- Department of Experimental Medicine, Sapienza University of Rome, Rome, Italy
Pancreatic ductal adenocarcinoma (PDAC) is the most aggressive and invasive type of pancreatic cancer and is expected to soon become the second leading cause of cancer-associated death. The high mortality rate is due to the clinical features that allow asymptomatic progression to advanced stages, a period when current therapeutic treatments have limited efficacy. To address these challenges, researchers are focused on identifying new molecular and circulating markers for early PDAC detection and precision medicine. In this mini-review, we report the most well-known and recently identified molecular and circulating biomarkers. This study aimed to emphasize the need for continued innovative research to develop diagnostic algorithms and therapies to improve the management of patients with PDAC.
1 Introduction
Pancreatic ductal adenocarcinoma (PDAC) remains one of the most challenging and devastating cancers and is characterized by late detection and aggressive biology that often leads to therapeutic failure (1). The number of pancreatic cancer cases has been increasing steadily, making it the second leading cause of cancer-related deaths, surpassing breast cancer and projected to soon surpass colorectal cancer (2, 3). The poor prognosis of PDAC is primarily attributed to the lack of effective early detection methods and inherent chemoresistance (1, 4–6). Due to the broad genetic heterogeneity and dense stromal environment of pancreatic tumors, current treatment options, including surgery, radiation, chemotherapy, immunotherapy, and targeted therapies, have yielded only marginal improvements in overall survival (4). To address these challenges, recent PDAC research has focused on a wide range of biomarkers, exploring their potential from multiple perspectives for early detection and improved disease management (7, 8). Some studies have specifically examined circulating tumor DNA (ctDNA) levels in plasma samples from patients with early-stage (I-III) pancreatic adenocarcinoma who are potential candidates for surgery. This review, however, centers on biomarkers for early disease detection, aiming to enhance strategies for the timely diagnosis of PDAC. We explore these emerging biomarkers and highlight their potential to transform PDAC diagnosis and improve patient outcomes through earlier therapeutic interventions.
2 Molecular markers
2.1 Key molecular biomarkers
Molecular markers play a crucial role in the diagnosis and prognosis of several cancers. Among these, key markers such as KRAS, TP53, SMAD4 and CDKN2A have been identified in PDAC (10). The complex genetic and molecular landscape of PDAC, including its heterogeneity and tumor microenvironment, further highlights the importance of these markers (11).
2.1.1 KRAS
Kirsten rat sarcoma viral oncogene homolog (KRAS) is the most well-known oncogene and has the highest mutation rate across all cancers (12). In PDAC, KRAS mutations serve as a major driver of the disease. These mutations are critical for PDAC development and progression, underscoring the importance of KRAS as a target for research and therapeutic interventions. In PDAC the most common alterations are predominantly found in codon 12, specifically G12D, G12V, and G12R (13, 14) and are often associated with poorer therapy response and overall survival (14). While G12D, G12V, and G12R mutations are the most frequent, G12R mutations have been linked to longer survival, particularly when not accompanied by PI3K pathway alterations (15). The development of small-molecule inhibitors targeting the KRAS pathway provides several treatment options for PDAC patients (16). Moreover, recent advancements have led to the development of irreversible covalent inhibitors for the KRAS-G12C variant, which have shown promise in clinical trials (17). Other strategies, such as targeting the MAPK signaling pathway to inhibit KRAS function, have also shown potential (18). Despite these advancements, challenges remain in developing small-molecule inhibitors that directly target KRAS, including the need for further optimization and the potential for resistance (19). However, the mutational status of KRAS may not be sufficient to identify tumors that are effectively dependent on it. Instead, the identification of specific KRAS-driven molecular biomarkers in certain PDAC subtypes may be more beneficial for tailoring targeted therapies (20). A subset of PDACs, known as KRAS-WT, exhibit a distinct molecular profile, including mutations in TP53 and BRAF, and are enriched with targetable alterations, suggesting the potential for expanded therapeutic options (21, 22).
2.1.2 TP53
The TP53 gene, which encodes the tumor suppressor protein p53, has garnered significant attention as a molecular marker in PDAC. Notably, the inactivation of TP53, particularly in combination with KRAS activation, has been shown to drive the development of PDAC (23). TP53 mutations are a common in this neoplasm, with a prevalence of 50-75% (24, 25). These mutations, especially gain-of-function mutations, play a significant role in promoting disease progression, including metastasis (24, 26). The presence of TP53 mutations is also associated with a poorer prognosis and resistance to antineoplastic therapies (27). Therefore, detecting and characterizing TP53 mutations in PDAC could be crucial for predicting patient outcomes and guiding treatment decisions. Missense mutations in TP53 have been found to enhance fibrosis and create an immunosuppressive microenvironment in PDAC tumor, potentially contributing to treatment resistance (28). Mutant TP53 can also alter the tumor microenvironment and immune responses, further promoting PDAC progression (26). Additionally, the introduction of wild-type TP53 has been found to increase the sensitivity of PDAC cells to various treatments, suggesting a potential therapeutic strategy (26).
2.1.3 SMAD4 (DPC4)
SMAD4, also known as DPC4 (Deleted in Pancreatic Cancer 4), belongs to a family of signal transduction proteins and it is considered a pivotal tumor suppressor gene involved in the TGF-β signaling pathway, which regulates cell growth, differentiation, and apoptosis. It is frequently inactivated in PDAC (29). Protein inactivation occurs either through deletion or mutation, making SMAD4 a critical molecular marker for this malignancy. SMAD4, has been suggested as a valuable diagnostic marker for PDAC, with higher expression compared to other cancers (30). Its loss is significantly associated with poor prognosis (31), and patients with SMAD4-expressing PDACs tend to have longer survival following surgical resection (32). The presence or absence of SMAD4 in PDAC significantly influences the response to specific therapies. SMAD4-positive tumors have been found to respond better to neoadjuvant therapy, with lower rates of lymph node metastasis (33, 34). SMAD4 status also affects the response to radiation therapy, with SMAD4-deleted PDAC showing worsened disease-free survival (35). Furthermore, SMAD4 loss has been associated with a response to neoadjuvant chemotherapy with the autophagy inhibitor hydroxychloroquine (36). These findings highlight the potential of SMAD4 as a marker for predicting treatment response and guiding personalized therapeutic strategies in PDAC.
2.1.4 CDKN2A (p16)
Another common genetic alteration in PDAC is the deletion or mutation of the CDKN2A gene, which encodes the p16 tumor suppressor protein. Studies have shown a higher frequency of deletions and mutations in this gene in PDAC cell lines compared to primary tumors (37, 38). These alterations, including frameshift, nonsense, and missense mutations, as well as homozygous deletions, are associated with the development and progression of the disease (6, 39). The presence of these alterations can also impact the biological behavior and clinical staging of PDAC, potentially aiding in prognosis (39). These findings underscore the importance of genetic and epigenetic alterations, including those in the CDKN2A gene, in the development and progression of this malignancy (40).
2.2 New potential biomarkers
A promising avenue in cancer research focuses on identifying specific molecular alterations unique to pancreatic ductal adenocarcinoma (PDAC). These include genetic mutations, epigenetic changes, and abnormal signaling pathways that may serve as targets for early detection and individualized treatment. Recent investigations have highlighted potential molecular biomarkers for PDAC, with the most promising candidates being BRCA1/2, MLL, as well as Gastrokine proteins and Nucleoporins. These emerging markers offer new opportunities for developing targeted therapies and improving diagnostic precision in PDAC management. Recent studies have identified promising therapeutic targets and diagnostic biomarkers in PDAC, including CCNB1, FHL, HLA-DPA1, and TUBB1. These markers may shed light on the underlying molecular mechanisms of pancreatic cancer and contribute to the development of more precise diagnostic tools and targeted treatments. Their role in cell cycle regulation, immune response, and tumor progression underscores their potential in enhancing early detection and therapeutic strategies, offering new hope for improving patient outcomes in this aggressive malignancy (41). However, further in-depth research is essential to validate the clinical utility of these markers and ensure their successful integration into standard diagnostic and therapeutic regimens. This step is critical to confirming their efficacy, reliability, and potential in improving patient outcomes, particularly in early detection and treatment personalization for PDAC.
2.2.1 BRCA1/2
The tumor suppressor genes BRCA1 and BRCA2 are involved in essential cellular functions necessary for cell replication and DNA synthesis, playing a crucial role in DNA repair and tumor suppression (31). BRCA1/2 mutations, present in approximately 10% of pancreatic cancer cases, have emerged as important biomarkers (42). Mutation in BRCA1 and BRCA2 are associated with an increased risk of pancreatic cancer (43–45). BRCA1 expression patterns in PDAC may serve as potential prognostic biomarkers, with cytosolic BRCA1 distribution linked to higher pathologic stage and potentially decreased recurrence-free survival (31). BRCA-associated pancreatic cancers have demonstrated greater sensitivity to platinum-based chemotherapy and PARP inhibitors, with patients showing partial responses to these treatments (45–47).
2.2.2 MLL
The Mixed-Lineage Leukemia (MLL) family of proteins, which includes several histone methyltransferases such as MLL1 (KMT2A), MLL2 (KMT2B), MLL3 (KMT2C), and MLL4 (KMT2D), plays a crucial role in regulating gene expression through histone modifications, particularly the methylation of histone H3 lysine 4 (H3K4). These modifications are critical for controlling chromatin structure and gene transcription, which are essential for normal cellular function and development (48). The role of the MLL family in the context of PDAC is attracting considerable interest due to its involvement in epigenetic regulation and cancer progression. However, the use of MLL as a molecular marker in PDAC remains a topic of ongoing debate, with some studies suggesting that mutations in MLL1, MLL2 and MLL3, are associated with improved survival (10). The MLL1-H3K4me3 axis has been implicated in regulating PD-L1 expression and immune evasion in pancreatic cancer, suggesting a potential role for MLL in this context as well (49).
2.2.3 Gastrokine proteins
Gastrokines (GKNs) are stomach-specific proteins with potential roles in gastric mucosal homeostasis and tumor suppression (50). The loss of GKN expression in various cancers, combined with its antiproliferative effects, suggests a tumor suppressor role for these proteins (50–52). GKN1 is abundantly expressed in normal gastric epithelium but is downregulated in gastric adenomas and carcinomas (52, 53). Additionally, overexpression of GKN1 in gastric cancer cells inhibits proliferation and induces cell death (52). Recent research has extended these findings to pancreatic cancer, showing that GKN proteins are expressed in early lesions of pancreatic intraepithelial neoplasm (PanIN), but are absent in healthy pancreas and invasive cancer (51). GKN1 and GKN2 appear to delay pancreatic carcinogenesis by promoting apoptosis and inhibiting proliferation in precursor lesions (51).
2.2.4 Nucleoporins (Nup170 and Nup160)
Nucleoporins (Nups) are proteins considered the building blocks of the nuclear pore complexes and have been linked to a multitude of cancers through nucleo-cytoplasmic cargo trafficking, cell division, signaling pathways, chromatin-related processes, and protein stability and degradation (54). Regarding PDAC, Nup 170 and Nup160, have been identified as potential molecular markers for the early diagnosis this neoplasm (55). Furthermore, the genetic inactivation of Nupr1, a key nucleoporin that mediates stress response in the pancreas, and is frequently upregulated in pancreatic cancer, has been shown to suppress malignant transformation and influence the development of different PDAC subtypes (56). These findings suggest that nucleoporins play a significant role in PDAC progression and may have potential as therapeutic targets.
2.2.5 CCNB1
The gene encoding the protein Cyclin B1 (CCNB1), an important cell cycle regulator involved in mitosis, has recently been identified as a potential prognostic biomarker in PDAC (57). Bioinformatics analyses of gene expression data revealed CCNB1 as a significantly upregulated gene in PDAC tissues (41, 57). CCNB1 was found to be involved in key cellular processes such as vesicle organization, lymphocyte activation, and adaptive immune responses (41). Validation studies confirmed the differential expression of CCNB1 in PDAC compared to normal tissues, supporting its potential as a diagnostic and prognostic marker (57). Additionally, the inhibition of Cyclin Dependent Kinase-1 (CDK-1), a protein that interacts with Cyclin B1, has shown promise as a novel therapeutic strategy against PDAC (58). These findings collectively underscore the potential of targeting CCNB1 and its associated proteins in the treatment of this neoplasm.
2.2.6 FHL
The Four and a Half LIM (FHL) family is a group of proteins characterized by the presence of LIM domains, which are double zinc finger motifs involved in protein-protein interactions. These proteins play critical roles in various cellular processes, including gene expression, cell adhesion, cytoskeletal organization, and signal transduction. FHL plays a significant role in PDAC progression and treatment resistance. In particular, FHL1 can act as both a tumor suppressor and an oncogenic protein, depending on its phosphorylation status and interactions with various signaling pathways (59). In PDAC, FHL1 expression is regulated by Immortalization-upregulated protein (IMUP) (60) and Nucleophosmin1 (NPM1) (61): depletion of IMUP increases FHL1 expression, while overexpression of NPM1 reduces it through promoter methylation (62). FHL2, in PDAC, has been shown to play a crucial role in cell survival, proliferation, and radio-resistance (63). Indeed, overexpression of FHL2 is associated with tumor metastasis in PDAC (64). In contrast, its depletion leads to reduced cell survival and increased apoptosis (63). Finally, FHL3 has also been reported to promote PDAC invasion and metastasis by inhibiting ubiquitin degradation of EMT-associated transcription factors (65). The findings highlights that these proteins may serve as promising therapeutic targets in PDAC.
2.2.7 HLA-DPA1
HLA-DPA1 belongs to the human leukocyte antigen (HLA) class II alpha chain paralogs and plays a central role in the immune system by presenting peptides derived from extracellular proteins. In PDAC, the expression of classical HLA class II antigens, including HLA-DPA1, has been associated with a poor prognosis (66). Similarly, high expression of HLA-G, a tolerogenic molecule implicated in tumor escape, has been linked to a poor prognosis in PDAC (66). These findings suggest a potential role for HLA-DPA1 in disease progression and the immune response, warranting further investigation.
2.2.8 TUBB1
TUBB1 Albahde (2020) found that TUBB1, a member of the tubulin superfamily, is overexpressed in PDAC, with its downregulation leading to reduced cellular proliferation, invasiveness, and tumor growth (67). Recent bioinformatics analyses of gene expression data have revealed several hub genes, including TUBB1, as potential biomarkers in PDAC (41). These genes are involved in various cellular processes, such as vesicle organization, lymphocyte activation, and adaptive immune responses. Collectively, these findings suggest that TUBB1 may serve as a potential therapeutic target and prognostic marker in PDAC.
3 Circulating biomarkers
Over the past decade, a wide range of potential circulating biomarkers has been described for PDAC, with serum being the most utilized matrix due to its ease of collection and non-invasive nature. Despite their routine use, current biomarkers such as CA19-9 and CEA exhibit significant limitations in sensitivity and specificity. Consequently, their clinical application is confined to monitoring treatment response and recurrence in diagnosed patients, and they remain insufficient for screening or early diagnosis, which is a critical challenge. To streamline current research findings, we provide an updated review of established PDAC biomarkers.
3.1 Currently used biomarker
3.1.2 CA19-9
Carbohydrate antigen 19-9 (CA19-9), also known as sialyl Lewis a (sLea), is currently the only widely used and approved biomarker for the pancreatic cancer diagnosis and monitoring (68, 69). However, CA19-9 has several limitations, such as yielding false negatives in patients with a Lewis blood type-negative phenotype and false positives in patients with obstructive jaundice (70). Moreover, CA19-9 is not tumor-type-specific, as its elevation can be observed in various malignancies, including those originating from the colorectum, gastric system, lung, breast, and liver, as well as in pancreatic neuroendocrine tumors (71). However, with the new frontiers of scientific research, despite its limitations, CA19-9 is finding new diagnostic roles, especially when used in combination with other biomarkers (71). In this regard, in PDAC, CA242 serum levels highly correlate with CA19-9 (72, 73), however, its complementary role to CA19-9 in the early diagnosis of PDAC remains unclear.
One of the most intriguing new functions of CA19-9 lies in its ability to accelerate pancreatic cancer progression by glycosylating proteins, binding to E-selectin, enhancing angiogenesis, and mediating the immunological response. This renders CA19-9 an attractive therapeutic target for cancer. Currently, therapeutic strategies employing CA19-9 to treat pancreatic cancer include using this molecule to develop specific anti-CA19-9 monoclonal antibodies that enable antibody-dependent cell-mediated cytotoxicity (ADCC) (71, 74). Additionally, CA19-9 can be targeted for drug delivery by binding it to nanoparticles (75). Another approach involves disrupting its biosynthesis by interfering with FUT3, a fucosyltransferase involved in the synthesis of Lewis antigens, and other enzymes (76). Therefore, targeting CA19-9 may offer novel therapeutic options, as interrupting CA19-9 could hinder the development and progression of PDAC.
3.1.3 CEA
Carcinoembryonic antigen (CEA) is a glycoprotein typically produced by normal cells during embryonic development and tends to increase in presence of inflammatory processes or in tumors of the gastrointestinal tract. Due to its lower sensitivity and specificity compared to CA19-9 in early diagnosis of PDAC (77, 78), this biomarker cannot be relied upon for solitary diagnostic use. However, it may help to discriminate between benign and malignant pancreatic lesions when used in combination with CA19-9 (79), particularly in cases of advanced PDAC (80).
3.2 New potential circulating biomarkers
Due to the aggressive nature and rapid progression of PDAC, early diagnosis presents a significant challenge. The swift development of the tumor underscores the urgent need for effective biomarkers that can facilitate timely detection and improve patient outcomes. As a result, research efforts are increasingly focused on identifying novel biomarkers for early detection. Several molecules, either alone or in combination with conventional markers, have shown significant potential in improving the early diagnosis of this neoplasm. Among the circulating molecules currently under investigation, matrix metalloproteinases (MMPs) and their tissue inhibitors (TIMP-1), growth differentiation factor 15 (GDF15), and protein induced by vitamin K absence (PIVKA II) have garnered particular attention for their potential in enhancing diagnostic sensitivity and specificity compared to traditional markers. These promising biomarkers could play a crucial role in early diagnosis and personalized therapy of PDAC.
3.2.1 MMPs
Matrix metalloproteinases (MMPs) are a family of zinc-dependent endopeptidases crucial for the remodeling of the extracellular matrix (ECM) (81). Initially thought to promote pre-metastatic development by degrading the ECM and basement membranes, MMPs also release soluble factors for cellular recruitment and express growth factor receptors in metastatic cells. Recent studies reveal that MMPs play a role in all stages of tumor progression, influencing signaling pathways, cytokine regulation, tumor growth, neo-angiogenesis, and cancer spread (82). During PC progression, MMPs drive both tumor growth and metastasis, making changes in their expression important surrogate markers. Pharmacological treatment or genetic ablation has been shown to reduce MMP levels, suggesting their potential anti-tumorigenic effects, which are currently under study (83). Regarding their role as biomarkers in Pancreatic cancer, combining MMP-7 with CA19-9 in periampullary carcinoma patients achieved a 100% positive predictive value (84). Additionally, Kahlert et al. found that serum levels of MMP-7 and MMP-12 are strong classifiers for diagnosing Pancreatic cancer compared to healthy controls (85).
3.2.2 TIMP-1
Tissue inhibitors of metalloproteinases (TIMPs) play a crucial role in regulating extracellular matrix (ECM) turnover, tissue remodeling, and cellular behavior through interactions with MMPs. Maintaining this balance is critical, as disruptions can signal cancer progression (86). Among the TIMPs, TIMP-1 is notably linked to cancer, influencing cell proliferation, apoptosis, and differentiation. TIMP-1 also interacts with activated neutrophils via CD63, activating the ERK signaling pathway and promoting the formation of neutrophil extracellular traps (NETs) (87). In PDAC, TIMP-1 is linked to gemcitabine (GEM) resistance, although the impact of TIMP-1 downregulation in combination with GEM treatment remains unclear (88).
One of the most intriguing aspects of TIMP-1 lies in its ability to interact with activated neutrophils by binding to its receptor CD63. This interaction activates ERK, and the noncanonical TIMP1/CD63/ERK signaling axis induces the formation of NETs (87). This process accelerates tumorigenesis, progression, and therapy resistance in PDAC, where NETs frequently co-localize with high TIMP-1 expression (87, 89). In PDAC patients, TIMP-1 levels correlate with NET markers such as DNA-bound myeloperoxidase, suggesting an enhancement of prognostic accuracy when combined with CA19-9 (87). Additionally, TIMP-1 plasma levels are associated with the monocyte activation marker CD163, indicating that these proteins, when combined, could serve as powerful prognostic indicators (90).
3.2.3 GDF15
Growth differentiation factor-15 (GDF-15), also known as MIC-1, belongs to the TGF-β superfamily and is typically undetectable under normal physiological conditions but its expression increases during cellular stress and pathological states (91). Originally identified in macrophages, elevated GDF-15 levels have been linked to various malignancies, making it a potential biomarker for disease (92–94). In response to stress signals, GDF-15 is released as a dimer through a specific secretory pathway, binds to the extracellular matrix, and undergoes cleavage into its active forms (95). It binds with high affinity to the GDNF family receptor α-like (GFRAL), which forms a complex with the tyrosine kinase co-receptor RET, activating signaling cascades involving the AKT, ERK1/2, and PLC γ pathways, but not the SMAD pathway (96). Elevated GDF-15 levels also influence immunological checkpoints, such as PD-1/PD-L1 (91), contributing to tumor development and suggesting its potential as a diagnostic marker for PDAC (96).
GDF-15 has also been implicated in the development of chemoresistance in PDAC. High levels of GDF-15 have been associated with resistance to commonly used chemotherapy drugs for PDAC, such as gemcitabine (GEM). This resistance mechanism can hinder the effectiveness of chemotherapy and worsen patient outcomes. Targeting GDF-15 signaling pathways has emerged as a potential therapeutic strategy for PDAC. Inhibition of GDF-15 or its downstream effectors may help inhibit tumor growth, metastasis, and chemoresistance in PDAC. Research efforts are underway to develop GDF-15-targeted therapies for the treatment of PDAC. Further research into the molecular mechanisms underlying GDF-15’s role in PDAC is needed to develop effective targeted therapies and biomarker-based diagnostic approaches (97).
3.2.4 PIVKA II
Protein Induced by Vitamin K absence or Antagonist-II (PIVKA-II), also known as DCP (Des-γcarboxy prothrombin), is an abnormal form of prothrombin, the precursor of Coagulation Factor II, and its synthesis is Vitamin K-dependent. Ten glutamic acid residues in prothrombin undergo carboxylation by reduced Vitamin K; in its absence, PIVKA-II accumulates instead of maturing into prothrombin, thus indicating Vitamin K deficiency (98, 99). Initially identified with elevated levels in hepatocellular carcinoma (HCC), PIVKA-II has also been found in various gastrointestinal neoplasms, such as PDAC, colorectal cancer (CRC), and cholangiocarcinoma (98, 100). However, the mechanisms underlying PIVKA-II overexpression in these cancers, including HCC and PDAC, remain incompletely understood. While genetic mutations are not the primary cause, recent evidence points to factors such as hypoxia in the microenvironment, reduced gamma-glutamyl carboxylase activity, and altered Vitamin K metabolism contributing to PIVKA-II hyperproduction (101–104). Additional factors implicated include the downregulation of the VKORC1 gene in HCC and increased PARP-1 activity stimulating prothrombin gene transcription, potentially explaining elevated PIVKA-II in this cancer (105, 106). Although research has predominantly focused on HCC, similar mechanisms likely underlie PIVKA-II elevation in PDAC and other gastrointestinal cancers. Recent studies indicate PIVKA-II’s potential as a diagnostic biomarker in PDAC, showing promising diagnostic performance compared to CA19-9 and CEA (107). PIVKA-II not only helps distinguish benign and malignant pancreatic conditions but also aids in monitoring disease recurrence post-surgery, with levels decreasing following resection (108). Studies also suggest PIVKA-II may serve as a predictive factor for vascular invasion in PDAC (99). While its exact role in PDAC is still being elucidated, in vitro studies using PANC-1 cells suggest PIVKA-II secretion correlates with glucose-induced epithelial-mesenchymal transition (EMT), hinting at its role in disease aggressiveness (109). These observations have also been reinforced by a recent study that highlighted the association of elevated PIVKA-II levels with increased vimentin levels in the serum of PDAC patients (110). Taken together, these results open new frontiers in the study and characterization of this biomarker in pancreatic cancer. However, further research is needed to fully understand PIVKA-II’s implications and potential therapeutic targets.
Table 1 summarizes the most relevant evidence on Molecular and Circulating biomarkers in PDAC.
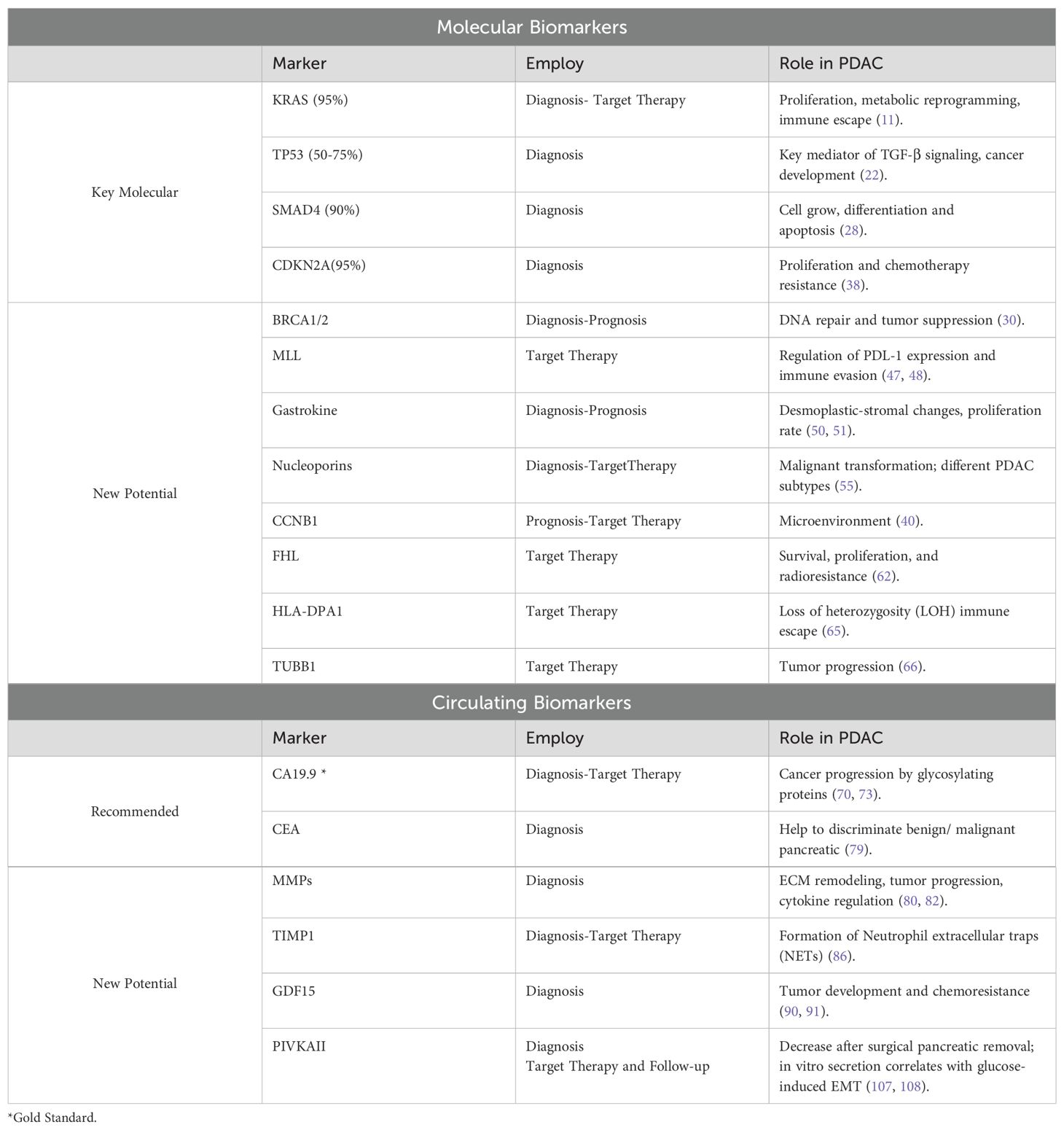
Table 1. A comprehensive overview of the clinical utility and established roles of molecular and circulating biomarkers in PDAC.
4 Conclusion
Pancreatic ductal adenocarcinoma (PDAC) remains a challenging malignancy with a poor prognosis, emphasizing the need for effective biomarkers to enhance diagnosis, treatment selection, and patient outcomes (46). Recent advances in molecular profiling have identified potential biomarkers for early diagnosis, prognosis, and targeted therapies (11, 111). Molecular markers such as KRAS and SMAD4, and novel circulating markers like GDF-15, PIVKA-II, and TIMP-1, show promise in enhancing diagnostic accuracy and prognostic evaluations. Additionally, genetic alterations in KRAS G12C, BRCA1/2, and HLA-DPA1 are emerging as predictive biomarkers for targeted treatments, including PARP inhibitors and immunotherapies (46, 112). As seen from the exploration of various emerging molecular and circulating biomarkers, research is increasingly focusing on proteins involved in the interplay between the tumor and its microenvironment. This shift underscores the importance of understanding the dynamic interactions within the tumor microenvironment, which are crucial in cancer progression and metastasis. As research advances, the integration of molecular and circulating biomarkers into clinical practice holds the potential to enable more personalized and effective treatment strategies for PDAC patients (46, 113, 114). In addition to the emerging biomarkers discussed, serum ctDNA levels have also shown great promise for diagnosing PDAC, detecting minimal residual disease (MRD), evaluating therapeutic response, and facilitating early detection of recurrence. ctDNA offers greater sensitivity and specificity compared to traditional antigen biomarkers, although standard clinical guidelines for its use are still under review and validation (9).
Finally, the development of sophisticated diagnostic algorithms incorporating these biomarkers could revolutionize PDAC management. Artificial intelligence (AI), combined with these algorithms, can analyze complex datasets by integrating molecular, genetic, and clinical information, resulting in more precise disease management. In conclusion, this mini-review underscores the importance of emerging molecular and circulating biomarkers, alongside the necessity of integrating them into diagnostic algorithms. Such integration aims to directly translate research findings into clinical practice, establishing a comprehensive approach to managing PDAC patients at all disease stages (Figure 1). Ongoing research and technological advancements will be essential to improving the early diagnosis and treatment of PDAC.
Figure 1. Features of PDAC Molecular and Circulating Biomarkers. The interplay between molecular and circulating biomarkers could represent a new strategy in the management of PDAC.
Author contributions
MM: Data curation, Project administration, Writing – original draft. AF: Data curation, Project administration, Writing – original draft. AA: Supervision, Writing – review & editing. EA: Supervision, Funding acquisition, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was funded by University of Rome la Sapienza, Ateneo 2022 n°AR22218167FCBC4B.
Acknowledgments
We are thankful to Valentina Viggiani, Giuseppina Gennarini. Francesca Cortese, and Barbara Colaprisca for their technical assistance and to D. Kanton for providing language help.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Orth M, M. P, Gerum S, Mayerle J, Schneider G, Belka C, et al. Pancreatic ductal adenocarcinoma: biological hallmarks, current status, and future perspectives of combined modality treatment approaches. Radiat Oncol. (2019) 14:141. doi: 10.1186/s13014-019-1345-6
2. Halbrook CJ, L. C, Pasca di Magliano M, Maitra A. Pancreatic cancer: Advances and challenges. Cell. (2023) 186:1729–54. doi: 10.1016/j.cell.2023.02.014
3. Badheeb M, A. A, Esmail A, Umoru G, Abboud K, Al-Najjar E, et al. Pancreatic tumorigenesis: precursors, genetic risk factors and screening. Curr Oncol. (2022) 29:8693–719. doi: 10.3390/curroncol29110686
4. Adamska A, D. A, Falasca M. Pancreatic ductal adenocarcinoma: current and evolving therapies. Int J Mol Sci. (2017) 18:1338. doi: 10.3390/ijms18071338
5. Falcone R, Davis PJ, Stain SC, Mousa SA. Emerging therapies for pancreatic ductal carcinoma. J Solid Tumors. (2016) 6:65. doi: 10.5430/jst.v6n1P65
6. Bartsch D, S. D, Tung WS, Kisker O, Wells SA Jr, Goodfellow PJ. Frequent mutations of CDKN2 in primary pancreatic adenocarcinomas. Genes Chromosomes Cancer. (1995) 14:189–95. doi: 10.1002/gcc.2870140306
7. Bugazia D, A.-N. E, Esmail A, Abdelrahim S, Abboud K, Abdelrahim A, et al. Pancreatic ductal adenocarcinoma: the latest on diagnosis, molecular profiling, and systemic treatments. Front Oncol. (2024) 14:1386699. doi: 10.3389/fonc.2024.1386699
8. Esmail A, Badheeb M, Abdelrahim M. Pancreatic Tumorigenesis: Precursors, Genetic Risk Factors and Screening [Internet]. Pancreatic Cancer- Updates in Pathogenesis, Diagnosis and Therapies. IntechOpen (2023). doi: 10.5772/intechopen.110887
9. Botta GP, Abdelrahim M, Drengler RL, Aushev VN, Esmail A, Laliotis G, et al. Association of personalized and tumor-informed ctDNA with patient survival outcomes in pancreatic adenocarcinoma. Oncologist. (2024) 29(10):859–69. doi: 10.1093/oncolo/oyae155
10. Aldyab M, E. JT, Parilla M, Lee H. Benign vs Malignant pancreatic lesions: Molecular insights to an ongoing debate. World J Gastrointest Surg. (2021) 13:406–18. doi: 10.4240/wjgs.v13.i5.406
11. Taherian M, W. H, Wang H. Pancreatic ductal adenocarcinoma: molecular pathology and predictive biomarkers. Cells. (2022) 11:3068. doi: 10.3390/cells11193068
12. Mondal K, P. M, Shenoy RP, Roychoudhury S. KRAS mutation subtypes and their association with other driver mutations in oncogenic pathways. Cells. (2024) 13:1221. doi: 10.3390/cells13141221
13. Stickler S, R. B, Hamilton G. Targeting KRAS in pancreatic cancer. Oncol Res. (2024) 32:799–805. doi: 10.32604/or.2024.045356
14. Nusrat F, K. A, Jain A, Jiang W, Lavu H, Yeo CJ, et al. The clinical implications of KRAS mutations and variant allele frequencies in pancreatic ductal adenocarcinoma. J Clin Med. (2024) 13:2103. doi: 10.3390/jcm13072103
15. Diehl JN, H. P, Ozkan-Dagliyan I, Goodwin CM, Howard SV, Cox AD, et al. Targeting the ERK mitogen-activated protein kinase cascade for the treatment of KRAS-mutant pancreatic cancer. Adv Cancer Res. (2022) 153:101–30. doi: 10.1016/bs.acr.2021.07.008
16. Dai M, C. S, Teng X, Chen K, Cheng W. KRAS as a key oncogene in the clinical precision diagnosis and treatment of pancreatic cancer. J Cancer. (2022) 13:3209–20. doi: 10.7150/jca.76695
17. Tang X, Ren H, Guo M, Qian J, Yang Y, Gu C. Review on circular RNAs and new insights into their roles in cancer. Comput Struct Biotechnol J. (2021) 19:910–28. doi: 10.1016/j.csbj.2021.01.018
18. Korzeniecki C, Priefer R. Targeting KRAS mutant cancers by preventing signaling transduction in the MAPK pathway. Eur J Med Chem. (2021) 211:113006. doi: 10.1016/j.ejmech.2020.113006
19. Chen H, S. J, Liu T, Ding K, Lu X. Small-molecule inhibitors directly targeting KRAS as anticancer therapeutics. J Med Chem. (2020) 63:14404–24. doi: 10.1021/acs.jmedchem.0c01312
20. Mottini C, C. L. Beyond the genomic mutation: rethinking the molecular biomarkers of K-RAS dependency in pancreatic cancers. Int J Mol Sci. (2020) 2020:21. doi: 10.3390/ijms21145023
21. Philip PA, A. I, Xiu J, Hall MJ, Hendifar AE, Lou E, et al. Molecular characterization of KRAS wild-type tumors in patients with pancreatic adenocarcinoma. Clin Cancer Res. (2022) 28:2704–14. doi: 10.1158/1078-0432.CCR-21-3581
22. Grinshpun A, Z. Y, Roszik J, Subbiah V, Hubert A. Beyond KRAS: practical molecular targets in pancreatic adenocarcinoma. Case Rep Oncol. (2019) 12:7–13. doi: 10.1159/000496018
23. Kim MP, L. X, Deng J, Zhang Y, Dai B, Allton KL, et al. Oncogenic KRAS Recruits an Expansive Transcriptional Network through Mutant p53 to Drive Pancreatic Cancer Metastasis. Cancer Discovery. (2021) 11:2094–111. doi: 10.1158/2159-8290.CD-20-1228
24. Morton JP, Timpson P, Karim SA, Ridgway RA, Athineos D, Doyle B, et al. Mutant p53 drives metastasis and overcomes growth arrest/senescence in pancreatic cancer. Proc Natl Acad Sci U.S.A. (2010) 107:246–51. doi: 10.1073/pnas.0908428107
25. Malvi D, V. F, Maloberti T, Sanza V, De Leo A, Fornelli A, et al. Molecular characterization of pancreatic ductal adenocarcinoma using a next-generation sequencing custom-designed multigene panel. Diagnostics (Basel). (2022) 12:1058. doi: 10.3390/diagnostics12051058
26. McCubrey JA, Y. L, Abrams SL, Steelman LS, Follo MY, Cocco L, et al. Effects of TP53 mutations and miRs on immune responses in the tumor microenvironment important in pancreatic cancer progression. Cells. (2022) 11:2155. doi: 10.3390/cells11142155
27. Kim SP, V. N, Zacharakis N, Krishna S, Yu Z, Gasmi B, et al. Adoptive cellular therapy with autologous tumor-infiltrating lymphocytes and T-cell receptor-engineered T cells targeting common p53 neoantigens in human solid tumors. Cancer Immunol Res. (2022) 10:932–46. doi: 10.1158/2326-6066.CIR-22-0040
28. Maddalena M, M. G, Nataraj NB, Shreberk-Shaked M, Hassin O, Mukherjee S, et al. TP53 missense mutations in PDAC are associated with enhanced fibrosis and an immunosuppressive microenvironment. Proc Natl Acad Sci U S A. (2021) 118:e2025631118. doi: 10.1073/pnas.2025631118
29. Zhao M, M. L, Deng CX. The role of TGF-β/SMAD4 signaling in cancer. Int J Biol Sci. (2018) 14:111–23. doi: 10.7150/ijbs.23230
30. Ali S, C. C, Little JV, Sequeira JH, Mosunjac MB, Siddiqui MT. The utility of SMAD4 as a diagnostic immunohistochemical marker for pancreatic adenocarcinoma, and its expression in other solid tumors. Diagn Cytopathol. (2007) 35:644–8. doi: 10.1002/dc.20715
31. Wang WQ, L. L, Xu HX, Wu CT, Xiang JF, Xu J, et al. Infiltrating immune cells and gene mutations in pancreatic ductal adenocarcinoma. Br J Surg. (2016) 103:1189–99. doi: 10.1002/bjs.10187
32. Tascilar M, S. H, Rosty C, Sohn T, Wilentz RE, Offerhaus GJ, et al. The SMAD4 protein and prognosis of pancreatic ductal adenocarcinoma. Clin Cancer Res. (2001) 7:4115–21.
33. Kassardjian A, Wang HL. SMAD4-expressing pancreatic ductal adenocarcinomas have better response to neoadjuvant therapy and significantly lower lymph node metastasis rates. Pancreas. (2020) 49(9):1153–60. doi: 10.1097/MPA.0000000000001636
34. Hsieh YY, L. T, Chou CJ, Chen HY, Lee KH, Yang PM. Integration of bioinformatics resources reveals the therapeutic benefits of gemcitabine and cell cycle intervention in SMAD4-deleted pancreatic ductal adenocarcinoma. Genes (Basel). (2019) 10:766. doi: 10.3390/genes10100766
35. Racu ML, B. D, Chaouche A, Zindy E, Navez J, Loi P, et al. SMAD4 positive pancreatic ductal adenocarcinomas are associated with better outcomes in patients receiving FOLFIRINOX-based neoadjuvant therapy. Cancers (Basel). (2023) 15:3765. doi: 10.3390/cancers15153765
36. Fei N, W. S, Ramanathan R, Hogg ME, Zureikat AH, Lotze MT, et al. SMAD4 loss is associated with response to neoadjuvant chemotherapy plus hydroxychloroquine in patients with pancreatic adenocarcinoma. Clin Transl Sci. (2021) 14:1822–9. doi: 10.1111/cts.13029
37. Huang L, G. T, Zhang SY, Klein-Szanto AJ, Chang H, Ruggeri BA. Deletion and mutation analyses of the P16/MTS-1 tumor suppressor gene in human ductal pancreatic cancer reveals a higher frequency of abnormalities in tumor-derived cell lines than in primary ductal adenocarcinomas. Cancer Res. (1996) 56:1137–41.
38. Qian Y, G. Y, Fan Z, Luo G, Huang Q, Deng S, et al. Molecular alterations and targeted therapy in pancreatic ductal adenocarcinoma. J Hematol Oncol. (2020) 13:130. doi: 10.1186/s13045-020-00958-3
39. Zhou J, Y. D, Zhang L, Wang J, Yao Q, Su Z, et al. Study on the relationship of alteration and expression of p16 gene to pancreatic carcinoma. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. (2000) 17:399–403.
40. Delpu Y, H. N, Lulka H, Sicard F, Selves J, Buscail L, et al. Genetic and epigenetic alterations in pancreatic carcinogenesis. Curr Genomics. (2011) 12:15–24. doi: 10.2174/138920211794520132
41. Giriyappagoudar M, V. B, Horakeri R, Vastrad C. Identification and interaction analysis of molecular markers in pancreatic ductal adenocarcinoma by bioinformatics and next-generation sequencing data analysis. Bioinform Biol Insights. (2023) 17:11779322231186719. doi: 10.1177/11779322231186719
42. Martinez-Useros J, Garcia-Foncillas J. Can molecular biomarkers change the paradigm of pancreatic cancer prognosis? BioMed Res Int. (2016) 2016:4873089. doi: 10.1155/2016/4873089
43. Lee YC, L. Y, Li CY. BRCA genes and related cancers: A meta-analysis from epidemiological cohort studies. Medicina (Kaunas). (2021) 57:905. doi: 10.3390/medicina57090905
44. McGarry JL, C. B, Kelly ME, Gallagher TK. Risk of pancreatic ductal adenocarcinoma associated with carriage of BRCA1 and/or BRCA2 mutation: A systematic review and meta-analysis. J Surg Oncol. (2022) 126:1028–37. doi: 10.1002/jso.26994
45. Lowery MA, K. D, Stadler ZK, Yu KH, Janjigian YY, Ludwig E, et al. An emerging entity: pancreatic adenocarcinoma associated with a known BRCA mutation: clinical descriptors, treatment implications, and future directions. Oncologist. (2011) 16:1397–402. doi: 10.1634/theoncologist.2011-0185
46. Brozos-Vázquez E, T.-F. M, Costa-Fraga N, García-Ortiz MV, Díaz-Lagares Á, Rodríguez-Ariza A, et al. Pancreatic cancer biomarkers: A pathway to advance in personalized treatment selection. Cancer Treat Rev. (2024) 125:102719. doi: 10.1016/j.ctrv.2024.102719
47. Golan T, K. Z, Epelbaum R, Devaud N, Dagan E, Holter S, et al. Overall survival and clinical characteristics of pancreatic cancer in BRCA mutation carriers. Br J Cancer. (2014) 111:1132–8. doi: 10.1038/bjc.2014.418
48. Jamali MB, Shi J. Unveiling the molecular landscape of pancreatic ductal adenocarcinoma: insights into the role of the COMPASS-like complex. Int J Mol Sci. (2024) 25:5069. doi: 10.3390/ijms25105069
49. Lu C, P. A, Shi H, Savage N, Waller JL, Sabbatini ME, et al. The MLL1-H3K4me3 axis-mediated PD-L1 expression and pancreatic cancer immune evasion. J Natl Cancer Inst. (2017) 109:djw283. doi: 10.1093/jnci/djw283
50. Menheniott TR, K. B, Giraud AS. Gastrokines: stomach-specific proteins with putative homeostatic and tumor suppressor roles. Am J Physiol Gastrointest Liver Physiol. (2013) 304:G109–21. doi: 10.1152/ajpgi.00374.2012
51. Steiner S, S. G, Reding T, Stopic M, Lenggenhager D, Ten Buren E, et al. De novo expression of gastrokines in pancreatic precursor lesions impede the development of pancreatic cancer. Oncogene. (2022) 41:1507–17. doi: 10.1038/s41388-022-02182-4
52. Yoon JH, S. J, Zhang C, Jin M, Kang YH, Nam SW, et al. Inactivation of the Gastrokine 1 gene in gastric adenomas and carcinomas. J Pathol. (2011) 223:618–25. doi: 10.1002/path.2838
53. Oien KA, M. F, Butler S, Ferrier RK, Downie I, Bryce S, et al. Gastrokine 1 is abundantly and specifically expressed in superficial gastric epithelium, down-regulated in gastric carcinoma, and shows high evolutionary conservation. J Pathol. (2004) 203:789–97. doi: 10.1002/path.1583
54. Jahangiri L. A mechanistic insight into cancer progression mediated by Nucleoporins. Cancer Genet. (2024) 286–287:35–42. doi: 10.1016/j.cancergen.2024.07.001
55. Shen Q, Y. M, Jia JK, Li WX, Tian YW, Xue HZ. Possible molecular markers for the diagnosis of pancreatic ductal adenocarcinoma. Med Sci Monit. (2018) 24:2368–76. doi: 10.12659/msm.906313
56. Cano CE, H. T, Garcia MN, Grasso D, Loncle C, Garcia S, et al. Genetic inactivation of Nupr1 acts as a dominant suppressor event in a two-hit model of pancreatic carcinogenesis. Gut. (2014) 63:984–95. doi: 10.1136/gutjnl-2013-305221
57. Zeng Y, Fan R. Identification and verification of CCNB1 as a potential prognostic biomarker by comprehensive analysis. Sci Rep. (2022) 12(1):16153. doi: 10.1038/s41598-022-20615-8
58. Wijnen R, P. C, Carbone D, Fiuji H, Avan A, Peters GJ, et al. Cyclin dependent kinase-1 (CDK-1) inhibition as a novel therapeutic strategy against pancreatic ductal adenocarcinoma (PDAC). Cancers (Basel). (2021) 13:4389. doi: 10.3390/cancers13174389
59. Wei X, Zhang H. Four and a half LIM domains protein 1 can be as a double-edged sword in cancer progression. Cancer Biol Med. (2020) 17(2):270–81. doi: 10.20892/j.issn.2095-3941.2019.0420
60. Wei R, Qi G, Zeng Z, Shen N, Wang Z, Shen H, et al. IMUP and GPRC5A: two newly identified risk score indicators in pancreatic ductal adenocarcinoma. Cancer Cell Int. (2021) 21:620. doi: 10.1186/s12935-021-02324-w
61. Karimi Dermani F, G. KS, Afshar S, Amini R. The potential role of nucleophosmin (NPM1) in the development of cancer. J Cell Physiol. (2021) 236:7832–52. doi: 10.1002/jcp.30406
62. Luo Q, P. Y, Fu Q, Zhang X, Zhou S, Yu P, et al. Immortalization-upregulated protein promotes pancreatic cancer progression by regulating NPM1/FHL1-mediated cell-cycle-checkpoint protein activity. Cell Biol Toxicol. (2023) 39:2069–87. doi: 10.1007/s10565-022-09695-4
63. Zienert E, E. I, Aust D, Cordes N. LIM-only protein FHL2 critically determines survival and radioresistance of pancreatic cancer cells. Cancer Lett. (2015) 364:17–24. doi: 10.1016/j.canlet.2015.04.019
64. Verset L, F. L, Trépant AL, De Wever O, Demetter P. FHL2: a scaffold protein of carcinogenesis, tumour-stroma interactions and treatment response. Histol Histopathol. (2016) 31:469–78. doi: 10.14670/HH-11-709
65. Li P, C. G, Zhang Y, Shi J, Cai K, Zhen L, et al. FHL3 promotes pancreatic cancer invasion and metastasis through preventing the ubiquitination degradation of EMT associated transcription factors. Aging (Albany NY). (2020) 12:53–69. doi: 10.18632/aging.102564
66. Hiraoka N, I. Y, Hori S, Yamazaki-Itoh R, Naito C, Shimasaki M, et al. Expression of classical human leukocyte antigen class I antigens, HLA-E and HLA-G, is adversely prognostic in pancreatic cancer patients. Cancer Sci. (2020) 111:3057–70. doi: 10.1111/cas.14514
67. Albahde MAH, Z. P, Zhang Q, Li G, Wang W. Upregulated expression of TUBA1C predicts poor prognosis and promotes oncogenesis in pancreatic ductal adenocarcinoma via regulating the cell cycle. Front Oncol. (2020) 10:49. doi: 10.3389/fonc.2020.00049
68. O'Neill RS, Stoita A. Biomarkers in the diagnosis of pancreatic cancer: Are we closer to finding the golden ticket? World J Gastroenterol. (2021) 27(26):4045–87. doi: 10.3748/wjg.v27.i26.4045
69. Yang J, Xu R, Wang C, Qiu J, Ren B, You L. Early screening and diagnosis strategies of pancreatic cancer: a comprehensive review. Cancer Commun (Lond). (2021) 41(12):1257–74. doi: 10.1002/cac2.12204
70. Scarà S, B. P, Scatena R. CA 19-9: biochemical and clinical aspects. Adv Exp Med Biol. (2015) 867:247–60. doi: 10.1007/978-94-017-7215-0_15
71. Luo G, J. K, Deng S, Cheng H, Fan Z, Gong Y, et al. Roles of CA19-9 in pancreatic cancer: Biomarker, predictor and promoter. Biochim Biophys Acta Rev Cancer. (2021) 1875:188409. doi: 10.1016/j.bbcan.2020.188409
72. Dong D, J. L, Zhang L, Ma N, Zhang A, Zhou Y, et al. Periostin and CA242 as potential diagnostic serum biomarkers complementing CA19.9 in detecting pancreatic cancer. Cancer Sci. (2018) 109:2841–51. doi: 10.1111/cas.13712
73. Dou H, S. G, Zhang L. CA242 as a biomarker for pancreatic cancer and other diseases. Prog Mol Biol Transl Sci. (2019) 162:229–39. doi: 10.1016/bs.pmbts.2018.12.007
74. Kaji M, I. H, Kishimoto T, Omi M, Ishizu A, Kimura C, et al. E-selectin expression induced by pancreas-carcinoma-derived interleukin-1 alpha results in enhanced adhesion of pancreas-carcinoma cells to endothelial cells. Int J Cancer. (1995) 60:712–7. doi: 10.1002/ijc.2910600524
77. Xing H, W. J, Wang Y, Tong M, Hu H, Huang C, et al. Diagnostic value of CA 19-9 and carcinoembryonic antigen for pancreatic cancer: A meta-analysis. Gastroenterol Res Pract. (2018) 2018:8704751. doi: 10.1155/2018/8704751
78. Nazli O, B. A, Tansug T, Kir R, Kaymak E. The diagnostic importance of CEA and CA 19-9 for the early diagnosis of pancreatic carcinoma. Hepatogastroenterology. (2000) 47:1750–2.
79. Lei XF, J. S, Ye J, Qiao YL, Zhao GM, Li XH, et al. Application values of detection of serum CA199, CA242 and CA50 in the diagnosis of pancreatic cancer. J Biol Regul Homeost Agents. (2017) 31:383–8.
80. van Manen L, G. J, Putter H, Vahrmeijer AL, Swijnenburg RJ, Bonsing BA, et al. Elevated CEA and CA19-9 serum levels independently predict advanced pancreatic cancer at diagnosis. Biomarkers. (2020) 25:186–93. doi: 10.1080/1354750X.2020.1725786
81. Niland S, R. A, Eble JA. Matrix metalloproteinases shape the tumor microenvironment in cancer progression. Int J Mol Sci. (2021) 23:146. doi: 10.3390/ijms23010146
82. de Almeida LGN, Thode H, Eslambolchi Y, Chopra S, Young D, Gill S, et al. Matrix metalloproteinases: from molecular mechanisms to physiology, pathophysiology, and pharmacology. Pharmacol Rev. (2022) 74(3):712–68. doi: 10.1124/pharmrev.121.000349
83. Slapak EJ, D. J, Tekin C, Bijlsma MF, Spek CA. Matrix metalloproteases in pancreatic ductal adenocarcinoma: key drivers of disease progression? Biol (Basel). (2020) 9:80. doi: 10.3390/biology9040080
84. Kuhlmann KF, v.T. J, Boermeester MA, de Reuver PR, Tzvetanova ID, Offerhaus GJ, et al. Evaluation of matrix metalloproteinase 7 in plasma and pancreatic juice as a biomarker for pancreatic cancer. Cancer Epidemiol Biomarkers Prev. (2007) 16:886–91. doi: 10.1158/1055-9965.EPI-06-0779
85. Kahlert C, F. M, Musso G, Halama N, Keim S, Mazzone M, et al. Prognostic impact of a compartment-specific angiogenic marker profile in patients with pancreatic cancer. Oncotarget. (2014) 5:12978–89. doi: 10.18632/oncotarget.2651
86. Cabral-Pacheco GA, G.-V. I, Castruita-De la Rosa C, Ramirez-Acuña JM, Perez-Romero BA, Guerrero-Rodriguez JF, et al. The roles of matrix metalloproteinases and their inhibitors in human diseases. Int J Mol Sci. (2020) 21:9739. doi: 10.3390/ijms21249739
87. Schoeps B, E. C, Prokopchuk O, Böttcher J, Häußler D, Steiger K, et al. TIMP1 triggers neutrophil extracellular trap formation in pancreatic cancer. Cancer Res. (2021) 81:3568–79. doi: 10.1158/0008-5472.CAN-20-4125
89. Zhang Y, C. V, Riquelme Sanchez E, Dutta P, Quesada PR, Rakoski A, et al. Interleukin-17-induced neutrophil extracellular traps mediate resistance to checkpoint blockade in pancreatic cancer. J Exp Med. (2020) 217:e20190354. doi: 10.1084/jem.20190354
90. Eckfeld C, S. B, Häußler D, Frädrich J, Bayerl F, Böttcher JP, et al. TIMP-1 is a novel ligand of Amyloid Precursor Protein and triggers a proinflammatory phenotype in human monocytes. J Cell Biol. (2023) 222:e202206095. doi: 10.1083/jcb.202206095
91. Wischhusen J, M. I, Fridman WH. Growth/differentiation factor-15 (GDF-15): from biomarker to novel targetable immune checkpoint. Front Immunol. (2020) 11:951. doi: 10.3389/fimmu.2020.00951
92. Pavo N, Wurm R, Neuhold S, Adlbrecht C, Vila G, Strunk G, et al. GDF-15 is associated with cancer incidence in patients with type 2 diabetes. Clin Chem. (2016) 62:1612–20. doi: 10.1373/clinchem.2016.257212
93. Yin T, C. S, Chen X. RNPC1, an RNA-binding protein and a p53 target, regulates macrophage inhibitory cytokine-1 (MIC-1) expression through mRNA stability. J Biol Chem. (2013) 288:23680–6. doi: 10.1074/jbc.M113.480186
94. Guo M, Zhao H. Growth differentiation factor-15 may be a novel biomarker in pancreatic cancer: A review. Med (Baltimore). (2024) 103(6):e36594. doi: 10.1097/MD.0000000000036594
95. Wang X, Baek SJ, Eling TE. The diverse roles of nonsteroidal anti-inflammatory drug activated gene (NAG-1/GDF15) in cancer. Biochem Pharmacol. (2013) 85:597–606. doi: 10.1016/j.bcp.2012.11.025
96. Emmerson PJ, W. F, Du Y, et al. The metabolic effects of GDF15 are mediated by the orphan receptor GFRAL. Nat Med. (2017) 23:1215–9. doi: 10.1038/nm.4393
97. Ozkan H, Demirbaş S, Ibiş M, Akbal E, Köklü S. Diagnostic validity of serum macrophage inhibitor cytokine and tissue polypeptide-specific antigen in pancreatobiliary diseases. Pancreatology. (2011) 11:295–300. doi: 10.1159/000328963
98. Dong L, Q. X, Gao F, Wang K, Xu X. Protein induced by vitamin K absence or antagonist II: Experience to date and future directions. Biochim Biophys Acta Rev Cancer. (2023) 1878:189016. doi: 10.1016/j.bbcan.2023.189016
99. Yang Y, L. G, Zhang Y, Cui Y, Liu J. Protein induced by vitamin K absence II: A potential biomarker to differentiate pancreatic ductal adenocarcinoma from pancreatic benign lesions and predict vascular invasion. J Clin Med. (2023) 12:2769. doi: 10.3390/jcm12082769
100. Takano S, H. I, Watanabe S, Soda H, Nagata M, Hoshino I, et al. PIVKA-II-producing advanced gastric cancer. Int J Clin Oncol. (2004) 9:330–3. doi: 10.1007/s10147-004-0398-0
101. Murata K, S. H, Okano H, Oyamada T, Yasuda Y, Sakamoto A. Hypoxia-induced des-gamma-carboxy prothrombin production in hepatocellular carcinoma. Int J Oncol. (2010) 36:161–70.
102. Marrero JA. Screening tests for hepatocellular carcinoma. Clin Liver Dis. (2005) 9:235–51. doi: 10.1016/j.cld.2004.12.006
103. Dong R, W. N, Yang Y, Ma L, Du Q, Zhang W, et al. Review on vitamin K deficiency and its biomarkers: focus on the novel application of PIVKA-II in clinical practice. Clin Lab. (2018) 64:413–24. doi: 10.7754/Clin.Lab.2017.171020
104. Hao Z, J. D, Chen X, Schurgers LJ, Stafford DW, Tie JK. [amp]]gamma;-Glutamyl carboxylase mutations differentially affect the biological function of vitamin K-dependent proteins. Blood. (2021) 137:533–43. doi: 10.1182/blood.2020006329
105. Yang X, W. Z, Zandkarimi F, Liu Y, Duan S, Li Z, et al. Regulation of VKORC1L1 is critical for p53-mediated tumor suppression through vitamin K metabolism. Cell Metab. (2023) 35:1474–90. doi: 10.1016/j.cmet.2023.06.014
106. Taniguchi T, K. K, Nakagawa T, Tanaka H, Tanaka T, Tomonari T, et al. Poly-(ADP-ribose) polymerase-1 promotes prothrombin gene transcription and produces des-gamma-carboxy prothrombin in hepatocellular carcinoma. Digestion. (2017) 95:242–51. doi: 10.1159/000470837
107. Tartaglione S, P. I, Zarrillo SR, Granato T, Viggiani V, Manganaro L, et al. Protein Induced by Vitamin K Absence II (PIVKA-II) as a potential serological biomarker in pancreatic cancer: a pilot study. Biochem Med (Zagreb). (2019) 29:20707. doi: 10.11613/BM.2019.020707
108. Tartaglione S, M. P, Viggiani V, Chirletti P, Angeloni A, Anastasi E. PIVKA-II: A biomarker for diagnosing and monitoring patients with pancreatic adenocarcinoma. PloS One. (2021) 16:e0251656. doi: 10.1371/journal.pone.0251656
109. Farina A, T. S, Preziosi A, Mancini P, Angeloni A, Anastasi E. PANC-1 cell line as an experimental model for characterizing PIVKA-II production, distribution, and molecular mechanisms leading to protein release in PDAC. Int J Mol Sci. (2024) 25:3498. doi: 10.3390/ijms25063498
110. Farina A, V. V, Cortese F, Moretti M, Tartaglione S, Angeloni A, et al. Combined PIVKA II and vimentin-guided EMT tracking in pancreatic adenocarcinoma combined biomarker-guided EMT tracking in PDAC. Cancers (Basel). (2024) 16:2362. doi: 10.3390/cancers16132362
111. Bärthel S, F. C, Rad R, Theis FJ, Saur D. Single-cell profiling to explore pancreatic cancer heterogeneity, plasticity and response to therapy. Nat Cancer. (2023) 4:454–67. doi: 10.1038/s43018-023-00526-x
112. Jazieh KA, F. M, Diaz LA Jr. The clinical utility of biomarkers in the management of pancreatic adenocarcinoma. Semin Radiat Oncol. (2014) 24:67–76. doi: 10.1016/j.semradonc.2013.11.007
113. Hu ZI, O'Reilly EM. Therapeutic developments in pancreatic cancer. Nat Rev Gastroenterol Hepatol. (2024) 21(1):7–24. doi: 10.1038/s41575-023-00840-w
Keywords: pancreas, adenocarcinoma, markers, diagnosis, target therapy
Citation: Moretti M, Farina A, Angeloni A and Anastasi E (2024) Emerging horizons on molecular and circulating biomarkers in pancreatic adenocarcinoma. Front. Oncol. 14:1483306. doi: 10.3389/fonc.2024.1483306
Received: 19 August 2024; Accepted: 17 October 2024;
Published: 07 November 2024.
Edited by:
Terence Moyana, The Ottawa Hospital, CanadaReviewed by:
Abdullah Esmail, Houston Methodist Hospital, United StatesCopyright © 2024 Moretti, Farina, Angeloni and Anastasi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Emanuela Anastasi, ZW1hbnVlbGEuYW5hc3Rhc2lAdW5pcm9tYTEuaXQ=
†These authors have contributed equally to this work and share first authorship
 Marta Moretti
Marta Moretti Antonella Farina
Antonella Farina Antonio Angeloni
Antonio Angeloni Emanuela Anastasi
Emanuela Anastasi