- 1Department of Medicine, Keck School of Medicine of the University of Southern California, Los Angeles, CA, United States
- 2Department of Pathology, Caris Life Sciences, Phoenix, AZ, United States
- 3Department of Clinical and Translational Research, Caris Life Sciences, Phoenix, AZ, United States
- 4Department of Pathology, City of Hope National Cancer Center, Duarte, CA, United States
- 5Department of Pathology, Affiliated Pathologists Medical Group, Orange, CA, United States
- 6Department of Obstetrics and Gynecology, University of California Irvine School of Medicine, Irvine, CA, United States
The discovery of gene fusions involving Neuregulin-1 (NRG1) within solid tumors has important therapeutic implications, as they are being actively explored as targets for emerging ERBB/ERBB2/ERBB3-directed anti-cancer agents. NRG1 fusions are very uncommon across all tumor types and are infrequently documented in the medical literature. We report a female patient presenting with widespread peritoneal carcinomatosis diagnosed as high grade serous fallopian tube carcinoma, which harbored a previously undescribed MYH10::NRG1 fusion. Moreover, we queried the whole transcriptome sequencing results of neoplasms analyzed by a commercial laboratory (Caris Life Sciences) to determine the overall incidence of NRG1 fusions in carcinomas of the ovary, fallopian tube, and peritoneum (0.18%). Twenty-five additional tumors were found to demonstrate NRG1 fusions, including 20 new genes partners that had not been previously identified in gynecologic carcinomas. Overall, NRG1 fusion events are rare in ovarian, fallopian tube, and primary peritoneal carcinomas, but they may carry diagnostic significance in the context of borderline/low grade serous tumors, which demonstrated exclusively CLU::NRG1 fusions, and could have important predictive implications for response to ERBB/ERBB2/ERBB3-directed therapies.
Introduction
Neuregulin-1 gene (NRG1) exists as six distinct isoforms that modulate the Erb-B2 receptor tyrosine kinase (ERBB) receptor pathways to influence ERBB2/ERBB3 protein signaling via an epidermal growth factor (EGF) family protein (1). Fusions including NRG1, in which it typically occurs as the 3’ partner, disrupt the normal growth and differentiation processes in epithelial and other cell types, and instead constitutively activate the phosphatidylinositol 3-kinase (PI3K) and mitogen-activated protein kinase (MAPK) signaling pathways through an intact EGF domain (2). To date, several 5’ fusion genes partners have been identified, with the most frequent being CD74 and SLC3A2 (3). However, with new diagnostic methodologies, the list of gene partners continues to expand. NRG1 fusions have been detected across solid tumor types with an incidence of 0.2%, with the greatest occurrence rate being observed in cholangiocarcinoma, pancreatic ductal adenocarcinoma, and renal cell carcinoma (0.5% for each) (3).
In one study of 21,858 tumor specimens (3), 0.4% of ovarian, fallopian tube, and primary peritoneal carcinomas were found to harbor NRG1 fusions. Within these tumors, the gene was fused to SETD4, TSHZ2, and ZMYM2. Recent research has also reported RAB3IL1 and CLU as partners (4). While rare, appropriate identification of the NRG1 fusion is essential, as it likely represents the oncogenic driver and occurs mutually exclusive with other targetable genetic alterations (5). Because differences in testing methodology can influence reliable detection of clinically actionable fusions involving genes like NRG1 and NTRK1/2/3 (6), the American Society of Clinical Oncology has advocated for the use of RNA sequencing (7). Ultimately, the presence of an NRG1 fusion may be important, in that it could predict response treatment. Our study highlights clinical course of single patient who, to-date, has experienced a good response to traditional platinum-based chemotherapy utilized during the management of gynecologic malignancies. Moreover, our work expands the spectrum of reported fusions involving NRG1 and novel partner genes, ultimately considering the utility of ERBB/ERBB2/ERBB3-directed anti-cancer agents, such as afitinib, tarloxotinib, seribantumab, zenocutuzumab, and others, that are under investigation for the treatment of NRG1 fusion-positive tumors (8–10).
Methods
Paraffin-embedded tumor samples were analyzed by DNA (592-gene or whole exome) and RNA (whole transcriptome) sequencing at Caris Life Sciences (Phoenix, AZ), utilizing a customized Agilent SureSelect Human All Exon V7 bait panel (Santa Clara, CA) and Illumina NovaSeq technology (San Diego, CA). NRG1 fusions were identified within the samples if ≥3 junction reads were detected by RNA sequencing. A board-certified pathologist (M.G.E.) evaluated those specimens with NRG1 fusions to render a pathologic diagnosis. In order to evaluate differential gene expression present within the NRG1 fusion-positive cases, Gene Set Enrichment Analysis (GSEA) was carried out using whole transcriptome data and the Hallmark gene set collection from the Human Molecular Signatures Database (11, 12). Transcriptomic signatures of the NRG1 fusion-positive tumors were compared to those of previously tested low grade (728 samples) and high grade (7,818 samples) serous carcinomas of the ovary, fallopian tube, or peritoneum lacking an NRG1 fusion.
Results
Of the 14,395 samples obtained from ovarian, fallopian tube, or primary peritoneal carcinomas that were tested by whole transcriptome sequencing, 26 were shown to harbor NRG1 fusions (incidence of 0.18%) between 2019 and 2023. A diversity of gene partners were identified and most were novel: CLU (n=4), SARAF (n=2), SCAF4 (n=1), MUC16 (n=1), WFDC2 (n=1), NOTCH2 (n=1), APP (n=1), SPIDR (n=1), HGSNAT (n=1), SPINT2 (n=1), RBPMS (n=1), JAG1 (n=1), NRP2 (n=1), CHMP4C (n=1), CXADR (n=1), TMEM65 (n=1), INSR (n=1), ADAM9 (n=1), LDLR (n=1), TNFRSF12A (n=1), SPON1 (n=1), and MYH10 (n=1). NRG1 breakpoints were observed in either exon 2 (46%, n=12) or exon 6 (54%, n=14); exon 2 splicing preserves the gene’s immunoglobulin and EGF domains, but only the EGF domain remains intact with exon 6 splicing. All fusion-positive samples were diagnosed as borderline/low grade or high grade serous carcinoma, but one case harboring a RBPMS::NRG1 fusion was classified as clear cell carcinoma. Of note, the CLU::NRG1 fusion was only observed in borderline/low grade cases.
One high grade tumor was classified as tumor mutational burden (TMB)-high with >10 mutations/megabase (Muts/Mb). Additionally, three cases were notable for homologous recombination deficiency (HRD) as indicated by pathogenic variants in BRCA1 and/or BRCA2, or a high genomic scar score that combines genomic loss of heterozygous (gLOH) with large-scale state transitions (LST). Those tumors with HRD were all diagnosed as high grade serous carcinoma, specifically harboring fusions involving NRG1 and the JAG1, SPON1, and TNFRSF12A partner genes.
Pathogenic genetic changes in addition to the NRG1 fusions and HRD-associated mutations that were detected within the NRG1 fusion-positive tumors are summarized in Supplementary Table 1. GSEA pathway analysis of those 26 neoplasms demonstrated that the G2M checkpoint pathway, E2F transcription factors, and KRAS downstream signaling were upregulated compared to fusion-negative, low grade serous carcinomas of the ovary, fallopian tube, or peritoneum (Figure 1). Moreover, compared to low grade serous carcinoma, the cancers harboring NRG1 fusions demonstrated statistically lower expression of TNFA signaling genes and those associated with the epithelial-mesenchymal transition. There were no significant pathways enriched when comparing NRG1 fusion-positive tumors with fusion-negative high grade serous carcinomas.
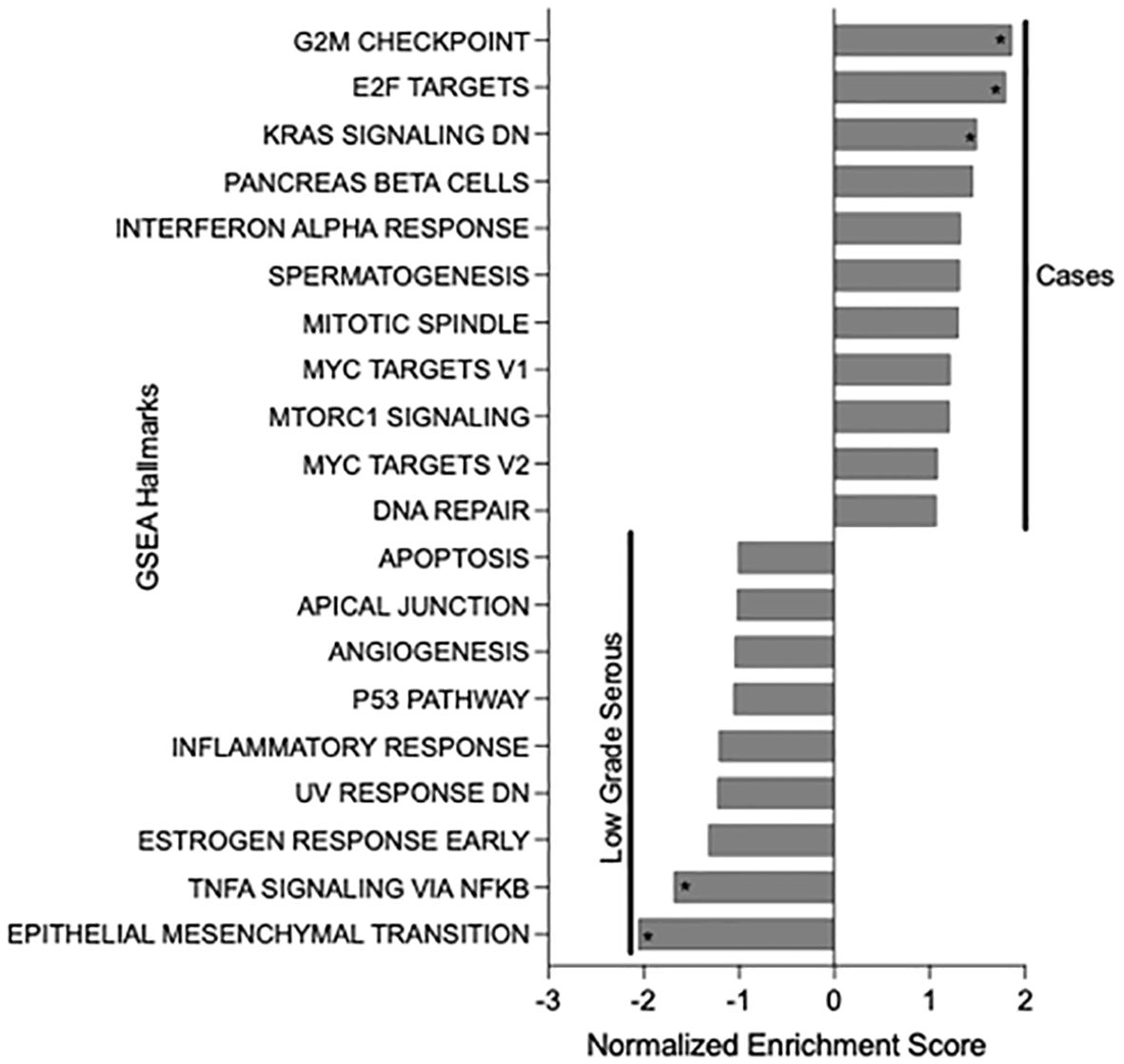
Figure 1. Gene Set Enrichment Analysis (GSEA) results of NRG1 fusion-positive tumor cases compared to low grade serous carcinomas of the female genital tract. Asterisks indicate statistically significance differences (p-value <0.05).
Case report
A 75-year-old G1P1 woman presented to the emergency department with a several-week history of fatigue and shortness of breath. In the prior several days, she had also developed a cough and abdominal bloating that were interfering with her daily life and had become alarming. Her past medical history included essential hypertension, and she endorsed an approximately 21-year smoking history. Upon evaluation, that patient was found to have decreased right-sided breath sounds and mild abdominal distention. Her laboratory studies were unremarkable with the exception of a CA-125 level of 357 units/milliliter. A chest radiograph demonstrated right lung opacities concerning for pneumonia and a right-sided pleural effusion. Thoracentesis was performed, and the patient was surprised to learn that microscopic examination of the pleural fluid revealed scattered nests of atypical cells arranged in papillary structures, demonstrating moderate amounts of eosinophilic cytoplasm and nuclei with prominent nucleoli, pleomorphism, and mitoses (Figure 2A). The cells were positive for PAX8, WT1, and ER by immunohistochemistry, consistent with high grade serous fallopian tube carcinoma. Whole transcriptome sequencing identified a MYH10 exon 3::NRG1 exon 2 fusion (Figure 2B), and exomic analysis revealed TP53 (p.K132R) and RET (p.C618Y) pathogenic single nucleotide variants, as well as a TMB of 3 Muts/Mb. Subsequent CT imaging demonstrated a large right pleural effusion and peritoneal carcinomatosis (Figure 3).
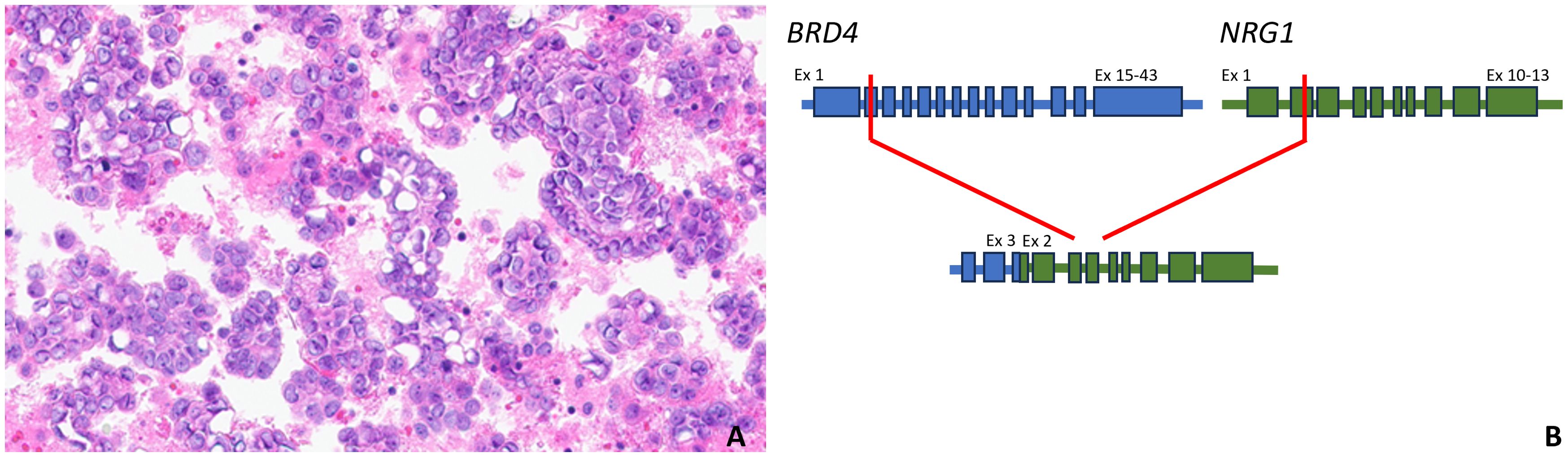
Figure 2. Hematoxylin and eosin-stained high grade serous carcinoma metastatic to pleural fluid harboring a MYH10 exon 3::NRG1 exon 2 fusion (400x magnification, (A); a pictorial representation of the MYH10::NRG1 gene fusion, which includes the intact immunoglobulin and EGF domains of NRG1. Ex, exon (B).
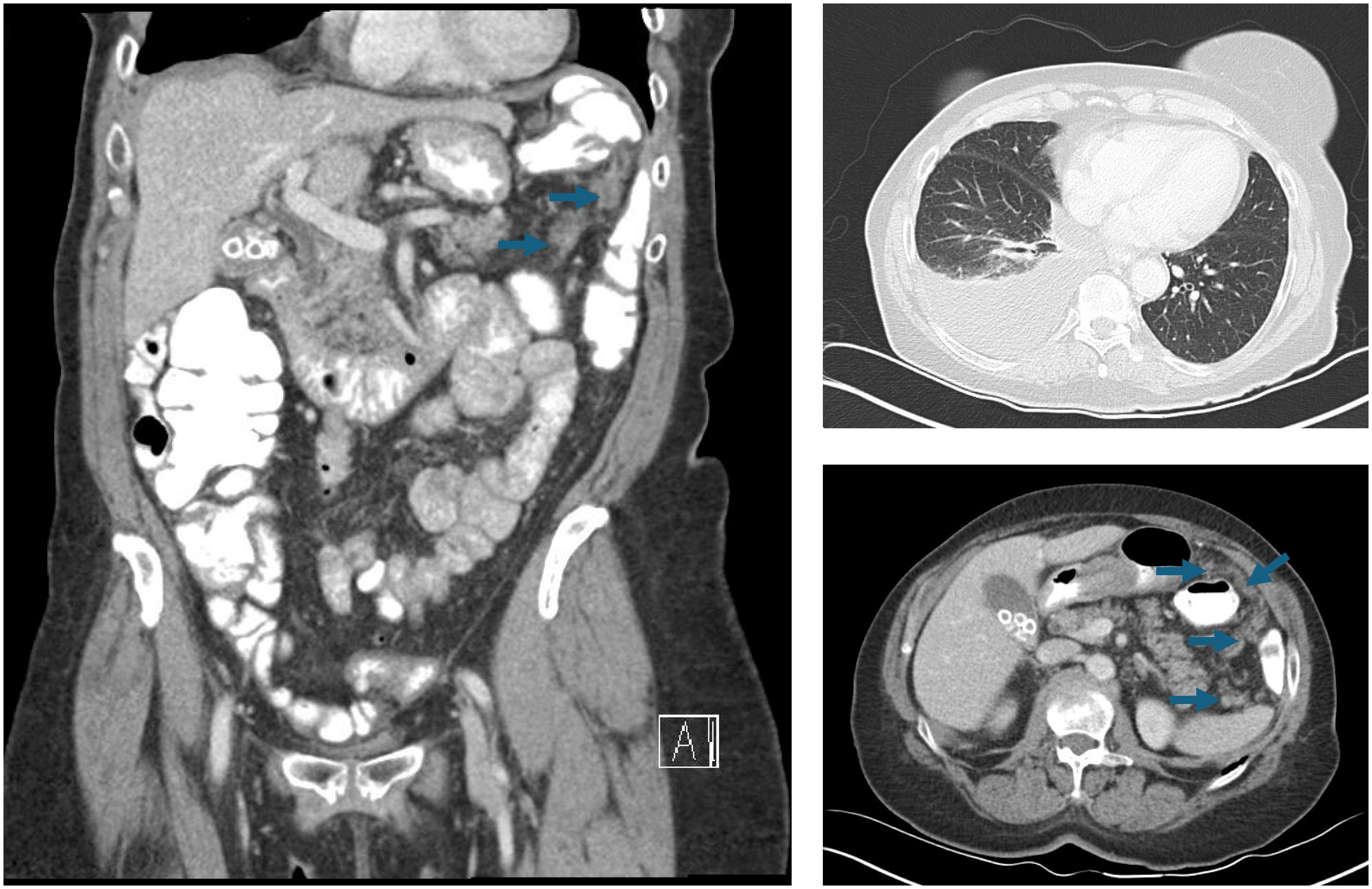
Figure 3. CT chest, abdomen, and pelvis imaging that reveals a right-sided pleural effusion and widespread peritoneal carcinomatosis (arrows).
The patient received paclitaxel and carboplatin neoadjuvant chemotherapy and developed severe abdominal pain, which indicated a large bowel obstruction for which she underwent a diverting colostomy. She received a total of 5 cycles of neoadjuvant chemotherapy prior to cytoreductive surgery that included a total abdominal hysterectomy with bilateral salpingo-oopherectomy, splenectomy, partial pancreatectomy, partial colectomy, omentectomy, cholecystectomy, appendectomy, tumor debulking and colostomy reversal. Residual high grade serous carcinoma was identified in numerous peritoneal implants, as well as within the bilateral ovaries and fallopian tubes, endometrium, cervix, parametria, pancreas, spleen, peri-appendiceal soft tissue, small intestinal mesentery, and omentum. The patient received 5 cycles of adjuvant paclitaxel and carboplatin with bevacizumab and remains on bevacizumab maintenance. Per the patient, the most challenging aspect of post-surgical treatment was persistent fatigue, which has improved significantly while receiving only bevacizumab. At last follow-up, 9 months following her final cycle of platinum-based chemotherapy, she was deemed disease-free by PET imaging. A summary of the patient’s clinical course is provided in Figure 4.
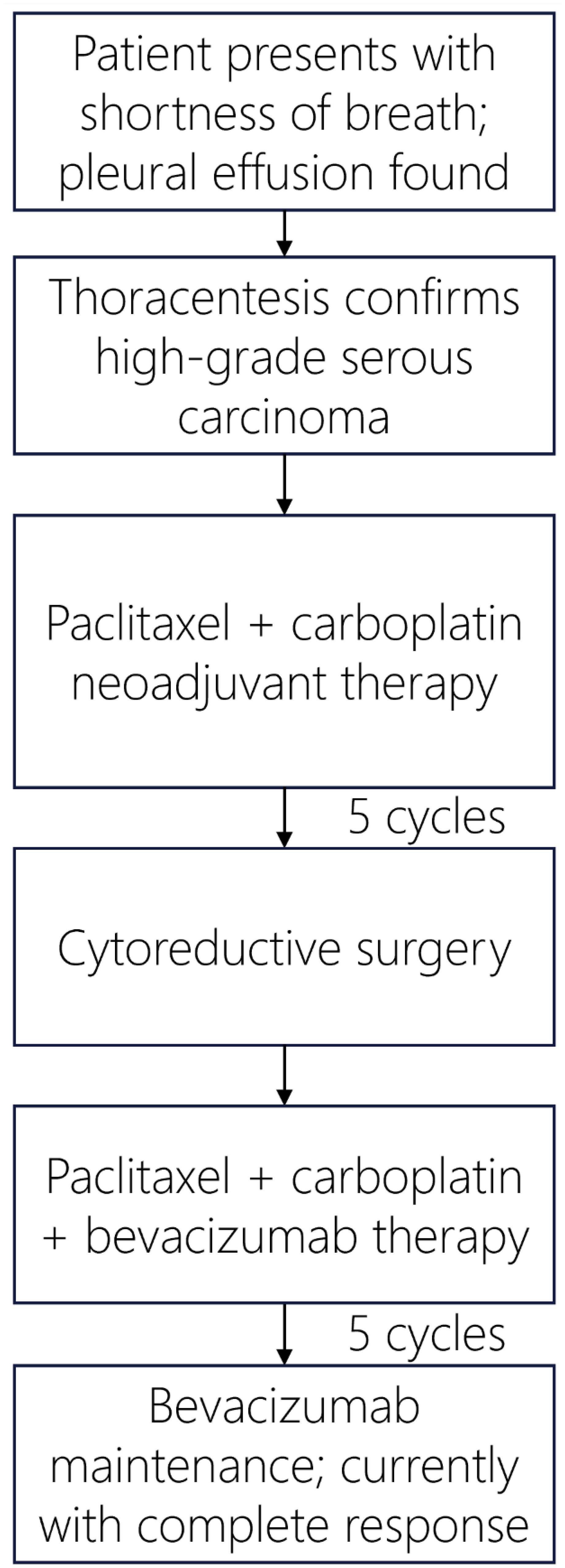
Figure 4. Patient’s treatment timeline for management of Stage IV high grade serous fallopian tube carcinoma.
Discussion
This patient’s tumor represents a high grade serous carcinoma harboring a previously unreported NRG1 fusion; the gene is spliced with MYH10 with an exon 2 breakpoint. Overall, our analysis demonstrates that such NRG1-rearranged gynecologic neoplasms are rare, with only 26 total examples identified within the total gynecologic tract cancer specimens that underwent whole transcriptome sequencing—an incidence of 0.18% among carcinomas of the ovary, fallopian tube, and peritoneum. Although uncommon, the list of genes fused with NRG1 is extensive, with our work identifying 20 rare partners. In some instances, the specific fusion could be diagnostic. In particular, CLU::NRG1 was only detected in cases demonstrating low grade/borderline histology. Beyond the novelty of these fusions, their gene products may have clinical significance with the advent of anti-ERBB/ERBB2/ERBB3 therapies. The efficacy of these drugs requires interaction with the immunoglobulin and EGF domains of NRG1 (13), which have been universally retained within the fusion proteins created upon rearrangement with the various partner genes.
Moreover, in keeping with prior studies (5), those neoplasms with NRG1 fusions described here demonstrated low TMB. Genetic changes responsible for HRD were indeed detected, although in a minority of the NRG fusion-positive samples (11.5%). While this proportion of cases is below the up to 50% prevalence of HRD reported in high grade serous carcinoma (14), these few cases could have been eligible for poly (ADP-ribose) polymerase (PARP) inhibition, which has demonstrated efficacy in treating gynecological tumors (15). Ultimately, the lack of HRD in our patient’s tumor rendered her ineligible for PARP inhibition. Whether the detected novel NRG1 fusion and co-occurring genetic alterations (involving RET and TP53) could influence her treatment options and prognosis remains unclear.
This reality stems from the limitations of our study. By focusing on a single case, the generalizability of our patient’s experience comes into question. Moreover, the lack of extensive follow-up data does not exclude a future change in her health status. Finally, utilization of testing from a single laboratory could introduce bias in detecting and reporting the NRG1 gene changes. Fusion confirmation could ultimately be achieved by alternative methodologies, including immunohistochemistry and fluorescence in situ hybridization directed against NRG1, although these techniques may not be specific to certain novel breakpoints and have limited commercial availability.
A comprehensive literature review summarizing diagnostic, therapeutic, and preventative strategies for treating tubal carcinomas by Colombi et al. advocates for surgical staging and debulking surgery when the patient’s physical condition allows, which would then be followed by systemic chemotherapy (16). And as would be indicated for managing our patient’s Stage IV tumor, the National Comprehensive Cancer Network (NCCN) guidelines recommend “platinum-based” regimen after cytoreductive surgical intervention (17). Similar to her personal experience, some research indicates that NRG1 fusion-positive tumors respond to such treatment. In a study of 19 individuals diagnosed with lung carcinoma, stable disease was noted in eight whom had received chemotherapy (47%). A partial response was observed in two (12%), and disease progression occurred in eight (41%) (18). No subjects demonstrated a response to anti-PD-L1 monotherapy or combination chemoimmunotherapy.
More encouraging are the recently developed anti-ERBB/ERBB2/ERBB3 targeted therapies for the treatment of carcinomas harboring NRG1 fusions, as would be considered for our patient if her tumor were to eventually recur. A multi-center alliance study compiling data from the management of NRG1 fusion-positive lung cancers documented a 25% overall response rate to afatinib, a pan-ERBB small molecular tyrosine kinase inhibitor (18). While progression-free survival (PFS) was only 2.8 months, the authors advocated for afitinib in the treatment of cancers with NRG1 fusions. A similar pan-ERBB inhibition approach is currently being explored with tarloxotinib for the management of NRG1 fusion-positive tumors (RAIN-701 trial, NCT03805841), the results of which are expected to be reported soon (19).
ERBB3-selective inhibitors, such as seribantumab, are also currently undergoing investigation. During a Phase II clinical trial (CRESTONE trial, NCT04383210), this anti-ERBB3 IgG2 monoclonal antibody produced an ORR of 33% across various tumor types harboring NRG1 fusions (20). A 92% percent disease control rate included two patients who demonstrated a complete response, thereby lending support for ERBB3-specific inhibition as a viable treatment strategy.
Recently, the United States Food and Drug Administration fast-tracked clinicals trials for a ERBB2/ERBB3 bispecific monoclonal antibody, zenocutuzumab. In a Phase I/II clinical trial (NCT02912949), this drug demonstrated excellent efficacy in treating NRG1 fusion-positive tumors, especially those arising from the pancreas (21). A partial response was documented in 42% the pancreatic tumors, with disease progression observed in only a single case. Overall, zenocutuzumab has demonstrated a preliminary ORR of 34% across solid tumor types (22). As a result, the simultaneous targeting of ERBB2 and ERBB3 could exist as a novel approach to treating NRG1 fusion-positive tumors. In the future, additional novel therapies are likely to be discovered, and proper assessment for changes in NRG1 will become necessary for treatment selection.
Conclusion
Our report of novel NRG1 gene rearrangements detected in serous carcinomas of the ovary, fallopian tube, and peritoneum indicates a small, but important prevalence of these fusions occurring within the female genital tract. While the knowledge gained from the experience of a single patient may be limited, it describes a successful treatment strategy including neoadjuvant/adjuvant platinum-based chemotherapy, cytoreductive surgery, and bevacizumab. Moreover, the patient’s novel NRG1 fusion highlights the potential role of anti-ERBB/ERBB2/ERBB3 therapies in managing similar tumors and illustrates the importance of diagnostic gene fusion testing.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Ethics statement
Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
Author contributions
AC: Data curation, Formal analysis, Investigation, Writing – original draft. ME: Conceptualization, Data curation, Formal analysis, Methodology, Supervision, Writing – review & editing. TA: Data curation, Formal analysis, Writing - review & editing. JR: Writing – review & editing. II: Writing – review & editing. MO: Writing – review & editing. JT: Conceptualization, Data curation, Formal analysis, Methodology, Supervision, Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Conflict of interest
TA, MO, and ME disclose a financial association with Caris Life Sciences full-time employment, travel/speaking expenses, stock/stock options.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2024.1472725/full#supplementary-material
References
1. Steinthorsdottir V, Stefansson H, Ghosh S, Birgisdottir B, Bjornsdottir S, Fasquel AC, et al. Multiple novel transcription initiation sites for NRG1. Gene. (2004) 342:97–105. doi: 10.1016/j.gene.2004.07.029
2. Fernandez-Cuesta L, Thomas RK. Molecular pathways: targeting NRG1 fusions in lung cancer. Clin Cancer Res. (2015) 21:1989–94. doi: 10.1158/1078-0432.CCR-14-0854
3. Jonna S, Feldman RA, Swensen J, Gatalica Z, Korn WM, Borghaei H, et al. Detection of NRG1 gene fusions in solid tumors. Clin Cancer Res. (2019) 25:4966–72. doi: 10.1158/1078-0432.CCR-19-0160
4. Nagasaka M, Ou SI. NRG1 and NRG2 fusion positive solid tumor Malignancies: a paradigm of ligand-fusion oncogenesis. Trends Cancer. (2022) 8:242–58. doi: 10.1016/j.trecan.2021.11.003
5. Severson E, Achyut BR, Nesline M, Pabla S, Previs RA, Kannan G, et al. RNA sequencing identifies novel NRG1 fusions in solid tumors that lack co-occurring oncogenic drivers. J Mol Diagn. (2023) 25:454–66. doi: 10.1016/j.jmoldx.2023.03.011
6. Solomon JP, Hechtman JF. Detection of NTRK fusions: merits and limitations of current diagnostic platforms. Cancer Res. (2019) 79:3163–8. doi: 10.1158/0008-5472.CAN-19-0372
7. Chakravarty D, Johnson A, Sklar J, Lindeman NI, Moore K, Ganesan S, et al. Somatic genomic testing in patients with metastatic or advanced cancer: ASCO Provisional Clinical Opinion. J Clin Oncol. (2022) 40:1231–58. doi: 10.1200/JCO.21.02767
8. Spigel D, Waqar SN, Burkard ME, Lin JJ, Chae YK, Socinski MA, et al. MO01.33 CRESTONE – clinical study of response to seribantumab in tumors with Neuregulin-1 (NRG1) fusions – a phase 2 study of the anti-HER3 mAb for advanced or metastatic solid tumors (NCT04383210). J Thorac Oncol. (2021) 16:S29–30. doi: 10.1016/j.jtho.2020.10.138
9. Carrizosa DR, Burkard ME, Elamin YY, Desai J, Gadgeel SM, Lin JJ, et al. CRESTONE: initial efficacy and safety of seribantumab in solid tumors harboring NRG1 fusions. J Clin Oncol. (2022) 40:3006–6. doi: 10.1200/JCO.2022.40.16_suppl.3006
10. Schram AM, Drilon AE, Macarulla T, O'Reilly EM, Rodon J, Wolpin BM, et al. A phase II basket study of MCLA-128, a bispecific antibody targeting the HER3 pathway, in NRG1 fusion-positive advanced solid tumors. J Clin Oncol. (2020) 38:TPS3654. doi: 10.1200/JCO.2020.38.15_suppl.TPS3654
11. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. (2005) 102:15545–50. doi: 10.1073/pnas.0506580102
12. Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. (2015) 1:417–25. doi: 10.1016/j.cels.2015.12.004
13. Jonna S, Feldman R, Ou SI, Nagasaka M, Swensen JJ, Korn WM, et al. Characterization of NRG1 gene fusion events in solid tumors. J Clin Oncol. (2020) 38:3113. doi: 10.1200/JCO.2020.38.15_suppl.3113
14. Mangogna A, Munari G, Pepe F, Maffii E, Giampaolino P, Ricci G, et al. Homologous recombination deficiency in ovarian cancer: from the biological rationale to current diagnostic approaches. J Pers Med. (2023) 13:284. doi: 10.3390/jpm13020284
15. O’Malley DM, Krivak TC, Kabil N, Munley J, Moore KN. PARP inhibitors in ovarian cancer: a review. Target Oncol. (2023) 18:471–503. doi: 10.1007/s11523-023-00970-w
16. Colombi I, D’Indinosante M, Lazzeri L, Zupi E, Pisaneschi S, Giusti M, et al. Tubal cancer clinical management: two exceptional scenarios and a review of the literature. J Clin Med. (2024) 13:5075. doi: 10.3390/jcm13175075
17. National Comprehensive Cancer Network. NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines®): Ovarian cancer [v.3.2024] (2024). Available online at: https://www.nccn.org/professionals/physician_gls/pdf/ovarian.pdf. (Accessed July 28, 2024)
18. Drilon A, Duruisseaux M, Han JY, Ito M, Falcon C, Yang SR, et al. Clinicopathologic features and response to therapy of NRG1 fusion-driven lung cancers: The eNRGy1 Global Multicenter Registry. J Clin Oncol. (2021) 39:2791–802. doi: 10.1200/JCO.20.03307
19. Liu SV. NRG1 fusions: biology to therapy. Lung Cancer. (2021) 158:25–8. doi: 10.1016/j.lungcan.2021.05.011
20. Thavaneswaran S, Chan WY, Asghari R, Grady JP, Deegan M, Jansen VM, et al. Clinical response to seribantumab, an anti-human epidermal growth factor receptor-3 immunoglobulin 2 monoclonal antibody, in a patient with metastatic pancreatic ductal adenocarcinoma harboring an NRG1 fusion. JCO Precis Oncol. (2022) 6:e2200263. doi: 10.1200/PO.22.00263
21. Xu C, Wang Q, Wang D, Wang W, Fang W, Li Z, et al. Expert consensus on the diagnosis and treatment of NRG1/2 gene fusion solid tumors. Glob Med Genet. (2024) 11:86–99. doi: 10.1055/s-0044-1781457
Keywords: NRG1 gene fusion, ovarian cancer, fallopian tube cancer, primary peritoneal cancer, serous carcinoma
Citation: Crymes A, Evans MG, Adeyelu T, Reid J, Ibe IO, Oberley MJ and Tseng JH (2024) Case report: High grade serous fallopian tube carcinoma with rare NRG1 gene fusion presenting as widespread peritoneal carcinomatosis. Front. Oncol. 14:1472725. doi: 10.3389/fonc.2024.1472725
Received: 29 July 2024; Accepted: 14 October 2024;
Published: 06 November 2024.
Edited by:
Kunqi Chen, Fujian Medical University, ChinaReviewed by:
Federica Perelli, Azienda USL Toscana Centro, ItalyMaria Prieto, WWF Spain, Spain
Natalie Danziger, Foundation Medicine Inc., United States
Copyright © 2024 Crymes, Evans, Adeyelu, Reid, Ibe, Oberley and Tseng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mark G. Evans, bWFya2V2YW5zMjAxMUBnbWFpbC5jb20=
 Anthony Crymes1
Anthony Crymes1 Mark G. Evans
Mark G. Evans Jack Reid
Jack Reid