- 1Division of Neonatology, Children’s Hospital of Philadelphia, Philadelphia, PA, United States
- 2Division of Neonatology, Children’s Hospital of Philadelphia, Perelman School of Medicine, Philadelphia, PA, United States
Neonatal meningitis is a devastating condition. Prognosis has not improved in decades, despite the advent of improved antimicrobial therapy and heightened index of suspicion among clinicians caring for affected infants. One in ten infants die from meningitis, and up to half of survivors develop significant lifelong complications, including seizures, impaired hearing and vision, and delayed or arrested development of such basic skills as talking and walking. At present, it is not possible to predict which infants will suffer poor outcomes. Early treatment is critical to promote more favorable outcomes, though diagnosis of meningitis in infants is technically challenging, time-intensive, and invasive. Profound neuronal injury has long been described in the setting of neonatal meningitis, as has elevated levels of many pro- and anti-inflammatory cytokines. Mechanisms of the host immune response that drive clearance of the offending organism and underlie brain injury due to meningitis are not well understood, however. In this review, we will discuss challenges in diagnosis, prognosis, and treatment of neonatal meningitis. We will highlight transcriptomic, proteomic, and metabolomic data that contribute to suggested mechanisms of inflammation and brain injury in this setting with a view toward fruitful areas for future investigation.
Introduction
Meningitis is a life-threatening disease, affecting 0.1–0.4 neonates per 1,000 live births, with a higher incidence in preterm and chronically hospitalized infants (1, 2). Approximately 10% of affected infants die, and 20–50% of survivors develop seizures, cognitive deficiencies, motor abnormalities, and hearing and visual impairments (3). Despite declines in mortality, morbidity has not improved since the 1970s.
Rapid initiation of appropriate broad-spectrum antimicrobial therapy in response to suspected neonatal meningitis is critical to optimize outcomes (4). Empiric therapy often chosen in the setting of suspected early-onset sepsis/meningitis ensures coverage of Group B Streptococcus (GBS), Listeria monocytogenes, and Gram-negative organisms, such as Escherichia coli (4, 5). Suspected late-onset infections are typically treated with even broader antimicrobial therapy to cover additional organisms in the nosocomial environment, including Pseudomonas aeruginosa and methicillin-resistant Staphylococcus aureus. Antibiotic therapy can only be narrowed if bacterial cultures reveal an offending organism, which hinges on swift, successful performance of a technically challenging, and sometimes risky, lumbar puncture (LP) prior to administration of antibiotics.
At a minimum, recommended therapy for meningitis lasts several weeks and may be longer based on organism or more extensive disease, such as ventriculitis or abscess formation (5). Infants with negative cultures despite a suspicion of meningitis are often managed conservatively and receive a full course of antibiotic therapy. In neonates, long-term antibiotic therapy is associated with emergence of resistant bacteria, superimposed fungal infections, and increased risk of necrotizing enterocolitis (6). Additional risks associated with long-term antibiotic therapy include need for central vascular access and its attendant complications (7). Thus, infants may sustain harm not only from meningitis but also from associated interventions.
Prognosis in the setting of neonatal meningitis is also fraught with difficulty. While outcomes are influenced by time to diagnosis and therapy, prognosis may also relate to virulence of the infecting pathogen. With our current understanding of meningitis, however, it is not possible to predict which infants will die, which infants will survive with disabilities, and which infants will survive with a normal neurodevelopmental outcome. Indeed, many infants still die, and many survivors still sustain lifelong morbidities, despite rapid initiation of appropriate antimicrobial therapy (8, 9).
Several adjuncts to antibiotic therapy have been proposed and tested to improve poor outcomes associated with meningitis. Steroid administration has not been shown to be beneficial as an adjunct to antimicrobial therapy in neonatal bacterial meningitis (10). While there is evidence for improved neurologic and auditory outcomes in pediatric Hemophilus influenzae B meningitis following steroid therapy, this pathogen is not a frequent cause of meningitis in neonates (11). The data surrounding benefit of steroids in pediatric or neonatal meningitis secondary to GBS, E. coli, Streptococcus pneumonia, and Neisseria meningitidis is unclear or poor, while long-term risks of exposing neonates to steroids are incompletely understood. Therefore, steroids are not recommended as adjunctive therapy in neonates evaluated for meningitis, unless there is strong suspicion for H. influenzae infection based on Gram stain or culture results. Trials of other adjunctive therapies, such as intravenous immunoglobulin and granulocyte-monocyte colony-stimulating factor, have also shown disappointing results.
To enhance outcomes, we must gain deeper insights into the pathophysiology of meningitis to identify diagnostic and prognostic tools and therapies that facilitate bacterial clearance but limit deleterious immune-mediated damage to brain tissue. Herein, we review challenges associated with the rapid and accurate diagnosis of neonatal meningitis. We discuss how a variety of large-scale datasets have extended our understanding of the host response to meningitis. Finally, we integrate the findings into a model to highlight new avenues for basic and translational investigations into the key immunologic pathways active in the setting of neonatal meningitis.
Diagnostic Challenges in Neonatal Meningitis
Culture of cerebrospinal fluid (CSF) is the traditional gold standard for diagnosis of bacterial meningitis. However, deciding when to perform LP to obtain and analyze CSF is challenging. Factors complicating this decision include the non-specific signs and symptoms of meningitis in the infant, cardiorespiratory instability that may preclude positioning of an infant for LP, and considerable practice variation (12–15). Meningitis is estimated to occur in approximately 1–2% of suspected cases of sepsis within the first 72 h of life, or early-onset sepsis, though the risk is limited almost entirely to symptomatic infants (16). The American Academy of Pediatrics policy statement on suspected or proven early-onset bacterial sepsis supports performing LP as part of the sepsis evaluation of the neonate with symptoms concerning for early-onset sepsis, but recommends a more limited evaluation in the asymptomatic neonate with sepsis risk factors (17). In contrast to early-onset sepsis, evaluations for late-onset sepsis (after the first 72 h of life) are almost always performed in response to concerning signs and symptoms. Several studies have noted discordance between blood culture and CSF culture results, with negative blood cultures in up to a third of infants with bacterial meningitis, highlighting that this diagnosis could be missed if LP is deferred or not performed (13–15). These findings strongly support obtaining CSF via LP prior to antibiotic administration in neonates evaluated for late-onset sepsis. In infants with positive blood cultures, an LP is essential to guide duration of therapy and provide prognostic information.
Interpretation of CSF results is frequently problematic. If LP is delayed, and infants are exposed to empiric broad-spectrum antibiotics, clinical yield of bacterial culture of CSF can be compromised (18–20). In these situations, clinicians rely on interpretation of CSF parameters, such as cell count, glucose, and protein levels to presumptively diagnose meningitis. However, there is considerable overlap in laboratory evaluation of CSF between groups of infected and uninfected infants, leading to difficulty in establishing cutoff levels that possess sufficient sensitivity and specificity for the diagnosis of bacterial meningitis (14, 15). Many other factors influence interpretation of these values in neonates, including gestational age, postnatal age, and trauma sustained during LP causing contamination of CSF with blood (12, 14, 15, 18, 21).
Furthermore, diagnostic markers may differ depending on the gestational maturity of the infant and may confound analyses unless proper controls matched for gestational age are included. Additional studies are warranted to understand how these markers vary during normal development and to better inform how they vary in the context of neonatal meningitis. Indeed, many studies have highlighted differences in the immune system of newborns and infants relative to that of children and adults (22, 23). The premature infant may be additionally immune-compromised compared to full-term infants, due, in part, to deficiencies in innate and adaptive immune system (22). Clinically, these deficiencies manifest as increased risk for coagulase-negative Staphylococcus spp., Staphylococcus aureus, and Candida spp. Of note, however, in an investigation of infants with sepsis, Smith et al. found no significant differences in key genes activated or repressed in the blood of infants of varying gestational ages (24). These data suggest that some key immune pathways activated upon serious bacterial infection may be similar between the preterm and the full-term infant.
Pathogen-Based Evaluation of CSF
A variety of approaches have been examined to improve the rapidity and fidelity of diagnosis of neonatal bacterial meningitis over conventional methods. These tests can be categorized broadly as: (1) identification of microbial signatures in CSF and (2) detection of a host response signature pathognomonic of sepsis and meningitis. There is much interest in development of bacterial nucleic acid-based polymerase chain reaction (PCR) assays for detection of common pathogens implicated in sepsis and meningitis (25). These tests have the potential advantage of rapid turnaround time compared to conventional microbiologic culture methods. Further, PCR-based testing has the ability to identify small amounts of nucleic acid signatures from non-viable bacteria, thereby improving diagnostic yield, especially in situations with low bacterial load, such as following antibiotic pretreatment (26). Investigators have studied conventional and real-time PCR methods, and several have employed broad-based bacterial PCR techniques directed against the 16S ribosomal RNA subunit that is conserved across bacterial species. However, in recent years, greater technical success has been noted with multiplex PCRs targeted to several common pathogens implicated in meningitis (27–29). Boriskin et al. reported the use of a focused microarray to detect and distinguish known genomic sequences of 13 viruses causing meningitis (30). Ben and colleagues employed a similar type of array to detect sequences of 20 common bacteria implicated in meningitis (31). Recently, a multiplex PCR assay has been approved by the FDA for detection of 14 pathogens, including Escherichia coli K1, Streptococcus agalactiae, Listeria monocytogenes, and Haemophilus influenzae (28, 29). Nonetheless, additional testing is required to determine the reliability of CSF PCR tests when compared with culture results in the management of bacterial meningitis in the clinical setting. To date, bacterial tests of sources other than CSF have shown disappointingly poor accuracy in diagnosis of meningitis (32).
Cytokine-Based Evaluation of CSF
The second approach to developing diagnostic tests is based on the premise that serious infections such as meningitis elicit a specific host immune and/or metabolic response that could represent a “host signature” pathognomonic of infection (33–36). Several studies of pediatric and neonatal meningitis have evaluated cytokine levels and other candidate biomarkers in CSF for their diagnostic utility (Table 1). In a prospective cohort of infants with suspected bacterial meningitis, Srinivasan et al. noted that interleukin-6 and interleukin-10 possessed the best area under the curve (AUC) in receiver operating characteristic analyses of multiple cytokines [Table 1 and Ref. (33)]. In a study of 140 subjects with pediatric meningitis, Ye et al. identified interleukin-6 alone and the ratio of CSF/blood interleukin-6 as useful diagnostic markers [Table 1 and Ref. (37)]. However, not all investigations of interleukin-6 have replicated this success. Mukai and colleagues noted poorer diagnostic accuracy with interleukin-6 compared to tumor necrosis factor-α, and Hsieh’s group demonstrated that interleukin-6 had good sensitivity but poorer specificity than interleukin-12 [Table 1 and Ref. (38, 39)]. Prasad et al. also noted that tumor necrosis factor-α and interleukin-8 provided excellent ability to discriminate bacterial meningitis, and outperformed interleukin-6, in their cohort of 87 pediatric patients [Table 1 and Ref. (40)]. Ours and an additional group concluded that, while tumor necrosis factor-α had good specificity for bacterial meningitis in infants and children, sensitivity was less promising [Table 1 and Ref. (41, 42)]. Challenges noted by investigators include the short half-life of many cytokines with only transient elevations in CSF in the setting of bacterial meningitis. The timing of LP in relation to onset of illness, therefore, becomes a crucial factor in interpretation of levels. As noted previously, however, it is often a challenge to obtain LPs early in the course of illness in unstable infants.
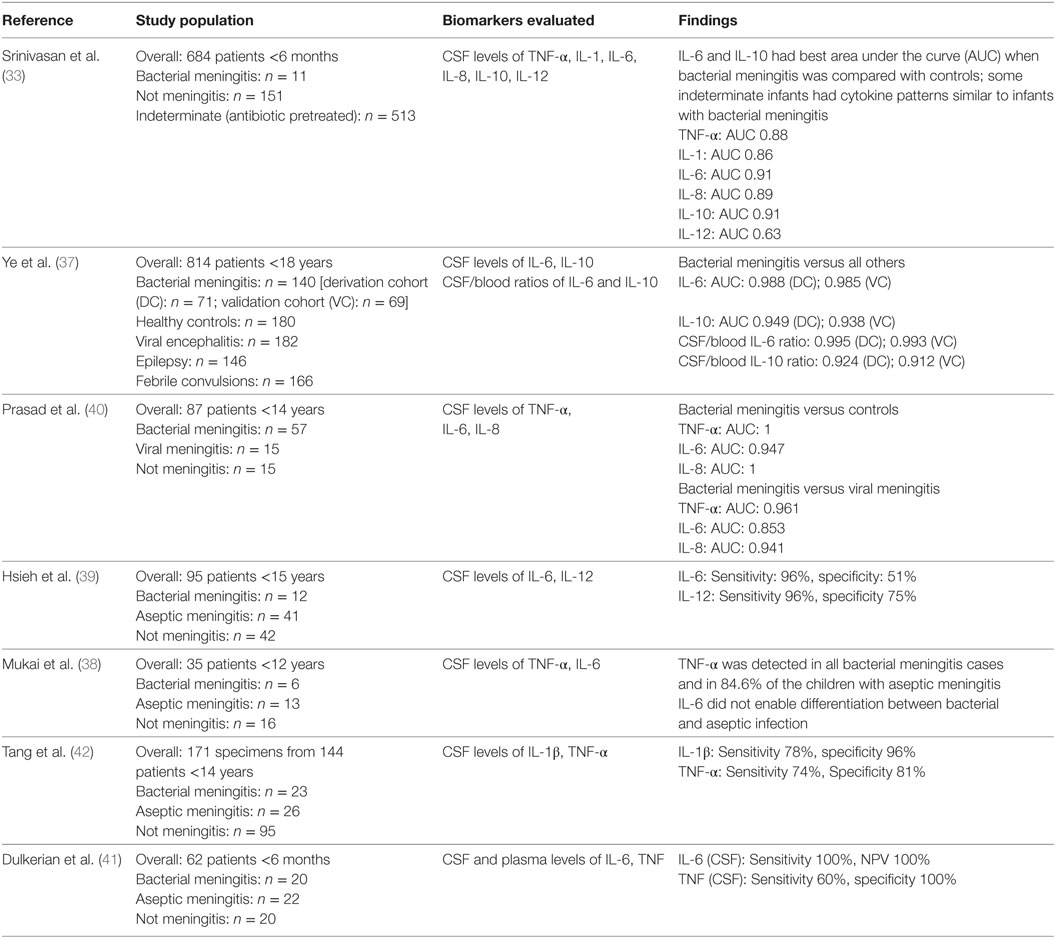
Table 1. Studies investigating diagnostic accuracy of cerebrospinal fluid (CSF) cytokines in pediatric and neonatal meningitis.
Biomarkers, or combinations of biomarkers, that demonstrate not only early but sustained elevations over the course of infection may be better candidates for diagnostic tests. Alterations in damage-associated molecular patterns (DAMPs) that aid in perpetuation of the inflammatory response may provide more stable measurement targets. S100-family proteins are DAMPs associated with innate immune activation (43). S100B showed early promise as a biomarker for meningitis due to its concentration in astrocytes and glia (44, 45). Further investigations, however, revealed that S100B in serum or CSF exhibited suboptimal sensitivity and specificity for bacterial meningitis. The highest, but still inconsistent, levels of S100B in CSF and serum were seen in viral encephalitis or in bacterial meningitis complicated by ventriculitis or obvious parenchymal abnormalities on brain imaging (44, 46). While many candidate cytokine and immunologic markers have shown promise as diagnostic tools in small studies, some have demonstrated conflicting findings across studies (Table 1). Thus, such markers have not been accepted into clinical practice as adjunctive tests for diagnosis of meningitis. These data emphasize the need for novel investigations into other classes of potential biomarkers to enhance diagnosis of meningitis beyond our current capability with clinical judgment and classical laboratory parameters.
Omics to Extend Knowledge of Bacterial Meningitis
Recently, investigators have embarked on exploratory approaches, taking advantage of advanced “omics” technologies to more deeply understand serious bacterial infection in infants, children, and adults (34–36, 47). Omics denotes the comprehensive investigation of any family of biologic molecules, including DNA (genomics), RNA transcripts (transcriptomics), proteins (proteomics), and metabolites (metabolomics), among others. In other words, omics are used to interrogate hundreds to thousands of molecules simultaneously from a body fluid or tissue, promoting generation of pathways or networks of molecules associated with presence or absence of a disease state, such as infection. Omics approaches can provide new insights into diagnosis, pathophysiology, risk stratification and prognostication, and potential therapeutic targets (48). Systems-level investigations into host–pathogen interactions involving many cells types have informed countless translational studies of complex human diseases. Bacterial meningitis is a complex infectious disease amenable to omics analyses to generate new hypotheses into mechanisms underlying inflammation and brain injury. A common obstacle to rigorous mechanistic studies of many human disorders, use of primary human tissue to study meningitis is generally not possible. CSF is an attractive source of information, as it communicates directly or indirectly with brain-resident cells, such as neurons, astrocytes, glia, and the blood–brain barrier. CSF is also in direct contact with infiltrating cells of the immune system recruited to the brain space in the setting of meningitis. Thus, CSF contains hosts of transcripts, proteins, and metabolites that, together, can paint a clearer picture of a disease process whose morbidity and mortality remain unacceptably high despite early recognition, definitive treatment with appropriate antibiotic therapy, and intensive care. Herein, we note advances made with model systems of human meningitis, but we will focus on transcriptomic, proteomic, and metabolomic work performed in human patients with bacterial meningitis.
Transcriptomics
Transcriptomics reveals genome-wide changes in gene expression by individual cells or populations of cells and often forms the backbone for gene-targeted analyses in animal models. Transcriptomics has evolved as an increasingly affordable and fruitful analysis, thanks in large part to several large, well-curated databases capable of organizing results into networks of known pathways. Numerous transcriptomic analyses in model systems of bacterial meningitis have utilized cultured human brain microvascular endothelial cells and cultured primary meningioma cells (49, 50). When cultured appropriately, such cells can recapitulate the blood–brain barrier in vitro. Both systems have been used to understand changes in host gene expression occurring upon contact with bacterial pathogens, such as N. meningitidis (51–53).
Animal models of meningitis have also provided great insight into the pathophysiology of neonatal meningitis, dating back to the first in vivo model in piglets to show disruption of the blood–brain barrier (54). Currently, rodent models of neonatal meningitis are often used to model human bacterial meningitis (55–58). One study by Coimbra and colleagues used a high-fidelity infant rat model of pneumococcal meningitis to separately profile the transcriptomes of the cortex and hippocampus in the early and late phases of disease (59). This model faithfully replicates much of the pathophysiology found in humans insofar as infant rats suffer similar cortical ischemia with resultant necrosis and hippocampal apoptosis as do human neonates who contract bacterial meningitis (60). Many genes associated type 1 inflammation and toll-like receptor activation, such as interleukin-6, interleukin-18, and STAT1, were increased in both the cortex and hippocampus of infant rats after induction of meningitis (59). Consistent with the distinct mechanisms of injury in the cortex and hippocampus, however, Coimbra et al. found differential regulation of genes associated with toll-like receptor signaling and innate immune activation, including CD14 and tumor necrosis factor-α, in each brain region. Supporting histologic findings, pro-apoptotic transcripts were upregulated selectively in the hippocampus, while increased expression of tropomyosin selectively in the cortex may reflect excess vasoconstriction and ischemia.
Another intriguing set of transcripts found to be upregulated in the cortex and hippocampus of infant rats with meningitis were several associated with complement activation, including C1q, C3, and C4 (59). In addition to a canonical role for complement in bacterial clearance, recent work has highlighted an additional role for complement in crosstalk with astrocytes and microglia to eliminate neuronal synapses (61–63). Indeed, a transcript indicative of activation of astrocytes (glial fibrillary acidic protein, or GFAP) was similarly upregulated in both the cortex and hippocampus of infant rats with meningitis. While clearance of synapses in the setting of bacterial meningitis has not been formally addressed, it is tempting to speculate that this may represent a critical mechanism of brain injury. Altogether, these data highlight the critical importance of animal models to our understanding of bacterial meningitis in human neonates. They also shed light on potential therapeutic targets as we gain greater knowledge of appropriate and maladaptive immune responses to bacterial meningitis.
While the transcriptome of brain tissue or CSF has not been examined in humans with meningitis, profiling of the transcriptome of whole blood in human patients with bacterial meningitis was performed by Lill and colleagues (64). Although the bulk of their study cohort was composed of adult patients, three neonates were included in the analysis: two with GBS meningitis and one with E. coli meningitis. Of note, clustering analysis of their data revealed that the neonates with meningitis had distinct whole blood transcriptomic profiles from the adults with meningitis. While adults with meningitis had a distinct whole blood transcriptome compared to adults without infection, the whole blood transcriptome of two out of three neonates most closely resembled that of control adults. This emphasizes the need for infant-specific studies of meningitis, as the neonatal and adult host responses to infection vary substantially (23, 65).
In the setting of bacterial meningitis, the most significantly changed individual and networks of transcripts in the blood were those associated with innate and adaptive immunity (64). Interestingly, type 2 immunity-associated genes, including interleukin-5 receptor, Fc-epsilon receptors, and markers of mast cell activation, were among the most upregulated in the blood of patients with bacterial meningitis relative to controls. These data support findings that type 2 immune responses are induced in humans and animal models with serious bacterial infection (66). In fact, type 2 immune signaling to macrophages drives better outcomes in animal models of sepsis and is associated with reduced severity of disease in humans with sepsis (66, 67). The data also are congruent with work to profile the inflammatory cytokine milieu in CSF of infants and children with bacterial meningitis, described above, showing type 2 cytokines interleukin-4 and interleukin-13 significantly upregulated in the CSF of infants and children with bacterial meningitis compared to controls (33, 68). Future studies will be needed to understand the significance of type 2 immunity in the setting of bacterial meningitis, especially in neonates who often exhibit blunted type 1 immune responses (65).
Notably, the study by Lill and colleagues (64) represents a first step to detect a signature of bacterial meningitis in the blood, outside of the brain space. Such lines of investigation may drive discovery and development of minimally invasive tools to diagnose bacterial meningitis, one day obviating the need for LP.
Proteomics
One caveat of transcriptomics is that some transcripts may not be translated into protein due to posttranscriptional modifications or regulation by non-coding RNA species. Transcriptomics also has limited clinical utility in the intensive care unit, as detection of mRNA transcripts can be expensive and time consuming. Proteomics offers the promise of novel biomarker discovery that may easily translate to clinical applications. Jesse and colleagues (69) applied the first large-scale proteomic approach to the study of bacterial and viral meningitis in adults. Analyzing CSF from adult patients with fluorescent two-dimensional difference gel electrophoresis, the authors aimed to find a protein signature of bacterial meningitis that differentiates it from viral meningitis and from controls. Six proteins were identified as candidate markers of bacterial meningitis in a pilot experiment and were subsequently validated in a larger cohort of patients. Interestingly, GFAP was one such protein marker specific to bacterial meningitis that was also identified by Coimbra et al. as an increased transcript in the setting of bacterial meningitis in infant rats (59). Activation of astrocytes and expression of GFAP can be induced by pro-inflammatory cytokines, such as interleukin-6 (70). Another protein marker of bacterial meningitis identified by Jesse et al. was Prostaglandin-H2 D-isomerase or prostaglandin D synthase (69). Recently, prostaglandin D synthase was implicated as a marker of potent T helper 2 cells that strongly elaborate type 2 cytokines, including interleukin-4, interleukin-5, and interleukin-13 (71). These data invoke the transcriptomic findings of Lill and colleagues, who showed increased levels of transcripts associated with type 2 immunity in the serum of patients with bacterial meningitis (64). As evidenced by this work, multiple omics modalities can be combined and used to link data across multiple studies and disciplines.
Using similar methodology to that of Jesse et al., Goonetilleke and colleagues performed comparative two-dimensional polyacrylamide gel electrophoresis to characterize the proteome of CSF from adult humans with pneumococcal meningitis (72). The focus of their analysis was to identify markers differentially regulated in survival versus death. Among the proteins relevant to the immune response, complement C3 was decreased in non-survivors, suggesting impaired complement-mediated bacterial clearance in the brain space or, possibly, impaired clearance of injured neurons by astrocytes and microglia (61–63), as discussed above. Another interesting finding was an increase in chitotriosidase in CSF of non-survivors. Chitotriosidase is associated with activation of macrophages, especially those exposed to type 2 inflammatory cytokines (73). As discussed above, there are several pieces of data supporting robust type 2 inflammation in bacterial meningitis, but whether excessive type 2 immune bias in the setting of bacterial meningitis is maladaptive and associated with poor outcomes remains to be investigated. The data of Goonetilleke et al. should be interpreted with some caution, as all patients with meningitis were coinfected with HIV in this study, but the interesting findings begin to address a critical need to predict not only diagnosis but also prognosis of bacterial meningitis. This is especially important in the neonatal population, whose potential disabilities due to meningitis are numerous and would benefit from early subspecialist involvement and intervention.
Cordeiro et al. presented the most recent proteomic data in the setting of meningitis, with a goal to differentiate among control subjects, those with pneumococcal meningitis, those with meningococcal meningitis, and those with enteroviral meningitis (74). The cohort consisted of adults and children, and CSF was analyzed using two-dimensional protein gel electrophoresis. The authors propose an algorithm to predict bacterial meningitis based on elevated apolipoprotein A1, also found to be elevated in bacterial meningitis by Song and colleagues (75), and complement C3, which participates in clearance of encapsulated microbes and in clearance of neuronal synapses (61–63). Once classified as bacterial meningitis, kininogen-1 was identified to discriminate between pneumococcal and meningococcal disease, though kininogen-1 is a pleiotropic effector molecule with possible, but poorly defined, effects on pro-inflammatory cytokine signaling, neurotransmission, and blood–brain barrier function (74). Overall, proteomic approaches to the study of bacterial meningitis are powerful and may lay the groundwork for future translational studies and clinical assays to improve rapidity and fidelity of diagnosis. While pediatric subjects were more represented in the study by Cordeiro and colleagues, there remains an absence of work dedicated to comprehensively addressing the proteome of neonates with bacterial meningitis.
Metabolomics
While approaches to simultaneously evaluate multitudes of metabolites have become available only recently, metabolic analysis of CSF is nearly a century old (76). Consistent with presumed cellular stress and high metabolic demand, elevated levels of lactate and decreased levels of glucose were among the first markers to be associated with meningitis. Since the original studies, lactate has been well validated as a highly accurate marker of bacterial meningitis (77). Additional investigations into the CSF metabolome of patients with meningitis have been performed, with a goal of discovering novel biomarkers of disease and uncovering aberrantly regulated pathways contributing to maladaptive inflammation and brain injury. Metabolomics offers an advantage over proteomics and transcriptomics in that metabolites are the final products of active enzymatic reactions in a cell or group of cells. In other words, metabolomics most accurately reflects the true cellular phenotype without having to consider posttranscriptional or posttranslational modifications that may add layers of complexity to transcriptomic or proteomic datasets, respectively. Challenges to pursuing metabolomic work, however, include expense, non-standardized modalities to acquire data, and still-evolving databases with which to compare and contextualize results.
Several studies of CSF in humans with bacterial meningitis employ proton nuclear magnetic resonance (NMR) spectroscopy to study a varied number of metabolites (78–80). Using NMR spectroscopy, Subramanian et al. investigated whether metabolites could distinguish CSF of children with tuberculous meningitis from CSF of children with non-tuberculous bacterial meningitis, viral meningitis, or no meningitis (79). One fascinating finding was detection of the metabolite cyclopropane specifically in CSF from children with tuberculous meningitis (79). Modifications to mycolic acids with cyclopropane in the cell wall of M. tuberculosis are critical for virulence (81). This piece of data highlights the potential of metabolomics to identify key pathways relevant to diagnosis and pathophysiology of bacterial meningitis.
In the study by Subramanian et al., other metabolites significantly elevated in CSF of those with bacterial meningitis relative to viral meningitis or controls included urea, creatine, alanine, citrate, pyruvate, acetoacetate, and beta-hydoxybutyrate (79). These data, particularly elevations in acetoacetate and beta-hydroxybutyrate, suggest increased ketosis and an increase in circulating free fatty acids that are subsequently metabolized into ketone bodies. Interestingly, poor outcomes in a cohort of adults with sepsis have been associated with increased levels of fatty acids in the serum (82). Further underscoring the importance of fatty acid oxidation in the immune response to infection, pro-inflammatory responses have been linked to activation of glycolytic pathways, while oxidation of fatty acids and ketosis has been linked to anti-inflammatory pathways in neonates with sepsis (22, 24). Altogether, these data may reflect inappropriately blunted immunity that may predispose to poor outcomes after serious bacterial infection, including sepsis and meningitis.
Overall, Subramanian et al. quantified twelve metabolites but distinguished non-tuberculous bacterial meningitis from controls with sensitivity and specificity of only 74 and 67%, respectively. When clinical variables were also considered, however, the model’s sensitivity and specificity rose to nearly 100%. These data speak to the value of combining multiple modalities in omics research, an approach recently used to create a high-fidelity clinical-metabolomic model of outcomes after bacterial sepsis in adult humans (82).
Coen et al. evaluated the CSF metabolome of mainly adult humans with bacterial, fungal, and viral meningitis, as compared to that of control adults and those with indwelling neurosurgical hardware (80). Of note, the cohort did include two infants with S. agalactiae (Group B Strep) meningitis. The group addressed a critical question relevant to the population in the NICU, as a small but significant proportion of infants born extremely prematurely will develop intraventricular hemorrhage and, subsequently, posthemorrhagic hydrocephalus. Using NMR spectroscopy to ascertain metabolic composition of CSF, Coen and colleagues were able to distinguish patients with bacterial meningitis from those with viral meningitis and those with no meningitis but with indwelling hardware. The most “influential” metabolites used to separate experimental groups were glucose and lactate, with additional contribution from beta-hydroxybutyrate, pyruvate, acetate, acetone, isoleucine, leucine, and valine. Of note, branched-chain amino acids, especially valine, have been shown to promote maturation, antigen presentation capability, and cytokine production of innate immune cells (83). The investigators were not able to distinguish CSF infected with Gram-positive organisms from CSF infected with Gram-negative organisms (80), in contrast to the findings of Lill and colleagues, who found unique changes in the plasma transcriptome of patients with bacterial meningitis due to pneumococcus relative to those with meningitis due to other bacterial organisms (64). This is perhaps due to small sample size, though it is feasible that the active metabolic pathways and mechanisms of cellular injury contributing to the CSF metabolome may not vary substantially among different microorganisms.
Although expensive, mass spectrometry-based approaches to metabolomics are alternatives to NMR spectroscopy and are capable of characterizing hundreds of metabolites from body fluids and tissues (84). No studies utilizing mass spectrometry-based metabolomics have been performed in patients, especially infants, with bacterial meningitis. To fill this gap in knowledge and complement known transcriptomic and proteomic data, it will be necessary to more comprehensively investigate the metabolome of CSF in model organisms and in human infants with bacterial meningitis.
Conclusion
Despite improvements in neonatal intensive care and timely administration of appropriate broad-spectrum antimicrobial agents, bacterial meningitis remains a major cause of morbidity and mortality in the neonatal period. Our understanding of mechanisms of neonatal brain injury due to meningitis remains limited. Although use of cytokines as biomarkers of meningitis has not achieved sufficient accuracy to be employed in clinical practice, these data complement a wealth of omics data from humans and model organisms with bacterial meningitis that now exist. Efforts to integrate these data sets and assemble pathway maps of genes, proteins, and metabolites altered in the setting of bacterial meningitis (Figure 1) will be of great benefit to advance the field. Subsequent approaches combining omics analyses of primary human tissue (e.g., CSF) with targeted studies in animal models will be required to understand the complex interplay among the numerous infiltrating and brain-resident cell types affected by bacterial meningitis. Such work can drive development of novel, rapid diagnostics and adjunctive therapies that may prevent the devastating, lifelong sequelae of bacterial meningitis in infants.
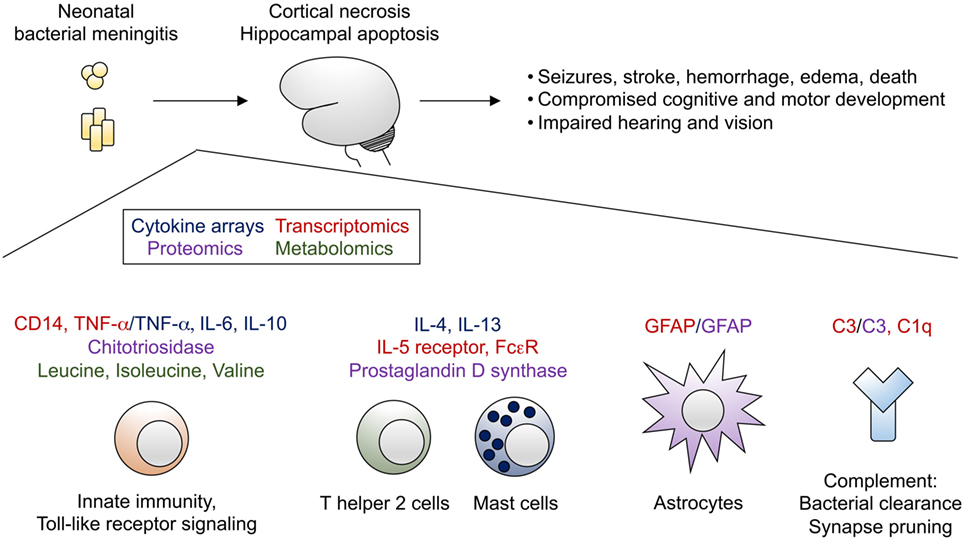
Figure 1. Bacterial meningitis of the neonate leads to profound brain injury, characterized by cortical necrosis and hippocampal apoptosis. Selected transcripts (highlighted in red), cytokines (highlighted in blue), other proteins (highlighted in purple), and metabolites (highlighted in green) identified from omics analyses have been integrated into a proposed model of immune activation in the setting of bacterial meningitis.
Author Contributions
SG, LS, and MH jointly conceived, wrote, and critically revised the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Okike IO, Johnson AP, Henderson KL, Blackburn RM, Muller-Pebody B, Ladhani SN, et al. Incidence, etiology, and outcome of bacterial meningitis in infants aged <90 days in the United kingdom and Republic of Ireland: prospective, enhanced, national population-based surveillance. Clin Infect Dis (2014) 59:e150–7. doi:10.1093/cid/ciu514
2. Lawn JE, Cousens S, Zupan J, Lancet Neonatal Survival Steering Team. 4 million neonatal deaths: when? where? why? Lancet (2005) 365:891–900. doi:10.1016/S0140-6736(05)71048-5
3. Mann K, Jackson MA. Meningitis. Pediatr Rev (2008) 29:417–29; quiz 430. doi:10.1542/pir.29-12-417
4. Heath PT, Okike IO. Neonatal bacterial meningitis: an update. Paediatr Child Health (2010) 20:526–30. doi:10.1016/j.paed.2010.04.001
5. American Academy of Pediatrics. Escherichia coli and other Gram-negative bacilli (septicemia and meningitis in neonates). In: Pickering LK, Baker CJ, Kimberlin DW, Long SS, editors. Red Book: 2012 Report of the Committee on Infectious Diseases. Elk Grove Village, IL: American Academy of Pediatrics (2012). p. 321–4.
6. Gritz EC, Bhandari V. The human neonatal gut microbiome: a brief review. Front Pediatr (2015) 3:17. doi:10.3389/fped.2015.00017
7. Coffin SE, Zaoutis TE. Healthcare-associated infections in the nursery. In: Remington JS, Klein JO, Wilson CB, Maldonado YA, editors. Infectious Diseases of the Fetus and Newborn. 7th ed. Philadelphia, PA: W.B. Saunders (2011). p. 1126–43.
8. Tan J, Kan J, Qiu G, Zhao D, Ren F, Luo Z, et al. Clinical prognosis in neonatal bacterial meningitis: the role of cerebrospinal fluid protein. PLoS One (2015) 10:e0141620. doi:10.1371/journal.pone.0141620
9. Janowski AB, Newland JG. From the microbiome to the central nervous system, an update on the epidemiology and pathogenesis of bacterial meningitis in childhood. F1000Res (2017) 6:86. doi:10.12688/f1000research.8533.1
10. Heath PT, Okike IO, Oeser C. Neonatal meningitis: can we do better? In: Curtis N, Finn A, Pollard AJ, editors. Hot Topics in Infection and Immunity in Children VIII. New York: Springer (2011). p. 11–24.
11. Brouwer MC, McIntyre P, Prasad K. Corticosteroids for acute bacterial meningitis. Cochrane Database Syst Rev (2015) CD004405. doi:10.1002/14651858.CD004405.pub5
12. Srinivasan L, Harris MC, Shah SS. Lumbar puncture in the neonate: challenges in decision making and interpretation. Semin Perinatol (2012) 36:445–53. doi:10.1053/j.semperi.2012.06.007
13. Stoll BJ, Hansen N, Fanaroff AA, Wright LL, Carlo WA, Ehrenkranz RA, et al. To tap or not to tap: high likelihood of meningitis without sepsis among very low birth weight infants. Pediatrics (2004) 113:1181–6. doi:10.1542/peds.113.5.1181
14. Garges HP, Moody MA, Cotten CM, Smith PB, Tiffany KF, Lenfestey R, et al. Neonatal meningitis: what is the correlation among cerebrospinal fluid cultures, blood cultures, and cerebrospinal fluid parameters? Pediatrics (2006) 117:1094–100. doi:10.1542/peds.2005-1132
15. Smith PB, Garges HP, Cotton CM, Walsh TJ, Clark RH, Benjamin DK. Meningitis in preterm neonates: importance of cerebrospinal fluid parameters. Am J Perinatol (2008) 25:421–6. doi:10.1055/s-0028-1083839
16. Ray B, Mangalore J, Harikumar C, Tuladhar A. Is lumbar puncture necessary for evaluation of early neonatal sepsis? Arch Dis Child (2006) 91:1033–5. doi:10.1136/adc.2006.105106
17. Polin RA; The Committee on Fetus and Newborn. Management of neonates with suspected or proven early-onset bacterial sepsis. Pediatrics (2012) 129:1006–15. doi:10.1542/peds.2012-0541
18. Srinivasan L, Shah SS, Padula MA, Abbasi S, McGowan KL, Harris MC. Cerebrospinal fluid reference ranges in term and preterm infants in the neonatal intensive care unit. J Pediatr (2012) 161:729–34. doi:10.1016/j.jpeds.2012.03.051
19. Kanegaye JT, Soliemanzadeh P, Bradley JS. Lumbar puncture in pediatric bacterial meningitis: defining the time interval for recovery of cerebrospinal fluid pathogens after parenteral antibiotic pretreatment. Pediatrics (2001) 108:1169–74. doi:10.1542/peds.110.3.651
20. Greenberg RG, Benjamin DK, Cohen-Wolkowiez M, Clark RH, Cotten CM, Laughon M, et al. Repeat lumbar punctures in infants with meningitis in the neonatal intensive care unit. J Perinatol (2011) 31:425–9. doi:10.1038/jp.2010.142
21. Srinivasan L, Shah SS, Abbasi S, Padula MA, Harris MC. Traumatic lumbar punctures in infants hospitalized in the neonatal intensive care unit. Pediatr Infect Dis J (2013) 32:1150–2. doi:10.1097/INF.0b013e31829862b7
22. Kan B, Razzaghian HR, Lavoie PM. An immunological perspective on neonatal sepsis. Trends Mol Med (2016) 22:290–302. doi:10.1016/j.molmed.2016.02.001
23. Levy O. Innate immunity of the newborn: basic mechanisms and clinical correlates. Nat Rev Immunol (2007) 7:379–90. doi:10.1038/nri2075
24. Smith CL, Dickinson P, Forster T, Craigon M, Ross A, Khondoker MR, et al. Identification of a human neonatal immune-metabolic network associated with bacterial infection. Nat Commun (2014) 5:4649. doi:10.1038/ncomms5649
25. Pammi M, Flores A, Versalovic J. Molecular assays for the diagnosis of sepsis in neonates. Cochrane Database Syst Rev (2015) 2:CD011926. doi:10.1002/14651858.CD011926.pub2
26. Srinivasan L, Pisapia JM, Shah SS, Halpern CH, Harris MC. Can broad-range 16S ribosomal ribonucleic acid gene polymerase chain reactions improve the diagnosis of bacterial meningitis? A systematic review and meta-analysis. Ann Emerg Med (2012) 60:609.e–20.e. doi:10.1016/j.annemergmed.2012.05.040
27. Chiba N, Murayama SY, Morozumi M, Nakayama E, Okada T, Iwata S, et al. Rapid detection of eight causative pathogens for the diagnosis of bacterial meningitis by real-time PCR. J Infect Chemother (2009) 15:92–8. doi:10.1007/s10156-009-0670-3
28. Arora HS, Asmar BI, Salimnia H, Agarwal P, Chawla S, Abdel-Haq N. Enhanced identification of group B Streptococcus and Escherichia coli in young infants with meningitis using the biofire filmarray meningitis/encephalitis panel. Pediatr Infect Dis J (2017):1–12. doi:10.1097/INF.0000000000001551
29. Graf EH, Farquharson MV, Cárdenas AM. Comparative evaluation of the FilmArray meningitis/encephalitis molecular panel in a pediatric population. Diagn Microbiol Infect Dis (2017) 87:92–4. doi:10.1016/j.diagmicrobio.2016.09.022
30. Boriskin YS, Rice PS, Stabler RA, Hinds J, Al-Ghusein H, Vass K, et al. DNA microarrays for virus detection in cases of central nervous system infection. J Clin Microbiol (2004) 42:5811–8. doi:10.1128/JCM.42.12.5811-5818.2004
31. Ben RJ, Kung S, Chang FY, Lu JJ, Feng NH, Hsieh YD. Rapid diagnosis of bacterial meningitis using a microarray. J Formos Med Assoc (2008) 107:448–53. doi:10.1016/S0929-6646(08)60152-7
32. Morrissey SM, Nielsen M, Ryan L, Dhanhani AH, Meehan M, McDermott S, et al. Group B streptococcal PCR testing in comparison to culture for diagnosis of late onset bacteraemia and meningitis in infants aged 7–90 days: a multi-centre diagnostic accuracy study. Eur J Clin Microbiol Infect Dis (2017):1–8. doi:10.1007/s10096-017-2938-3
33. Srinivasan L, Kilpatrick L, Shah SS, Abbasi S, Harris MC. Cerebrospinal fluid cytokines in the diagnosis of bacterial meningitis in infants. Pediatr Res (2016) 80:566–72. doi:10.1038/pr.2016.117
34. Mahajan P, Kuppermann N, Mejias A, Suarez N, Chaussabel D, Casper TC, et al. Association of RNA biosignatures with bacterial infections in febrile infants aged 60 days or younger. JAMA (2016) 316:846–57. doi:10.1001/jama.2016.9207
35. Wong HR, Cvijanovich NZ, Anas N, Allen GL, Thomas NJ, Bigham MT, et al. Developing a clinically feasible personalized medicine approach to pediatric septic shock. Am J Respir Crit Care Med (2015) 191:309–15. doi:10.1164/rccm.201410-1864OC
36. Langley RJ, Tipper JL, Bruse S, Baron RM, Tsalik EL, Huntley J, et al. Integrative “Omic” analysis of experimental bacteremia identifies a metabolic signature that distinguishes human sepsis from systemic inflammatory response syndromes. Am J Respir Crit Care Med (2014) 190:445–55. doi:10.1164/rccm.201404-0624OC
37. Ye Q, Shao WX, Shang SQ, Shen HQ, Chen XJ, Tang YM, et al. Clinical value of assessing cytokine levels for the differential diagnosis of bacterial meningitis in a pediatric population. Medicine (2016) 95:e3222. doi:10.1097/MD.0000000000003222
38. Mukai AO, Krebs VL, Bertoli CJ, Okay TS. TNF-α and IL-6 in the diagnosis of bacterial and aseptic meningitis in children. Pediatr Neurol (2006) 34:25–9. doi:10.1016/j.pediatrneurol.2005.06.003
39. Hsieh CC, Lu JH, Chen SJ, Lan CC, Chow WC, Tang RB. Cerebrospinal fluid levels of interleukin-6 and interleukin-12 in children with meningitis. Childs Nerv Syst (2009) 25:461–5. doi:10.1007/s00381-008-0715-4
40. Prasad R, Kapoor R, Srivastava R, Mishra OP, Singh TB. Cerebrospinal fluid TNF-α, IL-6, and IL-8 in children with bacterial meningitis. Pediatr Neurol (2014) 50:60–5. doi:10.1016/j.pediatrneurol.2013.08.016
41. Dulkerian SJ, Kilpatrick L, Costarino AT Jr, McCawley L, Fein J, Corcoran L, et al. Cytokine elevations in infants with bacterial and aseptic meningitis. J Pediatr (1995) 126:872–6. doi:10.1016/S0022-3476(95)70199-0
42. Tang RB, Lee BH, Chung RL, Chen SJ, Wong TT. Interleukin-1β and tumor necrosis factor-α in cerebrospinal fluid of children with bacterial meningitis. Childs Nerv Syst (2001) 17:453–6. doi:10.1007/s003810000422
43. Foell D, Wittkowski H, Vogl T, Roth J. S100 proteins expressed in phagocytes: a novel group of damage-associated molecular pattern molecules. J Leukoc Biol (2007) 81:28–37. doi:10.1189/jlb.0306170
44. Gazzolo D, Grutzfeld D, Michetti F, Toesca A, Lituania M, Bruschettini M, et al. Increased S100B in cerebrospinal fluid of infants with bacterial meningitis: relationship to brain damage and routine cerebrospinal fluid findings. Clin Chem (2004) 50:941–4. doi:10.1373/clinchem.2003.021048
45. Undén J, Christensson B, Bellner J, Alling C, Romner B. Serum S100B levels in patients with cerebral and extracerebral infectious disease. Scand J Infect Dis (2009) 36:10–3. doi:10.1080/00365540310017294
46. Peng QL, Tao SH, Yu N, Zhou XZ, Peng YZ, Fu N. Elevated levels of cerebrospinal fluid S100B are associated with brain injury and unfavorable outcomes in children with central nervous system infections. Int J Neurosci (2016) 127:1–9. doi:10.3109/00207454.2015.1135334
47. Ko ER, Yang WE, McClain MT, Woods CW, Ginsburg GS, Tsalik EL. What was old is new again: using the host response to diagnose infectious disease. Expert Rev Mol Diagn (2015) 15:1143–58. doi:10.1586/14737159.2015.1059278
48. Sweeney TE, Wong HR. Risk stratification and prognosis in sepsis. Clin Chest Med (2016) 37:209–18. doi:10.1016/j.ccm.2016.01.003
49. Weksler BB, Subileau EA, Perrière N, Charneau P, Holloway K, Leveque M, et al. Blood-brain barrier-specific properties of a human adult brain endothelial cell line. FASEB J (2005) 19:1872–4. doi:10.1096/fj.04-3458fje
50. Hardy SJ, Christodoulides M, Weller RO, Heckels JE. Interactions of Neisseria meningitidis with cells of the human meninges. Mol Microbiol (2000) 36:817–29. doi:10.1046/j.1365-2958.2000.01923.x
51. Bonnah RA, Muckenthaler MU, Carlson H, Minana B, Enns CA, Hentze MW, et al. Expression of epithelial cell iron-related genes upon infection by Neisseria meningitidis. Cell Microbiol (2004) 6:473–84. doi:10.1111/j.1462-5822.2004.00376.x
52. Schubert-Unkmeir A, Sokolova O, Panzner U, Eigenthaler M, Frosch M. Gene expression pattern in human brain endothelial cells in response to Neisseria meningitidis. Infect Immun (2007) 75:899–914. doi:10.1128/IAI.01508-06
53. Wells DB, Tighe PJ, Wooldridge KG, Robinson K, Ala’ Aldeen DA. Differential gene expression during meningeal-meningococcal interaction: evidence for self-defense and early release of cytokines and chemokines. Infect Immun (2001) 69:2718–22. doi:10.1128/IAI.69.4.2718-2722.2001
54. Temesvári P, Ábrahám CS, Speer CP, Kovács J, Megyeri P. Escherichia coli 0111 B4 lipopolysaccharide given intracisternally induces blood-brain barrier opening during experimental neonatal meningitis in piglets. Pediatr Res (1993) 34:182–6. doi:10.1203/00006450-199308000-00016
55. Mook-Kanamori B, Geldhoff M, Troost D, van der Poll T, van de Beek D. Characterization of a pneumococcal meningitis mouse model. BMC Infect Dis (2012) 12:71. doi:10.1186/1471-2334-12-71
56. Grandgirard D, Steiner O, Täuber MG, Leib SL. An infant mouse model of brain damage in pneumococcal meningitis. Acta Neuropathol (2007) 114:609–17. doi:10.1007/s00401-007-0304-8
57. Chen Y, Imai H, Ito A, Saito N. Novel modified method for injection into the cerebrospinal fluid via the cerebellomedullary cistern in mice. Acta Neurobiol Exp (Wars) (2013) 73:304–11.
58. Barichello T, Pereira JS, Savi GD, Generoso JS, Cipriano AL, Silvestre C, et al. A kinetic study of the cytokine/chemokines levels and disruption of blood-brain barrier in infant rats after pneumococcal meningitis. J Neuroimmunol (2011) 233:12–7. doi:10.1016/j.jneuroim.2010.10.035
59. Coimbra RS, Voisin V, de Saizieu AB, Lindberg RL, Wittwer M, Leppert D, et al. Gene expression in cortex and hippocampus during acute pneumococcal meningitis. BMC Biol (2006) 4:15. doi:10.1186/1741-7007-4-15
60. Grandgirard D, Leib SL. Meningitis in neonates: bench to bedside. Clin Perinatol (2010) 37:655–76. doi:10.1016/j.clp.2010.05.004
61. Stevens B, Allen NJ, Vazquez LE, Howell GR, Christopherson KS, Nouri N, et al. The classical complement cascade mediates CNS synapse elimination. Cell (2007) 131:1164–78. doi:10.1016/j.cell.2007.10.036
62. Schafer DP, Lehrman EK, Kautzman AG, Koyama R, Mardinly AR, Yamasaki R, et al. Microglia sculpt postnatal neural circuits in an activity and complement-dependent manner. Neuron (2012) 74:691–705. doi:10.1016/j.neuron.2012.03.026
63. Bialas AR, Stevens B. TGF-β signaling regulates neuronal C1q expression and developmental synaptic refinement. Nat Neurosci (2013) 16:1773–82. doi:10.1038/nn.3560
64. Lill M, Kõks S, Soomets U, Schalkwyk LC, Fernandes C, Lutsar I, et al. Peripheral blood RNA gene expression profiling in patients with bacterial meningitis. Front Neurosci (2013) 7:33. doi:10.3389/fnins.2013.00033
65. Adkins B, Leclerc C, Marshall-Clarke S. Neonatal adaptive immunity comes of age. Nat Rev Immunol (2004) 4:553–64. doi:10.1038/nri1394
66. Linch SN, Danielson ET, Kelly AM, Tamakawa RA, Lee JJ, Gold JA. Interleukin 5 is protective during sepsis in an eosinophil-independent manner. Am J Respir Crit Care Med (2012) 186:246–54. doi:10.1164/rccm.201201-0134OC
67. Gondorf F, Berbudi A, Buerfent BC, Ajendra J, Bloemker D, Specht S, et al. Chronic filarial infection provides protection against bacterial sepsis by functionally reprogramming macrophages. PLoS Pathog (2015) 11:e1004616. doi:10.1371/journal.ppat.1004616
68. Asano T, Ichiki K, Koizumi S, Kaizu K, Hatori T, Fujino O, et al. IL-17 is elevated in cerebrospinal fluids in bacterial meningitis in children. Cytokine (2010) 51:101–6. doi:10.1016/j.cyto.2010.03.001
69. Jesse S, Steinacker P, Lehnert S, Sdzuj M, Cepek L, Tumani H, et al. A proteomic approach for the diagnosis of bacterial meningitis. PLoS One (2010) 5:e10079. doi:10.1371/journal.pone.0010079
70. Hol EM, Pekny M. Glial fibrillary acidic protein (GFAP) and the astrocyte intermediate filament system in diseases of the central nervous system. Curr Opin Cell Biol (2015) 32:121–30. doi:10.1016/j.ceb.2015.02.004
71. Mitson-Salazar A, Yin Y, Wansley DL, Young M, Bolan H, Arceo S, et al. Hematopoietic prostaglandin D synthase defines a proeosinophilic pathogenic effector human TH2 cell subpopulation with enhanced function. J Allergy ClinImmunol (2016) 137:907.e–18.e. doi:10.1016/j.jaci.2015.08.007
72. Goonetilleke UR, Scarborough M, Ward SA, Gordon SB. Proteomic analysis of cerebrospinal fluid in pneumococcal meningitis reveals potential biomarkers associated with survival. J Infect Dis (2010) 202:542–50. doi:10.1086/654819
73. Di Rosa M, Malaguarnera G, De Gregorio C, Drago F, Malaguarnera L. Evaluation of CHI3L-1 and CHIT-1 expression in differentiated and polarized macrophages. Inflammation (2013) 36:482–92. doi:10.1007/s10753-012-9569-8
74. Cordeiro AP, Silva Pereira RA, Chapeaurouge A, Coimbra CS, Perales J, Oliveira G, et al. Comparative proteomics of cerebrospinal fluid reveals a predictive model for differential diagnosis of pneumococcal, meningococcal, and enteroviral meningitis, and novel putative therapeutic targets. BMC Genomics (2015) 16(Suppl 5):S11. doi:10.1186/1471-2164-16-S5-S11
75. Song H, Seishima M, Saito K, Maeda S, Takemura M, Noma A, et al. Apo A-I and apo E concentrations in cerebrospinal fluids of patients with acute meningitis. Ann Clin Biochem (1998) 35(Pt 3):408–14. doi:10.1177/000456329803500310
76. De Sanctis AG, Killian JA, Garcia T. Lactic acid of spinal fluid in meningitis: practical diagnostic and prognostic value. Am J Dis Child (1933) 46:239–49. doi:10.1001/archpedi.1933.01960020002001
77. Huy NT, Thao NT, Diep DT, Kikuchi M, Zamora J, Hirayama K. Cerebrospinal fluid lactate concentration to distinguish bacterial from aseptic meningitis: a systemic review and meta-analysis. Crit Care (2010) 14:R240. doi:10.1186/cc9395
78. Li Z, Du B, Li J, Zhang J, Zheng X, Jia H, et al. Cerebrospinal fluid metabolomic profiling in tuberculous and viral meningitis: screening potential markers for differential diagnosis. Clin Chim Acta (2017) 466:38–45. doi:10.1016/j.cca.2017.01.002
79. Subramanian A, Gupta A, Saxena S, Gupta A, Kumar R, Nigam A, et al. Proton MR CSF analysis and a new software as predictors for the differentiation of meningitis in children. NMR Biomed (2005) 18:213–25. doi:10.1002/nbm.944
80. Coen M, O’Sullivan M, Bubb WA, Kuchel PW, Sorrell T. Proton nuclear magnetic resonance-based metabonomics for rapid diagnosis of meningitis and ventriculitis. Clin Infect Dis (2005) 41:1582–90. doi:10.1086/497836
81. Glickman MS, Cox JS, Jacobs WR Jr. A novel mycolic acid cyclopropane synthetase is required for cording, persistence, and virulence of Mycobacterium tuberculosis. Mol Cell (2000) 5:717–27. doi:10.1016/S1097-2765(00)80250-6
82. Langley RJ, Tsalik EL, van Velkinburgh JC, Glickman SW, Rice BJ, Wang C, et al. An integrated clinico-metabolomic model improves prediction of death in sepsis. Sci Transl Med (2013) 5:195ra95. doi:10.1126/scitranslmed.3005893
83. Kakazu E, Kanno N, Ueno Y, Shimosegawa T. Extracellular branched-chain amino acids, especially valine, regulate maturation and function of monocyte-derived dendritic cells. J Immunol (2007) 179:7137–46. doi:10.4049/jimmunol.179.10.7137
Keywords: meningitis, neonatology, cytokines, metabolomics, proteomics, transcriptomics
Citation: Gordon SM, Srinivasan L and Harris MC (2017) Neonatal Meningitis: Overcoming Challenges in Diagnosis, Prognosis, and Treatment with Omics. Front. Pediatr. 5:139. doi: 10.3389/fped.2017.00139
Received: 25 March 2017; Accepted: 01 June 2017;
Published: 16 June 2017
Edited by:
Joseph M. Bliss, Women & Infants Hospital of Rhode Island, United StatesReviewed by:
Pascal M. Lavoie, BC Children’s Hospital Research Institute, CanadaRoland H. Hentschel, University Medical Center Freiburg, Germany
Copyright: © 2017 Gordon, Srinivasan and Harris. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mary Catherine Harris, aGFycmlzbUBlbWFpbC5jaG9wLmVkdQ==
 Scott M. Gordon
Scott M. Gordon Lakshmi Srinivasan
Lakshmi Srinivasan Mary Catherine Harris
Mary Catherine Harris