ruiz-Ojeda FJ, Gil-Campos M, Gil A. Immune-mediatet mechanisms of action of probiotics and synbiontic in treating pediatric intestinal diseases. Nutrients. (2018) 10:E42. doi: 10.3390/nu10010042
75. Lynch SV, Pedersen O. The human intestinal microbiome in health and disease. N Engl J Med. (2016) 375:2369–79. doi: 10.1056/NEJMra1600266
76. Man WH, de Steenhuijsen Piters WA, Bogaert D. The microbiota of the respiratory tract: gatekeeper to respiratory health. Nat Rev Microbiol. (2017) 15:259–70. doi: 10.1038/nrmicro.2017.14
77. Fyhrquist N, Ruokolainen L, Suomalainen A, Lehtimäki S, Veckman V, Vendelin J, et al. Acinetobacter species in the skin microbiota protect against allergic sensitization and inflammation. J Allergy Clin Immunol. (2014) 134:1301–9.e11. doi: 10.1016/j.jaci.2014.07.059
78. Sze MA, Dimitriu PA, Suzuki M, McDonough JE, Campbell JD, Brothers JF, et al. Host response to the lung microbiome in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. (2015) 192:438–45. doi: 10.1164/rccm.201502-0223OC
79. McAleer JP, Kolls JK. Contributions of the intestinal microbiome in lung immunity. Eur J Immunol. (2018) 48:39–49. doi: 10.1002/eji.201646721
80. Gollwitzer ES, Saglani S, Trompette A, Yadava K, Sherburn R, McCoy KD, et al. Lung microbiota promotes tolerance to allergens in neonates via PD-L1. Nat Med. (2014) 20:642–7. doi: 10.1038/nm.3568
81. Sabatel C, Radermecker C, Fievez L, Paulissen G, Chakarov S, Fernandes C, et al. Exposure to bacterial CpG DNA protects from airway allergic inflammation by expanding regulatory lung interstitial macrophages. Immunity. (2017) 46:457–73. doi: 10.1016/j.immuni.2017.02.016
82. Bradley CP, Teng F, Felix KM, Sano T, Naskar D, Block KE, et al. Segmented filamentous bacteria provoke lung autoimmunity by inducing gut-lung axis Th17 cells expressing dual TCRs. Cell Host Microbe. (2017) 22:697–704.e4. doi: 10.1016/j.chom.2017.10.007
83. Igartua C, Davenport ER, Gilad Y, Nicolae DL, Pinto J, Ober C. Host genetic variation in mucosal immunity pathways influences the upper airway microbiome. Microbiome. (2017) 5:16. doi: 10.1186/s40168-016-0227-5
84. Fonseca DM, Hand TW, Han SJ, Gerner MY, Glatman Zaretsky A, Byrd AL, et al. Microbiota-dependent sequelae of acute infection compromise tissue-specific immunity. Cell. (2015) 163:354–66. doi: 10.1016/j.cell.2015.08.030
85. Belkacem N, Serafini N, Wheeler R, Derrien M, Boucinha L, Couesnon A, et al. Lactobacillus paracasei feeding improves immune control of influenza infection in mice. PLoS ONE. (2017) 12:e0184976. doi: 10.1371/journal.pone.0184976
86. Shenoy MK, Iwai S, Lin DL, Worodria W, Ayakaka I, Byanyima P, et al. Immune response and mortality risk relate to distinct lung microbiomes in patients with HIV and pneumonia. Am J Respir Crit Care Med. (2017) 195:104–14. doi: 10.1164/rccm.201603-0523OC
87. Kanmani P, Clua P, Vizoso-Pinto MG, Rodriguez C, Alvarez S, Melnikov V, et al. Respiratory commensal bacteria Corynebacterium pseudodiphtheriticum improves resistance of infant mice to respiratory syncytial virus and Streptococcus pneumoniae superinfection. Front Microbiol. (2017) 8:1613. doi: 10.3389/fmicb.2017.01613
88. Weiss B, Bujanover Y, Yahav Y, Vilozni D, Fireman E, Efrati O. Probiotic supplementation affects pulmonary exacerbations in patients with cystic fibrosis: a pilot study. Pediatr Pulmonol. (2010) 45:536–40. doi: 10.1002/ppul.21138
89. Liu S, Hu P, Du X, Zhou T, Pei X. Lactobacillus rhamnosus GG supplementation for preventing respiratory infections in children: a meta-analysis of randomized, placebo-controlled trials. Indian Pediatr. (2013) 50:377–81. doi: 10.1007/s13312-013-0123-z
90. Elazab N, Mendy A, Gasana J, Vieira ER, Quizon A, Forno E. Probiotic administration in early life, atopy, and asthma: a meta-analysis of clinical trials. Pediatrics. (2013) 132:e666–76. doi: 10.1542/peds.2013-0246
91. Huang CF, Chie WC, Wang IJ. Efficacy of lactobacillus administration in school-age children with asthma: a randomized, placebo-controlled trial. Nutrients. (2018) 10:E1678. doi: 10.3390/nu10111678
92. Hojsak I, Abdović S, Szajewska H, Milosević M, Krznarić Z, Kolacek S. Lactobacillus GG in the prevention of nosocomial gastrointestinal and respiratory tract infections. Pediatrics. (2010) 125:e1171–7. doi: 10.1542/peds.2009-2568
93. Hojsak I, Snovak N, Abdović S, Szajewska H, Misak Z, Kolacek S. Lactobacillus GG in the prevention of gastrointestinal and respiratory tract infections in children who attend day care centers: a randomized, double-blind, placebo-controlled trial. Clin Nutr. (2010) 29:312–6. doi: 10.1016/j.clnu.2009.09.008
94. Du X, Wang L, Wu S, Yuan L, Tang S, Xiang Y, et al. Efficacy of probiotic supplementary therapy for asthma, allergic rhinitis, and wheeze: a meta-analysis of randomized controlled trials. Allergy Asthma Proc. (2019) 40:250–60. doi: 10.2500/aap.2019.40.4227
95. Wei X1, Jiang P, Liu J, Sun R, Zhu L. Association between probiotic supplementation and asthma incidence in infants: a meta-analysis of randomized controlled trials. J Asthma. (2019) 2019:1–12. doi: 10.1080/02770903.2018.1561893
96. Ozen M, Kocabas Sandal G, Dinleyici EC. Probiotics for the prevention of pediatric upper respiratory tract infections: a systematic review. Expert Opin Biol Ther. (2015) 15:9–20. doi: 10.1517/14712598.2015.980233
97. Wang Y, Li X, Ge T, Xiao Y, Liao Y, Cui Y, et al. Probiotics for prevention and treatment of respiratory tract infections in children: a systematic review and meta-analysis of randomized controlled trials. Medicine. (2016) 95:e4509. doi: 10.1097/MD.0000000000004509
98. Araujo GV, Oliveira Junior MH, Peixoto DM, Sarinho ES. Probiotics for the treatment of upper and lower respiratory-tract infections in children: systematic review based on randomized clinical trials. J Pediatr. (2015) 91:413–27. doi: 10.1016/j.jped.2015.03.002
Keywords: microbiota, microbiome, lung, airway, asthma, infections, immunity, probiotics
Citation: Pulvirenti G, Parisi GF, Giallongo A, Papale M, Manti S, Savasta S, Licari A, Marseglia GL and Leonardi S (2019) Lower Airway Microbiota. Front. Pediatr. 7:393. doi: 10.3389/fped.2019.00393
Received: 06 May 2019; Accepted: 12 September 2019;
Published: 27 September 2019.
Edited by:
Claudio Pignata, University of Naples Federico II, ItalyReviewed by:
Emily K. Cope, Northern Arizona University, United StatesDaniele Zama, Sant'Orsola-Malpighi Polyclinic, Italy
Copyright © 2019 Pulvirenti, Parisi, Giallongo, Papale, Manti, Savasta, Licari, Marseglia and Leonardi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Giuseppe Fabio Parisi, Z2l1c2VwcGVwYXJpc2k4OEBob3RtYWlsLml0
†Co-first authors
 Giulio Pulvirenti
Giulio Pulvirenti Giuseppe Fabio Parisi
Giuseppe Fabio Parisi Alessandro Giallongo1
Alessandro Giallongo1 Sara Manti
Sara Manti Salvatore Savasta
Salvatore Savasta Gian Luigi Marseglia
Gian Luigi Marseglia Salvatore Leonardi
Salvatore Leonardi