
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Parasitol., 28 April 2023
Sec. Molecular Cellular Parasitology
Volume 2 - 2023 | https://doi.org/10.3389/fpara.2023.1076759
 Seynabou D. Sene1†
Seynabou D. Sene1† Mariama N. Pouye1†
Mariama N. Pouye1† Rafael Miyazawa Martins2
Rafael Miyazawa Martins2 Fatoumata Diallo1
Fatoumata Diallo1 Khadidiatou Mangou1
Khadidiatou Mangou1 Amy K. Bei1,3
Amy K. Bei1,3 Alioune Barry4
Alioune Barry4 Oumar Faye5
Oumar Faye5 Oumar Ndiaye5
Oumar Ndiaye5 Ousmane Faye5
Ousmane Faye5 Amadou A. Sall5
Amadou A. Sall5 Jose-Juan Lopez-Rubio2
Jose-Juan Lopez-Rubio2 Alassane Mbengue1*
Alassane Mbengue1*The emergence of artemisinin partial resistance (ART-r) in Plasmodium falciparum malaria parasites has substantially compromised the efficacy of antimalarial treatments across southeast Asia (SE Asia). The spread of ART-r within the African continent could jeopardize past progress made in reducing worldwide malaria burden. A clinical index malaria case was identified in Kaolack, Senegal with persistent fever after complete artesunate-amodiaquine (ASAQ) treatment. Fifteen malaria-infected blood samples were collected by Institut Pasteur Dakar’s Senegalese sentinel surveillance system, from different healthcare centers surrounding the index case. We have identified one Plasmodium falciparum clinical isolate carrying R515K mutation in the artemisinin resistance gene PfKelch13. CRISPR-Cas9 genome editing was carried out and transgenic Pf3D7Pfkelch13R515K was tested for in vitro standard Ring-stage Survival Assay (RSA0-3hpi). Gene editing has confirmed that PfKelch13R515K drove increased in vitro RSA0-3hpi value. In this article, we report the functional significance of PfKelch13R515K mutation in an African context.
Artemisinin (ART) and its derivatives are the cornerstone of malaria case management for which no replacement is currently available on the market. The introduction of WHO recommended Artemisinin-based Combination Therapy (ACTs) as first-line treatment for uncomplicated malaria cases has partially contributed to the notable reduction of global malaria (WHO, 2015; World Health Organization. (2016)). ART has a short life and a rapid killing action for Plasmodium asexual blood stage parasites (Intharabut et al., 2019). Upon treatment, the rings are quickly eliminated and infected erythrocytes removed from the bloodstream, preventing sequestration (Mbengue et al., 2012). ART-r causes delayed parasite clearance upon 3 days of ACT treatment and increased in vitro RSA0-3hpi value. The parasite gene PfKelch13 (Pf3D7_1343700) discovered through a candidate gene approach is a primary marker of ART-r (Noedl et al., 2008; Ariey et al., 2014; Miotto et al., 2015). PfKelch13 mutations associated with ART-r were characterized in South East Asia (SEA) Plasmodium falciparum clinical isolates (Mbengue et al., 2015; Miotto et al., 2015; Mok et al., 2015; Tun et al., 2015). Since 2008, ART-r has been reported in India and a province in China (Huang et al., 2015; Das et al., 2018). In Africa where the malaria burden remains high, ART-r is a serious threat that has been documented in Rwanda and Uganda (Uwimana et al., 2020; Balikagala et al., 2021). As for chloroquine, ART-r will, if not contained, be a threat to malaria disease control plans in Sub-Saharan Africa (Trape et al., 1998; Lubell et al., 2014; Mukherjee et al., 2017; Sutherland et al., 2017). Clustered Regularly Interspaced Palindromic Repeat-Cas9 (CRISPR-Cas9) genome editing has been successfully applied in P. falciparum to confirm in vitro the function of key polymorphisms or alleles associated with resistance to antimalarials (Ghorbal et al., 2014; Lee and Fidock, 2014; Mbengue et al., 2015).
We found in Senegal, P. falciparum isolates with PfKelch13R515K an ART-r associated variant found in SEA upon ACT treatment (WWARN K13 Genotype-Phenotype Study Group, 2019). Transgenic lines were generated using CRISPR-Cas9 technology in the African background laboratory-adapted strain P. falciparum 3D7. Phenotypic assays of the transgenic Pf3D7Kelch13R515K line were performed following established RSA0-3hpi protocol (Witkowski et al., 2013).
Blood samples were collected by the IPD-4S in November 2018 (Surveillance des fièvres, 2015). The IPD-4S network is an important surveillance system initially built to strengthen influenza sentinel surveillance in partnership with the Senegalese Ministry of Health since 2012. This network of researchers, medical doctors, and nurses is implemented in all 14 regions of Senegal. Among its top priorities, the IPD-4S network also helps to routinely identify unusual health events to provide a rapid and appropriate medical response to the communities. Between 2017 and 2018 a surveillance program in response to a Dengue outbreak was carried out (Dieng et al., 2021). For this investigation, all declared PfRDT (SD Bioline malaria AG P.F) positive samples from the surrounding index case (ID 316443) area were collected from Ndoffane and other health care centers from the same region Kaolack as well as in the neighborhood region Diourbel. Malaria-positive samples were collected following the IPD-4S infection transmission control plan. Fifteen malaria-positive samples were collected, and 2 ml of venous blood was then shipped at 4°C to IPD the following day. Clinical information of the 18-year-old girl at day 9 post ACT treatment (ID 316443) is presented in Table 1b. DNA from the erythrocyte pellet was extracted using Quick-gDNA Blood MiniPrep kit from ZYMO research following the manufacturer’s instructions. Nested primers were designed to amplify the propeller domain of Kelch13. Full-length PfKelch13 was also amplified from biological replicates. Chromatograms of all PCR products were analyzed and the multiplicity of infection rate was determined using msp1 and msp2 typing protocol (Figure 1 and Supplementary Figure 1). Human malaria genius typing was done using a light cycler and LightMix modular Plasmodium genus (Malaria) Cat # 53-0694-96 and 40-0694-24 respectively (TIB BioMol). All primers are listed in Supplementary files. (Supplementary Tables 2A, B).
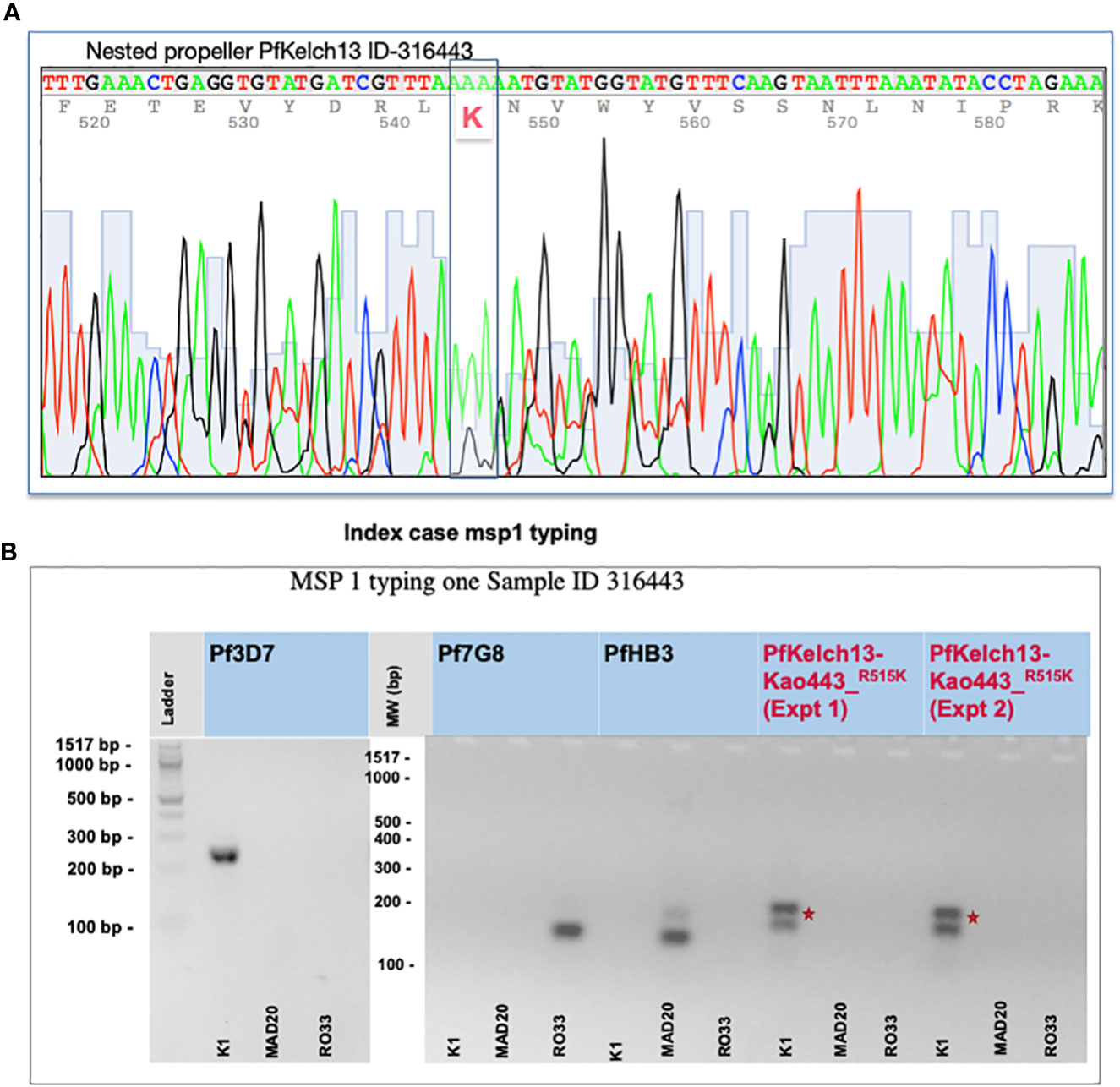
Figure 1 Identification of the R515K mutation and complexity of malaria infection. (A) PfKelch13 amplification from the original DNA batch was done. A major peak of Arginine (R codon AAA) and a minor peak of K (AGA) were detected at position 515 in the chromatogram indicating a mixed multi-clonal malaria parasite infection. (B) msp1 typing shows multiple clonal falciparum infections in the index case (ID 316443). gDNA from 3D7, Pf7G8, and PfHB3 strains were used as monogenic control for the complexity of the infected rate. Primers are provided in Supplementary Table 1. Experiments-1 and 2 represent biological replicates. *Shows the position of the nucleotide change before and after crispr cas.
The two-plasmid approach was employed to express Cas9, sgRNA, and a donor template (Ghorbal et al., 2014). SpCas9 was delivered on the pUF1 plasmid, which also contains a yeast dihydroorotate dehydrogenase (ydhodh) expression cassette which confers resistance to PfDHODH inhibitors such as DSM1. The sgRNA and the donor DNA template for homologous recombination repair were placed in the same plasmid pL7 (here pL7-238, have already cloned with the seed so we only need to clone the donor DNA). pL7 also expresses human dihydrofolate reductase (hdhfr) allowing positive selection with WR99210. The donor DNA to be used as a template to repair the double-strand breaks (DSBs) generated by Cas9 was designed from the kelch13 gene of Plasmodium falciparum 3D7 (PlasmoDB ID PF3D7_1343700). Donor’s DNA includes the homologous regions (HR1 and HR2) flanking the region of interest (ROI). The ROI carries the desired mutation and an additional modification (defined here as shield mutations) at the Cas9-target site. The shield’s mutations are silent but abolish recognition by Cas9, thereby protecting the modified locus from repeated cleavage. Additional silent mutations spanning the gap between the shield mutations and the desired modification can be introduced to help drive the repair event beyond the mutation-of-interest. Homology regions with plasmid and restriction sites were added for cloning. Donor DNA (synthetic DNA by Integrated DNA technologies) was cloned in pL7 plasmid at SpeI-AflII sites using Infusion cloning technology (Takara-Bio) (Supplementary Figure 2A).
P. falciparum asexual blood-stage parasites 3D7 wild-type were cultured in A+ human red blood cells (RBCs) in RPMI-1640 culture medium containing 25 mM Hepes + l-glutamine, supplemented with 10% Albumax II (Gibco Life Technologies), Human Sera (HS), hypoxanthine (C.C.Pro GmbH) and gentamicin (Sigma). Parasites were maintained at 37°C in 5% O2, 5% CO2, and 90% N2. Cultures were monitored by blood smears fixed in methanol, stained with Giemsa, and viewed by light microscopy. Synchronous cultures were obtained by sorbitol treatment. Prior to transfection, 50 μg of each plasmid circular DNA (pUF1-Cas9 and recombinant plasmid pL7) were ethanol-precipitated and resuspended in 30 μl of Tris-EDTA. The DNA precipitated plasmids were co-transfected into 100 μl rings stages parasites at 4.87% parasitemia and 270 μl cytomix, by electroporation using the Bio-Rad GenePulse Xcell™ electroporator, at 310 V, with a resistance of 950 μF and a transfection time of less than 10s.
Drug pressure was applied 15–20 h after transfection: WR99210 for pL7-238_Insert was used at 2.5 nM and DMS1 for pUF1-Cas9 was used at 1.5 μM. Media and drugs were renewed every 24 h for the first 5 days, then every other day for a week, and twice a week until parasites are visually detected by microscopy. Parasites came back during the third week post-transfection.
To test PfKelch13 single nucleotide integration, genomic DNA (gDNA) of bulk culture for each transfection (Pf3D7Kelch13R515K) was extracted from infected RBCs using the Mini NucleoSpin Blood QuickPure kit (MACHEREY-NAGEL). The high-fidelity polymerase PfuUltra II Fusion HS DNA polymerase was used for PCR amplification to detect the integration of locus. PCR conditions are as follows: 95°C for 2 min, followed by 30 cycles of 95°C for 30 s, 48°C for 20 s, 62°C for 15 s, and a final extension cycle of 72°C for 3 min. PCR products were migrated on 1% agarose gel for 20 min at 100V. NucleoSpin Gel and PCR Clean-up kit (MACHEREY-NAGEL) were used to extract amplified DNA at the expected size. Sequencing of the purified PCR products was done by Eurofins Genomics (TubeSeq service). All primers used in this study are listed in an additional file (Supplementary Table 1). Bulk-edited cultures were cloned via limiting dilution. For in vitro malaria culture synchronous culture of trophozoites stages was used to start the cloning dilution. A 1/1000 dilution was parasitemia checked by microscopy in a 2% hematocrit flask. Parasites were gassed and incubated at 37°C, in a static condition. Genomic DNA from selected clones after serial dilutions of transgenic lines were then amplified and sequence aligned with Pf3D7Kelch13WT parental sequence (Supplementary Figure 2B).
In vitro RSA0-3hpi were conducted on very early ring-stage parasites (0-3 hours post-invasion; hpi) as previously described (Witkowski et al., 2013). Pf3D7Kelch13R515K clones 1 and 4 were randomly picked and came back from sequencing with correct SNP integration in the Art-r gene marker (Supplementary Figure 2B). RSA0-3hpi level of transgenic parasite clones were studied. Parental laboratory strains Pf3D7Kelch13WT and PfNF54Kelch13C580Y were used as negative and positive controls respectively. In vitro RSA0-3hpi ring parasites were subjected to sorbitol treatment to eliminate remaining schizonts. Synchronized 0–3hpi rings were next adjusted to 0.5% parasitemia and 2% hematocrit in 1 mL volumes (in 48-well plates), and exposed to DHA (700nM) or 0.1% of its solvent dimethyl sulfoxide (DMSO) as previously described. Duplicate wells were established for each parasite line ± drug. Following wash out, parasites were maintained in complemented drug-free medium in an incubator condition for an additional 66h.
Parasite viability was assessed by microscopic examination of Giemsa-stained thin blood smears by counting viable parasites (Supplementary File 2C). A measure of 2 μL of the pellet was then used for each smear. Parasitemia was calculated from a total of at least 10,000 erythrocytes per assay. Slides were read from the two duplicate wells per assay. Percent survival was calculated as the parasitemia in the drug-treated sample divided by the parasitemia in the untreated sample ×100. The assay was done three times to confirm the results. The Mean (SD) of RSA0-3hpi based on three replicates was done.
PfKelch13 polymorphism was studied using the DNA of malaria-infected blood received to investigate a malaria index case reported with a fever persistence upon 3 days ACT treatment (ASAQ) in Kaolack, a low malaria endemic region in Senegal. A total of 15 malaria PfRDT-positive samples were collected including the index case. Sanger sequencing for PfKelch13 was carried out with both a nested PCR and full-length amplification methods. Table 1a shows the parasite ART-r gene marker distribution. The index case was infected by parasites carrying a major arginine (AAA coding for K) peak at position 515 of PfKelch13. Both the chromatogram and the msp1 typing confirm a mixed infection with a second clone carrying a low peak of wild-type PfKelch13 (AGA coding for R) (Figures 1A, B and Supplementary Figure 1). Plasmo typing assay shows 14 out of 15 patients had P. falciparum infections and 3 of the 15 had P. malariae co-infections (Supplementary Table 1B). None of the three remaining human malaria parasites (P. ovale, P. knowlesi, and P. vivax) were detected.
The lack of viable parasites (shipped to IPD a day after sample collection) to be culture-adapted and phenotypically tested was a limiting factor to further investigating ART-r phenotype. We next assessed an in vitro phenotypic assay to study the functional relevance of PfKelch13R515K.
pL7-2386R515K plasmid construct was used to generate a transgenic line (Supplementary Figure 2A). The resulting bulk cultures were cloned by serial limiting dilution (Supplementary Figure 2B). Individual isolated clones were sequenced to verify the creation of the R515K mutation. Pf3D7Kelch13R515K clone sequence is displayed and aligned with its parental Pf3D7 sequence (Figure 2A). The most frequent SEA ART-r mutation (KelchC580Y) was used as a positive control line also transfected in an African background P. falciparum counterpart PfNF54 (Dondorp et al., 2012; Mbengue et al., 2015; Miotto et al., 2015; Mok et al., 2015; Tun et al., 2015; Bhattacharjee et al., 2018).
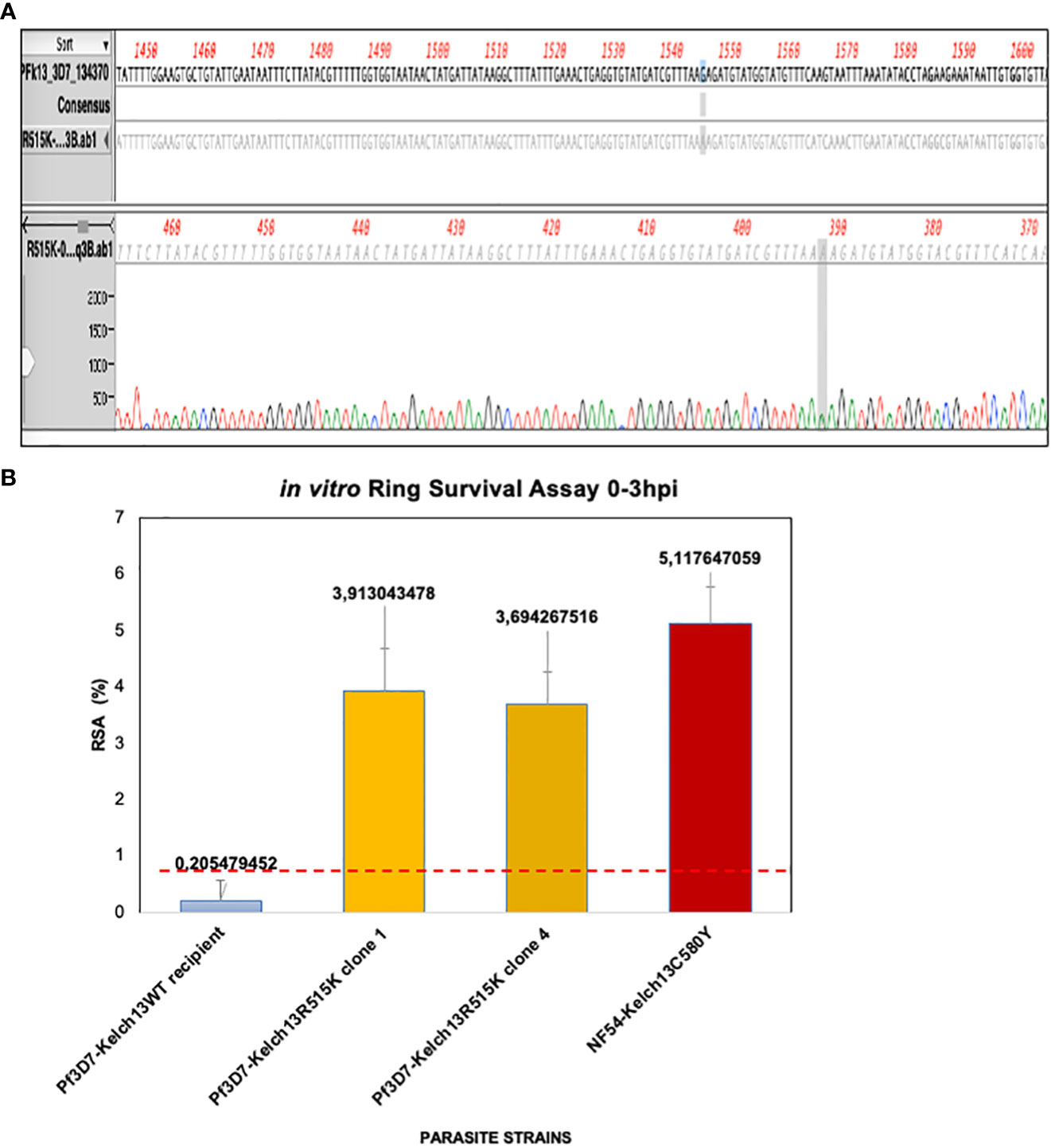
Figure 2 PfKelch13R515K induces increased in vitro RSA0-3hpi value. (A) Transgenic lines screening. Sequence confirmation of the generated Pf3D7PfKelch13R515K shows a single nucleotide substitution in the ART-r gene marker when compared to the parental Pf3D7 laboratory line. (B) PfKelch13R515K induces increased RSA0-3hpi value. Parasite lines were highly synchronized and grown for one hour under 700nM DHA for 6 hours. Parasitemia was counted by microscopy and RSA level was estimated as survival rate compared to DMSO control lines. The panel shows the level of in vitro ART-r. NF54-Kelch13C580Y shows displaying increased RSA0-3hpi value (5.12%) while Pf3D7Kelch13WT remain sensitive to DHA (0.21%). Two Pf3D7Kelch13R515K clones were used. Error bars of the median of the three biological replicates RSA0-3hpi for each line is shown. *Shows the position of the nucleotide change before and after crispr cas.
To validate PfKelch13 R515K mutation as a cause of the fever persistence in the malaria index case reported in Kaolack, we carried out in vitro drug sensitivity assay using the standard RSA0-3hpi protocol (Witkowski et al., 2013). Pf3D7Kelch13R515K showed an increased RSA0-3hpi value (Figure 2B). Pf3D7Kelch13R515K clones 1 and 4 showed a median RSA of 3.9% and 3.7% respectively while the parental Pf3D7 was sensitive to DHA (median RSA0-3hpi = 0.2%). The parasitemia assessed by microscopy showed a 5.12% RSA0-3hpi value for the positive control NF54Kelch13C580Y (Supplementary Figure 2C). In conclusion, our findings demonstrate that 3D7PfKelch13R515K results in an increased in vitro RSA0-3hpi value.
The emergence and spread of artemisinin partial resistance in Africa is a major public health concern. There is an urgent need to strengthen ART-r surveillance systems in sub-Saharan Africa. The molecular tool for ART-r provides evidence that PfKelch13 is under selection in African background circulating isolates.
Our finding does show the first appearance of PfKelch13R515K associated with fever persistence upon ACTs treatment in Senegal. PfKelch13R515K mutation is among the mutations shown to be clinically associated with slow parasite clearance. The functional relevance of R515K was studied using an African parasite strain Pf3D7. Pf3D7-Kelch13R515K mutation confers in vitro ART-r. Our work is to our knowledge the first PfKelch13R515K in vitro phenotypic validation. The presence of R515K mutation raises the possibility that this mutation can be used to assist in the detection of artemisinin partial resistance in Africa. We here determined the power of including genome editing in ART-r molecular tracking and case management in low-middle income countries where sample collection, storage, and transportation to research institutions are often challenging. Our findings suggest a potential risk of discrete artemisinin partial resistance spread across Africa and highlight the need to perform well-designed large-scale surveys.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
The studies involving human participants were reviewed and approved by RÉSEAU 4S Institut Pasteur Dakar. Written informed consent to participate in this study was provided by the participants’ legal guardian/next of kin. Written informed consent was obtained from the individual(s), and minor(s)’ legal guardian/next of kin, for the publication of any potentially identifiable images or data included in this article.
This manuscript was written by SS, AM. Lab work was done by the G4 unit at Institut Pasteur Dakar, Senegal. SS did the Crispr-cas9 constructs and in vitro RSA testings under J-JL-R supervision at University of Montpellier supervised. All authors contributed to the article and approved the submitted version.
This work was supported by the Crick African Network (CAN) which receives its funding from the UK’s Global Challenges Research Fund (MR/P028071/1), and by the Francis Crick Institute which receives its core funding from Cancer Research UK (FC1001647), the UK Medical Research Council (FC1001647), and the Wellcome Trust (FC1001647). SDS was supported by the French Embassy in Senegal through obtaining a scholarship from the Cooperation and Cultural Action Service of the French Embassy in Dakar. J-JL-R and RM were supported by #ANR-11-LABX-0024-01“ParaFrap” and Fondation pour la Recherche Médicale (DEQ2018033199). We did receive funds from the G4 group funding (G45267, Malaria Experimental Genetic Approaches and Vaccines) from the Institut Pasteur de Paris and Agence Universitaire de la Francophonie (AUF). For the purpose of Open Access, the author has applied a CC by public copyright license to any Author Accepted Manuscript version arising from this submission.
We are very grateful to the researchers at LPHI (Laboratory of Pathogen Host Interactions) in Montpellier and this team for their collaboration. We thank Kai Wengelnik, Prince Nyarko, Pratima Gurung, Ana Gomes, Diane-Ethna Mbang-Benet, Vincent, and Guitard (LPHI). We also thank all the workers at Arboviruses and Haemorrhagic Fever Viruses Unit, Virology Department and Epidemiology, Clinical Research and Data Science Unit, IPD for access to samples, and clinical and demographic data. We convey special thanks to Laty Gaye Thiam (G4-Malaria Experimental Genetic Approaches and Vaccines, Pôle Immunophysiopathologie et Maladies Infectieuses, Institut Pasteur Dakar) for the helpful review of the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpara.2023.1076759/full#supplementary-material
Ariey F., Witkowski B., Amaratunga C., Beghain J., Langlois A.C., Khim N., et al. (2013). A molecular marker of artemisinin-resistant plasmodium falciparum malaria. Nature 505 (7481), 50–55. doi: 10.1038/nature12876
World malaria report 2021. Available at: https://www.who.int/teams/global-malaria-programme/reports/world-malaria-report-2021.
Balikagala B., Fukuda N., Ikeda M., Katuro O. T., Tachibana S. I., Yamauchi M., et al. (2021). Evidence of artemisinin-resistant malaria in Africa. New Engl. J. Med. 385 (13), 1163–1171. doi: 10.1056/NEJMoa2101746
Bhattacharjee S., Coppens I., Mbengue A., Suresh N., Ghorbal M., Slouka Z., et al. (2018). Remodeling of the malaria parasite and host human red cell by vesicle amplification that induces artemisinin resistance. Blood 131 (21), 1234–1247. doi: 10.1182/blood-2017-11-814665
Das S., Saha B., Hati A. K., Roy S. (2018). Evidence of artemisinin-resistant Plasmodium falciparum malaria in Eastern India. New Engl. J. Med. 379, 1962–1964. doi: 10.1056/NEJMc1713777
Dieng I., Ndione M.H.D., Fall C., Diagne M.M., Diop M., Gaye A., et al. (2021). Multifoci and multiserotypes circulation of dengue virus in Senegal between 2017 and 2018. BMC Infect. Dis. 21 (1), 1–11. doi: 10.1186/s12879-021-06580-z
Dondorp A. M., Nosten F., Yi P., Das D., Hanpithakpong W., Lee S. J., Ringwald P., et al, et al. (2012). Artemisinin resistance in plasmodium falciparum malaria. N Engl. J. Med. 361, 455–467. doi: 10.1056/NEJMoa0808859
Ghorbal M., Gorman M., Macpherson C.R., Martins R.M., Scherf A., Lopez-Rubio J-J., et al. (2014). Genome editing in the human malaria parasite plasmodium falciparum using the CRISPR-Cas9 system. Nat. Biotechnol. 32 (8), 819–821. doi: 10.1038/nbt.2925
Huang F., Takala-Harrison S., Jacob C. G., Liu H., Sun X., Yang H., et al. (2015). A single mutation in K13 predominates in southern China and is associated with delayed clearance of plasmodium falciparum following artemisinin treatment. J. Infect. Dis. 212 (10), 1629–1635. doi: 10.1093/infdis/jiv249
Intharabut B., Kingston H. W., Srinamon K., Ashley E. A., Imwong M., Dhorda M., et al. (2019). Artemisinin resistance and stage dependency of parasite clearance in falciparum malaria. J. Infect. Dis. 219 (9), 1483–1489. doi: 10.1093/infdis/jiy673
Lee M. C., Fidock D. A. (2014). CRISPR-mediated genome editing of plasmodium falciparum malaria parasites. Genome Med. 6, 63. doi: 10.1186/s13073-014-0063-9
Lubell Y., Dondorp A., Guérin P.J., Drake T., Meek S., Ashley E., et al. (2014). Artemisinin resistance–modelling the potential human and economic costs. Malar J. 13, 452. doi: 10.1186/1475-2875-13-452
Mbengue A., Bhattacharjee S., Pandharkar T., Liu H., Estiu G., Stahelin R. V., et al. (2015). A molecular mechanism of artemisinin resistance in plasmodium falciparum malaria. Nature 520 (7549), 683–687. doi: 10.1038/nature14412
Mbengue A., Yam X. Y., Braun-Breton C. (2012). Human erythrocyte remodelling during plasmodium falciparum malaria parasite growth and egress. Br. J. Haematol 157, 171–179. doi: 10.1111/j.1365-2141.2012.09044.x
Miotto O., Amato R., Ashley E.A., MacInnis B., Almagro-Garcia J., Amaratunga C., et al. (2015). Genetic architecture of artemisinin-resistant plasmodium falciparum. Nat. Genet. 47 (3), 226–234. doi: 10.1038/ng.3189
Mok S., Ashley E. A., Ferreira P. E., Zhu L., Lin Z., Yeo T., et al. (2015). Population transcriptomics of human malaria parasites reveals the mechanism of artemisinin resistance. Sci. 347 (6220), 431–435. doi: 10.1126/science.1260403
Mukherjee A., Bopp S., Magistrado P., Wong W., Daniels R., Demas A., et al. (2017). Artemisinin resistance without pfkelch13 mutations in plasmodium falciparum isolates from Cambodia. Malar J. 16 (1), 195. doi: 10.1186/s12936-017-1845-5
Noedl H., Se Y., Schaecher K., Smith B. L., Socheat D., Fukuda M. M., et al. (2008). Evidence of artemisinin-resistant malaria in Western Cambodia. New Engl. J. Med. 359 (24), 2619–2620. doi: 10.1056/NEJMc0805011
Surveillance des fièvres (2015). Surveillance sentinelle syndromique au sénégal (Réseau 4S) (Senegal: Institut Pasteur de Dakar). Available at: https://www.pasteur.sn/fr/recherche-et-sante-pubique/epidemiologie/surveillance-des-fievres-surveillance-sentinelle-syndromique-au-senegal-reseau-4s.
Sutherland C. J., Lansdell P., Sanders M., Muwanguzi J., van Schalkwyk D.A., Kaur H., et al. (2017). pfk13-independent treatment failure in four imported cases of plasmodium falciparum malaria treated with artemether-lumefantrine in the UniteKingdom. Antimicrob Agents Chemother. 61(3):e02382-16. doi: 10.1128/AAC.02382-16
Trape J. F., Pison G., Preziosi MP., Enel C., Desgrées du Loû A., Delaunay V., et al. (1998). Impact of chloroquine resistance on malaria mortality. Comptes Rendus l’Academie Des. Sci. - Serie III 321 (8), 689–697. doi: 10.1016/s0764-4469(98)80009-7
Tun K. M., Imwong M., Lwin K. M., Win A. A., Hlaing T. M., Hlaing T., et al. (2015). Spread of artemisinin-resistant plasmodium falciparum in Myanmar: A cross-sectional survey of the K13 molecular marker. Lancet Infect. Dis. 15, 415–421. doi: 10.1016/S1473-3099(15)70032-0
Uwimana A., Legrand E., Stokes B.H., Ndikumana J.L.M., Warsame M., Umulisa N., et al. (2020). Emergence and clonal expansion of in vitro artemisinin-resistant plasmodium falciparum kelch13 R561H mutant parasites in Rwanda. Nat. Med. 26 (4), 1602–1608. doi: 10.1038/s41591-020-1005-2
World Health Organization (2016). Artemisinin andartemisinin-based combination therapy resistance: status report (No. WHO/HTM/GMP/2016.11). World Health Organization.
Witkowski B., Amaratunga C., Khim N., Sreng S., Chim P., Kim S., et al. (2013). Novel phenotypic assays for the detection of artemisinin-resistant plasmodium falciparum malaria in Cambodia: in-vitro and ex-vivo drug-response studies. Lancet Infect. Dis. 13, 1043–1049. doi: 10.1016/S1473-3099(13)70252-4
Keywords: P. falciparum, artemisinin-resistance PfKelch13, genetic variation, CRISPR-Cas9, genomic surveillance system, Senegal
Citation: Sene SD, Pouye MN, Martins RM, Diallo F, Mangou K, Bei AK, Barry A, Faye O, Ndiaye O, Faye O, Sall AA, Lopez-Rubio J-J and Mbengue A (2023) Identification of an in vitro artemisinin-resistant Plasmodium falciparum kelch13 R515K mutant parasite in Senegal. Front. Parasitol. 2:1076759. doi: 10.3389/fpara.2023.1076759
Received: 21 October 2022; Accepted: 16 March 2023;
Published: 28 April 2023.
Edited by:
Anastasios D. Tsaousis, University of Kent, United KingdomReviewed by:
Liwang Cui, University of South Florida, United StatesCopyright © 2023 Sene, Pouye, Martins, Diallo, Mangou, Bei, Barry, Faye, Ndiaye, Faye, Sall, Lopez-Rubio and Mbengue. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alassane Mbengue, YWxhc3NhbmUubWJlbmd1ZUBwYXN0ZXVyLnNu
†These authors share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.