- 1Analysis of Circulating Tumor Cells, Laboratory of Analytical Chemistry, Department of Chemistry, University of Athens, Athens, Greece
- 2Department of Pathology, Onassis Cardiac Surgery Center, Athens, Greece
- 3Department of Medical Oncology, University General Hospital of Alexandroupolis, Alexandroupolis, Greece
- 4Oncology Unit, Metropolitan Hospital, Athens, Greece
- 5Department of Clinical Therapeutics, School of Medicine, Alexandra Hospital, National and Kapodistrian University of Athens, Athens, Greece
- 6Third Department of Medical Oncology, Hygeia Hospital, Athens, Greece
- 7Section of Medical Oncology, Department of Internal Medicine, Faculty of Medicine, Attikon University Hospital, Athens, Greece
- 8Oncology Unit, 2nd Department of Surgery, School of Medicine, Aretaieio Hospital, National and Kapodistrian University of Athens, Athens, Greece
Introduction: Detection of PIK3CA mutations in primary tumors and liquid biopsy samples is of increasing importance for treatment decisions and therapy resistance in many types of cancer. The aim of the present study was to directly compare the efficacy of a relatively inexpensive ultrasensitive real-time PCR with the well-established and highly sensitive technology of ddPCR for the detection of the three most common hotspot mutations of PIK3CA, in exons 9 and 20, that are all of clinical importance in various types of cancer.
Patients and methods: We analyzed 42 gDNAs from primary tumors (FFPEs), 29 plasma-cfDNA samples, and 29 paired CTC-derived gDNAs, all from patients with ER+ metastatic breast cancer, and plasma from 10 healthy donors. The same blood draws were used for CTC isolation using EpCAM beads for positive immunomagnetic enrichment. All FFPEs and plasma-cfDNA samples were analyzed in parallel for PIK3CA mutations by ultrasensitive real-time PCR assay and droplet digital PCR.
Results: In gDNAs from FFPEs, using ultrasensitive real-time PCR, the p.E545K mutation was detected in 22/42(52.4%), and the p.E542K and p.H1047R mutations were detected in 14/42(33.3%) and 16/42(38.1%), respectively. Using ddPCR, the p.E545K mutation was detected in 22/42(52.4%), p.E542K in 17/42(40.5%), and p.H1047R in 19/42(45.2%) samples, revealing a concordance between the two methodologies of 81%, 78.6% and 78.6% for each mutation respectively. In plasma-cfDNA, using ultrasensitive real-time PCR, the p.E545K mutation was detected in 11/29(38%) and both p.E542K and p.H1047R mutations in 2/29(6.9%).In the same plasma-cfDNA samples using ddPCR, p.E545K was detected in 1/29(3.5%), p.E542K in 2/29(6.9%), and p.H1047R in 3/29(10.5%) samples, revealing a concordance of 65.5%,100% and 93.1% for each mutation respectively. In paired CTC-derived gDNAs p.E545K was detected in 11/29(38%), p.E542K in 3/29(10.3%), and p.H1047R in 7/29(24.1%) samples.
Conclusions: This low-cost, high-throughput and ultrasensitive real-time PCR assay provides accurate and specific detection of PIK3CA hotspot mutations in liquid biopsy samples and gives similar results to ddPCR. This assay can be performed in labs where digital PCR instrumentation is not available. In CTC-derived gDNA and paired plasma-cfDNA, PIK3CA mutations detected were not identical, revealing that CTC and plasma-cfDNA give complementary information.
1 Introduction
PI3K (Phosphoinositide 3-kinase) signaling is deregulated in a variety of cancers (1–4). The three main PIK3CA hotspot mutations, exon 9 p.E545K and p.E542K and exon 20 p.H1047R, are detected approximately in 40% of hormone receptor positive (HR+) breast cancer, mainly in the helical and kinase domains of the PIK3CA gene (5, 6). Beyond breast cancer, PIK3CA mutation detection is also very important in other types of cancer like lung (7), colorectal (4, 8), anal squamous cell carcinoma (9) and pancreatic cancer (10).
In HR+ metastatic breast cancer patients with lack of human epidermal growth factor receptor-2 (HER2) overexpression and/or amplification, the widely accepted therapy is the combination of endocrine therapy (ET) with cyclin-dependent kinases (CDK)4/6 inhibitors (11). Therapeutics targeting specific driver mutations in PIK3CA have revolutionized the treatment of breast cancer in recent years (6, 12, 13). Alpelisib, a PIK3 inhibitor, has been approved by both the U.S. Food and Drug Administration (FDA) and the European Medicines Agency (EMA) for use in combination with fulvestrantin patients with HR+, HER2-negative (HER2-), PIK3CA-mutated, advanced or metastatic breast cancer following progression on or after treatment with an endocrine-based regimen (14–17). Moreover, triplet therapy with palbociclib, taselisib, and fulvestrant has been reported to have a positive effect in patients with heavily pretreated PIK3CA-mutantER-positive (ER+)/HER2- advanced breast cancer (18). There are several factors that ultimately result in resistance to PI3K inhibitors such as a) inactivation or loss of phosphatase and tensin homolog (PTEN) activity, b) mutations and amplification of PI3K, c) drug-related toxicities and d) various resistance mechanisms (19).
Liquid Biopsy analysis (LBx), is minimally invasive and enables longitudinal follow-up for cancer patients in real time (20–22) through the analysis of circulating tumor cells (CTCs), plasma-cell-free DNA (cfDNA) and circulating tumor DNA (ctDNA (22, 23). LBx is very important for predicting disease progression or relapse, as well as monitoring response to treatment in breast cancer (24, 25), and minimal residual disease (MRD) detection (26–28). It is highly important to note that the implementation of LBx in clinical practice requires extensive standardization and analytical validation of protocols in every step of processing (24). There is now a variety of commercially available assays for PIK3CA mutation detection mainly based on PCR-based and next-generation sequencing (NGS) methodologies (29–34). Detection of PIK3CA mutations in CTCs has been reported in a few studies so far (35–39). This is a very challenging and demanding procedure, since CTC are usually detected at very low numbers in circulation, and moreover are highly heterogeneous (21–23). Detection of PIK3CA mutations in plasma-cfDNA has shown that ctDNA analysis can capture most of the mutations found in tissue biopsy, and the level of concordance ranges from 72.5% to 100%, depending on the different techniques used and the timing of tissue biopsy (37).
A plethora of research papers indicate that CTCs and plasma-cfDNA are complementary and both should be evaluated in parallel to gain more detailed information on the mutational landscape of a patient’s tumor. It was reported that a combination of CTCs and ctDNA evaluation for PIK3CA mutations in colorectal cancer is necessary in the clinical setting to achieve optimal surveillance of the course of disease and selection of the appropriate treatment (40). Another study comparing PIK3CA mutations in CTCs and primary tumors, revealed that CTCs can exhibit heterogeneity within a single patient, and may acquire additional genomic characteristics that differ from those of the primary tumor (36). Thus, a comprehensive liquid biopsy analysis that incorporates information from both ctDNA and CTCs, can be crucial for the selection of the appropriate treatment (37, 41–43). A relatively recent pilot study investigating PIK3CA mutations in ctDNA, CTCs, and extracellular vesicles (EVs), found consistent mutational profiles of EVs with CTCs, suggesting that EVs may have been released by CTCs (44).
Our team has reported the development of methodologies for the detection of hotspot mutations in exons 9 and 20 of the PIK3CA gene in primary tumors (45), and CTCs and ctDNA based on the combination of allele-specific priming, asymmetric PCR (ARMS-PCR), and melting curve analysis (35). Using this assay we analyzed plasma-cfDNA and paired CTCs as well as gDNAs isolated from CellSearch® cartridges for two hotspot PIK3CA mutations (p.E545K and p.H1047R) (38). We further developed highly sensitive methodologies for the detection of PIK3CA hotspot mutations based on Nuclease-Assisted Minor Allele Enrichment Using Overlapping Probes-Assisted Amplification-Refractory Mutation System (NAPA assay) (46) and a dual-Drop-off droplet digital PCR (ddPCR) Assay for the simultaneous detection of ten hotspot PIK3CA mutations (47).
The aim of the present study was to directly compare our previously described ultrasensitive real-time PCR assay (35) for the detection of three hotspot PIK3CA mutations with ddPCR in primary tumors and plasma-cfDNA, and directly compare the PIK3CA mutational status in CTC-derived gDNAs and paired plasma-cfDNA samples.
2 Materials and methods
2.1 Sample collection and processing
Forty-two FFPE samples from ER+/HER2- metastatic breast cancer patients, collected in the period of 2020-2022, were obtained from the Department of Pathology, Onassis Cardiac Surgery Center. gDNA was isolated from FFPEs using the QIAamp DNA FFPE Tissue Kit (Qiagen, Hilden, Germany), according to the manufacturer’s instructions. In parallel, peripheral blood (PB) in EDTA (10mL) was prospectively collected from 29 patients with ER+/HER2-metastatic breast cancer and 10 healthy donors. PB samples were collected in participating clinical centers under the Operational Program Competitiveness, Entrepreneurship and Innovation, under the call RESEARCH-CREATE-INNOVATE (project code: T1RCI-02935). All patients gave their informed consent and the study was approved by the Ethic committees from all participating institutions. Plasma was obtained by two consecutive centrifugations (530xg for 10min at room temperature followed by a second centrifugation at 2000xg for 10min) within 2-4h and was further stored at −70°C until analyzed. Plasma-cfDNA was extracted from 2mL of plasma using the QIAamp DSP cNA Kit (Qiagen, Hilden, Germany), and cfDNA was eluted in 30μL elution buffer (38). CTC-derived gDNA was obtained from identical blood draws as previously described (39). gDNA from CTCs was isolated using the QIAamp DNA Micro Kit, Qiagen (Qiagen, Hilden, Germany), according to the manufacturer’s instructions. DNA quantification in all samples was performed using the NanoDrop™ 1000 Spectrophotometer (ThermoFisher Scientific). All experimental procedures were performed in different rooms, dedicated labware and areas to avoid contamination. All preparation steps for the ddPCR setup were performed in a dedicated pre-PCR room and a PCR hood dedicated for the preparation of ddPCR reactions.
2.2 PIK3CA mutation analysis using the ultrasensitive real-time PCR assay
The ultrasensitive real-time PCR PIK3CA assay detects three PIK3CA hotspot mutations (exon 9 p.E542K, p.E545K and exon 20 p.H1047R) as single-plex assays. The ultra sensitive real-time PCR assay is based on a combination of allele-specific, asymmetric rapid PCR (ARMS-PCR) and melting curve analysis. The analytical validation of the assay for two of these mutations (p.E545K and p.H1047R) was previously performed in detail reporting a limit of detection (LOD) of 0.05% (35). The assay was further extended to include the PIK3CA exon 9 p.E542K mutation and was analytically validated in the same way showing a similar LOD in the LightCycler® 2.0 (IVD instrument, Roche Diagnostics, Germany) (results not shown).The interpretation of the results is performed by melting curve analysis as previously reported; the melting probe specific for each individual mutation provides a different melting temperature for its binding to the mutant allele as compared to its binding to the WT allele. PCR conditions and melting analysis protocols for each exon are described in detail in (35). Up to 50 ng/PCR DNA input is used. All samples were analyzed in the LightCycler® 2. Primers and probes sequences are given in detail in Supplementary Table S3 (35).
2.3 PIK3CA mutation analysis using droplet digital PCR
Droplet digital PCR was performed in the QX200 AutoDG ddPCR System (Bio-Rad, USA). Using ddPCR all 42 FFPEs and 29 plasma-cfDNA samples were analyzed for the same PIK3CA mutations (E545K, E542K and H1047R). We used PrimePCR™ ddPCR™ Mutation Detection Assay Kits: PIK3CA WT for p.E542K, p.E545K, p.H1047R and PIK3CA p.E542K (Bio-Rad: #1863131), PIK3CA p.E545K (Bio-Rad: #1863132), PIK3CA p.H1047R, Human (Bio-Rad: #1863133), according to the manufacturer’s instructions. The LOD for each mutation using ddPCR is ~0.1%. We set the cut-off depending on the number of positive droplets that were detected in the healthy donors control group in each mutation channel. For PIK3CA p.E545K and p.H1047R the cut-off was 2 positive droplets (0.4 cop/μL) while for p.E542K this cut-off was 3 positive droplets (0.2 cop/μL). PCR of 10 ng/μl DNA input in all cases was performed in the C1000™ Touch Thermal Cycler (95°C/10 min, 40 cycles of 94°C/30s, and 55°C/1 min with a final stage at 98°C/10 min). Finally, all samples were analyzed on a Bio-Rad QX200 droplet reader.
3 Results
3.1 Detection of PIK3CA hotspot mutations in primary tumors
The ultrasensitive real-time PCR PIK3CA assay was used to detect three PIK3CA hotspot mutations in 42 gDNA samples isolated from FFPEs. Characteristic real-time PCR melting graphs for these mutations are shown in Figure 1A. The p.E545K mutation was detected in 22/42(52.4%) samples, the p.E542K mutation in 14/42(33.3%) samples, and the p.H1047R mutation in 16/42(38.1%) samples (Figure 2A).
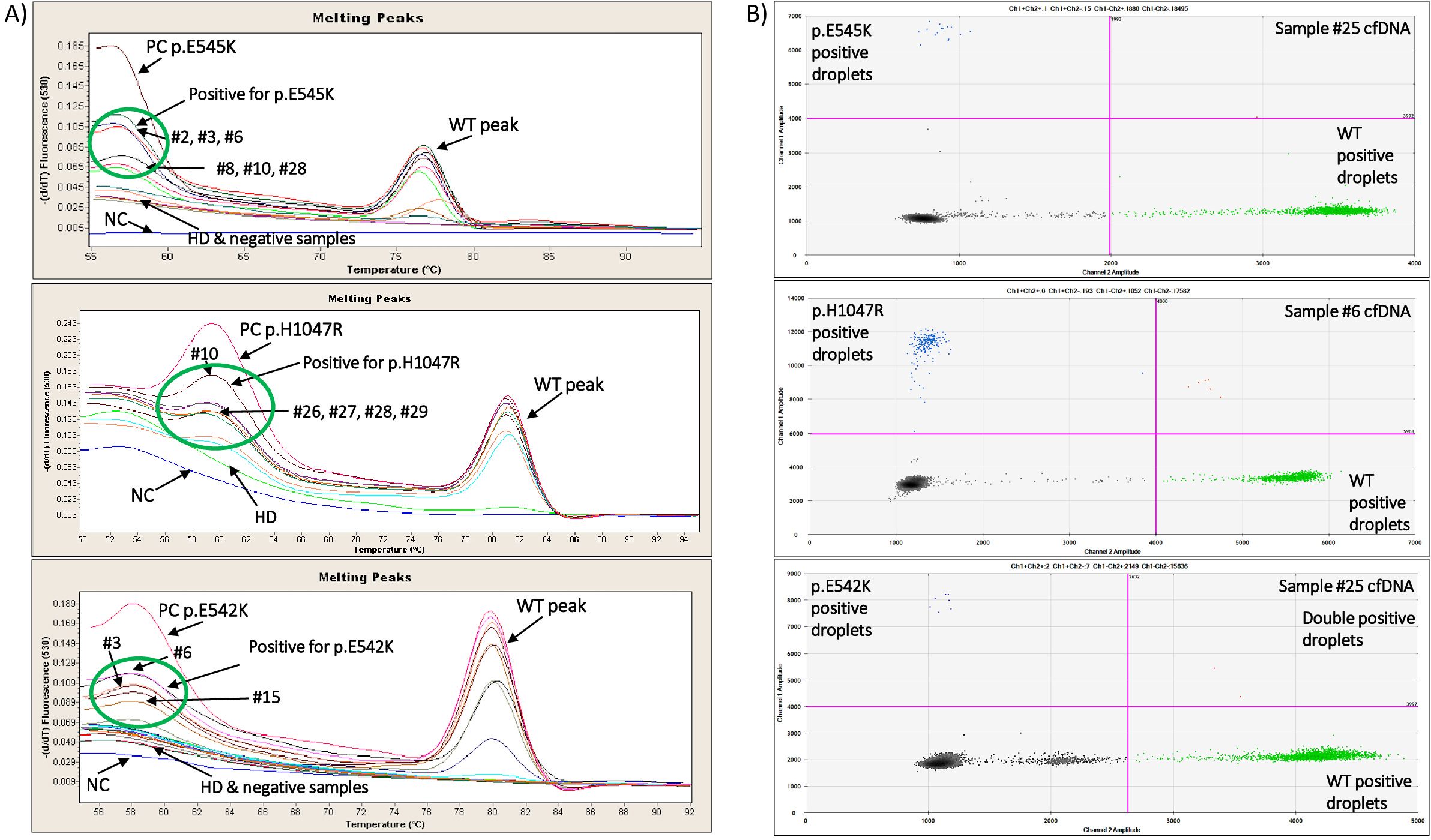
Figure 1. (A) Characteristic real-time PCR melting graphs for PIK3CA p.E545K, p.H1047R and p.E542K mutations of positive, negative and HD gDNA samples. (B) Characteristic ddPCR 2D dot-plots for PIK3CA p.E545K, p.H1047R and p.E542K mutations of 3 plasma cfDNA samples.
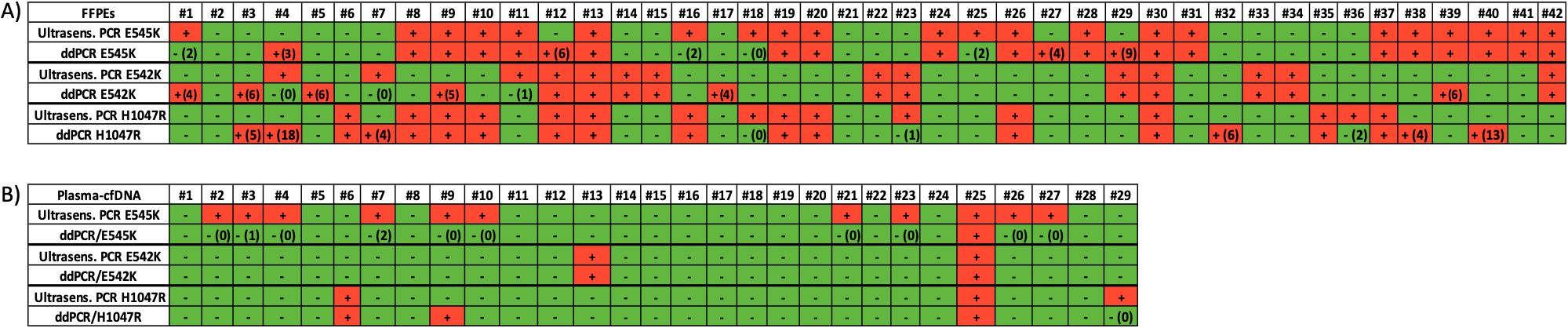
Figure 2. (A) Analysis of PIK3CA hotspot mutations in primary tumors (FFPEs gDNA). Direct comparison between the ultrasensitive real-time PIK3CA PCR assay with the ddPCR mutation test kit (Bio-Rad). Green: no mutation detected, red: mutation detected, ddPCR (#): number of droplets, (B) Analysis of PIK3CA hotspot mutations in plasma-cfDNA. Direct comparison between the ultrasensitive real-time PIK3CA PCR assay with the ddPCR mutation test kit (Bio-Rad). Green: no mutation detected, red: mutation detected, ddPCR (#): number of droplets.
We further analyzed the same gDNAs (FFPEs) using the ddPCR PIK3CA mutation test kit (Bio-Rad) for the detection of p.E545K, p.E542K and p.H1047R mutations. Characteristic ddPCR 2D dot plots for these mutations are shown in Figure 1B. Using the ddPCR assay, we detected p.E545K mutation in 22/42(52.4%) samples (Figure 2A). The concordance between these two methodologies for p.E545K PIK3CA mutation was 81% (k= 0.618, p<0.001) (Table 1). Similarly, both methods detected p.E542K mutation in 14/42 (33.3%) samples. As shown in Figure 2A, for PIK3CA p.E542K mutation the ddPCR assay detected three more positive samples than the ultrasensitive real-time PCR PIK3CA assay, resulting in a concordance of 78.6% (k=0.542, p<0.001). As for the p.H1047R mutation, 16/42(38.1%) samples were found positive by both methods while 20/42 samples were found negative (Figure 2A) and discrepancies were detected in nine samples leading to a concordance of 78.6% with statistically significant difference (k=0.562, p<0.001), (Table 1).
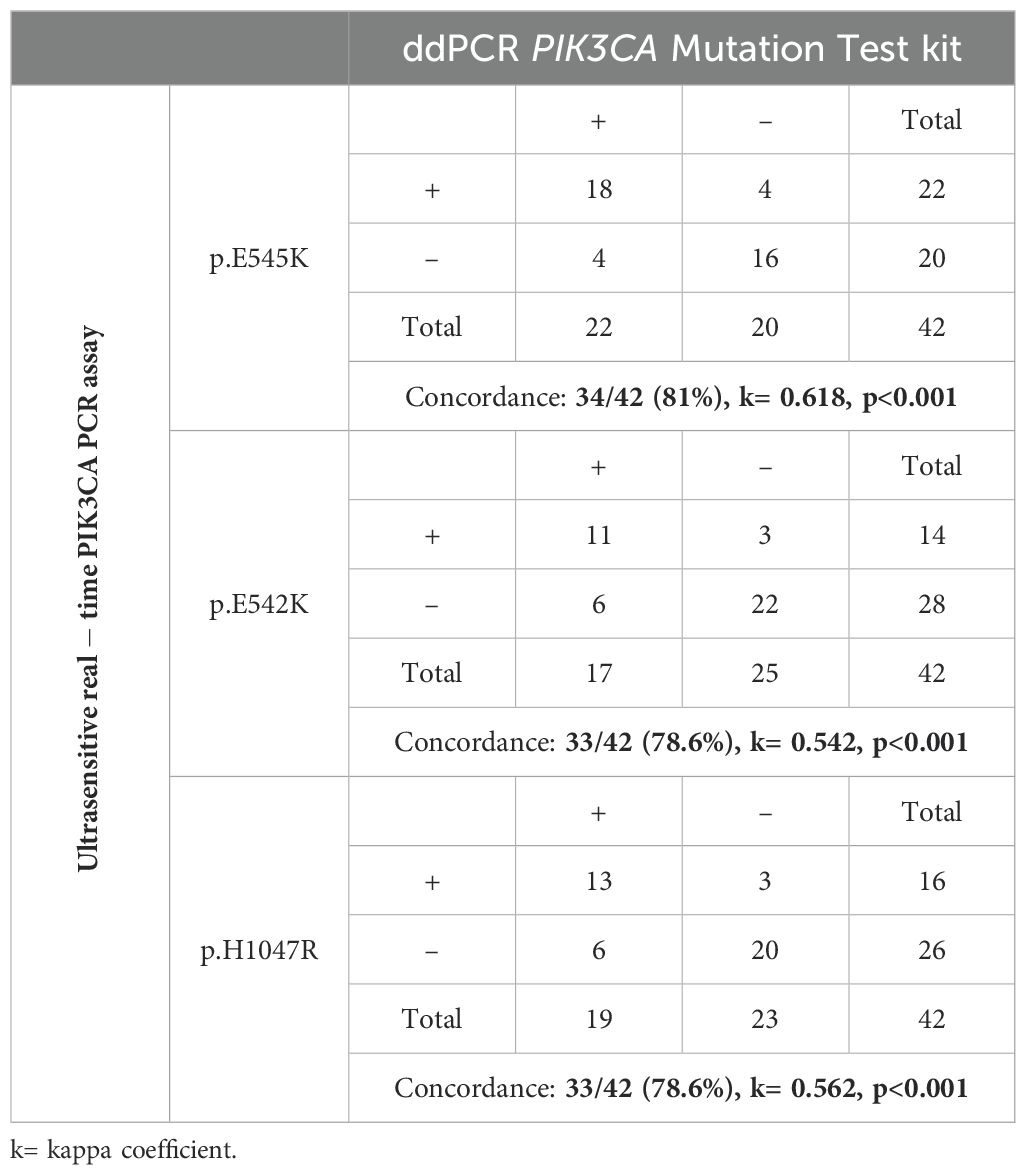
Table 1. Direct comparison between the ultrasensitive real-time PIK3CA PCR assay and the ddPCR PIK3CA mutation test for PIK3CA p.E545K, p.E542K and p.H1047R mutations in 42 primary tumor samples (FFPEs).
3.2 Detection of PIK3CA hotspot mutations in plasma-cfDNA
Using the ultrasensitive real-time PCR assay, in plasma-cfDNA, at least one PIK3CA mutation was detected in 14/29(48.3%) samples analyzed. Specifically, the p.E545K mutation was detected in 11/29(38%) samples, and bothp.H1047R and p.E542K mutation in 2/29(6.9%) samples (Figure 2B).
We further performed a direct comparison of the ultrasensitive real-time PCR PIK3CA assay with ddPCR for PIK3CA mutation analysis using the same 29 plasma-cfDNA samples. Using ddPCR, the p.E545K mutation was detected in 1/29(3.5%) samples, the p.E542K mutation in 2/29(6.9%) samples, and the p.H1047R mutation in 3/29(10.5%) samples (Figure 2B).The results of p.H1047R and p.E542K revealed an excellent concordance for both mutations (Table 2). Among these 29 plasma-cfDNA samples, eleven were found to be positive for p.E545K mutation with the ultrasensitive real-time PCR PIK3CA assay while only one of them was found positive with the ddPCR PIK3CA mutation test. However, these ten samples that were found positive by the ultrasensitive real-time PCR and negative by ddPCR had a very low number of positive droplets (0-2 positive droplets). These discrepancies between ultrasensitive real-time PCR and ddPCR found in plasma-cfDNA samples could be due to sampling errors, that occur at very low analyte concentrations (Poisson distribution effect) especially in plasma-cfDNA (Figure 2B).
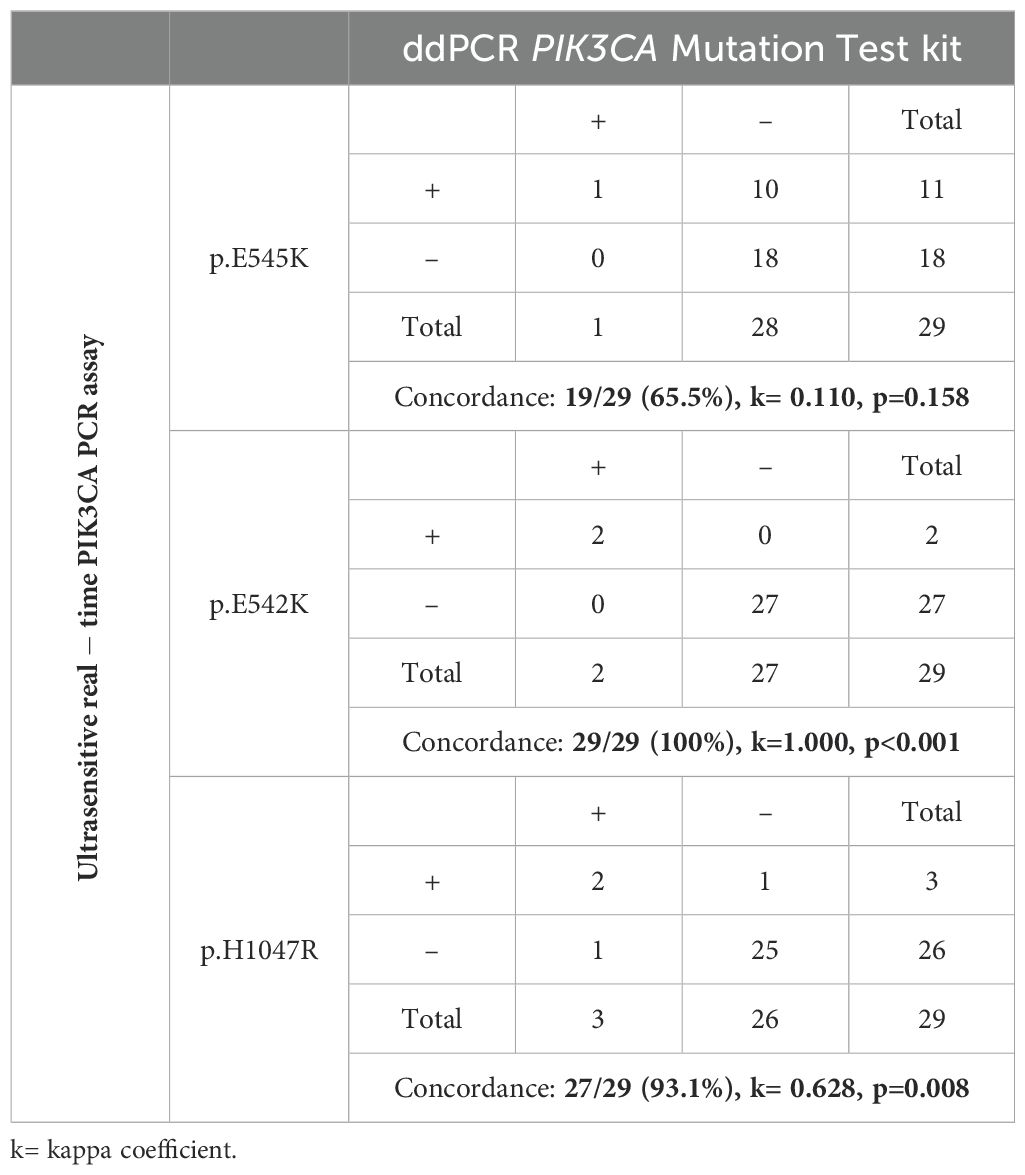
Table 2. Direct comparison between the ultrasensitive real-time PIK3CA PCR assay and the ddPCR PIK3CA Mutation Test for PIK3CA p.E545K, p.E542K and p.H1047R mutations in 29 plasma-cfDNA samples.
3.3 Detection of PIK3CA hotspot mutations in paired CTC-derived gDNAs
Using the ultrasensitive real-time PCR PIK3CA assay we analyzed PIK3CA mutations in paired CTC-derived gDNAs. The p.E545K mutation was detected in 11/29 (38%) samples, the p.E542K mutation in 3/29 (10.3%) samples and the p.H1047R mutation in 7/29 (24.1%) samples. All results for 29 plasma-cfDNA and paired CTC-derived gDNAs are summarized in Figure 3. As can be seen, mutation percentages are slightly higher in CTCs than plasma-cfDNA as expected (38). From 29 patients with ER+/HER2- breast cancer, five were found to be positive for p.E545K mutation both in plasma-cfDNA and paired CTC gDNA and one for p.H1047R (Figure 3). Nine patients were negative for these PIK3CA mutations in both types of samples. Concordance rates between plasma-cfDNA and paired CTC-derived gDNA was 45.5% for PIK3CA p.E545K mutation. It has been observed that in some cases, the genetic mutations found in CTC-derived gDNAs do not match those detected in the plasma-cfDNA. Similarly, there are instances where plasma-cfDNA mutations are not present in CTC-derived gDNAs. Mutation rates for PIK3CA p.E542K and p.H1047R are very low as the number of positive samples were relatively low.
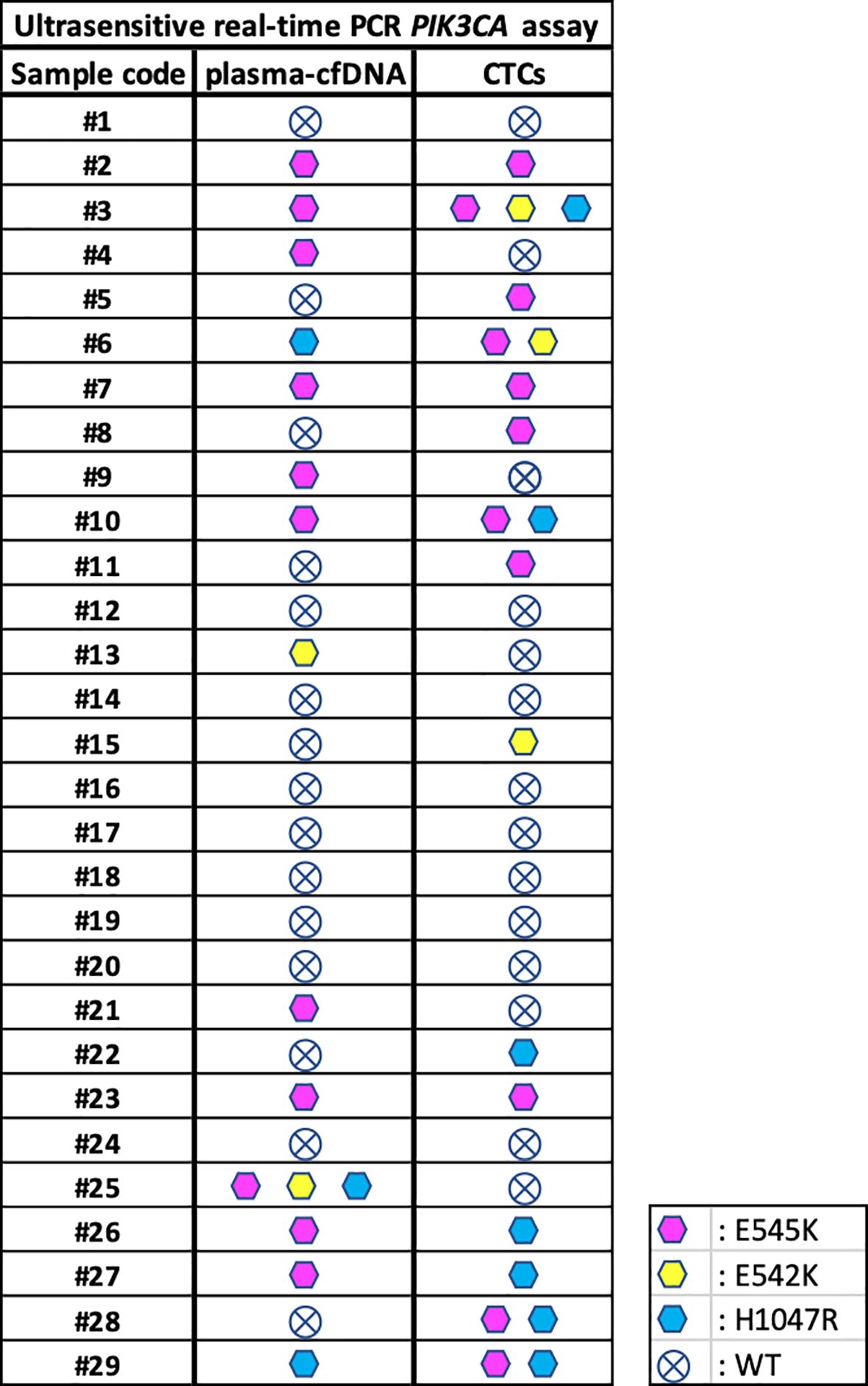
Figure 3. Direct comparison of PIK3CA mutations detected in plasma-cfDNA and paired CTC-derived gDNAs using the ultrasensitive real-time PCR assay.
4 Discussion
Since PIK3CA mutations are present in multiple types of cancer, the reliable detection of these hotspot mutations is a highly important tool to guide treatment, especially since alpelicib is widely used in the treatment of ER+/HER2- metastatic breast cancer (18). Quick, reliable and inexpensive identification of PIK3CA mutations is crucial for determining which breast cancer patients will benefit from these specific treatments (48). The presence of PIK3CA mutations in exon 9 was found to be associated with a more favorable outcome in some medical conditions. On the other hand, the presence of PIK3CA mutations in exon 20, was found to be linked to a relatively poor prognosis, indicating a higher likelihood of negative outcomes (49, 50).
In this study we directly compared the efficacy of a relatively inexpensive ultrasensitive real-time PCR with the now well-established and highly sensitive technology of ddPCR for the detection of the three most common hotspot mutations of PIK3CA, in exons 9 and 20, that are all of clinical importance in various types of cancer. According to our results, this simple and ultrasensitive real-time PCR assay gives similar results to ddPCR, can be performed in labs where digital PCR instrumentation is not available and provides accurate and specific detection of PIK3CA hotspot mutations in liquid biopsy samples.
The high positivity for PIK3CA mutations in primary tumors FFPE samples, detected by this assay is in agreement with previously reported studies (35, 47). In FFPEs our direct comparison study between the ultrasensitive real-time PCR assay with ddPCR for p.E545K, revealed a relatively good agreement, while the detection of p.E542K and p.H1047R was also in good agreement between the two assays. Using ddPCR three more samples were detected as positive for the p.E542K and p.H1047R in FFPEs. When classic real-time PCR was directly compared with ddPCR for the detection of PIK3CA mutations in FFPEs of Head and Neck Squamous Cell Carcinoma patients, it was reported that ddPCR was superior in terms of sensitivity in the PIK3CA mutation assessment in FFPE samples (51).
In plasma-cfDNA, the rates of PIK3CA positiveness usually reported are relatively low. Using the ultrasensitive real-time PCR PIK3CA assay ten plasma-cfDNA samples were found positive for the p.E545K mutation while they were found negative by ddPCR. These ten samples that are reported as negative using ddPCR for the PIK3CA p.E545K mutation were found to have between 0 to 2 droplets, indicating the absence of positive events, since the p.E545K positive copies detected are extremely low to be called as positive events. On the other hand, two plasma-cfDNA samples were found positive for the PIK3CAp.E542K mutation with both assays leading to complete agreement. Moreover, two plasma-cfDNA samples were found positive for p.H1047R mutation by the ultrasensitive real-time PCR PIK3CA assay, while by ddPCR p.H1047R mutation was detected in three plasma-cfDNA, revealing a strong agreement for this mutation too.
Analysis of PIK3CA mutations in CTC-derived gDNAs showed a higher percentage of PIK3CA hotspot mutations than in plasma-cfDNA, especially for p.H1047R. Moreover, we observed that when we compared mutations identified in CTCs and plasma-cfDNA, PIK3CA mutations were detected in CTCs but were not detectable in plasma-cfDNA in some peripheral blood samples, whereas in other blood samples, PIK3CA mutations were detected in plasma-cfDNA but were not detected in CTCs. Our results indicate that CTC and plasma-cfDNA give complementary information, especially at very low analyte concentrations, and that evaluating both CTCs and plasma-cfDNA analysis is necessary to ensure optimal disease surveillance and appropriate treatment selection in the clinical setting. We have recently shown that comprehensive liquid biopsy analysis including the analysis of PIK3CA mutations in CTCs and ctDNA is a very informative tool for the early detection of minimal residual disease in breast cancer (52). Based on a relatively small number of patients another study has come to the conclusion that combined analysis of CTCs and ctDNA may offer a new approach for monitoring of disease progression and to direct therapy in patients with advanced MBC, at a time when they are coming towards the end of other treatment options (53). Molecular analysis, performed both in CTCs and plasma-cfDNA, provides results that reflect certain subpopulations of the primary tumor as well as cells forming metastases and should be considered an important form to monitor the development of the disease and its status over time, particularly for its clinical impact in guiding drug selection. We have already shown in a previous study that the prevalence of detectable mutations of PIK3CA was higher in CTC-derived DNA than in the corresponding plasma-cfDNA (54).
ddPCR is considered as the next generation of PCR technology, since it offers absolute quantification of nucleic acid target sequences without the need for external calibration curves. However digital PCR is still very expensive in terms of both instrumentation and reagents, and is still not available in most molecular diagnostic labs worldwide. Real-time PCR still offers numerous benefits, including established protocols, well-known data analysis techniques, and availability of necessary instrumentation worldwide. Additionally, it boasts a significant low experiment cost, high sample throughputs, less consumables and a wide dynamic range for detection. Real-time PCR in combination with melting curve analysis is highly effective for mutational screening. This methodology is highly sensitive and provides quick and accurate results, making it a suitable option for cases where absolute quantification is not necessary. In our hands ultrasensitive real-time PCR for PIK3CA mutations showed similar sensitivity to ddPCR, however the limited number of clinical samples and the relatively small number of healthy controls analyzed, restricts the conclusions made. In CTC-derived gDNA and paired plasma-cfDNA, PIK3CA mutations detected were not identical, revealing that CTC and plasma-cfDNA give complementary information.
In conclusion, molecular diagnostic applications for PIK3CA mutation screening in liquid biopsy samples that require high sensitivity and precision can greatly benefit from the presented reliable, low-cost and high-throughput ultrasensitive real-time PCR assay.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics statement
The studies involving humans were approved by Ethic committees from all participating institutions. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
Author contributions
SS: Data curation, Writing – original draft, Writing – review & editing. LK: Resources, Writing – review & editing. IB: Resources, Writing – review & editing. HL: Resources, Writing – review & editing. APa: Resources, Writing – review & editing. FZ: Resources, Writing – review & editing. ER: Resources, Writing – review & editing. SK: Resources, Writing – review & editing. APs: Resources, Writing – review & editing. CP: Resources, Writing – review & editing. AM: Conceptualization, Methodology, Writing – review & editing. EL: Conceptualization, Methodology, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This study has been financially supported by the European Union and Greek national funds through the Operational Program Competitiveness, Entrepreneurship and Innovation, under the call RESEARCH-CREATE-INNOVATE (project code: T1RCI-02935).
Acknowledgments
We would like to thank all patients who participated in this study for providing the blood samples.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Glaviano A, Foo ASC, Lam HY, Yap KCH, Jacot W, Jones RH, et al. PI3K/AKT/mTOR signaling transduction pathway and targeted therapies in cancer. Mol Cancer. (2023) 22:138. doi: 10.1186/s12943-023-01827-6
2. Herberts C, Murtha AJ, Fu S, Wang G, Schönlau E, Xue H, et al. Activating AKT1 and PIK3CA mutations in metastatic castration-resistant prostate cancer. Eur Urol. (2020) 78:834–44. doi: 10.1016/j.eururo.2020.04.058
3. Heavey S, O’Byrne KJ, Gately K. Strategies for co-targeting the PI3K/AKT/mTOR pathway in NSCLC. Cancer Treat Rev. (2014) 40:445–56. doi: 10.1016/j.ctrv.2013.08.006
4. Voutsadakis IA. KRAS mutated colorectal cancers with or without PIK3CA mutations: Clinical and molecular profiles inform current and future therapeutics. Crit Rev Oncol Hematol. (2023) 186:103987. doi: 10.1016/j.critrevonc.2023.103987
5. Bachman KE, Argani P, Samuels Y, Silliman N, Ptak J, Szabo S, et al. The PIK3CA gene is mutated with high frequency in human breast cancers. Cancer Biol Ther. (2004) 3:772–5. doi: 10.4161/cbt.3.8.994
6. Cerma K, Piacentini F, Moscetti L, Barbolini M, Canino F, Tornincasa A, et al. Targeting PI3K/AKT/mTOR pathway in breast cancer: from biology to clinical challenges. Biomedicines. (2023) 11:109. doi: 10.3390/biomedicines11010109
7. Daher S, Zer A, Tschernichovsky R, Yacobi R, Barshack I, Tsabari S, et al. Driver mutation characteristics of phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha (PIK3CA) in advanced non-small cell lung cancer. Lung Cancer. (2023) 178:229–36. doi: 10.1016/j.lungcan.2023.02.023
8. Chen M, Mashima T, Oishi T, Muramatsu Y, Seto Y, Takamatsu M, et al. APC/PIK3CA mutations and β-catenin status predict tankyrase inhibitor sensitivity of patient-derived colorectal cancer cells. Br J Cancer. (2023) 130:151–62. doi: 10.1038/s41416-023-02484-8
9. Hamza A, Masliah-Planchon J, Neuzillet C, Lefèvre JH, Svrcek M, Vacher S, et al. Pathogenic alterations in PIK3CA and KMT2C are frequent and independent prognostic factors in anal squamous cell carcinoma treated with salvage abdominoperineal resection. Int J cancer. (2024) 154:504–15. doi: 10.1002/ijc.v154.3
10. Pflüger MJ, Jamouss KT, Afghani E, Lim SJ, Rodriguez Franco S, Mayo H, et al. Predictive ability of pancreatic cyst fluid biomarkers: A systematic review and meta-analysis. Pancreatology. (2023) 23:868–77. doi: 10.1016/j.pan.2023.05.005
11. Cardoso F, Paluch-Shimon S, Senkus E, Curigliano G, Aapro MS, André F, et al. 5th ESO-ESMO international consensus guidelines for advanced breast cancer (ABC 5). Ann Oncol Off J Eur Soc Med Oncol. (2020) 31:1623–49. doi: 10.1016/j.annonc.2020.09.010
12. Waarts MR, Stonestrom AJ, Park YC, Levine RL. Targeting mutations in cancer. J Clin Invest. (2022) 132:e154943. doi: 10.1172/JCI154943
13. Swain SM, Shastry M, Hamilton E. Targeting HER2-positive breast cancer: advances and future directions. Nat Rev Drug Discovery. (2022) 22:101–26. doi: 10.1038/s41573-022-00579-0
14. Markham A. Alpelisib: first global approval. Drugs. (2019) 79:1249–53. doi: 10.1007/s40265-019-01161-6
15. André F, Ciruelos E, Rubovszky G, Campone M, Loibl S, Rugo HS, et al. Alpelisib for PIK3CA -mutated, hormone receptor–positive advanced breast cancer. N Engl J Med. (2019) 380:1929–40. doi: 10.1056/NEJMoa1813904
16. de Godoy BLV, Moschetta-Pinheiro MG, Chuffa LG de A, Pondé NF, Reiter RJ, Colombo J, et al. Synergistic actions of Alpelisib and Melatonin in breast cancer cell lines with PIK3CA gene mutation. Life Sci. (2023) 324:121708. doi: 10.1016/j.lfs.2023.121708
17. André F, Ciruelos EM, Juric D, Loibl S, Campone M, Mayer IA, et al. Alpelisib plus fulvestrant for PIK3CA-mutated, hormone receptor-positive, human epidermal growth factor receptor-2-negative advanced breast cancer: final overall survival results from SOLAR-1. Ann Oncol Off J Eur Soc Med Oncol. (2021) 32:208–17. doi: 10.1016/J.ANNONC.2020.11.011
18. Pascual J, Lim JSJ, Macpherson IR, Armstrong AC, Ring A, Okines AFC, et al. Triplet therapy with palbociclib, taselisib, and fulvestrant in PIK3CA-mutant breast cancer and doublet palbociclib and taselisib in pathway-mutant solid cancers. Cancer Discovery. (2021) 11:92–107. doi: 10.1158/2159-8290.CD-20-0553
19. Mishra R, Patel H, Alanazi S, Kilroy MK, Garrett JT. PI3K inhibitors in cancer: clinical implications and adverse effects. Int J Mol Sci. (2021) 22:3464. doi: 10.3390/ijms22073464
20. Mazzitelli C, Santini D, Corradini AG, Zamagni C, Trerè D, Montanaro L, et al. Liquid biopsy in the management of breast cancer patients: where are we now and where are we going. Diagnostics. (2023) 13:1241. doi: 10.3390/diagnostics13071241
21. Alix-Panabières C, Pantel K. Liquid biopsy: from discovery to clinical application. Cancer Discovery. (2021) 11:858–73. doi: 10.1158/2159-8290.CD-20-1311
22. Lianidou E, Pantel K. Liquid biopsies. Genes Chromosomes Cancer. (2019) 58:219–32. doi: 10.1002/gcc.22695
23. Strati A, Markou A, Kyriakopoulou E, Lianidou E. Detection and molecular characterization of circulating tumour cells: challenges for the clinical setting. Cancers (Basel). (2023) 15:2185. doi: 10.3390/cancers15072185
24. Agostinetto E, Nader-Marta G, Ignatiadis M. Circulating tumor DNA in breast cancer: a biomarker for patient selection. Curr Opin Oncol. (2023) 35:426–35. doi: 10.1097/CCO.0000000000000964
25. Joosse SA, Pantel K. Circulating DNA and liquid biopsies in the management of patients with cancer. Cancer Res. (2022) 82:2213–5. doi: 10.1158/0008-5472.CAN-22-1405
26. Markou A, Tzanikou E, Lianidou E. The potential of liquid biopsy in the management of cancer patients. Semin Cancer Biol. (2022) 84:69–79. doi: 10.1016/j.semcancer.2022.03.013
27. Keller L, Belloum Y, Wikman H, Pantel K. Clinical relevance of blood-based ctDNA analysis: mutation detection and beyond. Br J Cancer. (2021) 124:345–58. doi: 10.1038/s41416-020-01047-5
28. Connal S, Cameron JM, Sala A, Brennan PM, Palmer DS, Palmer JD, et al. Liquid biopsies: the future of cancer early detection. J Transl Med. (2023) 21:118. doi: 10.1186/s12967-023-03960-8
29. Milbury CA, Creeden J, Yip WK, Smith DL, Pattani V, Maxwell K, et al. Clinical and analytical validation of FoundationOne®CDx, a comprehensive genomic profiling assay for solid tumors. PloS One. (2022) 17:e0264138. doi: 10.1371/journal.pone.0264138
30. PIK3CA Mutation Detection Test. DiaCarta, Inc. (2024). Available at: https://www.diacarta.com/products/qclamp-gene-mutation-detection-tests/pik3ca (Accessed January 29, 2024).
31. AmoyDxR PIK3CA Mutation Detection Kit - [AmoyDx]. Amoydiagnostics (2024). Available at: https://www.amoydiagnostics.com/products/amoydx-pik3ca-mutation-detection-kit (Accessed November 4, 2024).
32. PIK3CA Mutation Analysis Kit. EntroGen, Inc. (2024). Available at: https://entrogen.com/web3/pik3ca-mutation-analysis-kit/ (Accessed January 29, 2024).
33. FastPlexTM PIK3CA 11 Mutations PCR Detection Kit, Digital PCR Assay. PrecigenomeLLC. (2024). Available at: https://www.precigenome.com/product-page/fastplex-pik3ca-11-mutations-digital-pcr-detection-kit-dpcr-assay (Accessed January 29, 2024).
34. Schmidt C, Stöhr R, Dimitrova L, Beckmann MW, Rübner M, Fasching PA, et al. Quality-assured analysis of PIK3CA mutations in hormone receptor–positive/human epidermal growth factor receptor 2–negative breast cancer tissue. J Mol Diagnostics. (2024) 26:624–37. doi: 10.1016/j.jmoldx.2024.04.003
35. Markou A, Farkona S, Schiza C, Efstathiou T, Kounelis S, Malamos N, et al. PIK3CA mutational status in circulating tumor cells can change during disease recurrence or progression in patients with breast cancer. Clin Cancer Res. (2014) 20:5823–34. doi: 10.1158/1078-0432.CCR-14-0149
36. Pestrin M, Salvianti F, Galardi F, De Luca F, Turner N, Malorni L, et al. Heterogeneity of PIK3CA mutational status at the single cell level in circulating tumor cells from metastatic breast cancer patients. Mol Oncol. (2015) 9:749–57. doi: 10.1016/j.molonc.2014.12.001
37. Rossi G, Mu Z, Rademaker AW, Austin LK, Strickland KS, Costa RLB, et al. Cell-free DNA and circulating tumor cells: comprehensive liquid biopsy analysis in advanced breast cancer. Clin Cancer Res. (2018) 24:560–8. doi: 10.1158/1078-0432.CCR-17-2092
38. Tzanikou E, Markou A, Politaki E, Koutsopoulos A, Psyrri A, Mavroudis D, et al. PIK3CA hotspot mutations in circulating tumor cells and paired circulating tumor DNA in breast cancer: a direct comparison study. Mol Oncol. (2019) 13:2515–30. doi: 10.1002/1878-0261.12540
39. Chimonidou M, Strati A, Tzitzira A, Sotiropoulou G, Malamos N, Georgoulias V, et al. DNA methylation of tumor suppressor and metastasis suppressor genes in circulating tumor cells. (2011). doi: 10.1373/clinchem.2011.165902
40. Kidess-Sigal E, Liu HE, Triboulet MM, Che J, Ramani VC, Visser BC, et al. Enumeration and targeted analysis of KRAS, BRAF and PIK3CA mutations in CTCs captured by a label-free platform: Comparison to ctDNA and tissue in metastatic colorectal cancer. Oncotarget. (2016) 7:85349. doi: 10.18632/oncotarget.13350
41. Strati A, Zavridou M, Kallergi G, Politaki E, Kuske A, Gorges TM, et al. A comprehensive molecular analysis of in vivo isolated epCAM-positive circulating tumor cells in breast cancer. Clin Chem. (2021) 67:1395–405. doi: 10.1093/clinchem/hvab099
42. Park J, Chang ES, Kim JY, Chelakkot C, Sung M, Song JY, et al. c-MET-positive circulating tumor cells and cell-free DNA as independent prognostic factors in hormone receptor-positive/HER2-negative metastatic breast cancer. Breast Cancer Res. (2024) 26:13. doi: 10.1186/s13058-024-01768-y
43. Ge Z, Helmijr JCA, Jansen MPHM, Boor PPC, Noordam L, Peppelenbosch M, et al. Detection of oncogenic mutations in paired circulating tumor DNA and circulating tumor cells in patients with hepatocellular carcinoma. Transl Oncol. (2021) 14:101073. doi: 10.1016/j.tranon.2021.101073
44. Cardinali B, De Luca G, Tasso R, Coco S, Garuti A, Buzzatti G, et al. Targeting PIK3CA actionable mutations in the circulome: A proof of concept in metastatic breast cancer. Int J Mol Sci. (2022) 23:6320. doi: 10.3390/ijms23116320
45. Vorkas PA, Poumpouridou N, Agelaki S, Kroupis C, Georgoulias V, Lianidou ES. PIK3CA hotspot mutation scanning by a novel and highly sensitive high-resolution small amplicon melting analysis method. J Mol Diagn. (2010) 12:697–704. doi: 10.2353/jmoldx.2010.100008
46. Markou A, Tzanikou E, Ladas I, Makrigiorgos GM, Lianidou E. Nuclease-assisted minor allele enrichment using overlapping probes-assisted amplification-refractory mutation system: an approach for the improvement of amplification-refractory mutation system-polymerase chain reaction specificity in liquid biopsies. Anal Chem. (2019) 91:13105–11. doi: 10.1021/acs.analchem.9b03325
47. Stergiopoulou D, Smilkou S, Georgoulias V, Kaklamanis L, Lianidou E, Markou A. Development and validation of a novel dual-drop-off ddPCR assay for the simultaneous detection of ten hotspots PIK3CA mutations. Anal Chem. (2023) 95:14068–76. doi: 10.1021/acs.analchem.3c02692
48. Schneck H, Blassl C, Meier-Stiegen F, Neves RP, Janni W, Fehm T, et al. Analysing the mutational status of PIK3CA in circulating tumor cells from metastatic breast cancer patients. Mol Oncol. (2013) 7:976–86. doi: 10.1016/j.molonc.2013.07.007
49. Samuels Y, Waldman T. Oncogenic mutations of PIK3CA in human cancers. Curr Top Microbiol Immunol. (2010) 347:21. doi: 10.1016/J.MOLONC.2013.07.007
50. Ogino S, Nosho K, Kirkner GJ, Shima K, Irahara N, Kure S, et al. PIK3CA mutation is associated with poor prognosis among patients with curatively resected colon cancer. J Clin Oncol. (2009) 27:1477–84. doi: 10.1200/JCO.2008.18.6544
51. Borkowska EM, Barańska M, Kowalczyk M, Pietruszewska W. Detection of PIK3CA gene mutation in head and neck squamous cell carcinoma using droplet digital PCR and RT-qPCR. Biomolecules. (2021) 11:1477–84. doi: 10.3390/biom11060818
52. Stergiopoulou D, Markou A, Strati A, Zavridou M, Tzanikou E, Mastoraki S, et al. Comprehensive liquid biopsy analysis as a tool for the early detection of minimal residual disease in breast cancer. Sci Rep. (2023) 13:1258–8. doi: 10.1038/s41598-022-25400-1
53. Fernandez-Garcia D, Nteliopoulos G, Hastings RK, Rushton A, Page K, Allsopp RC, et al. Shallow WGS of individual CTCs identifies actionable targets for informing treatment decisions in metastatic breast cancer. Br J Cancer. (2022) 127:1858–64. doi: 10.1038/s41416-022-01962-9
54. Markou AN, Londra D, Stergiopoulou D, Vamvakaris I, Potaris K, Pateras IS, et al. Preoperative mutational analysis of circulating tumor cells (CTCs) and plasma-cfDNA provides complementary information for early prediction of relapse: A pilot study in early-stage non-small cell lung cancer. Cancers (Basel). (2023) 15:1877. doi: 10.3390/cancers15061877
Keywords: PIK3CA mutations, liquid biopsy, breast cancer, plasma cell-free DNA, circulating tumor cells, DNA melting curve analysis, ultrasensitive molecular assay, droplet digital PCR
Citation: Smilkou S, Kaklamanis L, Balgouranidou I, Linardou H, Papatheodoridi AM, Zagouri F, Razis E, Kakolyris S, Psyrri A, Papadimitriou C, Markou A and Lianidou E (2024) Direct comparison of an ultrasensitive real-time PCR assay with droplet digital PCR for the detection of PIK3CA hotspot mutations in primary tumors, plasma cell-free DNA and paired CTC-derived gDNAs. Front. Oncol. 14:1435559. doi: 10.3389/fonc.2024.1435559
Received: 20 May 2024; Accepted: 28 October 2024;
Published: 06 December 2024.
Edited by:
Chiara Nicolazzo, Sapienza University of Rome, ItalyReviewed by:
Eswari Dodagatta-Marri, University of California, San Francisco, United StatesChangming Lu, University of Alabama at Birmingham, United States
Copyright © 2024 Smilkou, Kaklamanis, Balgouranidou, Linardou, Papatheodoridi, Zagouri, Razis, Kakolyris, Psyrri, Papadimitriou, Markou and Lianidou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Evi Lianidou, bGlhbmlkb3VAY2hlbS51b2EuZ3I=; ZXZpbGlhbmlkb3VAZ21haWwuY29t
 Stavroula Smilkou
Stavroula Smilkou Loukas Kaklamanis2
Loukas Kaklamanis2 Helena Linardou
Helena Linardou Alkistis Maria Papatheodoridi
Alkistis Maria Papatheodoridi Flora Zagouri
Flora Zagouri Amanda Psyrri
Amanda Psyrri Christos Papadimitriou
Christos Papadimitriou Athina Markou
Athina Markou Evi Lianidou
Evi Lianidou