- 1Department of Neurosurgery, University of Oklahoma Health Sciences Center, Oklahoma City, OK, United States
- 2Department of Physiology, University of Oklahoma Health Sciences Center, Oklahoma City, OK, United States
- 3Department of Neurosurgery, Massachusetts General Hospital, Harvard Medical School, Boston, MA, United States
Introduction: Meningiomas are the most common primary central nervous system (CNS) tumor in adults, comprising one-third of all primary adult CNS tumors. Although several recent publications have identified molecular alterations in meningioma including characteristic mutations, copy number alterations, and gene expression signatures, our understanding of the drivers of meningioma recurrence is limited.
Objective: To identify gene expression signatures of 1p-22q-NF2- meningioma recurrence, with concurrent biallelic inactivation of NF2 and loss of chr1p that are heterogenous but enriched for recurrent meningiomas.
Methods: Transcriptomic alterations present in recurrent versus primary 1p-22q-NF2- meningiomas were identified using RNA sequencing (RNA-seq) data in a clinically annotated cohort.
Results: Recurrent 1p-22q-NF2- meningiomas were enriched for a newly identified GSTM1 null genotype compared to primary meningiomas that showed variable GSTM1 expression and independent external validation was performed.
Conclusions: The GSTM1 null genotype is a novel biomarker of 1p-22q-NF2- meningioma recurrence that resolves heterogeneity in existing meningioma subtypes and may be used to guide future clinical management decisions on extent of treatment to improve patient outcomes.
1 Introduction
Meningiomas are tumors that originate in the meninges and are the most common intracranial tumor type in adults, representing 39% of all primary adult central nervous system (CNS) tumors (1). The World Health Organization (WHO) classifies meningiomas into 15 subtypes and grades 1-3 based on histopathological features and specific molecular alterations (2). Alterations in NF2, moesin-ezrin-radixin like (MERLIN) tumor suppressor, are the most common genetic abnormality in meningioma, with up to 60% of sporadic meningiomas harboring them (3, 4). NF2 is mapped to chromosome 22 (chr22) and encodes merlin, an intracellular scaffold protein and tumor suppressor (5). Biallelic gene inactivation of NF2 resulting from chr22 monosomy and concurrent mutations in the remaining NF2 allele are characteristic alterations in multiple central and peripheral nervous system tumors in addition to meningioma including schwannomas and ependymomas (6). Allelic loss of chromosome 1p (chr1p) is the second most commonly observed chromosomal abnormality in meningiomas after deletion of chr22 (7, 8). Additional frequent genomic alterations in meningioma include mutations in TNF receptor associated factor 7 (TRAF7), KLF transcription factor 4 (KLF4), and phosphatidylInositol-4,5-bisphosphate 3-kinase catalytic subunit alpha (PIK3CA) (9, 10).
Recent advances in high throughput molecular profiling, including epigenetic, cytogenetic, and gene expression analyses, combined with computational modeling approaches have enabled enhanced classification of meningiomas into molecular subtypes that more accurately represent their clinical behavior than traditional histopathological evaluation (11, 12). A prominent molecular meningioma classification separates tumors into 3 subtypes: Type A meningiomas with TRAF7, KLF4, and/or AKT serine/threonine kinase 1 (AKT1) missense mutations and without significant chromosomal copy number alteration; type B meningiomas primarily distinguished by NF2 loss; and type C meningiomas with biallelic NF2 inactivation plus loss of chromosome 1p (13, 14). Different chr1p regions have been reported to be associated with meningioma development including 1p36, 1p34-1p32, 1p22, and 1p21.1-1p13 (15–18),, but the most frequent loss is in 1p34.1 (19, 20). Chromosome 1p loss leads to reduced expression of several genes including patched 2 (PTCH2) (21), AT-rich interaction domain 1A (ARID1A) (22, 23), alkaline phosphatase, biomineralization associated (ALPL) (24, 25), and cyclin dependent kinase inhibitor 2C (CDKN2C) (26).Type A and B meningiomas tend to follow a more benign course and type C are enriched for recurrent meningiomas. These subtypes better predicted tumor recurrence than WHO grading, the current clinical standard for predicting tumor recurrence. However, type C meningiomas with a higher recurrence risk remain a heterogenous group, with one-half recurring within 5 years and the other half exhibiting up to 5 years of recurrence free survival (13, 14). Another study divided meningiomas into four stable molecular groups (MG1-4) with type C meningiomas separated into two most clinically aggressive groups: hypermetabolic (MG3) and proliferative (MG4) (12, 13). The most frequent chromosomal abnormality in both MG3 and MG4 was the loss of both chr22q and chr1p (27). Novel somatic driver events identified in these meningiomas were chromatin remodeling and mutations in epigenetic regulators lysine demethylase 6A (KDM6A), chromodomain helicase DNA binding protein 2 (CHD2), and tumor suppressor phosphatase and tensin homolog (PTEN).
Finally, another subclassification of type C meningiomas based on transcriptomic data identifies A1 and A2 tumors distinguished by a 34-gene expression risk signature, with the latter group having a higher proportion of recurrent tumors (27). For this work, 13 bulk RNA-seq datasets were combined to create a dimension-reduced reference landscape of 1,298 meningiomas (28). The resulting reference map exhibited multiple clusters of tumors, indicating multiple RNA-seq-based meningioma subtypes, some of which were associated with distinct time to recurrence. The most striking differences of time to recurrence was seen between subclusters A1 and A2 within cluster A. While both A1 and A2 harbored meningiomas with biallelic inactivation of NF2 and loss of chr1p, subcluster A2 contained a much larger population of patients with poor outcomes compared to subcluster A1 (28). Interestingly, while A1 and A2 can be distinguished based on 34-gene expression risk signature, no single specific causative genetic alteration was identified (29).
Here, we directly compare transcriptional profiles of recurrent and primary 1p-22q-NF2- meningiomas, building on recent molecular advances in meningioma classification by focusing on this subtype enriched for recurrent tumors. The most differentially expressed gene (DEG) was glutathione S-transferase mu 1 (GSTM1), encoding a member of the mu class of cytosolic glutathione S-transferase (GST) family of metabolic isozymes responsible for detoxification of a broad range of substances including environmental toxins, drugs, and carcinogens. GSTs are divided into three major protein families with multiple subclasses in each family (30). Intensive study of GSTM1 has found frequent genetic polymorphisms, including complete biallelic deletion of the GSTM1 loci (31, 32).
Our results demonstrated that five out of twelve primary meningiomas had no expression of GSTM1, suggesting that these patients’ meningioma might reoccur in the future. The comparison of GSTM1-negative (GSTM1-) and GSTM1-positive (GSTM1+) transcriptomes revealed that GSTM1- meningiomas were characterized by higher expression of genes specific for positive regulation of cell motility, locomotion and angiogenesis, while transcriptomes of GSTM1+ tumors were enriched for transmembrane transport and system process genes. Taken together, our results present evidence that GSTM1 expression was completely absent in all recurrent meningiomas due to the GSTM1 null genotype resulting from inherited gene deletion and/or somatic deletion of chr1p, where GSTM1 is mapped.
Overall, existing work has largely focused on classifying meningiomas based on molecular alterations within primary tumors to predict the clinical presentation and course of the subtypes. This has transformed our understanding of the spectrum of meningioma clinical behavior but has not identified definitive biomarkers of meningioma recurrence, which are required in order to enable personalized medicine approaches to escalate treatment in aggressive meningiomas with the aim to delay recurrence and improve patient outcomes. This study identifies GSTM1 as a new biomarker of meningioma recurrence that builds on existing molecular subtypes to improve our ability to predict and manage recurrent meningiomas.
2 Materials and methods
2.1 Patients and sample collection
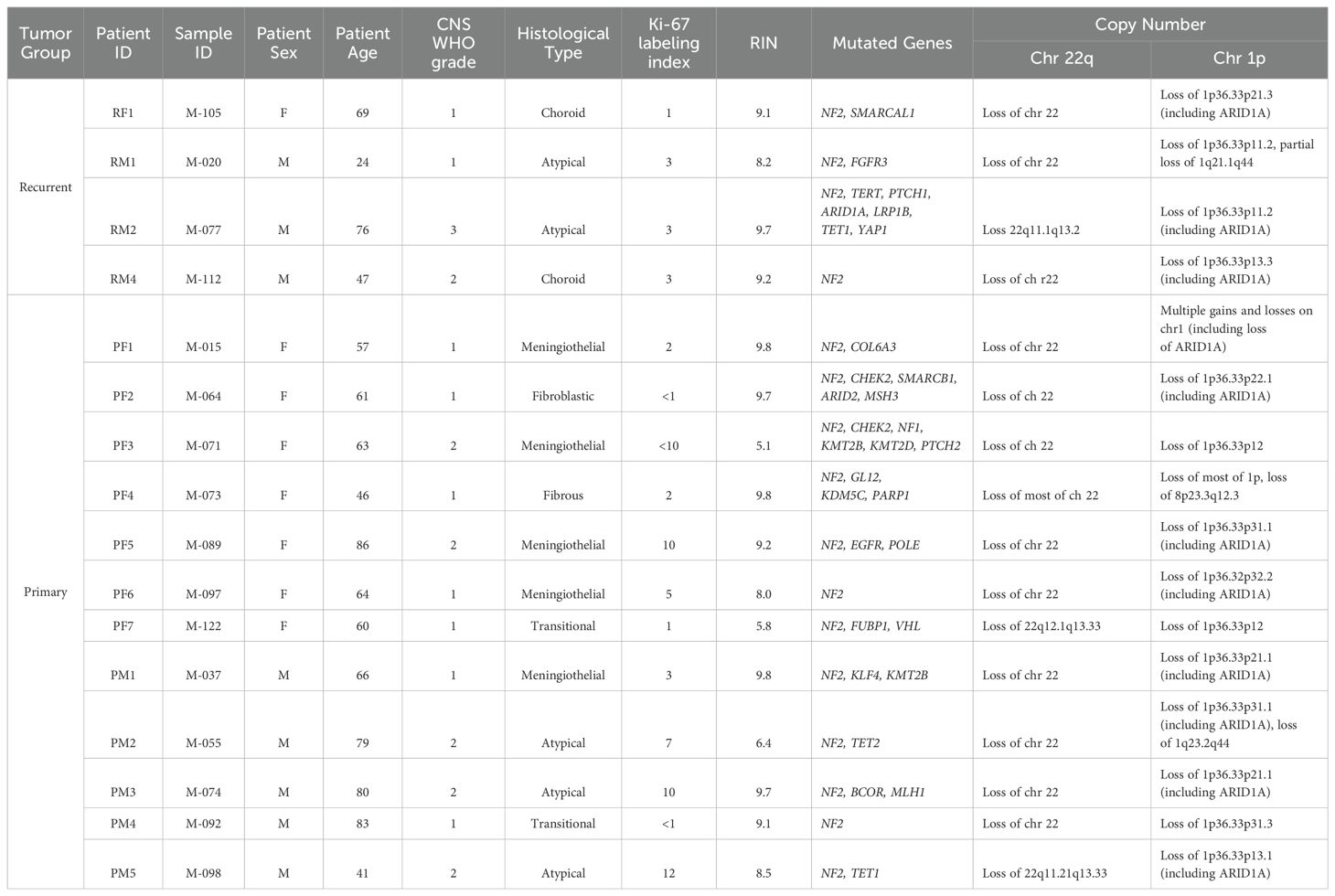
All procedures were approved by the institutional review board (IRB) of the University of Oklahoma Health Sciences Center (OUHSC) (IRB protocol number 10195). Sixteen samples from 16 patients (one sample/patient) diagnosed with meningioma at University of Oklahoma Medical Center were included in the study. All patients provided written informed consent for participation in the study. Clinical information including patient demographics, clinical course, and neuropathology was collected by retrospective chart review.
2.2 Histopathologic grading and genetic profiling
Following routine pathology processing, resected meningiomas were assigned a histopathologic grade according to the revised 4th edition of the WHO Classification of Tumors of the CNS (2). Ki-67 immunostaining was performed on at least one block in all cases. All samples were analyzed, graded, and independently confirmed by two staff neuropathologists. For genetic profiling of each tumor, specimens were sent to the Mayo Clinic Laboratories. Somatic mutations and gene rearrangements were examined by the NONCP panel. Copy number imbalances and loss of heterozygosity were estimated by a CMART panel. For RNA extraction, resected tissues were immediately submerged in RNAlater® Solution, kept at room temperature for 24 hours, and stored frozen long term (Fisher Scientific, AM7023).
2.3 RNA-seq and differential expression analysis
GST gene mRNA levels were calculated relative to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA levels measured by RNA-seq for GSTM genes and multiplex RT-qPCR for GSTT genes. Total RNA was extracted from tumors saved in RNAlater® Solution (Fisher Scientific, AM7023) with the RNeasy Plus mini kit (QIAGEN, 74136) with QIAshredder (QIAGEN, 79656). Preparation of cDNA libraries and sequencing was conducted by Novogene Co., LTD (Beijing, China). Significant DEGs were defined as those that had both an absolute log2FoldChange ≥ 1 as well as a false discovery rate adjusted p-value ≤ 0.05 for each comparison independently. Gene expression levels are expressed as Fragments Per Kilobase of transcript per Million mapped reads (FPKM). Pearson correlation analysis of the FPKM values for GSTM1 vs. GSTM2 expression was performed in GraphPad (v10.4) with p <0.05 considered statistically significant.
2.4 Quantitative PCR
Total tumor RNA was used to measure gene mRNA levels by real-time qPCR. Reverse transcription and cDNA amplification were performed in one tube using qScript™ XLT One-Step RT-qPCR ToughMix®, Low ROX™ (VWR Quanta Biosciences™, 95134) on an Applied Biosystems 7500 Fast Real-Time PCR System (Fisher Scientific). Sample reactions were run in 3-6 replicates. Each mRNA analysis was run in a DuPlex PCR reaction with GAPDH as an internal control. Standard curves for each gene were run to verify the linear range of amplification. Input RNA was kept under 200 ng per reaction to stay within the linear range for GAPDH levels. Gene expression levels for all genes of interest were determined by comparative ΔCT experiment runs, analyzed using the 7500 Software v2.3 and calculated as Relative Quantity (RQ). Shown expression values represent RQ multiplied by 1000. Pearson correlation analysis of RQ values for GSTM1 vs. GSTM4 expression was performed in GraphPad (v10.4) with p <0.05 considered statistically significant. Primers and Probes sequences used:
GSTT1-Fwd: TCCTTACTGGTCCTCACATCTC
GSTT1-Rev: GGCCTTCGAAGACTTGGC
GSTT1 Probe: ATGCATCAGCTCCGTGATGGCTAC (FAM – BHQ1)
GSTT4-Fwd: TCATCACCGGGAACCAAATC
GSTT4-Rev: GGAGCTGTTGAGGAAGACATTAT
GSTT4 Probe: TGGTGGAGATGATGCAGCCCAT (FAM – BHQ1)
GAPDH-Fwd: GGTGTGAACCATGAGAAGTATGA
GAPDH-Rev: GAGTCCTTCCACGATACCAAAG
GAPDH Probe: AGATCATCAGCAATGCCTCCTGCA (VIC-TAMRA)
2.5 Determination of GST(x) genes copy number by real-time PCR
Genomic DNA was extracted from tumor tissues with the DNeasy Blood and Tissue kit (Qiagen, 69506). Exon/intron junction or intron regions from genes of interest were detected by a duplex qPCR (FAM/VIC TaqMan® assay) for simultaneous detection of each gene of interest paired with apolipoprotein B (APOB) as a 2-copy standard reference gene. Standard curves were obtained by serial dilutions of a plasmid containing amplicon sequences of all genes of interest (GSTT1, GSTT4, GSTM1, GSTM2) and APOB sequence. The number of gene copies per cell (i.e. diploid genome) was determined by multiplying the ratio GST(x)/APOB by two. Primers and Probes sequences used:
GSTT1-Fwd: TCCTTACTGGTCCTCACATCTC
GSTT1-Rev: GGCCTTCGAAGACTTGGC
GSTT1 Probe: ATGCATCAGCTCCGTGATGGCTAC (FAM-BHQ1)
GSTT4-Fwd: TCATCACCGGGAACCAAATC
GSTT4-Rev: GGAGCTGTTGAGGAAGACATTAT
GSTT4 Probe: TGGTGGAGATGATGCAGCCCAT (FAM-BHQ1)
GSTM1-Fwd: CTGGGCATGATCTGCTACAA
GSTM1-Rev: TGTGCAGGAATGCAAGAGT
GSTM1 Probe: AGTGAGCTGCATCTGACAGAGTTTGG (FAM-BHQ1)
GSTM2-Fwd: CAAACTCTGCTATGACCCAGAT
GSTM2-Rev: AAGACCAAGAACTCACCAGAAG
GSTM2 Probe: CCTTTCCCTGCAGAGTTTGTGTCCA (FAM-BHQ1)
H-ApoB Fwd: TGAAGGTGGAGGACATTCCTCTA
H-ApoB Rev: CTGGAATTGCGATTTCTGGTAA
H-ApoB Probe: CGAGAATCACCCTGCCAGACTTCCGT (VIC-TAMRA)
3 Results
3.1 Clinical cohort characteristics
We built a cohort of sixteen patients with confirmed 1p-22q-NF2- meningioma samples for our analysis. Clinical and pathological features are outlined in (Table 1). The primary meningioma group consisted of twelve patients, seven females (PF1 to PF7) and five males (PM1 to PM5). The recurrent meningioma patients included 1 female (RF1) and 3 males (RM1, RM2, and RM4). RM3 was analyzed and excluded from further analysis because it did not pass RNA quality control (RNA Integrity Number (RIN) <3). The median RIN for the included samples was 9.2 (Table 1). The median age at the time of surgery for recurrent tumors was 58 and the median time to recurrence after the initial surgery was 12 years.
3.2 Transcriptomic signatures in recurrent versus primary meningiomas
To evaluate differences in the transcriptome of recurrent versus primary 1p-22q-NF2- meningiomas, we performed a differential comparison using RNA-seq data. We found that a vast majority of genes (11,951) were expressed in both primary and recurrent tumors, while relatively small number of genes (<8.3%) were expressed exclusively in primary (586 genes) or recurrent (496 genes) meningiomas (Figure 1A; Supplementary File 1). Nevertheless, the cohort showed a trend towards separation by recurrence status using this set of differentially expressed genes (DEGs) in a principal component analysis (PCA) plot (Figure 1B) and with hierarchical clustering (Supplementary Figure S1) and high Pearson correlation coefficient (Supplementary Figure S2).The most significant DEG was GSTM1 (Figure 1C) that was downregulated in recurrent tumors. These results suggest that GSTM1 under expression may be a biomarker of recurrence in 1p-22q-NF2- meningiomas, which are difficult to prognosticate currently. Other significant DEGs included zinc finger protein 536 (ZNF536) under expression (encoding a highly conserved transcription factor shown to negatively regulate neuronal differentiation) (33), AC005392.2 and LINC00485 long intergenic non-protein coding RNA overexpression, and KIAA2012 overexpression in recurrent meningiomas (encoding an uncharacterized protein of unknown function highly expressed in excitatory neurons, choroid plexus, and other ciliated cell types) (proteinatlas.org). Gene set enrichment analyses across three gene set databases (GO, KEGG, and Reactome) overall showed overexpression of cell structure and signaling pathways and under expression of developmental pathways in recurrent meningiomas (Figure 1D; Supplementary Figure S3). Interestingly, KEGG database analysis failed to identify any significantly enriched pathways in recurrent meningiomas while primary tumors were significantly enriched in “cAMP signaling pathway”, “Notch signaling pathway” and “protein digestion and absorption,” underscoring the value of a multi-database pathway analysis.
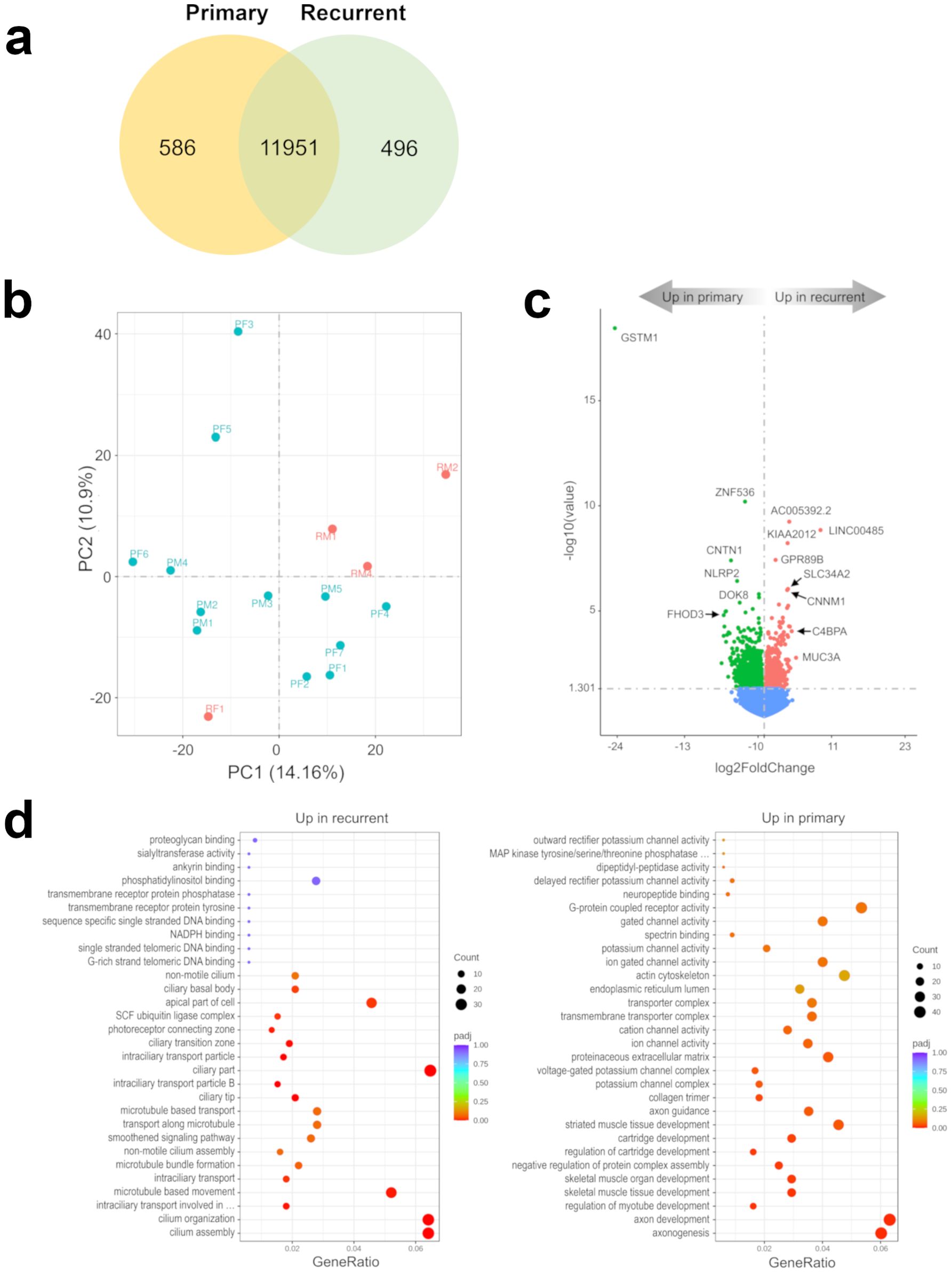
Figure 1. Recurrent 1p-22q-NF2- meningiomas have distinct transcriptomic markers. (A) Venn diagram showing the overlap of DEGs in recurrent and primary meningiomas by RNA-seq analysis. Common and tumor type-specific genes are depicted by numbers inside the corresponding circles. (B) PCA plot of recurrent (red dots) and primary (aquamarine dots) meningiomas using RNA-seq data. (C) Volcano plot displaying significant DEGs in recurrent vs. primary meningiomas. (D) Gene-set enrichment analyses of recurrent vs. primary meningiomas showing pathways that are upregulated and downregulated in recurrent meningioma. Bubbles represent pathways and gene ratios are percentages of significant genes of all genes in a pathway.
3.3 Transcriptomic signatures in GSTM1- versus GSTM1+ meningiomas
All recurrent tumors demonstrated the absence of GSTM1 expression and seven of 12 primary meningiomas showed GSTM1 expression (Supplementary File 2). Next, we compared mRNA expression profiles of GSTM1- and GSTM1+ subgroups to characterize differences between meningiomas with and without our marker of recurrence. There were 516 DEGs upregulated and 499 downregulated in GSTM1- meningiomas (Figure 2A) and the primary tumors with GSTM1 expression showed a trend towards clustering together (Figure 2B). Apart from expected differences in GSTM1 expression, GSTM1- meningiomas showed downregulation of anterior gradient 2, protein disulphide isomerase family member ((AGR2) encoding a member of the disulfide isomerase family of endoplasmic reticulum proteins important for oxidative protein folding) (34), we also found upregulation of microtubule associated protein 1 light chain 3 Gamma ((MAP1LC3C) encoding a member of the microtubule-associated family of proteins that are essential in the formation of autophagosomes and lysosomal degradation of cargo), neuropeptide Y receptor Y6 (NPY6R regulator of the growth hormone axis and body composition) (35), and NEDD4 E3 ubiquitin protein ligase ((NEDD4) encoding an E3 ubiquitin ligase enzyme that targets proteins for ubiquitination) as shown in Figure 2C. It was observed that meningiomas in this cohort harbored either GSMT1 or MAP1LC3C downregulation only and there were no samples with under expression of both (Figure 3). It has been shown that NEDD4 is frequently overexpressed in multiple human cancers and primarily functions as an oncogene in various malignancies (36), which aligns with its upregulation in recurrent meningiomas.
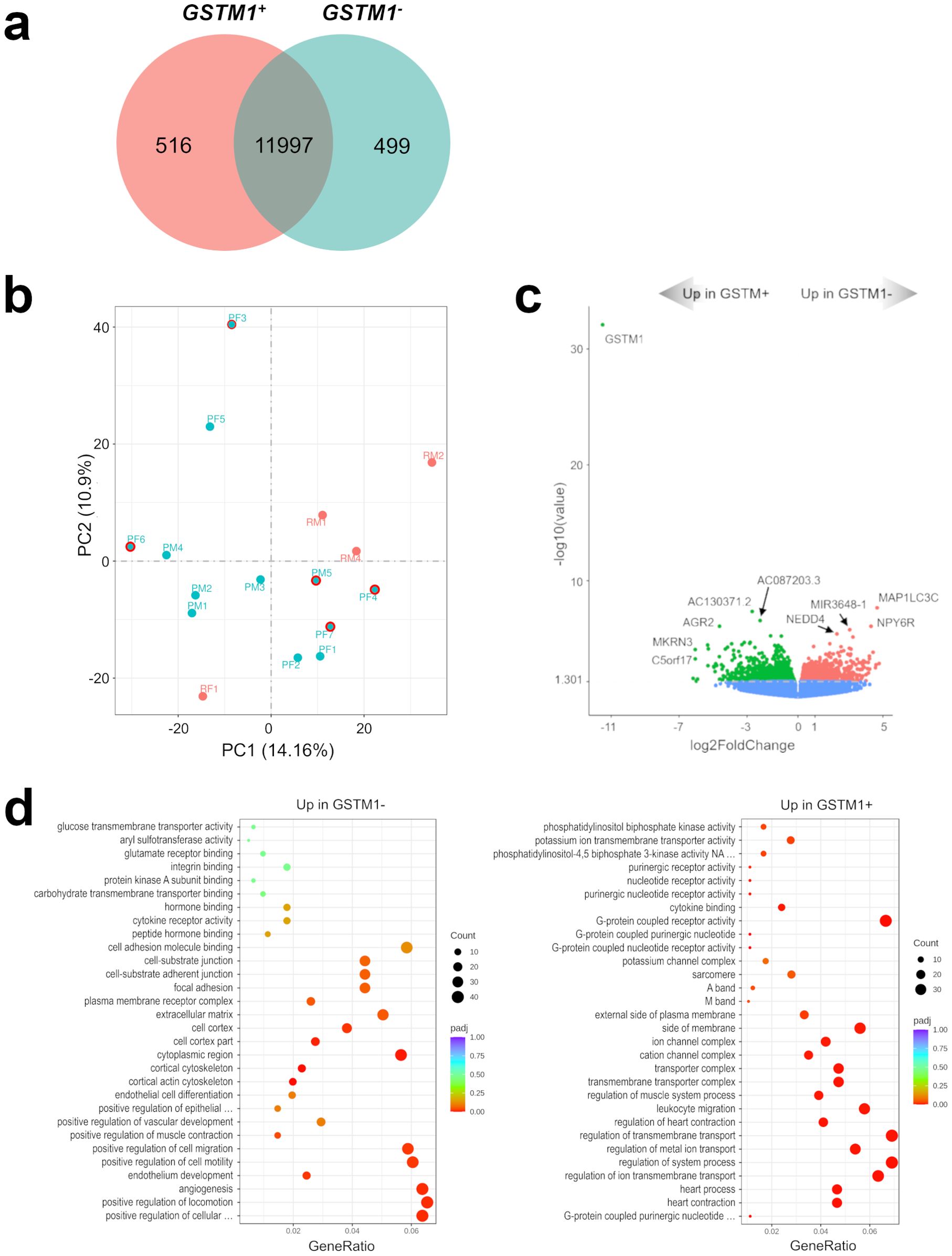
Figure 2. Characterization of the GSTM1- 1p-22q-NF2- meningioma transcriptome. (A) Venn diagram showing the overlap of DEGs in GSTM1+ and GSTM1- meningiomas by RNA-seq analysis. Common and tumor type-specific genes are depicted by numbers inside the corresponding circles. (B) PCA plot of RNA-seq analysis in GSTM1+ and GSTM1- meningiomas. As in Figure 1B, all recurrent meningiomas are indicated by red dots and all primary tumors are indicated by aquamarine dots. In addition, primary tumors without GSTM1 expression have a red outline. (C) Volcano plot displaying significant overexpressed and under expressed DEGs in GSTM1- meningiomas. (D) Gene-set enrichment analysis (GO) showing pathways upregulated and downregulated in GSTM1- meningiomas. Bubbles represent pathways and gene ratios are percentages of significant genes of all genes in a pathway.
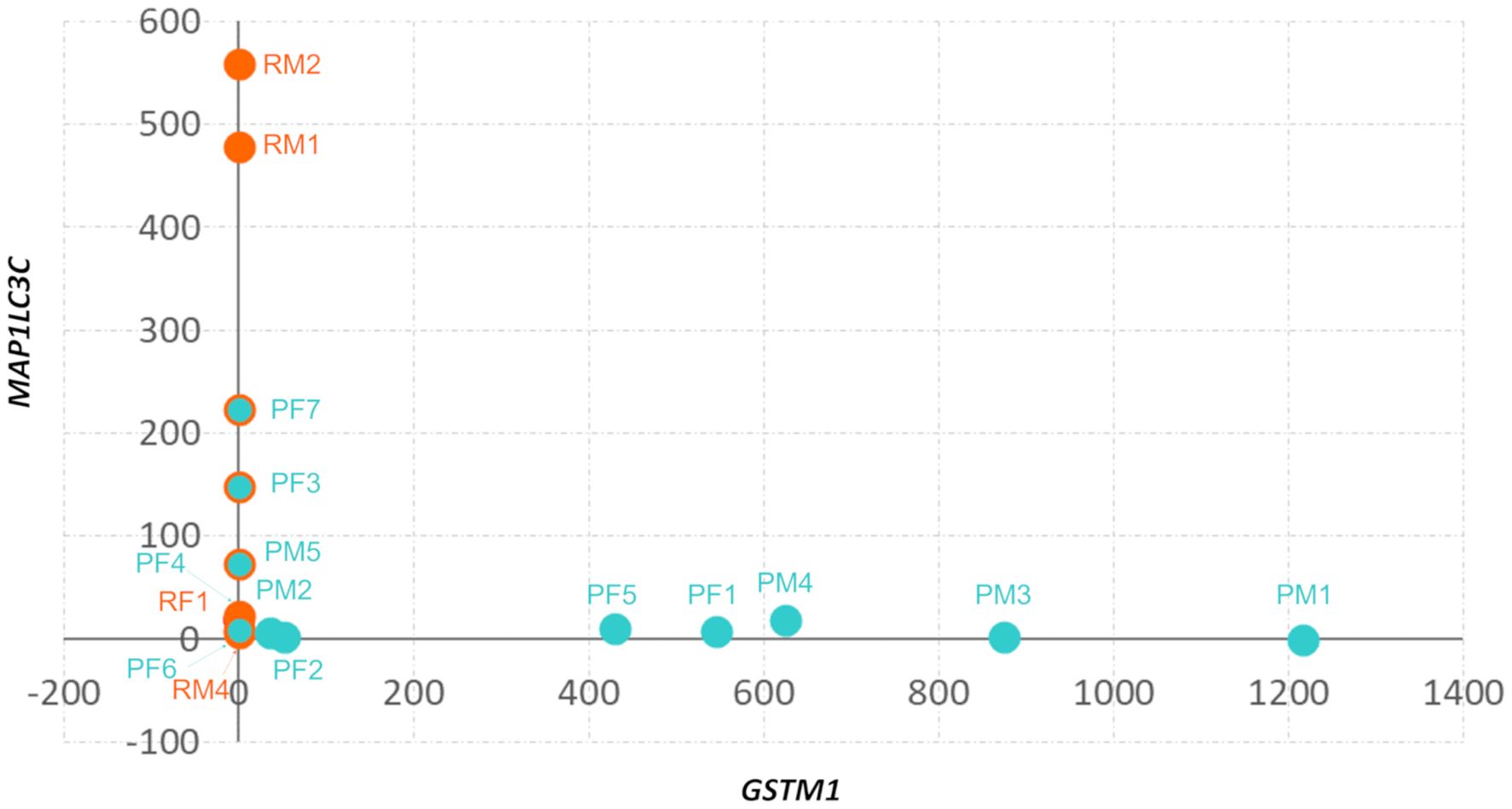
Figure 3. Divergent expression of GSTM1 and MAPLC3C in 1p-22q-NF2- meningiomas. Scatter plot of GSTM1 and MAP1LC3C expression in 1p-22q-NF2- meningiomas. Recurrent meningiomas are indicated by red dots, primary tumors are indicated by aquamarine dots, and primary tumors without GSTM1 expression have a red outline.
Gene set enrichment analyses revealed that pathways upregulated in GSTM1- meningiomas were related to cell-to-cell interaction and angiogenesis while downregulated pathways were related to transmembrane transport and intracellular signaling (Figure 2D; Supplementary Figure S4).
3.4 GSTM1 null genotype in meningiomas with GSTM1 downregulation
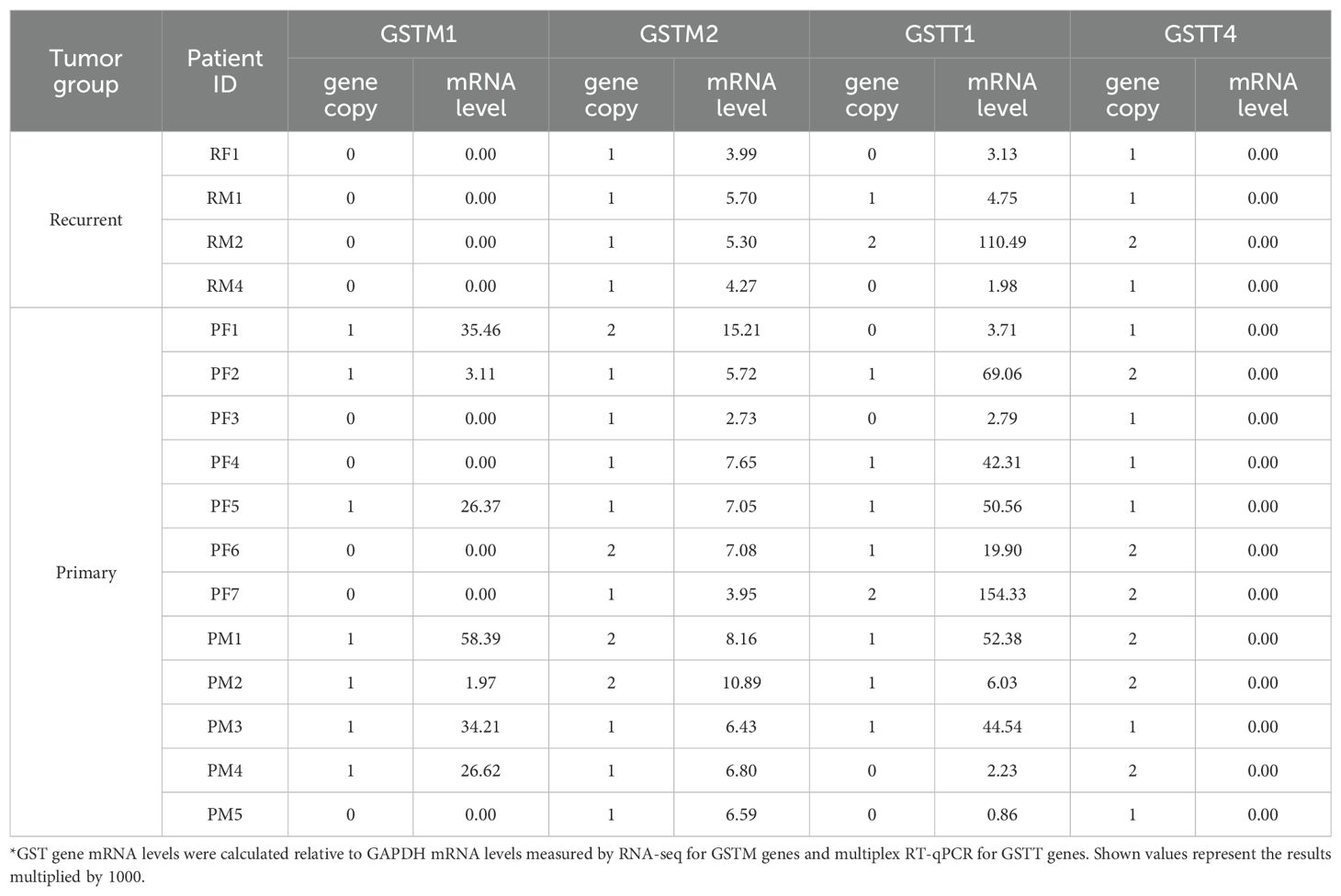
The expression of GSTM1 on chromosome 1p13 (Figure 4A) is downregulated in recurrent 1p-22q-NF2- meningiomas leading to a GSTM1 null genotype. To assess possible gene polymorphism as an explanation for differences in GSTM1 expression, we evaluated GSTM1 copy number variations (CNV) by multiplex quantitative polymerase chain reaction (qPCR). We also measured GSTM2 CNV due to its proximity on chr1p and lack of reported polymorphism (37). All meningiomas with GSTM1 under expression also showed copy number losses of GSTM1 (Table 2). Four meningiomas were GSTM2 homozygous at a copy number and none were recurrent tumors. GSTM2 homozygous tumors also displayed a two-fold increase in GSTM2 expression compared to tumors with one copy (Figure 4B, left), suggesting a proportional increase in expression to gene copy. Additionally, there was a significant positive correlation between GSTM1 and GSTM2 expression (Pearson r = 0.5021, 95% confidence interval = 0.0085 – 0.7999, p = 0.0475) (Figure 4B, right). This also aligns with the observation that GSTM1- meningiomas had significantly lower GSTM2 expression versus GSTM1+ meningiomas (Supplementary File 1).
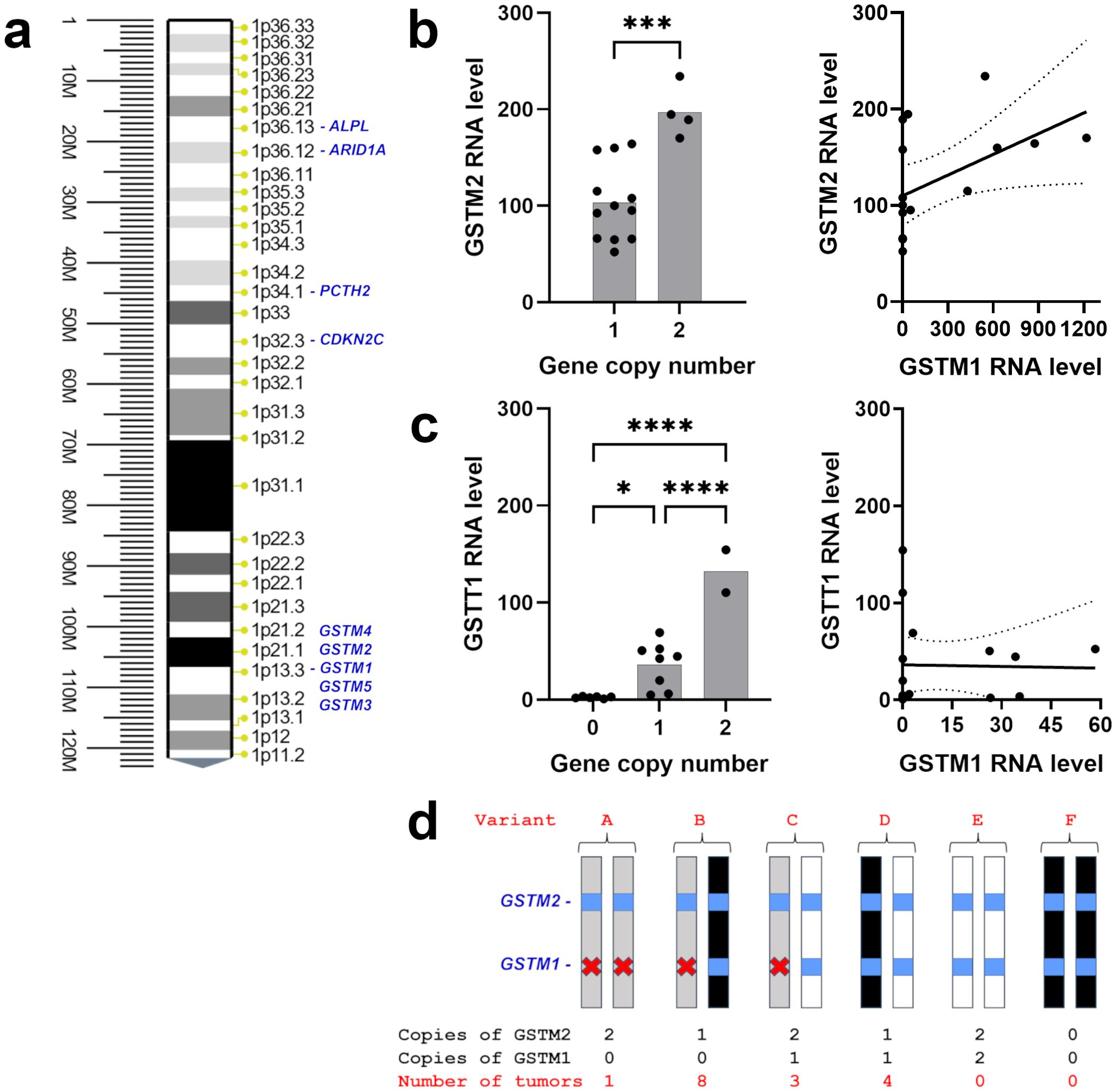
Figure 4. GST(x) gene expression and copy number in 1p-22q-NF2- meningiomas. (A) Chr1p position of GSTM1 and other related genes or genes of interest in meningioma. (B) (left) Scatter plot of GSTM2 expression level and copy number. (right) Correlation of GSTM2 expression with GSTM1 expression plotted with linear regression line (solid) and 95% confidence interval (dotted lines). r=0.5021, p=0.0475. (C) (left) Scatter plot of GSTT1 expression level and gene copy number. The y-axis displays relative quantity (RQ) multiplied by 1000. (right) Correlation of GSTT1 expression with GSTM1 expression plotted with linear regression line (solid) and 95% confidence interval (dotted lines). r=-0.0232, p=0.9321. (D) Schematic representation of GSTM1 and GSTM2 genotypes in 1p-22q-NF2- meningiomas and the number of tumors in each group. White bars depict GSTM1 present haplotype, black bars indicate somatic chr1p deletion in meningiomas, GSTM1 and GSTM2 genes are shown as blue squares, gray bars represent GSTM1 null haplotypes, and deletion of GSTM1 is indicated with a red x. *p<0.05; ***p<0.001; ****p<0.0001.
Recurrent GSTM1 null genotype meningiomas harbored either a) zero copies of GSTM1 and two copies of GSTM2, indicating that GSMT1 deletions in this patient were congenital (variant A), or b) were GSTM2 hemizygous (variant B), suggesting that the loss of one GSTM1 copy was inherited while another copy of the gene was lost in these tumors due to somatic deletion of chr1p (Figure 4D). Accordingly, recurrent meningiomas have inactivation of both copies of the gene either due to GSTM1 or 1p13 deletions. GSTM1 hemizygous non-recurrent meningiomas either had two GSTM2 alleles or were hemizygous for GSTM2, where loss of a GSTM1 copy was inherited (variant C) or lost due to somatic chr1p deletion (variant D). No meningiomas were homozygous for the wild type allele of GSTM1 (variant E) or had both GSTM1 and GSTM2 somatic deletions (variant F).
3.5 Meningioma recurrence is independent of GSTT1 expression
We also evaluated GSTT1 polymorphism in our cohort as a gene mapping to cytogenetic band 22q11.23 which is near NF2 (22q12.2) that is also frequently deleted in various cancers (31, 38). It is important to note that GSTT1 is not included in the primary human genome assembly GRCCh38.p14 but is found in NT_187633 Chromosome 22 Reference GRCh38.p14 ALT_REF_LOCI_1 assembly. GSTT1, mapping to 22q11.23, copy number was heterogenous in recurrent tumors with two tumors having no GSTT1, one having one copy, and one having two copies (Table 2) as well as being heterogenous in non-recurrent meningiomas. We next examined CNV of GSTT4, which maps only 30 kilobases apart from GSTT1 and does not display genetic polymorphism (37). Although none of 16 meningiomas displayed significant expression of GSTT4, nine tumors were GSTT4 hemizygous, while the rest were homozygous (Table 2). Taken together with the fact that GSTT4 is not usually deleted in humans, these results suggest that GSTT4 hemizygous meningiomas lost a copy of the gene together with somatic deletion of chr22. Accordingly, the loss of another allele in tumors with GSTT1 null genotype was inherited. GSTT1 expression levels were measured by reverse transcription (RT) qPCR using GAPDH gene as an internal control. The analysis revealed that GSTT1 expressed in a dose dependent manner, with the highest levels of expression in meningiomas with two copies of GSTT1 (Figure 4C, left). Further, there was no correlation between GSTM1 and GSTT1 expression (Pearson r = -0.0232, 95% confidence interval = -0.5130 – 0.4780, p = 0.9321) (Figure 4C, right). Overall, GSTT1 loss does not appear to be a biomarker of meningioma recurrence.
3.6 External validation of GSTM1 as a biomarker of recurrent meningioma
With the availability of public datasets in the gene expression omnibus (GEO), we evaluated meningioma expression studies to determine if we could find evidence of GSTM1 under expression in recurrent meningiomas. We identified five studies that clearly identified which tumors were from recurrent meningiomas. Lee et. al., 2010 (39), Clark et. al., 2013 (9), and Patel et. al., 2019 (13), each had expression data from 10-12 recurrent meningiomas and 58-145 primary tumors. Comparing expressions between the recurrent and primary tumors found no GSTM1 under expression (Geo2R default analysis, adjusted p=0.233-0.970). In the remaining studies, we found significant differences in GSTM1 expression between primary and recurrent tumors. For Schmidt et. al., 2016 (40), dataset, we were able to compare 31 recurrent and 23 primary tumors. Default Geo2R analysis found that there was a 98% decrease in GSTM1 expression (adjusted p=0.00365) in the recurrent tumors. Similarly, when we analyzed the Cimino et. al., 2019 (41), dataset consisting of 5 recurrent and 3 primary tumors, we found a 3.4-fold decrease in GSTM1 expression (adjusted p=0.00716) in the recurrent tumors. The main limitation of the public GEO datasets is that we could not determine which of the recurrent meningiomas were 1p-22q-NF2-. None the less, GSTM1 under expression is found in recurrent meningiomas from independent datasets, externally validating GSTM1- as a biomarker for 1p-22q-NF2-meningioma recurrence.
4 Discussion
Meningiomas are the most common primary brain tumors, but we are currently limited in our ability to prevent recurrence or to identify tumors that may recur due to the lack of robust molecular biomarkers of recurrence. This work extends beyond classification and subclassification of primary meningiomas to characterize a distinct transcriptomic biomarker that is enriched in recurrent 1p-22q-NF2- meningiomas. These meningiomas show the highest rate of recurrence using existing molecular classification but are still heterogenous with a substantial subset that do not recur. Our results demonstrate that 1p-22q-NF2- meningiomas without expression of GSTM1 comprise the recurrent 1p-22q-NF2- meningiomas compared to those that express GSTM1, thereby addressing the heterogeneity in this existing molecular classification scheme.
These observations are in agreement with numerous previous studies, emphasizing GSTM1 null genotype as a factor modulating the risk of developing cancer (42). For example, the GSTM1 null genotype was associated with an increased risk of cervical cancer in Indian and Chinese populations (43). Moreover, a study comprising of 2500 individuals demonstrated that environmental risk factors significantly increase susceptibility to head and neck cancer in GSTM1 null individuals (44). Importantly, colorectal cancer patients undergoing chemotherapy had higher survival rates even with only one copy of GSTM1 (45). Taken together, these results imply that there is an association between cancer progression and GSTM1 gene copy number, which is in general agreement with our observation that the expression of other genes from GST family (GSTM2 and GSTT1) is also copy number dependent (Figure 4). Interestingly, there is a report that GSTM2 expression may increase to compensate for the loss of GSTM1 (46). However, we found no evidence for that mechanism in this study since GSTM2 mRNA was positively correlated with GSTM1 mRNA (Figure 4B, right, Table 2). Thus, loss of GSTM1’s ability to detoxify electrophiles generated by xenobiotic-induced reactive oxygen species, along with the lack of compensation by other GST genes, increases cancer susceptibility and prompts further malignization of tumors, including meningiomas.
The association of GSTM1 and GSTT1 genetic polymorphism with the development of meningiomas has been previously examined. Whereas the GSTT1 null genotype was significantly associated with the risk of meningioma, no significant effect of GSTM1 genotypes on susceptibility was identified (47, 48). These results contradict our current findings of all recurrent meningiomas in our cohort displaying a GSTM1 null genotype, with no difference in GSTT1 genotypes in our study. Discrepancy with our results may be explained by our study focusing on the differences between recurrent and primary 1p-22q-NF2- meningiomas. Another difference between the studies was that the majority of meningiomas in our group with GSTM1 null genotype had only one copy of the gene lost due to inherited deletion of the gene, while another copy may have been lost due to somatic chr1p deletion. Whether a somatic chr1p deletion contained present or null allele remains to be determined by further investigations, which will require the analysis of germline GSTM1 copy numbers and should include a larger cohort of patients. Future experiments are needed to examine the existence of a significant relationship between GSTM1 copy numbers and meningioma development.
The comparison of recurrent and primary meningiomas transcriptomes in a gene-set enrichment analysis revealed that recurrent meningiomas were characterized by overexpression of gene sets involved in cell structure and signaling and under expression of developmental pathways. Additionally, a gene-set enrichment analysis of GSTM1 null meningiomas versus GSTM1 expressing meningiomas identified cell-to-cell interaction and angiogenesis pathways upregulated as well as transmembrane transport and intracellular signaling pathways downregulated in GSTM1- meningiomas. The most significant DEG in GSTM1 null meningiomas was upregulation of MAP1LC3C (Figure 2), which is located on chr1q43, and is required for the formation of autophagosomal membranes for autophagy-related processes and cell homeostasis (49), but is not routinely deleted in meningiomas (18). MAP1LC3C is present in all vertebrates except rodents, making the generation of MAP1LC3C-deficient mice impossible. MAP1LC3C has been shown to play a tumor-suppressing role in breast cancer (50) and renal clear cell carcinoma (51) development. Future experiments may offer a better understanding of the molecular mechanism of MAP1LC3C function in tumor cells and test whether it plays a positive role in meningioma development.
Another DEG upregulated in GSTM1- meningiomas compared to GSTM1+ tumors is NEDD4 (Figure 2), which encodes a member of HECT family of E3 ubiquitin ligases. NEDD4 is an evolutionarily conserved protein highly expressed in the early embryonic brain (52) where it promotes the development of neural dendrites (53), and results in neonatal lethality with targeted deletion (54), suggesting that NEDD4 is involved in neurodevelopment including meninges growth and organization. Additionally, NEDD4 is a proto-oncogene that downregulates PTEN expression, thereby impacting the PI3K/AKT/mTOR pathway that is implicated in multiple types of cancers (55). It will be important for future studies to further evaluate these additional pathways and DEGs to identify potential additional biomarkers of meningioma progression beyond GSTM1 as well as potential novel therapeutic targets.
In addition to detoxification, GSTM1 has been shown to modulate inflammation though NF-kB, GM-CSF (granulocyte–macrophage colony stimulating factor) and CCL2 (chemokine [C–C motif] ligands 2) signaling (56, 57). GSTM1 can also activate STAT3 signaling, a key modulator of multiple cellular processes including proliferation, apoptosis, and metastasis, which is altered in multiple types of cancers (56, 58, 59). The family of GSTs have also been demonstrated to affect signal transduction mechanisms involved in cell proliferation and apoptosis though modulation of c-Jun-N-terminal kinases (JNKs) and apoptosis signal-regulating kinase (ASK1) (57). Loss of NF2 leads to disruption of Merlin, a key tumor suppressor that targets multiple signaling pathways such as mTORC1 (cell growth and metabolism in response to environmental factors), Ras/Rac/PAK (cellular proliferation, differentiation, and transformation), and CRL4-DCAF (DNA damage response and mitotic exit) (60, 61). Thus, we can begin to build synergistic models between GSTM1 and NF2 signaling. Loss of GSTM1 leads to accumulation of intracellular toxins, which could be enhanced through dysregulated mTOC1 signaling. The toxins could then lead to DNA damage, worsened by disruption of CRL4, which allows the cell to escape apoptosis via STAT3, JNKs, and/or ASK1 signaling. Further, proliferation signals from STAT3 and the Ras/Rac/PAK pathways would be pro-oncogenic. While this model is partially supported by the increases in MAP1LC3C and NEDD4 expression in the GSTM1- meningiomas, future studies will be needed to investigate the various components of this model at both the mRNA and protein level.
The results of our study extend beyond understanding tumor development for the novel purpose of identifying a biomarker of tumor recurrence, which is crucial for informing clinical treatment decisions. A biomarker of meningioma recurrence has the potential to shape personalized medicine approaches in the future by allowing clinicians to match the extent of initial meningioma treatment to the risk for later recurrence based on the presence or absence of a GSTM1 null genotype. One postulated explanation for the relationship between GSTM1 under expression and meningioma recurrence is that GSTM1 function is necessary to remove substances secreted by meningiomas that stimulate their growth and without this host factor present meningioma growth is relatively less controlled. This work is limited to transcriptomic analysis and further validation is required at the protein expression level. Another likely limitation of this pilot study is that the examination of meningioma recurrence was limited to 1p-22q-NF2- tumors and did not include analysis of other meningioma subtypes, including 14q-22q- group tumors. Since measuring GSTM1 expression in additional tumor subtypes will allow us to determine if it is a common biomarker for recurrent meningiomas, we plan to analyze 14q-22q- and other subtypes of meningiomas in our future follow-up studies. Follow-up studies are required to evaluate GSTM1 as a molecular biomarker of recurrence prospectively and to assess its impact on treatment decisions and patient outcomes.
Data availability statement
The original contributions presented in the study are publicly available. This data can be found here: GSE283616. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE283616.
Ethics statement
The studies involving humans were approved by University of Oklahoma Health Sciences Center Institutional Review Board Protocol #10195. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
Author contributions
AJ: Conceptualization, Investigation, Writing – original draft, Writing – review & editing. ET: Conceptualization, Investigation, Methodology, Writing – original draft, Writing – review & editing. KP: Investigation, Methodology, Writing – review & editing. JZ: Writing – review & editing. AB: Investigation, Resources, Writing – review & editing. CG: Investigation, Resources, Writing – review & editing. SH: Investigation, Resources, Writing – review & editing. TS: Investigation, Resources, Writing – review & editing. YL: Investigation, Methodology, Writing – review & editing. GD: Investigation, Resources, Writing – review & editing. AT: Investigation, Methodology, Resources, Writing – review & editing. PJ: Investigation, Resources, Writing – review & editing. ID: Conceptualization, Resources, Supervision, Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Acknowledgments
The authors thank Jo Elle G. Peterson and Kar-Ming Fung, board-certified neuropathologists within the Department of Pathology at OU Health for providing pathology reports of resected meningiomas.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2024.1506708/full#supplementary-material
Supplementary Figure 1 | Hierarchical clustering of DEGs in groups between recurrent (orange planks) and primary (pink planks) meningioma samples. Columns corresponding to each meningioma are indicated at the bottom. Green and red colors indicate low and high relative mRNA expression levels, respectively.
Supplementary Figure 2 | Heat map of Pearson correlation between all tested meningioma samples. The change of the square (R2) value of the correlation coefficient of Pearson is indicated by the change of the blue color. Each grid in the figure represents the correlation between two samples; different colors represent correlation between samples. The deeper color indicates a bigger R2 value and a higher correlation between samples.
Supplementary Figure 3 | Bubble plot of Reactome (A) and KEGG (B) enrichment analysis of signaling pathways upregulated in recurrent (left panel) or primary (right panel) meningiomas. Each bubble represents a pathway. Gene ratio (x-axis) is the percentage of significant genes over the total genes in each pathway.
Supplementary Figure 4 | Bubble plot of Reactome (A) and KEGG (B) enrichment analysis of signaling pathways upregulated in recurrent (left panel) or primary (right panel) GSTM1- meningiomas. Each bubble represents a pathway. Gene ratio (x-axis) is the percentage of significant genes over the total genes in each pathway.
Supplementary File 1 | Recurrent meningioma vs. Primary meningioma differently expressed genes (DEG).
Supplementary File 2 | GSTM+ meningioma vs. GSTM- meningioma differently expressed genes (DEG).
References
1. Low JT, Ostrom QT, Cioffi G, Neff C, Waite KA, Kruchko C, et al. Primary brain and other central nervous system tumors in the United States (2014-2018): A summary of the cbtrus statistical report for clinicians. Neurooncol Pract. (2022) 9:165–82. doi: 10.1093/nop/npac015
2. Louis DN, Perry A, Wesseling P, Brat DJ, Cree IA, Figarella-Branger D, et al. The 2021 who classification of tumors of the central nervous system: A summary. Neuro Oncol. (2021) 23:1231–51. doi: 10.1093/neuonc/noab106
3. Gutmann DH, Giordano MJ, Fishback AS, Guha A. Loss of merlin expression in sporadic meningiomas, ependymomas and schwannomas. Neurology. (1997) 49:267–70. doi: 10.1212/wnl.49.1.267
4. Ruttledge MH, Sarrazin J, Rangaratnam S, Phelan CM, Twist E, Merel P, et al. Evidence for the complete inactivation of the nf2 gene in the majority of sporadic meningiomas. Nat Genet. (1994) 6:180–4. doi: 10.1038/ng0294-180
5. Rouleau GA, Merel P, Lutchman M, Sanson M, Zucman J, Marineau C, et al. Alteration in a new gene encoding a putative membrane-organizing protein causes neuro-fibromatosis type 2. Nature. (1993) 363:515–21. doi: 10.1038/363515a0
6. Petrilli AM, Fernández-Valle C. Role of Merlin/NF2 inactivation in tumor biology. Oncogene. (2016) 35:537–48. doi: 10.1038/onc.2015.125
7. Lindblom A, Ruttledge M, Collins VP, Nordenskjold M, Dumanski JP. Chromosomal deletions in anaplastic meningiomas suggest multiple regions outside chromosome 22 as important in tumor progression. Int J Cancer. (1994) 56:354–7. doi: 10.1002/ijc.2910560310
8. Simon M, von Deimling A, Larson JJ, Wellenreuther R, Kaskel P, Waha A, et al. Allelic losses on chromosomes 14, 10, and 1 in atypical and Malignant meningiomas: A genetic model of meningioma progression. Cancer Res. (1995) 55:4696–701.
9. Clark VE, Erson-Omay EZ, Serin A, Yin J, Cotney J, Ozduman K, et al. Genomic analysis of non-NF2 meningiomas reveals mutations in TRAF7, KLF4, AKT1, and SMO. Science. (2013) 339:1077–80. doi: 10.1126/science.1233009
10. Clark VE, Harmanci AS, Bai H, Youngblood MW, Lee TI, Baranoski JF, et al. Recurrent somatic mutations in POLR2A define a distinct subset of meningiomas. Nat Genet. (2016) 48:1253–9. doi: 10.1038/ng.3651
11. Choudhury A, Magill ST, Eaton CD, Prager BC, Chen WC, Cady MA, et al. Meningioma DNA methylation groups identify biological drivers and therapeutic vulnerabilities. Nat Genet. (2022) 54:649–59. doi: 10.1038/s41588-022-01061-8
12. Choudhury A, Chen WC, Lucas CG, Bayley JC, Harmanci AS, Maas SLN, et al. Hypermitotic meningiomas harbor DNA methylation subgroups with distinct biological and clinical features. Neuro Oncol. (2023) 25:520–30. doi: 10.1093/neuonc/noac224
13. Patel AJ, Wan YW, Al-Ouran R, Revelli JP, Cardenas MF, Oneissi M, et al. Molecular profiling predicts meningioma recurrence and reveals loss of dream complex repression in aggressive tumors. Proc Natl Acad Sci United States America. (2019) 116:21715–26. doi: 10.1073/pnas.1912858116
14. Bayley JC, Hadley CC, Harmanci AO, Harmanci AS, Klisch TJ, Patel AJ. Multiple approaches converge on three biological subtypes of meningioma and extract new insights from published studies. Sci Adv. (2022) 8:eabm6247. doi: 10.1126/sciadv.abm6247
15. Sulman EP, Dumanski JP, White PS, Zhao H, Maris JM, Mathiesen T, et al. Identification of a consistent region of allelic loss on 1p32 in meningiomas: correlation with increased morbidity. Cancer Res. (1998) 58:3226–30.
16. Leone PE, Bello MJ, de Campos JM, Vaquero J, Sarasa JL, Pestana A, et al. NF2 gene mutations and allelic status of 1p, 14q and 22q in sporadic meningiomas. Oncogene. (1999) 18:2231–9. doi: 10.1038/sj.onc.1202531
17. Bostrom J, Muhlbauer A, Reifenberger G. Deletion mapping of the short arm of chromosome 1 identifies a common region of deletion distal to D1s496 in human meningiomas. Acta Neuropathol. (1997) 94:479–85. doi: 10.1007/s004010050736
18. Bello MJ, de Campos JM, Vaquero J, Kusak ME, Sarasa JL, Rey JA. High-resolution analysis of chromosome arm 1p alterations in meningioma. Cancer Genet Cytogenet. (2000) 120:30–6. doi: 10.1016/s0165-4608(99)00249-6
19. Sulman EP, White PS, Brodeur GM. Genomic annotation of the meningioma tumor suppressor locus on chromosome 1p34. Oncogene. (2004) 23:1014–20. doi: 10.1038/sj.onc.1206623
20. Hansson CM, Buckley PG, Grigelioniene G, Piotrowski A, Hellstrom AR, Mantripragada K, et al. Comprehensive genetic and epigenetic analysis of sporadic meningioma for macro-mutations on 22q and micro-mutations within the NF2 locus. BMC Genomics. (2007) 8:16. doi: 10.1186/1471-2164-8-16
21. Tsitsikov EN, Hameed S, Tavakol SA, Stephens TM, Tsytsykova AV, Garman L, et al. Specific gene expression signatures of low grade meningiomas. Front Oncol. (2023) 13:1126550. doi: 10.3389/fonc.2023.1126550
22. Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol. (1994) 2:28–36.
23. Abedalthagafi MS, Bi WL, Merrill PH, Gibson WJ, Rose MF, Du Z, et al. Arid1a and tert promoter mutations in dedifferentiated meningioma. Cancer Genet. (2015) 208:345–50. doi: 10.1016/j.cancergen.2015.03.005
24. Muller P, Henn W, Niedermayer I, Ketter R, Feiden W, Steudel WI, et al. Deletion of chromosome 1p and loss of expression of alkaline phosphatase indicate progression of meningiomas. Clin Cancer Res. (1999) 5:3569–77.
25. Niedermayer I, Feiden W, Henn W, Steilen-Gimbel H, Steudel WI, Zang KD. Loss of alkaline phosphatase activity in meningiomas: A rapid histochemical technique indicating progression-associated deletion of a putative tumor suppressor gene on the distal part of the short arm of chromosome 1. J Neuropathol Exp Neurol. (1997) 56:879–86. doi: 10.1097/00005072-199708000-00006
26. Bostrom J, Meyer-Puttlitz B, Wolter M, Blaschke B, Weber RG, Lichter P, et al. Alterations of the tumor suppressor genes CDKN2A (P16(INK4A)), P14(ARF), CDKN2B (P15(INK4B)), and CDKN2C (P18(INK4C)) in atypical and anaplastic meningiomas. Am J Pathol. (2001) 159:661–9. doi: 10.1016/S0002-9440(10)61737-3
27. Nassiri F, Liu J, Patil V, Mamatjan Y, Wang JZ, Hugh-White R, et al. A clinically applicable integrative molecular classification of meningiomas. Nature. (2021) 597:119–25. doi: 10.1038/s41586-021-03850-3
28. Thirimanne HN, Almiron-Bonnin D, Nuechterlein N, Arora S, Jensen M, Parada CA, et al. Meningioma transcriptomic landscape demonstrates novel subtypes with regional associated biology and patient outcome. Cell Genom. (2024) 4:100566. doi: 10.1016/j.xgen.2024.100566
29. Chen WC, Choudhury A, Youngblood MW, Polley MC, Lucas CG, Mirchia K, et al. Targeted gene expression profiling predicts meningioma outcomes and radiotherapy responses. Nat Med. (2023) 29:3067–76. doi: 10.1038/s41591-023-02586-z
30. Oakley A. Glutathione transferases: A structural perspective. Drug Metab Rev. (2011) 43:138–51. doi: 10.3109/03602532.2011.558093
31. Nakanishi G, Bertagnolli LS, Pita-Oliveira M, Scudeler MM, Torres-Loureiro S, Almeida-Dantas T, et al. Gstm1 and gstt1 polymorphisms in healthy volunteers - a worldwide systematic review. Drug Metab Rev. (2022) 54:37–45. doi: 10.1080/03602532.2022.2036996
32. Allocati N, Masulli M, Di Ilio C, Federici L. Glutathione transferases: substrates, inihibitors and pro-drugs in cancer and neurodegenerative diseases. Oncogenesis. (2018) 7:8. doi: 10.1038/s41389-017-0025-3
33. Qin Z, Ren F, Xu X, Ren Y, Li H, Wang Y, et al. Znf536, a novel zinc finger protein specifically expressed in the brain, negatively regulates neuron differentiation by repressing retinoic acid-induced gene transcription. Mol Cell Biol. (2009) 29:3633–43. doi: 10.1128/MCB.00362-09
34. Jach D, Cheng Y, Prica F, Dumartin L, Crnogorac-Jurcevic T. From development to cancer - an ever-increasing role of AGR2. Am J Cancer Res. (2021) 11:5249–62.
35. Yulyaningsih E, Loh K, Lin S, Lau J, Zhang L, Shi Y, et al. Pancreatic polypeptide controls energy homeostasis via Npy6r signaling in the suprachiasmatic nucleus in mice. Cell Metab. (2014) 19:58–72. doi: 10.1016/j.cmet.2013.11.019
36. Tian X, Chen Y, Peng Z, Lin Q, Sun A. Nedd4 E3 ubiquitin ligases: promising biomarkers and therapeutic targets for cancer. Biochem Pharmacol. (2023) 214:115641. doi: 10.1016/j.bcp.2023.115641
37. Hayes JD, Strange RC. Glutathione S-transferase polymorphisms and their biological consequences. Pharmacology. (2000) 61:154–66. doi: 10.1159/000028396
38. Buchard A, Sanchez JJ, Dalhoff K, Morling N. Multiplex PCR detection of GSTM1, GSTT1, and GSTP1 gene variants: simultaneously detecting GSTM1 and GSTT1 gene copy number and the allelic status of the GSTP1 Ile105Val genetic variant. J Mol Diagn. (2007) 9:612–7. doi: 10.2353/jmoldx.2007.070030
39. Lee Y, Liu J, Patel S, Cloughesy T, Lai A, Farooqi H, et al. Genomic landscape of meningiomas. Brain Pathol. (2010) 20:751–62. doi: 10.1111/j.1750-3639.2009.00356.x
40. Schmidt M, Mock A, Jungk C, Sahm F, Ull AT, Warta R, et al. Transcriptomic analysis of aggressive meningiomas identifies PTTG1 and LEPR as prognostic biomarkers independent of who grade. Oncotarget. (2016) 7:14551–68. doi: 10.18632/oncotarget.7396
41. Cimino PJ, Yoda RA, Wirsching HG, Warrick JI, Dorschner MO, Ferreira M. Genomic profiling of anaplastic meningioma identifies recurrent genetic alterations with relevance to lower-grade meningioma. Neuropathol Appl Neurobiol. (2019) 45:179–82. doi: 10.1111/nan.12487
42. Di Pietro G, Magno LA, Rios-Santos F. Glutathione S-transferases: an overview in cancer research. Expert Opin Drug Metab Toxicol. (2010) 6:153–70. doi: 10.1517/17425250903427980
43. Ye J, Mu YY, Wang J, He XF. Individual effects of GSTM1 and GSTT1 polymorphisms on cervical or ovarian cancer risk: an updated meta-analysis. Front Genet. (2022) 13:1074570. doi: 10.3389/fgene.2022.1074570
44. Katiyar T, Yadav V, Maurya SS, Ruwali M, Singh M, Hasan F, et al. Interaction of glutathione-S-transferase genotypes with environmental risk factors in determining susceptibility to head and neck cancer and treatment response and survival outcome. Environ Mol Mutagen. (2020) 61:574–84. doi: 10.1002/em.22362
45. Funke S, Timofeeva M, Risch A, Hoffmeister M, Stegmaier C, Seiler CM, et al. Genetic polymorphisms in gst genes and survival of colorectal cancer patients treated with chemotherapy. Pharmacogenomics. (2010) 11:33–41. doi: 10.2217/pgs.09.132
46. Bhattacharjee P, Paul S, Banerjee M, Patra D, Banerjee P, Ghoshal N, et al. Functional compensation of glutathione S-transferase M1 (Gstm1) null by another GST superfamily member, GSTM2. Sci Rep. (2013) 3:2704. doi: 10.1038/srep02704
47. De Roos AJ, Rothman N, Inskip PD, Linet MS, Shapiro WR, Selker RG, et al. Genetic polymorphisms in GSTM1, -P1, -T1, and CYP2E1 and the risk of adult brain tumors. Cancer Epidemiol Biomarkers Prev. (2003) 12:14–22.
48. Elexpuru-Camiruaga J, Buxton N, Kandula V, Dias PS, Campbell D, McIntosh J, et al. Susceptibility to astrocytoma and meningioma: influence of allelism at glutathione S-transferase (GSTT1 and GSTM1) and cytochrome P-450 (CYP2D6) loci. Cancer Res. (1995) 55:4237–9.
49. von Muhlinen N, Akutsu M, Ravenhill BJ, Foeglein A, Bloor S, Rutherford TJ, et al. LC3C, bound selectively by a noncanonical LIR motif in NDP52, is required for antibacterial autophagy. Mol Cell. (2012) 48:329–42. doi: 10.1016/j.molcel.2012.08.024
50. Bell ES, Coelho PP, Ratcliffe CDH, Rajadurai CV, Peschard P, Vaillancourt R, et al. LC3C-mediated autophagy selectively regulates the met RTK and HGF-stimulated migration and invasion. Cell Rep. (2019) 29:4053–68.e6. doi: 10.1016/j.celrep.2019.11.063
51. Mikhaylova O, Stratton Y, Hall D, Kellner E, Ehmer B, Drew AF, et al. VHL-regulated MiR-204 suppresses tumor growth through inhibition of LC3B-mediated autophagy in renal clear cell carcinoma. Cancer Cell. (2012) 21:532–46. doi: 10.1016/j.ccr.2012.02.019
52. Kumar S, Harvey KF, Kinoshita M, Copeland NG, Noda M, Jenkins NA. cDNA cloning, expression analysis, and mapping of the mouse Nedd4 gene. Genomics. (1997) 40:435–43. doi: 10.1006/geno.1996.4582
53. Staub O, Gautschi I, Ishikawa T, Breitschopf K, Ciechanover A, Schild L, et al. Regulation of stability and function of the epithelial Na+ Channel (ENaC) by ubiquitination. EMBO J. (1997) 16:6325–36. doi: 10.1093/emboj/16.21.6325
54. Cao XR, Lill NL, Boase N, Shi PP, Croucher DR, Shan H, et al. Nedd4 controls animal growth by regulating IGF-1 signaling. Sci Signal. (2008) 1:ra5. doi: 10.1126/scisignal.1160940
55. Chen C, Matesic LE. The Nedd4-like family of E3 ubiquitin ligases and cancer. Cancer Metastasis Rev. (2007) 26:587–604. doi: 10.1007/s10555-007-9091-x
56. Huang YC, Lai JC, Peng PH, Wei KC, Wu KJ. Chromatin accessibility analysis identifies GSTM1 as a prognostic marker in human glioblastoma patients. Clin Epigenet. (2021) 13:201. doi: 10.1186/s13148-021-01181-8
57. Kano SI, Choi EY, Dohi E, Agarwal S, Chang DJ, Wilson AM, et al. Glutathione S-transferases promote proinflammatory astrocyte-microglia communication during brain inflammation. Sci Signal. (2019) 12(569):eaar2124. doi: 10.1126/scisignal.aar2124
58. Pham MH, Zada G, Mosich GM, Chen TC, Giannotta SL, Wang K, et al. Molecular genetics of meningiomas: A systematic review of the current literature and potential basis for future treatment paradigms. Neurosurg Focus. (2011) 30:E7. doi: 10.3171/2011.2.FOCUS1117
59. Shi D, Tao J, Man S, Zhang N, Ma L, Guo L, et al. Structure, function, signaling pathways and clinical therapeutics: the translational potential of stat3 as a target for cancer therapy. Biochim Biophys Acta Rev Cancer. (2024) 1879:189207. doi: 10.1016/j.bbcan.2024.189207
60. Look A, Lonser RR. Inherited genetic syndromes and meningiomas. Handb Clin Neurol. (2020) 169:121–9. doi: 10.1016/B978-0-12-804280-9.00007-X
Keywords: meningioma, recurrent, CNS tumors, transcriptome profiling, gene expression, NF2, GSTM1
Citation: Johnson AC, Tsitsikov EN, Phan KP, Zuccato JA, Bauer AM, Graffeo CS, Hameed S, Stephens TM, Liu Y, Dunn GP, Tsytsykova AV, Jones PS and Dunn IF (2024) GSTM1 null genotype underpins recurrence of NF2 meningiomas. Front. Oncol. 14:1506708. doi: 10.3389/fonc.2024.1506708
Received: 06 October 2024; Accepted: 25 November 2024;
Published: 12 December 2024.
Edited by:
David D. Tran, University of Southern California, United StatesReviewed by:
Kristin Huntoon, University of Arizona, United StatesSoma Sengupta, University of North Carolina at Chapel Hill, United States
Akash Patel, Baylor College of Medicine, United States
Copyright © 2024 Johnson, Tsitsikov, Phan, Zuccato, Bauer, Graffeo, Hameed, Stephens, Liu, Dunn, Tsytsykova, Jones and Dunn. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ian F. Dunn, aWFuLWR1bm5Ab3Voc2MuZWR1
 Anthony C. Johnson
Anthony C. Johnson Erdyni N. Tsitsikov3
Erdyni N. Tsitsikov3 Jeffrey A. Zuccato
Jeffrey A. Zuccato Andrew M. Bauer
Andrew M. Bauer Sanaa Hameed
Sanaa Hameed Tressie M. Stephens
Tressie M. Stephens Yufeng Liu
Yufeng Liu Ian F. Dunn
Ian F. Dunn