- 1Department of Hematology, Jingjiang People’s Hospital Affiliated to Yangzhou University, Taizhou, Jiangsu, China
- 2National Clinical Research Center for Hematologic Diseases, Jiangsu Institute of Hematology, the First Affiliated Hospital of Soochow University, Soochow University, Suzhou, China
- 3Changshu Hospital Affiliated to Soochow University, Changshu No.1 People’s Hospital, Suzhou, Jiangsu, China
- 4Department of Hematology, The Affiliated Zhangjiagang Hospital of Soochow University, Suzhou, Jiangsu, China
- 5Institute of Blood and Marrow Transplantation, Collaborative Innovation Center of Hematology, Soochow University, Suzhou, China
- 6Department of Hematology, People’s Hospital of Xinghua City, Taizhou, Jiangsu, China
- 7Department of Hematology, Suzhou Jsuniwell Medical Laboratory, Suzhou, China
- 8Department of Hematology, Huai’an Hospital Affiliated to Xuzhou Medical College and Huai’an Second People’s Hospital, Huai’an, Jiangsu, China
The gene ETV6 has been confirmed to be a genetic susceptibility gene for thrombocytopenia and leukemia. Here, we report a long-chain noncoding RNA AC010198.2 as a novel fusion partner of ETV6, showing a karyotype of del(12)(p13p11), with poor prognosis in a post-MPN AML that has never been reported, which may be an vital initial event in the transformation of MPN to AML and deterioration of disease.
Introduction
The ETV6 gene, located at chromosome 12p13, belongs to the ETS family of transcription factors, which share a conserved 80 amino acid DNA-binding domain, the ETS domain (1). It encodes an essential transcriptional repressor that is abundantly expressed in hematopoietic stem and progenitor cells (HSPCs). It is critical for the maintenance of HSPCs and the formation of platelets (2, 3), and has been shown to be closely associated with inherited thrombocytopenia and leukemia predisposition (3, 4). ETV6 variants include insertion/deletion mutations and structural variants (deletions, rearrangements) that may lead to truncation and loss of function of ETV6, ultimately contributing to thrombocytopenia and leukemia (4). It has been reported that ETV6 has over 30 partner genes (Supplementary Table 1), such as PDGFRB (1), RUNX1 (5), ACSL6 (6), and non-protein coding RNA such as LINC02260 (7), and is one of the most common translocation genes in acute lymphoblastic leukemia (ALL) and other hematological disorders such as acute myeloid leukemia (AML) and myelodysplastic syndrome (MDS) (1). Functional investigations of ETV6 variants have shown that they are associated with disrupted expression of genes involved in platelet production and platelet cytoskeleton dynamics, including the CDC42 and RHOA218 genes, but the mechanisms driving leukemia remain unclear (4). Recent studies have shown that ETV6 is required to repress inflammatory gene expression in HSPCs and this mechanism may be critical for maintaining HSPC function (8). Here, we report a long-chain non-coding RNA AC010198.2 as a novel fusion partner of ETV6, showing a karyotype of del(12)(p13p11), with poor prognosis in a post-myeloproliferative neoplasm Myeloproliferative Neoplasms (MPN) AML that has never been reported.
A 61-year-old male patient was diagnosed with myelofibrosis in 2018 and treated with hydroxyurea in a local hospital. In June 2020, he was admitted to the First Affiliated Hospital of Soochow University for a bone marrow (BM) examination which further confirmed the diagnosis of myelofibrosis with CALR exon9 gene mutation and karyotype of 48, XY, +Y, +21[10]. He was instructed to take ruxolitinib 20mg twice a day for treatment and was then lost to follow-up.
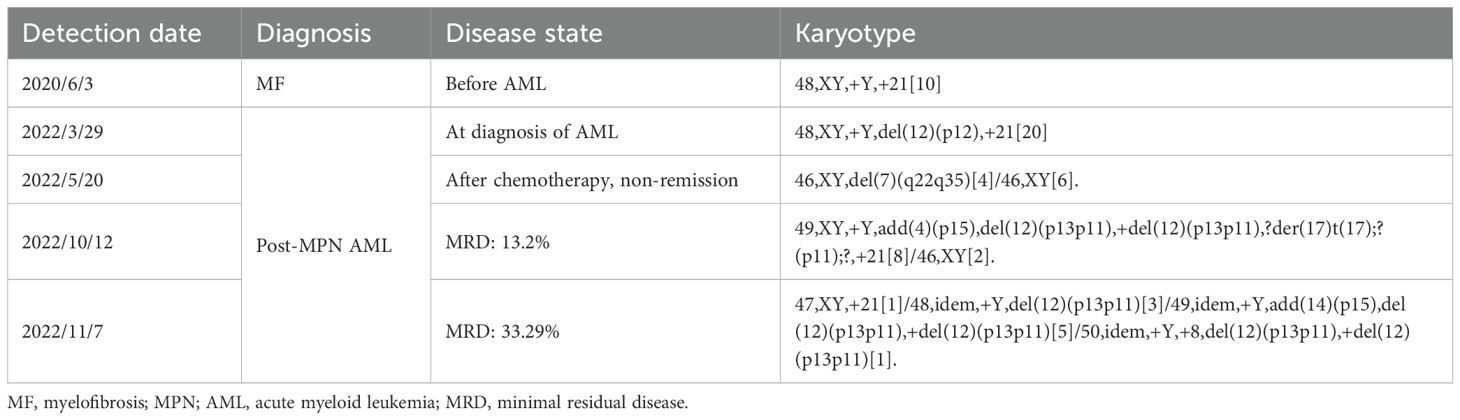
On 29 March 2022, the patient was referred to a local hospital for routine peripheral blood (PB) tests indicating a white blood cell count of 111.7×10^9/L with neutrophils 45.70×10^9/L, hemoglobin 8.6 g/dL, and thrombocytes 167×10^9/L. PB smear showed 33% of primitive monocytes and 19% myeloblasts. BM aspiration revealed 20% primitive monocytes with intracytoplasmic Auer bodies. Flow cytometry for BM cells indicated 27.55% of abnormal myeloblasts. Conventional R-band karyotype analysis suggested a karyotype of 48, XY, +Y, del(12)(p12), +21[20]. The BCR-ABL fusion gene and MPN-related gene (CALR, JAK2, MPL) mutations were negative. He was diagnosed with post-MPN AML and treated with HA (harringtonine and cytarabine) in combination with hydroxyurea and ruxolitinib. The first course of treatment failed to achieve remission, and next-generation sequencing (NGS) showed SRSF2, SETBP1, RUNX1, and IDH2 gene mutations which are reported to be enriched in post-MPN AML (9). Complete remission was still not achieved after the second D-HA (decitabine, homoharringtonine, and cytarabine) regimen immediately after the first course of treatment, and the gene mutations were persistently positive with a 1.23% relative expression level of the WT1 gene and a karyotype of 46, XY, del(7)(q22q35)[4]/46, XY[6]. A third course of azacitidine in combination with a venetoclax regimen resulted in a minimal residual disease (MRD) of 2.1×10-4 but there was a rapid relapse in a short time later with more complex chromosomal abnormalities of 49, XY, +Y, add(4)(p15), del(12)(p13p11), +del(12)(p13p11),?der(17)t(17);?(p11);?, +21[8]/46, XY[2] found on 12 October 2022. Soon thereafter the karyotype got even more complicated and was found to be 47, XY, +21[1]/48, idem, +Y, del(12)(p13p11)[3]/49, idem, +Y, add(14)(p15), del(12)(p13p11), +del(12)(p13p11)[5]/50, idem, +Y, +8, del(12)(p13p11), +del(12)(p13p11)[1]. This was detected on 7 November 2022. Furthermore, the relative expression level of WT1 increased to 44.08%. Targeted RNA sequencing showed a novel fusion composed of exons 1 to 3 of ETV6 and exon 3 of the AC010198.2 (Figure 1) that mapped to chromosome 12p13.2 and chromosome 12p11.21, respectively. AC010198.2 is a long-chain non-coding RNA, a type of lncRNA, which contains three non-coding exons and does not encode any proteins. The ETV6-AC010198.2 transcript is not expected to produce a chimeric protein but results in a truncated or possibly unproductive ETV6 protein that contributes to the inactivation of the ETV6 gene and thus promotes disease progression.

Figure 1. Laboratory examinations during the progression of the disease. (A) R-band karyotyping shows 47, XY, +21[1]/48, idem, +Y, del(12)(p13p11)[3]/49, idem, +Y, add(14)(p15), del(12)(p13p11), +del(12)(p13p11)[5]/50, idem, +Y, +8, del(12)(p13p11), and +del(12)(p13p11)[1]. (B) Schematic of the ETV6::AC010198.2 fusion breakpoint identified in this case. (C) A Circos plot. (D) RT-PCR for the ETV6-AC010198.2 fusion transcript with ETV6-F and AC010198.2-R primers and the expected product size was 382 bp. Lane a: 2000bp DNA ladder marker. Lane B: patient sample. Lane C: blank control. (E) Sanger sequencing of the PCR product confirmed the fusion between exon3 of the ETV6 gene and exon3 of the AC010198.2 gene.
Chromosomal karyotype is a highly significant predictor of leukemic progression in MPNs. A number of chromosomal abnormalities, including i(17q), inv(3)/3q21, monosomy 7, 12p-/12p11.2, 11q-/11q23, and autosomal trisomy (excluding +8/+9), have been reported to be associated with a poorer prognosis and a nearly two-fold increased risk of leukemic transformation compared to other traditionally unfavorable karyotypes. The median overall survival for patients with these abnormalities is only 1.2 years (9, 10). This patient had autosomal trisomy +21 at the MPN stage and transformed into AML upon the appearance of 12p-, followed by progressively complicated karyotypes (Table 1), which resulted in rapid disease deterioration and poor treatment response. It is notable that 12p- was consistently positive during disease progression, with the exception of transient negativity observed following the initial treatment. This suggests that 12p- and the resulting fusion ETV6::AC010198.2 is an important factor contributing to genome instability and plays a pivotal role in the leukemic progression.
A thorough review of the extant literature reveals a paucity of studies addressing the AC010198.2 lncRNA. A single study found that high expression of AC010198.2 was associated with the sensitivity of cervical cancer cells to cisplatin (11). However, ongoing research continues to investigate the pathogenic mechanisms of the ETV6 gene in hematological disorders. In view of the considerable number of ETV6 partner genes, some researchers have broadly classified these fusion genes into three groups: 1) protein tyrosine kinases, 2) transcription factors, and 3) “unproductive” fusions, i.e., fusion that do not seem to produce a meaningful fusion protein (1). The most prevalent fusion, ETV6::RUNX1, has been shown to promote ALL by impeding hematopoietic reconstitution and lymphocyte differentiation through the expression of fusion proteins (5). The ETV6::ACSL6 fusion gene, which occurs almost exclusively in AML, is not expected to produce fusion proteins. However, it has been demonstrated to trigger an increase in the transcription of the encoded inflammatory factors (e.g., IL-3) adjacent to and distal to the breakpoint through the translocation and activation of a super-enhancer located at the ETV6 gene, which ultimately leads to eosinophilia (12). The novel fusion gene ETV6::AC010198.2 reported in this study is similar to ETV6::ACSL6 in that it fails to produce fusion proteins. It is hypothesized that they may share analogous pathogenic mechanisms that stimulate the expression of neighboring genes under the super-enhancer of ETV6. Since this is the first international report, its pathogenic mechanism needs to be further verified.
In summary, we identified a novel ETV6 fusion gene transcript that does not express a functional protein in a post-MPN AML patient with del(12)(12p13p11) which may be a vital initial event in the transformation of MPN to AML and deterioration of disease.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
The studies involving humans were approved by the First Affiliated Hospital of Soochow University. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation in this study was provided by the participants’ legal guardians/next of kin. Written informed consent was obtained from the individual(s), and minor(s)’ legal guardian/next of kin, for the publication of any potentially identifiable images or data included in this article.
Author contributions
JL: Writing – original draft. ZW: Writing – original draft. GY: Writing – original draft. QZ: Writing – original draft. MW: Writing – original draft. LW: Writing – original draft. LC: Writing – review & editing. HC: Writing – review & editing. XJ: Writing – review & editing. CL: Writing – review & editing. TL: Writing – review & editing. QH: Writing – review & editing. ZL: Writing – original draft. SZ: Writing – original draft. ZZ: Writing – review & editing. JP: Writing – review & editing. JC: Writing – review & editing. SC: Writing – review & editing. MS: Writing – review & editing. XZ: Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported in part by grants from Administration of Traditional Chinese Medicine of Jiangsu Province (MS2022131), the National Key R&D Program of China (2022YFC2502701), the National Natural Science Foundation of China (82170158, 82100175, 82004187 and 82060810), the Open Project of Jiangsu Biobank of Clinical Resources (TC2022B008, CY202304), Huai’an Health Commission Research General Project (HAWJ2024017).
Acknowledgments
The sample was from Jiangsu Biobank of Clinical Resources.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2025.1495182/full#supplementary-material
References
1. Bohlander SK. ETV6: a versatile player in leukemogenesis. Semin Cancer Biol. (2005) 15:162–74. doi: 10.1016/j.semcancer.2005.01.008
2. Hock H, Meade E, Medeiros S, Schindler JW, Valk PJ, Fujiwara Y, et al. Tel/Etv6 is an essential and selective regulator of adult hematopoietic stem cell survival. Genes Dev. (2004) 18:2336–41. doi: 10.1101/gad.1239604
3. Hock H, Shimamura A. ETV6 in hematopoiesis and leukemia predisposition. Semin Hematol. (2017) 54:98–104. doi: 10.1053/j.seminhematol.2017.04.005
4. Klco JM, Mullighan CG. Advances in germline predisposition to acute leukaemias and myeloid neoplasms. Nat Rev Cancer. (2021) 21:122–37. doi: 10.1038/s41568-020-00315-z
5. Eldeeb M, Konturek-Ciesla A, Zhang Q, Kharazi S, Tingvall-Gustafsson J, Ungerbäck J, et al. Ontogeny shapes the ability of ETV6::RUNX1 to enhance hematopoietic stem cell self-renewal and disrupt early lymphopoiesis. Leukemia. (2024) 38:455–9. doi: 10.1038/s41375-024-02149-2
6. Zhang T, Wang Q, Xu Y, Wang M, Ma Z, Pan J, et al. ETV6::ACSL6 fusion gene in myeloid Malignancies with eosinophilia: a report of two cases with t(5;12) or normal karyotype. Leuk Lymphoma. (2023) 64:225–9. doi: 10.1080/10428194.2022.2136948
7. Zhang L, Wang M, Wang Z, Zeng Z, Wen L, Xu Y, et al. Identification of a novel ETV6 truncated fusion gene in myeloproliferative neoplasm, unclassifiable with t(4;12)(q12;p13). Ann Hematol. (2020) 99:2445–7. doi: 10.1007/s00277-020-04207-y
8. Bloom M, Oak N, Baskin-Doerfler R, Feng R, Iacobucci I, Baviskar P, et al. ETV6 represses inflammatory response genes and regulates HSPC function during stress hematopoiesis in mice. Blood Adv. (2023) 7:5608–23. doi: 10.1182/bloodadvances.2022009313
9. Dunbar AJ, Rampal RK, Levine R. Leukemia secondary to myeloproliferative neoplasms. Blood. (2020) 136:61–70. doi: 10.1182/blood.2019000943
10. Tefferi A, Nicolosi M, Mudireddy M, Lasho TL, Gangat N, Begna KH, et al. Revised cytogenetic risk stratification in primary myelofibrosis: analysis based on 1002 informative patients. Leukemia. (2018) 32:1189–99. doi: 10.1038/s41375-018-0018-z
11. Lv H, Jin S, Zou B, Liang Y, Xie J, Wu S. Analyzing the whole-transcriptome profiles of ncRNAs and predicting the competing endogenous RNA networks in cervical cancer cell lines with cisplatin resistance. Cancer Cell Int. (2021) 21:532. doi: 10.1186/s12935-021-02239-6
Keywords: AC010198.2, novel, MPN, AML, ETV6
Citation: Lu J, Yang G, Zhang Q, Wang M, Wang L, Chen L, Chen H, Jiang X, Liu C, Lin T, Han Q, Liu Z, Zhen S, Zeng Z, Pan J, Cen J, Chen S, Wang Z, Zhang X and Sun M (2025) A novel truncated fusion gene ETV6::AC010198.2 in post-MPN acute myeloid leukemia caused by del(12)(p13p11). Front. Oncol. 15:1495182. doi: 10.3389/fonc.2025.1495182
Received: 12 September 2024; Accepted: 16 January 2025;
Published: 11 February 2025.
Edited by:
Mario Tiribelli, University of Udine, ItalyReviewed by:
Pora Kim, University of Texas Health Science Center at Houston, United StatesHarsh Goel, All India Institute of Medical Sciences, India
Copyright © 2025 Lu, Yang, Zhang, Wang, Wang, Chen, Chen, Jiang, Liu, Lin, Han, Liu, Zhen, Zeng, Pan, Cen, Chen, Wang, Zhang and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Miao Sun, MTA2NjEwOTM2QHFxLmNvbQ==; Xingxia Zhang, enh4MjAwNDk5QDEyNi5jb20=; Zheng Wang, d2FuZzExNnpoZW5nQDE2My5jb20=
†These authors have contributed equally to this work
 Jiao Lu
Jiao Lu Guanqun Yang3†
Guanqun Yang3† Qiaoyan Han
Qiaoyan Han Zhao Zeng
Zhao Zeng Jinlan Pan
Jinlan Pan Suning Chen
Suning Chen Zheng Wang
Zheng Wang