
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Oncol. , 17 July 2024
Sec. Gastrointestinal Cancers: Colorectal Cancer
Volume 14 - 2024 | https://doi.org/10.3389/fonc.2024.1416806
This article is part of the Research Topic Regulation of Gastrointestinal (GI) Physiology by Gut-Microbiota Interactions in Healthy and Disease Conditions View all articles
Colorectal cancer (CRC) is one of the most common gastrointestinal malignancies in the world. With the rapid pace of life and changes in diet structure, the incidence and mortality of CRC increase year by year posing a serious threat to human health. As the most complex and largest microecosystem in the human body, intestinal microecology is closely related to CRC. It is an important factor that affects and participates in the occurrence and development of CRC. Advances in next-generation sequencing technology and metagenomics have provided new insights into the ecology of gut microbes. It also helps to link intestinal flora with CRC, and the relationship between intestinal flora and CRC can be continuously understood from different levels. This paper summarizes the relationship between intestinal flora and CRC and its potential role in the diagnosis of CRC providing evidence for early screening and treatment of CRC.
Colorectal cancer (CRC), which includes cancer of the colon and rectum, has a high incidence and mortality rate, which seriously affects people’s physical health. According to the latest epidemiological data in 2020, the number of new cases of CRC in the world is approximately 1.9 million, and 935,000 deaths, ranking as the third and second among all malignant tumors (1). Both the incidence and mortality of CRC are still increasing year by year. It is estimated that by 2030, the global burden of CRC is expected to increase by 60%, with more than 2.2 million new cases and 1.1 million cancer deaths (2).
The incidence of CRC is on the rise in developing countries, while it has stabilized or declined in developed countries where the incidence remains high (2, 3). This is due to the early implementation of population-specific CRC early screening and lifestyle management in developed countries. As we all know, the occurrence and development of CRC mostly follow the “adenoma–cancer” sequence, and the whole development process generally lasts 5–10 years (4). Timely detection of precancerous lesions and intervention treatment can effectively prevent the occurrence of CRC and improve the prognosis (4).
The increasing number of CRC cases over the past decade indicates that in addition to genetic factors, unhealthy dietary habits, such as high intake of red meat, processed meats, refined grains, sugary beverages, desserts and potatoes (5); lack of fiber intake; and a high-fat diet, are also important factors in the development of CRC (6). In addition, intestinal flora is also a specific environmental risk factor for CRC (7). Relevant clinical studies have found that the intestinal flora colonized in the colorectal directly or indirectly participate in the body’s digestion and absorption, substance metabolism, immune regulation, and other physiological processes (8). Moreover, it can maintain the stability of intestinal microecosystem (8). The status of intestinal flora is closely related to the occurrence and development of CRC. A large number of animal models and clinical studies have proven that changes in the composition and quantity of intestinal flora promote the occurrence and development of CRC (9).
In recent years, the research on intestinal microecology has gradually increased, and more and more researchers have paid attention to this field. Studies on the relationship between intestinal flora and CRC are also increasing. This paper reviewed the correlation between intestinal flora and CRC, as well as the application of intestinal microecology in CRC screening and diagnosis to provide evidence for early screening and treatment of CRC.
The gut microbiome refers to the microbes that live in a person’s gastrointestinal tract. There are approximately 100 trillion symbiotic bacteria in the normal human gut (10). This large and abundant microflora interacts with the human body to form the intestinal microecology. The number of microbes in the gut is approximately three times the total number of human cells (11). The human intestinal flora first appears in the fetus during the second trimester (12, 13), the abundance and diversity increased in the neonatal period (14, 15), and the α and β diversity of the flora forms and stabilizes in early childhood (16, 17). Although the core groups of human intestinal microbes are very similar, there are great differences in the relative content and strain types among different individuals.
Advances in research on the composition and metabolism of the human microbiome suggest that the gut microbiome has a significant impact on human health. Under normal physiological conditions, the intestinal flora coexists with the host in a stable state. The metabolism and transformation of substances in human body cannot be separated from the biological action of intestinal flora. Intestinal flora is involved in maintaining intestinal homeostasis, and plays an important role in digestion and absorption, substance metabolism, inhibition of pathogen invasion and colonization, regulation of immune response, and guarantee of intestinal mucosal barrier integrity (18–21), with high diversity, stability, resistance and adaptability (22).
Studies have shown that the composition of human gut microbiota is affected by many factors, mainly environmental factors. In healthy people, the intestinal microenvironment is in a stable state due to the mutual restriction of various microflora. However, when the intestinal microecosystem is affected by many factors, including diet, environment and host genes, its number, location, and proportion will change, which will cause the imbalance of intestinal microflora. It is mainly manifested in the changes of microecosystem composition, bacterial bioactivity, and location in different parts of the human body (23). The imbalance of intestinal flora will cause an increase in the number of pathogenic bacteria such as enterotoxigenic Bacteroides fragilis (ETBF), Escherichia coli (E. coli), and Clostridium difficile (24). At the same time, probiotics (such as Bifidobacterium, Lactobacillus, and Bacteroides) will decrease (24), which will lead to the occurrence of diseases. Studies have found that cardiovascular disease (25), diabetes (26), Alzheimer’s disease (27, 28), CRC (20), obesity (29, 30) and many other diseases are closely related to the imbalance of human intestinal flora. The overall classification of intestinal flora is as follows (Table 1):
The intestinal microecosystem is mainly composed of intestinal bacteria, viruses, and their metabolites, which interact with the host organism. Under normal physiological conditions, the intestinal microecosystem is in dynamic balance. When various factors disrupt the balance, the intestinal flora will change significantly. The main manifestation is that the number of pathogenic bacteria increases, and the beneficial bacteria decreases. This can lead to pathological changes in the host intestine and even cancerous changes (10). The intestinal tract plays an increasingly significant role in CRC, so intestinal flora has become the focus of many CRC-related studies (10).
Recent studies have shown that intestinal flora is involved in the occurrence and development of CRC (33, 34) and even in individual response to anti-cancer drugs (9). There is evidence in early animal studies to support the role of gut flora in the development of CRC. In 1967, the first study of gut microbiota mediating the carcinogenic effect of cysteine in germ-free mice was published (35), and it was found that gut microbiota played a crucial role in regulating the carcinogenic effect of cysteine on conventional rats, while cysteine failed to cause cancer in germ-free rats. In addition, a study published in 2017 showed that transferring stool from CRC patients to mice promoted intestinal cell proliferation in germ-free mice and promoted tumor formation in conventional mice given azomethane-induced colon cancer (36).
In human studies, metagenomics has been used to study the role of intestinal flora in CRC. Studies have found that intestinal flora imbalance exists in patients with rectal cancer (20, 37, 38), the reduction of symbiotic bacterial species, and the increase in harmful bacterial population (39). The intestinal flora associated with CRC patients was different from that of the healthy control group (33, 40–43), which was mainly reflected in the difference in bacterial composition and diversity.
There are significant changes in specific bacterial populations between the right colon (ascending, proximal, transverse colon) and the left colon (descending, sigmoid, distal colon) in patients with CRC, and this may affect the mucosal immune response (44). Chen et al. (42) found that compared with healthy subjects, the composition of mucosa-associated bacteria in CRC patients was significantly different. Fusobacterium, Porphyromonas, Peptostreptococcus, Gemella, Mogibacterium and Klebsiella have been found in CRC. It was enriched in patients, but decreased in non-bacteria Feacalibacterium, Blautia, Lachnospira, Bifidobacterium, and Anaerostipes (42). Changes in the composition of the microbiota in the mucosa of patients with CRC are not limited to cancer tissue (45). There are also differences between distal and proximal cancers (45). Bacterial diversity was reduced in stool samples from CRC patients compared to healthy individuals and was dominant in samples of intestinal mucosa.
In addition, it was found that the microbiota of cancer tissues showed lower diversity compared to non-cancer normal tissues (42). Similarly, a meta-analysis that included eight population-controlled studies from Europe, the United States, and East Asia found that 29 bacterial species had increased abundance in the fecal flora of patients with CRC (24), such as Fusobacterium, Porphyromonas, Parvimonas, Peptostreptococcus, Gemella, Prevotella, and Solobacterium, and eight other species with no genome reference sequence, among others (24). Bacterial species commonly associated with CRC development include Fusobacterium nucleatum (Fn), E.coli, Bacteroides fragile, Streptococcus bovis/Streptococcus acidophilus, Clostridium septicus, Enterococcus faecalis, and anaerobic Peptostreptococcus. A recent study found that there is heterogeneity in the flora of colorectal tumors or precancerous adenomas (46). The composition and abundance of some CRC-associated pathogens (including Clostridium, Bacteroides, Parvoomonas, and Prevotella) are different in different parts of the same tumor (46). This heterogeneity was significantly associated with CRC-related genetic changes (KRAS mutations and microsatellite instability) in the adenomato-cancer sequence (46).
Intestinal fungal flora is also a major component of human intestinal flora. Some studies have revealed the characteristics of intestinal fungal flora and pathogenic fungi in the stage of CRC development (47, 48). In addition, it has been proven that the interference of fungal flora can also promote the occurrence of CRC. Adult intestinal fungi are mainly composed of 10 genera from the phyla comycetes (70%) and basidiomycetes (30%). Intestinal fungi affect the host immune system by recognizing Toll-like receptors, C-type lectin receptors, galactolectin 3, NOD-like receptors, and NKp30, and have direct or indirect effects on CRC (49). Caspase recruitment domain containing protein 9 (CARD9) is the adapter protein of CARD expression in myeloid cells, which can activate the nuclear factor κB pathway through fungal surface receptors, and plays an important role in antifungal immunity (50). Wang et al. (50) found that in the mouse model of azomethane oxide and sodium glucan sulfate, mice with the deletion of the adapter protein CARD9 were more likely to suffer from colitis-related colon cancer. Fungal dysregulation can induce the accumulation of myeloid-derived inhibitory cells and promote the occurrence of colon cancer. The ability of macrophages lacking CARD9 to kill fungi was impaired resulting in an increase in intestinal fungi and a change in the composition of the fungal community. The enrichment of Candida tropicalis can induce mouse bone marrow cells to be myeloid-derived suppressor cells, which has a strong inhibitory effect on CD8+ and CD4+ T cells, thus promoting colon cancer. Antifungal treatment with fluconazole can reduce the accumulation of myeloid-derived inhibitory cells and inhibit colon cancer in CARD9-deficient mice. An increase in intestinal fungi, particularly Candida tropicalis, in colon cancer patients was found to be positively correlated with levels of myeloid-derived suppressor cells within the tumor (50). This reveals the important role of fungal dysregulation in the pathogenesis of colon cancer and its mechanism. Coker et al. (7) identified the characteristics of CRC-related intestinal fungi in the further study of CRC intestinal flora. CRC intestinal fungi was imbalanced, Basidiomycetes/Ascomycetes ratio increased, Malassezobacteria increased, and Yeast and Pneumosporobacteria decreased. The composition of fungi varies specifically in CRC, which is reflected in the enrichment of six genera. Among them, Rhododendron and Malassezia of Basidiomycetes and Acremonium of Ascomycetes are considered as opportunistic pathogens or potential carcinogens. In CRC, both the co-occurrence of intestinal fungi and the mutual repulsion of bacteria–fungi were increased, and the positive interaction between Proteobacteria and Ascomycetes became mutually exclusive in CRC suggesting that intestinal fungal disorders may be involved in the occurrence of CRC.
Although existing studies have found that the intestinal flora of CRC patients is unbalanced compared with healthy people, even some studies have confirmed that changes in intestinal flora can directly lead to the occurrence of CRC. But the specific role of which flora in the progression of CRC is still unclear. Further studies have proposed various models, such as the “Alpha-bug” (51) model and the “Driver–passenger” (52, 53) model, to illustrate the mechanisms by which gut microbiota contributes to the development of CRC. Some specific human intestinal symbiotic bacteria directly or indirectly affect intestinal mucosal epithelial cells to cause genetic mutations, and these intestinal bacteria are defined as “Alpha-bug.” The “Driver–passenger” model suggests that intestinal flora-induced CRC was related to changes in intestinal microenvironment. First, some specific intestinal symbiotic bacteria drive DNA damage in epithelial cells, and these intestinal symbiotic bacteria are defined as “driver.” Next, the intestinal microenvironment changes during the cancer process, which facilitates the proliferation of other bacteria (opportunistic pathogens or probiotics), which are defined as “passengers,” such as Fn, Streptococcus bovis, and Roxella. The key mechanisms of intestinal microbiota induction of CRC include genotoxins and virulence factors, intestinal microbial metabolites, inflammatory pathways, oxidative stress, and antioxidant defense regulation (54).
Although the gut microbiota is different, there are several individual bacteria associated with CRC. In recent years, the function and molecular mechanism of several specific bacteria, such as ETBF, Fn, Parvimonasmicros, and Prevotella, have been studied. Chicken hemolytic streptococcus, E. coli, and Peptostreptococcus may also stimulate CRC. Examples of pathogenic bacteria affecting the occurrence and development of CRC and their relationship with CRC are shown in Table 2 (55–67):
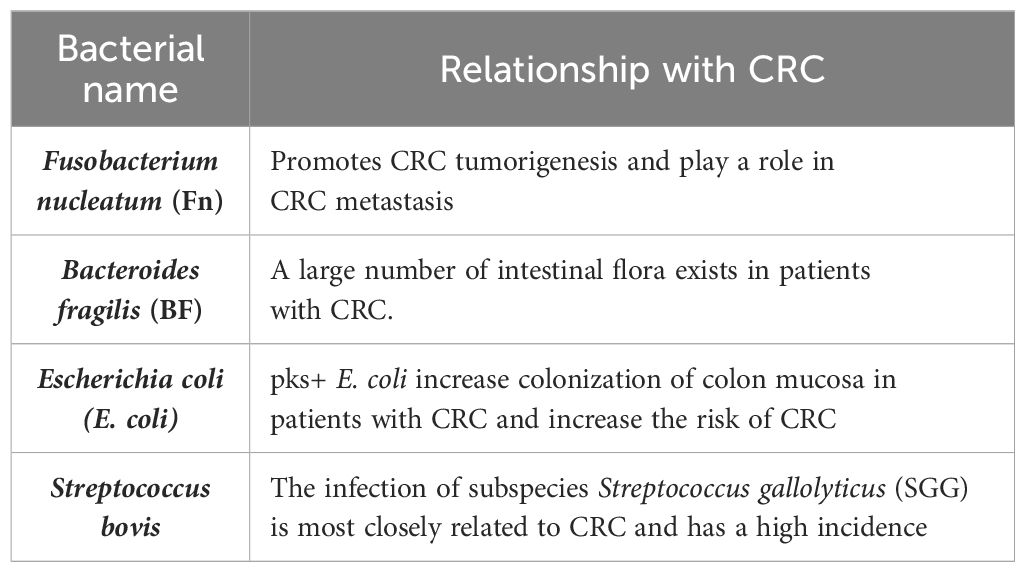
Table 2 The main pathogenic bacteria affecting the occurrence and development of CRC and their relationship with CRC.
Fn, belonging to Bacteroideaceae and Clostridium, is a Gram-negative spore free anaerobic bacterium. It extensively colonizes the symbiotic flora of human oral cavity and intestinal mucosa, and is a bridge microorganism in digestive tract flora. Changes in its number can cause microecological imbalance, which is closely related to the microecological environment of intestinal flora. Fn can promote cell proliferation, induce inflammatory response, inhibit host immune function, and disturb intestinal microenvironment balance and other mechanisms affecting the occurrence, development, and prognosis of colorectal diseases. An in-depth understanding of the pathogenic mechanism of Fn on CRC is helpful to provide a basis for clinical prevention and treatment of CRC.
Through PCR and 16S rDNA sequence analysis, a number of previous studies have found that Fn, as a pathogenic bacterium, is significantly enriched in feces and tissues of patients with CRC (68, 69). Yeoh et al. (70) analyzed 3,157 intestinal metagenomes from 16 populations around the world to study the distribution of Clostridium and its potential CRC-related genomic characteristics and found that the prevalence, relative abundance, and diversity of multiple known and unknown Clostridium groups in southern Chinese populations were higher than those in other regions, regardless of whether they had CRC. Besides Fn, Fusobacterium varium, and other Fusobacterium groups are also enriched in CRC.
Fn may affect multiple stages of CRC progression (71–73), such as promoting proliferation and metabolism, remodeling the immune microenvironment, promoting metastasis, and chemotherapy resistance (74). Regarding the influence of Fn on the progression of CRC, Rubinstein et al. (75) proposed a “two-hit” model for the occurrence of CRC, with somatic mutations as the first “hit” and Fn as the second “hit,” which aggravated the progression of cancer after benign cells became cancerous. The model extends the “adenomato-cancer” model and identifies microorganisms, such as Fn, as promoters of cancer. Li et al. (76) found that Fn can promote the progression of CRC through the Wnt/β-Catenin signaling pathway activated by Cdk5. In addition, Fn has been shown to be associated with CpG island methylation phenotype (CIMP) and microsatellite instability (MSI) in CRC (68, 77).
Xu et al. (58) showed that the abundance of Fn was higher in feces and tumors of patients with metastatic CRC, and the abundance of Fn increased in stage IV compared with that in stage I. It was also proven that Fn could promote the metastasis of CRC through the miR-1322/CCL20 axis and M2 polarization. Recent studies have also supported the role of Fn in promoting lymph node metastasis and lung metastasis in CRC (57).
Chemotherapy failure is the main reason for recurrence and poor prognosis of patients with CRC. Intestinal microflora plays a role in chemotherapy resistance of patients with CRC. Recent studies have also demonstrated that Fn is associated with chemotherapy resistance in CRC patients (78, 79). Due to several unique aspects of Fn on CRC, it can be used as a potential biomarker for the early detection of CRC (80, 81).
Fn forms intestinal flora together with other microflora of viruses, bacteria, fungi and other microorganisms in intestinal microenvironment. All microorganisms are combined in a certain proportion, which both restrict and depend on each other, and jointly maintain the stability of human microecosystem. The enrichment of Fn in CRC patients also reflects that the microecological balance of intestinal microflora in patients is broken. As a member of the intestinal microflora, Fn can affect the intestinal microecological environment. The enriched Fn in the intestinal tract plays multiple roles in the occurrence and development of CRC, which makes it expected to become a therapeutic target of the disease and provide a new strategy for CRC prevention and treatment.
Bacteroides fragilis (BF) is a common Gram-negative obligate anaerobic bacterium in the human gut. Some BF can secrete a toxin called BF toxin (BFT). According to the synthesis and secretion of BF enterotoxin, it can be divided into ETBF and non-enterotoxigenic BF (NTBF). BFT is the only recognized virulence factor of ETBF, and early studies have found that BFT can cause inflammatory diarrhea in children and adults (82).
However, in recent years, there has been increasing evidence that BFT is associated with the development of CRC (60), and studies have demonstrated increased ETBF colonization in CRC patients (61, 62). Colonization of ETBF in colitis-induced CRC mouse models increased the number of tumors (83), while in Apc Min/+ CRC mouse models, it promoted the development of colorectal adenomas (84) further confirming its carcinogenic potential. Recently, Liu et al. (85) found that ETBF increased the dryness of CRC by upregulating the expression of Nanog and Sox2, and found that ETBF significantly increased JMJD2B by activating the TLR4 pathway. Gao et al. (86) found that ETBF promoted intestinal inflammation and malignancies by inhibiting exosomal miRNA. In the colitis CRC model, WT-ETBF promoted polyp formation in the AOM/DSS mouse model and increased the incidence of colitis in BALB/c mice (83).
E. coli is a very common but small number of Gram-negative facultative anaerobes in the distal gastrointestinal tract. In addition to being an important member of the normal gut microbial community in humans and other mammals, E. coli also contains many pathogenic forms that can cause a wide range of diseases. At least six different pathogens cause intestinal disorders, such as diarrhea or dysentery, while others cause parenteral infections, including urinary tract infections and meningitis (87). The prevalence of pks+ E. coli was higher in CRC patients (14.7%) than in healthy people (4.3%) (88). Increased colonization of the colon mucosa by pks+ E. coli in CRC patients (89, 90) suggests its role in CRC. Various previous studies have also confirmed the link between E. coli and CRC (88, 91–93). In E. coli, the B2 strain has a certain “genotoxicity,” which carries a conserved pathogenic gene “polyketide synthase (pks)” island, which can produce a small molecular toxic substance colibactin. It leads to eukaryotic DNA damage, cell cycle arrest, mutation, and chromosome instability (94). In a recent study, Pleguezuelos-Manzano et al. (91) used human intestinal organoids to demonstrate that pks+ E. coli induces a CRC-associated mutational signature caused by exposure to CoPEC rich in human CRC tumors and metastases. Salesse et al. (95) found that colicin-producing E. coli (CoPEC) induced CRC in a mouse model of CRC lacking genetic susceptibility. Studies have also shown that in both humans and mice, CoPEC infection is associated with a reduction in tumor-infiltrating T lymphocytes leading to tumor resistance to immunotherapy (96). In addition, a new study is the first to link a Western-style diet to a specific cause of cancer (63). Analysis of two prospective cohort studies in the United States has shown that a Western-style diet increases CRC risk through carcinogenic pks+ E. coli (63).
Streptococcus bovis is a Gram-positive bacterium associated with endocarditis (97). Streptococcus bovis, also known as the Bovis/equine Streptococcus complex (SBSEC), includes many closely related subspecies, such as SGG, Streptococcus macedonicus, Streptococcus infantilus, and others (98).
Using mice genetically predisposed to CRC, Aymeric et al. (99) found that SGG colonization was 1,000 times higher in tumor-bearing mice than in normal mice. After tracking and detecting NF-kB and interleukin-8 (IL-8), Abdulamir et al. (62) found that SGG may promote the synthesis of cyclooxygenase-2 (COX-2) through these two factors. It can induce inflammation, resist apoptosis, and induce cancer through angiogenesis, thus proving that SGG has a certain correlation with CRC. Due to their powerful mutability, these molecules can participate in tumor formation by modifying cellular DNA. SGG infection promotes the deterioration of the colon epithelium, while the physiological status of the colon is also changed, including reduced intestinal mucus and increased permeability of the epithelial cells. These physiological changes can break the balance of intestinal microecology and make the host vulnerable to infection by pathogenic bacteria. A recent study found that SGG-expressed type VII secretion system (T7SS) can promote the adhesion of SGG to colon cells and enhance the intestinal colonization of SGG, thus mediating the promotion of SGG to colon tumors (100).
Although previous studies have confirmed that Streptococcus bovis is associated with CRC, the mechanism of its involvement in the occurrence of CRC has not been fully clarified. Studies have shown that Streptococcus bovis may induce inhibitory immunity by recruitment of tumor-infiltrating CD11b + TLR-4 + cells, thereby contributing to the development of CRC (101).
The 5-year survival rate for early-stage CRC can reach 90%, but the 5-year survival rate for metastatic disease rapidly drops below 15% (102). Effective screening for precancerous lesions or biomarkers of cancer can significantly reduce CRC-related mortality (103). At present, colonoscopy is the most effective means of diagnosis, but because the biggest problem of colonoscopy is invasive examination, and requires tedious intestinal preparation, in addition to the risk of perforation, it is not suitable for early screening of large samples of people (104). The immunofecal occult blood test (FIT) is a non-invasive, simple, and cost-effective method for CRC screening, but it often produces false-positive results and is less sensitive to advanced adenomas (105, 106). Multi-target fecal DNA testing is also very effective for CRC detection (sensitivity 92.3%), but the false-positive rate is higher than that of FIT (107), and the test is expensive. Therefore, it is necessary to develop a new non-invasive, simple, and effective method for CRC screening. Studies have reported associations between bacterial markers and clinical outcomes increasing the possibility of using these microbial markers for treatment and prognosis (19). Relevant studies have demonstrated the association between certain specific bacteria in intestinal flora and the diagnosis of CRC (19). Multiple studies have found that people with CRC have significantly different types and numbers of gut microbiota than healthy people (10). Exploring the intestinal microecosystem composition of CRC patients can provide a new means of screening for CRC. Currently, the difference in intestinal flora between CRC patients and healthy people has been used as one of the screening methods for CRC (19). The use of intestinal flora as a diagnostic and prognostic indicator for CRC needs further exploration to provide new ideas for the diagnosis and treatment of CRC by intestinal biota in the future.
There are differences in the abundance of gut microbiota among CRC patients, adenoma patients, and healthy people (108). Microbial markers can be used for microbial diagnosis of CRC. In recent years, large-scale studies of fecal metagenomes have identified microbial signatures that can predict CRC in different populations. Metagenome-based studies on the relationship between intestinal flora and CRC are shown in Table 3.
Recent studies have also found that CRC patients have the same strain of Fn in their CRC and mouth (120). Oral microbiota may also be used as a biomarker to screen for CRC and polyps (120–122).
In addition to bacteria, the imbalance of fungal flora in the gut is also closely related to CRC. Fungal flora can also be used for early diagnosis of CRC patients (47, 106). Its use in combination with bacterial biomarkers can be more accurate in clinical diagnosis of CRC. Recently, several studies have used machine learning-based meta-sequencing analyses of gut bacterial DNA to detect differences in gut microbiota between healthy individuals and CRC patients and have used this information for the development of CRC diagnostic models (123–125).
Gut microbial metabolites are important factors that link gut microbiota to CRC (126, 127). The relationship between intestinal flora and its metabolites and the occurrence and development of CRC is shown in Figure 1: How the species of bacteria and their metabolites affect the progression of CRC has been of great concern. Acetaldehyde-producing bacteria, sulfate-reducing bacteria, and 7α-dehydroxylating bacteria are thought to be one of the likely major contributors to CRC risk because their metabolites have colonic and tumor-inducing toxicity, including acetaldehyde, hydrogen sulfide, and secondary bile acids and so on (128). On the other hand, bacterial metabolites that may reduce the risk of CRC include butyrate and other short-chain fatty acids, as well as conjugated linoleic acid (128). Moreover, many studies have reported varying levels of microbial metabolites in stool samples from patients with CRC, including unsaturated fatty acids, ursodeoxycholic acid, lower levels of butyric acid, and higher levels of acetic acid (129). One study reported that in patients with precancerous polyps, the abundance of genes involved in amino acid and sulfur metabolism increased, while the abundance of genes involved in methane metabolism decreased relatively (43). By integrating the results of serum metabolomics and fecal metagenomic sequencing, Chen et al. (130) identified a group of serum metabolites (GMSM) of CRC and adenoma patients closely related to the gut microbiota, and established a GMSM-based model. The model was more effective than the clinical marker carcinoembryonic antigen (CEA) in differentiating CRC and adenoma patients from healthy individuals. Coker et al. (131) performed metabolomic and metagenomic analyses of stool samples from 386 subjects and found that metabolite markers (20 metabolites) could be used to diagnose CRC (AUC = 0.80) and that combining intestinal bacteria with metabolite markers improved their diagnostic ability (AUC = 0.94). CRC biomarkers based on intestinal flora and its metabolites are a promising method for CRC screening and diagnosis, which has the advantages of being non-invasive, sensitive, specific, and inexpensive. Compared with traditional CRC screening and diagnosis methods (such as fecal occult blood test, colonoscopy, serological testing), it has higher acceptability and accuracy.
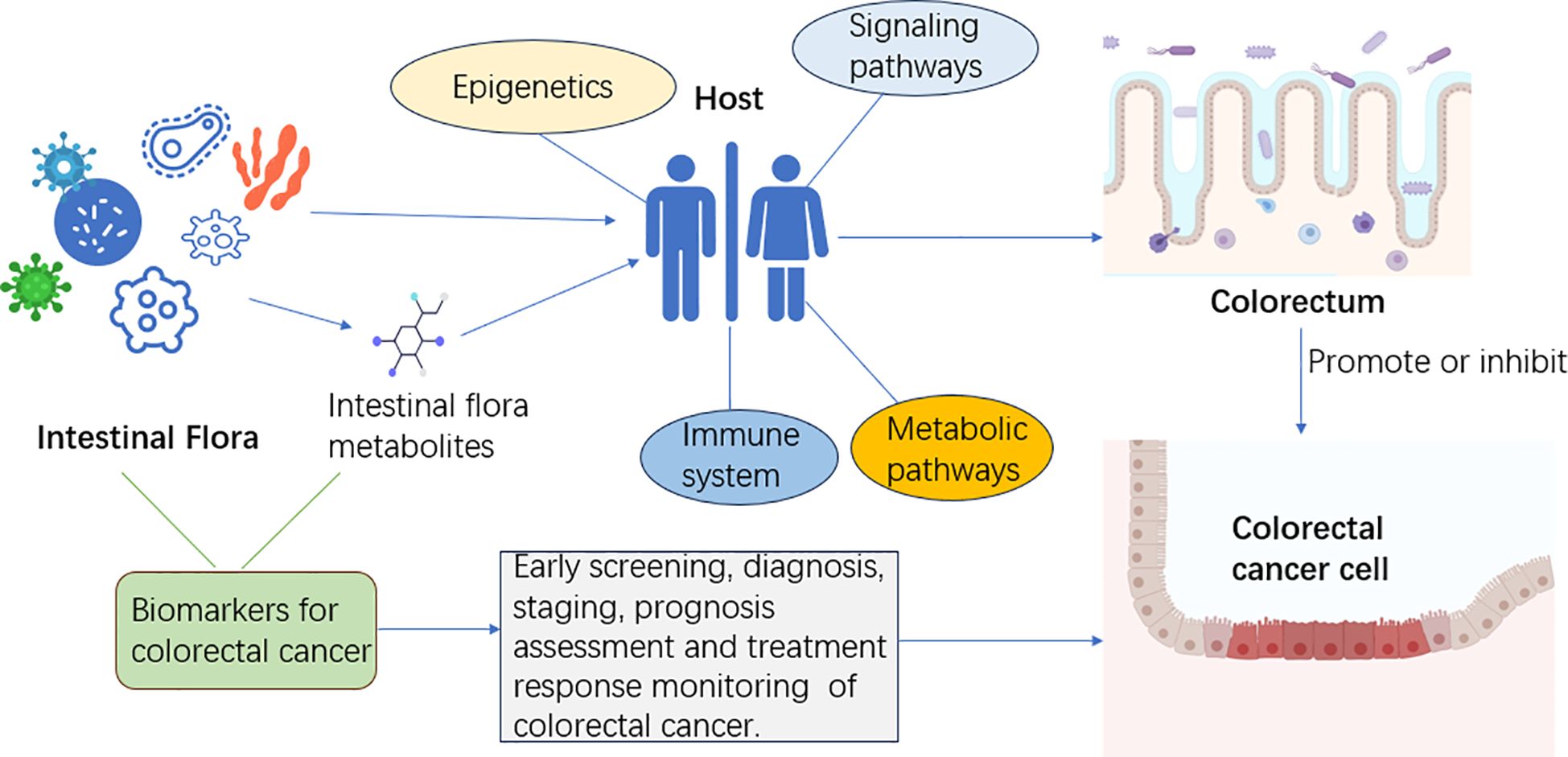
Figure 1 The gut microbiota and its metabolites influence the host’s immune system, metabolic pathways, epigenetics, and signaling pathways. Intestinal flora can interact with CRC cells through their metabolites. Intestinal flora and its metabolites can be used as biomarkers of CRC for early screening, diagnosis, staging, prognosis assessment, and treatment response monitoring.
According to epidemiological surveys, up to 80% of CRC incidence in Western populations can be attributed to dietary factors (128). People whose diets are high in animal fats, red and processed meats, and low in unrefined grains, dietary fiber, and vegetables have a higher chance of developing CRC (128). In recent years, with the in-depth study of intestinal flora, diet can have a huge impact on the structure and function of intestinal flora. These microorganisms ferment dietary components in an anaerobic manner that are not fully digested and absorbed by the upper gastrointestinal tract. Metabolites and antigens produced by the gut microbiota may play an important role in influencing CRC risk through their interactions with host metabolism and immunity (128).
A large number of experimental data show that high-fat diet (HFD) can significantly change the composition of intestinal flora and participate in the occurrence and development of CRC. Most studies have observed that high-fat diet has increased the proportion of Firmicutes and Bacteroides, decreased the diversity of intestinal flora, and increased the abundance of Desulfovibrio and Ruminococcus (103). The abundance of Prevotellaceae was reduced (132). A 10-day study (128) found that a short-term high-fat diet altered the composition of human gut microbes. A 6-month trial in a Chinese young population found (133) that compared with a low-fat diet, a high-fat diet reduced the diversity of intestinal flora, reduced the abundance of Faecalibacterium, and increased the abundance of Alistipes and Bacteroides.
High-fat diet cannot only cause structural changes in intestinal flora but also have an important effect on intestinal flora metabolism and produce a large number of secondary bile acids, fatty acids, and sulfide derivatives. When dietary fat enters the intestine, the liver and pancreas secrete large amounts of bile acids and lipases, respectively, to help further digest dietary fat. The primary bile acids secreted by the liver will be further ingested by the intestinal flora to produce secondary bile acids. Excessive secondary bile acids may promote the development of colorectal tumors. Secondary bile acids can cause DNA damage of intestinal epithelial cells, exacerbate inflammatory response, and promote the proliferation of intestinal Lgr5 (+) intestinal stem cells, thus promoting the occurrence and development of CRC (134). To establish a healthy diet, to better maintain the structural balance of intestinal flora, to a certain extent, help prevent or treat CRC.
CRC is one of the most common malignant tumors of the digestive tract. As an important influencing factor of CRC, the relationship between intestinal flora and CRC has been attracting attention. This study systematically and comprehensively described the different classification and corresponding physiological functions of intestinal flora, the research progress of the correlation between intestinal flora and CRC, the main pathogenic bacteria associated with CRC and their pathogenic mechanisms, and the value of intestinal flora in the diagnosis of CRC. At the same time, it summarized the influence of bad lifestyle on intestinal flora and its role in the occurrence and development of CRC. It lays a foundation for how to better maintain the health of intestinal microecology and apply intestinal microecology to the early prevention and screening of CRC in the future. Although the relevant studies on intestinal flora and CRC are gradually increasing, the clinical understanding of the relationship between intestinal microecology and CRC is still limited. Therefore, it is necessary to further study the correlation between intestinal flora and CRC in the future, correctly understand the role of intestinal flora, and deeply explore its mechanism of action on the occurrence and development of CRC, and finally provide new ideas and methods for the prevention, diagnosis, and treatment of CRC.
XW: Writing – original draft, Writing – review & editing. QZ: Conceptualization, Investigation, Writing – original draft, Writing – review & editing. RX: Investigation, Software, Writing – review & editing. ZH: Supervision, Writing – review & editing. XL: Supervision, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The Natural Science Foundation of Liaoning Province of China (2022-MS-320) funded this study.
The authors highly appreciate all the members who were involved in the present study. The graphs and tables in this article use elements from PowerPoint and the Med peer platform.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. (2021) 71:209–49. doi: 10.3322/caac.21660
2. Arnold M, Sierra MS, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global patterns and trends in colorectal cancer incidence and mortality. Gut. (2017) 66:683–91. doi: 10.1136/gutjnl-2015-310912
3. Cao W, Chen HD, Yu YW, Li N, Chen WQ. Changing profiles of cancer burden worldwide and in China: a secondary analysis of the global cancer statistics 2020. Chin Med J (Engl). (2021) 134:783–91. doi: 10.1097/CM9.0000000000001474
4. Dekker E, Tanis PJ, Vleugels JLA, Kasi PM, Wallace MB. Colorectal cancer. Lancet. (2019) 394:1467–80. doi: 10.1016/S0140-6736(19)32319-0
5. Tabung FK, Brown LS, Fung TT. Dietary patterns and colorectal cancer risk: A review of 17 years of evidence (2000-2016). Curr Colorectal Cancer Rep. (2017) 13:440–54. doi: 10.1007/s11888-017-0390-5
6. Jeon J, Du M, Schoen RE, Hoffmeister M, Newcomb PA, Berndt SI, et al. Determining risk of colorectal cancer and starting age of screening based on lifestyle, environmental, and genetic factors. Gastroenterology. (2018) 154:2152–2164 e19. doi: 10.1053/j.gastro.2018.02.021
7. Coker OO, Nakatsu G, Dai RZ, Wu WKK, Wong SH, Ng SC, et al. Enteric fungal microbiota dysbiosis and ecological alterations in colorectal cancer. Gut. (2019) 68:654–62. doi: 10.1136/gutjnl-2018-317178
8. Cheng Y, Ling Z, Li L. The intestinal microbiota and colorectal cancer. Front Immunol. (2020) 11:615056. doi: 10.3389/fimmu.2020.615056
9. Wong SH, Yu J. Gut microbiota in colorectal cancer: mechanisms of action and clinical applications. Nat Rev Gastroenterol Hepatol. (2019) 16:690–704. doi: 10.1038/s41575-019-0209-8
10. Zhang X, Zhao S, Song X, Jia J, Zhang Z, Zhou H, et al. Inhibition effect of glycyrrhiza polysaccharide (GCP) on tumor growth through regulation of the gut microbiota composition. J Pharmacol Sci. (2018) 137:324–32. doi: 10.1016/j.jphs.2018.03.006
11. Sender R, Fuchs S, Milo R. Revised estimates for the number of human and bacteria cells in the body. PloS Biol. (2016) 14:e1002533. doi: 10.1371/journal.pbio.1002533
12. Stinson LF, Boyce MC, Payne MS, Keelan JA. The not-so-sterile womb: evidence that the human fetus is exposed to bacteria prior to birth. Front Microbiol. (2019) 10:1124. doi: 10.3389/fmicb.2019.01124
13. La Rosa PS, Warner BB, Zhou Y, Zhou Y, Weinstock GM, Sodergren E, et al. Patterned progression of bacterial populations in the premature infant gut. Proc Natl Acad Sci U.S.A. (2014) 111:12522–7. doi: 10.1073/pnas.1409497111
14. Dominguez-Bello MG, Costello EK, Contreras M, Contreras M, Magris M, Hidalgo G, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci U.S.A. (2010) 107:11971–5. doi: 10.1073/pnas.1002601107
15. Palmer C, Bik EM, Digiulio DB, Relman DA, Brown PO. Development of the human infant intestinal microbiota. PloS Biol. (2007) 5:e177. doi: 10.1371/journal.pbio.0050177
16. Stewart CJ, Ajami NJ, O'brien JL, Hutchinson DS, Smith DP, Wong MC, et al. Temporal development of the gut microbiome in early childhood from the TEDDY study. Nature. (2018) 562:583–8. doi: 10.1038/s41586-018-0617-x
17. Fujimura KE, Sitarik AR, Havstad S, Lin DL, Levan S, Fadrosh D, et al. Neonatal gut microbiota associates with childhood multisensitized atopy and T cell differentiation. Nat Med. (2016) 22:1187–91. doi: 10.1038/nm.4176
18. Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, et al. Diversity of the human intestinal microbial flora. Science. (2005) 308:1635–8. doi: 10.1126/science.1110591
19. Tap J, Mondot S, Levenez F, Pelletier E, Caron C, Furet JP, et al. Towards the human intestinal microbiota phylogenetic core. Environ Microbiol. (2009) 11:2574–84. doi: 10.1111/j.1462-2920.2009.01982.x
20. Fan X, Jin Y, Chen G, Ma X, Zhang L. Gut microbiota dysbiosis drives the development of colorectal cancer. Digestion. (2021) 102:508–15. doi: 10.1159/000508328
21. Lucas C, Barnich N, Nguyen HTT. Microbiota, inflammation and colorectal cancer. Int J Mol Sci. (2017) 18. doi: 10.3390/ijms18061310
22. Meng C, Bai C, Brown TD, et al. Human gut microbiota and gastrointestinal cancer. Genomics Proteomics Bioinf. (2018) 16:33–49. doi: 10.1016/j.gpb.2017.06.002
23. Brennan CA, Garrett WS. Gut microbiota, inflammation, and colorectal cancer. Annu Rev Microbiol. (2016) 70:395–411. doi: 10.1146/annurev-micro-102215-095513
24. Wirbel J, Pyl PT, Kartal E, Zych K, Kashani A, Milanese A, et al. Meta-analysis of fecal metagenomes reveals global microbial signatures that are specific for colorectal cancer. Nat Med. (2019) 25:679–89. doi: 10.1038/s41591-019-0406-6
25. Witkowski M, Weeks TL, Hazen SL. Gut microbiota and cardiovascular disease. Circ Res. (2020) 127:553–70. doi: 10.1161/CIRCRESAHA.120.316242
26. Gurung M, Li Z, You H, Rodrigues R, Jump DB, Morgun A, et al. Role of gut microbiota in type 2 diabetes pathophysiology. EBioMedicine. (2020) 51. doi: 10.1016/j.ebiom.2019.11.051
27. Chen C, Liao J, Xia Y, Liu X, Jones R, Haran J, et al. Gut microbiota regulate Alzheimer’s disease pathologies and cognitive disorders via PUFA-associated neuroinflammation. Gut. (2022) 71:2233–52. doi: 10.1136/gutjnl-2021-326269
28. Cryan JF, O'riordan KJ, Cowan CSM, Sandhu KV, Bastiaanssen TFS, Boehme M, et al. The microbiota-gut-brain axis. Physiol Rev. (2019) 99:1877–2013. doi: 10.1152/physrev.00018.2018
29. Torres-Fuentes C, Schellekens H, Dinan TG, Cryan JF. The microbiota–gut–brain axis in obesity. Lancet Gastroenterol Hepatol. (2017) 2:747–56. doi: 10.1016/S2468-1253(17)30147-4
30. Liu B-N, Liu X-T, Liang Z-H, Wang JH. Gut microbiota in obesity. World J Gastroenterol. (2021) 27:3837–50. doi: 10.3748/wjg.v27.i25.3837
31. Keku TO, Dulal S, Deveaux A, Jovov B, Han X. The gastrointestinal microbiota and colorectal cancer. Am J Physiol Gastrointest Liver Physiol. (2015) 308:G351–63. doi: 10.1152/ajpgi.00360.2012
32. Jin M, Qian Z, Yin J, Xu W, Zhou X. The role of intestinal microbiota in cardiovascular disease. J Cell Mol Med. (2019) 23:2343–50. doi: 10.1111/jcmm.14195
33. Liu J, Qi M, Qiu C, Wang F, Xie S, Zhao J, et al. Integrative analysis of the mouse fecal microbiome and metabolome reveal dynamic phenotypes in the development of colorectal cancer. Front Microbiol. (2022) 13:1021325. doi: 10.3389/fmicb.2022.1021325
34. Peters BA, Dominianni C, Shapiro JA, Church TR, Wu J, Miller G, et al. The gut microbiota in conventional and serrated precursors of colorectal cancer. Microbiome. (2016) 4:69. doi: 10.1186/s40168-016-0218-6
35. Laqueur GL, Mcdaniel EG, Matsumoto H. Tumor induction in germfree rats with methylazoxymethanol (MAM) and synthetic MAM acetate. J Natl Cancer Inst. (1967) 39:355–71.
36. Wong SH, Zhao L, Zhang X, Nakatsu G, Han J, Xu W, et al. Gavage of fecal samples from patients with colorectal cancer promotes intestinal carcinogenesis in germ-free and conventional mice. Gastroenterology. (2017) 153:1621–1633 e6. doi: 10.1053/j.gastro.2017.08.022
37. Gagniere J, Raisch J, Veziant J, Barnich N, Bonnet R, Buc E, et al. Gut microbiota imbalance and colorectal cancer. World J Gastroenterol. (2016) 22:501–18. doi: 10.3748/wjg.v22.i2.501
38. Wu N, Yang X, Zhang R, Li J, Xiao X, Hu Y, et al. Dysbiosis signature of fecal microbiota in colorectal cancer patients. Microb Ecol. (2013) 66:462–70. doi: 10.1007/s00248-013-0245-9
39. Kumar A, Ali A, Kapardar RK, Dar GM, Nimisha, Nimisha Apurva, et al. Implication of gut microbes and its metabolites in colorectal cancer. J Cancer Res Clin Oncol. (2023) 149:441–65. doi: 10.1007/s00432-022-04422-2
40. Gao Z, Guo B, Gao R, Zhu Q, Qin H. Microbiota disbiosis is associated with colorectal cancer. Front Microbiol. (2015) 6:20. doi: 10.3389/fmicb.2015.00020
41. Lu Y, Chen J, Zheng J, Hu G, Wang J, Huang C, et al. Mucosal adherent bacterial dysbiosis in patients with colorectal adenomas. Sci Rep. (2016) 6:26337. doi: 10.1038/srep26337
42. Chen W, Liu F, Ling Z, Tong X, Xiang C. Human intestinal lumen and mucosa-associated microbiota in patients with colorectal cancer. PloS One. (2012) 7:e39743. doi: 10.1371/journal.pone.0039743
43. Yachida S, Mizutani S, Shiroma H, Shiba S, Nakajima T, Sakamoto T, et al. Metagenomic and metabolomic analyses reveal distinct stage-specific phenotypes of the gut microbiota in colorectal cancer. Nat Med. (2019) 25:968–76. doi: 10.1038/s41591-019-0458-7
44. Saffarian A, Mulet C, Regnault B, Amiot A, Tran-Van-Nhieu J, Ravel J, et al. Crypt- and mucosa-associated core microbiotas in humans and their alteration in colon cancer patients. Mbio. (2019) 10. doi: 10.1128/mBio.01315-19
45. Flemer B, Lynch DB, Brown JM, Jeffery IB, Ryan FJ, Claesson MJ, et al. Tumour-associated and non-tumour-associated microbiota in colorectal cancer. Gut. (2017) 66:633–43. doi: 10.1136/gutjnl-2015-309595
46. Liu W, Zhang X, Xu H, Li S, Lau HC, Chen Q, et al. Microbial community heterogeneity within colorectal neoplasia and its correlation with colorectal carcinogenesis. Gastroenterology. (2021) 160:2395–408. doi: 10.1053/j.gastro.2021.02.020
47. Lin Y, Lau HC, Liu Y, Kang X, Wang Y, Ting NL, et al. Altered mycobiota signatures and enriched pathogenic aspergillus rambellii are associated with colorectal cancer based on multicohort fecal metagenomic analyses. Gastroenterology. (2022) 163:908–21. doi: 10.1053/j.gastro.2022.06.038
48. Qin X, Gu Y, Liu T, Wang C, Zhong W, Wang B, et al. Gut mycobiome: A promising target for colorectal cancer. Biochim Biophys Acta Rev Cancer. (2021) 1875:188489. doi: 10.1016/j.bbcan.2020.188489
49. Richard ML, Sokol H. The gut mycobiota: insights into analysis, environmental interactions and role in gastrointestinal diseases. Nat Rev Gastroenterol Hepatol. (2019) 16:331–45. doi: 10.1038/s41575-019-0121-2
50. Wang T, Fan C, Yao A, Xu X, Zheng G, You Y, et al. The adaptor protein CARD9 protects against colon cancer by restricting mycobiota-mediated expansion of myeloid-derived suppressor cells. Immunity. (2018) 49:504–14. doi: 10.1016/j.immuni.2018.08.018
51. Sears CL, Pardoll DM. Perspective: alpha-bugs, their microbial partners, and the link to colon cancer. J Infect Dis. (2011) 203:306–11. doi: 10.1093/jinfdis/jiq061
52. Tjalsma H, Boleij A, Marchesi JR, Dutilh BE. A bacterial driver-passenger model for colorectal cancer: beyond the usual suspects. Nat Rev Microbiol. (2012) 10:575–82. doi: 10.1038/nrmicro2819
53. Avril M, Depaolo RW. "Driver-passenger" bacteria and their metabolites in the pathogenesis of colorectal cancer. Gut Microbes. (2021) 13:1941710. doi: 10.1080/19490976.2021.1941710
54. Pandey H, Tang DWT, Wong SH, Lal D. Gut microbiota in colorectal cancer: biological role and therapeutic opportunities. Cancers (Basel). (2023) 15. doi: 10.3390/cancers15030866
55. Zhang Y, Zhang L, Zheng S, Li M, Xu C, Jia D, et al. Fusobacterium nucleatum promotes colorectal cancer cells adhesion to endothelial cells and facilitates extravasation and metastasis by inducing ALPK1/NF-kappaB/ICAM1 axis. Gut Microbes. (2022) 14:2038852. doi: 10.1080/19490976.2022.2038852
56. Lu X, Xu Q, Tong Y, Zhang Z, Dun G, Feng Y, et al. Long non-coding RNA EVADR induced by Fusobacterium nucleatum infection promotes colorectal cancer metastasis. Cell Rep. (2022) 40:111127. doi: 10.1016/j.celrep.2022.111127
57. Chen S, Su T, Zhang Y, Lee A, He J, Ge Q, et al. Fusobacterium nucleatum promotes colorectal cancer metastasis by modulating KRT7-AS/KRT7. Gut Microbes. (2020) 11:511–25. doi: 10.1080/19490976.2019.1695494
58. Xu C, Fan L, Lin Y, Shen W, Qi Y, Zhang Y, et al. Fusobacterium nucleatum promotes colorectal cancer metastasis through miR-1322/CCL20 axis and M2 polarization. Gut Microbes. (2021) 13:1980347. doi: 10.1080/19490976.2021.1980347
59. Chen Y, Chen Y, Zhang J, Cao P, Su W, Deng Y, et al. Fusobacterium nucleatum Promotes Metastasis in Colorectal Cancer by Activating Autophagy Signaling via the Upregulation of CARD3 Expression. Theranostics. (2020) 10:323–39. doi: 10.7150/thno.38870
60. Si H, Yang Q, Hu H, Ding C, Wang H, Lin X. Colorectal cancer occurrence and treatment based on changes in intestinal flora. Semin Cancer Biol. (2021) 70:3–10. doi: 10.1016/j.semcancer.2020.05.004
61. Zamani S, Taslimi R, Sarabi A, Jasemi S, Sechi LA, Feizabadi MM. Enterotoxigenic bacteroides fragilis: A possible etiological candidate for bacterially-induced colorectal precancerous and cancerous lesions. Front Cell Infect Microbiol. (2019) 9:449. doi: 10.3389/fcimb.2019.00449
62. Wang T, Cai G, Qiu Y, Fei N, Zhang M, Pang X, et al. Structural segregation of gut microbiota between colorectal cancer patients and healthy volunteers. ISME J. (2012) 6:320–9. doi: 10.1038/ismej.2011.109
63. Arima K, Zhong R, Ugai T, Zhao M, Haruki K, Akimoto N, et al. Western-Style Diet, pks Island-Carrying Escherichia coli, and Colorectal Cancer: Analyses From Two Large Prospective Cohort Studies. Gastroenterology. (2022) 163:862–74. doi: 10.1053/j.gastro.2022.06.054
64. Butt J, Jenab M, Willhauck-Fleckenstein M, Michel A, Pawlita M, Kyrø C, et al. Prospective evaluation of antibody response to Streptococcus gallolyticus and risk of colorectal cancer. Int J Cancer. (2018) 143:245–52. doi: 10.1002/ijc.31283
65. Pasquereau-Kotula E, Martins M, Aymeric L, Dramsi S. Significance of Streptococcus gallolyticus subsp. gallolyticus Association With Colorectal Cancer. Front Microbiol. (2018) 9:614. doi: 10.3389/fmicb.2018.00614
66. Corredoira J, Grau I, Garcia-Rodriguez JF, Alonso-Garcia P, Garcia-Pais MJ, Rabuñal R, et al. The clinical epidemiology and Malignancies associated with Streptococcus bovis biotypes in 506 cases of bloodstream infections. J Infect. (2015) 71:317–25. doi: 10.1016/j.jinf.2015.05.005
67. Andres-Franch M, Galiana A, Sanchez-Hellin V, Ochoa E, Hernandez-Illan E, Lopez-Garcia P, et al. Streptococcus gallolyticus infection in colorectal cancer and association with biological and clinical factors. PloS One. (2017) 12:e0174305. doi: 10.1371/journal.pone.0174305
68. Ito M, Kanno S, Nosho K, Sukawa Y, Mitsuhashi K, Kurihara H, et al. Association of Fusobacterium nucleatum with clinical and molecular features in colorectal serrated pathway. Int J Cancer. (2015) 137:1258–68. doi: 10.1002/ijc.29488
69. De Carvalho AC, De Mattos Pereira L, Datorre JG, Dos Santos W, Berardinelli GN, Matsushita MM, et al. Microbiota profile and impact of fusobacterium nucleatum in colorectal cancer patients of barretos cancer hospital. Front Oncol. (2019) 9:813. doi: 10.3389/fonc.2019.00813
70. Yeoh YK, Chen Z, Wong MCS, Hui M, Yu J, Ng SC, et al. Southern Chinese populations harbour non-nucleatum Fusobacteria possessing homologues of the colorectal cancer-associated FadA virulence factor. Gut. (2020) 69:1998–2007. doi: 10.1136/gutjnl-2019-319635
71. Brennan CA, Garrett WS. Fusobacterium nucleatum - symbiont, opportunist and oncobacterium. Nat Rev Microbiol. (2019) 17:156–66. doi: 10.1038/s41579-018-0129-6
72. Yang Y, Weng W, Peng J, Hong L, Yang L, Toiyama Y, et al. Fusobacterium nucleatum increases proliferation of colorectal cancer cells and tumor development in mice by activating toll-like receptor 4 signaling to nuclear factor-kappaB, and up-regulating expression of microRNA-21. Gastroenterology. (2017) 152:851–866 e24. doi: 10.1053/j.gastro.2016.11.018
73. Li R, Shen J, Xu Y. Fusobacterium nucleatum and colorectal cancer. Infect Drug Resist. (2022) 15:1115–20. doi: 10.2147/IDR.S357922
74. Wang N, Fang JY. Fusobacterium nucleatum, a key pathogenic factor and microbial biomarker for colorectal cancer. Trends Microbiol. (2023) 31:159–72. doi: 10.1016/j.tim.2022.08.010
75. Rubinstein MR, Baik JE, Lagana SM, Han RP, Raab WJ, Sahoo D, et al. Fusobacterium nucleatum promotes colorectal cancer by inducing Wnt/beta-catenin modulator Annexin A1. EMBO Rep. (2019) 20. doi: 10.15252/embr.201847638
76. Li X, Huang J, Yu T, Fang X, Lou L, Xin S, et al. Fusobacterium nucleatum promotes the progression of colorectal cancer through cdk5-activated wnt/beta-catenin signaling. Front Microbiol. (2020) 11:545251. doi: 10.3389/fmicb.2020.545251
77. Hamada T, Zhang X, Mima K, Bullman S, Sukawa Y, Nowak JA, et al. Fusobacterium nucleatum in colorectal cancer relates to immune response differentially by tumor microsatellite instability status. Cancer Immunol Res. (2018) 6:1327–36. doi: 10.1158/2326-6066.CIR-18-0174
78. Liu Y, Baba Y, Ishimoto T, Tsutsuki H, Zhang T, Nomoto D, et al. Fusobacterium nucleatum confers chemoresistance by modulating autophagy in oesophageal squamous cell carcinoma. Br J Cancer. (2021) 124:963–74. doi: 10.1038/s41416-020-01198-5
79. Yu T, Guo F, Yu Y, Sun T, Ma D, Han J, et al. Fusobacterium nucleatum promotes chemoresistance to colorectal cancer by modulating autophagy. Cell. (2017) 170:548–563 e16. doi: 10.1016/j.cell.2017.07.008
80. Yamaoka Y, Suehiro Y, Hashimoto S, Hoshida T, Fujimoto M, Watanabe M, et al. Fusobacterium nucleatum as a prognostic marker of colorectal cancer in a Japanese population. J Gastroenterol. (2018) 53:517–24. doi: 10.1007/s00535-017-1382-6
81. Peng BJ, Cao CY, Li W, Zhou YJ, Zhang Y, Nie YQ, et al. Diagnostic performance of intestinal fusobacterium nucleatum in colorectal cancer: A meta-analysis. Chin Med J (Engl). (2018) 131:1349–56. doi: 10.4103/0366-6999.232814
82. Sears CL. Enterotoxigenic Bacteroides fragilis: a rogue among symbiotes. Clin Microbiol Rev. (2009) 22:349–69. doi: 10.1128/CMR.00053-08
83. Hwang S, Lee CG, Jo M, Park CO, Gwon SY, Hwang S, et al. Enterotoxigenic Bacteroides fragilis infection exacerbates tumorigenesis in AOM/DSS mouse model. Int J Med Sci. (2020) 17:145–52. doi: 10.7150/ijms.38371
84. Geis AL, Fan H, Wu X, Wu S, Huso DL, Wolfe JL, et al. Regulatory T-cell response to enterotoxigenic bacteroides fragilis colonization triggers IL17-dependent colon carcinogenesis. Cancer Discovery. (2015) 5:1098–109. doi: 10.1158/2159-8290.CD-15-0447
85. Liu QQ, Li CM, Fu LN, Wang HL, Tan J, Wang YQ, et al. Enterotoxigenic Bacteroides fragilis induces the stemness in colorectal cancer via upregulating histone demethylase JMJD2B. Gut Microbes. (2020) 12:1788900. doi: 10.1080/19490976.2020.1788900
86. Cao Y, Wang Z, Yan Y, Ji L, He J, Xuan B, et al. Enterotoxigenic bacteroidesfragilis promotes intestinal inflammation and Malignancy by inhibiting exosome-packaged miR-149-3p. Gastroenterology. (2021) 161:1552–1566 e12. doi: 10.1053/j.gastro.2021.08.003
87. Kaper JB, Nataro JP, Mobley HL. Pathogenic escherichia coli. Nat Rev Microbiol. (2004) 2:123–40. doi: 10.1038/nrmicro818
88. Iyadorai T, Mariappan V, Vellasamy KM, Wanyiri JW, Roslani AC, Lee GK, et al. Prevalence and association of pks+ Escherichia coli with colorectal cancer in patients at the University Malaya Medical Centre, Malaysia. PloS One. (2020) 15:e0228217. doi: 10.1371/journal.pone.0228217
89. Dejea CM, Fathi P, Craig JM, Boleij A, Taddese R, Geis AL, et al. Patients with familial adenomatous polyposis harbor colonic biofilms containing tumorigenic bacteria. Science. (2018) 359:592–7. doi: 10.1126/science.aah3648
90. Bonnet M, Buc E, Sauvanet P, Darcha C, Dubois D, Pereira B, et al. Colonization of the human gut by E. coli and colorectal cancer risk. . Clin Cancer Res. (2014) 20:859–67. doi: 10.1158/1078-0432.CCR-13-1343
91. Pleguezuelos-Manzano C, Puschhof J, Rosendahl Huber A, van Hoeck A, Wood HM, Nomburg J, et al. Mutational signature in colorectal cancer caused by genotoxic pks(+) E. coli. Nat. (2020) 580:269–73. doi: 10.1038/s41586-020-2080-8
92. Wassenaar TME. coli and colorectal cancer: a complex relationship that deserves a critical mindset. Crit Rev Microbiol. (2018) 44:619–32. doi: 10.1080/1040841X.2018.1481013
93. Piciocchi A, Germinario E, Garcia Etxebarria K, Rossi S, Sanchez-Mete L, Porowska B, et al. Association of polygenic risk score and bacterial toxins at screening colonoscopy with colorectal cancer progression: A multicenter case-control study. Toxins (Basel). (2021) 13. doi: 10.3390/toxins13080569
94. Wang Y, Fu K. Genotoxins: the mechanistic links between escherichia coli and colorectal cancer. Cancers (Basel). (2023) 15. doi: 10.3390/cancers15041152
95. Salesse L, Lucas C, Hoang MHT, Sauvanet P, Rezard A, Rosenstiel P, et al. Colibactin-producing escherichia coli induce the formation of invasive carcinomas in a chronic inflammation-associated mouse model. Cancers (Basel). (2021) 13. doi: 10.3390/cancers13092060
96. Lopes A, Billard E, Casse AH, Villéger R, Veziant J, Roche G, et al. Colibactin-positive Escherichia coli induce a procarcinogenic immune environment leading to immunotherapy resistance in colorectal cancer. Int J Cancer. (2020) 146:3147–59. doi: 10.1002/ijc.32920
97. Corredoira J, Alonso MP, Coira A, Casariego E, Arias C, Alonso D, et al. Characteristics of Streptococcus bovis endocarditis and its differences with Streptococcus viridans endocarditis. Eur J Clin Microbiol Infect Dis. (2008) 27:285–91. doi: 10.1007/s10096-007-0441-y
98. Jans C, Meile L, Lacroix C, Stevens MJ. Genomics, evolution, and molecular epidemiology of the Streptococcus bovis/Streptococcus equinus complex (SBSEC). Infect Genet Evol. (2015) 33:419–36. doi: 10.1016/j.meegid.2014.09.017
99. Aymeric L, Donnadieu F, Mulet C, du Merle L, Nigro G, Saffarian A, et al. Colorectal cancer specific conditions promote Streptococcus gallolyticus gut colonization. Proc Natl Acad Sci U.S.A. (2018) 115:E283–91. doi: 10.1073/pnas.1715112115
100. Taylor JC, Gao X, Xu J, Holder M, Petrosino J, Kumar R, et al. A type VII secretion system of Streptococcus gallolyticus subsp. gallolyticus contributes to gut colonization and the development of colon tumors. PloS Pathog. (2021) 17:e1009182. doi: 10.1371/journal.ppat.1009182
101. Deng Q, Wang C, Yu K, Wang Y, Yang Q, Zhang J, et al. Streptococcus bovis Contributes to the Development of Colorectal Cancer via Recruiting CD11b(+)TLR-4(+) Cells. Med Sci Monit. (2020) 26:e921886. doi: 10.12659/MSM.921886
102. Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2021. CA Cancer J Clin. (2021) 71:7–33. doi: 10.3322/caac.21654
103. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. (2019) 69:7–34. doi: 10.3322/caac.21551
104. Kim SY, Kim HS, Park HJ. Adverse events related to colonoscopy: Global trends and future challenges. World J Gastroenterol. (2019) 25:190–204. doi: 10.3748/wjg.v25.i2.190
105. Imperiale TF, Gruber RN, Stump TE, Emmett TW, Monahan PO. Performance characteristics of fecal immunochemical tests for colorectal cancer and advanced adenomatous polyps: A systematic review and meta-analysis. Ann Intern Med. (2019) 170:319–29. doi: 10.7326/M18-2390
106. Liu NN, Jiao N, Tan JC, Wang Z, Wu D, Wang AJ, et al. Multi-kingdom microbiota analyses identify bacterial-fungal interactions and biomarkers of colorectal cancer across cohorts. Nat Microbiol. (2022) 7:238–50. doi: 10.1038/s41564-021-01030-7
107. Kisiel JB, Limburg PJ. Colorectal cancer screening with the multitarget stool DNA test. Am J Gastroenterol. (2020) 115:1737–40. doi: 10.14309/ajg.0000000000000968
108. Amitay EL, Krilaviciute A, Brenner H. Systematic review: Gut microbiota in fecal samples and detection of colorectal neoplasms. Gut Microbes. (2018) 9:293–307. doi: 10.1080/19490976.2018.1445957
109. Thomas AM, Manghi P, Asnicar F, Pasolli E, Armanini F, Zolfo M, et al. Metagenomic analysis of colorectal cancer datasets identifies cross-cohort microbial diagnostic signatures and a link with choline degradation. Nat Med. (2019) 25:667–78. doi: 10.1038/s41591-019-0405-7
110. Shah MS, Desantis TZ, Weinmaier T, et al. Leveraging sequence-based faecal microbial community survey data to identify a composite biomarker for colorectal cancer. Gut. (2018) 67:882–91. doi: 10.1136/gutjnl-2016-313189
111. Yu J, Feng Q, Wong SH, Zhang D, Liang QY, Qin Y, et al. Metagenomic analysis of faecal microbiome as a tool towards targeted non-invasive biomarkers for colorectal cancer. Gut. (2017) 66:70–8. doi: 10.1136/gutjnl-2015-309800
112. Osman MA, Neoh HM, Ab Mutalib NS, Chin SF, Mazlan L, Raja Ali RA, et al. Parvimonas micra, Peptostreptococcus stomatis, Fusobacterium nucleatum and Akkermansia muciniphila as a four-bacteria biomarker panel of colorectal cancer. Sci Rep. (2021) 11:2925. doi: 10.1038/s41598-021-82465-0
113. Liu K, Yang X, Zeng M, Yuan Y, Sun J, He P, et al. The Role of Fecal Fusobacterium nucleatum and pks(+) Escherichia coli as Early Diagnostic Markers of Colorectal Cancer. Dis Markers. (2021) 2021:1171239. doi: 10.1155/2021/1171239
114. Eklof V, Lofgren-Burstrom A, Zingmark C, Edin S, Larsson P, Karling P, et al. Cancer-associated fecal microbial markers in colorectal cancer detection. Int J Cancer. (2017) 141:2528–36. doi: 10.1002/ijc.31011
115. Liang Q, Chiu J, Chen Y, Huang Y, Higashimori A, Fang J, et al. Fecal bacteria act as novel biomarkers for noninvasive diagnosis of colorectal cancer. Clin Cancer Res. (2017) 23:2061–70. doi: 10.1158/1078-0432.CCR-16-1599
116. Wong SH, Kwong TNY, Chow TC, Luk AKC, Dai RZW, Nakatsu G, et al. Quantitation of faecal Fusobacterium improves faecal immunochemical test in detecting advanced colorectal neoplasia. Gut. (2017) 66:1441–8. doi: 10.1136/gutjnl-2016-312766
117. Guo S, Li L, Xu B, Li M, Zeng Q, Xiao H, et al. A simple and novel fecal biomarker for colorectal cancer: ratio of fusobacterium nucleatum to probiotics populations, based on their antagonistic effect. Clin Chem. (2018) 64:1327–37. doi: 10.1373/clinchem.2018.289728
118. Liang JQ, Li T, Nakatsu G, Chen YX, Yau TO, Chu E, et al. A novel faecal Lachnoclostridium marker for the non-invasive diagnosis of colorectal adenoma and cancer. Gut. (2020) 69:1248–57. doi: 10.1136/gutjnl-2019-318532
119. Dai Z, Coker OO, Nakatsu G, Wu WKK, Zhao L, Chen Z, et al. Multi-cohort analysis of colorectal cancer metagenome identified altered bacteria across populations and universal bacterial markers. Microbiome. (2018) 6:70. doi: 10.1186/s40168-018-0451-2
120. Komiya Y, Shimomura Y, Higurashi T, Sugi Y, Arimoto J, Umezawa S, et al. Patients with colorectal cancer have identical strains of Fusobacterium nucleatum in their colorectal cancer and oral cavity. Gut. (2019) 68:1335–7. doi: 10.1136/gutjnl-2018-316661
121. Flemer B, Warren RD, Barrett MP, Cisek K, Das A, Jeffery IB, et al. The oral microbiota in colorectal cancer is distinctive and predictive. Gut. (2018) 67:1454–63. doi: 10.1136/gutjnl-2017-314814
122. Zhang S, Kong C, Yang Y, Cai S, Li X, Cai G, et al. Human oral microbiome dysbiosis as a novel non-invasive biomarker in detection of colorectal cancer. Theranostics. (2020) 10:11595–606. doi: 10.7150/thno.49515
123. Konishi Y, Okumura S, Matsumoto T, Itatani Y, Nishiyama T, Okazaki Y, et al. Development and evaluation of a colorectal cancer screening method using machine learning-based gut microbiota analysis. Cancer Med. (2022) 11:3194–206. doi: 10.1002/cam4.4671
124. Zhang SL, Cheng LS, Zhang ZY, Sun HT, Li JJ. Untangling determinants of gut microbiota and tumor immunologic status through a multi-omics approach in colorectal cancer. Pharmacol Res. (2023) 188:106633. doi: 10.1016/j.phrs.2022.106633
125. Ai L, Tian H, Chen Z, Chen H, Xu J, Fang JY. Systematic evaluation of supervised classifiers for fecal microbiota-based prediction of colorectal cancer. Oncotarget. (2017) 8:9546–56. doi: 10.18632/oncotarget.v8i6
126. Louis P, Hold GL, Flint HJ. The gut microbiota, bacterial metabolites and colorectal cancer. Nat Rev Microbiol. (2014) 12:661–72. doi: 10.1038/nrmicro3344
127. Yang Y, Misra BB, Liang L, Bi D, Weng W, Wu W, et al. Integrated microbiome and metabolome analysis reveals a novel interplay between commensal bacteria and metabolites in colorectal cancer. Theranostics. (2019) 9:4101–14. doi: 10.7150/thno.35186
128. David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. (2014) 505:559–63. doi: 10.1038/nature12820
129. Weir TL, Manter DK, Sheflin AM, Barnett BA, Heuberger AL, Ryan EP, et al. Stool microbiome and metabolome differences between colorectal cancer patients and healthy adults. PloS One. (2013) 8:e70803. doi: 10.1371/journal.pone.0070803
130. Chen F, Dai X, Zhou C-C, Li KX, Zhang YJ, Lou XY, et al. Integrated analysis of the faecal metagenome and serum metabolome reveals the role of gut microbiome-associated metabolites in the detection of colorectal cancer and adenoma. Gut. (2022) 71:1315–25. doi: 10.1136/gutjnl-2020-323476
131. Coker OO, Liu C, Wu WKK, Wong SH, Jia W, Sung JJY, et al. Altered gut metabolites and microbiota interactions are implicated in colorectal carcinogenesis and can be non-invasive diagnostic biomarkers. Microbiome. (2022) 10:35. doi: 10.1186/s40168-021-01208-5
132. Tong Y, Gao H, Qi Q, Liu X, Li J, Gao J, et al. High fat diet, gut microbiome and gastrointestinal cancer. Theranostics. (2021) 11:5889–910. doi: 10.7150/thno.56157
133. Wan Y, Wang F, Yuan J, Li J, Jiang D, Zhang J, et al. Effects of dietary fat on gut microbiota and faecal metabolites, and their relationship with cardiometabolic risk factors: a 6-month randomised controlled-feeding trial. Gut. (2019) 68:1417–29. doi: 10.1136/gutjnl-2018-317609
Keywords: intestinal flora, colorectal cancer, early screening, biomarker, correlation
Citation: Wang X, Zhang Q, Xu R, Li X and Hong Z (2024) Research progress on the correlation between intestinal flora and colorectal cancer. Front. Oncol. 14:1416806. doi: 10.3389/fonc.2024.1416806
Received: 13 April 2024; Accepted: 24 June 2024;
Published: 17 July 2024.
Edited by:
Adil Bhat, University of California, Los Angeles, United StatesReviewed by:
Muhammad Azhar Nisar, Tulane University, United StatesCopyright © 2024 Wang, Zhang, Xu, Li and Hong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaofeng Li, bHhmX2NoZW5AZG11LmVkdS5jbg==; Zhijun Hong, aG9uZ3poaWp1bkBmaXJzdGhvc3AtZG11LmNvbQ==
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.