- 1Center for Childhood Cancer, Abigail Wexner Research Institute at Nationwide Children’s Hospital, Columbus, OH, United States
- 2Department of Microbiology, Immunology and Cell Biology, West Virginia University, Morgantown, WV, United States
- 3Department of Biology, College of Science, Mathematics, and Technology, Wenzhou-Kean University, Wenzhou, China
- 4Wenzhou Municipal Key Laboratory for Applied Biomedical and Biopharmaceutical Informatics, Wenzhou-Kean University, Wenzhou, China
- 5Zhejiang Bioinformatics International Science and Technology Cooperation Center, Wenzhou-Kean University, Wenzhou, China
- 6Shenzhen Huayuan Biological Science Research Institute, Shenzhen Huayuan Biotechnology Co. Ltd., Shenzhen, China
- 7Center for Molecular Medicine and Genetics, School of Medicine, Wayne State University, Detroit, MI, United States
Radiotherapy (RT) serves as one of the key adjuvant treatments in management of breast cancer. Nevertheless, RT has two major problems: side effects and radioresistance. Given that patients respond differently to RT, it is imperative to understand the molecular mechanisms underlying these differences. Two-thirds of human genes do not encode proteins, as we have realized from genome-scale studies conducted after the advent of the genomic era; nevertheless, molecular understanding of breast cancer to date has been attained almost entirely based on protein-coding genes and their pathways. Long non-coding RNAs (lncRNAs) are a poorly understood but abundant class of human genes that yield functional non-protein-coding RNA transcripts. Here, we canvass the field to seek evidence for the hypothesis that lncRNAs contribute to radioresistance in breast cancer. RT-responsive lncRNAs ranging from “classical” lncRNAs discovered at the dawn of the post-genomic era (such as HOTAIR, NEAT1, and CCAT), to long intergenic lncRNAs such as LINC00511 and LINC02582, antisense lncRNAs such as AFAP-AS1 and FGD5-AS1, and pseudogene transcripts such as DUXAP8 were found during our screen of the literature. Radiation-related pathways modulated by these lncRNAs include DNA damage repair, cell cycle, cancer stem cells phenotype and apoptosis. Thus, providing a clear picture of these lncRNAs’ underlying RT-relevant molecular mechanisms should help improve overall survival and optimize the best radiation dose for each individual patient. Moreover, in healthy humans, lncRNAs show greater natural expression variation than protein-coding genes, even across individuals, alluding to their exceptional potential for targeting in truly personalized, precision medicine.
1 The intersection of radiotherapy and long non-coding RNAs in breast cancer
Radiotherapy (RT) serves as one of the adjuvant treatment modalities in the control of many malignancies including breast cancer. The other modalities are chemotherapy, hormone therapy, immunotherapy, and targeted therapy (1–3). Along with surgery and chemotherapy, RT significantly reduces the risk of recurrence and improves overall survival in breast cancer patients (4, 5).
However, breast cancer is heterogeneous in terms of its genetics and clinical characteristics, and this heterogeneity is classified through distinct subtypes. While 5-10% of breast cancers are due to inherited disease causative alleles and mutations in the BRCA1 and BRCA2 genes (6, 7), the vast majority of cases have a more complex etiology that is not confined to genetics and does not involve major single-gene risk factors. Breast cancer is traditionally classified into four groups based on immunohistochemical expression of hormone receptors: estrogen receptor positive (ER+), progesterone receptor positive (PR+), human epidermal growth factor receptor positive (HER2+), and triple-negative (TNBC), characterized by the absence of any of the above receptors. However, in light of a deeper understanding of breast cancer biology at the molecular level and by considering gene expression profiles, breast cancer can be stratified into four primary groups as luminal A, luminal B, HER-2, basal, and also a normal-breast-like group which closely resembles luminal A subtype (8, 9). While research over the past decade has focused primarily on protein-coding genes, the importance of non-coding regions of the genome cannot be underestimated.
Beyond protein-coding genes, non-coding RNAs (ncRNAs) constitute a significant portion of the human transcriptome (99% of all RNAs in a cell (10), and encoded by >65% of all genes), providing a promising horizon to capture key regulators in the cancer gene networks. It is well documented in the literature that long non-coding RNAs (lncRNAs), as a subset of ncRNAs, play a major role in the pathogenesis of breast cancer (11). In our previous work, we demonstrated that estrogen-induced/repressed lncRNAs serve as key oncogenes or tumor suppressors in breast cancer (12). Nevertheless, little remains known about how lncRNAs are functionally pertinent to radiation response, motivating this review. Beyond breast cancer, mounting evidence points to a direct functional association between Y chromosome-encoded lncRNAs and radiation sensitivity in male non-small cell lung cancer (NSCLC) (13). In our comprehensive exploration, we seize the opportunity to provide a briefing about radiotherapy in breast cancer management to accommodate readers unacquainted with this specialized field. Additionally, we recap how the cell reacts to the radiation at the molecular level and what the consequences of the exposure are: apoptosis, necrosis, necroptosis, pyroptosis, ferroptosis, cuproptosis, autophagy, senescence, and mitotic catastrophe (14). MicroRNAs in breast cancer are already well-characterized; this review hence will not pursue that topic. Instead, we delve into the world of lncRNAs due to their important role in the genome, offering a concise introduction for those seeking general information. Next, we tabulated the results of PubMed search for full-text articles with the following search terms:{“radioresistant” and/or “radiosensitive”} and “breast cancer” and (“lncrna” and/or “lincrna”). Finally, we reiterate how parallel development of chemical modifiers and technological breakthroughs across the wide spectrum of clinical approaches, from imaging technology to high-throughput omics, and with special relevance to the role of lncRNAs in radiosensitivity and resistance, impacts the field of RT in a way that will empower personalized medicine.
1.1 Understanding radiotherapy in breast cancer: protocols, challenges, and molecular insights
1.1.1 Current standards for breast cancer radiotherapy
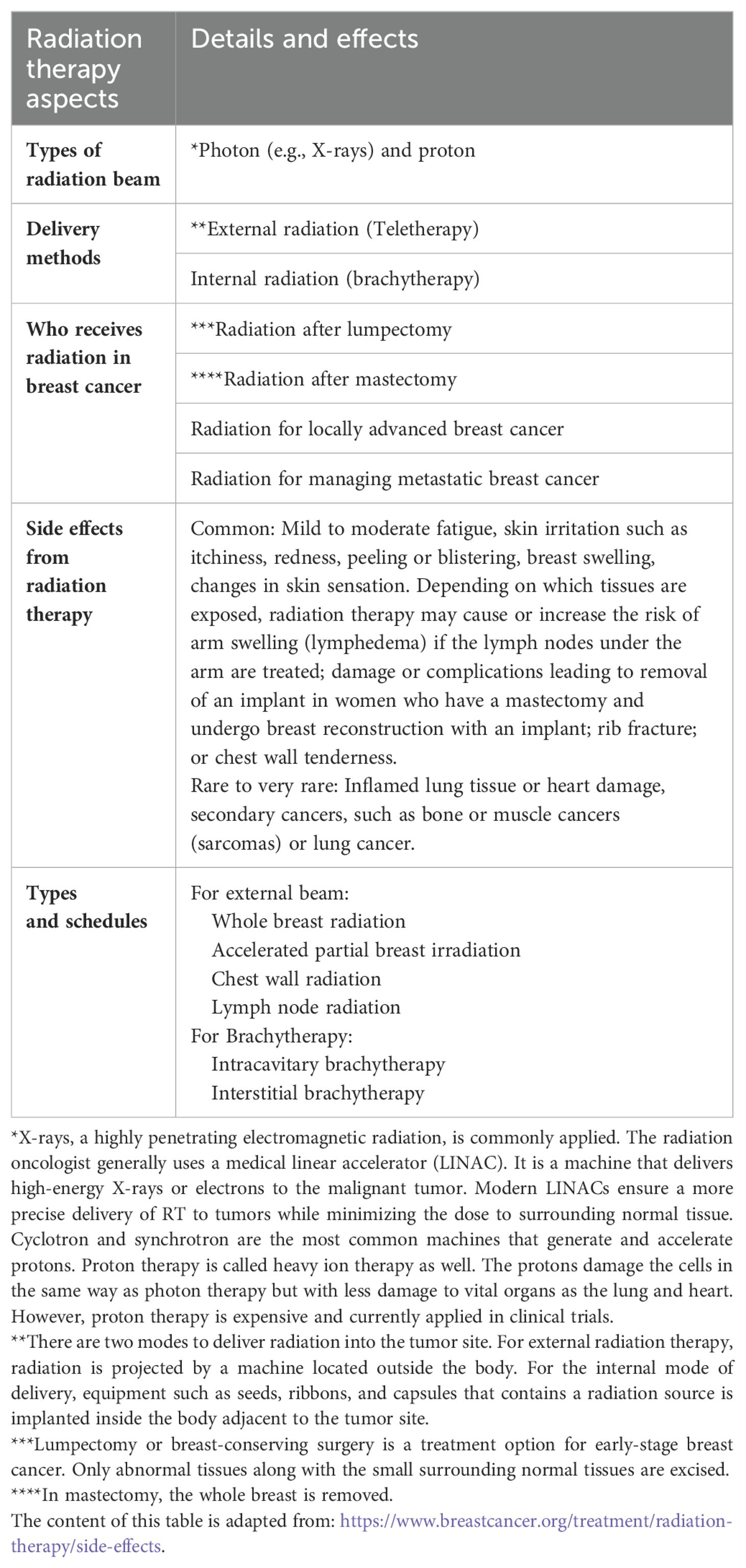
Various radiotherapy protocols are available for RT of breast cancer patients. The standard of care is to deliver a total radiation dose of 50 Gray in 25 fractions of 2.0 Gray over 5 weeks. Nevertheless, the current guidelines of the American Society for Radiation Oncology (ASTRO) recommend a hypofractionated (or accelerated) regimen for whole-breast radiation therapy in which patients receive the same total dose fractionated over a shorter period of time (15). A radiation oncologist selects the appropriate fractionation schedules according to the clinical features and histopathologic characteristics of patients (16). Table 1 summarizes the guiding principles of RT for breast cancer.
1.1.2 Challenges in breast cancer radiotherapy: side effects and radioresistance
Though highly effective, RT as an approach to treating cancer has two problems: side effects and radioresistance. The most common short-term side effects are fatigue, swelling in the breast, skin changes in the treated area similar to a sunburn (redness, skin peeling, darkening of the skin), or other serious long-term complications as lymphedema, shoulder stiffness, brachial plexopathy, predisposition to rib fracture and angiosarcoma, a rare type of cancer (17). Locoregional recurrence following RT could be attributed to intrinsic radioresistance or the development of de novo resistance features in a particular subpopulation (18). Moreover, different molecular subtypes of breast cancer do not respond equally well to RT, in which luminal cancers, in particular, luminal subtype A, benefit the most compared to HER2-positive and TNBC (19).
1.1.3 Cellular and molecular responses to radiotherapy in breast cancer
In the context of RT, cell fates and molecular events play a crucial role in the response to treatment. RT harnesses the intense energy of photons to target cancerous cells that persist after surgery, potentially aiding in the cure of metastases. Mechanisms of action include but are not limited to: direct, by induction of DNA damage and subsequent cell death; or indirect, by targeting tumor microenvironment through vascular damage or modulation of anti-tumor immune responses (20–22).
Following delivery at the tumor site, high-energy ionizing radiation directly breaks DNA, and also decomposes water into free radicals (water radiolysis) thus indirectly damaging DNA. Various DNA lesions occur as a result, including: base damage, single-strand breaks (SSBs), double-strand breaks (DSBs), and intra as well as inter DNA crosslinks. As an aftermath of severe damage, cell death occurs. Mitotic catastrophe and mitotic death, apoptosis, necrosis, senescence, autophagy, and necroptosis and ferroptosis are among the currently recognized types of radiation-induced cell death (4). Nevertheless, cancerous cells are not the only cells that are affected by RT, since RT does not distinguish between cell types. These side effects of RT occur systematically and locally on the normal cells nearby the tumor site that is being treated. Due to the proximity of the lung, heart, and contralateral healthy breast to cancerous breast tissue, and also blood exposure to RT, complications can arise (23). Moreover, each tumor resides in a field of non-cancerous cells, such as stromal cells which could govern radiotherapy responsiveness. The dynamic crosstalk of these neighboring cells with tumors within the tumor microenvironment makes them a potential modulator on radiation response. The key types of stromal cells include immune cells, cancer-associated fibroblasts (CAF), extracellular matrix, endothelial cells, and adipocytes. They promote radioresistance by adopting cancer stem cell properties, providing the secreted pro-survival factors, affecting metabolites and oxygen availability, angiogenesis, and immunomodulatory effects. Hence, besides the tumor-intrinsic factors, the tumor microenvironment and host immune system are notable factors affecting RT response (24).
Upon irradiation-induced damage, DNA damage responses (DDRs) are invoked. The key proteins Ataxia-Telangiectasia Mutated (ATM) and Ataxia-Telangiectasia and Rad3-Related protein (ATR) sense the damage and trigger the DNA damage response. Depending on the cell cycle stage, DNA double-strand lesions are repaired by non-homologous end joining (NHEJ) and homologous recombination (HR). The same protective DDRs safeguard tumor cells against radiation-induced cell death. Thus, radiation-induced DNA lesions and protective DDRs are, essentially, double-edged swords: they act in a way that develops, in parallel, dysfunctional normal tissue and cell damages as well as radioresistant features. Hence, a major consideration in RT is how to tip this balance in favor of the patient, by therapeutic approaches that minimize toxicity in normal tissues (e.g., by the development of mechanistically-driven radioprotectors, or techniques to more precisely deliver radiation to tumor site) and to better sensitize tumor cells to radiation (e.g., by the development of radiosensitizers, or targeting DDR signaling pathways to heighten tumor radiosensitization).
While radiation can damage DNA irreparably by affecting cell organelles, cell membrane properties, signal transduction, tumor cell phenotype, and the tumor immune response (25), cancerous cells can still evade lethal DNA damage by activating efficient DNA repair mechanisms. A growing list of lncRNAs facilitates various steps of the DNA repair from the detection of DNA lesions to activating signaling pathways that initiate DNA repair processes (26). The lncRNA GUARDIN is noteworthy for its association with BRCA1 and BARD1, which participate in a variety of DNA damage response pathways (DDRs) (27). This is merely the proverbial “tip of the iceberg.” There are definitely many other lncRNAs interacting with DNA damage repair and hence radiation response genes. Identification of any radiation-induced or suppressed lncRNAs would have practical implications for sensitizing cancer cells to RT. Emerging evidence unequivocally indicates that there is a huge reservoir of yet-unknown oncogenes and tumor suppressors in lncRNA data, including specifically in breast cancer (12), and undoubtedly many of these will be relevant to radiation response.
Considering the marked differences between patients in response to RT, and the associated toxicity and side effects, it is of utmost importance to decipher the complex molecular networks that are responsible for these differences and for the mechanisms leading to radioresistance and treatment failure. Providing a clear picture of the underlying molecular mechanisms could help improve overall survival and to optimize the best radiation dose for any individual, in a clear application of personalized medicine. Accordingly, the present review aims to summarize all the efforts of the last two decades (~2000-2022) to unravel radiation-induced pathways with a focus on breast cancer, specifically on the role of lncRNAs therein.
1.2 LncRNAs: emerging players in genome regulation
1.2.1 Unraveling the genomic landscape: LncRNAs in focus
Completion of the Human Genome Project (HGP) in 2003 spurred the emergence of disciplines and projects aimed at elucidating genome functionality. The Encyclopedia of DNA Elements (ENCODE) and the Functional Annotation of the Mammalian Genome (FANTOM) consortia, as HGP successors, demonstrated that 80% of the genome is functional (28) and that mammals have more non-coding RNA genes than protein-coding genes (29), respectively. The number contrasts with the mere 1.5% of the genome that is occupied by exons of protein-coding genes. Formerly considered to be “junk DNA,” the non-coding part of the genome is now widely understood to contain regulatory elements and non-coding RNA genes. Much of this non-coding DNA is transcribed into non-coding RNAs (ncRNAs), including micro and macro noncoding RNAs. Among these ncRNAs, lncRNAs represent the most prevalent and functionally diverse class. They have been increasingly highlighted as transcripts with emerging roles in crucial aspects of biological processes. It is estimated that they are pervasively transcribed from 15,000 to 80,000 distinct loci in the human genome (30–32). LncRNAs are defined as transcripts of more than 500 nucleotides in length, lacking open reading frames, and transcribed by RNA Polymerase II. They can interact with genomic DNA, RNA partners, as well as with RNA binding proteins. LncRNAs are involved in target-specific as well as in global regulatory mechanisms, and are ubiquitously transcribed (10).
1.2.2 LncRNAs: architects of evolution and cellular identity
The numbers of lncRNAs, unlike protein-coding genes, correlate with species complexity during evolution, and approximately 60-75% of human lncRNAs are primate-specific (32–34). They may have appeared and increased in their numbers in ancestral mammalian species soon after the mammalian radiation, conferring higher levels of complexity and gene regulation (32, 35) that may have even led to the development of evolutionarily new organs (for example, chimpanzees have entire organs – such as the vomeronasal organ – that are absent in humans, whereas human and gorilla RNAs in the brain show highly discrepant splicing (36)). While the knowledge of primary amino acid sequence and homology modeling greatly enhanced the ability to predict two or three-dimensional structures of proteins and hence their function, this has not been the case for lncRNAs since most RNA structural biology has historically focused on ribosomal RNAs and other “classical” short RNAs (37). Despite the challenges associated with in silico prediction methods for deciphering structural/primary sequence correlations, the concept of a “modular RNA regulatory code” arose due to evidence that secondary structures in lncRNAs remain conserved in orthologs between mammalian species despite an absence of sequence conservation (38). The latter makes lncRNAs “invisible” to sequence homology-based bioinformatic tools used for protein-coding genes. It is therefore possible to predict lncRNAs’ modes of action by mapping higher-order structural features and functional networks rather than their primary sequences. Genetic experiments could help determine whether their regulatory function is modularity-dependent and/or independent (38). Furthermore, we expect artificial intelligence (AI) to make a measurable impact both in the area of predicting lncRNA secondary structure and in detecting cryptic cross-species lncRNA homologies.
As a result of the evolutionary plasticity of these sequences, it is increasingly posited that the genome was liberated from the rigidity of mechanisms driven by highly conserved coding genes, allowing for new functions to be developed. In contrast to mRNAs, lncRNAs usually exhibit restricted and tissue-specific expression patterns (32, 39) and are often cell-type-specific, which suggests they are involved in cell state and developmental pathway regulation (40). In addition, most are located in the nucleus, but a significant proportion is in the cytoplasm (10). The cytoplasmic proportion is estimated to be 75% in human and Drosophila cells (41). Consistent with these results, in Drosophila models, RNA FISH revealed that more lncRNAs were found in the cytoplasm (40%) than in the nucleus (4%) (42).
Similarly to mRNAs, most cytoplasmic lncRNAs are spliced, polyadenylated, and 7-methylguanosine-capped. Based on the genomic positions of lncRNA-encoding loci relative to nearby or overlapping protein-coding genes, lncRNA genes can be classified as intergenic, antisense, intronic, and/or overlapping in other ways, relative to those coding genes (10).
1.2.3 Functional characteristics of LncRNAs
LncRNAs regulate gene expression at multiple levels, including chromatin organization, transcriptional and post-transcriptional regulation (Figure 1).
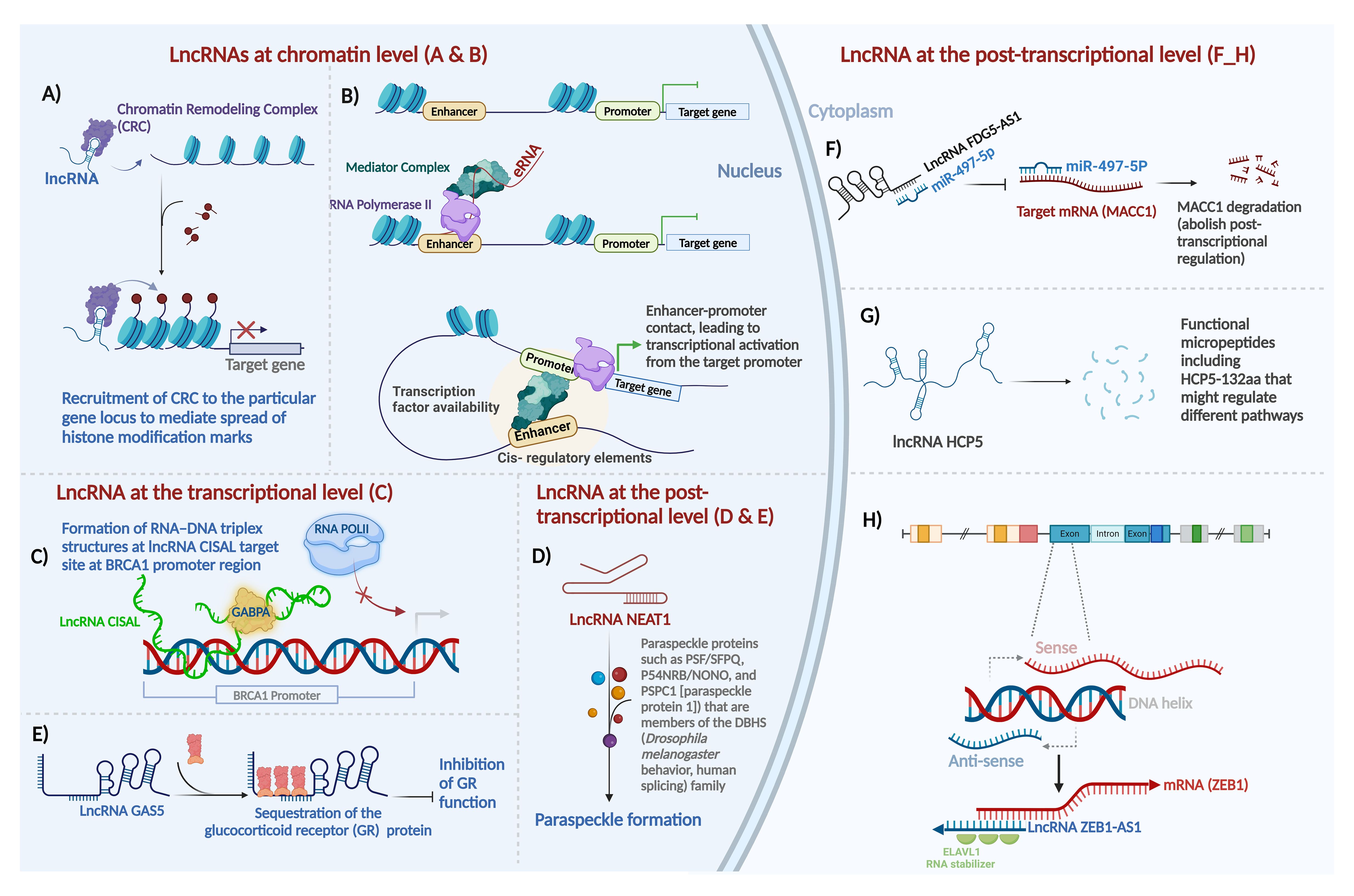
Figure 1. Major mechanisms of action of long non-coding RNAs in mammalian systems. Gene expression is regulated at three distinct levels - epigenetic (chromatin), transcriptional, and post-transcriptional - by lncRNAs as shown here and as detailed in the main text. (A) guide lncRNA, (B) enhancer RNA, (C) forming a triplex structure with DNA sequence at regulatory regions and interaction with transcription factors to regulate gene expression, (D) lncRNA as scaffold, (E) lncRNA as decoy, (F) lncRNA as sponge for miRNAs, (G) lncRNA-encoded protein, and (H) mRNA:lncRNA hybridization in post-transcriptional sense-antisense pairs. Most of these effects can be classified as epigenetic and transcriptional regulation, as illustrated in the nucleus on the left, or post-transcriptional mechanisms in the cytoplasm, on the right. For details about lncRNA function at the chromatin level, the transcriptional level, and the post-transcriptional level refer to the main text. Created with BioRender.com.
LncRNAs at the chromatin level: LncRNAs can modulate gene expression through deposition of epigenetic modifiers at particular regions of the genome by acting either in cis or in trans. For the purposes of this review, we define “cis” regulation as that occurring within the same locus, for example when an antisense lncRNA regulates its counterpart sense mRNA transcribed from the opposite direction in the same locus (regardless of allele specificity), and we define “trans” regulation as any scenario where the lncRNA and its target are encoded at different loci (43). Exemplifying this regulatory modality, the lncRNA HOX antisense intergenic RNA (HOTAIR), among the first oncogenic lncRNAs identified in primary and metastatic breast tumors (44), serves as a platform for the assembly of two distinct histone modifiers. Through the recruitment of Polycomb Repressive Complex 2 (PRC2) and LSD1–CoREST, lncRNA HOTAIR facilitates H3K27 trimethylation (45) and H3K4 demethylation (46), respectively, to repress gene expression (Figure 1A). In a contrarian fashion, the upstream master lncRNA of the inflammatory chemokine locus (UMLILO) activates expression of its target gene through recruitment of activating factors, such as the WD repeat-containing protein 5 (WDR5)–mixed lineage leukemia protein 1 (MLL1) (WDR5–MLL) complex, to the target promoter enabling their H3K4Me3 epigenetic priming (47). In addition to recruiting epigenetic modifiers, lncRNAs can induce chromatin loop formation to reformat the genome. For instance, ncRNA-a7, member of a class of lncRNAs involved in long-range transcriptional activation through the association with the Mediator complex, forms a co-activator complex to form a loop with its target locus (Figure 1B) (48).
LncRNAs at the transcriptional level: LncRNAs also function positively and negatively - depending on the specific lncRNA, the target/s, and the cellular and network contexts - as transcriptional regulators in many cases. LncRNAs can directly interact with transcription factors along the genomic DNA to induce or suppress transcription. The mechanisms of actions include but are not limited to several key modalities. By preventing transcription initiation complex formation at promoters, lncRNA may suppress transcription through RNA–DNA triplex formation. The regulatory outcome in this case is that lncRNAs sequester transcription factors from their cognate DNA-binding sites. For instance, through direct binding to the BRCA1 (breast cancer early onset 1, the first breast cancer gene that was ever discovered) promoter and forming a tertiary structure, cisplatin-sensitivity-associated lncRNA (CISAL) sequesters the BRCA1-activating transcription factor GABPA away from downstream regulatory regions (Figure 1C) (49). LncRNAs also activate transcription by recruiting transcription factors to the targeted promoters or acting as transcription factor co-activators. LncRNA DLEU1 (deleted in lymphocytic leukemia 1) acts as the coactivator for hypoxia inducible factor 1 subunit alpha (HIF-1α) to induce expression of cytoskeleton associated protein 2 (CKAP2) and consequently pro-tumor activities in breast cancer (50). Additionally, a number of lncRNAs regulate transcription by controlling nucleocytoplasmic transport of transcription factors. For instance, the lncRNA non-coding repressor of NFAT (NRON) inhibits nuclear translocation of dephosphorylated nuclear factor of activated T cells (NFAT) trans-activator by interacting with importin-beta family members (51).
LncRNAs as regulators at the post-transcriptional level: Post-transcriptionally, lncRNAs exhibit distinct mechanisms, which are diverse and protean in their versatility - in contrast to the mono-mechanistic nature of microRNAs, which act almost solely as post-transcriptional suppressors.
LncRNAs serve as architectural scaffolds for assembling proteins to enable biological events, such as the formation of key nuclear subcompartments, in particular paraspeckles, containing the lncRNA NEAT1 (Figure 1D) (52), as well as determinants of chromosome structure in the interphase nucleus, including long-range interchromosomal interactions (53), regulators of telomere activity such as TERRA (telomeric repeat-containing RNA) (54); and essential accessories of the actin cytoskeleton (55).
LncRNA-protein interactions comprise an additional important aspect of lncRNA-mediated gene regulation. LncRNAs act through direct functional interactions with specific proteins, forming lncRNA ribonucleoprotein complexes (lncRNPs), such as the complex that allows the BRCA1 protein in breast cancer to function upon activation by a direct-binder primate-specific lncRNA. For instance, as a p53-responsive lncRNA, GUARDIN acts as a binding platform joining the breast cancer early onset protein BRCA1 and its partner BARD1, which cooperate to stimulate cell proliferation and survival (27). Another example of an lncRNA that interacts with proteins to accomplish its function in breast cancer is the lncRNA LINP. It contributes to radioresistance in TNBC by stabilizing Ku80 and DNA-PKcs complexes after double-strand DNA breaks (DSB) (56).
One important subtype of lncRNAs that act through lncRNP formation is lncRNA which directly binds transcription factor proteins. In this context, lncRNAs directly interact with transcription factors and serve as their co-activators or co-repressors. For instance, the glucocorticoid response element (GRE)-like element ribomimic sequence, found within the lncRNA growth arrest-specific 5 (GAS5), mimics the consensus genomic-DNA binding site motif that GR recognizes, and as a result, the element serves as a decoy to repress glucocorticoid receptor (GR) by titrating bioavailable GR molecules out of the pool that is available to bind to GR-responsive promoters (Figure 1E) (57). Through such protein binding that leads to inactivation or sequestration, lncRNAs inhibit the function of proteins.
MicroRNAs can bind lncRNAs based on Watson-Crick sequence complementarity. As a result of sponging microRNAs, lncRNAs may rescue the half-life, stability, or translation of the mRNAs cognate to the affected microRNAs. LncRNAs act as competing endogenous RNAs (ceRNAs) if they bind miRNAs that are hence prevented from binding the mRNAs that they would otherwise be suppressing. For instance, the lncRNA FYVE RhoGEF and PH Domain containing 5 antisense RNA 1 (FGD5-AS1) promotes the radioresistance of breast cancer cells in an FGD5-independent manner, by sponging miR-497-5p, which in turn results in the upregulation of mir-497-5p’s target mRNA, metastasis-associated in colon cancer 1 (MACC1) (Figure 1F) (58). MACC1 was first discovered in colon cancer (59). As an oncogene, it plays an important role in tumor invasion and metastasis in a wide range of solid tumors, primarily by regulating genes that contribute to the epithelial-mesenchymal transition (EMT) (60).
In addition to their multiple functional modalities that we have thus far discussed, lncRNAs often contain cryptic short open reading frames (ORFs) that can encode micropeptides, allowing the lncRNAs to be bifunctional through distinct RNA-based and peptide-based roles. Despite early assumptions that most lncRNAs are genuinely noncoding and lack even short ORFs that could be translated by ribosomes to yield peptides, protein-coding capabilities of a small but reproducible subset of lncRNAs were discovered by us (61) and subsequently confirmed by numerous groups. For example, in breast cancer, the LINC00908-encoded polypeptide ASRPS (a small regulatory peptide of STAT3) (62), the lncRNA MAGI2-AS3-encoded polypeptide (63), and the lncRNA HCP5-encoded peptide (64) have been proven to have functional roles in breast cancer pathogenesis (Figure 1G).
Direct relationships of lncRNAs to mRNAs in ceRNA networks are well-documented (65). Antisense lncRNAs generally regulate sense protein-coding transcripts in two ways: cis and trans (29). A major and frequent mechanism of lncRNA post-transcriptional action is antisense regulation of cognate protein-coding mRNAs encoded on the opposite strand of the same locus; this can be positive or negative, depending on the specific sense-antisense pair (29, 66). Sense/antisense, coding/noncoding, mRNA/lncRNA pairs are a prevalent phenomenon in the human transcriptome with demonstrated functional importance in breast cancer. For instance, lncRNA ZEB1-AS1 positively regulates ZEB1 expression. By binding to embryonic lethal vision-like protein 1 (ELAVL1), ZEB1-AS1 stabilizes ZEB1 mRNA, facilitating the progression of TNBC (Figure 1H) (67). Breast cancer literature already demonstrates the prevalence of sense/antisense gene pair regulation by lncRNAs in mRNA/lncRNA pairs, such as PDCD4/lncRNA PDCD4-AS1 (68), ZNRD1/lncRNA ZNRD1-AS1 (69), HMMR/lncRNA HMMR-AS1 (70), HYOU1/lncRNA HYOU1-AS (71), and HIF-1α/lncRNA HIF-1α-AS (72).
Beside overlapping-gene regulation by antisense lncRNAs, they can regulate genes in trans and at distant loci. Certain lncRNAs partially pair with target mRNA 3’UTRs through their Alu elements and activate STAU-mediated mRNA decay (73); these Alu-Alu sense-antisense mechanisms are repeat-mediated and, unlike more common antisense regulatory modalities, generally involve transcripts from different loci, rather than from opposite strands of the same locus.
Keeping with the broad theme of sequence-mediated regulation of downstream effectors of lncRNAs, pseudogene transcripts are another abundant class of lncRNAs and have been implicated in the regulation of mRNA transcripts of the pseudogenes’ parental genes, through a protean variety of versatile mechanisms which include but are not limited to epigenetic feedback to the parental gene promoter as well as ceRNA networks dependent on the pseudogenes’ sponging of the miRNA regulators of the parental genes. Pseudogene-derived functional lncRNAs are now well-documented, including as fundamental regulators of the expression of the cognate protein-coding genes (74, 75). Recent studies suggest that pseudogenes regulate gene expression, in part, by being processed into short interfering RNAs that regulate coding genes, as well as, in other cases, by acting as microRNA decoys to regulate tumor suppressors and oncogenes (76). The lncRNA DUXAP8, transcribed from a pseudogene, modulates both the P3K/AKT/mTOR pathway and the EZH2-E-cadherin/RHOB pathways to exert its role in inducing breast cancer radioresistance (77).
2 Long non-coding RNAs of known functional relevance to radiation response in breast cancer
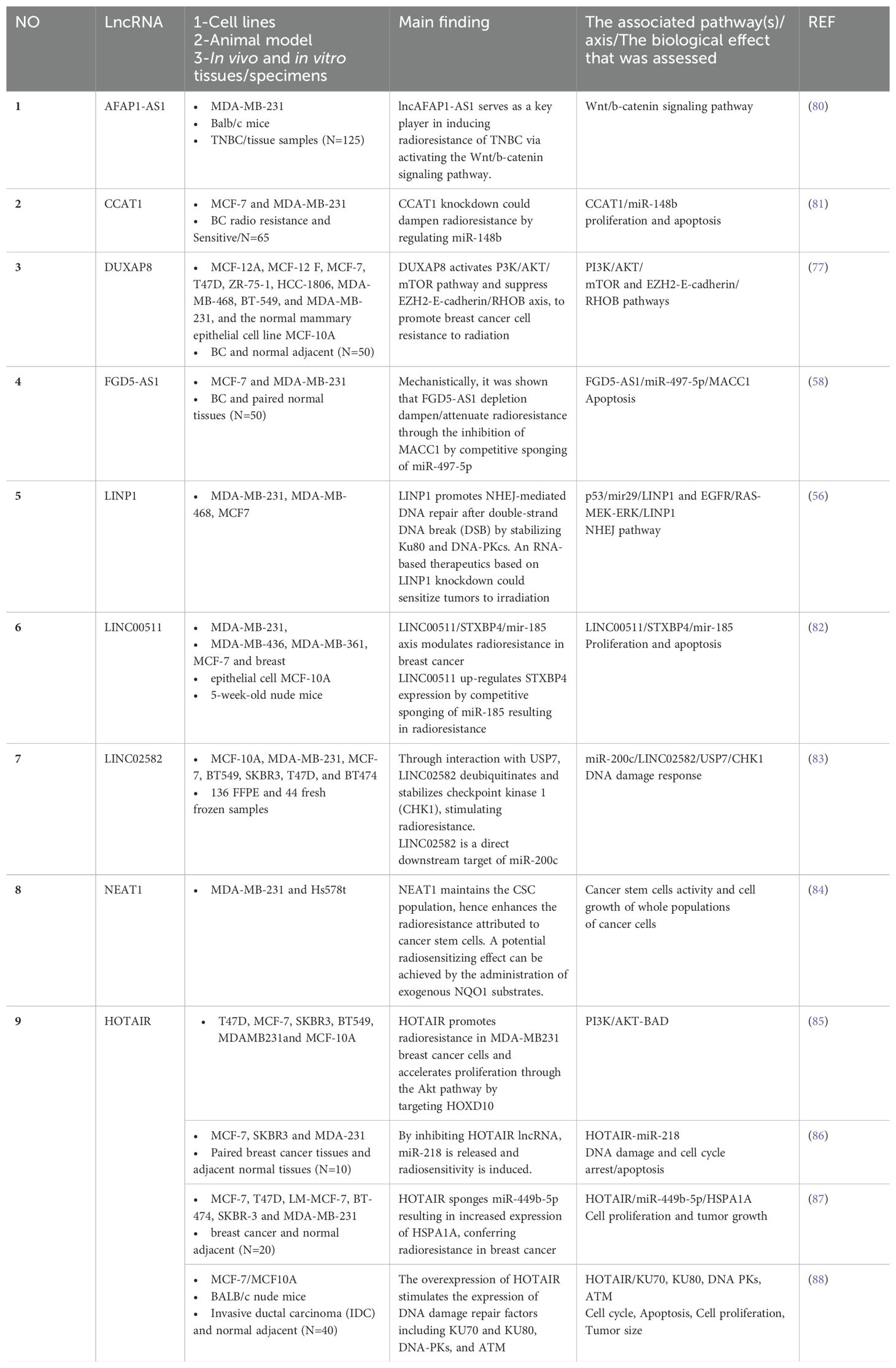
The role of lncRNAs in cancer drug resistance is now well-established (78). The emerging evidence suggests the involvement of lncRNA in RT response. Dissecting the underlying mechanisms in radioresponsiveness can provide predictive biomarkers of radioresponsiveness and identify functional molecules in radiation response pathways that will contribute to the development of targeted radiotherapies (79). Table 2 summarizes the key literature of the past 20 years on lncRNAs involved in radiation response in breast cancer.
Among these are classical (discovered during the early era of the field and now well-understood) lncRNAs such as CCAT1 (81), NEAT1 (84), and HOTAIR (85–88). The aberrant expression of HOTAIR (89), NEAT1 (90), and CCAT1 (91) is implicated in breast cancer pathogenesis. Additionally, a number of gene-desert long intergenic non-coding RNAs (lincRNAs) including LINC00511 (82) and LINC02582 (83) have been discovered during this screen. Furthermore, there are several lncRNAs highlighted in the literature relevant to RT whose genomic position provides immediate clues to their function and thereby facilitates specific therapeutic targeting or rescue approaches, including AFAP-AS1 (80) and FGD5-AS1 (58). A number of cancers are pathoetiologically linked to the lncRNA actin filament-associated protein 1 antisense RNA1 (lncAFAP1-AS1). Its second exon overlaps with exons 14-16 of the AFAP1 gene on 4p16.1. Via AFAP1-dependent and independent activities, it affects the signaling pathways involved in migratory potential and metastatic activities including PI3K/AKT, Wnt/b-catenin, EGFR/AKT, PTEN/pAKT, and RhoA/Rac2. In general, AFAP1-AS1 is considered an oncogenic lncRNA. AFAP1-independent mechanisms mediate most of the effects exerted by this lncRNA during carcinogenesis (92, 93). It has also been shown that AFAP1-AS1 induces EMT through the influence of Wnt/b-catenin signaling, not only in TNBC cells (94), but also in tongue squamous cell carcinoma (95), osteosarcoma (96), colon cancer (97), and cervical cancer (98). There is now evidence that AFAP1-AS1 is involved in promoting TNBC radioresistance via activation of the Wnt/b-catenin pathway (80).
The FGD5-AS1 lncRNA is antisense to the FGD5 gene whose direct role in breast cancer (99) gives us an immediate clue concerning the potential use of this RNA as a target for frontline therapy. We showed nearly two decades ago that antisense lncRNAs are key regulators in cancer models (29). Various cancers have abnormally high FGD5-AS1 expression that correlates with lymph node metastasis, tumor invasion, survival time, and recurrence rate. FGD5-AS1 stimulates cancer cell proliferation, metastasis, invasion, and chemoresistance both in vitro and in vivo by competing with microRNAs (including miR-5590-3p, miR-129-5p, miR-196a-5p, and miR-142-5p), leading to the mRNA’s stability and hence cell growth (100). Its relevance to radiation response through the FGD5-AS1/miR-497-5p/MACC1 axis (58) suggests that it is potentially a target in radiation sensitization. Therefore, FGD5-AS1 is an extraordinary sponge and sink for at least five different microRNAs that, in its absence, may downregulate the FGD5 mRNA. Furthermore, here for the first time we canvassed the literature to show that the expressed pseudogenes such as DUXAP8 are relevant not just to breast cancer (101) but to radiotherapy against breast cancer (77).
Therefore, we presented evidence that these dispersedly studied RT-associated lncRNAs are involved in the same biological processes including but not limited to DNA damage repair, cell cycle, cancer stem cells phenotype, and apoptosis.
3 Development of non-invasive radio-responsiveness biomarkers from the circulating lncRNAome
The primary source that drives a molecular signature with the potential to predict or assess the prognosis of radio-responsiveness and adjust to the optimum radiation intensity for each patient is the breast tumor itself. Nonetheless, liquid biopsies (blood, urine, saliva, etc.) have long proven to be a useful surrogate reservoir for biomarker investigation. Blood, by having the advantage of being collected noninvasively, mirrors valuable information to assess a condition such as radiation response for a period of time, provided that the markers arising out of the tumor are expressed and correlated with the tumor properties. This helps continuous monitoring of patients over course of (pre-, on- or post-) treatment with the promising application for a metastatic disease where the metastatic site might not be detectable or accessible for examination.
Whole blood, and specific fractions such as peripheral blood mononuclear cells (PBMC), cell-free DNA, or exosomes, are all repositories of valuable biological data (102). Exosomes are nanosized vesicles, produced by all cell types and shed outside of the cells, that are an integral component of biofluids and hence are collected in any liquid biopsy. These vesicles contain DNA, RNA, protein, and metabolites which reflect the ongoing disease processes. These cargos correlate and change with disease conditions and hence are potential biomarkers that can track disease progression (103). Collectively, these information layers can be data-mined to define the best representative signature for accurate diagnosis, prognosis, and prediction, here in the context of radiation response (104). The ultimate goal of this effort is to eventually derive a “radiation fingerprint” capable of predicting a patient’s radiation response and adjusting the best radiation dose for any individual patient.
Currently, there are no clinically validated biomarkers to reliably guide optimal RT strategies. However, circular RNAs, including non-coding circRNAs, have emerged as stable and useful biomarkers in numerous types of cancer (105). Leveraging blood-derived biomarkers from non-invasive liquid biopsies allows real-time assessment of patient response. Given their advantages of early detection, noninvasiveness, and cost-effectiveness, developing circRNA biomarkers for radioresistance and radiosensitivity in breast cancer is a promising research avenue (106). Although efforts are underway, these biomarkers still require refinement and validation due to challenges related to sensitivity and specificity.
Breast cancer is complex and heterogeneous, with different pathway activations leading to varied oncogenic drivers even within the same subtype. These differences affect tumor responses to RT, and gene signatures may not fully capture this complexity. Validating gene signatures is challenging due to variations in treatment regimens, patient subtypes, and study designs. Considering the low expression level of lncRNAs in general, as well as differences in RNA extraction methods and gene expression analysis methods, also complicates the validation process. Translating lab research into clinical tests requires standard procedures, cost-effectiveness, and proof of improvement over existing practices. These challenges limit the clinical use of gene signatures in breast cancer treatment. Nonetheless, the scientific community remains optimistic about the impact of personalized medicine on breast cancer treatment in the coming decades (106).
Figure 2 depicts an integrated approach using the combination of all modalities to find the best informative model that could be applied individually to direct the RT.
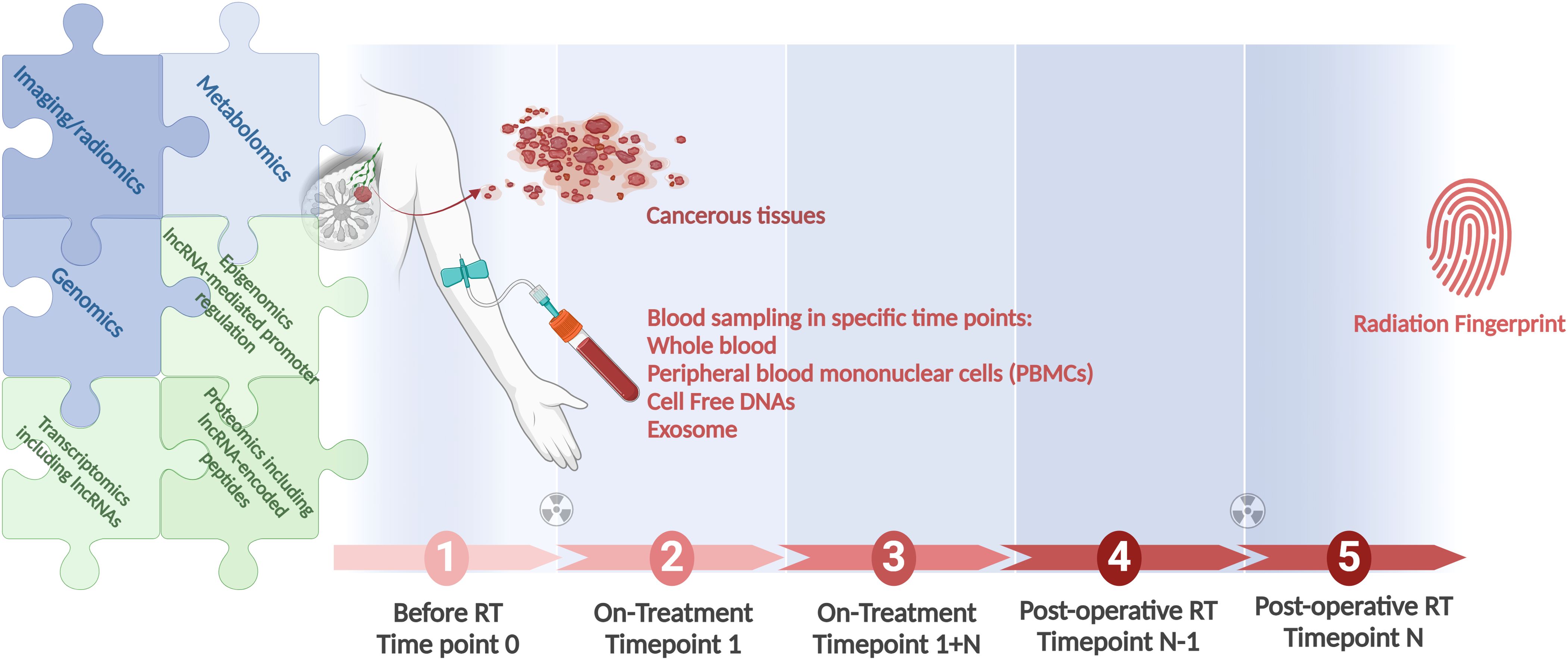
Figure 2. A workflow for integration of all clinical modules in RT, as genomics, lncRNA-inclusive transcriptomics, and radiomics, to identify the most informative radiation responsiveness signature. Longitudinal studies conducted in large cohorts provide the foundation. Associating patient genomics data with radiation response is at the focus of field radiation genomics. Areas dependent on lncRNAs, in view of these transcripts’ abundance, function, and prevalence, include epigenomics, transcriptomics, and proteomics. Correlating radiomics features with patient molecular profiles is the focus of the growing field of imaging genomics. Radiogenomics is a term that should be applicable for both the molecular and the imaging aspects of the field. Created with BioRender.com.
4 Radiosensitizers and radioprotectors as potential regulators and targets of lncRNAs
Technological improvements in RT, including conformal radiotherapy (CRT), intensity-modulated radiotherapy (IMRT), image-guided radiotherapy (IGRT), and proton‐beam radiotherapy (PBT), have improved RT efficacy by precisely targeting the tumor and minimizing the collateral normal tissue damage. To make cancerous cells more vulnerable to RT and reduce the associated toxicity on normal cells, radiosensitizers and radioprotectors, respectively, have been developed (107).
Based on their structure, radiosensitizers are categorized into small molecules, macromolecules, and nanomaterials. A comprehensive list of radiosensitizers, and of the mechanisms through which radiosensitizers boost radiation response, has been developed (108). In general, they sensitize the tumors by enhancing ionizing radiation energy deposition, catalyzing reactive oxygen species (ROS) generation and subsequently reinforcing radiation’s damaging effect on biomolecules, and modulating the tumor microenvironment (TME) (108).
Radioprotectors, which are commonly antioxidants, minimize the deleterious effect of radiation when applied before or shortly after radiation therapy. To be adopted in clinical settings, radioprotectors should not possess protective effects on cancerous cells nor toxic effects on normal tissues and need to be conveniently administrated as well (109). Examples of radioprotectors include antioxidant compounds such as glutathione, lipoic acid, and the antioxidant vitamins A, C, and E, nitroxides, cysteine and cysteamine, melatonin, and novel radioprotectors as tetracyclines and fluoroquinolones. The most widely used US Food and Drug Administration (FDA) approved radioprotector is Amifostine (WR-2721), a systemically effective radiation countermeasure. However, the associated side effects have limited their application in all oncologic settings and highlight the unmet need for the development of novel protective compounds. A list of natural radioprotectors has recently been tabulated (110). A database of radiosensitizers and radioprotectors in which users can browse typical information of a desired compound has been reported as well (111). A curated database of validated radioprotectors is also available (112).
In view of the compelling evidence that lncRNAs contribute to radioresistance, as we have demonstrated here in the context of breast cancer, targeting them along with radiotherapy could potentially increase treatment effectiveness. An in-depth exploration of lncRNA-mediated regulation of radiosensitivity has highlighted potential radiation-related signaling pathways in various cancers (104, 113). In brief they exert their influence through intricate molecular mechanisms, including DNA damage repair, cell cycle regulation, apoptosis, modulation of cancer stem cells, EMT, and autophagy. However, additional basic and clinical studies are needed to understand the intricate interactions between lncRNAs and signaling molecules that affect radiosensitivity. Advancing the field necessitates the incorporation of both small and long RNAs in medium- to high-throughput screenings, aiming to replicate the effects of radioprotectors, thus underscoring the need for continued research in this area.
5 Personalized radiation oncology and how it relates to lncRNAomics
An ideal state of the field of radiation oncology could be envisaged when the right radiation dose could be applied for the right patient with maximum desired impacts and minimal side effects. This is a part of personalized medicine, where therapeutic plan, including radiation schedules, will be tailored with genetics and phenotypic features of each individual patient to achieve the most beneficial outcome. Up until now, the one-size-fits-all approach has been adopted and dose protocols have therefore been applied uniformly for all patients. Hence, RT-related side effects and resistance remain major challenges on the way of treating breast cancer. Due to the integral contribution of genetic background to individual radiation response as well as to the essential nature of medical imaging in clinical decision-making, here we discuss the complementary role of these frontiers in further detail.
Genetic background, along with clinical characteristics of patients such as histopathology and tumor grade, progression, and drug response profiles, holds useful clues for understanding the observed disparities in radioresponsiveness (114). The advent of high-throughput next-generation sequencing has paved the way to decipher this complexity at the genomic, transcriptomic, and epigenomic levels. In 2009, a Radiogenomics Consortium (RGC) was established to facilitate and promote multi-center collaboration of researchers linking genetic variants with response to radiation therapy.
A molecular radiosensitivity index (RSI) was developed to predict RT therapeutic benefit in two independent breast cancer datasets totaling 503 patients (115). The gene expression profile of 10 genes (AR, cJun, STAT1, PKC, RelA, cABL, SUMO1, CDK1, HDAC1, and IRF1) was assessed using a linear regression algorithm (115). Clonogenic survival assays coupled with gene expression subsequently facilitated the generation of a human breast cancer-specific radiosensitivity signature (Radiotype DX) with the potential to predict locoregional recurrence and personalized RT (116). Given that this signature is assessed in various solid tumors and is independent of molecular subtype in predicting local recurrence, it potentially could be adopted in various clinical contexts (117). Integration of genomics with RT decision-making in the clinic, in a retrospective cohort study, yielded a genome-based model for adjusting radiotherapy dose (GARD score) applicable to multiple solid tumors including breast cancer (118). That study was a pioneering attempt towards the individualization of RT dose to tumor radiosensitivity using a genomically guided approach on a large scale.
To advance radiogenomics research, larger cohort studies and multi-center collaborations are necessary and candidate signatures should be validated in independent datasets. Single-cell approaches have the potential to help advance RT because they provide higher resolution with clues to detect intrinsic or induced radioresistance subpopulations. Since radiation is an exogenous intervention affecting gene expression networks, the epigenome is another informative layer for accurate characterization of the underlying mechanisms for radioresistance with the potential to be developed as biomarkers of radioresponse or druggable targets to overcome radioresistance.
The same holistic approach could be applied to mine other clinical features of patients which have been mirrored in imaging and pathology metrics. The clinical images produced by mammography, magnetic resonance imaging (MRI), and ultrasound conceivably embrace many hidden quantitative trends undetectable by humans. Describing an image by its quantitative features, notably using the strengths of sophisticated modeling by AI, provides a chance to combine and mine multi-dimensional information to develop a signature that could be routinely utilized to support clinical decision-making and predict overall survival. This is the aim of the relatively new research field of radiomics, where imaging signatures are extracted manually by predefined features (feature-based radiomics) or identified and generated from the underlying data (deep learning-based radiomics) (119). The essential steps for extracting quantitative image features can be deconstructed into four general tasks: 1- image segmentation or determination of the region of interest; 2- image processing, in order to adjust image features as pixels or intensity to make feature extraction between images with minimum error; 3- feature extraction (calculation of features as a final processing step), per the available guidelines of the Image Biomarker Standardization Initiative (IBSI) that suggest a consensus to report the extracted feature metrics and offer a consensus for standardized feature calculations from all radiomic feature matrices; 4- feature selection/dimension reduction (120). Future studies should examine whether non-coding genomic signatures are associated with medical imaging features by analyzing textural information with AI techniques.
Approximately 70-75% of human lncRNAs are primate-specific (33, 121), hence they provide a promising new line of therapy that will have more direct outcomes with fewer side effects. LncRNAs are new members of regulatory networks, as nodes that have evolved recently during evolution. They have not yet had the time to accrete too many new edges for that evolutionary reason. Accordingly, lncRNAs can be efficiently targeted without devastating downstream effects, given that side effects of drugs are often due to disrupting other components of the same complex and more evolutionarily ancient networks that a drug target is within. The distinctive features of lncRNAs identify them as advantageous targets for personalized medicine in radiation oncology and beyond. LncRNAs often exhibit high levels of variation in expression among individuals than protein-coding genes (122–124). As a result of these profiles, tailored therapies can be developed for each patient, based on their unique lncRNA expression profile (125). Moreover, they demonstrate lower expression levels, yet their expression is more specific to particular diseases, tissues, and cell types when compared to proteins or small RNAs (123, 126). This particularity has sparked an increasing interest in the capabilities of them as candidates for targeted and personalized treatments, offering reduced on-target toxicity to healthy cells and tissues.
However, despite the development of pan-cancer genomic radiosensitivity signatures and of signatures that provide predictions of radiation response in a range of different cancers, the development of assays that incorporate lncRNA signatures is still at the nascent (106). Regardless of the underlying reason, many proposed signatures for identifying intrinsic radiosensitivity show minimal to no overlap in their gene sets. LncRNAs serve as key regulators in numerous signaling pathways and act as central hubs in cellular processes. Therefore, comparing lncRNAs across not only different samples of same tumor type, but also different cancers have the potential to reveal more consistent gene expression profiles. Due to their dysregulation in multiple tumor types by considering their cancer type-specific functions, they may be ideal next-generation targets as tumor-agnostic RNA-based therapeutics (127). Due to the limited therapeutic targets available in breast cancer, obvious limitations of today’s surgical, radiation, and chemotherapy options, and a particular paucity of treatment options in TNBC – where numerous lncRNAs are directly implicated in the pathogenesis (12), lncRNAs as a class of targets merit further in-depth investigation.
6 Conclusions and future perspectives
The profound impact of RT in improving the overall survival of breast cancer patients has been frequently evidenced by large-cohort studies (4, 5). However, continual observation of radiation side effects and radioresistance in breast cancer patients implicates that we have not yet arrived at our sought-after destination in radiation oncology: a state where all patients receive their own individualized radiation protocols which hold the maximum benefit and the most advantageous risk-to-benefits ratio. Since genetic background and molecular tumor heterogeneity are among the major contributors to patients’ RT response, in this review - to evaluate where we stand in the field of radiation oncology - we undertook a comprehensive search of peer-reviewed literature referenced in PubMed and Google Scholar, in order to identify studies with a focus on dissecting the molecular basis of radioresistance and radiosensitivity in breast cancer. As we have summarized, the radiation-related pathways modulated by these lncRNAs include but are not limited to DNA damage repair, cell cycle, cancer stem cells phenotype and apoptosis. The development of sophisticated imaging technologies and radio-modifier compounds have positively reshaped RT. However, the advent of big data through the integration of ‘omics approaches will allow the field to more precisely define the most informative predictors.
Immune cells contribute fundamentally to the tumor microenvironment. Due to the challenges related to modeling immunity in vitro, immune system-related lncRNAs discovered from cell culture-based studies may not portray the bona fide players in the field. The advent of single-cell and third-generation sequencing is expected to yield additional insights on the subtypes of and the differences in tumor immune infiltration. Hence, more in vivo work on radiosensitivity- and radioresistance-associated lncRNAs, including in the tumor microenvironment context, is needed in animal models. However, just as in in vitro models, lncRNAs pose challenges in animal models. The fact that the majority of human lncRNA genes have no homologs outside of primates hinders performing endogenous loss of function experiments in rodents. However, humanized mice containing primate-specific lncRNA genes can serve as an alternative. Furthermore, human organoid systems that can replace animal models will ultimately be necessary.
The field of RT, from the discovery of X−rays by Wilhelm Conrad Röntgen in 1895 (128) to the modern era of its unequivocal application in clinics, has witnessed, and fundamentally grown through, the seminal discoveries by grand scientists as Nikola Tesla, Mihajlo Idvorski Pupin, and Maria Sklodowska-Curie on its path (129). The parallel technological breakthroughs in other clinical modules complementary to the field of radiation oncology indicate that we are expecting a treasure trove of data, with the potential to be integrated to eventually prescribe individualized radiation schedules to improve patients’ overall survival. Genomics and radiomics are prominent contributors to this field. Neither Wilhelm Roentgen’s discovery of X-rays, nor Mendel’s foundational determination of the principles of inheritance, could have possibly envisaged the incredible impact of their discoveries in diverse clinical modules and on this type of scale.
LncRNAomics offers promising improvements in cancer treatment. Their high variability in expression among individuals and tissue-specificity render them prime candidates for exploitation in precision medicine. LncRNAs can serve as biomarkers for diagnosis and prognosis, helping identify patients who may benefit from specific radiotherapy protocols. Targeting lncRNAs involved in DNA damage repair can enhance radiotherapy effectiveness. Combining lncRNAomics with imaging and clinical data can lead to more personalized treatment plans. Clinical trials focusing on integrating lncRNAomics and radiation oncology are crucial for developing new, effective treatment protocols. To move towards clinical trials, therapeutic lncRNAs are first selected and validated in vitro. As lncRNAs could be therapeutically targeted by antisense oligonucleotides before (ASOs), siRNAs or small molecules this process is followed by identifying the most effective molecules. Subsequent studies then focus on confirming their efficacy and safety in appropriate disease models.
Future research in radiation oncology and lncRNAomics faces challenges and opportunities. Technological advancements like hybrid MRI and PET scans are crucial for precise tumor targeting but are costly and require specialized training. The main challenge in lncRNA research is the limited knowledge derived from a small number of studied lncRNAs, which hinders our grasp of their mechanisms, functions, and structures. Overcoming the hurdle of delivering oligonucleotides to solid tumors remains a critical goal, despite significant progress in the field (125). As a result of refinements in AI and high-level statistical modeling, the implementation of molecular-guided treatment strategies at the resolution of one individual person has emerged as a final and reachable goal of precision medicine. Breast cancer is one of the leading cancers from the viewpoints of incidence, mortality, and cost; therefore, it will enormously benefit from incorporating a functional understanding of the role of lncRNA genes – the majority of human genes and a huge but still poorly understood pool of oncogenes, tumor suppressors, and radioresponse modifiers – into all treatment modules, from diagnosis and prediction response to designing rational therapeutic strategies.
Author contributions
FY: Conceptualization, Supervision, Visualization, Writing – original draft, Writing – review & editing. IM: Conceptualization, Supervision, Writing – review & editing. LL: Conceptualization, Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. Leonard Lipovich, PhD, was funded by the Wenzhou-Kean University faculty start-up research grant, the Wenzhou Municipal Key Laboratory for Applied Biomedical and the Biopharmaceutical Informatics (WB20211227000125), and the Zhejiang Bioinformatics International Science and Technology Cooperation Center at Wenzhou-Kean University (WB20210429000008). Ivan Martinez, PhD, was supported by Research Scholar Grants RSG-24-1039619-01-RMC and DOI 1039619 from the American Cancer Society.
Conflict of interest
Author LL has been collaborating with the Shenzhen Huayuan Biological Science Research Institute since 2019. This academic-industry research collaboration does not entail any past or present appointments at or compensation by the company, does not entail consulting, and is not remunerated. The research reported in this paper is completely outside the scope of this collaboration. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Chew HK. Adjuvant therapy for breast cancer: who should get what? West J Med. (2001) 174:284–7. doi: 10.1136/ewjm.174.4.284
2. Becker D, Hershman DL. Chapter 8 - adjuvant therapy for elderly patients with breast, colon, and lung cancer. In: Naeim A, Reuben DB, Ganz PA, editors. Management of Cancer in the Older Patient W.B. Saunders, Philadelphia (2012). p. 79–88. doi: 10.1016/B978-1-4377-1398-5.10008-6
3. Abbas Z, Rehman S. An overview of cancer treatment modalities. In: Shahzad HN, editor, Neoplasm vol. 6 . IntechOpen, Rijeka (2018). https://doi.org/10.5772/intechopen.76558.
4. Darby S, McGale P, Correa C, Taylor C, Arriagada R, Clarke M, et al. Effect of radiotherapy after breast-conserving surgery on 10-year recurrence and 15-year breast cancer death: meta-analysis of individual patient data for 10,801 women in 17 randomised trials. Lancet (London England). (2011) 378:1707–16. doi: 10.1016/S0140-6736(11)61629-2
5. Fisher B, Anderson S, Bryant J, Margolese RG, Deutsch M, Fisher ER, et al. Twenty-year follow-up of a randomized trial comparing total mastectomy, lumpectomy, and lumpectomy plus irradiation for the treatment of invasive breast cancer. N Engl J Med. (2002) 347:1233–41. doi: 10.1056/NEJMoa022152
6. Welcsh PL, King M-C. BRCA1 and BRCA2 and the genetics of breast and ovarian cancer. Hum Mol Genet. (2001) 10:705–13. doi: 10.1093/hmg/10.7.705
7. Friedman LS, Ostermeyer EA, Szabo CI, Dowd P, Lynch ED, Rowell SE, et al. Confirmation of BRCA1 by analysis of germline mutations linked to breast and ovarian cancer in ten families. Nat Genet. (1994) 8:399–404. doi: 10.1038/ng1294-399
8. Malhotra GK, Zhao X, Band H, Band V. Histological, molecular and functional subtypes of breast cancers. Cancer Biol Ther. (2010) 10:955–60. doi: 10.4161/cbt.10.10.13879
9. Anderson WF, Rosenberg PS, Prat A, Perou CM, Sherman ME. How many etiological subtypes of breast cancer: two, three, four, or more? JNCI J Natl Cancer Inst. (2014) 106:dju165. doi: 10.1093/jnci/dju165
10. Mattick JS, Amaral PP, Carninci P, Carpenter S, Chang HY, Chen L-L, et al. Long non-coding RNAs: definitions, functions, challenges and recommendations. Nat Rev Mol Cell Biol. (2023) 24(6):430–47. doi: 10.1038/s41580-022-00566-8
11. Su J, Deng L, Wang Y-D. Roles and mechanisms of long non-coding RNAs in breast cancer. Int J Mol Sci. (2023) 24(1). doi: 10.3390/ijms24010089
12. Lin C-Y, Kleinbrink EL, Dachet F, Cai J, Ju D, Goldstone A, et al. Primate-specific oestrogen-responsive long non-coding RNAs regulate proliferation and viability of human breast cancer cells. Open Biol. (2016) 6(12). doi: 10.1098/rsob.150262
13. Brownmiller T, Juric JA, Ivey AD, Harvey BM, Westemeier ES, Winters MT, et al. Y chromosome lncRNA are involved in radiation response of male non-small cell lung cancer cells. Cancer Res. (2020) 80:4046–57. doi: 10.1158/0008-5472.CAN-19-4032
14. Chen H, Han Z, Luo Q, Wang Y, Li Q, Zhou L, et al. Radiotherapy modulates tumor cell fate decisions: a review. Radiat Oncol. (2022) 17:196. doi: 10.1186/s13014-022-02171-7
15. Smith BD, Bellon JR, Blitzblau R, Freedman G, Haffty B, Hahn C, et al. Radiation therapy for the whole breast: Executive summary of an American Society for Radiation Oncology (ASTRO) evidence-based guideline. Pract Radiat Oncol. (2018) 8:145–52. doi: 10.1016/j.prro.2018.01.012
16. Kamran SC, Mouw KW. Applying precision oncology principles in radiation oncology. JCO Precis Oncol. (2018) 2. doi: 10.1200/PO.18.00034
17. Janssen S, Käsmann L, Fahlbusch FB, Rades D, Vordermark D. Side effects of radiotherapy in breast cancer patients. Strahlentherapie und Onkol. (2018) 194:136–42. doi: 10.1007/s00066-017-1197-7
18. Gray M, Turnbull AK, Ward C, Meehan J, Martínez-Pérez C, Bonello M, et al. Development and characterisation of acquired radioresistant breast cancer cell lines. Radiat Oncol. (2019) 14:64. doi: 10.1186/s13014-019-1268-2
19. Langlands FE, Horgan K, Dodwell DD, Smith L. Breast cancer subtypes: response to radiotherapy and potential radiosensitisation. Br J Radiol. (2013) 86:20120601. doi: 10.1259/bjr.20120601
20. Desouky O, Ding N, Zhou G. Targeted and non-targeted effects of ionizing radiation. J Radiat Res Appl Sci. (2015) 8:247–54. doi: 10.1016/j.jrras.2015.03.003
21. Sia J, Szmyd R, Hau E, Gee HE. Molecular mechanisms of radiation-induced cancer cell death: A primer. Front Cell Dev Biol. (2020) 8:41. doi: 10.3389/fcell.2020.00041
22. Kumari S, Mukherjee S, Sinha D, Abdisalaam S, Krishnan S, Asaithamby A. Immunomodulatory effects of radiotherapy. Int J Mol Sci. (2020) 21. doi: 10.3390/ijms21218151
23. Brownlee Z, Garg R, Listo M, Zavitsanos P, Wazer DE, Huber KE. Late complications of radiation therapy for breast cancer: Evolution in techniques and risk over time. Gland Surg. (2018) 7:371–8. doi: 10.21037/gs
24. Krisnawan VE, Stanley JA, Schwarz JK, Denardo DG. Tumor microenvironment as a regulator of radiation therapy: New insights into stromalmediated radioresistance. Cancers (Basel). (2020) 12:1–25. doi: 10.3390/cancers12102916
25. Wang J-S, Wang H-J, Qian H-L. Biological effects of radiation on cancer cells. Mil Med Res. (2018) 5:20. doi: 10.1186/s40779-018-0167-4
26. Dianatpour A, Ghafouri-Fard S. The role of long non coding RNAs in the repair of DNA double strand breaks. Int J Mol Cell Med. (2017) 6:1–12.
27. Hu WL, Jin L, Xu A, Wang YF, Thorne RF, Zhang XD, et al. GUARDIN is a p53-responsive long non-coding RNA that is essential for genomic stability. Nat Cell Biol. (2018) 20:492–502. doi: 10.1038/s41556-018-0066-7
28. Dunham I, Kundaje A, Aldred SF, Collins PJ, Davis CA, Doyle F, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. (2012) 489:57–74. doi: 10.1038/nature11247
29. Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, et al. Antisense transcription in the mammalian transcriptome. Science. (2005) 309:1564–6. doi: 10.1126/science.1112009
30. Iyer MK, Niknafs YS, Malik R, Singhal U, Sahu A, Hosono Y, et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. (2015) 47:199–208. doi: 10.1038/ng.3192
31. Hon C-C, Ramilowski JA, Harshbarger J, Bertin N, Rackham OJL, Gough J, et al. An atlas of human long non-coding RNAs with accurate 5’ ends. Nature. (2017) 543:199–204. doi: 10.1038/nature21374
32. Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. (2012) 22:1775–89. doi: 10.1101/gr.132159.111
33. Washietl S, Kellis M, Garber M. Evolutionary dynamics and tissue specificity of human long noncoding RNAs in six mammals. Genome Res. (2014) 24:616–28. doi: 10.1101/gr.165035.113
34. Necsulea A, Kaessmann H. Evolutionary dynamics of coding and non-coding transcriptomes. Nat Rev Genet. (2014) 15:734–48. doi: 10.1038/nrg3802
35. Taft RJ, Pheasant M, Mattick JS. The relationship between non-protein-coding DNA and eukaryotic complexity. Bioessays. (2007) 29:288–99. doi: 10.1002/bies.20544
36. Lipovich L, Hou Z-C, Jia H, Sinkler C, McGowen M, Sterner KN, et al. High-throughput RNA sequencing reveals structural differences of orthologous brain-expressed genes between western lowland gorillas and humans. J Comp Neurol. (2016) 524:288–308. doi: 10.1002/cne.23843
37. Graf J, Kretz M. From structure to function: Route to understanding lncRNA mechanism. BioEssays : News Rev molecular Cell Dev Biol United States;. (2020) 42:e2000027. doi: 10.1002/bies.202000027
38. Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature. (2012) 482:339–46. doi: 10.1038/nature10887
39. Gloss BS, Dinger ME. The specificity of long noncoding RNA expression. Biochim Biophys Acta - Gene Regul Mech. (2016) 1859:16–22. https://www.sciencedirect.com/science/article/pii/S1874939915001741.
40. Flynn RA, Chang HY. Long noncoding RNAs in cell-fate programming and reprogramming. Cell Stem Cell. (2014) 14:752–61. doi: 10.1016/j.stem.2014.05.014
41. Benoit Bouvrette LP, Cody NAL, Bergalet J, Lefebvre FA, Diot C, Wang X, et al. CeFra-seq reveals broad asymmetric mRNA and noncoding RNA distribution profiles in Drosophila and human cells. RNA. (2018) 24:98–113. doi: 10.1261/rna.063172.117
42. Wilk R, Hu J, Blotsky D, Krause HM. Diverse and pervasive subcellular distributions for both coding and long noncoding RNAs. Genes Dev. (2016) 30:594–609. doi: 10.1101/gad.276931.115
43. Wood EJ, Chin-Inmanu K, Jia H, Lipovich L. Sense-antisense gene pairs: sequence, transcription, and structure are not conserved between human and mouse. Front Genet. (2013) 4:183. doi: 10.3389/fgene.2013.00183
44. Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. (2010) 464:1071–6. doi: 10.1038/nature08975
45. Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. (2007) 129:1311–23. doi: 10.1016/j.cell.2007.05.022
46. Tsai M-C, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science. (2010) 329:689–93. doi: 10.1126/science.1192002
47. Fanucchi S, Fok ET, Dalla E, Shibayama Y, Börner K, Chang EY, et al. Immune genes are primed for robust transcription by proximal long noncoding RNAs located in nuclear compartments. Nat Genet. (2019) 51:138–50. doi: 10.1038/s41588-018-0298-2
48. Lai F, Orom UA, Cesaroni M, Beringer M, Taatjes DJ, Blobel GA, et al. Activating RNAs associate with Mediator to enhance chromatin architecture and transcription. Nature. (2013) 494:497–501. doi: 10.1038/nature11884
49. Fan S, Tian T, Lv X, Lei X, Yang Z, Liu M, et al. lncRNA CISAL inhibits BRCA1 transcription by forming a tertiary structure at its promoter. iScience. (2020) 23:100835. doi: 10.1016/j.isci.2020.100835
50. Ma HN, Chen HJ, Liu JQ, Li WT. Long non-coding RNA DLEU1 promotes Malignancy of breast cancer by acting as an indispensable coactivator for HIF-1α-induced transcription of CKAP2. Cell Death Dis [Internet]. (2022) 13:625. doi: 10.1038/s41419-022-04880-z
51. Willingham AT, Orth AP, Batalov S, Peters EC, Wen BG, Aza-Blanc P, et al. A strategy for probing the function of noncoding RNAs finds a repressor of NFAT. Science. (2005) 309:1570–3. doi: 10.1126/science.1115901
52. Fox AH, Lam YW, Leung AKL, Lyon CE, Andersen J, Mann M, et al. Paraspeckles: a novel nuclear domain. Curr Biol. (2002) 12:13–25. doi: 10.1016/S0960-9822(01)00632-7
53. Maass PG, Barutcu AR, Weiner CL, Rinn JL. Inter-chromosomal contact properties in live-cell imaging and in hi-C. Mol Cell. (2018) 69:1039–45. doi: 10.1016/j.molcel.2018.02.007
54. Lalonde M, Chartrand P. TERRA, a multifaceted regulator of telomerase activity at telomeres. J Mol Biol. (2020) 432:4232–43. doi: 10.1016/j.jmb.2020.02.004
55. García-Padilla C, Muñoz-Gallardo MDM, Lozano-Velasco E, Castillo-Casas JM, Caño-Carrillo S, García-López V, et al. New Insights into the Roles of lncRNAs as Modulators of Cytoskeleton Architecture and Their Implications in Cellular Homeostasis and in Tumorigenesis. Non-coding RNA. (2022) 8. doi: 10.3390/ncrna8020028
56. Zhang Y, He Q, Hu Z, Feng Y, Fan L, Tang Z, et al. Long noncoding RNA LINP1 regulates repair of DNA double-strand breaks in triple-negative breast cancer. Nat Struct Mol Biol. (2016) 23:522–30. doi: 10.1038/nsmb.3211
57. Goustin AS, Thepsuwan P, Kosir MA, Lipovich L. The growth-arrest-specific (GAS)-5 long non-coding RNA: A fascinating lncRNA widely expressed in cancers. Non-coding RNA. (2019) 5. doi: 10.3390/ncrna5030046
58. Li J, Lei C, Chen B, Zhu Q. LncRNA FGD5-AS1 facilitates the radioresistance of breast cancer cells by enhancing MACC1 expression through competitively sponging miR-497-5p. Front Oncol. (2021) 11:671853. doi: 10.3389/fonc.2021.671853
59. Stein U, Walther W, Arlt F, Schwabe H, Smith J, Fichtner I, et al. MACC1, a newly identified key regulator of HGF-MET signaling, predicts colon cancer metastasis. Nat Med. (2009) 15:59–67. doi: 10.1038/nm.1889
60. Radhakrishnan H, Walther W, Zincke F, Kobelt D, Imbastari F, Erdem M, et al. MACC1-the first decade of a key metastasis molecule from gene discovery to clinical translation. Cancer Metastasis Rev. (2018) 37:805–20. doi: 10.1007/s10555-018-9771-8
61. Bánfai B, Jia H, Khatun J, Wood E, Risk B, Gundling WEJ, et al. Long noncoding RNAs are rarely translated in two human cell lines. Genome Res. (2012) 22:1646–57. doi: 10.1101/gr.134767.111
62. Wang Y, Wu S, Zhu X, Zhang L, Deng J, Li F, et al. LncRNA-encoded polypeptide ASRPS inhibits triple-negative breast cancer angiogenesis. J Exp Med. (2019) 217:e20190950. doi: 10.1084/jem.20190950
63. Zhang Z, Yi Y, Wang Z, Zhang H, Zhao Y, He R, et al. LncRNA MAGI2-AS3-encoded polypeptide restrains the proliferation and migration of breast cancer cells. Mol Biotechnol. (2023) 66(6):1409–23. doi: 10.1007/s12033-023-00801-3
64. Tong X, Yu Z, Xing J, Liu H, Zhou S, Huang Y, et al. LncRNA HCP5-encoded protein regulates ferroptosis to promote the progression of triple-negative breast cancer. Cancers (Basel). (2023) 15. doi: 10.3390/cancers15061880
65. Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. (2011) 146:353–8. doi: 10.1016/j.cell.2011.07.014
66. Engström PG, Suzuki H, Ninomiya N, Akalin A, Sessa L, Lavorgna G, et al. Complex Loci in human and mouse genomes. PloS Genet. (2006) 2:e47. doi: 10.1371/journal.pgen.0020047
67. Luo N, Zhang K, Li X, Hu Y. ZEB1 induced-upregulation of long noncoding RNA ZEB1-AS1 facilitates the progression of triple negative breast cancer by binding with ELAVL1 to maintain the stability of ZEB1 mRNA. J Cell Biochem. (2020) 121:4176–87. doi: 10.1002/jcb.29572
68. Jadaliha M, Gholamalamdari O, Tang W, Zhang Y, Petracovici A, Hao Q, et al. A natural antisense lncRNA controls breast cancer progression by promoting tumor suppressor gene mRNA stability. PloS Genet. (2018) 14:e1007802. doi: 10.1371/journal.pgen.1007802
69. Kim HW, Jeong D, Ham J, Kim H, Ji HW, Choi EH, et al. ZNRD1 and its antisense long noncoding RNA ZNRD1-AS1 are oppositely regulated by cold atmospheric plasma in breast cancer cells. Oxid Med Cell Longev. (2020) 2020:9490567. doi: 10.1155/2020/9490567
70. Liu W, Ma J, Cheng Y, Zhang H, Luo W, Zhang H. HMMR antisense RNA 1, a novel long noncoding RNA, regulates the progression of basal-like breast cancer cells. Breast Cancer (Dove Med Press. (2016) 8:223–9. doi: 10.2147/BCTT
71. Hao A, Wang Y, Zhang X, Li J, Li Y, Li D, et al. Long non-coding antisense RNA HYOU1-AS is essential to human breast cancer development through competitive binding hnRNPA1 to promote HYOU1 expression. Biochim Biophys Acta - Mol Cell Res. (2021) 1868:118951. https://www.sciencedirect.com/science/article/pii/S0167488921000057.
72. Zheng F, Chen J, Zhang X, Wang Z, Chen J, Lin X, et al. The HIF-1α antisense long non-coding RNA drives a positive feedback loop of HIF-1α mediated transactivation and glycolysis. Nat Commun. (2021) 12:1341. doi: 10.1038/s41467-021-21535-3
73. Gong C, Maquat LE. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3’ UTRs via Alu elements. Nature. (2011) 470:284–8. doi: 10.1038/nature09701
74. Milligan MJ, Harvey E, Yu A, Morgan AL, Smith DL, Zhang E, et al. Global intersection of long non-coding RNAs with processed and unprocessed pseudogenes in the human genome. Front Genet. (2016) 7:26. doi: 10.3389/fgene.2016.00026
75. Choo S-W, Zhong Y, Sendler E, Goustin A-S, Cai J, Ju D, et al. Estrogen distinctly regulates transcription and translation of lncRNAs and pseudogenes in breast cancer cells. Genomics. (2022) 114:110421. https://www.sciencedirect.com/science/article/pii/S0888754322001665.
76. Pink RC, Wicks K, Caley DP, Punch EK, Jacobs L, Carter DRF. Pseudogenes: pseudo-functional or key regulators in health and disease? RNA. (2011) 17:792–8. doi: 10.1261/rna.2658311
77. Lei C, Li S, Fan Y, Hua L, Pan Q, Li Y, et al. LncRNA DUXAP8 induces breast cancer radioresistance by modulating the PI3K/AKT/mTOR pathway and the EZH2-E-cadherin/RHOB pathway. Cancer Biol Ther. (2022) 23:1–13. doi: 10.1080/15384047.2022.2132008
78. He J, Zhu S, Liang X, Zhang Q, Luo X, Liu C, et al. LncRNA as a multifunctional regulator in cancer multi-drug resistance. Mol Biol Rep. (2021) 48:1–15. doi: 10.1007/s11033-021-06603-7
79. Xie Y, Han J, Xie K, Gou Q. LncRNAs as biomarkers for predicting radioresistance and survival in cancer: a meta-analysis. Sci Rep. (2022) 12:18494. doi: 10.1038/s41598-022-21785-1
80. Bi Z, Li Q, Dinglin X, Xu Y, You K, Hong H, et al. Nanoparticles (NPs)-meditated lncRNA AFAP1-AS1 silencing to block wnt/β-catenin signaling pathway for synergistic reversal of radioresistance and effective cancer radiotherapy. Adv Sci (Weinheim Baden-Wurttemberg Ger. (2020) 7:2000915. doi: 10.1002/advs.202000915
81. Lai Y, Chen Y, Lin Y, Ye L. Down-regulation of LncRNA CCAT1 enhances radiosensitivity via regulating miR-148b in breast cancer. Cell Biol Int. (2018) 42:227–36. doi: 10.1002/cbin.10890
82. Liu L, Zhu Y, Liu A-M, Feng Y, Chen Y. Long noncoding RNA LINC00511 involves in breast cancer recurrence and radioresistance by regulating STXBP4 expression via miR-185. Eur Rev Med Pharmacol Sci. (2019) 23:7457–68. doi: 10.26355/eurrev_201909_18855
83. Wang B, Zheng J, Li R, Tian Y, Lin J, Liang Y, et al. Long noncoding RNA LINC02582 acts downstream of miR-200c to promote radioresistance through CHK1 in breast cancer cells. Cell Death Dis. (2019) 10:764. doi: 10.1038/s41419-019-1996-0
84. Lin L-C, Lee H-T, Chien P-J, Huang Y-H, Chang M-Y, Lee Y-C, et al. NAD(P)H:quinone oxidoreductase 1 determines radiosensitivity of triple negative breast cancer cells and is controlled by long non-coding RNA NEAT1. Int J Med Sci. (2020) 17:2214–24. doi: 10.7150/ijms.45706
85. Zhou Y, Wang C, Liu X, Wu C, Yin H. Long non-coding RNA HOTAIR enhances radioresistance in MDA-MB231 breast cancer cells. Oncol Lett. (2017) 13:1143–8. doi: 10.3892/ol.2017.5587
86. Hu X, Ding D, Zhang J, Cui J. Knockdown of lncRNA HOTAIR sensitizes breast cancer cells to ionizing radiation through activating miR-218. Biosci Rep. (2019) 39. doi: 10.1042/BSR20181038
87. Zhang S, Wang B, Xiao H, Dong J, Li Y, Zhu C, et al. LncRNA HOTAIR enhances breast cancer radioresistance through facilitating HSPA1A expression via sequestering miR-449b-5p. Thorac cancer. (2020) 11:1801–16. doi: 10.1111/1759-7714.13450
88. Qian L, Fei Q, Zhang H, Qiu M, Zhang B, Wang Q, et al. lncRNA HOTAIR promotes DNA repair and radioresistance of breast cancer via EZH2. DNA Cell Biol. (2020) 39(12):2166–73. doi: 10.1089/dna.2020.5771
89. Cantile M, Di Bonito M, Cerrone M, Collina F, De Laurentiis M, Botti G. Long non-coding RNA HOTAIR in breast cancer therapy. Cancers. (2020) 12. doi: 10.3390/cancers12051197
90. Knutsen E, Harris AL, Perander M. Expression and functions of long non-coding RNA NEAT1 and isoforms in breast cancer. Br J Cancer. (2022) 126:551–61. doi: 10.1038/s41416-021-01588-3
91. Selem NA, Youness RA, Gad MZ. What is beyond LncRNAs in breast cancer: A special focus on colon cancer-associated Transcript-1 (CCAT-1). Non-coding RNA Res. (2021) 6:174–86. https://www.sciencedirect.com/science/article/pii/S2468054021000445.
92. Ghafouri-Fard S, Khoshbakht T, Hussen BM, Taheri M, Mokhtari M. A review on the role of AFAP1-AS1 in the pathoetiology of cancer. Front Oncol. (2021) 11:777849. doi: 10.3389/fonc.2021.777849
93. Ji D, Zhong X, Jiang X, Leng K, Xu Y, Li Z, et al. The role of long non-coding RNA AFAP1-AS1 in human Malignant tumors. Pathol Res Pract. (2018) 214:1524–31. doi: 10.1016/j.prp.2018.08.014
94. Zhang K, Liu P, Tang H, Xie X, Kong Y, Song C, et al. AFAP1-AS1 promotes epithelial-mesenchymal transition and tumorigenesis through wnt/β-catenin signaling pathway in triple-negative breast cancer. Front Pharmacol. (2018) 9:1248. doi: 10.3389/fphar.2018.01248
95. Wang Z-Y, Hu M, Dai M-H, Xiong J, Zhang S, Wu H-J, et al. Upregulation of the long non-coding RNA AFAP1-AS1 affects the proliferation, invasion and survival of tongue squamous cell carcinoma via the Wnt/β-catenin signaling pathway. Mol Cancer. (2018) 17:3. doi: 10.1186/s12943-017-0752-2
96. Li R, Liu S, Li Y, Tang Q, Xie Y, Zhai R. Long noncoding RNA AFAP1−AS1 enhances cell proliferation and invasion in osteosarcoma through regulating miR−4695−5p/TCF4−β−catenin signaling. Mol Med Rep. (2018) 18:1616–22. doi: 10.3892/mmr
97. Bo H, Fan L, Li J, Liu Z, Zhang S, Shi L, et al. High expression of lncRNA AFAP1-AS1 promotes the progression of colon cancer and predicts poor prognosis. J Cancer. (2018) 9:4677–83. doi: 10.7150/jca.26461
98. Bo H, Fan L, Gong Z, Liu Z, Shi L, Guo C, et al. Upregulation and hypomethylation of lncRNA AFAP1−AS1 predicts a poor prognosis and promotes the migration and invasion of cervical cancer. Oncol Rep. (2019) 41:2431–9. doi: 10.3892/or
99. Fang K, Xu Z-J, Jiang S-X, Tang D-S, Yan C-S, Deng Y-Y, et al. lncRNA FGD5−AS1 promotes breast cancer progression by regulating the hsa−miR−195−5p/NUAK2 axis. Mol Med Rep. (2021) 23. doi: 10.3892/mmr
100. He N, Xiang L, Chen L, Tong H, Wang K, Zhao J, et al. The role of long non-coding RNA FGD5-AS1 in cancer. Bioengineered. (2022) 13:11026–41. doi: 10.1080/21655979.2022.2067292
101. Wang Y, Jiang X, Zhang D, Zhao Y, Han X, Zhu L, et al. LncRNA DUXAP8 as a prognostic biomarker for various cancers: A meta-analysis and bioinformatics analysis. Front Genet. (2022) 13:907774. doi: 10.3389/fgene.2022.907774
102. Lone SN, Nisar S, Masoodi T, Singh M, Rizwan A, Hashem S, et al. Liquid biopsy: a step closer to transform diagnosis, prognosis and future of cancer treatments. Mol Cancer. (2022) 21:79. doi: 10.1186/s12943-022-01543-7
103. Wang X, Tian L, Lu J, Ng IO-L. Exosomes and cancer - Diagnostic and prognostic biomarkers and therapeutic vehicle. Oncogenesis. (2022) 11:54. doi: 10.1038/s41389-022-00431-5
104. Xie Y, Han J, Xie K, Gou Q. LncRNAs as biomarkers for predicting radioresistance and survival in cancer: a meta-analysis. Sci Rep. (2022) 12:18494. doi: 10.1038/s41598-022-21785-1
105. Zhang Z, Yang T, Xiao J. Circular RNAs: promising biomarkers for human diseases. EBioMedicine. (2018) 34:267–74. doi: 10.1016/j.ebiom.2018.07.036
106. Meehan J, Gray M, Martínez-Pérez C, Kay C, Pang LY, Fraser JA, et al. Precision medicine and the role of biomarkers of radiotherapy response in breast cancer. Front Oncol. (2020) 10:628. doi: 10.3389/fonc.2020.00628
107. Citrin DE. Radiation modifiers. Hematol Oncol Clin North Am. (2019) 33:1041–55. doi: 10.1016/j.hoc.2019.08.004
108. Gong L, Zhang Y, Liu C, Zhang M, Han S. Application of radiosensitizers in cancer radiotherapy. Int J Nanomedicine. (2021) 16:1083–102. doi: 10.2147/IJN.S290438
109. Raviraj J, Bokkasam VK, Kumar VS, Reddy US, Suman V. Radiosensitizers, radioprotectors, and radiation mitigators. Indian J Dent Res Off Publ Indian Soc Dent Res. (2014) 25:83–90. doi: 10.4103/0970-9290.131142
110. Kuruba V, Gollapalli P. Natural radioprotectors and their impact on cancer drug discovery. Radiat Oncol J. (2018) 36:265–75. doi: 10.3857/roj.2018.00381
111. Dongre P, Joshi A. A systematic organization of bioinformatics database of radiosensitizers and radioprotectors. J Radiat Cancer Res. (2018) 9:102. doi: 10.4103/jrcr.jrcr_5_18
112. Aliper AM, Bozdaganyan ME, Sarkisova VA, Veviorsky AP, Ozerov IV, Orekhov PS, et al. Radioprotectors. org: An open database of known and predicted radioprotectors. Aging (Albany NY). (2020) 12:15741–55. doi: 10.18632/aging.v12i15
113. Zhu J, Chen S, Yang B, Mao W, Yang X, Cai J. Molecular mechanisms of lncRNAs in regulating cancer cell radiosensitivity. Biosci Rep. (2019) 39. doi: 10.1042/BSR20190590
114. Shui L, Ren H, Yang X, Li J, Chen Z, Yi C, et al. The era of radiogenomics in precision medicine: an emerging approach to support diagnosis, treatment decisions, and prognostication in oncology. Front Oncol. (2021) 10. doi: 10.3389/fonc.2020.570465
115. Eschrich SA, Fulp WJ, Pawitan Y, Foekens JA, Smid M, Martens JWM, et al. Validation of a radiosensitivity molecular signature in breast cancer. Clin Cancer Res. (2012) 18:5134–43. doi: 10.1158/1078-0432.CCR-12-0891
116. Speers C, Zhao S, Liu M, Bartelink H, Pierce LJ, Feng FY. Development and validation of a novel radiosensitivity signature in human breast cancer. Clin Cancer Res. (2015) 21:3667–77. doi: 10.1158/1078-0432.CCR-14-2898
117. Kunkler I. Genomic-adjusted radiation dose. Lancet Oncol. (2017) 18:e128. doi: 10.1016/S1470-2045(17)30090-6
118. Scott JG, Berglund A, Schell MJ, Mihaylov I, Fulp WJ, Yue B, et al. A genome-based model for adjusting radiotherapy dose (GARD): a retrospective, cohort-based study. Lancet Oncol. (2017) 18:202–11. doi: 10.1016/S1470-2045(16)30648-9
119. Lohmann P, Bousabarah K, Hoevels M, Treuer H. Radiomics in radiation oncology—basics, methods, and limitations. Strahlentherapie und Onkol. (2020) 196:848–55. doi: 10.1007/s00066-020-01663-3
120. van Timmeren JE, Cester D, Tanadini-Lang S, Alkadhi H, Baessler B. Radiomics in medical imaging—“how-to” guide and critical reflection. Insights Imaging. (2020) 11:91. doi: 10.1186/s13244-020-00887-2
121. Necsulea A, Soumillon M, Warnefors M, Liechti A, Daish T, Zeller U, et al. The evolution of lncRNA repertoires and expression patterns in tetrapods. Nature. (2014) 505:635–40. doi: 10.1038/nature12943
122. de Goede OM, Nachun DC, Ferraro NM, Gloudemans MJ, Rao AS, Smail C, et al. Population-scale tissue transcriptomics maps long non-coding RNAs to complex disease. Cell. (2021) 184:2633–48. doi: 10.1016/j.cell.2021.03.050
123. Kornienko AE, Dotter CP, Guenzl PM, Gisslinger H, Gisslinger B, Cleary C, et al. Long non-coding RNAs display higher natural expression variation than protein-coding genes in healthy humans. Genome Biol. (2016) 17:14. doi: 10.1186/s13059-016-0873-8
124. Melé M, Ferreira PG, Reverter F, DeLuca DS, Monlong J, Sammeth M, et al. Human genomics. The human transcriptome across tissues and individuals. Science. (2015) 348:660–5. doi: 10.1126/science.aaa0355
125. Coan M, Haefliger S, Ounzain S, Johnson R. Targeting and engineering long non-coding RNAs for cancer therapy. Nat Rev Genet. (2024) 25:578–95. doi: 10.1038/s41576-024-00693-2
126. Unfried JP, Ulitsky I. Substoichiometric action of long noncoding RNAs. Nat Cell Biol. (2022) 24:608–15. doi: 10.1038/s41556-022-00911-1
127. Chen B, Dragomir MP, Yang C, Li Q, Horst D, Calin GA. Targeting non-coding RNAs to overcome cancer therapy resistance. Signal Transduct Target Ther. (2022) 7:121. doi: 10.1038/s41392-022-00975-3
128. Tubiana M. [Wilhelm conrad röntgen and the discovery of X-rays]. Bull Acad Natl Med. (1996) 180:97–108.
Keywords: breast neoplasms, long non-coding RNAs, non-coding RNAs, precision medicine, radioresistance, radiosensitivity, radiotherapy, genomics
Citation: Yazarlou F, Martinez I and Lipovich L (2024) Radiotherapy and breast cancer: finally, an lncRNA perspective on radiosensitivity and radioresistance. Front. Oncol. 14:1437542. doi: 10.3389/fonc.2024.1437542
Received: 23 May 2024; Accepted: 01 August 2024;
Published: 13 September 2024.
Edited by:
Brian D. Adams, Brain Institute of America, United StatesReviewed by:
Zhuofei Bi, Sun Yat-sen University, ChinaFrancesco P. Cammarata, National Research Council (CNR), Italy
Copyright © 2024 Yazarlou, Martinez and Lipovich. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fatemeh Yazarlou, RmF0ZW1laC5ZYXphcmxvdUBuYXRpb253aWRlY2hpbGRyZW5zLm9yZw==; Leonard Lipovich, TExJUE9WSUNAV0tVLkVEVS5DTg==
 Fatemeh Yazarlou
Fatemeh Yazarlou Ivan Martinez
Ivan Martinez Leonard Lipovich
Leonard Lipovich