- Department of Pediatrics, Shandong Provincial Hospital Affiliated to Shandong University, Jinan, China
Long non-coding RNA (lncRNA) has been associated with human diseases. To study the role of lncRNA in the pathogenic mechanism of acute fulminant myocarditis (AFM), we used a microarray to analyze lncRNA and messenger RNA (mRNA) expression in leukocyte samples from AFM patients and normal children. In total, using a 2/0.5-fold change and P < 0.05 as the cutoff criteria, we found that 3,101 lncRNAs and 2,170 mRNAs were differentially expressed in AFM patients. Quantitative real-time polymerase chain reaction (RT-qPCR) analysis was used to verify the microarray data. Eight differentially expressed molecules were randomly selected, including 3 upregulated lncRNAs, 3 downregulated lncRNAs, and 2 upregulated mRNAs. Among them, 7 expression profiles were consistent with the microarray results. Gene Ontology enrichment and Kyoto encyclopedia of genes and genomes (KEGG) pathway analysis were used to investigate the biological functions of these genes. Establishment of a lncRNA-mRNA co-expression network and lncRNA target predication were performed to study the molecular interactions of these molecules. Our study is the first to use microarrays to examine the lncRNA and mRNA expression profiles associated with AFM, and the results indicate that the immune system plays an important role in AFM. These findings may provide a new perspective for the pathogenesis, diagnosis, and therapy of AFM.
Introduction
Acute fulminant myocarditis (AFM) is an inflammatory process that occurs in the myocardium and causes acute-onset heart failure (1). It is a type of myocarditis that can arise quickly, progress rapidly and lead to sudden cardiac death, with mortality rates as high as 50–70% (2). At present, many studies have aimed to determine the clinical features of AFM, including the clinical presentation, diagnosis, treatment, and outcome (3–6). However, few studies have examined the pathogenic mechanism of AFM, except for myocarditis. Previous records have confirmed the relationship between myocarditis and the immune system. On the one hand, some moieties, such as Toll-like receptors (TLRs) (7), midkine (8), and STATs (9), can activate inflammatory responses to conserve the host protective system. On the other hand, when an antigen interacts with the variable region of the T cell receptor, acquired immunity will be activated (10). Data have shown that myocarditis is closely related to signaling pathways, such as NF-κB (11), AKT/caspase-3 (12), IL-1β (13), MAPK (14), and TLR-4/NF-κB p65 (15). However, little attention has been focused on the molecular mechanism of the immune system of AFM patients.
Long non-coding RNAs (lncRNAs) are defined as transcripts of more than 200 nucleotides that are not translated into proteins (16), including antisense, intronic, intergenic, pseudogene, and retrotransposon transcripts (16). LncRNAs participate in various developmental processes, acting as signals, decoys, guides, and scaffolds in epigenetic, transcriptional, or post-transcriptional regulation (17). At present, an increasing number of lncRNAs are emerging as having roles in cardiovascular diseases (18). Some lncRNAs are involved in cardiovascular system diseases, including atherosclerosis (19), heart failure (20), and arrhythmia (21). However, data related to myocarditis are scarce, and only two studies have focused on lncRNAs and cardiac inflammation (22, 23). We aim to provide further information on lncRNAs and AFM to examine the potential pathogenicity of lncRNAs in AFM patients. In addition, microarrays have been regarded as a useful tool in transcriptome gene expression profiling. They have been broadly used to investigate the pathobiology of diverse forms of diseases (24, 25).
To determine whether lncRNAs of peripheral leukocytes might correlate with AFM, we used a microarray to analyze the dysregulated profiles of lncRNAs and mRNAs in leukocytes of children with AFM and healthy children.
Materials and Methods
Patients and Samples
We recruited children (aged 4 months−10 years) with AFM in this study based on the following criteria (26): sudden onset of disease, obvious initial symptoms of viral infection (especially severe fatigue and poor appetite), rapidly emerging severe hemodynamic dysfunction, serious myocardial injury, and a diffuse decrease in ventricular wall motion. The exclusion criteria included coronary heart disease, viral pneumonia, sepsis myocarditis, common acute myocarditis, AFM caused by autoimmune disease, toxic drug effects, or drug allergies. The controls consisted of healthy children.
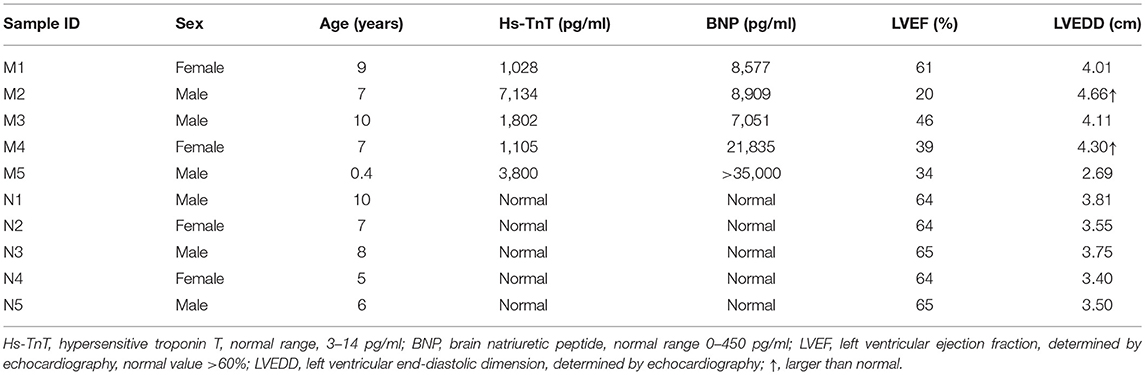
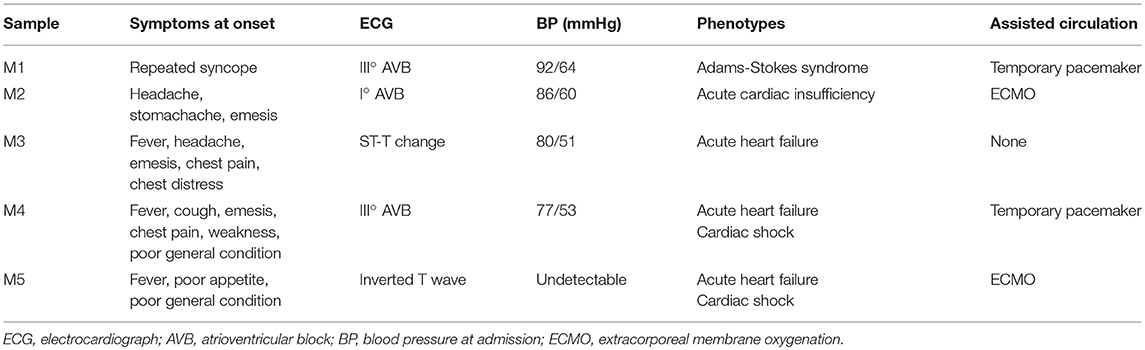
Blood samples (3 ml) were collected into EDTA anticoagulant tubes. Leukocytes were isolated within 4 h and were immediately frozen at −80°C with 1 ml of RNeasy Total RNA Isolation Kit reagent (Qiagen, GmBH, Germany). A total of 10 children from Shandong Provincial Hospital Affiliated to Shandong University (May 2018 to February 2019) were included in this study, including 5 controls and 5 patients. The clinical characteristics of patients and controls are presented in Tables 1, 2. All patients were diagnosed with AFM using cardiac magnetic resonance imaging. Three AFM samples (M1, M2, M3) and three control samples (N1, N2, N3) were used for microarray analysis.
RNA Extraction
Total RNA was extracted from isolated leukocytes using the RNeasy Total RNA Isolation Kit (Qiagen, GmBH, Germany) according to the manufacturer's protocols. The RNA integrity coefficient (RIN) was determined by an Agilent Bioanalyzer 2,100 (Agilent Technologies, Santa Clara, CA, US).
Microarray Analysis
RNA samples were used to generate biotinylated cDNA targets with the Sino Human ceRNA array V3.0 (Shanghai Sinomics Corporation, Shanghai, China) (27). Upon hybridization of the biotinylated cRNA targets, an Agilent Microarray Scanner (Agilent Technologies) was used for slide scanning. Data were extracted with Feature Extraction software 10.7 (Agilent Technologies). Raw data were normalized using the Quantile algorithm in the R package “limma.” Data analysis was conducted at Sinotech Genomics Corporation according to the protocol specified by Agilent Technologies. A fold change cutoff of 2 was adopted.
qRT-PCR Validation
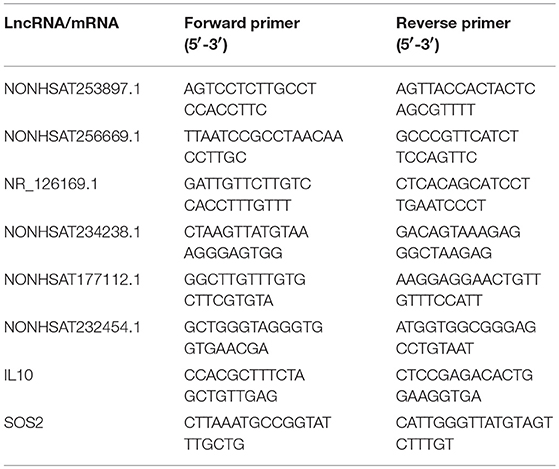
Quantitative real-time polymerase chain reaction (qRT-PCR) analysis with a LightCycler 480 (Roche, Shanghai, China) system was used to verify the microarray data. Extracted RNA was subjected to cDNA synthesis with the PrimeScriptTM RT reagent Kit (Takara, Beijing, China). qRT-PCR was conducted with TB GreenTM Premix Ex TaqTM II (Takara) as directed by the manufacturer. The relative expression levels were determined by the 2−ΔΔCt method. Primer sequences used in the qRT-PCR analysis of lncRNA were shown in Table 3.
GO and KEGG Pathway Analysis
Gene Ontology (GO) analysis was performed to determine the biological significance of genes in unique or representative maps of differentially expressed genes (28). KEGG pathway analysis was used to predict the underlying biological functions of dysregulated lncRNAs in pathways (29). GO and KEGG pathway analysis were performed for annotation of genes as a whole network, and differentially expressed genes were analyzed using Fisher's exact test in the R package “cluster profiler.” GO categories and pathways with P < 0.05 in Fisher's exact test were selected.
Co-expression Network Analysis
We predicted the co-expression relationship between lncRNAs and mRNAs according to the dynamic changes in the gene expression signal values to investigate the relationship between lncRNAs and mRNAs. Through the co-expression network, we analyzed regulatory ability of genes and determined the core regulatory genes. The co-expression network was constructed using Cytoscape.
Cis/trans lncRNA Target Prediction
We predicted the cis and trans targets of lncRNAs. Cis target gene prediction involved identification of mRNAs genes located within 10 kb upstream or downstream of the lncRNA as the target gene of the lncRNA. Trans target gene prediction was based on the principle of complementary sequence pairing. Blast alignment was used to obtain mRNAs that were complementary with lncRNAs. Then, RNA plex software was used to calculate the thermodynamic parameters of lncRNAs that were complementary with mRNAs, and sequences with e ≤30 were selected.
Statistical Analysis
Statistically significant differences between groups were estimated by the Mann-Whitney U test using SPSS 25.0; P < 0.05 was considered statistically significant.
Results
Differential Expression of lncRNAs and mRNAs
Three samples each from the AFM patient and healthy control groups were analyzed using Sino Human ceRNA Array V3.0 microarray hybridization. Volcano plot analysis was used to assess variations in lncRNA and mRNA expression between these two populations (Figures 1A,B). Moreover, hierarchical clustering was used to distinguish AFM patients from healthy children based on gene expression data (Figures 1C,D). In total, using a 2/0.5-fold change and P < 0.05 as the cutoff criteria, 3,101 lncRNAs displayed differential expression in AFM patients, including 1,645 upregulated lncRNAs, and 1,456 downregulated lncRNAs. The top of 10 upregulated and downregulated lncRNAs are shown in Table 4. The distribution of differentially expressed lncRNAs on human chromosomes is shown in Figure 1E. In addition, a total of 2,170 mRNAs were dysregulated; among them, 733 were upregulated, and 1,437 were downregulated.
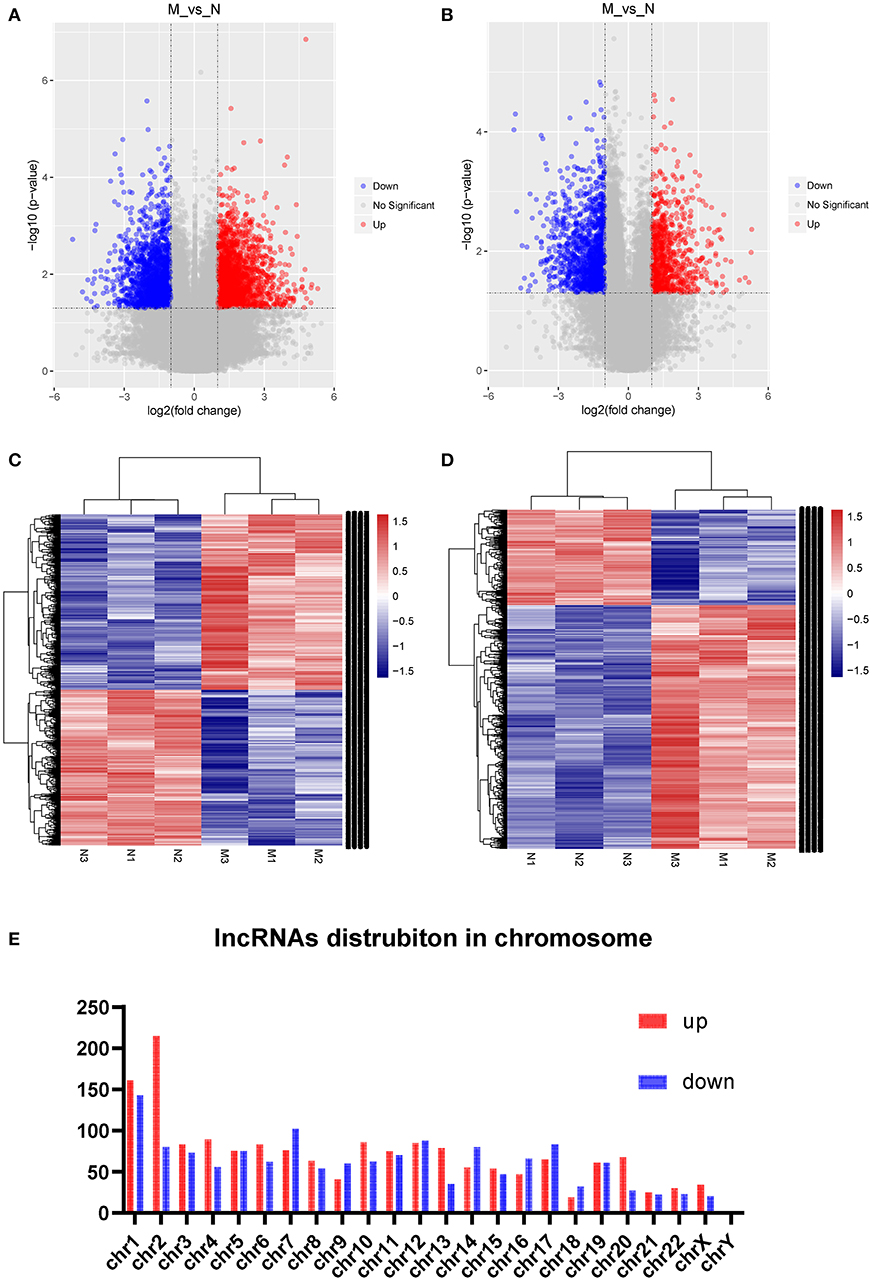
Figure 1. Expression profile of lncRNAs and mRNAs in AFM patients. Volcano plots were used to distinguish differentially expressed lncRNAs (A) and mRNAs (B). Hierarchical clustering analysis of differentially expressed lncRNAs (C) and mRNAs (D). Distribution of dysregulated lncRNAs in human chromosomes (E). Red and blue represent upregulated and downregulated expression, respectively. M indicates the experimental group; N indicates the normal group.
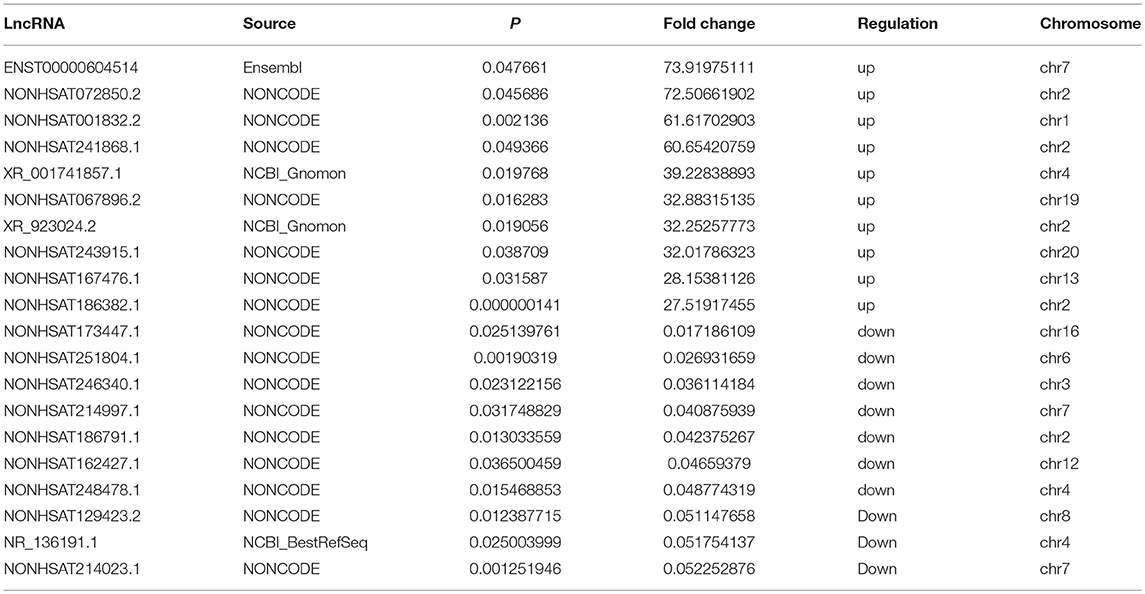
Table 4. Top 10 upregulated and downregulated (fold change ≥2 and P < 0.05) lncRNAs in AFM patients.
Validation by qRT-PCR
To confirm the microarray data, we randomly selected 3 upregulated lncRNAs (NONHSAT253897.1, NONHSAT177112.1, and NONHSAT234238.1), 3 downregulated lncRNAs (NONHSAT256669.1, NR_126169.1, and NONHSAT232454.1), and 2 mRNAs (IL10 and SOS2), for qRT-PCR analysis (n = 10) (Figure 2). The results of 5 lncRNAs (NONHSAT253897.1, NONHSAT177112.1, NONHSAT256669.1, NR_126169.1, and NONHSAT232454.1) and 2 mRNAs (IL10 and SOS2) were consistent with the microarray data. The results for NONHSAT234238.1 were inconsistent with the trend sown by the microarray data.
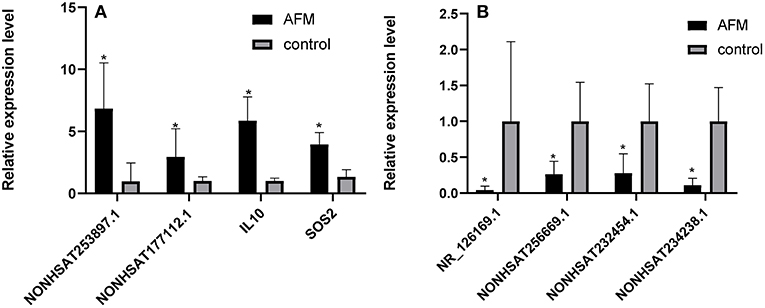
Figure 2. Comparison of the relative RNA expression levels between AFM patients and normal children. The expression of 2 upregulated mRNAs (A) and 3 downregulated and 1 upregulated lncRNA (B) were validated using the 2−ΔΔCt method. The data are displayed as the mean ± SD, and samples of the two groups were compared using the Mann-Whitney U test. *P < 0.05; AFM acute fulminant myocarditis; lncRNAs long non-coding RNAs.
GO and Pathway Analysis
We performed GO analysis to determine the potential biological role of the dysregulated lncRNAs. The GO enrichment terms of the 30 top cis- (Figure 3A) and trans-acting lncRNAs (Figure 3D) were determined. The most prominent GO terms were T cell activation and T cell receptor complex for cis-acting lncRNAs and pyramidal neuron development, nuclear membrane, and transmembrane receptor protein serine/threonine kinase activity for trans-acting lncRNAs. According to the KEGG classification, the immune system and signal transduction were notable pathways. We chose the 10 top KEGG pathways (Figures 3C,D) within the immune system and signal transduction pathways are notable in Supplementary Figure 1; the notable pathways included hematopoietic cell lineage, complement and coagulation cascades, antigen processing, and presentation, T cell receptor signaling pathway, and Jak-STAT signaling pathway.
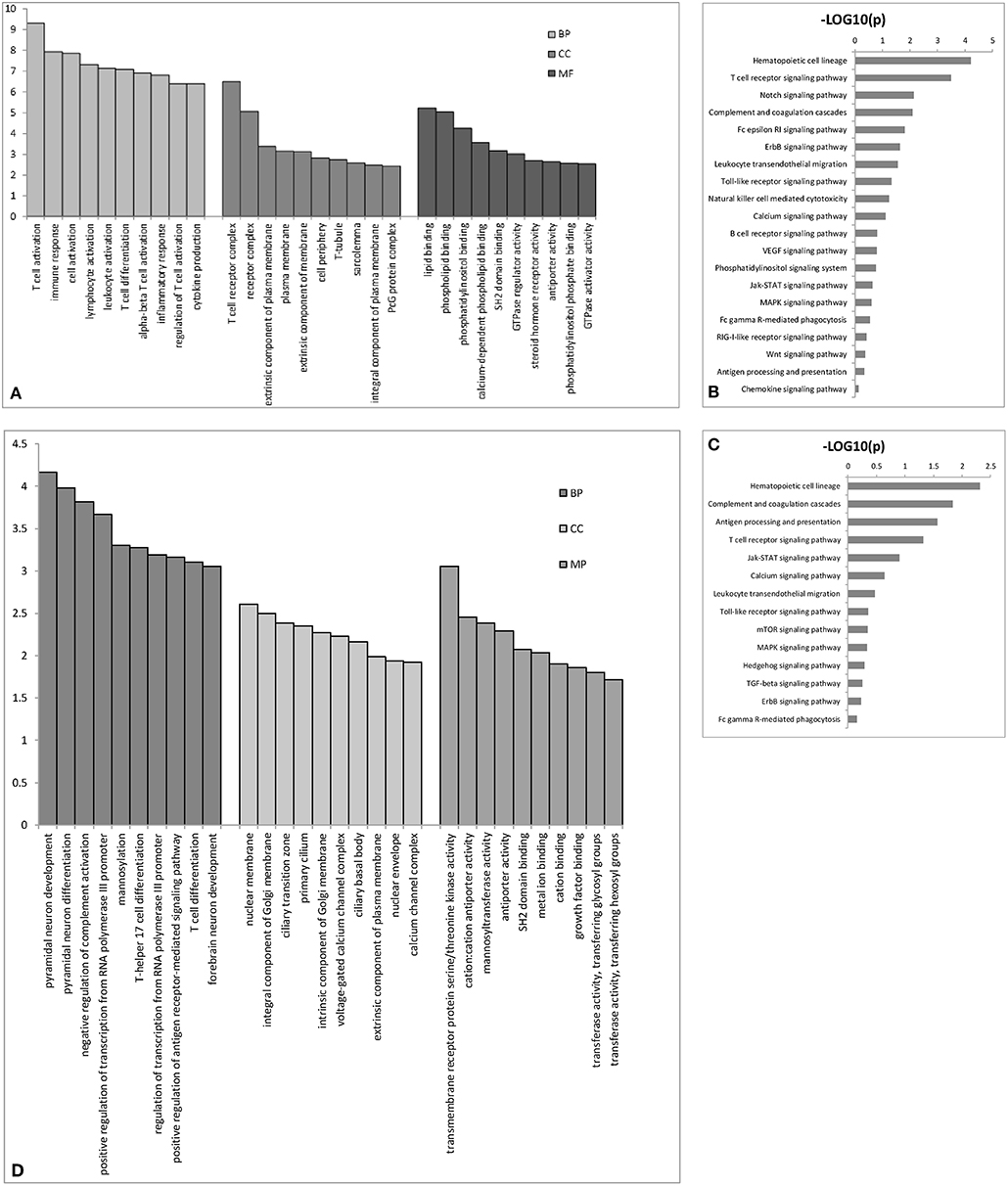
Figure 3. The top 30 GO terms for cis- (A) or trans-acting lncRNAs (D). The top 10 KEGG pathways for cis- (B) or trans-acting lncRNAs (C). BP biological process; CC cellular component; MP molecular function.
lncRNA-mRNA Co-expression Network
To predict gene function, we constructed an lncRNA-mRNA co-expression network and performed pathway analyses (Figure 4). We identified a total of 95 lncRNAs and 20 mRNAs in the T cell receptor and Jak-STAT signaling pathways (Pearson's coefficient >0.95). The co-expression network was composed of 115 network nodes and 186 connections. Each mRNA was associated with 1 to 17 lncRNAs, and each lncRNA was associated with 1–31 mRNAs.
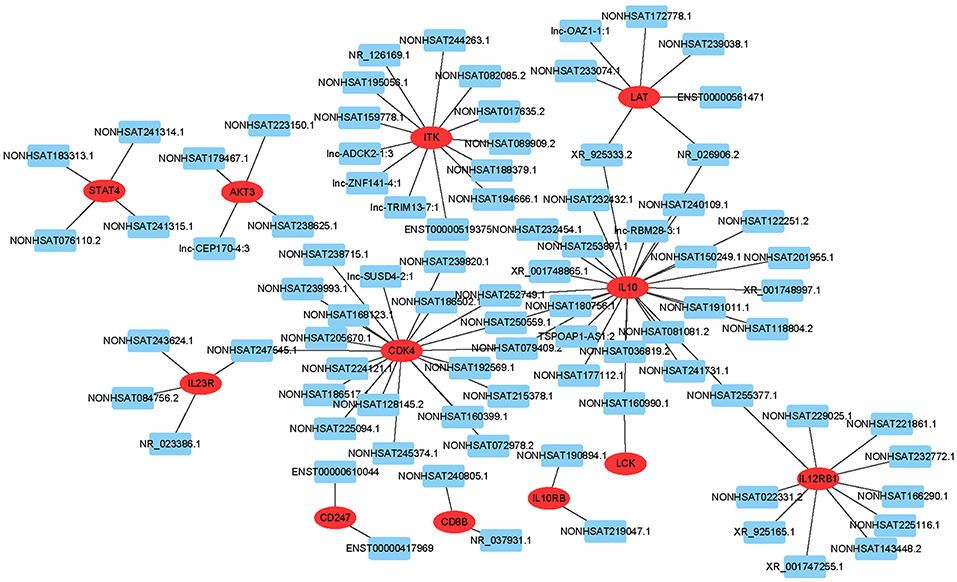
Figure 4. lncRNA-mRNA network analysis of the T cell receptor and Jak-STAT signaling pathways (Pearson's coefficient > 0.95). Red ovals represent mRNAs, blue rectangles represent lncRNAs, a line represents the correlation.
Cis/trans lncRNA Target Prediction
Target prediction was performed for differentially expressed lncRNAs to investigate whether they can regulate genes and to determine the signaling pathways associated with AFM. We performed partial lncRNA target prediction in the T cell receptor and Jak-STAT signaling pathways (Figure 5). The principle of cis target gene prediction is that the function of the lncRNA is related to the protein-coding genes adjacent to its location. The basic principle of trans target gene prediction is that the lncRNA is a distant transcriptional activator or repressor of the target. The data showed that most lncRNAs acted in a cis manner. This information may aid in determining the functional mechanism of AFM.
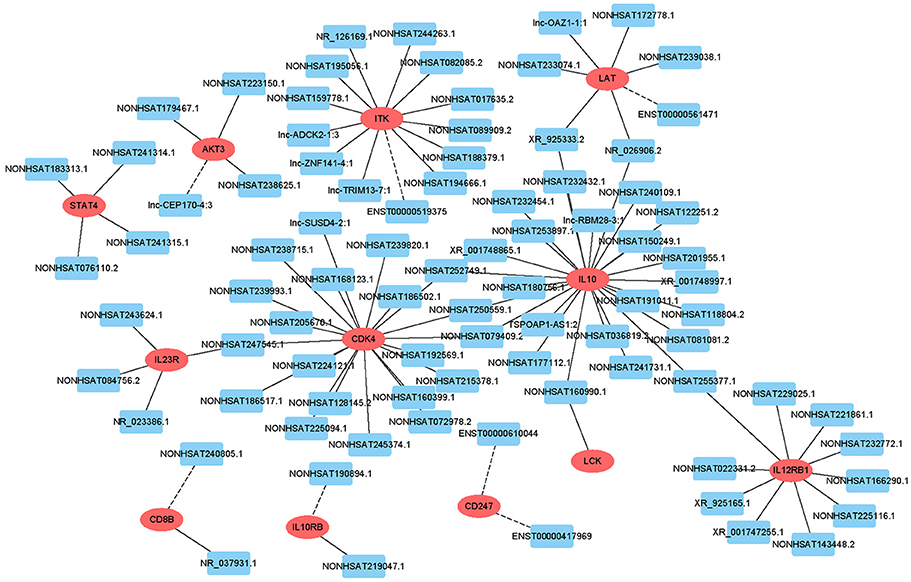
Figure 5. Cis/trans lncRNA target prediction in the T cell receptor and Jak-STAT signaling pathways. Red ovals represent mRNAs, blue rectangles represent lncRNAs. The solid line indicates a trans lncRNA, and the dotted line indicates a cis lncRNA.
Discussion
AFM is characterized by acute and severe inflammation and global myocardium injury (30). The etiology and pathophysiological mechanism of AFM are unknown because of its complexity. Moreover, most of the present research focuses on the diagnosis and treatment of AFM, and the molecular mechanism of AFM is rarely reported. Many researchers believe that AFM is closely related to the immune system, which provides a platform for the study of the pathophysiological mechanisms of AFM.
LncRNAs including antisense, intronic, intergenic, pseudogene, and retrotransposon transcripts, play a vital role in biological processes (16), acting as signals, decoys, guides and scaffolds in epigenetic, transcriptional, or post-transcriptional regulation, and these molecules are emerging as dominating regulators of gene expression in the immune system. At present, most descriptions of the function of lncRNAs include modulation of the target genomic loci in a cis or in trans manner by binding to target DNA based on recognition of specific chromatin features or as an RNA-DNA heteroduplex or RNA-DNA-DNA triplex (31). LncRNAs can also function through RNA-RNA interactions. They can act as “sponges” for miRNA (32) or act by N6-methyladenosine to modify introns to form a secondary structure (33). Many studies have focused on the roles of lncRNAs in the immune system, but only a few studies have demonstrated that lncRNAs are associated with AFM. Zhang et al. (22) studied the relationship between lncRNAs and myocardial inflammation for the first time. They suggested that lncRNA TUG1 inhibits apoptosis and the inflammatory response in lipopolysaccharide (LPS)-treated H9c2 cells by downregulating mir-29b. Zhang et al. (23) demonstrated that silencing lncRNA CHRF protects H9c2 cells against LPD-induced injury via upregulation of mir-221. These two studies discussed the relationship between lncRNA and microRNA but did not examine mRNA. However, research on the transcriptional regulation of myocarditis has mainly focused on the role of mRNAs and their translated proteins.
Our study showed differential expression profiles of lncRNAs and mRNAs in children with AFM and healthy controls. We found 1,645 upregulated lncRNAs, 1,456 downregulated lncRNAs, 733 upregulated mRNAs, and 1,437 downregulated mRNAs. To verify the accuracy of the microarray, we randomly selected 8 molecules for qRT-PCR, including 3 upregulated lncRNAs (NONHSAT253897.1, NONHSAT177112.1, and NONHSAT234238.1), 3 downregulated lncRNAs (NONHSAT256669.1, NR_126169.1, and NONHSAT232454.1), and 2 upregulated mRNAs (IL-10 and SOS2). Among them, 7 molecules showed the same upregulation or downregulation trends of lncRNAs in the AFM and healthy groups. Therefore, the results from the qPCR analysis coincided with the microarray data.
Next, we used GO and KEGG analysis to determine the potential biological functions of differentially expressed lncRNAs and mRNAs. The most notable cellular processes were immune processes, including T cell activation, immune response, T cell receptor complex, negative regulation of complement activation, T-helper 17 cell differentiation, and T cell differentiation for GO terms, and those for the KEGG pathways included the complement and coagulation cascades, antigen processing and presentation, the T cell receptor signaling pathway, the Jak-STAT signaling pathway, the TLR signaling pathway, and the MAPK signaling pathway. These data further indicate the accuracy of the microarray analysis and provide more potential biological functions of lncRNAs and mRNAs related to AFM.
Furthermore, we constructed an lncRNA-mRNA co-expression network in the T cell receptor and Jak-STAT signaling pathways to obtain additional information on lncRNAs and mRNAs. We compared differentially correlated lncRNAs and mRNAs from the leukocytes of AFM patients and healthy children, which will provide a better understanding of the pathogenic mechanism of AFM.
In conclusion, the data indicate that mutual regulation between lncRNAs and mRNAs may be involved in the pathogenic process of AFM, and the results provide essential information to identify AFM.
Our study had some limitations. First, due to the low incidence of fulminant myocarditis, the sample size of the microarray analysis and that used to verify the results was small. Second, the subsequent functional verification needs to be further improved.
Data Availability
The raw data supporting the conclusions of this manuscript will be made available by the authors, without undue reservation, to any qualified researcher.
Ethics Statement
This study was conducted in accordance with the recommendations and guidelines of the Ethics Committee of Shandong Provincial Hospital Affiliated to Shandong University. All study participants and their parents gave written informed consent in accordance with the Declaration of Helsinki. The protocol was approved by the Ethics Committee of Shandong Provincial Hospital Affiliated to Shandong University.
Author Contributions
QL designed the study and performed the experiments. YK, LZ, and HJ performed the experiments, analyzed the data, and wrote the manuscript. BH and DJ supervised the experiments.
Funding
This study was supported by the Science and Technology Development Plan of Jinan City (No. 201805020), the Taishan scholars (No. ts201511099), and National Natural Science Foundation of China (No. 8187020860).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fped.2019.00283/full#supplementary-material
Supplementary Figure 1. KEGG classification of lncRNAs in patients with AFM. Immune system and signal transduction are notable in the figure.
References
1. Gupta S, Markham DW, Drazner MH, Mammen PP. Fulminant myocarditis. Nat Clin Pract Cardiovasc Med. (2008) 5:693–706. doi: 10.1038/ncpcardio1331
2. Wang D, Li S, Jiang J, Yan J, Zhao C, Wang Y, et al. Chinese society of cardiology expert consensus statement on the diagnosis and treatment of adult fulminant myocarditis. Sci China Life Sci. (2018) 62:187–202. doi: 10.1007/s11427-018-9385-3
3. Ammirati E, Veronese G, Cipriani M, Moroni F, Garascia A, Brambatti M, et al. Acute and fulminant myocarditis: a pragmatic clinical approach to diagnosis and treatment. Curr Cardiol Rep. (2018) 20:114. doi: 10.1007/s11886-018-1054-z
4. Callan PD, Baltabaeva A, Kamal M, Wong J, Lane R, Robertus JL, et al. Acute fulminant necrotizing eosinophilic myocarditis: early diagnosis and treatment. ESC Heart Fail. (2017) 4:660–4. doi: 10.1002/ehf2.12146
5. Wang Q, Pan W, Shen L, Wang X, Xu S, Chen R, et al. Clinical features and prognosis in Chinese patients with acute fulminant myocarditis. Acta Cardiol. (2012) 67:571–6. doi: 10.1080/AC.67.5.2174132
6. Xiong H, Xia B, Zhu J, Li B, Huang W. Clinical outcomes in pediatric patients hospitalized with fulminant myocarditis requiring extracorporeal membrane oxygenation: a meta-analysis. Pediatr Cardiol. (2017) 38:209–14. doi: 10.1007/s00246-016-1517-1
7. Marsland BJ, Nembrini C, Grun K, Reissmann R, Kurrer M, Leipner C, et al. TLR ligands act directly upon T cells to restore proliferation in the absence of protein kinase C-theta signaling and promote autoimmune myocarditis. J Immunol. (2007) 178:3466–73. doi: 10.4049/jimmunol.178.6.3466
8. Weckbach LT, Grabmaier U, Uhl A, Gess S, Boehm F, Zehrer A, et al. Midkine drives cardiac inflammation by promoting neutrophil trafficking and NETosis in myocarditis. J Exp Med. (2019) 216:350–68. doi: 10.1084/jem.20181102
9. Gonnella PA, Del Nido PJ, McGowan FX. Oral tolerization with cardiac myosin peptide. (614-629) ameliorates experimental autoimmune myocarditis: role of STAT 6 genes in BALB/CJ mice. J Clin Immunol. (2009) 29:434–43. doi: 10.1007/s10875-009-9290-z
10. Tavares PS, Rocon-Albuquerque R, Leite-Moreira AF. Innate immune receptor activation in viral myocarditis: pathophysiologic implications. Rev Port Cardiol. (2010) 29:57–78. doi: 10.1038/ki.2010.111
11. Xue YL, Zhang SX, Zheng CF, Li YF, Zhang LH, Hao YF, et al. Silencing of STAT4 protects against autoimmune myocarditis by regulating Th1/Th2 immune response via inactivation of the NF-κB pathway in rats. Inflammation. (2019). doi: 10.1007/s10753-019-00978-3. [Epub ahead of print].
12. Wang X, Li M, Yu Y, Liu G, Yu Y, Zou Y, et al. FTY720 alleviates coxsackievirus B3-induced myocarditis and inhibits viral replication through regulating sphingosine 1-phosphate receptors and AKT/caspase-3 pathways. J Cell Physiol. (2019) 234:18029–40. doi: 10.1002/jcp.28434
13. Kraft L, Erdenesukh T, Sauter M, Tschope C, Klingel K. Blocking the IL-1beta signalling pathway prevents chronic viral myocarditis and cardiac remodeling. Basic Res Cardiol. (2019) 114:11. doi: 10.1007/s00395-019-0719-0
14. Zheng C, Wu SM, Lian H, Lin YZ, Zhuang R, Thapa S, et al. Low-intensity pulsed ultrasound attenuates cardiac inflammation of CVB3-induced viral myocarditis via regulation of caveolin-1 and MAPK pathways. J Cell Mol Med. (2019) 23:1963–75. doi: 10.1111/jcmm.14098
15. Liu T, Zhang M, Niu H, Liu J, Ruilian M, Wang Y, et al. Astragalus polysaccharide from Astragalus Melittin ameliorates inflammation via suppressing the activation of TLR-4/NF-kappaB p65 signal pathway and protects mice from CVB3-induced virus myocarditis. Int J Biol Macromol. (2019) 126:179–86. doi: 10.1016/j.ijbiomac.2018.12.207
16. Kopp F, Mendell JT. Functional classification and experimental dissection of long non-coding RNAs. Cell. (2018) 172:393–407. doi: 10.1016/j.cell.2018.01.011
17. Ma H, Hao Y, Dong X, Gong Q, Chen J, Zhang J, et al. Molecular mechanisms and function prediction of long non-coding RNA. Scient World J. (2012) 2012:541786. doi: 10.1100/2012/541786
18. Coelho-Lima J, Spyridopoulos I. Non-coding RNA regulation of T cell biology: implications for age-associated cardiovascular diseases. Exp Gerontol. (2018) 109:38–46. doi: 10.1016/j.exger.2017.06.014
19. Cremer S, Michalik KM, Fischer A, Pfisterer L, Jae N, Winter C, et al. Hematopoietic deficiency of the long non-coding RNA MALAT1 promotes atherosclerosis and plaque inflammation. Circulation. (2018) 139:1320–34. doi: 10.1161/CIRCULATIONAHA.117.029015
20. Boeckel JN, Perret MF, Glaser SF, Seeger T, Heumuller AW, Chen W, et al. Identification and regulation of the long non-coding RNA Heat2 in heart failure. J Mol Cell Cardiol. (2018) 126:13–22. doi: 10.1016/j.yjmcc.2018.11.004
21. Li Z, Wang XM, Wang WZ, Du JJ, Wei JQ, Zhang Y, et al. Altered long non-coding RNA expression profile in rabbit atria with atrial fibrillation: TCONS_00075467 modulates atrial electrical remodeling by sponging miR-328 to regulate CACNA1C. J Mol Cell Cardiol. (2017) 108:73–85. doi: 10.1016/j.yjmcc.2017.05.009
22. Zhang H, Li H, Ge A, Guo E, Liu S, Zhang L. Long non-coding RNA TUG1 inhibits apoptosis and inflammatory response in LPS-treated H9c2 cells by down-regulation of miR-29b. Biomed Pharmacother. (2018) 101:663–9. doi: 10.1016/j.biopha.2018.02.129
23. Zhang L, Wang L, Guo E, Qi Y. Silence of lncRNA CHRF protects H9c2 cells against lipopolysaccharide-induced injury via up-regulating microRNA-221. Exp Mol Pathol. (2019) 107:43–50. doi: 10.1016/j.yexmp.2019.01.010
24. Kierzek R, Turner DH, Kierzek E. Microarrays for identifying binding sites and probing structure of RNAs. Nucleic Acids Res. (2015) 43:1–12. doi: 10.1093/nar/gku1303
25. Mantione KJ, Kream RM, Kuzelova H, Ptacek R, Raboch J, Samuel JM, et al. Comparing bioinformatic gene expression profiling methods: microarray and RNA-Seq. Med Sci Monit Basic Res. (2014) 20:138–42. doi: 10.12659/MSMBR.892101
26. Sun D, Ding H, Zhao C, Li Y, Wang J, Yan J, et al. Value of SOFA, APACHE IV and SAPS II scoring systems in predicting short-term mortality in patients with acute myocarditis. Oncotarget. (2017) 8:63073–83. doi: 10.18632/oncotarget.18634
27. Zhang H, Zhu L, Bai M, Liu Y, Zhan Y, Deng T, et al. Exosomal circRNA derived from gastric tumor promotes white adipose browning by targeting the miR-133/PRDM16 pathway. Int J Cancer. (2018) 144:2501–15. doi: 10.1002/ijc.31977
28. Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. (2009) 4:44–57. doi: 10.1038/nprot.2008.211
29. Khatri P, Sirota M, Butte AJ. Ten years of pathway analysis: current approaches and outstanding challenges. PLoS Comput Biol. (2012) 8:e1002375. doi: 10.1371/journal.pcbi.1002375
30. Zhang J, Hang W, Hui R, Zhao Q, Desai SS. China's treatment regimen for fulminant myocarditis is bringing wonderful achievement to the world. Sci China Life Sci. (2019) 62:282–4. doi: 10.1007/s11427-018-9445-2
31. Chen YG, Satpathy AT, Chang HY. Gene regulation in the immune system by long non-coding RNAs. Nat Immunol. (2017) 18:962–72. doi: 10.1038/ni.3771
32. Dykes IM, Emanueli C. Transcriptional and post-transcriptional gene regulation by long non-coding RNA. Genomics Proteomics Bioinformat. (2017) 15:177–86. doi: 10.1016/j.gpb.2016.12.005
Keywords: acute fulminant myocarditis, long non-coding RNAs, microarray, messenger RNA, functional analysis
Citation: Liu Q, Kong Y, Han B, Jiang D, Jia H and Zhang L (2019) Long Non-coding RNA Expression Profile and Functional Analysis in Children With Acute Fulminant Myocarditis. Front. Pediatr. 7:283. doi: 10.3389/fped.2019.00283
Received: 04 April 2019; Accepted: 25 June 2019;
Published: 11 July 2019.
Edited by:
Fu Lijun, Shanghai Children's Medical Center, ChinaReviewed by:
Bing He, Wuhan University, ChinaGiuseppe Limongelli, Second University of Naples, Italy
Copyright © 2019 Liu, Kong, Han, Jiang, Jia and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bo Han, aGFuYm8zNSYjeDAwMDQwOzE2My5jb20=
 Qingqing Liu
Qingqing Liu Yaru Kong
Yaru Kong Bo Han
Bo Han