
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
BRIEF RESEARCH REPORT article
Front. Oncol. , 25 March 2025
Sec. Hematologic Malignancies
Volume 15 - 2025 | https://doi.org/10.3389/fonc.2025.1552928
 Jialong Liu1†
Jialong Liu1† Yaqing Feng2*†
Yaqing Feng2*† Yanfang Zhang2
Yanfang Zhang2 Yingnan Xiao1
Yingnan Xiao1 Xi Liu2
Xi Liu2 Tingting Xiao1
Tingting Xiao1 Junyan Zou1
Junyan Zou1 Kai Fan2
Kai Fan2 Lisha Lu1,3
Lisha Lu1,3 Xiaoxia Yang2
Xiaoxia Yang2 Jinying Gong1,3*
Jinying Gong1,3*A novel fusion gene NRF1::PDGFRA was identified in a patient with myeloid/lymphoid neoplasms with eosinophilia and tyrosine kinase gene fusions (MLN-TK), harboring the chromosome abnormality t(4;7)(q12;q32). This represents the first reported case of the NRF1::PDGFRA fusion gene, and the ninth PDGFRA-associated fusion gene identified in MLN-TK. The fusion event led to the constitutive activation of the PDGFRA kinase, resulting in uncontrolled eosinophil proliferation and potentially contributing to the occurrence of cerebral infarction. Our study indicates treatment with low-dose imatinib effectively alleviates the symptoms associated with NRF1::PDGFRA gene fusion.
The World Health Organization (WHO) 2022 classification and the International Consensus Classification of Myeloid and Lymphoid Neoplasms (ICC-MLN) define a distinct subcategory of myeloid neoplasms as “myeloid/lymphoid neoplasms with eosinophilia and tyrosine kinase gene fusions” (MLN-TK), which are driven by rearrangements/fusion genes involving PDGFRA, PDGFRB, FGFR1, JAK2, ABL1, or FLT3 (1–3). Notably, myeloid and lymphoid neoplasms with eosinophilia and PDGFRA rearrangements are recognized as a distinct entity within the section of MLN-TK (1, 4). At the time of writing, all fusion partners of PDGFRA that have been described in MLN-TK, including FIP1L1, BCR, ETV6, KIF5B, CDK5RAP2, STRN, TNKS2, FOXP1, and AKAP9 (4–13). The FIP1L1 gene is the most frequent fusion partner, followed by the BCR gene, with other partner genes being infrequent. In this study, we describe the identification of NRF1 as a novel and rare fusion partner of PDGFRA in an adult patient with MLN-TK, and provide a detailed account of the patient’s clinical course following treatment with low-dose imatinib.
Chromosomal analysis was performed by examining short-term cultures of bone marrow (BM) according to standard conventional cytogenetic protocols. Fresh bone marrow was collected from each patient and cultured for 24 h (in RPMI 1640 medium, 20% calf serum) without any growth factors. A methanol–glacial acetic acid fixation method was used for obtaining metaphase cells, with G-banding and R-banding performed. Analysis was performed using an Ikaros automated scanning system (Metasystems, Germany). At least 20 cells in metaphase were analyzed in each case. Karyotype descriptions are based on the International System for Human Cytogenomic Nomenclature (ISCN 2020).
BM of this patient was collected and processed following standard cytogenetic protocols. For interphase fluorescence in situ hybridization (FISH), the slides containing fixed cells were incubated in 2× SSC at 37°C for 30 minutes, rinsed with deionized water, and subsequently dehydrated through graded ethanol series (70%, 85%, and 100%). After air drying, slides were denatured at 83°C for 5 minutes in the water bath, hybridized with PDGFRA Tricolor Rearrangement Probe (HealthCare, China) at 37°C for 16 hours using the hybridizer (Abbott, USA), stained with DAPI, and analyzed under the fluorescence microscope (Zeiss, Germany). For metaphase FISH, the labeled slides from chromosome analysis should undergo de-oiling and destaining before incubation in 2× SSC buffer.
Total RNA was extracted from diagnostic BM using TRIzol, following the manufacturer’s protocol. 500-1000 ng of RNA was Poly(A)-based mRNA enrichment using VAHTS mRNA Capture Beads 2.0 (Vazyme, China). The preparation of cDNA library using VAHTS Universal V8 RNA-seq Library Prep Kit for Illumina (Vazyme, China). Finally, Illumina Novaseq 6000 sequencing platform was used for library sequencing. Raw FASTQ data were quality-checked with Fastp, and fusion gene candidates were identified using Arriba (v2.2.0). Hot gene pairs were reviewed with an in-house database and manually verified in Integrative Genomics Viewer (v2.16.2).
Total RNA was reverse transcribed into cDNA using the High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher, USA) according to the manufacturer’s instructions. RT-qPCR was performed in a 96-well plate (20 µL/well) containing TaqMan gene Expression Master Mix, primers (500 nM), probe (250 nM), and cDNA template. The primers and probe sequences are provided in Supplementary Figure 1E. Thermal cycling was as follows: 50°C for 2 min, 95°C for 10 min, followed by 45 cycles of 95°C for 15 sec and 60°C for 1 min. cABL was used as a housekeeping gene to confirm the amplifiability and quality of the cDNA.
A 27-year-old male patient with a history of recurrent pruritus and irritative cough was admitted to the hospital complaining about the persistent eosinophilia and recurrent episodes of dizziness. Six months earlier, he had experienced a sudden onset of left-sided limb weakness and dysarthria. At that time, Cranial computed tomography (CT) demonstrated a wedge-shaped hyperintense lesion at the right frontotemporal-parietal junction, indicative of a cerebral infarction. His peripheral blood exhibited a mildly elevated white blood cell count (18.42×109/L) with marked eosinophilia (43.2%), and thrombocytopenia (73×109/L). No hepatosplenomegaly was detected upon physical examination. Cerebrospinal fluid was normal.
Bone marrow (BM) morphology showed marked granulocytic hyperplasia with an increase in cytoplasmic granules within granulocytes and a heightened proportion of eosinophils (Supplementary Figure 1A). BM biopsy indicated extremely active marrow proliferation (>90%) with an increased proportion of granulocytes, and noticeable eosinophil and megakaryocyte proliferation. Flow cytometry revealed a low proportion of myeloid blasts with no significant phenotypic abnormalities, and an increased proportion of eosinophils (Supplementary Figure 1B).
Conventional chromosome analysis using G-banding and R-banding demonstrate a male karyotype with an apparently balanced t(4;7)(q12; q32) observed in 16/20 BM cells examined (Figure 1B, Supplementary Figure 1C). Interphase FISH analysis using the PDGFRA Tricolor Rearrangement Probe (Figure 1A) revealed a 71% positive signal (Supplementary Figure 1D). One overlapping green/orange/aqua signal on the normal chromosome 4 (pink arrows), one overlapping green/orange signal on the abnormal chromosome 7 (orange arrows), and one separate aqua signal on the abnormal chromosome 4 (purple arrows) were detected by metaphase FISH (Figure 1C). Normal chromosome 7 had no signal (yellow arrows).
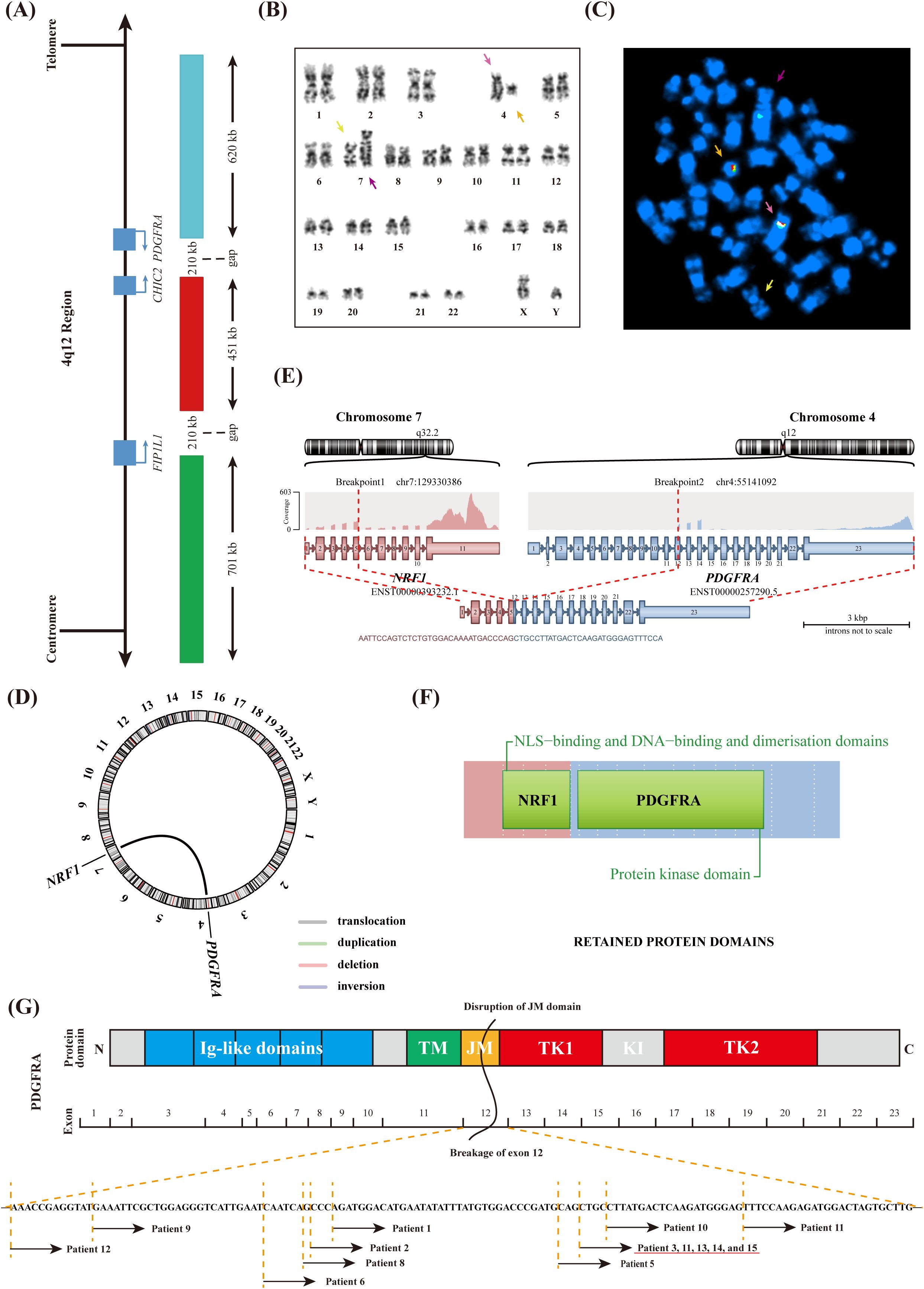
Figure 1. Molecular Genetics analyses of BM cells. (A) Schematic representation of the 4q12 region indicating relevant genes and the PDGFRA Tricolor Rearrangement Probe. (B) Karyotype showing abnormal chromosomes identified by G-banding. (C) Metaphase FISH hybridized with PDGFRA Tricolor Rearrangement Probe. (D) Identification of a novel NRF1::PDGFRA fusion gene by RNA sequencing. (E) Schematic representation of the recombination between the NRF1 and PDGFRA genes. (F) The domain of the NRF1-PDGFRA fusion protein. (G) Schematic representation of the PDGFRA protein domains and the break positions within exon 12 of PDGFRA in MLN-TK patients.
The abnormal FISH signal in this case indicated that PDGFRA was cleaved and that the telomere proximal site of chromosome 4q was translocated to chromosome 7q, which had not been previously reported. Consequently, the RNA sequencing analysis was conducted to identify the unknown fusion partner, and the NRF1 (Nuclear Respiratory Factor 1; Gene ID: 4899) was confirmed at the RNA level (Figures 1D, E). Quantitative analysis revealed that NRF1::PDGFRA transcript exhibited a higher expression level (210.72%). Based on the above results, we confirmed the in-frame fusion between the intact exon 5 of NRF1 and the truncated exon 12 of PDGFRA. The resulting fusion protein comprised the nuclear localization signal (NLS)-binding and DNA-binding and dimerization domains of NRF1, along with the protein kinase domain of PDGFRA (Figure 1F).
The patient was diagnosed with MLN-TK and initiated treatment with imatinib at a daily dose of 100 mg. After three days, the patient’s pruritus significantly improved. After two weeks, the cough had fully resolved, and dizziness had largely subsided. Follow-up complete blood count results revealed a white blood cell count of 5.25×109/L, an eosinophil percentage of 3.6%, and a platelet count of 172×109/L, indicating the achievement of complete hematologic remission. After three months of treatment, both chromosome analysis and fluorescence in situ hybridization (FISH) analysis were negative, confirming the achievement of complete cytogenetic remission. Additionally, quantitative analysis of the NRF1::PDGFRA fusion gene demonstrated a substantial reduction from 210.72% to 0.11%, suggesting partial molecular remission. Magnetic resonance imaging (MRI) scans revealed a marked decrease in high signal intensity within the right frontal-temporal-parietal junction cortex compared to prior imaging. However, residual patchy high signal lesions persisted in the cortical region, accompanied by localized gliosis and mild atrophic changes (Supplementary Figure 1F). During the course of imatinib therapy, the patient experienced mild (grade 1) myalgia in the limbs, which spontaneously resolved after approximately one month, likely attributed to the imatinib treatment.
MLN-TK encompasses a wide range of histological types, including MPN, MDS, MDS/MPN, AML, and MPAL, as well as B or T lymphocytic leukemia/lymphoma (ALL). Extramedullary disease is common. Although eosinophilia is a frequent and significant feature, it may be absent in some cases (1). In the reported cohort of 135 MLN-TK patients, blast phase was primary in about 70% of patients, with a lower relative frequency of 16% in patients with PDGFRA/PDGFRB fusion genes, and only 6% had secondary blast phase after a median of 87 months, likely because >90% of patients achieved persistent complete hematologic, complete cytogenetic (PDGFRB), and complete molecular (FIP1L1::PDGFRA) remissions after treatment with imatinib (3, 11).
NRF1 functions as a transcription factor that activates the expression of some key metabolic genes regulating cellular growth and nuclear genes required for respiration, heme biosynthesis, and mitochondrial DNA transcription and replication (14). The fusion partners of NRF1 that have been discovered through Next-Generation Sequencing (NGS) include MTMR1, BRAF, STRIP2, PPP2R5A and MKLN1, among others (15, 16). However, only the NRF1-BRAF fusion has been reported in two cases, associated with pleomorphic xanthoastrocytoma and urothelial carcinoma, with genomic breakpoints in NRF1 occurred within exon 5 and exon 10, respectively (17, 18). Regrettably, the functional alterations resulting from NRF1 rearrangements have scarcely been explored in the literature. Nevertheless, research has revealed that the knockdown of NRF1 in rats adversely impacts mitochondrial biogenesis and function, and the knockout of NRF1 in mice even resulting in embryonic lethality (19). The impact of NRF1 rearrangements remains to be further studied.
PDGFRA rearrangements typically activate receptor tyrosine kinases (RTKs), initiating a cascade of aberrant signaling pathways, which ultimately result in uncontrolled cell proliferation and inhibition of apoptosis (20). As shown in Figure 1G, PDGFRA consists of five extracellular immunoglobulin-like (Ig-like) domains, a transmembrane (TM) domain, a juxtamembrane (JM) domain, and a bipartite tyrosine kinase catalytic domain (TK1 and TK2), which are separated by a kinase insertion region (KI) (21). An unusual phenomenon has been observed in which the breakpoint in PDGFRA always occurs within exon 12 (Figure 1G, Table 1). Previous studies have demonstrated that exon 12 of PDGFRA encodes a portion of the JM domain, which is known to exert an autoinhibitory function in other receptor tyrosine kinases. Disruptions to this domain, including missense mutations, in-frame insertions, or in-frame deletions, can result in the constitutive activation of the corresponding tyrosine kinase (22). For this phenomenon, we believe it is related to the special position of exon 12 that encodes the JM domain. If the breakage position is before exon 12, the complete JM domain will inhibit the activity of the RTKs. Conversely, if the break occurs after exon 12, the disrupted TK domain leads to RTKs inactivation. Both scenarios do not result in disease phenotypes and thus remain undetected. Similar to other fusion tyrosine kinases, NRF1-PDGFRA is also likely a constitutively active tyrosine kinase that can transform hematopoietic cells both in vitro and in vivo, consistent with mechanisms observed in previous studies, although further investigation is necessary to substantiate the mechanism.
The MLN-TK with cerebral infarction is rarely reported in the literature. Cerebral infarction caused by eosinophilia may involve three mechanisms: (i) endocardial damage leading to mural thrombosis, embolism, and infarction; (ii) a hypercoagulable state promoting thrombosis through eosinophil release of various proteins; and (iii) insufficient perfusion due to eosinophil-induced vascular permeability, microcirculation disturbance, and oxygen deficiency (23–25). Although imaging studies in this patient did not reveal any large vessel abnormalities, Cranial MRI identified a wedge-shaped hyperintense lesion at the junction of the right frontal, temporal, and parietal lobes, which is consistent with the typical characteristics of a watershed infarction. This finding may indicate localized ischemic changes, potentially resulting from small vessel inflammation or a state of hypoperfusion. Notably, after imatinib treatment, the hyperintense cortical signal in the right frontal-temporal-parietal junction area significantly reduced, indicating a potential association between the brain infarction and eosinophilia in this patient.
In summary, this is the first report to describe the NRF1::PDGFRA fusion gene in MLN-TK, which exhibits highly sensitivity to low-dose imatinib. Additionally, the patient in this case exhibited a rare occurrence of brain infarction associated with eosinophilia. This discovery not only highlights a novel genetic alteration in MLN-TK but also provides a basis for further investigation into the role of this fusion gene in disease pathogenesis and treatment responsiveness.
The original contributions presented in the study are publicly available. This data can be found here: https://doi.org/10.6084/m9.figshare.28602737.
The studies involving humans were approved by Drug Clinical Trial Ethics Committee of the Third People’s Hospital of Datong City. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study. Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
JL: Conceptualization, Data curation, Formal analysis, Investigation, Validation, Writing – original draft, Writing – review & editing. YF: Conceptualization, Methodology, Validation, Writing – original draft, Writing – review & editing. YZ: Data curation, Writing – original draft. YX: Validation, Writing – original draft. XL: Validation, Writing – original draft. TX: Validation, Writing – original draft. JZ: Validation, Writing – original draft. KF: Validation, Writing – original draft. LL: Validation, Writing – original draft. XY: Validation, Writing – original draft. JG: Conceptualization, Methodology, Resources, Supervision, Validation, Writing – review & editing.
The author(s) declare that no financial support was received for the research and/or publication of this article.
We would like to thank all colleagues for technical assistance and clinical support from Tianjin Union Precision Medical Diagnostics Co. Ltd, Blood Diseases Hospital, Chinese Academy of Medical Sciences & Peking Union Medical College and the Third People’s Hospital of Datong City.
Authors JL, YX, TX, JZ, LL and JG were employed by the company Tianjin Union Precision Medical Diagnostics Co. Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2025.1552928/full#supplementary-material.
1. Khoury JD, Solary E, Abla O, Akkari Y, Alaggio R, Apperley JF, et al. The 5th edition of the World Health Organization Classification of Haematolymphoid Tumours: Myeloid and Histiocytic/Dendritic Neoplasms. Leukemia. (2022) 36:1703–19. doi: 10.1038/s41375-022-01613-1
2. Arber DA, Orazi A, Hasserjian RP, Borowitz MJ, Calvo KR, Kvasnicka HM, et al. International Consensus Classification of Myeloid Neoplasms and Acute Leukemias: integrating morphologic, clinical, and genomic data. Blood. (2022) 140:1200–28. doi: 10.1182/blood.2022015850
3. Metzgeroth G, Steiner L, Naumann N, Lubke J, Kreil S, Fabarius A, et al. Myeloid/lymphoid neoplasms with eosinophilia and tyrosine kinase gene fusions: reevaluation of the defining characteristics in a registry-based cohort. Leukemia. (2023) 37:1860–7. doi: 10.1038/s41375-023-01958-1
4. Kaur P, Khan WA. Myeloid/Lymphoid Neoplasms with Eosinophilia and Platelet Derived Growth Factor Receptor Alpha (PDGFRA) Rearrangement. Exon Publications (2022) 129–146. doi: 10.36255/exon-publications-leukemia-pdgfra-rearrangement
5. Cools J, DeAngelo DJ, Gotlib J, Stover EH, Legare RD, Cortes J, et al. A tyrosine kinase created by fusion of the PDGFRA and FIP1L1 genes as a therapeutic target of imatinib in idiopathic hypereosinophilic syndrome. N Engl J Med. (2003) 348:1201–14. doi: 10.1056/NEJMoa025217
6. Baxter EJ, Hochhaus A, Bolufer P, Reiter A, Fernandez JM, Senent L, et al. The t(4;22)(q12;q11) in atypical chronic myeloid leukaemia fuses BCR to PDGFRA. Hum Mol Genet. (2002) 11:1391–7. doi: 10.1093/hmg/11.12.1391
7. Trempat P, Villalva C, Laurent G, Armstrong F, Delsol G, Dastugue N, et al. Chronic myeloproliferative disorders with rearrangement of the platelet-derived growth factor alpha receptor: a new clinical target for STI571/Glivec. Oncogene. (2003) 22:5702–6. doi: 10.1038/sj.onc.1206543
8. Safley AM, Sebastian S, Collins TS, Tirado CA, Stenzel TT, Gong JZ, et al. Molecular and cytogenetic characterization of a novel translocation t(4;22) involving the breakpoint cluster region and platelet-derived growth factor receptor-alpha genes in a patient with atypical chronic myeloid leukemia. Genes Chromosomes Cancer. (2004) 40:44–50. doi: 10.1002/gcc.20014
9. Score J, Curtis C, Waghorn K, Stalder M, Jotterand M, Grand FH, et al. Identification of a novel imatinib responsive KIF5B-PDGFRA fusion gene following screening for PDGFRA overexpression in patients with hypereosinophilia. Leukemia. (2006) 20:827–32. doi: 10.1038/sj.leu.2404154
10. Walz C, Curtis C, Schnittger S, Schultheis B, Metzgeroth G, Schoch C, et al. Transient response to imatinib in a chronic eosinophilic leukemia associated with ins(9;4)(q33;q12q25) and a CDK5RAP2-PDGFRA fusion gene. Genes Chromosomes Cancer. (2006) 45:950–6. doi: 10.1002/gcc.20359
11. Curtis CE, Grand FH, Musto P, Clark A, Murphy J, Perla G, et al. Two novel imatinib-responsive PDGFRA fusion genes in chronic eosinophilic leukaemia. Br J Haematol. (2007) 138:77–81. doi: 10.1111/j.1365-2141.2007.06628.x
12. Sugimoto Y, Sada A, Shimokariya Y, Monma F, Ohishi K, Masuya M, et al. A novel FOXP1-PDGFRA fusion gene in myeloproliferative neoplasm with eosinophilia. Cancer Genet. (2015) 208:508–12. doi: 10.1016/j.cancergen.2015.07.001
13. Sahin Y, Pei J, Baldwin DA, Mansoor N, Koslosky L, Abdelmessieh P, et al. Acute myeloid leukemia with a novel AKAP9::PDGFRA fusion transformed from essential thrombocythemia: A case report and mini review. Leuk Res Rep. (2024) 21:100465. doi: 10.1016/j.lrr.2024.100465
14. Mulder H. Transcribing β-cell mitochondria in health and disease. Mol Metab. (2017) 6:1040–51. doi: 10.1016/j.molmet.2017.05.014
15. Huret JL, Dessen P, Bernheim A. Atlas of Genetics and Cytogenetics in Oncology and Haematology, updated. Nucleic Acids Res. (2001) 29:303–4. doi: 10.1093/nar/29.1.303
16. Kim P, Tan H, Liu J, Lee H, Jung H, Kumar H, et al. FusionGDB 2.0: fusion gene annotation updates aided by deep learning. Nucleic Acids Res. (2022) 50:D1221–D30. doi: 10.1093/nar/gkab1056
17. Phillips JJ, Gong H, Chen K, Joseph NM, van Ziffle J, Jin LW, et al. Activating NRF1-BRAF and ATG7-RAF1 fusions in anaplastic pleomorphic xanthoastrocytoma without BRAF p.V600E mutation. Acta Neuropathol. (2016) 132:757–60. doi: 10.1007/s00401-016-1616-3
18. Isaacson AL, Guseva NV, Bossler AD, Ma D. Urothelial carcinoma with an NRF1-BRAF rearrangement and response to targeted therapy. Cold Spring Harb Mol Case Stud. (2019) 5(3):a003848. doi: 10.1101/mcs.a003848
19. Mattingly KA, Ivanova MM, Riggs KA, Wickramasinghe NS, Barch MJ, Klinge CM. Estradiol stimulates transcription of nuclear respiratory factor-1 and increases mitochondrial biogenesis. Mol Endocrinol. (2008) 22:609–22. doi: 10.1210/me.2007-0029
20. Montor WR, Salas AROSE, Melo F. Receptor tyrosine kinases and downstream pathways as druggable targets for cancer treatment: the current arsenal of inhibitors. Mol Cancer. (2018) 17:1–18. doi: 10.1186/s12943-018-0792-2
21. Martinho O, Reis RM. Malignant gliomas: role of platelet-derived growth factor receptor A (PDGFRA). Tumors Cent Nervous System Volume 1: Gliomas: Glioblastoma (Part 1). (2011) 1:109–18. doi: 10.1007/978-94-007-0344-5_12
22. Gotlib J, Cools J, Malone JM 3rd, Schrier SL, Gilliland DG, Coutre SE. The FIP1L1-PDGFRalpha fusion tyrosine kinase in hypereosinophilic syndrome and chronic eosinophilic leukemia: implications for diagnosis, classification, and management. Blood. (2004) 103:2879–91. doi: 10.1182/blood-2003-06-1824
23. Jia Y, Zhao X, Li Y, Liu G, Wang B, Zhu X, et al. Case Report Eosinophilia complicated with cerebral infarction: a case report and literature review. Int J Clin Exp Med. (2023) 16:142–7.
24. Sethi HS, Schmidley JW. Cerebral infarcts in the setting of eosinophilia: three cases and a discussion. Arch Neurol. (2010) 67:1275–7. doi: 10.1001/archneurol.2010.256
Keywords: eosinophilia, PDGFRA rearrangement, NRF1, MLN-TK, gene fusion, imatinib
Citation: Liu J, Feng Y, Zhang Y, Xiao Y, Liu X, Xiao T, Zou J, Fan K, Lu L, Yang X and Gong J (2025) Identification of a novel NRF1::PDGFRA fusion in myeloid/lymphoid neoplasms with eosinophilia and tyrosine kinase gene fusions. Front. Oncol. 15:1552928. doi: 10.3389/fonc.2025.1552928
Received: 29 December 2024; Accepted: 07 March 2025;
Published: 25 March 2025.
Edited by:
Valentin Garcia-Gutierrez, Ramón y Cajal University Hospital, SpainReviewed by:
Teresa de Souza Fernandez, National Cancer Institute (INCA), BrazilCopyright © 2025 Liu, Feng, Zhang, Xiao, Liu, Xiao, Zou, Fan, Lu, Yang and Gong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jinying Gong, Z29uZ2ppbnlpbmdAaWhjYW1zLmFjLmNu; Yaqing Feng, MzQ0MjE4NzE2NkBxcS5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.