- 1Scuola Superiore Meridionale (SSM), Clinical and Translational Oncology, Naples, Italy
- 2Department of Medical Oncology, School of Medicine and Surgery, University of Naples Federico II, Naples, Italy
- 3Public Health Department, University of Naples Federico II, Naples, Italy
- 4Urology and Gynecology Department, Istituto Nazionale Tumori IRCCS, Fondazione G. Pascale, Naples, Italy
Objective: Preclinical studies have emphasized the potential connection between BRCA specific domains defects and the activity of Poly ADP-ribose polymerase inhibitors (PARPi). Nevertheless, real-world evidence regarding the impact of BRCA domain defects and mutations on PARPi efficacy are limited. The aim of his study was to evaluate the efficacy of PARPi in terms of progression free survival (PFS) according to BRCA domains defects and mutation types.
Methods: A retrospective analysis was performed among 79 BRCA mutated patients, diagnosed with advanced High-grade serous ovarian carcinoma (HGSOC) who received first- and second-line platinum- based chemotherapy followed by PARPi maintenance treatment. PFS was evaluated according to BRCA1 [Really Interesting Gene (RING), DNA Binding (DBD), Serine Cluster (SCD), BRCA1 C-terminal (BRCT)] and BRCA2 [RAD-51 Domain (RAD-51 BD), DBD] specific domain defects and mutation types [missense (MS), nonsense (NS), frameshift (FS), splicing (S), or large rearrangements (LR)].
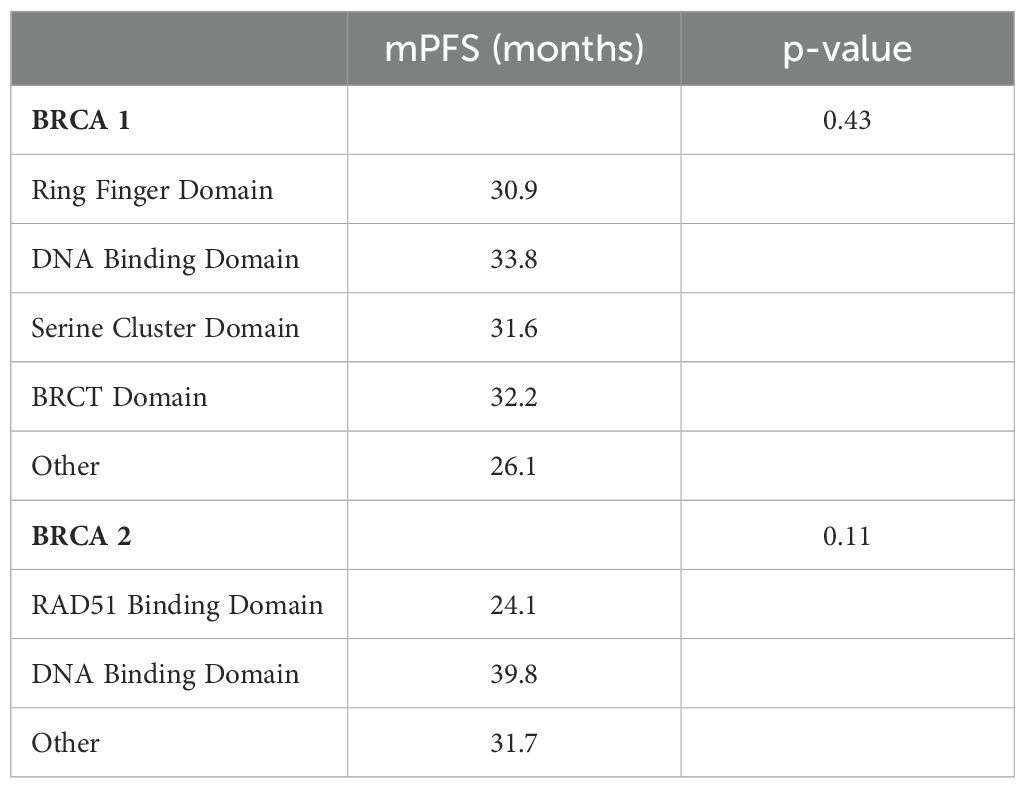
Results: After a median follow-up of 51 months, no significant difference in PFS was observed between the BRCA functional domains or mutation types in the BRCA1 and BRCA2 subgroups. Patients with BRCA2 DBD and RAD51-BD defects had the longest (39.8 months) and shortest (24.1 months) median PFS, respectively (p = 0.11). Additionally, patients with BRCA1 DBD defects had the greatest benefit (median PFS = 33.8 months) while those with BRCA1 RING domain mutations experienced the worst outcome (median PFS = 30.9 months (p = 0.43).
Conclusion: The efficacy of maintenance treatment with PARPi is independent by BRCA domain defects or mutation types. Patients DBD domain defects experienced numerically longer median PFS compared to those with other BRCA1/2 alterations.
Introduction
Ovarian cancer is a significant contributor to global cancer-related deaths, and it is the leading cause of death for gynecological malignancies (1, 2). Although platinum-based chemotherapy combinations are the primary treatment for epithelial ovarian cancer, recent years have seen the introduction of targeted therapies, including humanized monoclonal antibody against vascular endothelial growth factor (VEGF) bevacizumab, inhibitors of poly (ADP-ribose) polymerase (PARPi), and antifolate receptor alpha antibody drug conjugates, expanding the pharmacologic landscape.
High-grade serous ovarian carcinoma (HGSOC) is the most prevalent histological subtype of epithelial ovarian cancer and is associated with mutations in the breast cancer susceptibility genes 1 and 2 (BRCA1 and BRCA2) in approximately 25% of cases (3–6). BRCA1 and BRCA2 are tumor suppressor genes whose mutations have been historically associated with an increased lifetime risk of developing breast cancer (50 -80%) and ovarian cancer (30 - 50%) (7). BRCA1 and BRCA2 proteins are crucial interactors in the Homologous Recombination (HR) DNA double-strand break (DSB) repair system, which ensures genomic stability and DNA integrity during the S and G2 cell cycle phases (8, 9). BRCA1 also plays a role in other DNA repair pathways, including the error-prone non-homologous end-joining (NHEI) DSB repair system and single-strand annealing (SSA) (10). Ovarian cancer cells with mutations in BRCA1, BRCA2, or genes involved in the HR mechanism have an Homologous Recombination Deficiency (HRD) status, which renders them highly dependent on the DNA single-strand break (SSB) repair mechanism to maintain genomic stability. Notably, the DNA SSB recognition and repair process is primarily driven by chromatin - associated proteins, such as poly (ADP-ribose) polymerase, which promote the synthesis of poly (ADP- ribose) chains to recruit DNA repair factors to SSBs and ensure cell survival (11).
The inhibition of poly (ADP-ribose) polymerase (PARP) in BRCA-deficient ovarian cancer cells impairs the DNA damage response and induces cell apoptosis through a phenomenon known as “synthetic lethality.” (12). However, the range of PARP inhibition sensitivity extends from primary resistance to long-lasting responses. Interestingly, the degree of tumor sensitivity to PARP inhibition can be influenced by specific BRCA domain defects. BRCA1, for example, has several functional domains, including the amino terminal RING domain, which promotes BRCA1-BARD1 (BRCA1 Associated RING Domain protein 1) heterodimer formation and its E3 ubiquitin ligase activity (13). The BRCT domain mediates binding to phosphorylated proteins like CtBP-interacting protein (Ctlp) and ABRAXAS, promoting recruitment to DNA damage sites, DNA end resection, and G2/M checkpoint activation (14). The binding domain directly interacts with DNA regions, functioning as both a DNA damage sensor and repair promoter (15). BRCA2 has two functional domains: the RAD51-binding domain, which contains BRC repeats that bind to RAD51 and promote its recruitment to the double-strand break (DSB) to ultimately form RAD5 filaments on single-strand DNA (ssDNA), and the DNA binding domain, which is crucial for mediating BRCA2 interaction with both ssDNA and double-strand DNA (16). Preclinical studies have investigated the potential impact of BRCA-specific domain loss on treatment activity. In genetically engineered mice with two common BRCA1 frameshift mutations (BRCA1185delAG and BRCA15382insC), the loss of the BRCA1 ring domain was associated with resistance to both cisplatin and olaparib (17). Conversely, the expression of a BRCA2 variant lacking the DNA Binding and C-terminal domains resulted in defective BRCA2 architectural rearrangement, leading to increased PARPi sensitivity in murine cell lines (18).
Limited clinical data are available on the impact of BRCA-specific defects on PARPi efficacy. A post-hoc analysis of the PAOLA 1 trial showed that although the benefits of the combination regimen have been observed regardless of BRCA domains, mutations affecting the BRCA2 DNA-binding domain (DBD) are associated with increased sensitivity to platinum salts treatment and improved median Progression Free Survival (mPFS). Conversely, defects in the BRCA1 DBD were correlated to a worse clinical outcome, indicating reduced sensitivity to platinum salts treatment in this patient subgroup (19). Therefore, understanding the pattern of PARPi sensitivity based on BRCA domain defects may help predict the extent of PARP inhibition’s benefits and improve the clinical management of patients with HGSOC.
Methods
Patients’ selection
A retrospective, multicenter study of electronic health records from patients who were referred to the oncology departments of the University Hospital Federico II, IRCCS Fondazione Pascale, and AORN Cardarelli Hospital between April 2016 and September 2023 was undertaken. The study population comprised patients aged 18 years or older with an Eastern Cooperative Oncology Group Performance Status (ECOG PS) of 0-2 who were treated with platinum-based chemotherapy as first- or second-line treatment followed by maintenance therapy with PARPi. Eligible patients had a histological diagnosis of FIGO stage III or IV high-grade serous ovarian cancer (HGSOC) with a germline or somatic BRCA1 or BRCA2 mutation. The study included patients who underwent primary debulking surgery (PDS) or interval debulking surgery (IDS), regardless of the surgical outcome. PARPi treatment could be administered for up to 24 months (olaparib) or 36 months (niraparib) in patients with no evidence of disease (NED) in the first- line setting. In patients with residual disease and in the second-line setting, PARPi was administered until disease progression, death, or unacceptable toxicity. All patients treated with second-line PARPi received platinum-based first-line treatment, followed by a maintenance regimen with bevacizumab.
Genetic characteristics
Patients were included if they had tested positive for pathogenic germline or somatic mutations in BRCA1 or BRCA2 genes. Patients with BRCA1/2 variants of unknown significance (VUS) (n = 8) or those carrying mutations in any other genes involved in the Homologous Recombination Repair (HRR) were excluded from the analysis. The mutation records were classified based on the consultation of the two largest BRCA mutation databases, BRCA Exchange (20) and ClinVar (21), according to mutation type and BRCA protein functional domains. Pathogenic variants were classified as missense (MS), nonsense (NS), frameshift (FS), splicing (S), or large rearrangements (LR). The BRCA1 protein functional domains were defined as the RING Finger Domain (amino acids 8-96), the DNA Binding Domain (DBD, amino acids 452-1092), the Serine Cluster Domain (SCD, amino acids 1280-1524), the BRCA1 C-terminal (BRCT, amino acids 1646-1736 and 1760-1855). The BRCA2 protein functional domains were defined as the RAD51-Binding Domain (RAD-51 BD, amino acids 900-2000) and the DNA Binding Domain (DBD, amino acids 2459-3190). Any other mutation not involving those reported amino acid regions for BRCA1 or BRCA2 was classified as “Other.”
Statistical analysis
Recorded data were processed using Jamovi (22–24) software, version 2.3.28. The follow-up time was determined from the date of diagnosis to October 2023. Progression-free survival (PFS) was measured from the date of diagnosis to the time of disease progression. If disease progression did not occur, patients were censored at the date of their last follow-up. PFS was analyzed and visualized using the Kaplan–Meier method, and the comparison was made using the log-rank test. The Cox-regression model was used to evaluate hazard ratios (HR), which were expressed in 95% confidence intervals (95% CI). A significance level of 5% (p ≤ 0.05) was considered statistically significant. The study was approved by the Research Ethics Committee, and patients provided informed consent for the use of their clinical data and tumor samples (BiOnCam protocol).
Results
Cohort characteristics
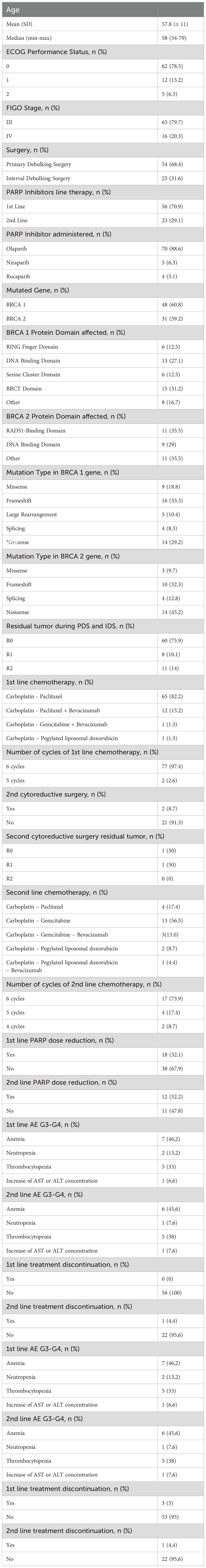
The baseline demographic and clinical characteristics of the 79 consecutive patients included in this study are presented in Table 1. The median age at diagnosis was 58 years, with a range of 34 to 79 years. Sixty- two (78%) patients were diagnosed with stage III HGSOC, while 17 (22%) had stage IV HGSOC. Of these patients, 54 (68%) underwent PDS, and 25 (32%) received IDS. Fifty-six (71%) patients received first-line platinum-based chemotherapy, followed by maintenance therapy with PARPi. Twenty-three (29%) patients received platinum-based chemotherapy as first-line treatment, followed by maintenance therapy with intravenous bevacizumab. All these patients experienced a platinum-sensitive disease recurrence or progression and received a second-line platinum-based chemotherapy followed by PARPi.
Distribution of mutations in BRCA1 and BRCA2
Figure 1 and Table 1 summarize the BRCA1 and BRCA2 mutation type and location. BRCA1 and BRCA2 mutations were detected in 48 (61%) and 31 (39%) patients, respectively. The BRCT (15 cases, 31%) and the DBD (13 cases, 27%) were the most common mutated domains in BRCA1-mutated HGSOC. Among patients with BRCA2-mutated HGSOC, 11 (35%) harbored a mutation in RAD51-BD, and 9 (29%) had a mutation in the DBD. Notably, a substantial number of mutations occurred outside the identified functional domains for BRCA1 (8 cases, 17%) and BRCA2 (11 cases, 35%) (Figure 1A). According to mutation type, FS (16 cases, 33%) were the most common alterations for BRCA1, and NS (14 cases, 45%) were the most frequent for BRCA2 (Figure 1B).
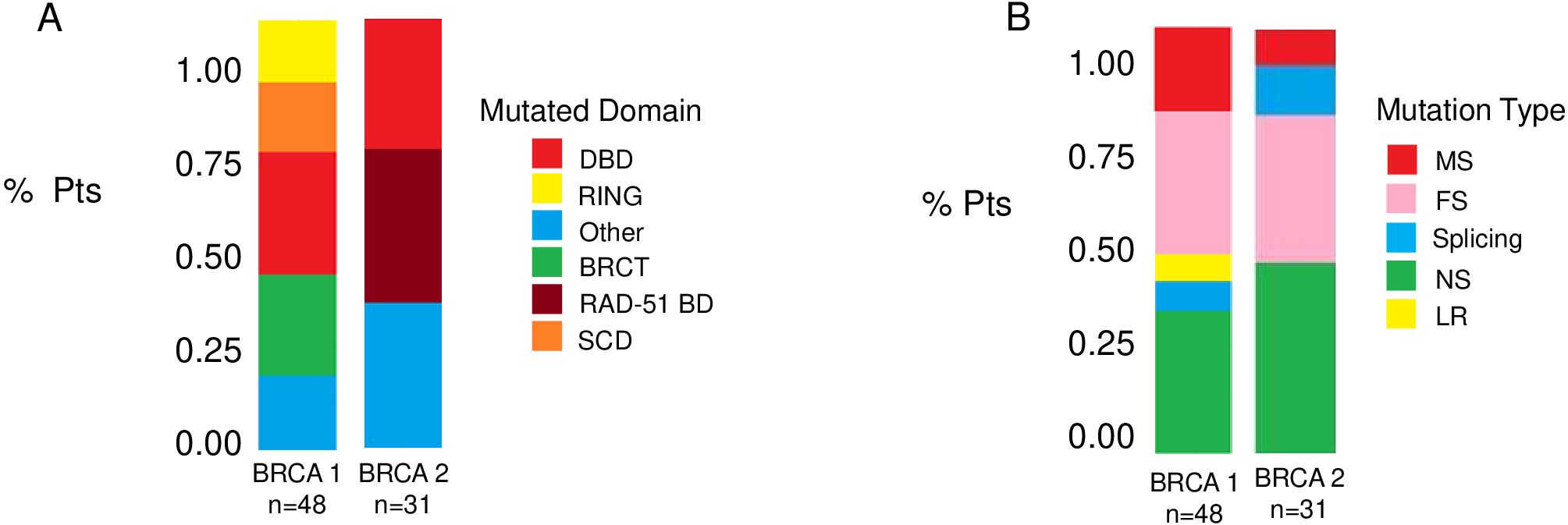
Figure 1. Distribution of BRCA1 and BRCA2 mutations. Bar plots show the frequencies of BRCA mutations according to BRCA specific domain defects (A) and to mutation type (B). BRCT, C-terminal domain of BRCA1; DBD, DNA-binding domain; RING, Really Interesting New Gene; RAD51 BD, RAD51-binding domain; SCD, serine cluster domain; MS, missense; FS, frameshift; MS, missense; LR, large rearrangement.
Progression-free survival
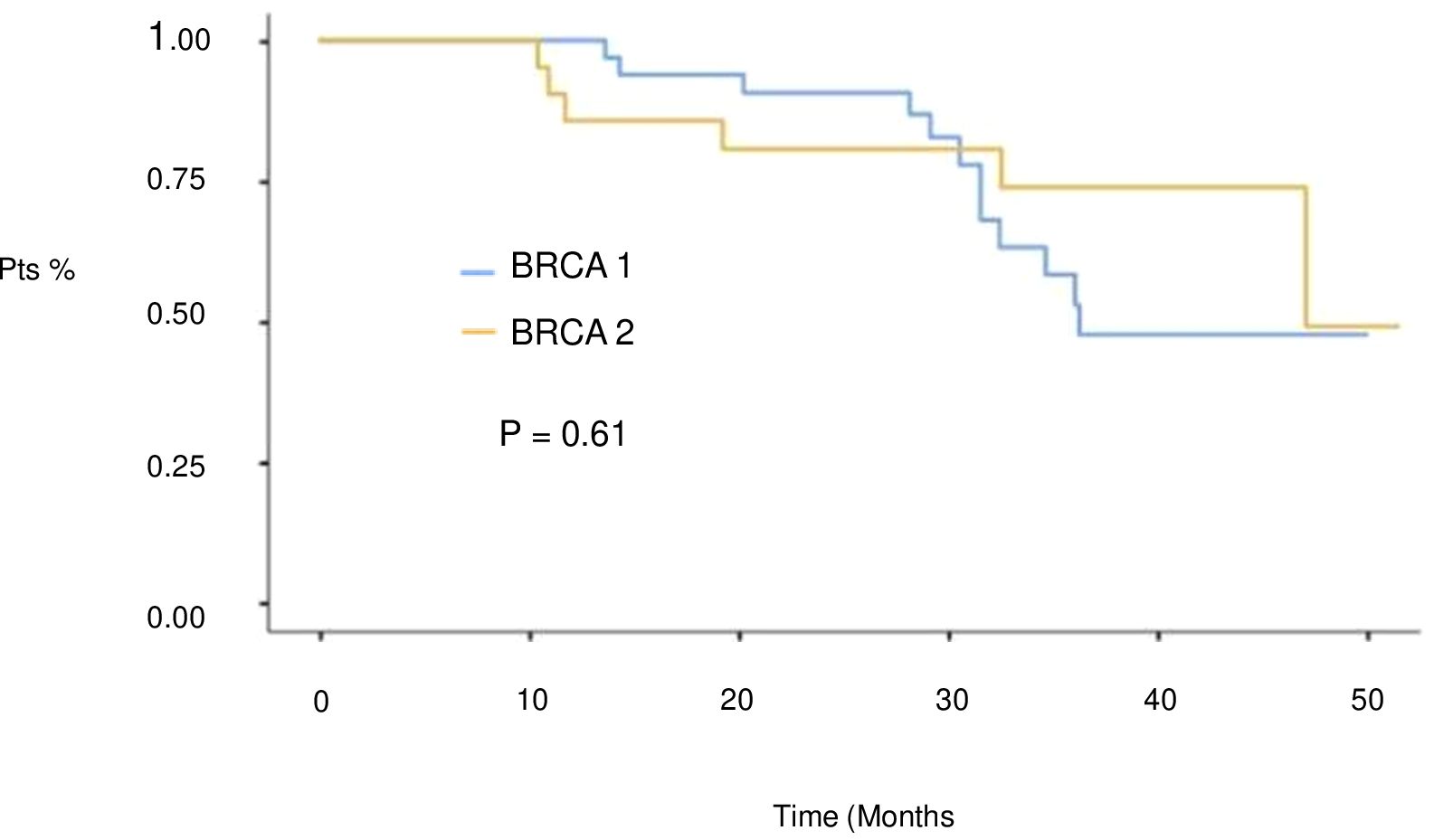
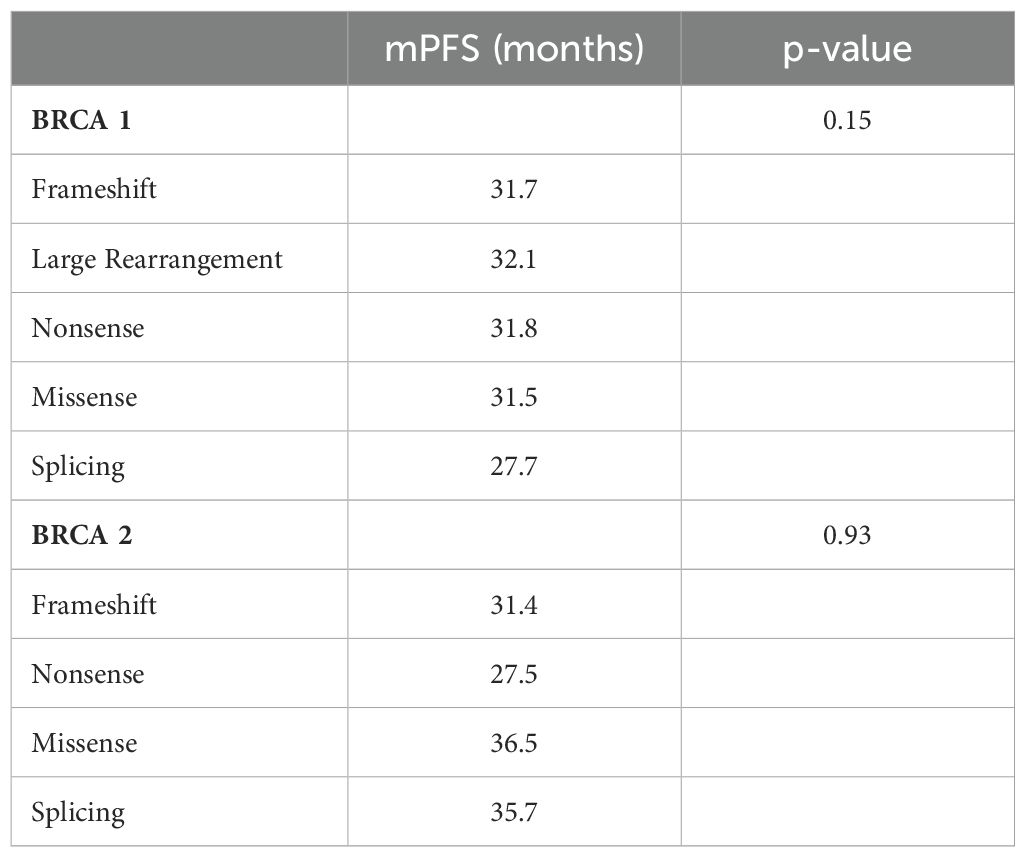
At a median follow-up of 51 months (IQR 17,4 months) no statistically significant difference in median PFS (mPFS) was observed between patients with BRCA1 or BRCA2 mutations who received first line platinum-based chemotherapy and PARPi (31.3 vs 30.1 months, respectively; hazard ration [HR] 0.77, 95% CI 0.29-2.07, p-value = 0.61) (Figure 2). Among patients with BRCA1-mutated HGSOC, those with defects in the DBD had the longest mPFS (33.8 months), while those with mutations in the Ring Finger Domain had the worst outcome (30.9 months). However, the magnitude of PARPi’s benefit across groups was not statistically significant (p-value = 0.43) (Figure 3A; Table 2A). In the BRCA2 cohort, patients with DBD mutations achieved a numerically longer, but not statistically significant, mPFS compared with those with mutations in RAD51-BD (39.8 months vs 24.1 months, p-value = 0.11) (Figure 3B; Table 2A). Additionally, PFS was analyzed according to the type of BRCA1/2 mutations. Among BRCA1 patients, those with LR mutations had the longest mPFS (33.2 months), while those with splicing mutations had the worst outcome (27.1 months; p-value = 0.15) (Figure 4A, Table 2B). Conversely, among BRCA2 patients, mPFS was longer in those with mutations in the MS domain (36.5 months), although no statistically significant correlation was observed (p-value = 0.93) (Figure 4B, Table 2B). A separate descriptive analysis was performed for patients treated with PARPi in the second-line setting. Results showed that patients with mutations in the BRCA1 Ring Finger Domain and BRCA2 DBD had the longest mPFS compared with other mutated protein domains. Furthermore, patients with FS mutations in BRCA1 and NS mutations in BRCA2 had improved outcomes based on mutation type classification. These findings were summarized in Supplementary Tables 1A, B.
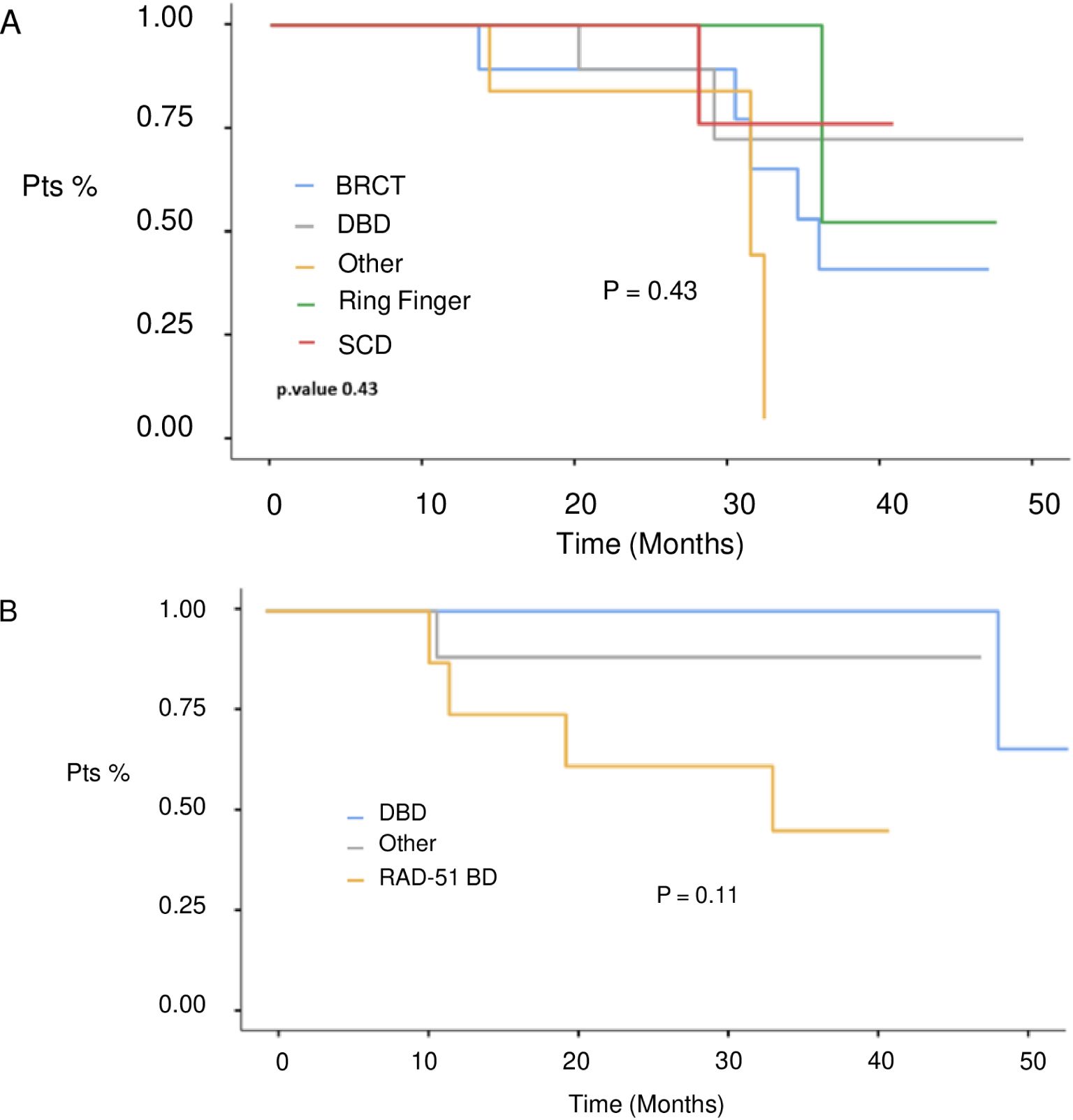
Figure 3. PFS in 1st line setting according to BRCA1 (A) and BRCA 2 (B) protein domains defects. BRCT, C-terminal domain of BRCA1; DBD, DNA-binding domain; RING, Really Interesting New Gene; RAD51 BD, RAD51-binding domain; SCD, serine cluster domain.
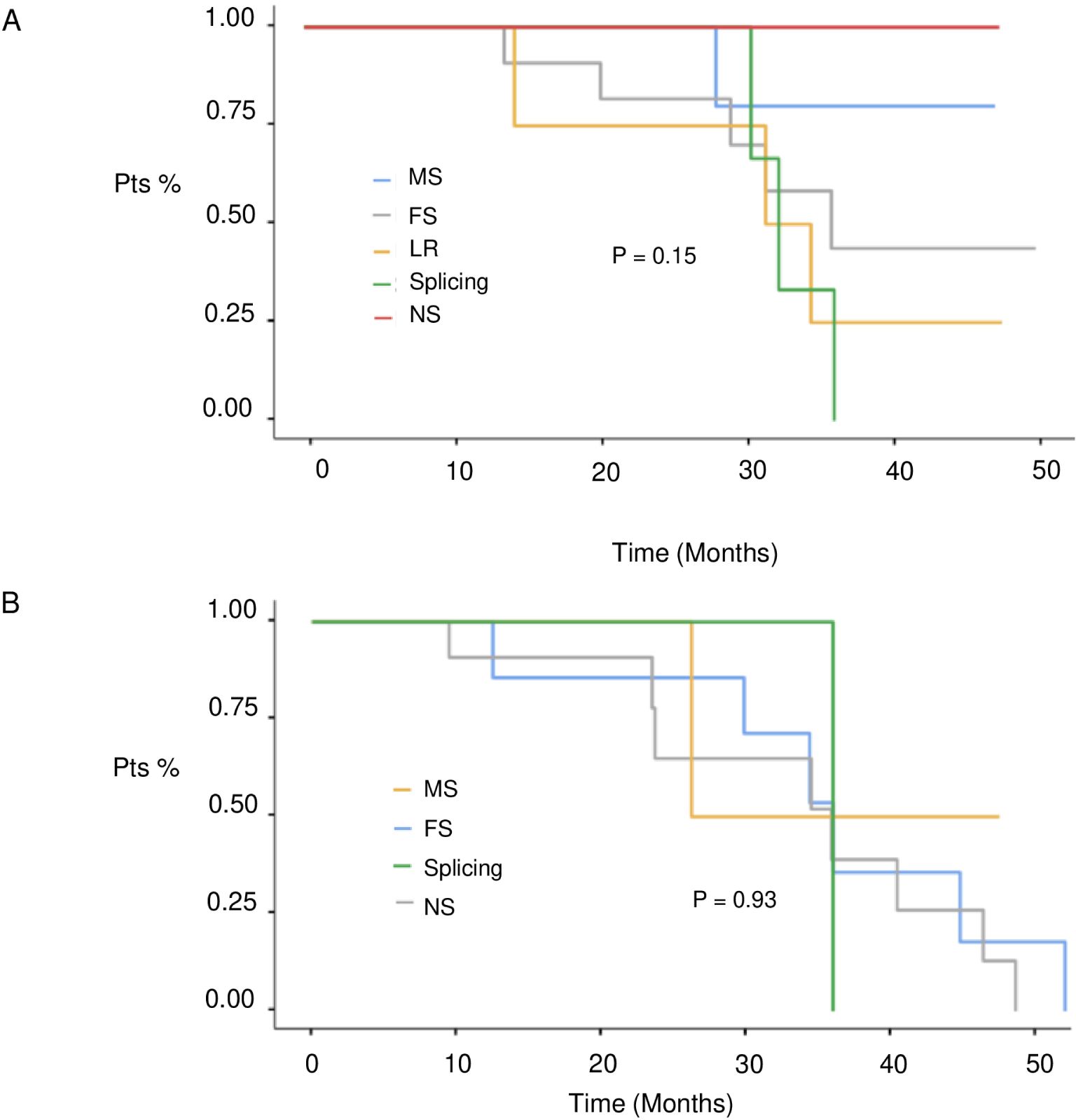
Figure 4. PFS in 1st line setting according to BRCA1 (A) and BRCA 2 (B) mutation type. MS, missense; FS, frameshift; MS, missense; LR, large rearrangement.
Discussion
PARP inhibitors have gained a prominent position in the ovarian cancer treatment algorithm and have become a complementary tool to platinum agents. However, although PARP inhibition in the OC maintenance regimen has shown remarkable results, it is important to consider several prognostic and predictive factors, such as BRCA status, Homologous Recombination Deficiency, and residual disease, as they may affect the benefit observed from PARP inhibition. In our retrospective study, we aimed to explorethe potential impact of BRCA domain defects and type of BRCA mutation on PARP inhibitor efficacy. Our analysis showed that the benefit in terms of PFS from PARP inhibitors was observed regardless of the type and location of BRCA1/2 mutations. Our results align with recent findings from a post-hoc analysis of the phase III PAOLA-1 trial, which evaluated the effectiveness of adding olaparib or placebo to bevacizumab as first-line maintenance therapy in patients newly diagnosed with HGSOC and Homologous Recombination Deficiency. Among patients with a BRCA mutation, the combination of olaparib and bevacizumab was found to be highly effective, regardless of specific domain defects or the type of mutations (19). Although not statistically significant, our study also revealed a varying degree of benefit according to specific mutations. Specifically, among patients with BRCA1 mutations, those with defects in the DBD achieved the highest PFS, while those with Ring Finger Domain mutations had the worst outcomes. These results align with previous preclinical and clinical evidence (19), which has suggested that RING domain-deficient BRCA1 may promote resistance to PARP inhibitors and platinum therapy in breast cancer cell lines harboring a hemizygous BRCA1 185delAG mutation (25). In particular, while initially sensitive to both PARPi and cisplatin, resistant clones emerged from native cells because of the increased expression of a RING domain deficient BRCA1 protein. Notably, this hypomorphic protein does not rely on the interaction with BAR D 1 for its activity, independently promoting RAD51 foci formation, DNA repair, and PARPi/platinum resistance (25). The potential impact of RING domain loss on PARPi activity has been further demonstrated in preclinical mouse models. In breast cancer chimeric mice carrying BRCA1185delAG and BRCA15382insC mutations, the activity of HRD targeted therapies, such as platinum agents and PARP inhibitors, has been evaluated. Intriguingly, mice expressing a BRCA1185delAG mutation, resulting in RING-less BRCA1 protein expression, developed PARPi resistance more rapidly than those harboring a BRCA15382insC mutation, suggesting that RING domain loss may serve as a potential marker of poor response to PARP inhibition (17). Conversely, consistent with our results, patients with defects in DBD enrolled in the PAOLA-1 trial were extremely sensitive to the olaparib plus bevacizumab combination (HR = 0.08; 95%CI [0.02, 0.28]) (19). In contrast, those randomized to the placebo group exhibited a poor response with a median PFS of 16 months compared to 19.9 months observed in patients with BRCT domains. Notably, the lack of benefit from platinum-based chemotherapy, which counteracts the impressive response to olaparib plus bevacizumab, may, at least in part, be related to the incomplete overlap between platinum and PARP resistance in these patients (19). Additionally, in the BRCA2 subgroup, patients harboring mutations in DBD achieved a median PFS longer than those displaying mutations in RAD51-BD. These results appear to be in line with preclinical evidence demonstrating a significant impact of DBD loss on cell survival and PARP inhibition (19). Specifically, clonogenic survival analyses upon treatment with DNA targeting agents such as olaparib, cisplatin, ionizing radiation, and mitomycin were conducted using mouse embryonic stem cells characterized by full length BRCA2 gene or variants lacking DBD (ΔDBD), CTD (ΔCTD), DBD and CTD (ΔDBDΔCTD). Intriguingly, DBD loss led to heightened sensitivity to ionizing radiation and olaparib, regardless of CTD expression. Unexpectedly, DBD loss did not hinder the formation of RAD51 foci, BRCA2 diffusion, or immobilization. On the other hand, in the FACS-based BRCA2-targeting assay, BRCA domain defects prevented HR activation and affected BRCA2 architectural plasticity and its interaction with binding partners (18). Additionally, the DBD region has been reported to be less affected by the reversal of pathogenic BRCA mutations, in contrast to the high prevalence of those described in the N-terminal domain. It is worth mentioning that pathogenic BRCA mutations located in the DBD appear to be less prone to effective reversal due to the surrounding highly conserved sequences that play a central role in the HR (26).
Our study is one of the first real-world experiences to describe the potential impact of BRCA domain defects on PARP activity. Similar results have been reported by P. Torres-Mozas et al. in a retrospective study involving 26 HGSOC patients treated with either first-line (35%), first relapse (47%), or ≥second-line (18%) PARPi. In line with our findings, patients with RING domain mutations showed a poor response to treatment, whereas those with mutations in the DBD domain of BRCA1 or BRCA2 genes demonstrated a remarkable response (27). Furthermore, a larger retrospective analysis conducted by Lorusso et al. on 122 patients with BRCA1 (60%) and BRCA2 (40%) mutations showed that patients with DBD alterations had no survival events compared to those with other BRCA domain defects (log rank p = 0.079). Additionally, missense mutations were associated with longer progression-free survival (2 years PFS 100%) compared to splicing or nonsense mutations (log rank p = 0.021 and p = 0.049, respectively) (28). These results are in contrast with our observations and the results from the PAOLA-1 trial showing a reduced sensitivity, although not significant, to bevacizumab plus olaparib in patients with missense or splicing mutations. Additionally, the specific domain defects rather than the mutation type may have driven the benefit since no data regarding the distribution of missense mut in DBD domain were provided.
Our study has several limitations, including its retrospective nature and the small cohort of patients included in the analysis. The majority of patients in our study received Olaparib, rather than Rucaparib or Niraparib, limiting any reliable evaluation of potential differences between the PARPi. Furthermore, the number of patients treated with PARPi at relapse was too small to perform any statistical evaluation beyond a descriptive analysis. Additionally, full data on side effects and treatment adherence that may affect PARPi efficacy were not available.
Conclusions
Our real-world dataset indicates that PARPi was effective regardless of BRCA specific domain defects or mutation type in patients with HGSOC who were treated with first-line platinum-based chemotherapy followed by PARPi maintenance. In terms of PFS, BRCA1 and BRCA2 DBD alterations were associated with the greatest benefits, while BRCA1 RING domain and BRCA1 splicing mutations showed the worst outcomes. This analysis provides intriguing evidence regarding the potential impact of BRCA specific domain defects on primary PARPi sensitivity. However, larger real-world datasets and collaborative studies are needed to further elucidate the impact of BRCA mut on predicting PARPi efficacy, in addition to platinum interval and HRD status.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics statement
The studies involving humans were approved by Comitato Etico “Università FEDERICO II- A.O.R.N A.Cardarelli”. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
Author contributions
RB: Writing – original draft, Conceptualization, Formal analysis, Investigation, Methodology, Resources, Writing – review & editing. GN: Writing – original draft, Conceptualization, Formal analysis, Investigation, Methodology, Resources, Writing – review & editing. ACa: Writing – original draft, Conceptualization, Resources. AL: Writing – original draft, Resources. FM: Supervision, Writing – review & editing. ACe: Writing – review & editing, Supervision. ML: Writing – review & editing, Supervision. FP: Data curation, Supervision, Writing – review & editing. JV: Writing – review & editing, Supervision. UM: Writing – review & editing, Data curation, Supervision. GT: Supervision, Writing – review & editing. MG: Writing – review & editing, Supervision. GA: Writing – review & editing, Supervision. SP: Writing – review & editing, Supervision. CD: Methodology, Project administration, Writing – review & editing, Conceptualization, Data curation, Formal Analysis, Investigation, Resources, Supervision, Validation, Visualization, Writing – original draft.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2024.1412807/full#supplementary-material
References
1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. (2021) 71:209–49. doi: 10.3322/caac.21660
2. Vergote I, González-Martín A, Ray-Coquard I, Harter P, Colombo N, Pujol P, et al. European experts consensus: BRCA/homologous recombination deficiency testing in first-line ovarian cancer. Ann Oncol. (2022) 33:276–87. doi: 10.1016/j.annonc.2021.11.013
3. Moschetta M, George A, Kaye SB, Banerjee S. BRCA somatic mutations and epigenetic BRCA modifications in serous ovarian cancer. Ann Oncol. (2016) 27:1449–55. doi: 10.1093/annonc/mdw142
4. Stewart C, Ralyea C, Lockwood S. Ovarian cancer: an integrated review. Semin Oncol Nurs. (2019) 35:151–6. doi: 10.1016/j.soncn.2019.02.001
5. González-Martín A, Harter P, Leary A, Lorusso D, Miller RE, Pothuri B, et al. Newly diagnosed and relapsed epithelial ovarian cancer: ESMO Clinical Practice Guideline for diagnosis, treatment and follow-up. Ann Oncol. (2023) 34:833–48. doi: 10.1016/j.annonc.2023.07.011
6. Moore K, Colombo N, Scambia G, Kim BG, Oaknin A, Friedlander M, et al. Maintenance olaparib in patients with newly diagnosed advanced ovarian cancer. N Engl J Med. (2018) 379:2495–505. doi: 10.1056/NEJMoa1810858
7. Casaubon JT, Kashyap S, Regan JP. BRCA1 and BRCA2 mutations. In: StatPearls [Internet]. (Treasure Island (FL): StatPearls Publishing) (2024).
8. González-Martín A, Pothuri B, Vergote I, DePont Christensen R, Graybill W, Mirza MR, et al. Niraparib in patients with newly diagnosed advanced ovarian cancer. N Engl J Med. (2019) 381:2391–402. doi: 10.1056/NEJMoa1910962
9. Ledermann JA, Oza AM, Lorusso D, Aghajanian C, Oaknin A, Dean A, et al. Rucaparib for patients with platinum-sensitive, recurrent ovarian carcinoma (ARIEL3): post-progression outcomes and updated safety results from a randomized, placebo-controlled, phase 3 trial. Lancet Oncol. (2020) 21:710–22. doi: 10.1016/S1470-2045(20)30061-9
10. Blasiak J. Single-strand annealing in cancer. Int J Mol Sci. (2021) 22(4):2167. doi: 10.3390/ijms22042167
11. Boussios S, Abson C, Moschetta M, Rassy E, Karathanasi A, Bhat T, et al. Poly (ADP-ribose) polymerase inhibitors: talazoparib in ovarian cancer and beyond. Drugs R D. (2020) 20:55–73. doi: 10.1007/s40268-020-00301-8
12. Wu Y, Xu S, Cheng S, Yang J, Wang Y. Clinical application of PARP inhibitors in ovarian cancer: from molecular mechanisms to the current status. J Ovarian Res. (2023) 16:6. doi: 10.1186/s13048-023-01094-5
13. Clark SL, Rodriguez AM, Snyder RR, Hankins GDV, Boehning D. Structure-function of the tumor suppressor BRCA1. Comput Struct Biotechnol J. (2012) 1(1):e201204005. doi: 10.5936/csbj.201204005
14. Wang B, Matsuoka S, Ballif BA, Zhang D, Smogorzewska A, Gygi SP, et al. Abraxas and RAP80 form a BRCA1 protein complex required for the DNA damage response. Science. (2007) 316:1194–8. doi: 10.1126/science.1139476
15. Mukherjee S, Abdisalaam S, Bhattacharya S, Srinivasan K, Sinha D, Asaithamby A. Mechanistic link between DNA damage sensing, repairing and signaling factors and immune signaling. Adv Protein Chem Struct Biol. (2019) 115:297–324. doi: 10.1016/bs.apcsb.2018.11.004
16. Bonilla B, Hengel SR, Grundy MK, Bernstein KA. RAD51 gene family structure and function. Annu Rev Genet. (2020) 54:25–46. doi: 10.1146/annurev-genet-021920-092410
17. Drost R, Dhillon KK, van der Gulden H, van der Heijden I, Brandsma I, Cruz C, et al. BRCA1185delAG tumors may acquire therapy resistance through expression of RING-less BRCA1. J Clin Invest. (2016) 126:2903–18. doi: 10.1172/JCI70196
18. Paul MW, Sidhu A, Liang Y, van Rossum-Fikkert SE, Odijk H, Zelensky AN, et al. Role of BRCA2 DNA-binding and C-terminal domain in its mobility and conformation in DNA repair. Elife. (2021) 10:e67926. doi: 10.7554/eLife.67926
19. Labidi-Galy SI, Rodrigues M, Sandoval JL, Kurtz JE, Heitz F, Mosconi AM, et al. Association of location of BRCA1 and BRCA2 mutations with benefit from olaparib and bevacizumab maintenance in high-grade ovarian cancer: phase III PAOLA-1/ENGOT-ov25 trial subgroup exploratory analysis. Ann Oncol. (2023) 34:152–62. doi: 10.1016/j.annonc.2022.11.003
20. Cline MS, Liao RG, Parsons MT, Paten B, Alquaddoomi F, Antoniou A, et al. BRCA Challenge: BRCA Exchange as a global resource for variants in BRCA1 and BRCA2. PloS Genet. (2018) 14:e1007752. doi: 10.1371/journal.pgen.1007752
21. Landrum MJ, Lee JM, Benson M, Brown G, Chao C, Chitipiralla S, et al. ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res. (2016) 44:D862–8. doi: 10.1093/nar/gkv1222
22. The jamovi project. jamovi (2022). Available at: https://www.jamovi.org.
23. Therneau TM. A package for survival analysis in R. R package (2020). Available at: https://cran.r-project.org/package=survival.
24. Borgan Ø. Modeling Survival Data: Extending the Cox Model Vol. 20. Therneau TM, Grambsch PM, editors. New York: Springer-Verlag (2000) p. 2053–4, ISBN: ISBN 0-387- 98784-3.
25. Wang Y, Krais JJ, Bernhardy AJ, Nicolas E, Cai KQ, Harrell MI, et al. RING domain-deficient BRCA1 promotes PARP inhibitor and platinum resistance. J Clin Invest. (2016) 126:3145–57. doi: 10.1172/JCI87033
26. Pettitt SJ, Frankum JR, Punta M, Lise S, Alexander J, Chen Y, et al. Clinical BRCA1/2 reversion analysis identifies hotspot mutations and predicted neoantigens associated with therapy resistance. Cancer Discovery. (2020) 10:1475–88. doi: 10.1158/2159-8290.CD-19-1485
27. Galvez Montosa F, Rodriguez Gonzalez CJ, Torres-Mozas P. 69P Relationship between BRCA genotype by location and duration of response to iPARP treatment: Real-life data. ESMO Open. (2023) 8:100849. doi: 10.1016/j.esmoop.2023.100849
Keywords: HGSOC, PARP inhibitors, BRCA mutation domain, resistance, ovarian cancer
Citation: Buonaiuto R, Neola G, Caltavituro A, Longobardi A, Mangiacotti FP, Cefaliello A, Lamia MR, Pepe F, Ventriglia J, Malapelle U, Troncone G, Giuliano M, Arpino G, Pignata S and De Angelis C (2024) Efficacy of PARP inhibitors in advanced high-grade serous ovarian cancer according to BRCA domain mutations and mutation type. Front. Oncol. 14:1412807. doi: 10.3389/fonc.2024.1412807
Received: 05 April 2024; Accepted: 22 August 2024;
Published: 09 September 2024.
Edited by:
Emanuele Perrone, Agostino Gemelli University Polyclinic (IRCCS), ItalyReviewed by:
Raffaella Ergasti, Agostino Gemelli University Polyclinic (IRCCS), ItalyRima Pathak, Tata Memorial Hospital, India
Copyright © 2024 Buonaiuto, Neola, Caltavituro, Longobardi, Mangiacotti, Cefaliello, Lamia, Pepe, Ventriglia, Malapelle, Troncone, Giuliano, Arpino, Pignata and De Angelis. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Carmine De Angelis, Y2FybWluZS5kZWFuZ2VsaXMxQHVuaW5hLml0
†These authors have contributed equally to this work
 Roberto Buonaiuto
Roberto Buonaiuto Giuseppe Neola
Giuseppe Neola Aldo Caltavituro
Aldo Caltavituro Alessandra Longobardi
Alessandra Longobardi Federica Pia Mangiacotti
Federica Pia Mangiacotti Amedeo Cefaliello
Amedeo Cefaliello Maria Rosaria Lamia
Maria Rosaria Lamia Francesco Pepe
Francesco Pepe Jole Ventriglia
Jole Ventriglia Umberto Malapelle
Umberto Malapelle Giancarlo Troncone
Giancarlo Troncone Mario Giuliano
Mario Giuliano Grazia Arpino
Grazia Arpino Sandro Pignata
Sandro Pignata Carmine De Angelis
Carmine De Angelis