- Department of Otolaryngology-Head and Neck Surgery, Shanghai General Hospital, Shanghai Jiaotong University School of Medicine, Shanghai, China
Head and neck cancer is the main cause of cancer death worldwide, with squamous cell carcinoma (HNSCC) being the second most frequent subtype. HNSCC poses significant health threats due to its high incidence and poor prognosis, underscoring the urgent need for advanced research. Histone modifications play a crucial role in the regulation of gene expression and influencing various biological processes. In the context of HNSCC, aberrant histone modifications are increasingly recognized as critical contributors to its development and pathologic progression. This review demonstrates the molecular mechanisms, by which histone modifications such as acetylation, methylation, phosphorylation, and ubiquitination, impact the pathogenesis of HNSCC. The dysregulation of histone-modifying enzymes, including histone acetyltransferases (HATs), histone deacetylases (HDACs), and histone methyltransferases (HMTs), is discussed for its role in altering chromatin structure and gene expression in HNSCC. Moreover, we will explore the potential of targeting histone modifications as a therapeutic strategy, highlighting current preclinical and clinical studies that investigate histone deacetylase inhibitors (HDIs) and other epigenetic drugs, referring to the completed and ongoing clinical trials on those medications.
1 Introduction
Exploring the pathogenesis of tumors to find effective treatments has always been a main theme of medical research. Head and neck squamous cell carcinoma (HNSCC) is one of the most common malignancies in the body, occurring mainly in the oral cavity, pharynx, and larynx. Globally, there are more than 600,000 new cases diagnosed each year, with a higher incidence in regions such as Southeast Asia and Central Europe. Characterized by high heterogeneity and mortality rate, about 60% of patients are already in the advanced stage when they seek medical help (1). It is well-known that genetic and epigenetic alterations are the main hallmarks of HNSCC tumorigenesis (Figure 1). Changes in the genetic material of the organism underlie all tumorigenesis, with gene mutations and chromosomal aberrations being the two primary aspects of genetic alteration. However, epigenetic changes are relatively complex, including DNA methylation, histone modifications, and the regulation of non-coding RNAs (2). Histone modification refers to the process of chemically modifying histones to alter their structure and function, they can activate or repress genes by changing the accessibility of DNA to transcription factors and other regulatory proteins, which is one of the most significant features of epigenetic changes. Histone modifications include methylation, acetylation, phosphorylation, ubiquitination, and many other forms (3) (Figure 2). Recent studies have highlighted the importance of specific histone modifications in HNSCC pathogenesis (4). This article provides a thorough overview of recent progress in comprehending the correlation between histone modification alterations and HNSCC. Certain modifications are deemed “classic” due to their early discovery, extensive examination, and frequent interactions, forming an intricate “histone code” capable of precisely regulating gene expression. By making these distinctions, our goal is to clarify the interactions among histone modifications, ultimately aiding in the discovery of more effective diagnostic markers and therapeutic targets for HNSCC.
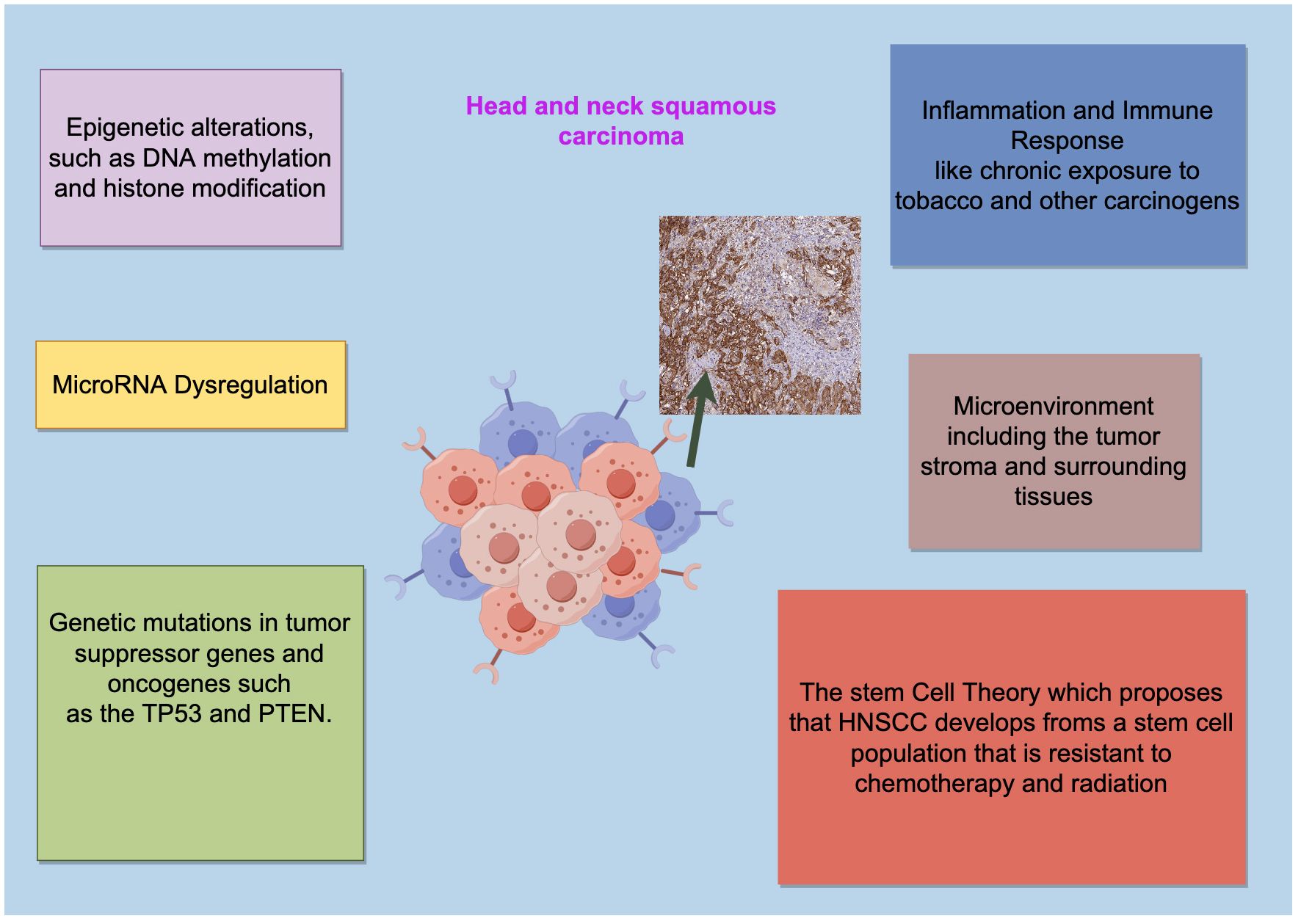
Figure 1 The pathogenesis of HNSCC is a multistep process. The pathogenesis of HNSCC is a complex multistep process influenced by multiple factors including epigenetic alterations, MicroRNA Dysregulation, genetic mutations in tumor suppressor genes and oncogenes and so on. Drawing using Figdraw tool.
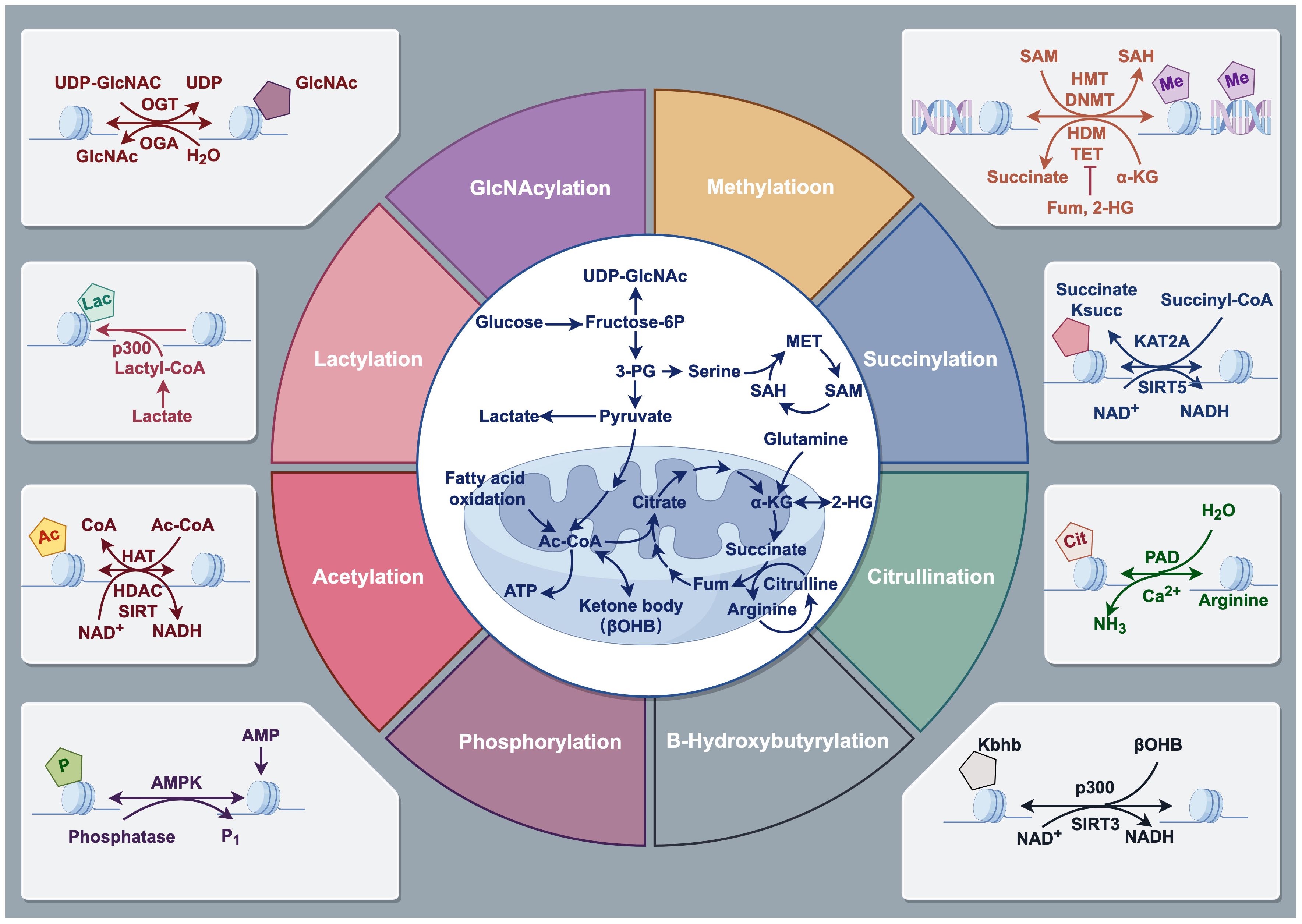
Figure 2 Types of Histone modifications. Histone modifications refer to the addition or removal of chemical groups, such as methyl, acetyl, phosphate, and ubiquitin, to specific amino acids on the histone protein tails. These modifications can alter the structure and function of chromatin, which is the complex of DNA and histones, and can either facilitate or inhibit gene expression. Histone modifications include methylation, acetylation, phosphorylation, ubiquitination, and many other forms. Drawing using Figdraw tool.
2 Classic histone modifications in HNSCC
2.1 Histone methylation
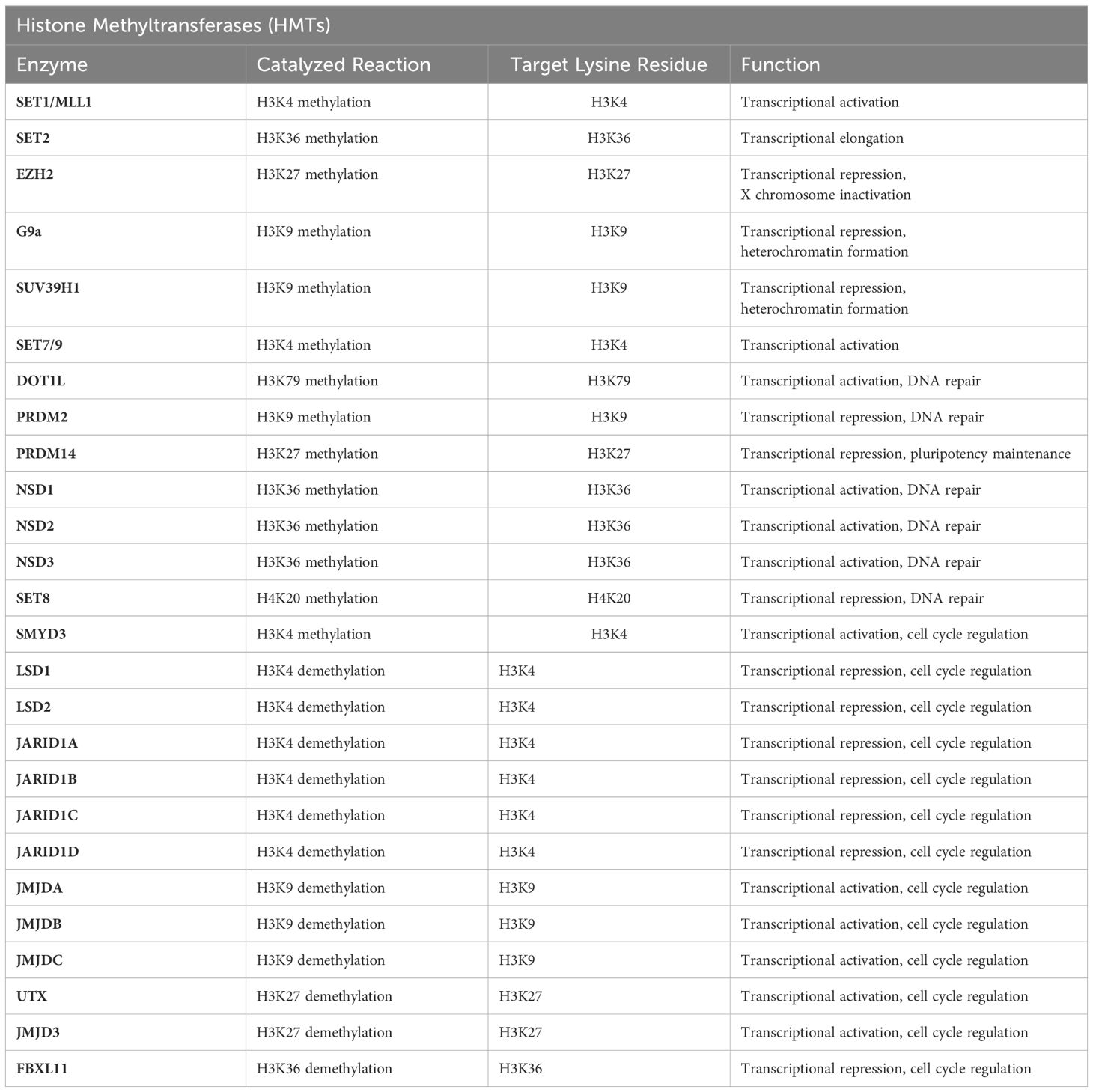
Histone methylation is a crucial post-translational modification in chromatin structure regulation, significantly impacting gene expression and cellular fate. This process predominantly involves the methylation of lysine and arginine residues on histones H3 and H4. Catalyzed by a series of specific enzymes, such as histone methyltransferases (HMTs) and histone demethylases (HDMs) (Table 1), histone methylation alters chromatin structure, thereby regulating gene activity. This modification is dynamic and reversible, for instance, H3K4me3 is typically associated with gene activation, whereas H3K27me3 correlates with gene silencing. These characteristics make histone methylation a pivotal mechanism in regulating gene expression and determining cellular fate (Figure 3).

Figure 3 Writers and erasers in Histone Methylation. Histone methylation is a critical epigenetic modification involved in the regulation of gene expression. It involves the addition of one, two, or three methyl groups to specific lysine or arginine residues on histone tails. This modification is dynamically controlled by two classes of enzymes: “writers” and “erasers”. Drawing using Figdraw tool.
In recent years, numerous studies have made remarkable achievements on the correlation between histone methylation and HNSCC. These investigations have revealed that antitumor immunity in HNSCC can be effectively induced by promoting DNA hypomethylation and derepression of retrotransposons through the induction of interferon response. Notably, the deactivation of histone H3K36 methyltransferase NSD1 leads to a depletion of H3K36 dimethylation (H3K36me2) and an elevation in H3K27 trimethylation (H3K27me3), which subsequently diminishes tumor immune infiltration (5). Through machine learning, another study screened 429 chromatin regulatory factors and discovered that histone methyltransferases enhance the stemness and migratory capabilities of tumor cells by upregulating SMYD3 and inducing H3K4 trimethylation (H3K4me3)-mediated HMGA2 expression (6). Current research suggests that approximately 20% of HNSCC cases exhibit reduced methylation of histone H3 at lysine 36 (H3K36me). This reduction is caused by mutations in the histone methyltransferase NSD1 or by lysine-to-methionine substitutions (H3K36M) in histone H3. Manipulation of the key epigenetic marker H3K27me3 can potentially play a supporting role in pharmacological treatment (7).
2.2 Histone acetylation imbalance
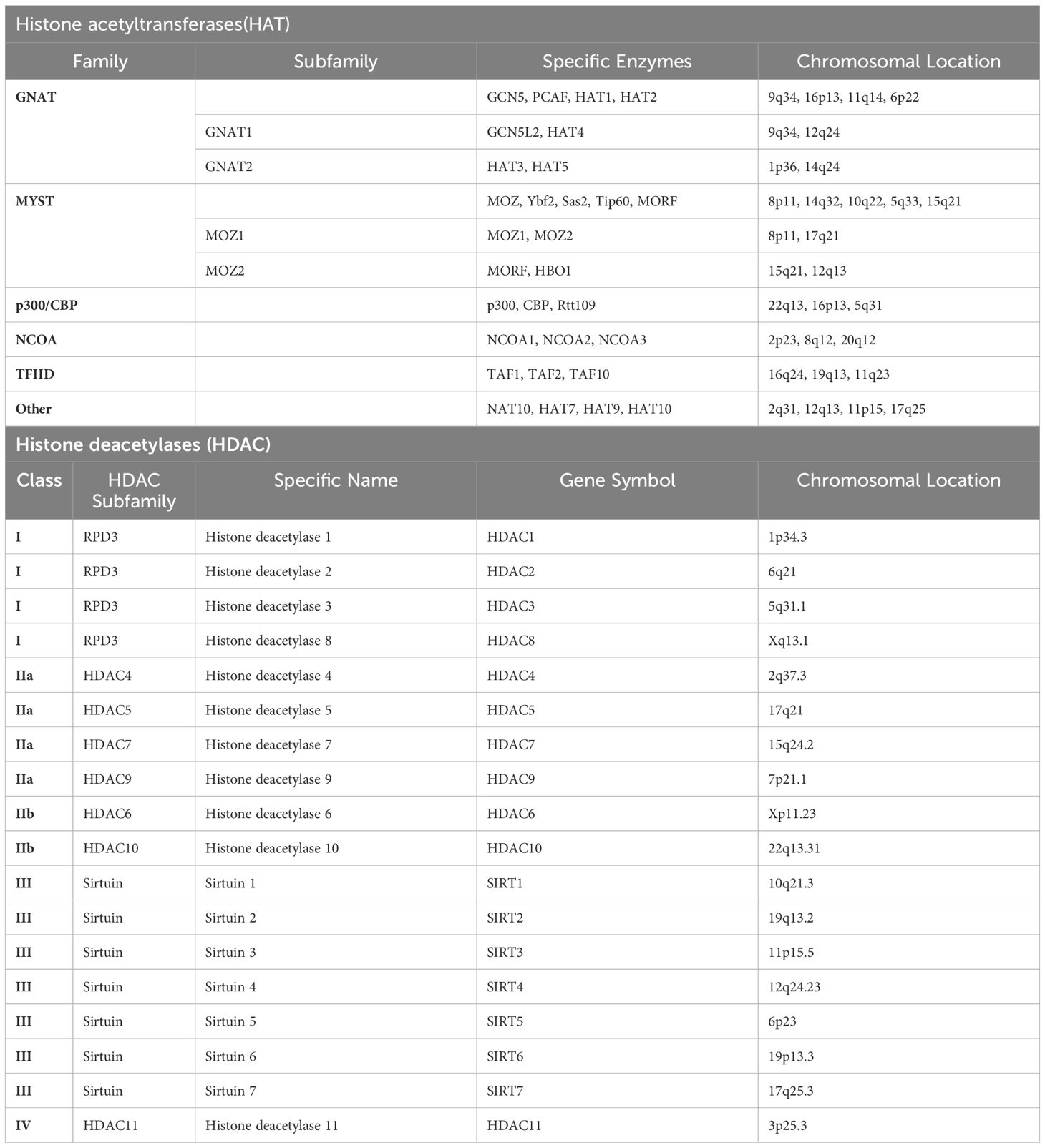
Histone acetylation is a process involves the addition of an acetyl group to the lysine residues in the histone proteins around which DNA is wound. This acetylation process is dynamic and catalyzed by histone acetyltransferases (HATs) (Table 2), specifically including B HAT and others primarily grouped into three families: GCN5/PCAF, CBP/p300, and the MYST family, which encompasses MOZ, MOF, TIP60, and HBO1. Among these, the MYST family stands as the largest subfamily of HATs, distinguished by a highly conserved MYST domain. This domain comprises a zinc finger and an acetyl-CoA binding motif. Additionally, HATs possess additional structures such as zinc fingers linked to plant homeodomains (seen in MORF and MOZ) and chromodomains (found in TIP60 and MOF). These structures contribute to the formation of protein complexes involved in both oncogenic and tumor suppressor activities. Another group of N-acetyltransferases (GNATs), represented by p300/CREB-binding protein (CBP), PCAF, HAT1, and GCN5, is associated with GCN5. Notably, GCN5 bears a bromodomain and catalyzes the acetylation of lysine residues on histones H2B, H3, and H4. The CBP/p300 subfamily comprises numerous subdomains capable of interacting with various proteins harboring disordered domains, such as p53 and NF-κB. The introduction of acetyl groups, which carry a negative charge, neutralizes the positive charge of lysine residues. This neutralization weakens the electrostatic attraction between histones and DNA. In contrast, histone deacetylases (HDACs) (Table 2) are capable of removing acetyl groups, thereby restoring the positive charge of histones, strengthening the binding between histones and DNA. This process results in a more condensed chromatin structure, ultimately inhibiting gene transcription. The balance between histone acetylation and deacetylation is crucial for the proper regulation of gene expression and is involved in numerous biological processes, including cell growth and development, DNA repair, and the response to environmental stimuli. Abnormalities in these processes can lead to diseases such as cancer and neurodegenerative disorders.
Most tumor cells exhibit lower levels of histone acetylation and maintain an acidic intracellular environment, with HNSCC serving as a prime example of this phenomenon (8). Through whole-exome sequencing of human HNSCC tumors (n=235), studies have confirmed that mutations in histone acetyltransferases CREBBP, EP300, or CASP8 are associated with poor prognosis after radiotherapy for HNSCC. This effect is achieved by inhibiting HAT function, thereby suppressing homologous recombination and increasing DNA damage (9). On the other hand, experimental evidence currently suggests that HDAC1 and HDAC2 can bind to ΔNp63α, a prognostic marker for HNSCC, forming an active transcriptional repressor complex that promotes tumor progression (10). Mounting evidence from similar studies converges to suggest that modulating histone dysregulation has emerged as a promising therapeutic strategy for HNSCC treatment. Several drugs have been shown to exert their therapeutic effects by targeting this pathway. Cetuximab (11), for instance, a monoclonal antibody that targets the epidermal growth factor receptor (EGFR), has been studied extensively in the treatment of HNSCC. A crucial aspect of Cetuximab’s pharmacological mechanism involves its ability to suppress tumor cell proliferation by modulating the activity of Acetyl-CoA Carboxylase (ACC). ACC is a metabolic enzyme, and its activity dysregulation is closely linked to the progression of various solid tumors. numerous studies indicate a possible correlation between alterations in acetylation status within tumor cells and Cetuximab therapy resistance. Modulating the activity of HDACs and HATs may potentially overcome Cetuximab resistance, thereby improving therapeutic efficacy (12). Similar compounds such as ACY-241 (13), and JQ1 (14), have also been found to exert their therapeutic effects by targeting this pathway, offering a new avenue for treatment exploration.
2.3 Histone phosphorylation
Histone phosphorylation is a post-translational modification (PTM) that plays a crucial role in regulating chromatin structure and gene expression. It involves the addition of a phosphate group to specific amino acid residues (usually serine, threonine, or tyrosine) on histone proteins. This process is catalyzed by protein kinases, typically utilizing ATP as the source of phosphate groups. Phosphorylation modification may reduce the binding affinity of histones to DNA, thereby facilitating DNA unwinding and the binding of transcription factors. This, in turn, stimulates gene transcription and expression. Research reveals that histone phosphorylation levels are abnormally elevated in cancer cells across multiple tumor types, a phenomenon closely correlated with cellular proliferation, metastasis, and invasion capabilities. For instance, studies have shown that histone H3 phosphorylation at serine 10 (H3S10ph) is significantly increased in various cancers, including breast cancer and colorectal cancer, and is associated with poor prognosis. Current research has revealed that in HNSCC, not only do histones themselves, such as H1 (15) and H3, H4 (16), undergo modifications, but specific amino acid residues like H3Ser10 (17) are also phosphorylated to regulate tumor progression. In HNSCC, abnormal histone phosphorylation activates various signaling pathways, including PI3K/Akt, MAPK, and JNK. The activation of these oncogenic signaling pathways and the inactivation of tumor suppressor pathways accelerate the progression of HNSCC. Tumor therapeutic drugs are developed based on these findings. For instance, PI-828 and PI-103 can block PI3K, affecting downstream signaling molecules such as Akt and mTOR, thereby inducing apoptosis in HNSCC cells, altering cell cycle regulation, and reducing invasiveness (18). Additionally, cetuximab has been reported to activate the epidermal growth factor receptor in HNSCC cell lines, regulating tyrosine 1173 hyperphosphorylation (19) and contributing to therapeutic effects.
2.4 Histone ubiquitylation
Ubiquitin, a highly conserved small protein, conjugates with protein substrates through a stepwise enzymatic cascade involving E1 ubiquitin-activating enzyme, E2 ubiquitin-conjugating enzyme, and E3 ubiquitin ligase. This process plays a crucial role in various cellular processes, including protein degradation, cell signaling, DNA repair, and cell cycle regulation. Defined as the covalent attachment of ubiquitin molecules to target proteins, ubiquitination stands as one of the most prevalent post-translational modifications within the proteome. Structurally, ubiquitin linkage can be classified as three patterns: monoubiquitination, polyubiquitination, and branched ubiquitination. Notably, each type of ubiquitination, alongside lysine ubiquitin linkage occurrences, triggers unique biological responses.
In HNSCC, the involvement of ubiquitin is particularly crucial as it impacts the stability and functionality of oncogenic proteins and tumor suppressor genes, ultimately influencing tumor development and progression. Several genome-wide studies have revealed common mutations/deletions in the CYLD and TRAF3 genes in HNSCC. CYLD, a crucial gene in clear K63 polyubiquitin and M1 linear ubiquitin chains, inhibits NFκB signaling activation at multiple distinct steps (20). Furthermore, TRAF3 serves as a negative regulator of both canonical and non-canonical NFκB pathways, and its deletion can enhance tumor stemness (21). Mutation analysis further indicates that SCFFBXW7, the most frequently mutated gene in HPV- HNSCC, promotes the stabilization of NFκB protein p100 by reducing its degradation through the influence of E3 ligase. This may potentially increase the inactive precursor induced by stimulation, thereby enhancing NFκB activity (22). In a study adopted customized siRNA and cDNA libraries for deubiquitinating enzymes, it was discovered that USP7 serves as the bona fide TAZ deubiquitinase in HNSCC. USP7 promotes cell proliferation, migration, in vitro invasion, and tumor growth by stabilizing TAZ. USP7 interacts with TAZ to deubiquitinate and stabilize it by selectively removing its K48-linked ubiquitin chains. Inhibition of USP7 significantly suppresses tumor growth in HNSCC xenografts and PDX models (23). Furthermore, ubiquitin-targeted therapies show a promising future in HNSCC. The development of isoform-specific E3 ligase inhibitors or DUB inhibitors could potentially minimize off-target effects and enhance therapeutic selectivity (24). Additionally, identifying novel ubiquitin-binding domains (UBDs) and elucidating their roles in HNSCC may contribute to the discovery of new drug targets.
3 Novel histone modifications in HNSCC
3.1 Histone succinylation
Histone succinylation is a recently discovered PTM that plays a crucial role in regulating chromatin structure, gene expression, and cellular processes. Succinylation is the covalent attachment of a succinyl group (CH3CH2CO-) to a specific amino acid side chain on a histone protein. This modification is carried out by enzymes called succinyltransferases, which transfer the succinyl group from succinyl-CoA to the target histone. Succinylation can occur on multiple histone proteins, including H2A, H3, and H4. Histone succinylation can impact the structure and function of chromatin. This modification may lead to chromatin relaxation, promoting transcriptional activity of genes, or regulating gene expression through other mechanisms. Research indicates that histone succinylation plays a significant role in the development and progression of HNSCC. The succinylation levels of histones H3K9, H3K14, and H3K56 are notably elevated in HNSCC tissues, correlating with tumor staging and prognosis (25). Another study suggests that histone H3K122 succinylation promotes HNSCC cell proliferation and invasion by regulating the NOTCH1 signaling pathway (26).
Given the significant role of histone succinylation in HNSCC, targeting this epigenetic modification has emerged as a new therapeutic strategy for HNSCC treatment. EPZ-6438, also known as Tazemetostat, is a small molecule drug belonging to the class of histone methyltransferase (EZH2) inhibitors. Its primary function is to inhibit the activity of the EZH2 enzyme. It has been found to target histone H3K27 succinylation (5), thereby inhibiting the proliferation and invasion of HNSCC cells and inducing apoptosis. Another study revealed that the histone deacetylase inhibitor SAHA (27) exerts its therapeutic effect by inhibiting the succinylation of histones H3K9 and H3K56 (28). These studies suggest that targeting histone succinylation represents a promising treatment strategy for HNSCC, which can be used alone or in combination with traditional chemotherapy drugs to enhance therapeutic outcomes and reduce side effects.
3.2 Histone lactylation
Histone lactylation involves the attachment of a lactyl group (composed of a hydroxyl and a carboxyl group) to the lysine residues of histone proteins. This process is catalyzed by enzymes known as histone lactylases, which belong to the Sirtuin protein family. Utilizing NAD+ as a cofactor, these enzymes facilitate the transfer of a lactyl group from β-alanine to the ϵ-amino group of lysine residues on histone proteins. Notably, this reaction is reversible, implying that the lactyl group can be detached by an enzyme designated as histone lactylase-removing enzyme (LLRE). In HNSCC, research on histone lactylation mainly focuses on three aspects: transcription regulation, DNA repair mechanisms, and cell cycle control. Studies have indicated the abnormal expression of various enzymes associated with histone lactylation in HNSCC. Specifically, the expression of histone acetyltransferase P300/CBP is significantly upregulated in HNSCC, whereas the expression of histone deacetylases HDAC1 and HDAC2 is notably downregulated (29). Furthermore, the expression levels of histone acetyltransferase P300/CBP and HDAC1 in HNSCC correlate with the malignancy and prognosis of the tumor (30).
Given the pivotal role of lactate in tumorigenesis and progression, research targeting histone lactylation is becoming a hot topic. GPR81, standing for G protein-coupled receptor 81 and also known as lactate receptor 1, is a G protein-coupled receptor reported to be widely expressed in various solid tumors, including HNSCC. GPR81 is intricately linked to tumor growth and metastasis. Silencing GPR81 has been shown to exert effective therapeutic effects (31).
3.3 Histone glycosylation
Histone glycosylation involves the addition of a carbohydrate moiety, specifically a sugar molecule, to the histone proteins. This modification is catalyzed by a family of enzymes called histone O-glycosyltransferases, which are responsible for adding a specific sugar molecule, such as N-acetylglucosamine (O-GlcNAc), to the histone proteins. Histone glycosylation occurs at specific serine and threonine residues on the histone proteins, which are recognized by the glycosyltransferases. Histone glycosylation modifications can be primarily categorized into two types: O-GlcNAzylation and ADP-ribosylation. O-GlcNAzylation refers to the process of attaching an N-acetylglucosamine group to the serine or threonine residues of a protein, whereas ADP-ribosylation involves the addition of an ADP-ribose group to the glutamic acid or aspartic acid residues of a protein. Studies have found that O-GlcNAc transferase (OGT)-mediated glycosylation modification at the H2BSer112 site of histone is significantly upregulated in various solid tumors such as HNSCC. Knockdown of OGT can inhibit tumor cell proliferation and induce apoptosis (32). Additionally, Lin et al. (33) found that the glycosyltransferase GALNT2 increased in HNSCC through immunohistochemistry, this increasement can regulates tumor growth via the EGFR/AKT signaling axis. OSMI-1, a cell-permeable O-GlcNAc transferase (OGT) inhibitor, inhibit protein O-GlcNAcylation without qualitatively altering N- or O-linked glycans on the cell surface. In a study targeting esophageal squamous cell carcinoma, it was determined that OSMI-1 can exert a therapeutic effect by regulating PD-1 in tumor cells (34), the above results suggesting that targeting OGT-mediated histone O-GlcNAcylation modification may be a promising anti-HNSCC strategy.
3.4 Histone β-hydroxybutyrylation
Histone β-hydroxybutyrylation (Kbhb) is the addition of a β-hydroxybutyryl group to the lysine residues of histone proteins. This modification is catalyzed by a family of enzymes called histone β-hydroxybutyryltransferases (H3BTs), which use β-hydroxybutyrate as a cosubstrate. This modification usually occurs on the acetyl groups of histones H3 and H4, leading to structural changes in the histones, which subsequently affect the state of chromatin and gene expression. Current research indicates a close relationship between Kbhb and metabolic pathways. The elevation of histone H3K9bhb levels during bodily hunger is associated with the upregulation of genes in hunger-responsive metabolic pathways, and p300-catalyzed histone Kbhb can directly activate transcription (35). Although nowadays research on Kbhb in the field of squamous cell carcinoma is limited, certain enzymes or genes in this pathway have been shown to impact HNSCC. For instance, HMGCS2, which encodes 3-hydroxy-3-methylglutaryl-CoA synthase (HMGCR), is a crucial enzyme in cholesterol biosynthesis and ketogenesis. HMGCS2 can reduce beta-hydroxybutyrate (β-OHB) levels, thereby decreasing histone kbhb modifications, highlighting a tight relationship between the two. A study analyzing HMGCS2 levels in patients with esophageal squamous cell carcinoma (ESCC) revealed significant downregulation in primary ESCC, correlating strongly with patient prognosis (36). Additionally, metformin, a well-known drug, has been proven to increase Kbhb levels in the body. It has been demonstrated to downregulate gene expression profiles associated with cancer stemness and epithelial-mesenchymal transition while increasing the expression of squamous differentiation genes, thus preventing the development of HNSCC (37). In summary, there is a certain correlation between Kbhb and HNSCC, which may hold significant importance in gene expression regulation, metabolic reprogramming, and potential disease treatments.
3.5 Other novel histone modifications in HNSCC
Several recent studies have shown that there are also multiple types of histone modifications that are associated with the development and treatment of HNSCC. Histone crotonylation, for instance, is the addition of a crotonyl group (-CH=CH-COO-) to the lysine residues of histone proteins. This modification is catalyzed by a family of enzymes called histone crotonyltransferases (KAT5/8), which use crotonyl-CoA as a cosubstrate. Histone crotonylation can activate or repress the transcription of specific genes in HNSCC by altering chromatin structure, thereby influencing disease progression (38). Through sequencing and enrichment analysis, it has been found that histone malonylation of HSP90AB1 mediates the onset and progression of oral squamous cell carcinoma (39). With understanding of histone modifications deepens, researchers have reported novel modification patterns like histone SUMOylation. SUMO (Small Ubiquitin-like MOdifier) is a small protein that is similar in structure to ubiquitin, a protein that plays a key role in protein degradation which is also known as sentrin or SMT3. Unlike ubiquitination, SUMOylation does not promote the degradation of its target proteins. Instead, SUMOylation has been shown to regulate various aspects such as protein stability, subcellular localization, protein interactions, and transcriptional regulation (40). Currently, researchers have intervened in the development of HNSCC by applying SUMO-1 inhibitors (41). As these studies progressed, they have improved our understanding of the mechanisms of HNSCC occurrence.
4 Summary and prospects
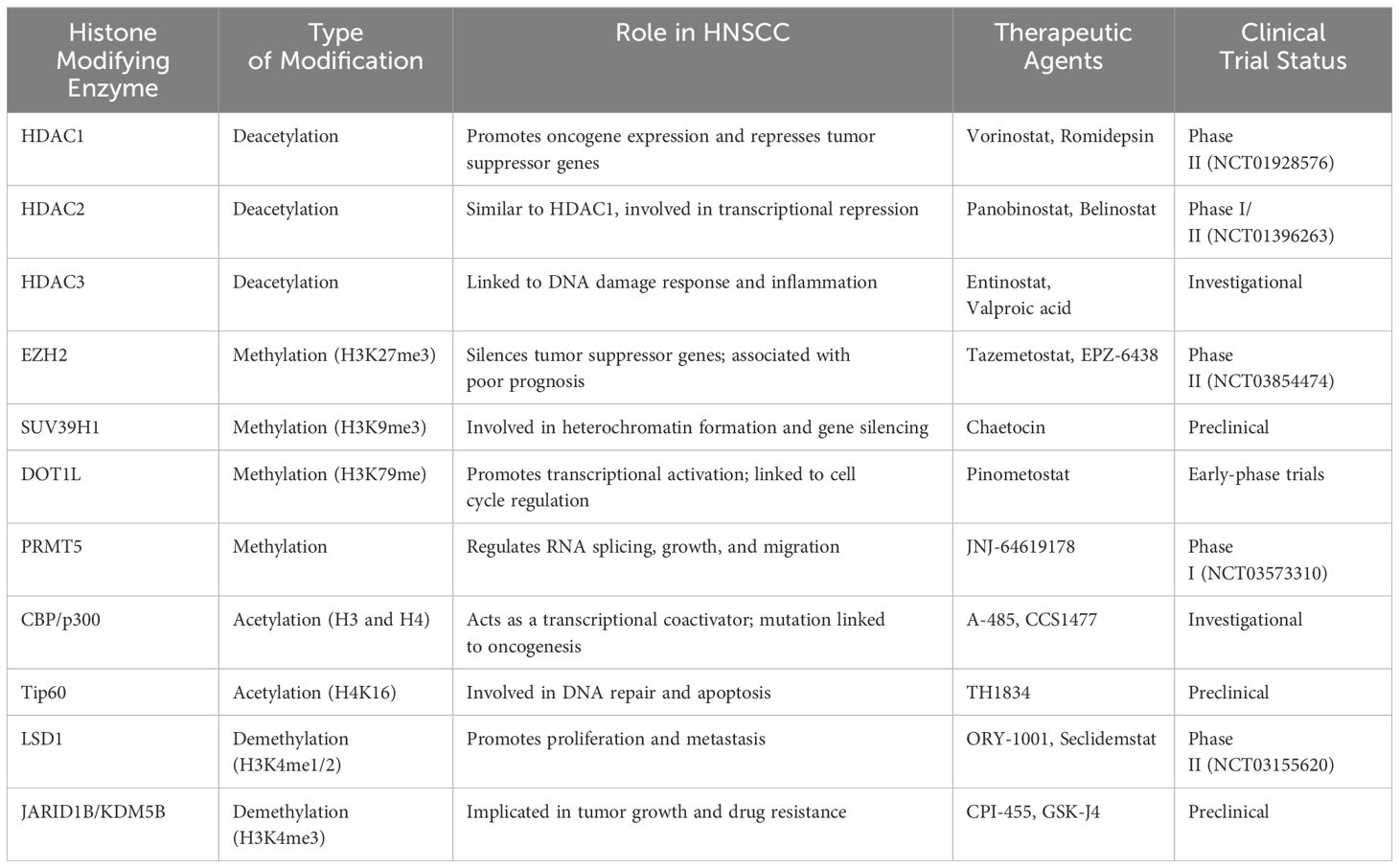
In summary, the role of histone modifications in the pathogenesis of HNSCC is increasingly recognized as a critical factor in tumor development and progression. Key findings from recent studies underscore the significant impact of histone acetylation, methylation, and other modifications on gene expression and cellular behavior in HNSCC. Recent advancements in molecular biology, genomics, and proteomics have broaden our insights on the mechanisms underlying HNSCC development. For instance, aberrant histone deacetylation and methylation have been linked to the repression of tumor suppressor genes and activation of oncogenes, contributing to the malignant phenotype. The significance of these findings lies in the potential for histone modifications to serve as therapeutic targets (Table 3), metaphorically operating as a covert puppeteer manipulating various tumorigenic events. This review delves into the profound impact of histone modifications in HNSCC and summarizes the latest research on targeting these modifications as a therapeutic strategy.
In HNSCC the equilibrium of histone modifications becomes disrupted, ultimately leading to altered gene expression patterns that drive tumorigenesis and disease progression (Figure 4). The primary results of this disruption can be categorized into three primary aspects: the transcriptional dysregulation of oncogenes and tumor suppressor genes mediated by abnormal histone modifications, the modulation of the stem-like properties of HNSCC tumor cells, and the perturbation of the tumor microenvironment. Despite the expanding numbers of research in this field, numerous gaps persist, primarily due to the intricate nature of histone post-translational modifications (PTMs). Recently, several histone modifications have emerged as potent prognostic indicators in HNSCC, offering valuable insights into patient survival and disease trajectory. For example, Trimethylation of histone H3 lysine 27 (H3K27me3) is associated with gene repression and is often found in the tumor suppressor gene loci. High levels of H3K27me3 have been correlated with poor prognosis and decreased overall survival in HNSCC patients. Ubiquitination of histone H2B lysine 5 (H2BK5ub) has been shown to play a critical role in immune evasion and tumor progression in HNSCC. High levels of H2BK5ub have been linked to increased tumor immune infiltration, which is associated with improved prognosis and better response to immunotherapy. Concurrently, research efforts focused on the development of drugs targeting specific histone-modifying enzymes are progressing, thereby carving new therapeutic avenues for HNSCC. Furthermore, the invention of transferase inhibitors, notably DNA methyltransferase inhibitors (DNMTis) and histone deacetylase inhibitors (HDACis), has enriched the therapeutic methods for HNSCC, by effectively targeting abnormal histone modifications and impeding tumor proliferation and metastasis.
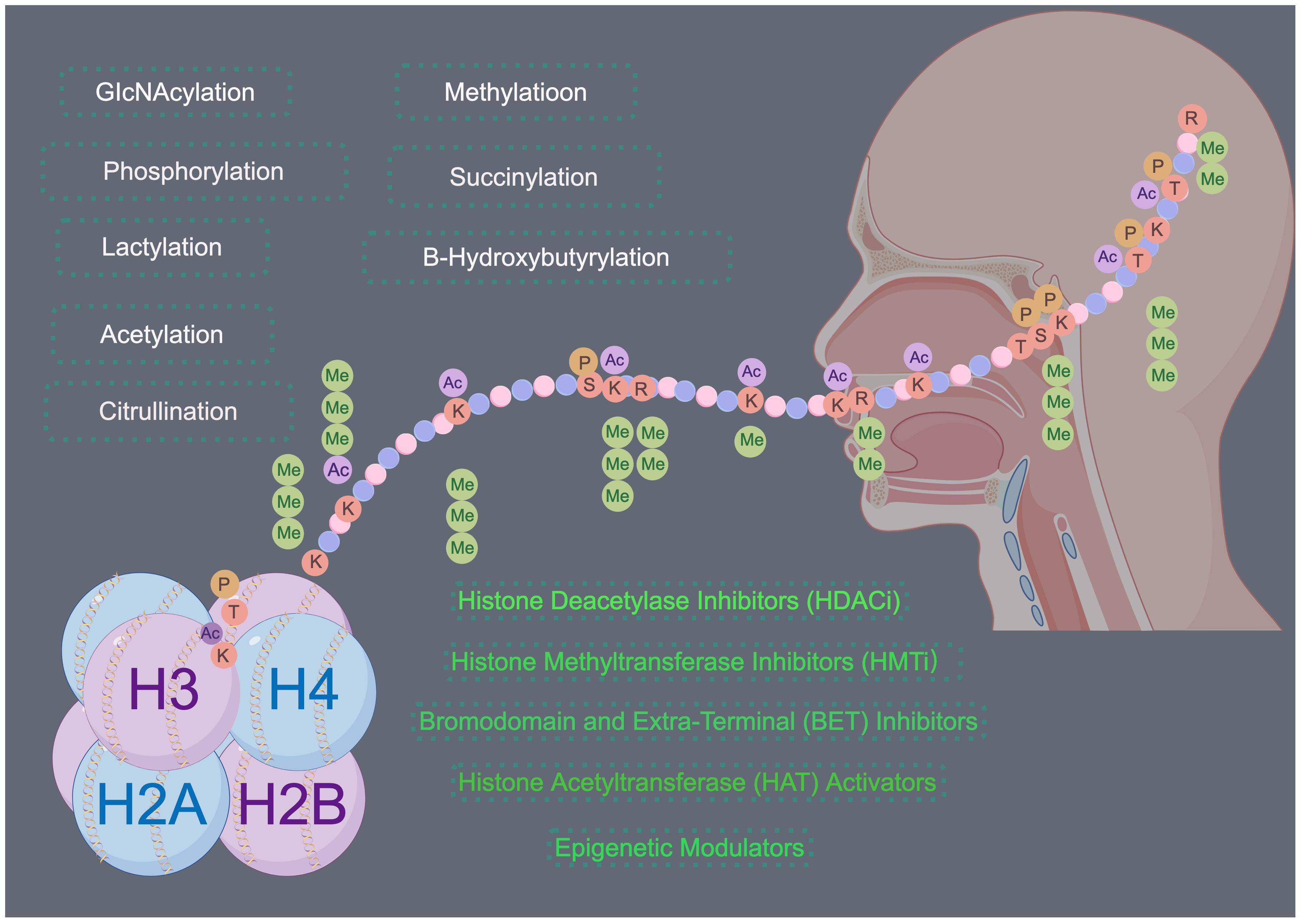
Figure 4 Histone modifications in HNSCC. Histone modifications play a significant role in the development and progression of HNSCC. Targeting these modifications through inhibition of histone-modifying enzymes or direct targeting of histone modifications offers a promising therapeutic strategy for the treatment of HNSCC. Drawing using Figdraw tool.
Future research should focus on several key areas. First, there is a need for comprehensive studies to map the global landscape of histone modifications in HNSCC. This includes identifying specific histone marks associated with different stages of tumorigenesis and their prognostic significance. Second, understanding the interplay between histone modifications and other epigenetic mechanisms, such as DNA methylation and non-coding RNAs, could provide deeper insights into the complex regulatory networks driving HNSCC. Moreover, clinical trials are necessary to evaluate the safety and efficacy of epigenetic therapies targeting histone modifications in HNSCC patients. Such trials should explore combination therapies, integrating epigenetic drugs with conventional treatments like chemotherapy, radiotherapy, and immunotherapy, to assess potential synergistic effects. We are optimistic that a deeper understanding of abnormal histone modifications in HNSCC will allow us to overcome current therapeutic limitations, and provide patients with more precise and personalized treatment options. This will vastly improve patient prognosis, giving renewed hope and energy to those fighting the disease. We eagerly await further research progress in this field.
Author contributions
PD: Resources, Writing – review & editing. WM: Conceptualization, Writing – original draft, Writing – review & editing. BW: Resources, Writing – review & editing. RH: Resources, Writing – review & editing. ZS: Resources, Writing – review & editing. MY: Resources, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work and related studies were supported by National Natural Science Foundation of China, Project No. 82101188.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Yongprayoon V, Wattanakul N, Khomate W, Apithanangsiri N, Kasitipradit T, Nantajit D, et al. Targeting BRD4: Potential therapeutic strategy for head and neck squamous cell carcinoma (Review). Oncol Rep. (2024) 51(6). doi: 10.3892/or.2024.8733
2. Cao J, Yan Q. Cancer epigenetics, tumor immunity, and immunotherapy. Trends Cancer. (2020) 6:580–92. doi: 10.1016/j.trecan.2020.02.003
3. Zhang Y, Sun Z, Jia J, Du T, Zhang N, Tang Y, et al. Overview of histone modification. Adv Exp Med Biol. (2021) 1283:1–16. doi: 10.1007/978-981-15-8104-5_1
4. Bajbouj K, Al-Ali A, Ramakrishnan RK, Saber-Ayad M, Hamid Q. Histone modification in NSCLC: molecular mechanisms and therapeutic targets. Int J Mol Sci. (2021) 22. doi: 10.3390/ijms222111701
5. Li Y, Goldberg EM, Chen X, Xu X, McGuire JT, Leuzzi G, et al. Histone methylation antagonism drives tumor immune evasion in squamous cell carcinomas. Mol Cell. (2022) 82:3901–18.e7. doi: 10.1016/j.molcel.2022.09.007
6. Yang Z, Liu F, Li Z, Liu N, Yao X, Zhou Y, et al. Histone lysine methyltransferase SMYD3 promotes oral squamous cell carcinoma tumorigenesis via H3K4me3-mediated HMGA2 transcription. Clin Epigenet. (2023) 15:92. doi: 10.1186/s13148-023-01506-9
7. Caeiro LD, Nakata Y, Borges RL, Zha M, Garcia-Martinez L, Bañuelos CP, et al. Methylation of histone H3 lysine 36 is a barrier for therapeutic interventions of head and neck squamous cell carcinoma. Genes Dev. (2024) 38:46–69. doi: 10.1101/gad.351408.123
8. Webber LP, Wagner VP, Curra M, Vargas PA, Meurer L, Carrard VC, et al. Hypoacetylation of acetyl-histone H3 (H3K9ac) as marker of poor prognosis in oral cancer. Histopathology. (2017) 71:278–86. doi: 10.1111/his.13218
9. Kumar M, Molkentine D, Molkentine J. CREBBP/EP300 mutation is associated with poor outcome in HNSCC and targetable with synthetic cytotoxicity. bioRxiv. (2020). doi: 10.1101/2020.04.10.028217
10. Ramsey MR, He L, Forster N, Ory B, Ellisen LW. Physical association of HDAC1 and HDAC2 with p63 mediates transcriptional repression and tumor maintenance in squamous cell carcinoma. Cancer Res. (2011) 71:4373–9. doi: 10.1158/0008-5472.CAN-11-0046
11. Leonard B, Brand TM, O'keefe RA, Lee ED, Zeng Y, Kemmer JD, et al. BET inhibition overcomes receptor tyrosine kinase–mediated cetuximab resistance in HNSCC. Cancer Res. (2018) 78:4331–43. doi: 10.1158/0008-5472.CAN-18-0459
12. Provencio M, Sánchez A. Therapeutic integration of new molecule-targeted therapies with radiotherapy in lung cancer. Transl Lung Cancer Res. (2014) 3:89–94. doi: 10.3978/j.issn.2218-6751.2014.03.06
13. Park SJ, Joo SH, Lee N, Jang WJ, Seo JH, Jeong CH. ACY-241, an HDAC6 inhibitor, overcomes erlotinib resistance in human pancreatic cancer cells by inducing autophagy. Arch Pharm Res. (2021) 44:1062–75. doi: 10.1007/s12272-021-01359-x
14. Cho HY, Lee SW, Jeon YH, Lee DH, Kim GW, Yoo J, et al. Combination of ACY-241 and JQ1 synergistically suppresses metastasis of HNSCC via regulation of MMP-2 and MMP-9. Int J Mol Sci. (2020) 21:6873. doi: 10.3390/ijms21186873
15. Saloura V, Vougiouklakis T, Bao R, Kim S, Baek S, Zewde M, et al. WHSC1 monomethylates histone H1 and induces stem-cell like features in squamous cell carcinoma of the head and neck. Neoplasia. (2020) 22:283–93. doi: 10.1016/j.neo.2020.05.002
16. Li H, Yang Z, Yang X, Zhang F, Wang J, Wu Z, et al. LINC01123 promotes immune escape by sponging miR-214-3p to regulate B7–H3 in head and neck squamous-cell carcinoma. Cell Death Dis. (2022) 13:109. doi: 10.1038/s41419-022-04542-0
17. Guan Z, Wang X-R, Zhu X-F, Huang X-f, Xu J, Wang L-h, et al. Aurora-A, a negative prognostic marker, increases migration and decreases radiosensitivity in cancer cells. Cancer Res. (2007) 67:10436–44. doi: 10.1158/0008-5472.CAN-07-1379
18. Simpson DR, Mell LK, Cohen EE. Targeting the PI3K/AKT/mTOR pathway in squamous cell carcinoma of the head and neck. Oral Oncol. (2015) 51:291–8. doi: 10.1016/j.oraloncology.2014.11.012
19. Mandic R, Rodgarkia-Dara CJ, Zhu L, Folz BJ, Bette M, Weihe E, et al. Treatment of HNSCC cell lines with the EGFR-specific inhibitor cetuximab (Erbitux®) results in paradox phosphorylation of tyrosine 1173 in the receptor. FEBS Lett. (2006) 580:4793–800. doi: 10.1016/j.febslet.2006.07.064
20. Squarize CH, Castilho RM, Sriuranpong V, Pinto DS Jr, Gutkind JS. Molecular cross-talk between the NFκB and STAT3 signaling pathways in head and neck squamous cell carcinoma. Neoplasia. (2006) 8:733–46. doi: 10.1593/neo.06274
21. Hajek M, Sewell A, Kaech S, Burtness B, Yarbrough WG. TRAF3/CYLD mutations identify a distinct subset of human papillomavirus-associated head and neck squamous cell carcinoma. Cancer. (2017) 123:1778–90. doi: 10.1002/cncr.30570
22. Davis RJ, Welcker M, Clurman BE. Tumor suppression by the Fbw7 ubiquitin ligase: mechanisms and opportunities. Cancer Cell. (2014) 26:455–64. doi: 10.1016/j.ccell.2014.09.013
23. Li J, Dai Y, Ge H, Guo S, Zhang W, Wang Y, et al. The deubiquitinase USP7 promotes HNSCC progression via deubiquitinating and stabilizing TAZ. Cell Death Dis. (2022) 13:677. doi: 10.1038/s41419-022-05113-z
24. Martinez VD, Vucic EA, Thu KL, Pikor LA, Lam S, Lam WL. Disruption of KEAP1/CUL3/RBX1 E3-ubiquitin ligase complex components by multiple genetic mechanisms: association with poor prognosis in head and neck cancer. Head Neck. (2015) 37:727–34. doi: 10.1002/hed.v37.5
25. Bommi PV, Ravindran S, Raychaudhuri P, Bagchi S. DDB2 regulates epithelial-to-mesenchymal transition (EMT) in oral/head and neck squamous cell carcinoma. Oncotarget. (2018) 9:34708. doi: 10.18632/oncotarget.v9i78
26. Ianni A, Kumari P, Tarighi S, Braun T, Vaquero A. SIRT7: a novel molecular target for personalized cancer treatment? Oncogene. (2024) 43(14):993–1006. doi: 10.1038/s41388-024-02976-8
27. Kumar B, Yadav A, Lang JC, Teknos TN, Kumar P. Suberoylanilide hydroxamic acid (SAHA) reverses chemoresistance in head and neck cancer cells by targeting cancer stem cells via the downregulation of nanog. Genes Cancer. (2015) 6:169. doi: 10.18632/genesandcancer.v6i3-4
28. Drogaris P, Villeneuve V, Pomiès C, Lee E-H, Bourdeau V, Bonneil É, et al. Histone deacetylase inhibitors globally enhance h3/h4 tail acetylation without affecting h3 lysine 56 acetylation. Sci Rep. (2012) 2:220. doi: 10.1038/srep00220
29. Bosic MM, Brasanac DC, Stojkovic-Filipovic JM, Zaletel IV, Gardner JM, Cirovic SL. Expression of p300 and p300/CBP associated factor (PCAF) in actinic keratosis and squamous cell carcinoma of the skin. Exp Mol Pathol. (2016) 100:378–85. doi: 10.1016/j.yexmp.2016.03.006
30. Zhao R, Chen K, Cao J, Yu H, Tian L, Liu M. A correlation analysis between HDAC1 over-expression and clinical features of laryngeal squamous cell carcinoma. Acta Oto-Laryngologica. (2016) 136:172–6. doi: 10.3109/00016489.2015.1101781
31. Zhang Y, Song H, Li M, Lu P. Histone lactylation bridges metabolic reprogramming and epigenetic rewiring in driving carcinogenesis: Oncometabolite fuels oncogenic transcription. Clin Trans Med. (2024) 14:e1614. doi: 10.1002/ctm2.1614
32. Özcan SY. A novel role for 5-hmC in the regulation of cancer testis gene expression in cancer and mesenchymal to epithelial transition. Turkey: Bilkent Universitesi (2014).
33. Lin M-C, Huang M-J, Liu C-H, Yang T-L, Huang M-C. GALNT2 enhances migration and invasion of oral squamous cell carcinoma by regulating EGFR glycosylation and activity. Oral Oncol. (2014) 50:478–84. doi: 10.1016/j.oraloncology.2014.02.003
34. Yuan Y, Wang L, Ge D, Cao B, Fan H, Xue L. Exosomal O-GlcNAc transferase from esophageal carcinoma stem cell promotes cancer immunosuppression through up-regulation of PD-1 in CD8+ T cells. Cancer Lett. (2021) 500:98–106. doi: 10.1016/j.canlet.2020.12.012
35. Zhang D, Xie Z, Chung D, Tang Z, Huang H, Dai L, et al. Metabolic regulation of gene expression by histone lysine β-hydroxybutyrylation. FASEB J. (2017) 31:755.2–.2. doi: 10.1096/fasebj.31.1_supplement.755.2
36. Tang H, Wu Y, Qin Y, Wang H, Jia Y, Yang S, et al. Predictive significance of HMGCS2 for prognosis in resected Chinese esophageal squamous cell carcinoma patients. OncoTargets Ther. (2017) 2553–60. doi: 10.2147/OTT
37. Wu X, Yeerna H, Goto Y, Ando T, Wu VH, Zhang X, et al. Metformin inhibits progression of head and neck squamous cell carcinoma by acting directly on carcinoma-initiating cells. Cancer Res. (2019) 79:4360–70. doi: 10.1158/0008-5472.CAN-18-3525
38. Jiang L, Yin X, Zhang H, Zhang X, Cao Z, Zhou M, et al. Development and validation of a prognostic signature based on the lysine crotonylation regulators in head and neck squamous cell carcinoma. BioMed Res Int. (2023) 2023. doi: 10.1155/2023/4444869
39. Yin X, Zhang H, Wei Z, Wang Y, Han S, Zhou M, et al. Large-scale identification of lysine crotonylation reveals its potential role in oral squamous cell carcinoma. Cancer Manage Res. (2023) 1165–79. doi: 10.2147/CMAR.S424422
40. Chen X, Zhang Y, Wang Q, Qin Y, Yang X, Xing Z, et al. The function of SUMOylation and its crucial roles in the development of neurological diseases. FASEB J. (2021) 35:e21510. doi: 10.1096/fj.202002702R
Keywords: head and neck squamous cell carcinoma (HNSCC), histone modification, methylation, acetylation, targeted therapies
Citation: Mao W, Wang B, Huang R, Sun Z, Yan M and Dong P (2024) Histone modifications in head and neck squamous cell carcinoma. Front. Oncol. 14:1427725. doi: 10.3389/fonc.2024.1427725
Received: 04 May 2024; Accepted: 10 June 2024;
Published: 25 June 2024.
Edited by:
Nerina Denaro, IRCCS Ca’ Granda Foundation Maggiore Policlinico Hospital, ItalyReviewed by:
Liu Shuli, Shanghai Jiao Tong University, ChinaJozsef Dudas, Innsbruck Medical University, Austria
Copyright © 2024 Mao, Wang, Huang, Sun, Yan and Dong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Minzhu Yan, eW16MTEyOUAxNjMuY29t; Pin Dong, ZHBzaGdoQHllYWgubmV0; ZG9uZ3BpbjY0QGFsaXl1bi5jb20=
 Wei Mao
Wei Mao Baoxin Wang
Baoxin Wang Pin Dong
Pin Dong