- 1Children’s Cancer Therapy Development Institute, Beaverton, OR, USA
- 2Faculty of the 2015 Pediatric Cancer Biology Nanocourse, Children’s Cancer Therapy Development Institute, Fort Collins, CO, USA
Limited research exists regarding the most aggressive forms of hepatoblastoma. Cell lines of the rare subtypes of hepatoblastoma with poor prognosis are not only difficult to attain but also challenging to characterize histologically. A community-driven approach to educating parents and families, regarding the need for donated tissue, is necessary for scientists to have access to resources for murine models and drug discovery. Herein, we describe the currently available resources, existing gaps in research, and the path to move forward for uniform cure of hepatoblastoma.
Introduction
Hepatoblastoma is the most common primary liver tumor diagnosed in childhood (1), with approximately 100 cases in the U.S. annually (2). Despite a high cure rate for those children whose tumor is resectable, there remains a group of children for whom a cure is out of reach.
The disease predominantly occurs in young children, from birth to 5 years of age (1). The histological subtypes of hepatoblastoma are fetal, embryonal, mixed epithelial–mesenchymal, and small cell undifferentiated (3). However, there is currently a lack of understanding regarding the origins and pathophysiology of these different subtypes of hepatoblastoma.
Clinically, the empirically driven advancements in postoperative chemotherapy and surgery, including the multidisciplinary approach set forth through the Pretreatment Extent of Disease guidelines (1), has improved outcomes for hepatoblastoma. These guidelines rely on a standardized staging system using imaging for detecting amount of tumor involvement (4). Despite these clinical advancements, the more aggressive forms of hepatoblastoma remain difficult to treat. Current treatments for aggressive forms of hepatoblastoma include doxorubicin, irinotecan (clinical trials), hepatic artery chemoembolization in addition to chemotherapy agents, as well as liver transplantation or partial resection with neoadjuvant chemotherapy (5).
Scientists and clinicians are now seeking non-chemotherapeutic treatments for patients with unresectable or metastatic tumor – treatments that directly target the molecular underpinnings of hepatoblastoma progression. For example, clinical trials regarding cixutumumab and pazopanib, monoclonal antibodies, and alisertib, a kinase inhibitor, have all been completed or are actively being investigated in phase 2 clinical trails for the treatment of refractory hepatoblastoma (6).
In order to find other targeted therapies, researchers need hepatoblastoma tissues and cell lines. There is an absence of diversity in hepatoblastoma cell lines for scientists and clinicians to use to better understand the disease. In this paper, we describe the need for more cell lines and murine models to advance the discovery of therapeutic targets for the more aggressive subtypes of hepatoblastoma.
Methods
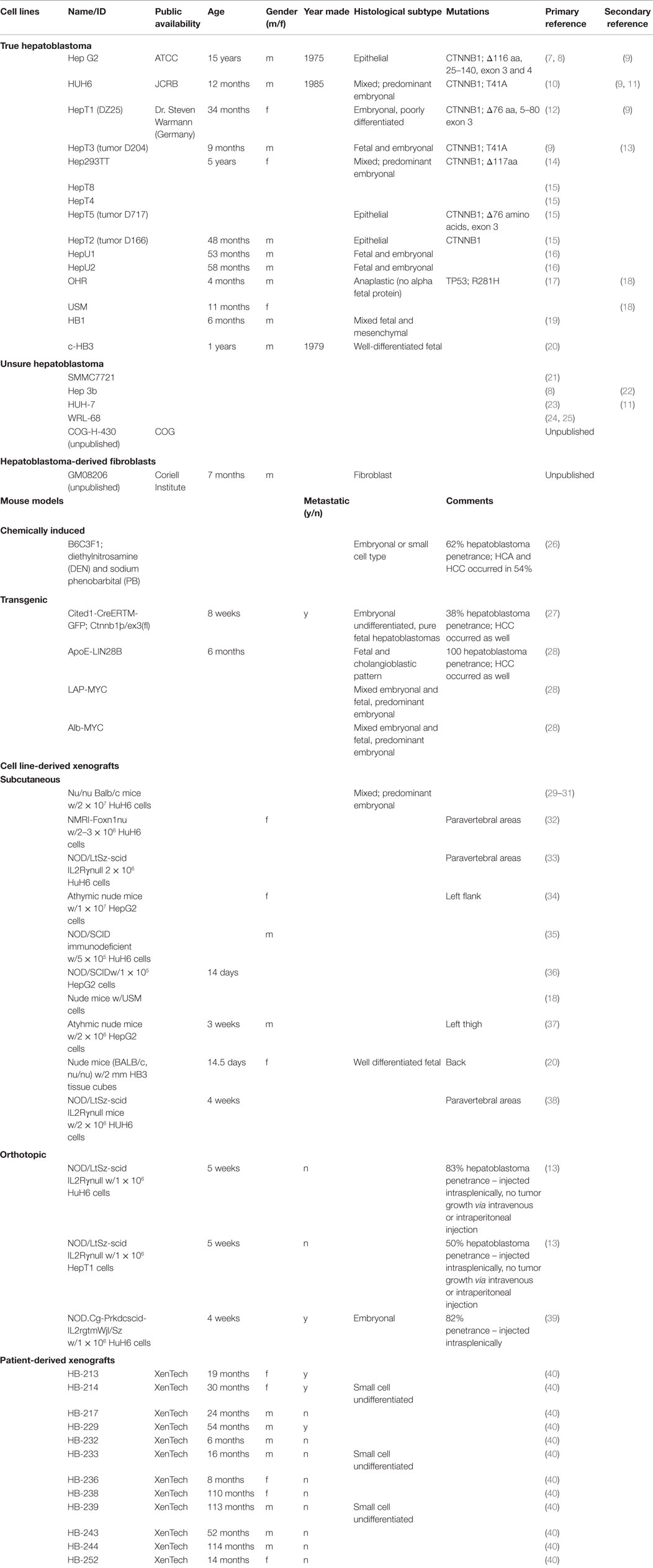
An extensive literature review of hepatoblastoma via PubMed was conducted to obtain information on research with unique hepatoblastoma cell lines and murine models. First, the authors found the number of distinct hepatoblastoma cell lines published in the literature. The search terms used were “‘hepatoblastoma’ [All Fields] and ‘cell line’ [MeSH Terms].” The search provided approximately 450 publications, all reviewed by the authors of this paper. Any publication that had a focus on hepatoblastoma was mentioned in Table 1. For each unique cell line, the authors attempted to find the primary article characterizing the cell line. Secondary publications using hepatoblastoma cell lines were also included in Table 1.
Manuscripts that had differing characterizations of certain cell lines were included in the “Unsure Hepatoblastoma” portion of Table 1.
Next, the authors reviewed literature via PubMed to find murine models of hepatoblastoma. The search terms used were “hepatoblastoma murine models.” This provided approximately 50 publications that the authors reviewed. All publications that studied murine models of hepatoblastoma were included in Table 1. The models were classified as chemically induced, transgenic, and cell-derived xenografts. Furthermore, the xenograft studies were sorted based on subcutaneous or orthotopic models.
Finally, the authors searched for patient-derived xenografts via PubMed and found no manuscript publications. The authors then searched the European Journal of Cancer using the term “hepatoblastoma xenograft” and found a published abstract using patient-derived xenografts, which is included in Table 1.
Results
Data regarding histological characterization and experimental murine models from only a few hepatoblastoma cell lines exist. These cell lines tend to have a favorable histology, leading to an underrepresentation of the high-risk subtypes (41).
Cell Lines
Fifteen hepatoblastoma cell lines are described in current literature (Table 1). Additionally, there are four cell lines that are potentially hepatoblastoma, but significant inconsistencies in the literature render the data obtained from these lines unreliable. Even among the confirmed hepatoblastoma cell lines, however, there are many documented instances in which cell lines were mistaken for hepatocellular carcinoma (7). A fibroblast cell line harvested from the liver of a Beckwith–Wiedemann syndrome patient with hepatoblastoma is described (Table 1).
Most confirmed cell lines are of the mixed histology subtype. However, there are no cell lines of the small cell undifferentiated subtype, which carries the worst prognosis (7).
Murine Models
Only one chemically induced murine model of hepatoblastoma has been reported (Table 1). Although four different transgenic murine models are described in the literature, these murine models were not specifically developed for the purpose of modeling hepatoblastoma. The transgenic murine models phenotypically express both hepatocellular carcinoma and hepatoblastoma (Table 1).
Ten unique cell line-derived subcutaneous xenografts and three cell line-derived orthotopic murine models of human hepatoblastoma exist (Table 1). These models primarily utilize the Hep G2 and HuH6 cells lines. Twelve unique patient-derived xenografts exist (Table 1).
Potential Genetic Targets for Aggressive Hepatoblastoma
Many studies have noted genetic mutations specific to histological subtypes of hepatoblastoma (42). Hepatoblastoma cells have shown gain of 2q, 1q, Xp, and Xq; loss of 4q, 2q, and 1q; and loss of heterozygosity of insulin growth factor 2 (5). Subtypes with increased Notch expression are of the fetal subtype and tend to have a better prognosis. Those with overexpression of the Wingless-type MMTV Integration Site Family pathway are of the small cell undifferentiated subtype and carry a less favorable prognosis (3). Additionally, the more aggressive forms of hepatoblastoma have telomerase reverse transcriptase promoter mutations (43). Blocking the Wingless-type MMTV Integration Site Family pathway using NK1R antagonists has been shown to slow the progression of hepatoblastoma cell growth in vitro (44). Hepatoblastoma cells show an increase in activity of the hedgehog pathway, and abnormal signaling has been linked to more malignant potential (45). Forkhead Box G1 is overexpressed in hepatoblastoma, specifically the more aggressive subtypes, when compared to the fetal subtype (46).
Discussion
In order to find targeted therapeutic options for hepatoblastoma, basic science studies need to be conducted. The few cell lines characterized and the inconsistencies in the literature on certain cell lines provide a major hurdle toward this goal. In addition, the availably of the cell lines is limited, which explains the narrow spectrum of cell lines used to derive xenografts from the already few hepatoblastoma cell lines. Additionally, diversity of histological subtypes is needed in order to find better treatment modalities for the more aggressive forms of hepatoblastoma. Interestingly, expression of fibroblasts enhances the growth of hepatoblastoma (47), which is why the hepatoblastoma-derived fibroblast cell line, GM08206 (Table 1), carries potential for more advanced studies. It is of important note that the majority of liver cells are aneuploid, which has been thought to protect the liver from chronic injury (48). Culturing surrounding normal liver tissue in addition to the tumor would provide insight into premalignant tissue field effect at the site of the tumor (46).
Certain repositories for hepatoblastoma are in the early stages of developing around the world, providing optimism for advancements in basic science research, and potentially leading to clinical trials for hepatoblastoma. The Children’s Oncology Group developed a Rare Tumor Committee that has lead to promising clinical trials for rare pediatric cancer (49), including a current clinical trial involving combination chemotherapy for different stages of hepatoblastoma. Although histological analysis is not used in the staging process, this trial presents the opportunity to provide awareness of hepatoblastoma and an opportunity to increase tissue donation. Currently, the Children’s Oncology group has one hepatoblastoma cell line (COG-H-430), not available on the open distribution list, but can be obtained with a materials transfer agreement (personal communication).
In addition to the Children’s Oncology Group, the Japanese Collection of Research Biosources hosts a cell bank that provided the cell lines for the majority of hepatoblastoma manuscripts in the literature review (9). However, currently only HUH6 is available for public distribution. Many published hepatoblastoma cell lines found in the literature review were not within the last decade, which could explain the difficulty in obtaining certain cell lines today.
Most importantly, many international groups, such as Childhood Liver Tumors Strategy Group and the Society for Pediatric Oncology and Hematology, have collaborated with Children’s Oncology Group and the Japanese Collection of Research Biosources, which initially led to the Pretreatment Extent of Disease guidelines (50). It is this type of collaboration that can result in an increase in cell lines and tissue-banking repositories. One example is the Childhood Liver Tumors Strategy Group, which runs a tissue bank for childhood liver tumors (51).
Recently, further collaboration has allowed for the Children’s Hepatic tumors International Collaboration, to obtain data on 1,605 hepatoblastoma patients, aimed at creating a database to identify prognostic factors for this rare pediatric cancer (52). One limitation to the database, mentioned by the authors, was the exclusion of histology due to the lack of international consensus in characterizing subtypes (52).
As more interaction among family members is made, newer registries, in addition to those previously mentioned, will continue to grow. The Macy Easom Foundation has committed to funding development of the Hepatoblastoma Registry, as well as the expense of administration, data compilation, and analysis (18).
Despite the current development of repositories, an increase in cell lines and murine models available for research purposes cannot progress unless methods are in place to increase awareness for tissue donation in hepatoblastoma. Both parents and treating physicians must be made aware of the need for hepatoblastoma tissue and the opportunity to support research via autopsy tissue donations. The decision, whether to make an autopsy tissue donation, is difficult, intensely personal, and unique for each family. The authors recognize the delicate balance between making parents aware of the need and opportunity while taking care to respect every family’s response and perspective.
A parent who wishes to arrange for an autopsy donation should not be burdened with making the arrangements. With parents’ consent, volunteers and professionals must be in place to make the necessary contacts and establish logistics of the donation. These arrangements may include contacting the treating physician, speaking with the local pathologist who will perform the autopsy, connecting the researcher who will receive the donated tissue with the pathologist, and arranging for transport of the body from the child’s home to the hospital (and return to the funeral home).
Grassroots communication and interaction among family members, caregivers, and others affected by a particular diagnosis has significantly influenced progress in some areas of pediatric cancer research. As an example, interaction among families affected by diffuse intrinsic pontine glioma (DIPG) in an online discussion group is considered by some to be the first step in raising a tide turning awareness in that community. The result was a promising therapeutic drug, panobinostat, for treatment (53).
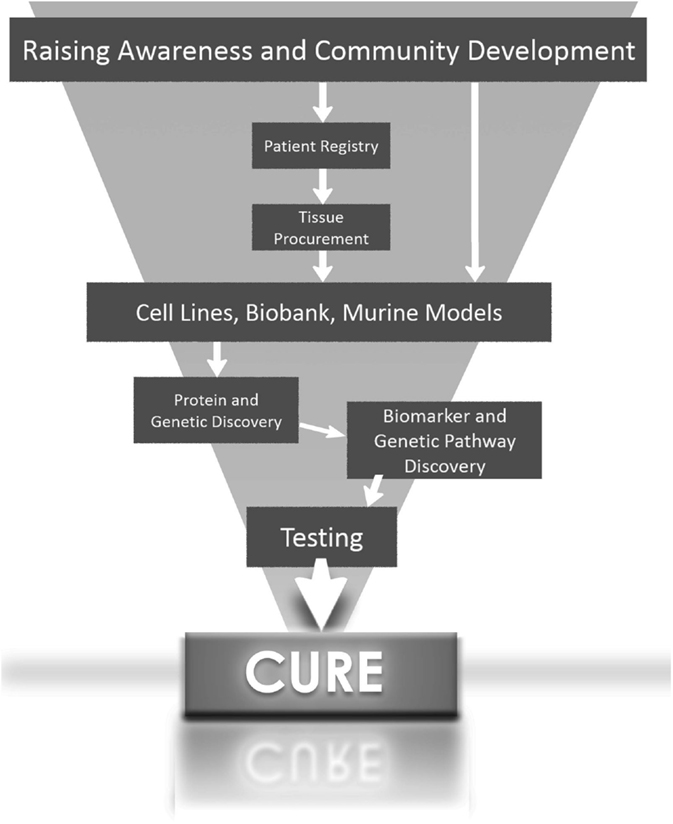
In summary, the greatest potential for the development of targeted therapy for aggressive forms of hepatoblastoma will come when scientists have access to hepatoblastoma cells lines and tissues with histological subtype diversity (Figure 1).
Author Contributions
RR, KS, RH, MSB, MB, and LH: contributed significantly to the acquisition, intellectual content, and final approval and are in agreement with all aspects of the work.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to the 2012 Nanocourse Group focused on hepatoblastoma and who provided a jump-start for this project: Deb Fuller, Zaahid Kaahn, Lisa Petke, and Jonathan Winegarden. We also appreciate the insight and analysis of Matthew Randolph, D.V.M. Finally, we are especially thankful to Charles Keller, M.D., for his commitment to reproducible, tangible research and his dedication to improving the outcomes for children diagnosed with hepatoblastoma.
References
1. Khaderi S, Guiteau J, Cotton RT, O’Mahony C, Rana A, Goss JA. Role of liver transplantation in the management of hepatoblastoma in the pediatric population. World J Transplant (2014) 4(4):294–8. doi: 10.5500/wjt.v4.i4.294
2. Johnson KJ, Williams KS, Ross JA, Krailo MD, Tomlinson GE, Malogolowkin MH, et al. Parental tobacco and alcohol use and risk of hepatoblastoma in offspring: a report from the children’s oncology group. Cancer Epidemiol Biomarkers Prev (2013) 22(10):1837–43. doi:10.1158/1055-9965.EPI-13-0432
3. Cairo S, Armengol C, De Reynies A, Wei Y, Thomas E, Renard CA, et al. Hepatic stem-like phenotype and interplay of Wnt/beta-catenin and Myc signaling in aggressive childhood liver cancer. Cancer Cell (2008) 14(6):471–84. doi:10.1016/j.ccr.2008.11.002
4. Pappo AS, Furman WL, Schultz KA, Ferrari A, Helman L, Krailo MD. Rare tumors in children: progress through collaboration. J Clin Oncol (2015) 33(27):3047–54. doi:10.1200/JCO.2014.59.3632
5. Venkatramani R, Furman WL, Fuchs J, Warmann SW, Malogolowkin MH. Current and future management strategies for relapsed or progressive hepatoblastoma. Paediatr Drugs (2012) 14(4):221–32. doi:10.2165/11597740-000000000-00000
6. Cixutumumab in Treating Patients with Relapsed or Refractory Solid Tumors clinicaltrials.gov (2015). Available from: https://clinicaltrials.gov/ct2/show/study/NCT00831844?term=hepatoblastoma&rank=9§=X70156
7. Lopez-Terrada D, Cheung SW, Finegold MJ, Knowles BB. Hep G2 is a hepatoblastoma-derived cell line. Hum Pathol (2009) 40(10):1512–5. doi:10.1016/j.humpath.2009.07.003
8. Zannis VI, Breslow JL, SanGiacomo TR, Aden DP, Knowles BB. Characterization of the major apolipoproteins secreted by two human hepatoma cell lines. Biochemistry (1981) 20(25):7089–96. doi:10.1021/bi00528a006
9. Koch A, Denkhaus D, Albrecht S, Leuschner I, von Schweinitz D, Pietsch T. Childhood hepatoblastomas frequently carry a mutated degradation targeting box of the beta-catenin gene. Cancer Res (1999) 59(2):269–73.
10. Tokiwa T, Doi I, Sato J. Preparation of single cell suspensions from hepatoma cells in culture. Acta Med Okayama (1975) 29(2):147–50.
11. Purcell R, Childs M, Maibach R, Miles C, Turner C, Zimmermann A, et al. HGF/c-Met related activation of beta-catenin in hepatoblastoma. J Exp Clin Cancer Res (2011) 30:96. doi:10.1186/1756-9966-30-96
12. Pietsch T, Fonatsch C, Albrecht S, Maschek H, Wolf HK, von Schweinitz D. Characterization of the continuous cell line HepT1 derived from a human hepatoblastoma. Lab Invest (1996) 74(4):809–18.
13. Ellerkamp V, Armeanu-Ebinger S, Wenz J, Warmann SW, Schafer J, Ruck P, et al. Successful establishment of an orthotopic hepatoblastoma in vivo model in NOD/LtSz-scid IL2Rgammanull mice. PLoS One (2011) 6(8):e23419. doi:10.1371/journal.pone.0023419
14. Chen TT, Rakheja D, Hung JY, Hornsby PJ, Tabaczewski P, Malogolowkin M, et al. Establishment and characterization of a cancer cell line derived from an aggressive childhood liver tumor. Pediatr Blood Cancer (2009) 53(6):1040–7. doi:10.1002/pbc.22187
15. Koch A, Waha A, Hartmann W, Hrychyk A, Schuller U, Waha A, et al. Elevated expression of Wnt antagonists is a common event in hepatoblastomas. Clin Cancer Res (2005) 11(12):4295–304. doi:10.1158/1078-0432.CCR-04-1162
16. Scheil S, Hagen S, Bruderlein S, Leuschner I, Behnisch W, Moller P. Two novel in vitro human hepatoblastoma models, HepU1 and HepU2, are highly characteristic of fetal-embryonal differentiation in hepatoblastoma. Int J Cancer (2003) 105(3):347–52. doi:10.1002/ijc.11082
17. Kanno S, Tsunoda Y, Shibusawa K, Ishikawa M, Okamoto S, Matsumura M, et al. [Establishment of a human hepatoblastoma (immature type) cell line OHR]. Hum Cell (1989) 2(2):211.
18. Ohnishi H, Kawamura M, Hanada R, Kaneko Y, Tsunoda Y, Hongo T, et al. Infrequent mutations of the TP53 gene and no amplification of the MDM2 gene in hepatoblastomas. Genes Chromosomes Cancer (1996) 15(3):187–90. doi:10.1002/(SICI)1098-2264(199603)15:3<187::AID-GCC8>3.0.CO;2-Z
19. Manchester KM, Warren DJ, Erlandson RA, Wheatley JM, La Quaglia MP. Establishment and characterization of a novel hepatoblastoma-derived cell line. J Pediatr Surg (1995) 30(4):553–8. doi:10.1016/0022-3468(95)90129-9
20. Hata Y, Uchino J, Sato K, Sasaki F, Une Y, Naito H, et al. Establishment of an experimental model of human hepatoblastoma. Cancer (1982) 50(1):97–101. doi:10.1002/1097-0142(19820701)50:1<97::AID-CNCR2820500118>3.0.CO;2-4
21. Feng X, Tang Z, Zheng Z. [Preliminary studies on the effects of tumor necrosis factor gene transfer on the growth of human hepatocellular carcinoma cells in nude mice]. Zhonghua Zhong Liu Za Zhi (1995) 17(3):167–9.
22. Delgado ER, Yang J, So J, Leimgruber S, Kahn M, Ishitani T, et al. Identification and characterization of a novel small-molecule inhibitor of beta-catenin signaling. Am J Pathol (2014) 184(7):2111–22. doi:10.1016/j.ajpath.2014.04.002
23. Nakabayashi H, Taketa K, Miyano K, Yamane T, Sato J. Growth of human hepatoma cells lines with differentiated functions in chemically defined medium. Cancer Res (1982) 42(9):3858–63.
24. Hsu IC, Tokiwa T, Bennett W, Metcalf RA, Welsh JA, Sun T, et al. p53 gene mutation and integrated hepatitis B viral DNA sequences in human liver cancer cell lines. Carcinogenesis (1993) 14(5):987–92. doi:10.1093/carcin/14.5.987
25. Falk PM, Sabater RT, Carballo DD Jr. Response of the human hepatic tissue cultures Hep-G2 and WRL-68 to cocaine. J Pharmacol Toxicol Methods (1995) 33(2):113–20. doi:10.1016/1056-8719(94)00065-C
26. Sakairi T, Kobayashi K, Goto K, Okada M, Kusakabe M, Tsuchiya T, et al. Greater expression of transforming growth factor alpha and proliferating cell nuclear antigen staining in mouse hepatoblastomas than hepatocellular carcinomas induced by a diethylnitrosamine-sodium phenobarbital regimen. Toxicol Pathol (2001) 29(4):479–82. doi:10.1080/01926230152499962
27. Mokkapati S, Niopek K, Huang L, Cunniff KJ, Ruteshouser EC, deCaestecker M, et al. Beta-catenin activation in a novel liver progenitor cell type is sufficient to cause hepatocellular carcinoma and hepatoblastoma. Cancer Res (2014) 74(16):4515–25. doi:10.1158/0008-5472.CAN-13-3275
28. Nguyen LH, Robinton DA, Seligson MT, Wu L, Li L, Rakheja D, et al. Lin28b is sufficient to drive liver cancer and necessary for its maintenance in murine models. Cancer Cell (2014) 26(2):248–61. doi:10.1016/j.ccr.2014.06.018
29. Berger M, Neth O, Ilmer M, Garnier A, Salinas-Martin MV, de Agustin Asencio JC, et al. Hepatoblastoma cells express truncated neurokinin-1 receptor and can be growth inhibited by aprepitant in vitro and in vivo. J Hepatol (2014) 60(5):985–94. doi:10.1016/j.jhep.2013.12.024
30. Wagner F, Henningsen B, Lederer C, Eichenmuller M, Godeke J, Muller-Hocker J, et al. Rapamycin blocks hepatoblastoma growth in vitro and in vivo implicating new treatment options in high-risk patients. Eur J Cancer (2012) 48(15):2442–50. doi:10.1016/j.ejca.2011.12.032
31. Cairo S, Wang Y, de Reynies A, Duroure K, Dahan J, Redon MJ, et al. Stem cell-like micro-RNA signature driven by Myc in aggressive liver cancer. Proc Natl Acad Sci U S A (2010) 107(47):20471–6. doi:10.1073/pnas.1009009107
32. Eicher C, Dewerth A, Thomale J, Ellerkamp V, Hildenbrand S, Warmann SW, et al. Effect of sorafenib combined with cytostatic agents on hepatoblastoma cell lines and xenografts. Br J Cancer (2013) 108(2):334–41. doi:10.1038/bjc.2012.539
33. Lieber J, Eicher C, Wenz J, Kirchner B, Warmann SW, Fuchs J, et al. The BH3 mimetic ABT-737 increases treatment efficiency of paclitaxel against hepatoblastoma. BMC Cancer (2011) 11:362. doi:10.1186/1471-2407-11-362
34. Marotta DE, Cao W, Wileyto EP, Li H, Corbin I, Rickter E, et al. Evaluation of bacteriochlorophyll-reconstituted low-density lipoprotein nanoparticles for photodynamic therapy efficacy in vivo. Nanomedicine (Lond) (2011) 6(3):475–87. doi:10.2217/nnm.11.8
35. Hayashi S, Fujita K, Matsumoto S, Akita M, Satomi A. Isolation and identification of cancer stem cells from a side population of a human hepatoblastoma cell line, HuH-6 clone-5. Pediatr Surg Int (2011) 27(1):9–16. doi:10.1007/s00383-010-2719-x
36. Mezzanotte L, Fazzina R, Michelini E, Tonelli R, Pession A, Branchini B, et al. In vivo bioluminescence imaging of murine xenograft cancer models with a red-shifted thermostable luciferase. Mol Imaging Biol (2010) 12(4):406–14. doi:10.1007/s11307-009-0291-3
37. Klein JL, Nguyen TH, Laroque P, Kopher KA, Williams JR, Wessels BW, et al. Yttrium-90 and iodine-131 radioimmunoglobulin therapy of an experimental human hepatoma. Cancer Res (1989) 49(22):6383–9.
38. Lieber J, Dewerth A, Wenz J, Kirchner B, Eicher C, Warmann SW, et al. Increased efficacy of CDDP in a xenograft model of hepatoblastoma using the apoptosis sensitizer ABT-737. Oncol Rep (2013) 29(2):646–52. doi:10.3892/or.2012.2150
39. Lieber J, Ellerkamp V, Vogt F, Wenz J, Warmann SW, Fuchs J, et al. BH3-mimetic drugs prevent tumour onset in an orthotopic mouse model of hepatoblastoma. Exp Cell Res (2014) 322(1):217–25. doi:10.1016/j.yexcr.2013.12.007
40. Fabre M, Nicolle D, Gorse A, Déas O, Mussini C, Brugières L, et al. A panel of pediatric liver cancer patient-derived xenografts to improve stratification of children with hepatoblastoma. In: Pieters R, editor. European Journal of Cancer. ENA 2014: EORTC-NCI-AACR Symposium; 2014; Badalona, Spain. Oxford: Elsevier (2014). 25 p.
41. Zhang Y, Zhang WL, Huang DS, Hong L, Wang YZ, Zhu X, et al. Clinical effectiveness of multimodality treatment on advanced pediatric hepatoblastoma. Eur Rev Med Pharmacol Sci (2014) 18(7):1018–26.
42. Lopez-Terrada D, Gunaratne PH, Adesina AM, Pulliam J, Hoang DM, Nguyen Y, et al. Histologic subtypes of hepatoblastoma are characterized by differential canonical Wnt and Notch pathway activation in DLK+ precursors. Hum Pathol (2009) 40(6):783–94. doi:10.1016/j.humpath.2008.07.022
43. Eichenmuller M, Trippel F, Kreuder M, Beck A, Schwarzmayr T, Haberle B, et al. The genomic landscape of hepatoblastoma and their progenies with HCC-like features. J Hepatol (2014) 61(6):1312–20. doi:10.1016/j.jhep.2014.08.009
44. Ilmer M, Garnier A, Vykoukal J, Alt E, von Schweinitz D, Kappler R, et al. Targeting the neurokinin-1 receptor compromises canonical Wnt signaling in hepatoblastoma. Mol Cancer Ther (2015) 14(12):2712–21. doi:10.1158/1535-7163.MCT-15-0206
45. Li YC, Deng YH, Guo ZH, Zhang MM, Zhu J, Pu CL, et al. Prognostic value of hedgehog signal component expressions in hepatoblastoma patients. Eur J Med Res (2010) 15(11):468–74.
47. Asada N, Tanaka Y, Hayashido Y, Toratani S, Kan M, Kitamoto M, et al. Expression of fibroblast growth factor receptor genes in human hepatoma-derived cell lines. In Vitro Cell Dev Biol Anim (2003) 39(7):321–8. doi:10.1290/1543-706X(2003)039<0321:EOFGFR>2.0.CO;2
48. Duncan AW, Hanlon Newell AE, Bi W, Finegold MJ, Olson SB, Beaudet AL, et al. Aneuploidy as a mechanism for stress-induced liver adaptation. J Clin Invest (2012) 122(9):3307–15. doi:10.1172/JCI64026
49. Kotecha RS, Kees UR, Cole CH, Gottardo NG. Rare childhood cancers – an increasing entity requiring the need for global consensus and collaboration. Cancer Med (2015) 4(6):819–24. doi:10.1002/cam4.426
50. Aronson DC, Czauderna P, Maibach R, Perilongo G, Morland B. The treatment of hepatoblastoma: its evolution and the current status as per the SIOPEL trials. J Indian Assoc Pediatr Surg (2014) 19(4):201–7. doi:10.4103/0971-9261.142001
51. Grotzer M. History SIOPEL (2016). Available from: http://www.siopel.org/?q=node/47
52. Czauderna P, Haeberle B, Hiyama E, Rangaswami A, Krailo M, Maibach R, et al. The Children’s Hepatic tumors International Collaboration (CHIC): novel global rare tumor database yields new prognostic factors in hepatoblastoma and becomes a research model. Eur J Cancer (2016) 52:92–101. doi:10.1016/j.ejca.2015.09.023
Keywords: hepatoblastoma, roadmap, xenograft models, cell lines, tissue procurement
Citation: Rikhi RR, Spady KK, Hoffman RI, Bateman MS, Bateman M and Howard LE (2016) Hepatoblastoma: A Need for Cell Lines and Tissue Banks to Develop Targeted Drug Therapies. Front. Pediatr. 4:22. doi: 10.3389/fped.2016.00022
Received: 02 February 2016; Accepted: 07 March 2016;
Published: 21 March 2016
Edited by:
Anat Erdreich-Epstein, Children’s Hospital Los Angeles and University of Southern California, USAReviewed by:
Steven G. Gray, Trinity College Dublin, IrelandJeffrey Toretsky, Georgetown University, USA
Copyright: © 2016 Rikhi, Spady, Hoffman, Bateman, Bateman and Howard. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rishi Raj Rikhi, cnJpa2hpJiN4MDAwNDA7bWVkLm1pYW1pLmVkdQ==
 Rishi Raj Rikhi
Rishi Raj Rikhi Kimberlee K. Spady
Kimberlee K. Spady Ruth I. Hoffman
Ruth I. Hoffman Michael S. Bateman
Michael S. Bateman Max Bateman
Max Bateman Lisa Easom Howard2
Lisa Easom Howard2