- 1Molecular Pharmacology and Therapeutics Graduate Program, University of Minnesota School of Medicine, Minneapolis, MN, United States
- 2Department of Obstetrics, Gynecology and Women’s Health, University of Minnesota School of Medicine, Minneapolis, MN, United States
- 3Department of Laboratory Medicine and Pathology, University of Minnesota School of Medicine, Minneapolis, MN, United States
Genomic analysis has played a significant role in the identification of driver mutations that are linked to disease progression and response to drug treatment in ovarian cancer. A prominent example is the stratification of epithelial ovarian cancer (EOC) patients with homologous recombination deficiency (HRD) characterized by mutations in DNA damage repair genes such as BRCA1/2 for treatment with PARP inhibitors. However, recent studies have shown that some epithelial ovarian tumors respond to PARP inhibitors irrespective of their HRD or BRCA mutation status. An exclusive focus on the genome overlooks the significant insight that can be gained from other biological analytes, including proteins, which carry out cellular functions. Proteogenomics is the integration of genomics, transcriptomics, epigenomics and proteomics data. This review paper provides novel insight into the role of proteogenomics as an analytical approach to identify predictive biomarkers of drug treatment response in epithelial ovarian cancer. Proteogenomic analysis can facilitate the identification of predictive biomarkers of drug treatment response, consequently greatly improving the stratification of patients with EOC for treatment towards a goal of personalized medicine.
1 Ovarian cancer pathology
Ovarian cancer is the second deadliest gynecological cancer globally with a 46% 5-year survival rate (1, 2, 5–7). In 2024, there will be an estimated 19,680 new cases of ovarian cancer and 12,740 ovarian cancer-related deaths in the United States (3). More than 70% of epithelial ovarian cancers (EOCs) are of the high grade serous (HGSOC) subtype (4).
The majority of HGSOC cases are diagnosed at an advanced stage due to the absence of specific symptoms and the lack of effective screening tests (5–7). Pelvic examination, positron-emission tomography, transvaginal ultrasound, magnetic resonance imaging (MRI), laparoscopy, and CA125 and HE4 protein biomarker levels measured using enzyme linked immunosorbent assay (ELISA) are utilized as screening tests for EOC (8–10). However, the sensitivity and specificity of these diagnostic tests do not reach the targets of >75% and 99.7%, respectively to achieve a positive predictive value (PPV) of 10% (11).
2 Current treatment modalities
The standard treatment for EOC is cytoreductive (debulking) surgery followed by a combination of platinum (cisplatin and carboplatin) and taxane (paclitaxel or docetaxel) based chemotherapy (3, 12–15). Most HGSOCs exhibit an initial high chemosensitivity; however, ~75% of patients relapse within 5 years after first-line treatment, resulting in a low 5-year survival rate of <50% (13, 15, 16). Other EOC subtypes, such as mucinous and clear cell, are highly resistant to chemotherapy. This increases the need for novel treatments with enhanced efficacy, such as targeted therapy and immunotherapy. Three classes of targeted therapies are currently used for the treatment of EOCs: anti-angiogenic agents, poly (ADP-ribose) polymerase inhibitors (PARPi) and antibody drug conjugates.
2.1 Targeted therapy: anti-angiogenesis drugs
In June 2018, the humanized monoclonal antibody against vascular endothelial growth factor (VEGF), bevacizumab (Avastin), became the first US FDA-approved antiangiogenic drug for the treatment of stage III and IV EOC after surgery (17–19). Bevacizumab binds to circulating VEGF-A to competitively prevent it from binding to its endothelial cell surface receptor (VEGFR), thereby inhibiting tumor angiogenesis (20).
National Comprehensive Cancer Network (NCCN) guidelines allow for the combination of bevacizumab in multiple clinical scenarios including combination with chemotherapy as primary adjuvant therapy, platinum-sensitive relapse, and platinum-resistant EOC. Additionally, bevacizumab can be given alone for platinum resistant disease (18). Combining chemotherapy with bevacizumab increases progression free survival (PFS) (20). Other antiangiogenic drugs such as aflibercept, anlotinib and cediranib are being investigated in clinical trials for the management of EOC (21–24).
2.2 Targeted therapy: PARP inhibitors
Poly (ADP-ribose) polymerase (PARP) proteins are a group of 17 glycosyl-transferase nuclear enzymes with roles in DNA repair via an enzymatic process termed PARylation (25, 26). As part of the DNA damage response, BRCA1 and 2 function within the homologous recombination repair (HRR) pathway. When these genes are mutated, DNA can be repaired via alternative pathways that involve PARP. Inhibition of PARP in homologous recombinant (HR) deficient cancer prevents DNA damage repair, resulting in cell death through synthetic lethality (18, 27–29).
Olaparib, niraparib and rucaparib are the three PARP inhibitors (PARPi) approved by the FDA for the treatment of advanced germline BRCA-mutated ovarian cancer. These PARPi are also approved for use as maintenance therapy for EOC. In 2014, olaparib became the first PARPi targeting PARPs 1-4 approved by the FDA as a monotherapy for the treatment of advanced germline BRCA-deficient ovarian cancer, and it was approved in 2017 as maintenance therapy for platinum-sensitive EOC (SOLO-1 and SOLO-2 clinical trials, respectively) (20, 30–36). Rucaparib targets most of the PARP enzymes (PARPs 1-4, 12, 15 and 16). In 2016, it became the second PARPi to receive FDA approval as maintenance therapy for advanced ovarian cancer (20, 30), and it is currently approved for the treatment of BRCAmut recurrent EOC based on data from the ARIEL 3 and 4 clinical trials (25, 37–39) Niraparib is another PARP1/2 inhibitor with FDA approval as maintenance therapy for recurrent EOC. Currently, niraparib is approved as maintenance therapy for platinum-sensitive recurrent ovarian cancer independent of BRCA1/2 mutation and HR status based on results from the PRIMA and NOVA clinical trials (40–43). Emerging PARPi under clinical investigation for the treatment of EOC include talozaparib, veliparib, pamiparib and fuzuloparib (44, 45, 62). Table 1 lists the clinical trials associated with the currently approved and emerging PARPi.
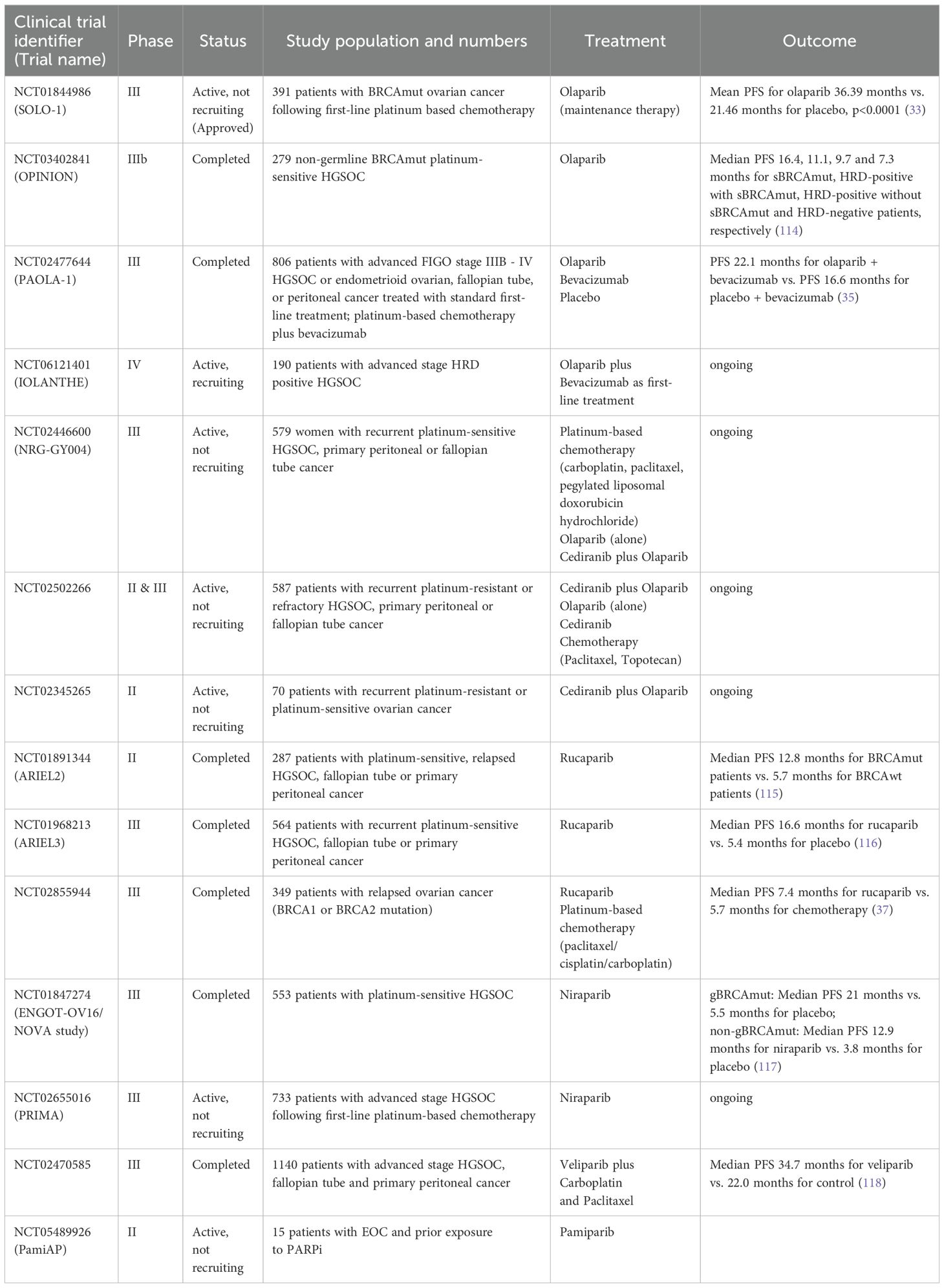
Table 1. Clinical trials of PARPi alone or in combination with anti-angiogenic drugs and/or chemotherapy.
2.3 Targeted therapy: antibody drug conjugates
Antibody drug conjugates (ADC) consist of a cytotoxic drug conjugated through a linker to a monoclonal antibody targeting specific tumor-associated antigens. ADCs use monoclonal antibodies to deliver cytotoxic drugs to cancer cells by selectively binding to a specific antigen (typically a cell surface receptor) overexpressed on the surface of cancer cells. ADCs have shown potential positive effects in recurrent and platinum-resistant ovarian cancer. Mirvetuximab soravtansine (MIRV) is the only FDA approved ADC for the treatment of folate receptor alpha (FRα) positive platinum-resistant epithelial ovarian cancer (46–50). It is comprised of an anti-FRα monoclonal antibody, a cytotoxic payload (maytansinoid, a microtubule inhibitor) and a cleavable disulfide linker. The SORAYA (51) and MIRASOL (52) trials demonstrated the efficacy of MIRV in platinum-resistant ovarian cancer with an increase in PFS when compared to chemotherapy alone. Ongoing trials for the treatment of platinum-resistant ovarian cancer include the use of other ADCs such as Farletuzumab ecteribulin (53), Raludotatug deruxtecan (54), Luveltamab tazevibulin (55), Anetumab ravtansine (56, 57) and Lifastuzumab vedotin (58).
2.4 Targeted therapy: immunotherapy
EOC tumors are “immunogenic tumors” that cause sporadic anti-tumor immune responses detected in the peripheral blood, ascites and tumors (59, 60). The tumor microenvironment (TME) is implicated in EOC prognosis (61, 62). Early studies demonstrated evidence of tumor-infiltrating lymphocytes (TILs), programmed death 1 (PD-1) and programmed death-ligand 1 (PD-L1) in EOC. TILs, PD-1 and PD-L1 correlate with improved EOC prognosis, which renders immunotherapy for ovarian cancer an area of great interest (60, 63). Some immunotherapies for EOC include immune checkpoint inhibitors (ICI), chimeric antigen receptor T (CAR-T) cells, vaccines and monoclonal antibodies (62). Nivolumab and Pembrolizumab are ICIs for PD-1 that have been tested for the treatment of relapsed platinum-resistant and advanced ovarian cancer, respectively. Avelumab, an anti-PD-L1 antibody, and Ipilimumab, an anti-CTLA-4 antibody, have been tested for the treatment of recurrent platinum-sensitive ovarian cancer (60, 63). Despite a justified rationale for the use of ICIs in EOC, the results from clinical trials have shown a limited clinical efficacy of ICIs; the median overall response rates (ORR) are 10-15% (64, 65). An ORR of 22.2% was achieved in a study of 12 patients with recurrent or metastatic EOC treated with the anti-PD-L1 inhibitor, Atezolizumab (65, 66). These mediocre ORRs are complicated by several adverse events experienced by up to 75% of patients, including fatigue, gastrointestinal, endocrine and dermatological events, and neurological, cardiological, pulmonary and renal toxicities (67, 68). The limited efficacy of immune checkpoint blockade in the setting of EOC could be due to several factors, including the compensatory upregulation of alternative immune checkpoints, low tumor mutational burden, loss of mutation-associated antigens, or the expression of multiple inhibitory receptors on the infiltrating T-cells (60, 61, 64, 66).
Additionally, immunotherapy has been added to chemotherapy in multiple large phase III trials of patients with primary and recurrent ovarian cancer. Unfortunately, across all of these studies, patients who received immunotherapy did not derive any benefit in terms of PFS and OS compared to those who received placebo. It is not known if there may be subsets within this population that could derive some benefit from immunotherapy — this is an area in need of further evaluation.
Several clinical trials are ongoing for ICIs combined with platinum-based chemotherapy, PARPi, anti-angiogenic agents and other biologic agents in recurrent EOC (Table 2). Of note, immunotherapy is not currently incorporated in routine clinical practice.
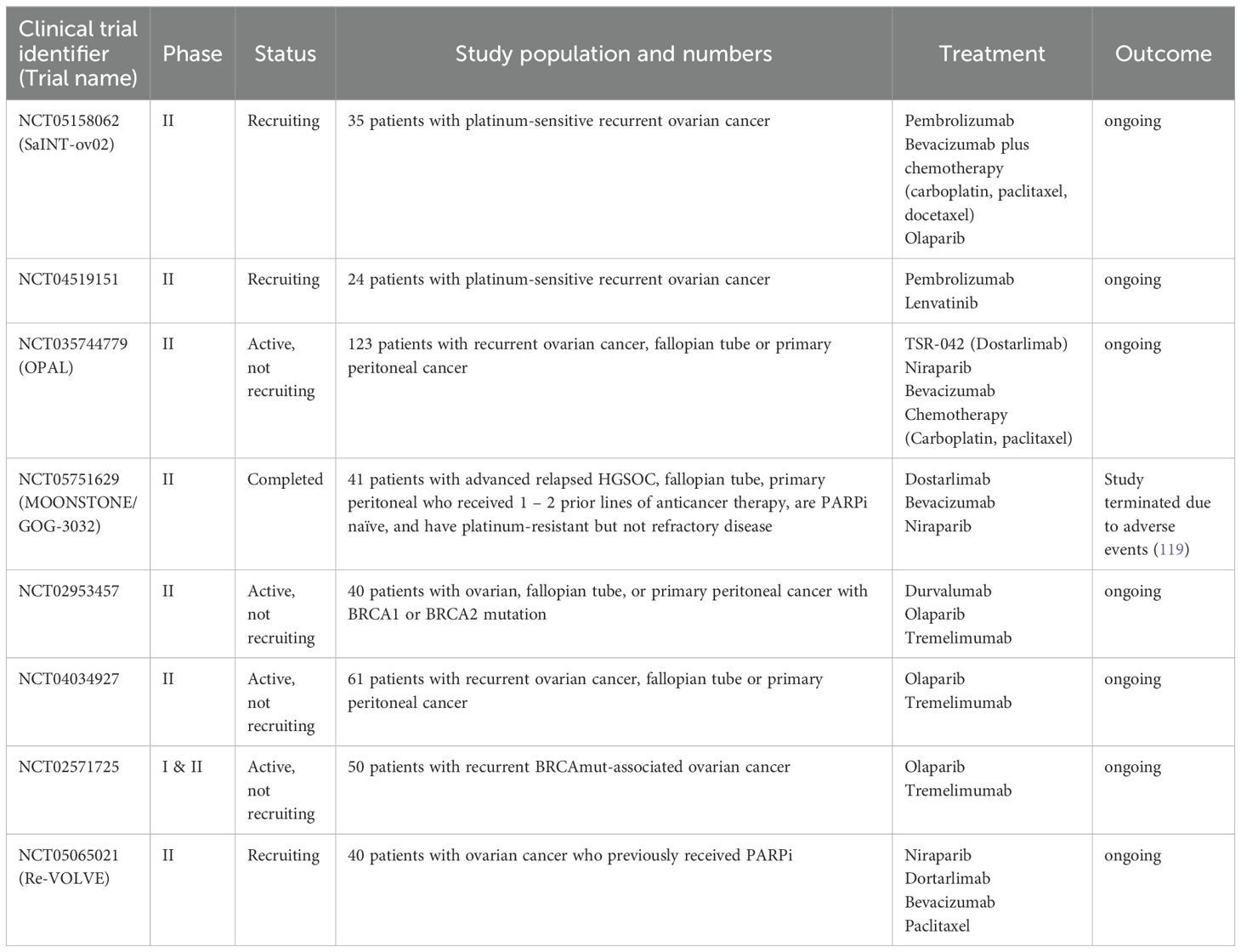
Table 2. Clinical trials of immune checkpoint inhibitors combined with PARPi or anti-angiogenic drugs and/or chemotherapy.
3 Treatment resistance and disease recurrence
Biomarkers are measured and evaluated as an indication of normal biological processes, disease state and pharmacological response to treatment (69). They can serve as tools to screen for, characterize, or diagnose disease, consequently enabling personalized drug treatment and the prediction of drug toxicity and adverse drug reactions (5). Predictive biomarkers measure the potential response or lack of response to a particular treatment, helping to identify patients who are likely to benefit from the treatment (69).
In EOC, >80% of patients respond to the first line of treatment, i.e. cytoreductive surgery and platinum-based chemotherapy, with approximately 80% having recurrence at different time intervals after chemotherapy (70–72). Patients having recurrence >6 months of standard treatment are considered platinum-sensitive. Recurrence during chemotherapy or within a month after treatment is referred to as chemotherapy-refractory ovarian cancer (73, 74). This recurrence is intrinsic or a form of primary resistance. Identifying this primary resistance using predictive biomarkers can help patients receive more effective alternative treatments.
Another form of recurrence occurs in patients who initially respond to chemotherapy but develop recurrence within 6 months of treatment. These patients’ tumors are referred to as platinum-resistant (73, 75). This type of acquired resistance occurs after exposure to drug treatment (76). Predictive biomarkers for acquired resistance can help determine when to change treatment. Less than 20% of clear cell, mucinous, endometrioid, and LGSOC tumors respond to standard treatment. Approximately 90% of patients with HGSOC respond to standard treatment, but some patients still relapse after treatment (74). This increases the need to understand the molecular basis of EOC for improved disease management (70).
Among the factors that influence drug resistance by decreasing the intracellular drug concentration are increased expression of drug efflux pumps (e.g., copper-transporter 2, which is involved in the efflux of platinum), overexpression of ATP-binding cassette (ABC) transporters (e.g., P-glycoprotein), and the decreased expression of drug influx transporters (77). P-glycoprotein is highly expressed in EOC patients with decreased PFS, therefore drug efflux pumps or drug transporters are candidate predictive biomarkers due to their ability to modulate EOC resistance (74). Solute-like-carrier (SLC) family proteins are drug influx transporters that increase intracellular accumulation of drugs. Lower expression of SLC22A5 and SLC31A1 has been observed in patients with HGSOC, whereas their elevated levels correlate with increased treatment response and survival rates (77). Dysregulation of autophagy and evasion of apoptosis also contribute to drug resistance (77). Hence, the proteins involved in these pathways represent feasible predictive biomarker candidates.
The activation of DNA damage repair pathways also plays a major role in ovarian cancer treatment resistance. DNA damaging agents such as platinum-based chemotherapies cause double strand breaks that are repaired by the HR pathway to increase cell survival. Several academic and commercial entities have developed tests to assess the genomic instability of tumor DNA as a biomarker of HR (73, 74, 78). Deleterious mutations in the BRCA1/2 tumor suppressor genes lead to error-prone non-homologous end joining (NHEJ), resulting in genome instability (73, 77, 79, 80). EOC patients with BRCA1 or BRCA2 mutations are sensitive to DNA-damaging agents. Other proteins such as RAD51, ATM, ATR and PALB2 are also involved in HR, and mutations in the genes encoding for these proteins increase the sensitivity of HGSOC patients to chemotherapy (81, 82). Most HGSOC patients with HR deficiency have an increased response rate to platinum-based therapy (83).
The combination of standard treatment with other chemotherapies or targeted therapy can help improve patient outcomes. Stratifying patients for specific drug treatments based on predictive biomarkers has the potential to increase drug response and reduce drug resistance in the context of personalized or precision medicine.
4 The role of predictive biomarkers in precision medicine-based treatment of EOC
Over the past decade, biomedical research has focused on understanding the disease state of individual patients to develop specific diagnosis and treatment options, forming the basis for precision medicine, which is known as personalized medicine (84). In 2015, U.S. President Barack Obama announced the launch of the Precision Medicine Initiative (PMI) to improve healthcare. The goal of the PMI was “to enable a new era of medicine through research, technology, and policies that empower patients, researchers and providers to work together toward the development of individualized care” (85, 86). The initiative focused on providing the most effective treatment for patients based on their genetic changes. The following year, the National Cancer Moonshot Initiative was launched by then Vice President Joe Biden to improve cancer prevention, diagnosis and treatment with the goal of ending cancer deaths (86, 87).
Precision medicine aims to use information from patients’ genes, proteins and the environment to identify early health indicators, detect disease stages, slow disease progression, and alter the health trajectories of patients through targeted treatments or lifestyle changes (84, 88, 89). In oncology, precision medicine has significantly improved cancer treatment by identifying the unique molecular characteristics of different cancers, allowing for the development of targeted therapies that can effectively target specific cancer types (90). An example is EOC patients with BRCA1/2 mutations who benefit from PARPi (91). Additionally, with precision medicine, patient response to drugs can be assessed using molecular assays to test the sensitivity of the tumor cells to current or emerging therapies (92). Precision medicine leads to better treatment outcomes and reduces the trial-and-error treatment approach, which has traditionally followed a “one-size fits all” modality (93).
Predictive biomarkers play an important role in cancer precision medicine whereby they serve as tools to predict treatment response. Genome-based biomarkers were the first widely used predictive biomarkers in cancer precision medicine focusing on genome sequencing to identify cancer-specific somatic and germline mutations to provide information on cancer susceptibility and treatment options (84, 94, 95). Individuals with germline mutations in specific oncogenes are at higher risk of developing cancer. These mutations include missense mutations, nonsense mutations, insertions or deletion and rearrangements (94).
5 Opportunities for predictive proteogenomic biomarkers in EOC precision medicine
Despite the promising potential of genome-based predictive biomarkers in precision medicine, several limitations remain, including the complexity and variability of these mutations, which can affect the accuracy and reliability of predictive biomarkers. These mutations can vary widely even within different areas of the same tumor, making it difficult to develop an effective treatment approach based on a single biopsy.
Genomics has provided fundamental insights into the disease mechanism of several cancer types; however, genomic data alone does not predict disease prognosis and treatment efficacy due to the indirect correlation between genotypes and phenotypes (96). Transcriptomics, proteomics, epigenetics and metabolomics can be utilized as biomarker tools in cancer precision medicine (95). Another analytical approach known as proteogenomics has recently gained importance in cancer precision medicine.
Proteogenomics is the integration of genomics, transcriptomics, epigenomics and mass spectrometry-based proteomics data, which aims to provide a comprehensive view of the molecular basis of disease processes (96, 97). By identifying novel protein-coding regions, post-translational modifications (PTMs), proteoforms, and single amino acid variations, proteogenomics enables the exploration of the functional consequences of genetic variations (98).
Proteogenomic analysis entails whole-genome sequencing (WGS) or whole exome sequencing (WES) to identify genetic variants and genomic mutations, RNA-seq to identify alternative splicing and non-coding RNAs, and mass spectrometry-based proteomic analysis to identify and quantify peptides (99). The peptides generated from proteomic analysis are mapped to a custom protein sequence database derived from the sample-specific genomic, transcriptomic or epigenomic data instead of a reference proteome database to identify possible novel proteins (100). The custom protein database can be generated using bioinformatics tools such as Galaxy-P, customProBD, sapFinder and PGA (98).
Proteogenomics has been applied in cancer research in an approach termed onco-proteogenomics to identify tumor-specific peptides. Onco-proteogenomics enables the validation of the translation of specific cancer-associated genomic mutations into proteins (phenotypes), and the identification of PTMs and altered signaling pathways involved in cancer development (98). The National Cancer Institute’s Clinical Proteomic Tumor Analysis Consortium (CPTAC) has published several landmark studies applying proteogenomics to characterize the molecular landscape of multiple cancer types, including breast, colorectal, pancreatic, lung, clear cell renal, and ovarian carcinoma (101–104). Onco-proteogenomics has a role in the identification and development of diagnostic, prognostic, and predictive biomarkers.
Liquid biopsies containing circulating tumor cells (CTCs) can be utilized as samples for onco-proteogenomic testing. CTCs contain molecular markers from patient tumors, which can be measured to assess drug treatment response (105). Given that the somatic mutational landscape of HGSOC remains stable with chemotherapy treatment, subjecting patients to rebiopsy at each recurrence time point would not be informative or practical due to patient discomfort and the financial cost (105, 106). In contrast, it is possible that the somatic proteogenomic landscape of HGSOC is altered during the course of chemotherapy treatment. Establishing proteogenomic biomarkers of treatment response using CTCs represents a novel, feasible, and minimally invasive approach.
Most molecularly targeted therapies target proteins in cancer cells. Examples of these targeted therapies include PARP inhibitors, kinase inhibitors and immunomodulatory proteins (102). Proteogenomics can help identify predictive biomarkers for the diagnosis and effective treatment of patients with EOC. PARPi are particularly effective in EOC patients with BRCA1/2 mutations or HRD. Proteogenomic analyses can help identify additional biomarkers beyond BRCA mutations, such as specific protein expression patterns or mutations in other DNA repair genes that can predict sensitivity to PARP inhibitors (107). This could expand the population of EOC patients who benefit from these therapies.
In ovarian cancer, proteogenomics has identified PTMs such as phosphorylation, acetylation and glycosylation as candidate predictive biomarkers of response to targeted therapies. A CPTAC study integrating the proteomic measurements of 174 HGSOC tumor samples with the genomic data generated by TCGA provided several novel insights into HGSOC biology, including the association between protein relative abundance and chromosomal instability, the impact of copy number alterations (CNA) on the proteome, the signaling pathways that diverse genome rearrangements coverage on, and the signaling pathways that are associated with short overall survival (108, 109). The proteogenomic and phosphoproteomic characterization of 83 HGSOC and normal fallopian tube tissues identified signaling pathways that differentiated HGSOC from normal tissues based on their HRD status (109, 110). These studies also demonstrated that decreased acetylation levels of Histone H4 Lysine12 and Lysine16 were associated with HRD in HGSOC (109, 110). These PTMs are candidate biomarkers for HRD, which creates the potential for HGSOC patients to be stratified for treatment with DNA repair pathway targeted therapies, including histone deacetylation (HDAC) inhibitors and PARPi (108, 110).
Resistance to platinum-based chemotherapy is a major challenge in EOC treatment. Proteogenomic studies can identify proteins and pathways associated with chemoresistance, such as alterations in the PI3K/AKT pathway, which could serve as predictive biomarkers (108). Proteogenomics also has potential utility in identifying predictive biomarkers of response to treatment with other targeted therapies that are used in the treatment of EOC. For example, predictive biomarkers such as PD-L1 expression, tumor infiltrating lymphocytes (CD8+, CD4+), tumor mutational burden and chemokines (CXCL 9,11,13) can be used to identify EOC patients that do or do not respond to ICI (111). Increased levels of chemokines and tumor infiltrating lymphocytes have been associated with increased response to immunotherapy (112, 113).
6 Conclusion and outlook
The proteogenomic molecular data from liquid biopsies of patients with EOC is a largely unexplored source of predictive biomarkers of treatment response. These samples can be obtained from patients before, during and after treatment to inform treatment decisions. CTCs contain information from the RNA, DNA, proteins and metabolites from the patient tumors and can therefore serve as valuable biospecimens for disease detection and monitoring treatment response.
In vitro diagnostic clinical tests can be developed and validated to measure these proteogenomic biomarkers with an intended use of predicting treatment response. This will enable patients to be stratified for treatment, consequently preventing them from being prescribed drug treatment regimens from which they could potentially not derive any benefit, which is incongruent with the goal of precision medicine. Proteogenomics is a valuable tool supporting personalized medicine efforts. Incorporating this powerful analytical method into translational research studies advances a goal of improving the overall survival of patients with EOC.
Author contributions
TP: Writing – original draft, Writing – review & editing. BE: Writing – original draft, Writing – review & editing. ST: Conceptualization, Supervision, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Feeney L, Harley IJ, McCluggage WG, Mullan PB, Beirne JP. Liquid biopsy in ovarian cancer: Catching the silent killer before it strikes. World J Clin Oncol. (2020) 11(11):868–89. doi: 10.5306/wjco.v11.i11.868
2. Ali AT, Al-Ani O, Al-Ani F. Epidemiology and risk factors for ovarian cancer. Prz Menopauzalny. (2023) 22(2):93–104. doi: 10.5114/pm.2023.128661
3. Stewart C, Ralyea C, Lockwood S. Ovarian cancer: An integrated review. Semin Oncol Nurs. (2019) 35(2):151–6. doi: 10.1016/j.soncn.2019.02.001
4. Lheureux S, Braunstein M, Oza AM. Epithelial ovarian cancer: Evolution of management in the era of precision medicine. CA Cancer J Clin. (2019) 69(4):280–304. doi: 10.3322/caac.21559
5. Bonifácio VDB. Ovarian cancer biomarkers: Moving forward in early detection. Adv Exp Med Biol. (2020) 1219:355–63. doi: 10.1007/978-3-030-34025-4_18
6. Forstner R. Early detection of ovarian cancer. Eur Radiol. (2020) 30(10):5370–3. doi: 10.1007/s00330-020-06937-z
7. Webb PM, Jordan SJ. Epidemiology of epithelial ovarian cancer. Best Pract Res Clin Obstet Gynaecol. (2017) 41:3–14. doi: 10.1016/j.bpobgyn.2016.08.006
8. Punzón-Jiménez P, Lago V, Domingo S, Simón C, Mas A. Molecular management of high-grade serous ovarian carcinoma. Int J Mol Sci. (2022) 23(22):13777. doi: 10.3390/ijms232213777
9. Xiao Y, Bi M, Guo H, Li M. Multi-omics approaches for biomarker discovery in early ovarian cancer diagnosis. EBioMedicine. (2022) 79:104001. doi: 10.1016/j.ebiom.2022.104001
10. Zhang M, Cheng S, Jin Y, Zhao Y, Wang Y. Roles of CA125 in diagnosis, prediction, and oncogenesis of ovarian cancer. Biochim Biophys Acta Rev Cancer. (2021) 1875(2):188503. doi: 10.1016/j.bbcan.2021.188503
11. Englisz A, Smycz-Kubańska M, Mielczarek-Palacz A. Sensitivity and specificity of selected biomarkers and their combinations in the diagnosis of ovarian cancer. Diagnostics (Basel). (2024) 14(9):949. doi: 10.3390/diagnostics14090949
12. An D, Banerjee S, Lee JM. Recent advancements of antiangiogenic combination therapies in ovarian cancer. Cancer Treat Rev. (2021) 98:102224. doi: 10.1016/j.ctrv.2021.102224
13. Arnaoutoglou C, Dampala K, Anthoulakis C, Papanikolaou EG, Tentas I, Dragoutsos G, et al. Epithelial ovarian cancer: A five year review. Medicina (Kaunas). (2023) 59(7):1183. doi: 10.3390/medicina59071183
14. Kuroki L, Guntupalli SR. Treatment of epithelial ovarian cancer. BMJ. (2020) 371:m3773. doi: 10.1136/bmj.m3773
15. Kossaï M, Leary A, Scoazec JY, Genestie C. Ovarian cancer: A heterogeneous disease. Pathobiology. (2018) 85(1-2):41–9. doi: 10.1159/000479006
16. Pergialiotis V, Sotiropoulou IM, Liatsou E, Liontos M, Frountzas M, Thomakos N, et al. Quality of life of ovarian cancer patients treated with combined platinum taxane chemotherapy: a systematic review of the literature. Support Care Cancer. (2022) 30(9):7147–57. doi: 10.1007/s00520-022-07053-y
17. Vetter MH, Hays JL. Use of targeted therapeutics in epithelial ovarian cancer: A review of current literature and future directions. Clin Ther. (2018) 40(3):361–71. doi: 10.1016/j.clinthera.2018.01.012
18. Gadducci A, Guarneri V, Peccatori FA, Ronzino G, Scandurra G, Zamagni C, et al. Current strategies for the targeted treatment of high-grade serous epithelial ovarian cancer and relevance of BRCA mutational status. J Ovarian Res. (2019) 12(1):9. doi: 10.1186/s13048-019-0484-6
19. Nag S, Aggarwal S, Rauthan A, Warrier N. Maintenance therapy for newly diagnosed epithelial ovarian cancer- a review. J Ovarian Res. (2022) 15(1):88. doi: 10.1186/s13048-022-01020-1
20. Huang X, Li XY, Shan WL, Chen Y, Zhu Q, Xia BR. Targeted therapy and immunotherapy: Diamonds in the rough in the treatment of epithelial ovarian cancer. Front Pharmacol. (2023) 14:1131342. doi: 10.3389/fphar.2023.1131342
21. García García Y, Marín Alcalá M, Martínez Vila C. Anti-angiogenic therapy for ovarian cancer. EJC Suppl. (2020) 15:77–86. doi: 10.1016/j.ejcsup.2020.02.003
22. Tew WP, Colombo N, Ray-Coquard I, Del Campo JM, Oza A, Pereira D, et al. Intravenous aflibercept in patients with platinum-resistant, advanced ovarian cancer: results of a randomized, double-blind, phase 2, parallel-arm study. Cancer. (2014) 120(3):335–43. doi: 10.1002/cncr.28406
23. Moroney JW, Sood AK, Coleman RL. Aflibercept in epithelial ovarian carcinoma. Future Oncol. (2009) 5(5):591–600. doi: 10.2217/fon.09.35
24. Grunewald T, Ledermann JA. Targeted therapies for ovarian cancer. Best Pract Res Clin Obstet Gynaecol. (2017) 41:139–52. doi: 10.1016/j.bpobgyn.2016.12.001
25. Loizzi V, Ranieri G, Laforgia M, Gadaleta CD, Gargano G, Kardhashi A, et al. PARP inhibitors and epithelial ovarian cancer: Molecular mechanisms, clinical development and future prospective. Oncol Lett. (2020) 20(4):90. doi: 10.3892/ol.2020.11951
26. Vanacker H, Harter P, Labidi-Galy SI, Banerjee S, Oaknin A, Lorusso D, et al. PARP-inhibitors in epithelial ovarian cancer: Actual positioning and future expectations. Cancer Treat Rev. (2021) 99:102255. doi: 10.1016/j.ctrv.2021.102255
27. Zheng F, Zhang Y, Chen S, Weng X, Rao Y, Fang H. Mechanism and current progress of poly ADP-ribose polymerase (PARP) inhibitors in the treatment of ovarian cancer. BioMed Pharmacother. (2020) 123:109661. doi: 10.1016/j.biopha.2019.109661
28. Rose M, Burgess JT, O'Byrne K, Richard DJ, Bolderson E. PARP inhibitors: Clinical relevance, mechanisms of action and tumor resistance. Front Cell Dev Biol. (2020) 8:564601. doi: 10.3389/fcell.2020.564601
29. Gadducci A, Guerrieri ME. PARP inhibitors alone and in combination with other biological agents in homologous recombination deficient epithelial ovarian cancer: From the basic research to the clinic. Crit Rev Oncol Hematol. (2017) 114:153–65. doi: 10.1016/j.critrevonc.2017.04.006
30. Luo L, Keyomarsi K. PARP inhibitors as single agents and in combination therapy: the most promising treatment strategies in clinical trials for BRCA-mutant ovarian and triple-negative breast cancers. Expert Opin Investig Drugs. (2022) 31(6):607–31. doi: 10.1080/13543784.2022.2067527
31. Banerjee S, Moore KN, Colombo N, Scambia G, Kim BG, Oaknin A, et al. Maintenance olaparib for patients with newly diagnosed advanced ovarian cancer and a BRCA mutation (SOLO1/GOG 3004): 5-year follow-up of a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol. (2021) 22(12):1721–31. doi: 10.1016/S1470-2045(21)00531-3
32. DiSilvestro P, Banerjee S, Colombo N, Scambia G, Kim BG, Oaknin A, et al. Overall survival with maintenance olaparib at a 7-year follow-up in patients with newly diagnosed advanced ovarian cancer and a BRCA mutation: The SOLO1/GOG 3004 trial. J Clin Oncol. (2023) 41(3):609–17. doi: 10.1200/JCO.22.01549
33. Moore K, Colombo N, Scambia G, Kim BG, Oaknin A, Friedlander M, et al. Maintenance olaparib in patients with newly diagnosed advanced ovarian cancer. N Engl J Med. (2018) 379(26):2495–505. doi: 10.1056/NEJMoa1810858
34. Poveda A, Floquet A, Ledermann JA, Asher R, Penson RT, Oza AM, et al. Olaparib tablets as maintenance therapy in patients with platinum-sensitive relapsed ovarian cancer and a BRCA1/2 mutation (SOLO2/ENGOT-Ov21): a final analysis of a double-blind, randomised, placebo-controlled, phase 3 trial. Lancet Oncol. (2021) 22(5):620–31. doi: 10.1016/S1470-2045(21)00073-5
35. Ray-Coquard I, Pautier P, Pignata S, Pérol D, González-Martín A, Berger R, et al. Olaparib plus bevacizumab as first-line maintenance in ovarian cancer. N Engl J Med. (2019) 381(25):2416–28. doi: 10.1056/NEJMoa1911361
36. Vergote I, Ray-Coquard I, Anderson DM, Cantuaria G, Colombo N, Garnier-Tixidre C, et al. Population-adjusted indirect treatment comparison of the SOLO1 and PAOLA-1/ENGOT-ov25 trials evaluating maintenance olaparib or bevacizumab or the combination of both in newly diagnosed, advanced BRCA-mutated ovarian cancer. Eur J Cancer. (2021) 157:415–23. doi: 10.1016/j.ejca.2021.08.023
37. Kristeleit R, Lisyanskaya A, Fedenko A, Dvorkin M, de Melo AC, Shparyk Y, et al. Rucaparib versus standard-of-care chemotherapy in patients with relapsed ovarian cancer and a deleterious BRCA1 or BRCA2 mutation (ARIEL4): an international, open-label, randomised, phase 3 trial. Lancet Oncol. (2022) 23(4):465–78. doi: 10.1016/S1470-2045(22)00122-X
38. O'Donnell DM. Rucaparib for BRCA1/2-mutated pretreated ovarian cancer: reflections from the ARIEL4 trial. Lancet Oncol. (2022) 23(4):440–1. doi: 10.1016/S1470-2045(22)00150-4
39. Clamp AR, Lorusso D, Oza AM, Aghajanian C, Oaknin A, Dean A, et al. Rucaparib maintenance treatment for recurrent ovarian carcinoma: the effects of progression-free interval and prior therapies on efficacy and safety in the randomized phase III trial ARIEL3. Int J Gynecol Cancer. (2021) 31(7):949–58. doi: 10.1136/ijgc-2020-002240
40. González-Martín A, Pothuri B, Vergote I, DePont Christensen R, Graybill W, Mirza MR, et al. Niraparib in patients with newly diagnosed advanced ovarian cancer. N Engl J Med. (2019) 381(25):2391–402. doi: 10.1056/NEJMoa1910962
41. Moore KN, Secord AA, Geller MA, Miller DS, Cloven N, Fleming GF, et al. Niraparib monotherapy for late-line treatment of ovarian cancer (QUADRA): a multicentre, open-label, single-arm, phase 2 trial. Lancet Oncol. (2019) 20(5):636–48. doi: 10.1016/S1470-2045(19)30029-4
42. O'Cearbhaill RE, Pérez-Fidalgo JA, Monk BJ, Tusquets I, Mccormick C, Fuentes J, et al. Efficacy of niraparib by time of surgery and postoperative residual disease status: A post hoc analysis of patients in the PRIMA/ENGOT-OV26/GOG-3012 study. Gynecol Oncol. (2022) 166(1):36–43. doi: 10.1016/j.ygyno.2022.04.012
43. Hardesty MM, Krivak TC, Wright GS, Hamilton E, Fleming EL, Belotte J, et al. OVARIO phase II trial of combination niraparib plus bevacizumab maintenance therapy in advanced ovarian cancer following first-line platinum-based chemotherapy with bevacizumab. Gynecol Oncol. (2022) 166(2):219–29. doi: 10.1016/j.ygyno.2022.05.020
44. Wu X, Zhu J, Wang J, Lin Z, Yin R, Sun W, et al. Pamiparib monotherapy for patients with germline BRCA1/2-mutated ovarian cancer previously treated with at least two lines of chemotherapy: A multicenter, open-label, phase II study. Clin Cancer Res. (2022) 28(4):653–61. doi: 10.1158/1078-0432.CCR-21-1186
45. Lickliter JD, Voskoboynik M, Mileshkin L, Gan HK, Kichenadasse G, Zhang K, et al. Phase 1A/1B dose-escalation and -expansion study to evaluate the safety, pharmacokinetics, food effects and antitumor activity of pamiparib in advanced solid tumours. Br J Cancer. (2022) 126(4):576–85. doi: 10.1038/s41416-021-01632-2
46. Moore KN, Martin LP, O'Malley DM, Matulonis UA, Konner JA, Vergote I, et al. A review of mirvetuximab soravtansine in the treatment of platinum-resistant ovarian cancer. Future Oncol. (2018) 14(2):123–36. doi: 10.2217/fon-2017-0379
47. Moore KN, O'Malley DM, Vergote I, Martin LP, Gonzalez-Martin A, Malek K, et al. Safety and activity findings from a phase 1b escalation study of mirvetuximab soravtansine, a folate receptor alpha (FRα)-targeting antibody-drug conjugate (ADC), in combination with carboplatin in patients with platinum-sensitive ovarian cancer. Gynecol Oncol. (2018) 151(1):46–52. doi: 10.1016/j.ygyno.2018.07.017
48. Moore KN, Borghaei H, O'Malley DM, Jeong W, Seward SM, Bauer TM, et al. Phase 1 dose-escalation study of mirvetuximab soravtansine (IMGN853), a folate receptor α-targeting antibody-drug conjugate, in patients with solid tumors. Cancer. (2017) 123(16):3080–7. doi: 10.1002/cncr.30736
49. Moore KN, Martin LP, O'Malley DM, Matulonis UA, Konner JA, Perez RP, et al. Safety and activity of mirvetuximab soravtansine (IMGN853), a folate receptor alpha-targeting antibody-drug conjugate, in platinum-resistant ovarian, fallopian tube, or primary peritoneal cancer: A phase I expansion study. J Clin Oncol. (2017) 35(10):1112–8. doi: 10.1200/JCO.2016.69.9538
50. Ponte JF, Ab O, Lanieri L, Lee J, Coccia J, Bartle LM, et al. Mirvetuximab soravtansine (IMGN853), a folate receptor alpha-targeting antibody-drug conjugate, potentiates the activity of standard of care therapeutics in ovarian cancer models. Neoplasia. (2016) 18(12):775–84. doi: 10.1016/j.neo.2016.11.002
51. Matulonis UA, Lorusso D, Oaknin A, Pignata S, Dean A, Denys H, et al. Efficacy and safety of mirvetuximab soravtansine in patients with platinum-resistant ovarian cancer with high folate receptor alpha expression: Results from the SORAYA study. J Clin Oncol. (2023) 41(13):2436–45. doi: 10.1200/JCO.22.01900
52. Moore KN, Angelergues A, Konecny GE, García Y, Banerjee S, Lorusso D, et al. Mirvetuximab soravtansine in FRα-positive, platinum-resistant ovarian cancer. N Engl J Med. (2023) 389(23):2162–74. doi: 10.1056/NEJMoa2309169
53. Herzog TJ, Pignata S, Ghamande SA, Rubio MJ, Fujiwara K, Vulsteke C, et al. Randomized phase II trial of farletuzumab plus chemotherapy versus placebo plus chemotherapy in low CA-125 platinum-sensitive ovarian cancer. Gynecol Oncol. (2023) 170:300–8. doi: 10.1016/j.ygyno.2023.01.003
54. Suzuki H, Nagase S, Saito C, Takatsuka A, Nagata M, Honda K, et al. Raludotatug deruxtecan, a CDH6-targeting antibody-drug conjugate with a DNA topoisomerase I inhibitor DXd, is efficacious in human ovarian and kidney cancer models. Mol Cancer Ther. (2024) 23(3):257–71. doi: 10.1158/1535-7163.MCT-23-0287
55. Li X, Zhou S, Abrahams CL, Krimm S, Smith J, Bajjuri K, et al. Discovery of STRO-002, a novel homogeneous ADC targeting folate receptor alpha, for the treatment of ovarian and endometrial cancers. Mol Cancer Ther. (2023) 22(2):155–67. doi: 10.1158/1535-7163.MCT-22-0322
56. Hassan R, Blumenschein GR, Moore KN, Santin AD, Kindler HL, Nemunaitis JJ, et al. First-in-Human, multicenter, phase I dose-escalation and expansion study of anti-mesothelin antibody-drug conjugate anetumab ravtansine in advanced or metastatic solid tumors. J Clin Oncol. (2020) 38(16):1824–35. doi: 10.1200/JCO.19.02085
57. Santin AD, Vergote I, González-Martín A, Moore K, Oaknin A, Romero I, et al. Safety and activity of anti-mesothelin antibody-drug conjugate anetumab ravtansine in combination with pegylated-liposomal doxorubicin in platinum-resistant ovarian cancer: multicenter, phase ib dose escalation and expansion study. Int J Gynecol Cancer. (2023) 33(4):562–70. doi: 10.1136/ijgc-2022-003927
58. Moore KN, Birrer MJ, Marsters J, Wang Y, Chio Y, Royer-Joo S, et al. Phase 1b study of anti-NaPi2b antibody-drug conjugate lifastuzumab vedotin (DNIB0600A) in patients with platinum-sensitive recurrent ovarian cancer. Gynecol Oncol. (2020) 158(3):631–9. doi: 10.1016/j.ygyno.2020.05.039
59. Palaia I, Tomao F, Sassu CM, Musacchio L, Benedetti Panici P. Immunotherapy for ovarian cancer: Recent advances and combination therapeutic approaches. Onco Targets Ther. (2020) 13:6109–29. doi: 10.2147/OTT.S205950
60. Rodriguez GM, Galpin KJC, McCloskey CW, Vanderhyden BC. The tumor microenvironment of epithelial ovarian cancer and its influence on response to immunotherapy. Cancers (Basel). (2018) 10(8):242. doi: 10.3390/cancers10080242
61. Macpherson AM, Barry SC, Ricciardelli C, Oehler MK. Epithelial ovarian cancer and the immune system: Biology, interactions, challenges and potential advances for immunotherapy. J Clin Med. (2020) 9(9):2967. doi: 10.3390/jcm9092967
62. Bund V, Azaïs H, Bibi-Triki S, Lecointre L, Betrian SB, Angeles MA, et al. Basics of immunotherapy for epithelial ovarian cancer. J Gynecol Obstet Hum Reprod. (2022) 51(2):102283. doi: 10.1016/j.jogoh.2021.102283
63. Hartnett EG, Knight J, Radolec M, Buckanovich RJ, Edwards RP, Vlad AM. Immunotherapy advances for epithelial ovarian cancer. Cancers (Basel). (2020) 12(12):3733. doi: 10.3390/cancers12123733
64. Odunsi K. Immunotherapy in ovarian cancer. Ann Oncol. (2017) 28(suppl_8):viii1–7. doi: 10.1093/annonc/mdx444
65. González-Martín A, Sánchez-Lorenzo L. Immunotherapy with checkpoint inhibitors in patients with ovarian cancer: Still promising? Cancer. (2019) 125 Suppl 24:4616–22. doi: 10.1002/cncr.32520
66. Liu JF, Gordon M, Veneris J, Braiteh F, Balmanoukian A, Eder JP, et al. Safety, clinical activity and biomarker assessments of atezolizumab from a phase I study in advanced/recurrent ovarian and uterine cancers. Gynecol Oncol. (2019) 154(2):314–22. doi: 10.1016/j.ygyno.2019.05.021
67. Maughan BL, Bailey E, Gill DM, Agarwal N. Incidence of immune-related adverse events with program death receptor-1- and program death receptor-1 ligand-directed therapies in genitourinary cancers. Front Oncol. (2017) 7:56. doi: 10.3389/fonc.2017.00056
68. Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, McDermott DF, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med. (2012) 366(26):2443–54. doi: 10.1056/NEJMoa1200690
69. Verdaguer H, Saurí T, Macarulla T. Predictive and prognostic biomarkers in personalized gastrointestinal cancer treatment. J Gastrointest Oncol. (2017) 8(3):405–17. doi: 10.21037/jgo.2016.11.15
70. Le Page C, Huntsman DG, Provencher DM, Mes-Masson AM. Predictive and prognostic protein biomarkers in epithelial ovarian cancer: recommendation for future studies. Cancers (Basel). (2010) 2(2):913–54. doi: 10.3390/cancers2020913
71. Ray-Coquard I, Mirza MR, Pignata S, Walther A, Romero I, du Bois A. Therapeutic options following second-line platinum-based chemotherapy in patients with recurrent ovarian cancer: Comparison of active surveillance and maintenance treatment. Cancer Treat Rev. (2020) 90:102107. doi: 10.1016/j.ctrv.2020.102107
72. Ogundipe OD, Olajubutu O, Adesina SK. Targeted drug conjugate systems for ovarian cancer chemotherapy. BioMed Pharmacother. (2023) 165:115151. doi: 10.1016/j.biopha.2023.115151
73. Havasi A, Cainap SS, Havasi AT, Cainap C. Ovarian cancer-insights into platinum resistance and overcoming it. Medicina (Kaunas). (2023) 59(3):544. doi: 10.3390/medicina59030544
74. Marchetti C, De Felice F, Romito A, Iacobelli V, Sassu CM, Corrado G, et al. Chemotherapy resistance in epithelial ovarian cancer: Mechanisms and emerging treatments. Semin Cancer Biol. (2021) 77:144–66. doi: 10.1016/j.semcancer.2021.08.011
75. Yang L, Xie HJ, Li YY, Wang X, Liu XX, Mai J. Molecular mechanisms of platinum−based chemotherapy resistance in ovarian cancer (Review). Oncol Rep. (2022) 47(4):82. doi: 10.3892/or.2022.8293
76. Freimund AE, Beach JA, Christie EL, Bowtell DDL. Mechanisms of drug resistance in high-grade serous ovarian cancer. Hematol Oncol Clin North Am. (2018) 32(6):983–96. doi: 10.1016/j.hoc.2018.07.007
77. Mehrotra M, Phadte P, Shenoy P, Chakraborty S, Gupta S, Ray P. Drug-resistant epithelial ovarian cancer: Current and future perspectives. Adv Exp Med Biol. (2024) 1452:65–96. doi: 10.1007/978-3-031-58311-7_4
78. Christinat Y, Ho L, Clément S, Genestie C, Sahouli J, Cinieri S, et al. Normalized LST is an efficient biomarker for homologous recombination deficiency and olaparib response in ovarian carcinoma. JCO Precis Oncol. (2023) 7:e2200555. doi: 10.1200/PO.22.00555
79. Stiegeler N, Garsed DW, Au-Yeung G, Bowtell DDL, Heinzelmann-Schwarz V, Zwimpfer TA. Homologous recombination proficient subtypes of high-grade serous ovarian cancer: treatment options for a poor prognosis group. Front Oncol. (2024) 14:1387281. doi: 10.3389/fonc.2024.1387281
80. Biegała Ł, Gajek A, Marczak A, Rogalska A. PARP inhibitor resistance in ovarian cancer: Underlying mechanisms and therapeutic approaches targeting the ATR/CHK1 pathway. Biochim Biophys Acta Rev Cancer. (2021) 1876(2):188633. doi: 10.1016/j.bbcan.2021.188633
81. Toh M, Ngeow J. Homologous recombination deficiency: Cancer predispositions and treatment implications. Oncologist. (2021) 26(9):e1526–37. doi: 10.1002/onco.13829
82. Mangogna A, Munari G, Pepe F, Maffii E, Giampaolino P, Ricci G, et al. Homologous recombination deficiency in ovarian cancer: from the biological rationale to current diagnostic approaches. J Pers Med. (2023) 13(2):284. doi: 10.3390/jpm13020284
83. Cordani N, Bianchi T, Ammoni LC, Cortinovis DL, Cazzaniga ME, Lissoni AA, et al. An overview of PARP resistance in ovarian cancer from a molecular and clinical perspective. Int J Mol Sci. (2023) 24(15):11890. doi: 10.3390/ijms241511890
84. Olivier M, Asmis R, Hawkins GA, Howard TD, Cox LA. The need for multi-omics biomarker signatures in precision medicine. Int J Mol Sci. (2019) 20(19):4781. doi: 10.3390/ijms20194781
85. Sankar PL, Parker LS. The precision medicine initiative's all of us research program: an agenda for research on its ethical, legal, and social issues. Genet Med. (2017) 19(7):743–50. doi: 10.1038/gim.2016.183
86. Brant JM, Mayer DK. Precision medicine: Accelerating the science to revolutionize cancer care. Clin J Oncol Nurs. (2017) 21(6):722–9. doi: 10.1188/17.CJON.722-729
87. Collins FS, Varmus H. A new initiative on precision medicine. N Engl J Med. (2015) 372(9):793–5. doi: 10.1056/NEJMp1500523
88. Ramaswami R, Bayer R, Galea S. Precision medicine from a public health perspective. Annu Rev Public Health. (2018) 39:153–68. doi: 10.1146/annurev-publhealth-040617-014158
89. Johnson KB, Wei WQ, Weeraratne D, Frisse ME, Misulis K, Rhee K, et al. Precision medicine, AI, and the future of personalized health care. Clin Transl Sci. (2021) 14(1):86–93. doi: 10.1111/cts.12884
90. Saeed RF, Awan UA, Saeed S, Mumtaz S, Akhtar N, Aslam S. Targeted therapy and personalized medicine. Cancer Treat Res. (2023) 185:177–205. doi: 10.1007/978-3-031-27156-4_10
91. Karki R, Pandya D, Elston RC, Ferlini C. Defining "mutation" and "polymorphism" in the era of personal genomics. BMC Med Genomics. (2015) 8:37. doi: 10.1186/s12920-015-0115-z
92. Singh T, Neal AS, Moatamed NA, Memarzadeh S. Exploring the potential of drug response assays for precision medicine in ovarian cancer. Int J Mol Sci. (2020) 22(1):305. doi: 10.3390/ijms22010305
93. Carrigan P, Krahn T. Impact of biomarkers on personalized medicine. Handb Exp Pharmacol. (2016) 232:285–311. doi: 10.1007/164_2015_24
94. Sarhadi VK, Armengol G. Molecular biomarkers in cancer. Biomolecules. (2022) 12(8):1021. doi: 10.3390/biom12081021
95. Tsimberidou AM, Fountzilas E, Nikanjam M, Kurzock R. Review of precision cancer medicine: Evolution of the treatment paradigm. Cancer Treat Rev. (2020) 86:102019. doi: 10.1016/j.ctrv.2020.102019
96. Joshi SK, Piehowski P, Liu T, Gosline SJC, McDermott JE, Druker BJ, et al. Mass spectrometry-based proteogenomics: New therapeutic opportunities for precision medicine. Annu Rev Pharmacol Toxicol. (2024) 64:455–79. doi: 10.1146/annurev-pharmtox-022723-113921
97. Savage SR, Yi X, Lei JT, Wen B, Zhao H, Liao Y, et al. Pan-cancer proteogenomics expands the landscape of therapeutic targets. Cell. (2024) 187(16):4389–4407.e15. doi: 10.1016/j.cell.2024.05.039
98. Ang MY, Low TY, Lee PY, Wan Mohamad Nazarie WF, Guryev V, Jamal R. Proteogenomics: From next-generation sequencing (NGS) and mass spectrometry-based proteomics to precision medicine. Clin Chim Acta. (2019) 498:38–46. doi: 10.1016/j.cca.2019.08.010
99. Aebersold R, Mann M. Mass-spectrometric exploration of proteome structure and function. Nature. (2016) 537(7620):347–55. doi: 10.1038/nature19949
100. Nesvizhskii AI. Proteogenomics: concepts, applications and computational strategies. Nat Methods. (2014) 11(11):1114–25. doi: 10.1038/nmeth.3144
101. Rivers RC, Kinsinger C, Boja ES, Hiltke T, Mesri M, Rodriquez H. Linking cancer genome to proteome: NCI's investment into proteogenomics. Proteomics. (2014) 14(23-24):2633–6. doi: 10.1002/pmic.201400193
102. Zhang B, Whiteaker JR, Hoofnagle AN, Baird GS, Rodland KD, Paulovich AG. Clinical potential of mass spectrometry-based proteogenomics. Nat Rev Clin Oncol. (2019) 16(4):256–68. doi: 10.1038/s41571-018-0135-7
103. Whiteaker JR, Halusa GN, Hoofnagle AN, Sharma V, MacLean B, Yan P, et al. Using the CPTAC assay portal to identify and implement highly characterized targeted proteomics assays. Methods Mol Biol. (2016) 1410:223–36. doi: 10.1007/978-1-4939-3524-6_13
104. Hu Y, Pan J, Shah P, Ao M, Thomas SN, Lui Y, et al. Integrated proteomic and glycoproteomic characterization of human high-grade serous ovarian carcinoma. Cell Rep. (2020) 33(3):108276. doi: 10.1016/j.celrep.2020.108276
105. Zhang YW, Gvozdenovic A, Aceto N. A molecular voyage: Multiomics insights into circulating tumor cells. Cancer Discovery. (2024) 14(6):920–33. doi: 10.1158/2159-8290.CD-24-0218
106. Nordgård O, Tjensvoll K, Gilje B, Søreide K. Circulating tumour cells and DNA as liquid biopsies in gastrointestinal cancer. Br J Surg. (2018) 105(2):e110–20. doi: 10.1002/bjs.10782
107. Huang KL, Li S, Mertins P, Cao S, Gunawardena HP, Ruggles KV, et al. Proteogenomic integration reveals therapeutic targets in breast cancer xenografts. Nat Commun. (2017) 8:14864. doi: 10.1038/ncomms14864
108. Zhang H, Liu T, Zhang Z, Payne SH, Zhang B, McDermott JE, et al. Integrated proteogenomic characterization of human high-grade serous ovarian cancer. Cell. (2016) 166(3):755–65. doi: 10.1016/j.cell.2016.05.069
109. Mani DR, Krug K, Zhang B, Satpathy S, Clauser KR, Ding L, et al. Cancer proteogenomics: current impact and future prospects. Nat Rev Cancer. (2022) 22(5):298–313. doi: 10.1038/s41568-022-00446-5
110. McDermott JE, Arshad OA, Petyuk VA, Fu Y, Gritsenko MA, Clauss TR, et al. Proteogenomic characterization of ovarian HGSC implicates mitotic kinases, replication stress in observed chromosomal instability. Cell Rep Med. (2020) 1(1):100004. doi: 10.1016/j.xcrm.2020.100004
111. Na JR, Liu Y, Fang K, Tan Y, Liang PP, Yan M, et al. Unraveling the potential biomarkers of immune checkpoint inhibitors in advanced ovarian cancer: a comprehensive review. Invest New Drugs. (2024) 42(6):728–38. doi: 10.1007/s10637-024-01478-4
112. Ding Y, Ye Z, Ding B, Feng S, Zhang Y, Shen Y. Identification of CXCL13 as a promising biomarker for immune checkpoint blockade therapy and PARP inhibitor therapy in ovarian cancer. Mol Biotechnol. (2024), 1–15. doi: 10.1007/s12033-024-01207-5
113. Lampert EJ, Zimmer A, Padget M, Cimino-Mathews A, Nair JR, Liu Y, et al. Combination of PARP inhibitor olaparib, and PD-L1 inhibitor durvalumab, in recurrent ovarian cancer: a proof-of-Concept phase II study. Clin Cancer Res. (2020) 26(16):4268–79. doi: 10.1158/1078-0432.CCR-20-0056
114. Poveda A, Lheureux S, Colombo N, Cibula D, Lindemann K, Weberpals J, et al. Olaparib maintenance monotherapy in platinum-sensitive relapsed ovarian cancer patients without a germline BRCA1/BRCA2 mutation: OPINION primary analysis. Gynecol Oncol. (2022) 164(3):498–504. doi: 10.1016/j.ygyno.2021.12.025
115. Swisher EM, Lin KK, Oza AM, Scott CL, Giordano H, Sun J, et al. Rucaparib in relapsed, platinum-sensitive high-grade ovarian carcinoma (ARIEL2 part 1): an international, multicentre, open-label, phase 2 trial. Lancet Oncol. (2017) 18(1):75–87. doi: 10.1016/S1470-2045(16)30559-9
116. Coleman RL, Oza AM, Lorusso D, Aghajanian C, Oaknin A, Dean A, et al. Rucaparib maintenance treatment for recurrent ovarian carcinoma after response to platinum therapy (ARIEL3): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet. (2017) 390(10106):1949–61. doi: 10.1016/S0140-6736(17)32440-6
117. Mirza MR, Monk BJ, Herrstedt J, Oza AM, Mahner S, Redondo A, et al. Niraparib maintenance therapy in platinum-sensitive, recurrent ovarian cancer. N Engl J Med. (2016) 375(22):2154–64. doi: 10.1056/NEJMoa1611310
118. Coleman RL, Fleming GF, Brady MF, Swisher EM, Steffensen KD, Friedlander M, et al. Veliparib with first-line chemotherapy and as maintenance therapy in ovarian cancer. N Engl J Med. (2019) 381(25):2403–15. doi: 10.1056/NEJMoa1909707
Keywords: ovarian cancer, PARP inhibitor, proteogenomics, immunotherapy, biomarkers, antibody drug conjugate (ADC)
Citation: Philips TJ, Erickson BK and Thomas SN (2025) Opportunities for predictive proteogenomic biomarkers of drug treatment sensitivity in epithelial ovarian cancer. Front. Oncol. 14:1503107. doi: 10.3389/fonc.2024.1503107
Received: 28 September 2024; Accepted: 12 December 2024;
Published: 07 January 2025.
Edited by:
Liang Xue, Pfizer, United StatesReviewed by:
Nicole James, Women & Infants Hospital of Rhode Island, United StatesCopyright © 2025 Philips, Erickson and Thomas. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Stefani N. Thomas, c3RlZmFuaXRAdW1uLmVkdQ==
 Trudy J. Philips
Trudy J. Philips Britt K. Erickson
Britt K. Erickson Stefani N. Thomas
Stefani N. Thomas